Abstract
Severe immunosuppression is a hallmark of Morbillivirus infections. To study the underlying mechanisms, we have developed a ferret model of canine distemper virus infection. The model reproduces all clinical signs of measles, but the lack of ferret-specific reagents has limited the characterization of the cellular immune response. Towards this, we cloned ferret cytokines and established semi-quantitative real-time PCR assays. To demonstrate the utility of these assays we compared the cytokine profiles elicited by lethal and non-lethal strains during the prodromal phase. We observed a general lack of cytokine induction in animals that later succumbed to the disease, whereas survivors mounted a robust and sustained response. The newly developed cytokine assays strengthen and expand the ferret model not only for Morbillivirus pathogenesis studies but also for several other human respiratory viruses including influenza and SARS.
Keywords: Morbillivirus, Canine distemper virus, Immunosuppression, Ferret cytokine mRNA quantification, Prediction of disease outcome
Introduction
Morbilliviruses are highly contagious respiratory pathogens that cause systemic disease and are accompanied by severe immunosuppression (Griffin, 2001, Murphy et al., 1999). Disease severity varies from moderate to severe for measles (MV) in humans and canine distemper virus (CDV) in dogs to usually lethal for CDV in most wild carnivores and rinderpest in cattle (Rima and Duprex, 2006). Regardless of the ultimate disease outcome, the infection is characterized by a dramatic decrease of white blood cells and an inhibition of lymphocyte proliferation within the first weeks after infection (Borrow and Oldstone, 1995, Griffin, 2001, von Messling et al., 2003). However, the knowledge regarding interactions between the host and the immune system preceding these events remains limited.
Since MV only infects humans and certain non-human primates, we have established a model based on the study of the closely related canine distemper virus (CDV) in ferrets, one of its natural hosts (von Messling et al., 2003). Ferrets are highly sensitive to the disease, and for the most part succumb to infection with a wild type virus. On the other hand, infection with attenuated CDV strains results in disease signs and progression that parallels MV infection in humans (Kauffman et al., 1982). Virus control and clearance in surviving animals usually occur within 3 to 4 weeks after inoculation and coincide with the development of neutralizing antibodies (von Messling et al., 2004, von Messling et al., 2003). The application of our model to study the immune response during the crucial early phase of CDV infection has so far been difficult as specific reagents to detect cytokine responses in ferrets are not readily available.
This lack of immunological reagents constitutes a major limitation for the use of ferrets in pathogen–host interaction studies, for which it is becoming increasingly attractive. Ferrets are naturally susceptible to several human respiratory pathogens and a number of gastro-intestinal bacterial diseases (Ball, 2006). They have long been recognized as a model for the efficacy assessment of new vaccines or treatment approaches against influenza and more recently severe acute respiratory syndrome (SARS), and they are also used to study prion diseases (Bartz et al., 1998, Maher and DeStefano, 2004, Ter Meulen et al., 2006).
The purpose of the present study was to establish mRNA-based cytokine assays for ferrets and to validate this new tool by comparing the early host response to lethal and non-lethal CDV infections. Towards this, we completed the sequences of ferret cytokines representing the different axes of the cellular immune response and developed semi-quantitative real-time PCR assays. We then evaluated cytokine mRNA expression levels at 3 and 7 days after inoculation and found that survival was associated with a robust and sustained response whereas animals that succumbed to the disease experienced a generalized shutoff of cytokine expression.
Results
Cloning and characterization of ferret cytokines
Since reagents for the assessment of the cellular immune response in ferrets are not commercially available, we set out to obtain the sequences of a ferret cytokine panel including those reflecting early innate immune response activation, tumor necrosis factor (TNF) α, interferon (IFN) α, and interleukin (IL) 6, those indicating Th1 polarization, IFNγ, IL2, and IL12p40, and the Th2 cytokines IL4 and IL10. As mustelids, ferrets are most closely related to other carnivores like dogs and cats (McKenna and Bell, 1997). The first set of primers was therefore based on canine and feline sequences available in GenBank. We initially obtained small fragments of ferret IFNα, IFNγ, IL2, IL4, IL6, and TNFα and used those sequences to measure cytokine mRNA induction using a gel-based assay (von Messling et al., 2006). To develop a robust and versatile real-time PCR-based assay that generates a representative cytokine profile, we included IL10 and IL12p40 and pursued the sequencing of larger parts of the open reading frames. This strategy has so far yielded complete open reading frames for IL2, IL4, IL6, IFNα (consensus sequence), and TNFα (GenBank accession nos. IL2 EF368206; IL4 EF368210; IL6 EF368209; IFNα EF368207; TNFα EF368211) and partial gene sequences for IL10 and IFNγ (> 95) and IL12p40 (∼ 60%; GenBank accession nos. IL10 EF368212; IFNγ EF368214; IL12p40 EF368213). When comparing the ferret cytokines with the respective canine and feline proteins, we observed a slightly higher overall amino acid identity between ferrets and dogs than between ferrets and cats (Table 1 ). This degree of similarity on the amino acid and nucleotide level (data not shown) confirms the identity of the obtained sequences.
Table 1.
Comparison of ferret cytokines with the respective canine and feline protein sequences available in GenBank
| Protein | % AA identity |
% AA identity |
|---|---|---|
| Ferret/Dog | Ferret/Cat | |
| IFNαa | 67 | 72 |
| IFNγb | 88 | 85 |
| TNFα | 95 | 92 |
| IL2 | 86 | 85 |
| IL4 | 83 | 77 |
| IL6 | 78 | 73 |
| IL10b | 93 | 89 |
| IL12p40b | 94 | 92 |
Values based on consensus sequence.
Values based on alignment of available gene sequence.
Early CDV immunosuppression is independent of the viral load
To establish and validate ferret cytokine real-time PCR assays, we compared samples from animals that succumbed to the disease with those originating from survivors. Different viruses were included to assure the identification of general characteristics of disease outcome rather than profiles associated with one individual strain. Samples included in the lethal group originated from animals infected with either 5804P-eGFP/H (von Messling et al., 2004) or A75eH (Rudd et al., 2006), whereas the survivor group consisted of individuals inoculated with the highly pathogenic but non-lethal strain 5804P-eGFP/uN (von Messling et al., 2004) or survivors of 5804P/A75eH chimeric viruses. These viruses, in which the envelope glycoproteins were exchanged between the two lethal strains, cause severe disease but only 60–80% mortality (unpublished results). In the case of 5804P-eGFP/uN, which carries an eGFP-containing additional transcription unit upstream of the N gene, the non-lethal phenotype is due to a slight delay in replication efficacy, whereas minimal incompatibilities between viral proteins originating from different strains are the likely cause of survival in the case of the 5804P/A75 chimeric viruses.
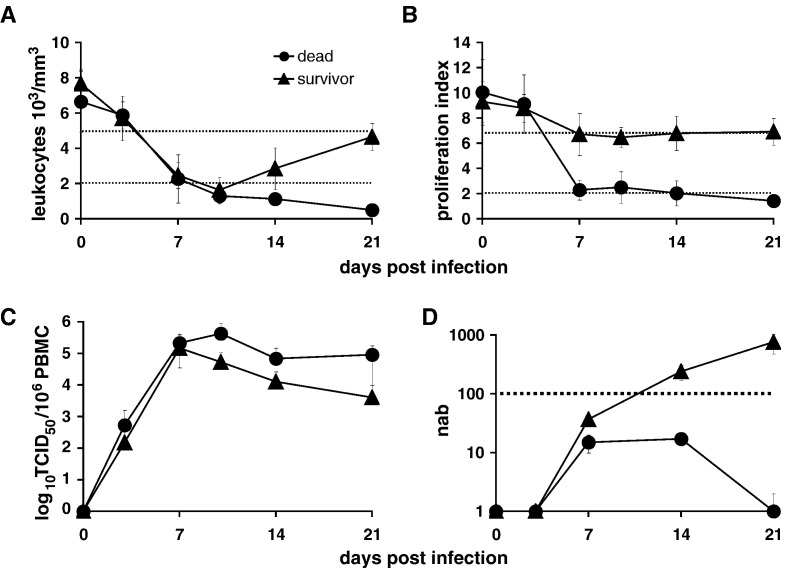
In animals that succumb to the disease, we observed a dramatic drop in leukocyte numbers and an almost complete loss of peripheral blood mononuclear cell (PBMC) proliferation activity upon phytohemagglutinin (PHA) stimulation between 3 and 7 days after inoculation, prior to the onset of disease signs (Figs. 1A and B, circles). In contrast, survivors experienced an equally severe but transient leukocyte drop, and the inhibition of lymphocyte proliferation was less pronounced (Figs. 1A and B, triangles). No marked differences in peripheral blood mononuclear cell (PBMC)-associated virus titers and neutralizing antibody levels against CDV were noted between the two groups at these early time points (Figs. 1C and D).
Fig. 1.
Clinical parameters in animals infected with lethal or non-lethal CDV strains. (A) Leukocyte numbers, (B) in vitro proliferation activity of lymphocytes, (C) cell-associated virus titers expressed in 50% log10 tissue culture infectious doses (TCID50) in PBMCs, and (D) neutralizing antibody titers over the first 21 days of the disease. Animals infected with the lethal strains 5804P-eGFP/H (n = 5) or A75eH (n = 6) are represented by circles, those infected with the non-lethal virus 5804P-eGFP/uN (n = 4) and the survivors of infections with chimeric 5804P/A75eH envelope exchange viruses (n = 4) by triangles. Days post-infection are indicated on the X axis, leukocyte number, proliferation activity, cell-associated virus titer, or neutralizing antibody titer on the Y axis. Error bars represent the standard deviation. The narrow dotted lines indicate cut-off values associated with moderate (upper line) and severe (lower line) immunosuppression. The wider dotted line represents the protective threshold of neutralizing antibodies.
Blood cell composition and infection levels at early disease stages are independent of disease outcome
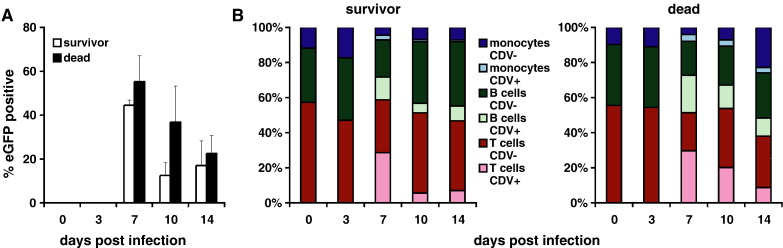
Because of their important role in CDV pathogenesis and easy accessibility, PBMCs are ideally suited to follow the systemic cytokine response over time in the same animal. We have previously shown that 5804P targets primarily T and B cells and that a lethal infection results in a relative reduction of the T cell population, beginning at 10 days post-infection (d.p.i.) (von Messling et al., 2006). To assure the comparability between samples from both groups, we determined infection levels and PBMC subpopulation composition at the different time points. Consistent with our previous results, we observed no infected cells or relevant changes in PBMC subpopulations at 3 d.p.i. (Figs. 2A and B, compare 0 and 3 d.p.i.). At 7 d.p.i., more than 40% of cells expressed eGFP (Fig. 2A, 7 d.p.i.), which could be further subdivided in 28–30% T cells, 13–21% B cells, and 3–4% monocytes regardless of the disease outcome (Fig. 2B, left and right panel). After the onset of clinical signs at 10 d.p.i., the two groups diverged similar to what was observed in the immunological parameters (Fig. 1).
Fig. 2.
Extent and PBMC subtype specificity of lethal and non-lethal CDV infection. (A) FACS analysis of the percentage of eGFP expression in PBMCs isolated from animals that succumbed (black bars) to or survived (white bars) the disease at 0 to 14 days after inoculation. Error bars represent the standard deviation. (B) Relative percentages of PBMC subtypes infected 0 to 14 days after infection with lethal or non-lethal viruses. T cells are shown in red, B cells are shown in green, and monocyte-lineage cells are shown in blue. Lighter shades represent eGFP-expressing cells; full shades represent negative cells. The means of the results from all animals assigned to the respective groups are shown.
CDV survival is associated with a robust cytokine response
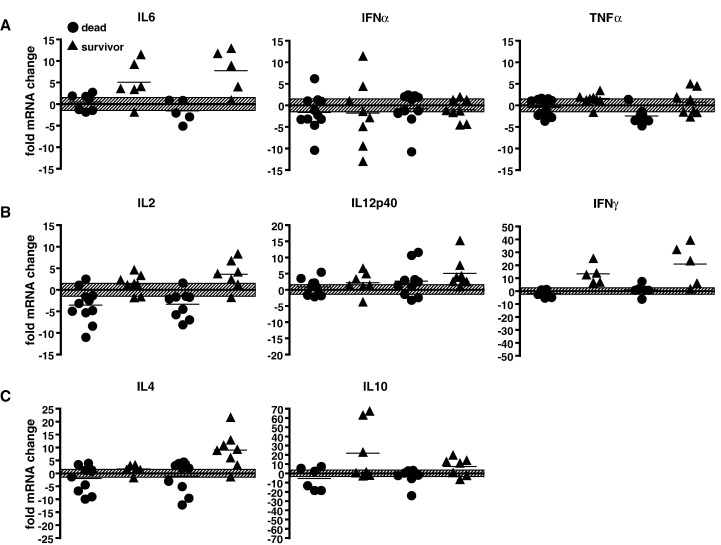
To determine if the newly established cytokine assays allow a discrimination of the two groups at early disease stages, when other assays fail to detect a difference, we compared the relative induction of the different cytokine mRNAs at 3 and 7 d.p.i. in animals that succumbed to the disease with survivors. The most striking finding was a generalized lack of cytokine upregulation in animals infected with a lethal strain at both time points (Figs. 3A–C, circles). For TNFα, IL6, and IL2, we even observed a reduction in cytokine mRNA levels below baseline in the majority of animals analyzed, suggesting an infection-induced shutdown of gene expression (Figs. 3A and B, circles). The only exception was IL12p40, which was upregulated in a small subset of the animals examined (Fig. 3B, middle panel, circles).
Fig. 3.
Comparison of cytokine responses in animals infected with lethal or non-lethal CDV strains. (A) Innate immunity, (B) Th1-type, and (C) Th2-type cytokine mRNA expression in both groups at 3 (D3) and 7 (D7) days post-infection. Animals infected with lethal strains (n = 11) are represented by circles, those infected with non-lethal viruses (survivors; n = 8) by triangles. The fold mRNA expression change compared to pre-infection levels of the same animal is indicated on the Y axis. Each samples was analyzed in triplicates and only samples where the variation among triplicates was less than 10% were included in the analysis, resulting in the variability in numbers of individuals plotted for the different cytokines. The black bar represents the mean of all the samples. The hatched zone represents threshold levels of 1.5 fold expression change, which is considered normal variability.
In contrast, the majority of animals infected with non-lethal viruses showed a robust upregulation of the innate immune response cytokine IL6 and the Th1-type cytokine IFNγ at 3 d.p.i., which was sustained through day seven (Figs. 3A and B, triangles). At the earlier time point, a broad range of IFNα expression levels was observed, and a subset of animals displayed a very strong IL10 response (Figs. 3A and C, D3, triangles). Seven days after inoculation, the Th1-type cytokines IL2 and IL12p40 and the Th2-type cytokines IL4 and IL10 were also strongly expressed (Figs. 3B and C, D7, triangles).
Discussion
The dramatic drop in leukocyte numbers and the loss of function in the remaining cells observed within the first week after infection indicate that Morbilliviruses interfere with the induction of an appropriate immune response at very early stages (Borrow and Oldstone, 1995, Schneider-Schaulies et al., 2001). To better understand the basis for host immune response control in our ferret model, we established semi-quantitative real-time RT-PCR assays based on ferret cytokine sequences and compared the response associated with lethal disease outcome or survival. We found that animals succumbing to the disease failed to mount a sustained response, while survivors showed initially a Th1 polarization that later transitioned into a Th2-biased response.
Cytokine responses to CDV have so far been mostly studied in dogs with naturally contracted disease at the time of euthanasia. The results represent thus the profile at the late stages of the acute disease phase or at the onset of neurologic disease after recovery from the acute infection. Interestingly, animals with severe signs of disease and viremia at the time of death displayed low or absent cytokine mRNA induction in blood-derived RNA, consistent with our initial findings in 5804P-infected ferrets and this study (Grone et al., 1998, von Messling et al., 2006). In contrast, individuals with mild or absent general signs of disease but neurologic manifestations mounted a broad response not only in the blood but also locally in the CNS (Grone et al., 1998, Markus et al., 2002). The role of the systemic and local immune response in the context of this delayed neurologic form of CDV remains to be investigated in ferrets.
The striking lack of any cytokine response in animals infected with a lethal virus as early as 3 d.p.i. indicates a viral interference with immune system activation at a time when the infection in PBMCs is barely detectable. In vitro, it has been demonstrated that minute amounts of MV envelope are suffice to induce an anergic state in exposed lymphocytes, but the exact interactions remain to be elucidated (Schlender et al., 1996). In addition, the non-essential protein V has been shown to inhibit activation of an anti-viral response by interfering with nuclear localization of STATs (Devaux et al., in press, Palosaari et al., 2003). It is possible that the shutdown of cytokine expression observed for the lethal CDV strains results from contacts between viral glycoproteins and immune cells in combination with a V and phosphoprotein-dependent inhibition of the induction of the anti-viral state in infected cells. Taken together our findings demonstrate that the disease outcome is decided within the first days of infection.
The Th1/Th2 switch observed in animals infected with non-lethal viruses mirrors previous reports of children naturally infected with MV, where IL2 and IFNγ were expressed during the acute phase of the disease, and the increase of IL4 coincided with antibody-mediated virus clearance (Griffin et al., 1994, Moss et al., 2002). In the same context, elevated IL10 levels were detected for weeks after the infection, which might be an important factor in the long-lasting immunosuppression associated with MV (Moss et al., 2002). In our study, ferrets infected with non-lethal viruses closely reproduce these findings, further supporting the value of this model for the characterization of the mechanisms underlying Morbillivirus-induced immunosuppression.
In addition to Morbillivirus pathogenesis studies, ferrets are a recognized model for the development of vaccines and anti-viral therapies against influenza, and more recently SARS (Maher and DeStefano, 2004, Ter Meulen et al., 2006). They are also used for transmission studies of different transmissible spongiforme encephalopathies (Bartz et al., 1998, Bartz et al., 1994) and parallel human disease manifestations for several bacterial diseases including Helicobacter-induced gastric ulcers, hemolytic uremic syndrome caused by Shiga toxin-producing Escherichia coli, and campylobacteriosis (O'Rourke and Lee, 2003, Woods et al., 2002). By comparing samples from animals with different disease outcome, we have demonstrated that our semi-quantitative real-time PCR assays are sufficiently sensitive to detect a very weak response and even a reduction from baseline levels as observed in animals that later succumbed to the disease. At the same time, the assays allow the quantification of the response across a much wider range than the previous gel-based approach (von Messling et al., 2006). This will enable the more detailed comparison of the cytokine responses to specifically attenuated viruses in future studies. Furthermore, this RNA-based assay can easily be applied to various tissues as well as bodily fluids, enabling the assessment of the local and systemic responses at the same time. It thus adds a versatile new tool to the study of host–pathogen interactions in this model.
Materials and methods
Ferret cytokine sequences
To obtain sequences encoding ferret IL2, 4, 6, 10, and 12p40, IFNα and γ, and TNFα, as well as GAPDH as a housekeeping gene, total RNA was isolated (RNeasy, Qiagen) from PBMCs of healthy animals stimulated with 5 μg/ml PHA (Sigma) for 24 h. The RNA was then reverse transcribed (Superscript III, Invitrogen) using a mixture of oligoT and random hexamer primers to maximize cDNA output. Primer pairs for the initial amplification of ferret cytokines were based on conserved regions in the respective genes of the phylogenetically related carnivores, dogs and cats, available in GenBank (canine sequences: IL2 AM238655; IL4 NM001003159; IL6 NM001003301; IL10 NM001003077; IL12p40 U49100; IFNα A33693; IFNγ NM001003174; TNFα Z70046; feline sequences: IL2 NM001043337; IL4 NM001043339; IL6 NM001009211; IL10 NM001009209; IL12p40 Y07762; IFNα DQ220469; IFNγ X86974; TNFα NM001009835). Once a partial ferret sequence was determined (von Messling et al., 2006), ferret specific primers were chosen to obtain increasing parts of the open reading frame either in combination additional canine/feline-based primers or by 3′ and 5′ RACE according to the manufacturer's instruction (GeneRacer RACE-Ready cDNA kit, Invitrogen). Primer pairs for the real-time PCR assay were designed using the Primer Quest program of Integrated DNA Technologies (Table 2 ). Protein alignments and the calculation of percent amino acid identity were performed using MegAlign (DNASTAR, Lasergene).
Table 2.
Primer pairs used in the cytokine real-time PCR assays
| Gene | Primers |
|
|---|---|---|
| Forward | Reverse | |
| IFNα | 5′-ATGCTCCTGCGACAAATGAGGAGA-3′ | 5′-TTCTGCAGCTGCTTGCTGTCAAAC-3′ |
| IFNγ | 5′-CCATCAAGGAAGACATGCTTGTCAGG-3′ | 5′-CTGGACCTGCAGATCATTCACAGGAA-3′ |
| TNFα | 5′-TGGAGCTGACAGACAACCAGCTAA-3′ | 5′-TGATGGTGTGGGTAAGGAGCACAT-3′ |
| IL2 | 5′-TGCTGCTGGACTTACAGTTGCTCT-3′ | 5′-CAATTCTGTGGCCTTCTTGGGCAT-3′ |
| IL4 | 5′-CGTTGAACATCCTCACAGCGAGAAAC-3′ | 5′-TTGCCATGTTCCTGAGGTTCCTGTGA-3′ |
| IL6 | 5′-CAAATGTGAAGACAGCAAGGAGGCA-3′ | 5′-TCTGAAACTCCTGAAGACCGGTAGTG-3′ |
| IL10 | 5′-TCCTTGCTGGAGGACTTTAAGGGT-3′ | 5′-TCCACCGCCTTGCTCTTATTCTCA-3′ |
| IL12p40 | 5′-ATCGAGGTTGTGGTGGGTGCTATT-3′ | 5′-TAGGTTCATGGGTGGGTCTGGTTT-3′ |
| GAPDH | 5′-AACATCATCCCTGCTTCCACTGGT-3′ | 5′-TGTTGAAGTCGCAGGAGACAACCT-3′ |
Viruses and animal experiments
The samples analyzed in this study originated from animals infected with either one of the lethal strains 5804P-eGFP/H (von Messling et al., 2004) and A75eH (Rudd et al., 2006), the non-lethal strain 5804P-eGFP/uN (von Messling et al., 2004), and survivors of infections with chimeric 5804P/A75eH envelope exchange viruses that cause 60–80% lethality (unpublished results).
Male ferrets over 16 weeks of age (Marshall Farms) were inoculated intranasally with 105 50% tissue culture infectious doses of the respective virus, and blood samples were collected twice during the first 2 weeks, and weekly thereafter. At each time point, the total white blood cell count, proliferation activity, cell-associated virus titers, and neutralizing antibody titers were determined as described previously (Rudd et al., 2006, von Messling et al., 2003). Flow cytometry analysis of the PBMC subpopulations was carried our following the protocol published previously (von Messling et al., 2004, von Messling et al., 2006).
For the cytokine mRNA quantification, 3 ml blood was collected in Na–EDTA-coated vacuum tubes (Vacutainer, Beckton Dickinson) immediately before and at 3 and 7 d.p.i. PBMCs were isolated by destroying the erythrocytes with ACK lysis buffer (150 mM NH4Cl, 10 mM KHCO3, 0.01 mM EDTA, pH 7.2–7.4), resuspended in RNAlater (Qiagen), and stored at − 20 °C. The RNA was purified using the RNeasy kit in combination with an on-the-column DNase treatment (Qiagen), and the yield was quantified by UV spectrophotometry.
Semi-quantitative real-time PCR assays
To assess cytokine expression, the cDNA produced from 10 ng RNA was combined with 0. 5 μM of each primer and the DyNAmo SYBR Green 2-Step qRT-PCR reaction mix (Finnzymes) to a 10 μl reaction according to the manufacturer's instructions. The real-time PCR was performed in a RG-3000A (Rotor-Gene), using the following protocol: denaturation at 95 °C for 15 min followed by 40 cycles of 95 °C for 5 s, 60 °C for 15 s, and 72 °C for 25 s, and a melting curve analysis to confirm reaction specificity. Each experiment was performed in triplicate, and only values that varied less than 10% were considered. The fold change ratios between experimental and control samples for each gene were calculated with GAPDH levels as a reference. Towards this the cycle at which GAPDH crosses the detection threshold (Ct) is subtracted from the Ct value of the gene of interest at each time point (ΔCt). The fold change is then calculated by subtracting the ΔCt values of the respective gene at day 0 from those obtained at subsequent days (ΔΔCt). To transform these values into absolute values, the formula: fold change = 2− ΔΔCt is used (Schmittgen et al., 2000).
Acknowledgments
We thank Roberto Cattaneo, Charles M. Dozois, and Alain Lamarre for comments on the manuscript and are grateful to all laboratory members for support and lively discussion. This work was supported by grants from the CIHR (MOP-66989), CFI (9488), and a CIHR New Investigator Award to V.v. M, NIH grant R01 A163476 to Roberto Cattaneo (V.v.M. co-investigator), and an Armand-Frappier Foundation scholarship to N.S.
References
- Ball R.S. Issues to consider for preparing ferrets as research subjects in the laboratory. ILAR J. 2006;47:348–357. doi: 10.1093/ilar.47.4.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz J.C., McKenzie D.I., Bessen R.A., Marsh R.F., Aiken J.M. Transmissible mink encephalopathy species barrier effect between ferret and mink: PrP gene and protein analysis. J. Gen. Virol. 1994;75(Pt. 11):2947–2953. doi: 10.1099/0022-1317-75-11-2947. [DOI] [PubMed] [Google Scholar]
- Bartz J.C., Marsh R.F., McKenzie D.I., Aiken J.M. The host range of chronic wasting disease is altered on passage in ferrets. Virology. 1998;251:297–301. doi: 10.1006/viro.1998.9427. [DOI] [PubMed] [Google Scholar]
- Borrow P., Oldstone M.B. Measles virus–mononuclear cell interactions. Curr. Top. Microbiol. Immunol. 1995;191:85–100. doi: 10.1007/978-3-642-78621-1_6. [DOI] [PubMed] [Google Scholar]
- Devaux, P., von Messling, V., Songsungthong, W., Springfeld, C., Cattaneo, R., in press. Tyrosine 110 in the measles virus phosphoprotein is required to block STAT1 phosphorylation. Virol. [DOI] [PubMed]
- Griffin D.E. Measles virus. In: Knipe D.M., Howley P.M., editors. fourth ed. vol. 1. Lippincott Williams and Wilkins; Philadelphia: 2001. pp. 1401–1441. (Fields Virology). (2 vols.) [Google Scholar]
- Griffin D.E., Ward B.J., Esolen L.M. Pathogenesis of measles virus infection: an hypothesis for altered immune responses. J. Infect. Dis. 1994;170(Suppl. 1):S24–S31. doi: 10.1093/infdis/170.supplement_1.s24. [DOI] [PubMed] [Google Scholar]
- Grone A., Frisk A.L., Baumgartner W. Cytokine mRNA expression in whole blood samples from dogs with natural canine distemper virus infection. Vet. Immunol. Immunopathol. 1998;65:11–27. doi: 10.1016/s0165-2427(98)00170-6. [DOI] [PubMed] [Google Scholar]
- Kauffman C.A., Bergman A.G., O'Connor R.P. Distemper virus infection in ferrets: an animal model of measles-induced immunosuppression. Clin. Exp. Immunol. 1982;47:617–625. [PMC free article] [PubMed] [Google Scholar]
- Maher J.A., DeStefano J. The ferret: an animal model to study influenza virus. Lab. Anim. (NY) 2004;33:50–53. doi: 10.1038/laban1004-50. [DOI] [PubMed] [Google Scholar]
- Markus S., Failing K., Baumgartner W. Increased expression of pro-inflammatory cytokines and lack of up-regulation of anti-inflammatory cytokines in early distemper CNS lesions. J. Neuroimmunol. 2002;125:30–41. doi: 10.1016/s0165-5728(02)00027-9. [DOI] [PubMed] [Google Scholar]
- McKenna M.C., Bell S.K. Columbia University Press; New York City, NY, USA: 1997. Classification of Mammals Above the Species Level. [Google Scholar]
- Moss W.J., Ryon J.J., Monze M., Griffin D.E. Differential regulation of interleukin (IL)-4, IL-5, and IL-10 during measles in Zambian children. J. Infect. Dis. 2002;186:879–887. doi: 10.1086/344230. [DOI] [PubMed] [Google Scholar]
- Murphy F., Gibbs E., Horzinek M., Studdert M. Veterinary Virology. 3rd ed. Academic Press; San Diego, CA: 1999. pp. 421–426. [Google Scholar]
- O'Rourke J.L., Lee A. Animal models of Helicobacter pylori infection and disease. Microbes Infect. 2003;5:741–748. doi: 10.1016/s1286-4579(03)00123-0. [DOI] [PubMed] [Google Scholar]
- Palosaari H., Parisien J.P., Rodriguez J.J., Ulane C.M., Horvath C.M. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 2003;77:7635–7644. doi: 10.1128/JVI.77.13.7635-7644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rima B.K., Duprex W.P. Morbilliviruses and human disease. J. Pathol. 2006;208:199–214. doi: 10.1002/path.1873. [DOI] [PubMed] [Google Scholar]
- Rudd P.A., Cattaneo R., von Messling V. Canine distemper virus uses both the anterograde and the hematogenous pathway for neuroinvasion. J. Virol. 2006;80:9361–9370. doi: 10.1128/JVI.01034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlender J., Schnorr J.J., Spielhoffer P., Cathomen T., Cattaneo R., Billeter M.A., ter Meulen V., Schneider-Schaulies S. Interaction of measles virus glycoproteins with the surface of uninfected peripheral blood lymphocytes induces immunosuppression in vitro. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13194–13199. doi: 10.1073/pnas.93.23.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T.D., Zakrajsek B.A., Mills A.G., Gorn V., Singer M.J., Reed M.W. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal. Biochem. 2000;285:194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
- Schneider-Schaulies S., Niewiesk S., Schneider-Schaulies J., ter Meulen V. Measles virus induced immunosuppression: targets and effector mechanisms. Curr. Mol. Med. 2001;1:163–181. doi: 10.2174/1566524013363960. [DOI] [PubMed] [Google Scholar]
- Ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E., Preiser W., Doerr H.W., Chow V.T., de Kruif J., Peiris J.S., Goudsmit J. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Messling V., Springfeld C., Devaux P., Cattaneo R. A ferret model of canine distemper virus virulence and immunosuppression. J. Virol. 2003;77:12579–12591. doi: 10.1128/JVI.77.23.12579-12591.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Messling V., Milosevic D., Cattaneo R. Tropism illuminated: lymphocyte-based pathways blazed by lethal Morbillivirus through the host immune system. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14216–14221. doi: 10.1073/pnas.0403597101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Messling V., Svitek N., Cattaneo R. Receptor (SLAM [CD150]) recognition and the V protein sustain swift lymphocyte-based invasion of mucosal tissue and lymphatic organs by a Morbillivirus. J. Virol. 2006;80:6084–6092. doi: 10.1128/JVI.00357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods J.B., Schmitt C.K., Darnell S.C., Meysick K.C., O'Brien A.D. Ferrets as a model system for renal disease secondary to intestinal infection with Escherichia coli O157:H7 and other Shiga toxin-producing E. coli. J. Infect. Dis. 2002;185:550–554. doi: 10.1086/338633. [DOI] [PubMed] [Google Scholar]