Abstract
Pancreatic duct epithelial cells (PDECs) have been shown to express calcium activated chloride channels (CaCCs) and there is evidence for their involvement in fluid secretion from these cells. The molecular identity of the CaCC in PDECs remains unknown. Recently, the bestrophin family of proteins have been proposed as a potential molecular candidate for CaCCs. Expression of bestrophins is strongly correlated with the function of CaCCs in a variety of tissues. In the present study, the expression of bestrophins has been investigated in the cystic fibrosis pancreatic duct cell line, CFPAC-1. Iodide efflux analysis was used to characterise native CaCCs in CFPAC-1 cell monolayers. Efflux was induced with the addition of UTP (100 μm, 10.2 ± 1.5 nmol min−1), which was blocked by the chloride channel blockers niflumic acid (81%) and DIDS (90%). The UTP-stimulated iodide efflux was shown to be Ca2+ dependent and cAMP independent. RT-PCR analysis of RNA isolated from CFPAC-1 cells demonstrated positive identification of all four human bestrophin mRNAs. Western blot of CFPAC-1 cell protein isolates with antibodies specific to human bestrophin 1 (hBest1) showed that hBest1 protein was expressed in this cell line. HBest1 was present on the cell surface, demonstrated using biotinylation and confocal imaging, as well as in the cytoplasm. SiRNA-mediated silencing of hBest1 in CFPAC-1 cells reduced the UTP-stimulated iodide efflux by around 40%. This study provides evidence that the bestrophins are expressed in pancreatic duct cells and, more specifically, that hBest1 plays a role in the CaCCs found in these cells.
Pancreatic duct epithelial cells (PDECs) secrete a bicarbonate rich fluid that forms the basis of pancreatic juice. This bicarbonate rich fluid helps move digestive enzymes secreted by the pancreatic acini towards the gut and also neutralises the acidic chyme entering the duodenum from the stomach (Argent & Case, 1994). The production of this bicarbonate rich secretion is controlled by a number of different transport proteins and ion channels (for a review see Steward et al. 2005). The movement of bicarbonate through the PDEC to the lumen of the duct is thought to be in part regulated by two chloride channels, the cystic fibrosis transmembrane conductance regulator (CFTR) chloride channel and the calcium-activated chloride channel (CaCC). CFTR is a protein kinase A (PKA) regulated channel that belongs to the family of ATP-binding cassette (ABC) transporters (Sheppard & Welsh, 1999). Cystic fibrosis (CF) is the commonest cause of pancreatic insufficiency in Caucasian children and affects about 1 in 2500 of the population (Durie & Forstner, 1989). CF is associated with defective trafficking and/or function of the CFTR chloride channel, which is highly expressed on the proximal ducts of the pancreas (Marino et al. 1991; Trezise et al. 1993). In CF, dysfunction of CFTR reduces pancreatic duct bicarbonate and fluid secretions leading to precipitation of proteins within the duct lumen that eventually block and destroy the gland.
Activation of alternative chloride conductances has been proposed as a way to treat CF-related organ disease. One of the main candidates for developing such a therapy is the CaCC. CaCCs have been identified in PDECs from several animal species including mouse (Winpenny et al. 1995), rat and guinea pig (Gray et al. 2002). They have also been identified in both freshly isolated human PDECs and in immortalised cell lines of duct origin (Winpenny et al. 1998). Despite this functional data, the molecular identity of the CaCCs in PDECs is unknown.
Several proteins have emerged as possible candidates for the CaCCs in PDECs. Since the first member of the CLCA family was cloned from the bovine trachea (bCLCA1)(Cunningham et al. 1995), several other members have emerged over a variety of different species, including four members of human origin, hCLCA 1–4 (for a review of the CLCA family see Loewen & Forsyth, 2005). However, RT-PCR data from a human pancreatic duct cell line, HPAF, suggested that CLCA1 and 2 were not present in pancreatic duct cells (Fong et al. 2003). Furthermore, it has been suggested that the CLCA proteins do not structurally represent an integral chloride channel and are secreted from the cell surface (Gibson et al. 2005; Elble et al. 2006).
The bestrophin protein family consists of four human homologues, hBest1–4 (Sun et al. 2001; Tsunenari et al. 2003) and have recently been proposed as a candidate for CaCCs. HBest1 is the product of the gene VMD2, which is the gene that is mutated in the disease Best's macular dystrophy (BMD) (Marquardt et al. 1998). Members of the bestrophin family have also been identified in other species including Drosophila (Sun et al. 2001; Tavsanli et al. 2001), C. elegans (Sun et al. 2001; Tavsanli et al. 2001), Xenopus laevis (Qu et al. 2003) and mouse (Bakall et al. 2003; Qu et al. 2004). The bestrophins are members of the RFP-TM gene family (Stohr et al. 2002), which is characterised by a conserved 350–400 amino acid region that contains an arginine (R), phenylalanine (F) and proline (P) motif. HBest1 is a 585 amino acid protein that has a 68 kDa molecular mass (Petrukhin et al. 1998) and the bestrophin protein is thought to contain four transmembrane spanning domains (Tsunenari et al. 2003). Analysis of all four human bestrophins show that they share 55–66% sequence identity at the N-terminal end of the protein but very little homology is seen at the C-terminus (Tsunenari et al. 2003). Initial studies suggested that the bestrophins were chloride ion channels (Sun et al. 2001) and identified a Ca2+-sensitive component of bestrophin currents when expressed in HEK 293 cells (Sun et al. 2002; Fischmeister & Hartzell, 2005). Studies on mouse bestrophin 2 (mBest2) identified residues that when mutated augmented chloride channel activity and were suggested to be responsible for permeability and conduction of chloride by the channel (Qu et al. 2004; Qu & Hartzell, 2004). More recently, several investigations have demonstrated that bestrophins are expressed in cells and tissues that would usually be associated with measurable CaCCs, including olfactory neurons and epithelial cells (Barro Soria et al. 2006; Pifferi et al. 2006). Furthermore, studies using gene silencing techniques have related the silencing of bestrophin to reduction in functional CaCCs (Barro Soria et al. 2006; Chien et al. 2006).
In the present study we have investigated the expression of bestrophins, in particular hBest1, in a human pancreatic duct epithelial cell line, CFPAC-1. CFPAC-1 cells contain the most prevalent mutation found in CF patients, F508del-CFTR, which prevents CFTR trafficking to the cell surface, causing a loss of the predominant apical membrane Cl− conductance in pancreatic ducts (Schoumacher et al. 1990). Our data show that bestrophins are expressed in PDECs and that hBest1 is involved in calcium-mediated iodide efflux and is therefore a potential candidate for the CaCC in PDECs.
Methods
Cell culture
CFPAC-1 cells were obtained from the ATCC and used between passages 25 and 60. Culture medium consisted of Iscove's modified Dulbecco's medium, supplemented with 10% fetal calf serum (FCS), 5 mm l-glutamine and 1% penicillin–streptomycin. Spontaneously arising human retinal pigmented epithelial (ARPE-19) cells (Dunn et al. 1996) were grown in a 1 : 1 mixture of Dulbecco's modified Eagle's medium (DMEM)–Ham's F12 nutrient mixture supplemented with 10% FCS, 1% penicillin–streptomycin, 2.5 mm l-glutamate and 1.2 g l−1 sodium bicarbonate. All cells were cultured at 37°C in a humidified atmosphere of 95% O2–5% CO2. Following trypsin treatment, cells were seeded on coverslips for immunocytochemistry, on tissue culture dishes for RNA isolation and in six-well plates for iodide efflux experiments. For cell surface biotinylation, CFPAC-1 cells were grown to confluence in T75 tissue culture flasks.
Iodide efflux assay
Iodide efflux assays were carried out as previously published (Winpenny et al. 1998). Briefly, cells were incubated with loading buffer (containing in mm: 136 NaI, 3 KNO3, 2 Ca(NO3)2, 20 Hepes and 11 glucose, adjusted to pH 7.4 using 1 m NaOH) for 1 h at room temperature (24°C), washed with efflux buffer to remove excess iodide (containing the same constituents as loading buffer except 136 NaNO3 instead of NaI) and the cells washed sequentially with 1 ml of efflux buffer at 1 min intervals for 15 min. The washes were collected in 12-well plates for analysis. The samples were analysed using an iodide sensitive electrode (Russell pH Ltd, Auchtermuchty, UK) and the amount of iodide in each sample of efflux buffer determined. The experimental data were quantified using a spreadsheet created in Microsoft Excel, similar to that described by (Lansdell et al. 1998; Hughes et al. 2004). Essentially, we convert the iodide-selective electrode readings (mV) into iodide content (nmol min−1) using a calibration curve of NaI solutions from 1 μm to 1 mm. Test chemicals, for example ionomycin, were added to the efflux buffer from time point six until the end of the experiment. Thirty minutes after starting to incubate cells with loading buffer, 4,4′-diisothiocyanatostilbene-2,2′-disulfonic acid (DIDS), niflumic acid (NA) and BAPTA-AM were added to the efflux buffer; these agents were present in the extracellular solution until the end of the experiment. All iodide efflux experiments were performed at room temperature (24°C), similar to other groups that have used this method (Lansdell et al. 1998; Chappe et al. 2003).
RNA isolation and reverse-transcription polymerase chain reaction (RT-PCR)
Total RNA was isolated using the RNeasy spin-column method (Qiagen, Crawley, UK). RNA was quantified by assessing optical density at 260 and 280 nm. The isolates were stabilised using RNasin RNase inhibitor (Promega) and stored at −80°C. RT-PCR was performed using the Qiagen One-Step RT-PCR kit. All primers were designed using the OligoPerfect primer designer (Invitrogen) (see Table 1). RT was performed at 50°C for 30 min and Taq polymerase activated at 95°C for 15 min. The product was then subject to 40 rounds of three step cycling, consisting of denaturation (94°C for 45 s), annealing (58°C for 45 s) and extension (72°C for 1 min). The final elongation step was performed at 72°C for 10 min. The products were stored at 4°C, until required. β-Actin or GAPDH was used as a positive control and the negative control consisted of the PCR reaction without the addition of the template. Products were separated by electrophoresis on a 0.8% agarose gel (containing 0.01% ethidium bromide) in Tris-borate-EDTA buffer. The products were visualised using a UV transilluminator (UVP Bio-Doc-It system). Bands were cut out of the gel and the products extracted using Qiagen gel extraction kit, and sequenced by the John Innes Genome Laboratory (Norwich Research Park, Norwich, UK).
Table 1.
Bestrophin and control primer sequences used in the RT-PCR amplification reactions
| Gene | Sense (5′–3′) | Antisense (5′–3′) | Expected product size (bp) | GenBank accession |
|---|---|---|---|---|
| hBest1 5′ | 304-GTATTGCGACAGCTACATCCAG-327 | 794-ATCCAGTCGTAGGCATACAGGT-771 | 488 | NM 004183 |
| hBest1 mid | 771-ACCTGTATGCCTACGACTGGAT-794 | 1233-CTGGAACTCCATCTCCTCTTTG-1210 | 460 | NM 004183 |
| hBest1 3′ | 1105-GGACATGTACTGGAATAAGCCC-1128 | 1514-GTGTCTGGGGCACTGTAGTC-1493 | 407 | NM 004183 |
| hBest2 | 443-AGTTTGAAAACCTGAACTCATCCTA-469 | 865-AGAAAGTTGGTCTCAAAGTCATCAT-839 | 420 | NM 017682 |
| hBest3 | 833-ACAAGTGACAGCTCCATGTTCTTAC-859 | 1350-TTTTAGAAAGGTATCACCAGGGTCT-1324 | 515 | NM 032735 |
| hBest4 | 957-TTTGAGACAAATCAGCTCATAGACC-983 | 1568-TCTTCTCTTTCAAGTTCTGTCCCTA-1544 | 609 | NM 153274 |
| β-Actin | 260-GCATCCTCACCCTGAAGTAC-281 | 717-TTCTCCTTAATGTCACGCAC-697 | 455 | NM 001101 |
| GAPDH | 627-ACCACAGTCCATGCCATCAC-648 | 1080-TCCACCACCCTGTTGCTGTA-1059 | 451 | NM 002046 |
PCR primers for hBest1, 2, 3, 4, β-actin and GAPDH that have been used in this study. Primers were designed using the OligoPerfect™ primer design programme by Invitrogen.
Western blot analysis of protein
Protein was isolated from CFPAC-1 cells and ARPE-19 cells using lysis buffer containing 150 mm NaCl, 50 mm Tris, 1.5 mm MgCl2, 10% glycerol and 1% Triton X-100 (pH 7.5) supplemented with complete protease inhibitor (Roche) and protein phosphatase inhibitors (Calbiochem). Protein concentration was determined using the Bradford assay. Protein was separated on a 10% SDS–polyacrylamide gel, transferred to PVDF membrane, which was blocked with 5% low fat milk and incubated with primary antibody overnight at 4°C. Full details of all the primary antibodies used in this study are listed in Table 2. The membrane was incubated with secondary antibody (HRP-conjugated Affinipure goat anti-rabbit or goat anti-mouse, Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) for 2 h at room temperature and the HRP activated using SuperSignal Pico substrate (Pierce Biotechnology, Inc., Rockford, IL, USA). Protein was detected on radiography film using a Xograph system (Xograph Healthcare, Tetbury, UK, compact 4). Membranes were stripped by washing with water for 5 min, incubating with 0.2 m NaOH for 5 min, washing with water for 5 min, then blocking with milk for 1 h. All subsequent stages were carried out as described above. Peptide block was carried out by incubating the primary antibody overnight in a 20× excess of blocking peptide at 4°C. The mixture was applied to the Western blot as per the primary antibody and the membrane processed as normal. The difference in band intensity was quantified using Volocity software (Improvision, Coventry, UK) and analysed using GraphPad Prism (GraphPad Software Inc., San Diego, CA, USA) using β-actin as the standard for each blot.
Table 2.
Antibodies used in Western blotting and confocal microscopy analysis
| Antibody | Supplier | Animal raised in | Target/Function | Peptide location | Sequence antibody raised against |
|---|---|---|---|---|---|
| Bst112 | Fg | RP | hBest1 | aa 445–464 | WKLKAVDAFKSAPLYQRPGY |
| Bst121 | Fg | RP | hBest1 | aa 566–585 | TLKDHMDPYWALENRDEAHS |
| BstMM | Ab | MM | hBest1 | aa 568–585 | KDHMDPYWALENRDEAHS |
| β-Actin | Ab | RP | β-actin | — | Target sequence between aa 1–100 |
| Pan cadherin | Ab | MM | Pan cadherin/plasma membrane marker | aa 883–902 | DYDYLNDWGPRFKKLADMYGGGDD |
| Calnexin | Ab | MM | Calnexin ER marker | — | Target sequence not divulged by supplier |
| GM130 | Ab | MM | Golgi marker | — | Target sequence not divulged by supplier |
| Clathrin | Ab | MM | Vesicular marker | — | Target sequence not divulged by supplier |
| LAMP-1 | Ab | MM | Lysosomal marker | — | Target sequence not divulged by supplier |
| ZO-1 | BD | MM | Tight junction marker | aa 1048–1247 | Target sequence not divulged by supplier |
Ab, Abcam; Fg, Fabgennix; BD, BD Biosciences; RP, rabbit polyclonal; MM, mouse monoclonal.
Biotinylation
Cell surface protein was separated using a Cell Surface Protein Isolation Kit (Pierce) according to the manufacturer's instructions. Briefly, CFPAC-1 cells were treated with biotin solution for 30 min, the solution quenched and the cells scraped from the base of the flask and centrifuged at 604g for 3 min. The cells were washed with Tris buffered saline (TBS) then treated with 500 μl lysis buffer for 30 min. The lysate was centrifuged (15800g for 2 min at 4°C) and the supernatant applied to an avidin column and incubated at 24°C for 1 h. The column was washed four times with wash buffer and the protein collected using SDS sample buffer. The cell surface proteins were then subject to Western blotting as for whole cell protein.
Immunocytochemistry
Cells were fixed for 15 min with 4% paraformaldehyde. The cells were incubated with primary antibody (see Table 2– made up in 1% BSA–PBS) at room temperature for 1 h. The cells were then incubated with secondary antibody (Alexafluor 488 goat anti-rabbit or Alexafluor 688 goat anti-mouse were purchased from Molecular Probes/Invitrogen, UK) for 45 min at room temperature. Cells were treated with 4 μg ml−1 DAPI for 3 min and mounted onto slides using Hydromount treated with 1,4-diazabicyclo(2,2,2)octane (DABCO). The slides were visualised with a Zeiss LSM 510 Meta confocal microscope (Henry Wellcome Laboratory for Cell Imaging, University of East Anglia, Norwich). Double immunostaining to determine colocalisation involved the incubation of a second set of primary and secondary antibodies, which were applied as for the first set of antibodies. Colocalisation between two proteins was determined using Volocity colocalisation software (Improvision).
Cell surface immunostaining
Live cells were washed in PBS and treated with primary antibody (mouse monoclonal, BstMM) for 30 min at 24°C, and then subject to a gentle fix with 0.5% paraformaldehyde for 2 min to stabilise the cells. Cells were treated with the secondary antibody (Alexofluor 488 goat anti-mouse) for 30 min then fixed with 4% paraformaldehyde for 15 min. Cells were analysed using the confocal microscope.
SiRNA-mediated gene knockdown
SiRNAs designed to knockdown hBest1 were purchased from Ambion (Austin, TX, USA). The Qiagen siRNA starter kit contained negative and positive siRNA controls. The negative control was a non-silencing siRNA with no known homology to mammalian genes. The positive control was a siRNA targeting MAPK1. The manufacture states that both control siRNAs have been thoroughly tested and validated; however, we also validated the control siRNAs by real-time quantitative PCR and Western blot analysis (see Results). SiRNAs were transfected into CFPAC-1 cells using the Amaxa electroporation method (Amaxa Inc., Walkersville, MD, USA). 1 × 106 CFPAC-1 cells were used per reaction. The cells were mixed with 100 μl nucleofector V solution and 30 nM siRNA and subject to electroporation using the A-33 programme. Cells were gently mixed with culture medium and seeded in six-well plates. RNA was isolated from the cells using the RNeasy mini spin column (Qiagen) method and subject to reverse transcription using TaqMan RT-PCR kit (Applied Biosystems, Foster City, CA, USA). Real time PCR was carried out using primers specific for hBest1, MAPK (positive control) and GAPDH (negative control). Sybr Green Jumpstart Taq Readymix (Sigma) was used to perform real time PCR analysis using the following programme: 95°C for 2 min, followed by 40 rounds of 95°C for 15 s and 60°C for 1 min (iQ5 thermocycler, Bio-Rad). Results were analysed using a Microsoft Excel spreadsheet kindly provided by Dr Stuart Rushworth, University of East Anglia, Norwich. Protein knockdown was assessed using Western blot as described in the previous section. The amount of protein knockdown was determined using Volocity software to quantify the reduction of band intensity in siRNA treated samples.
Statistics
Values are given as means ±s.e.m., n represents the number of experimental replicates. Iodide efflux data was statistically analysed using the two-way ANOVA with Bonferroni's post hoc test and significance was taken as P≤ 0.05. Real time PCR results were analysed using Student's one-tailed paired t test. The level of significance is indicated thus: *P < 0.05, **P < 0.01 and ***P < 0.001. Analysis was performed using GraphPad Prism version 4.00 for Windows package.
Results
Iodide efflux analysis of native CaCCs in CFPAC-1 cells
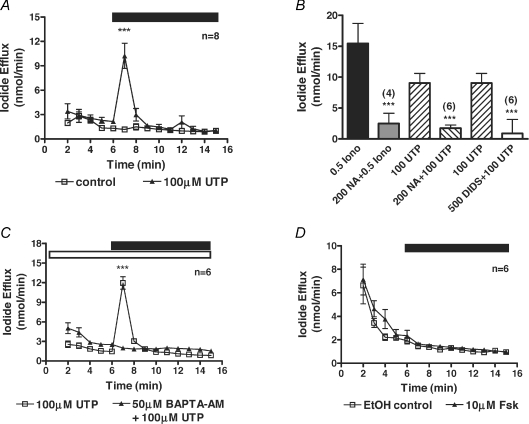
Initially, using the patch-clamp technique, we attempted to record whole-cell currents from CFPAC-1 cells. Despite repeated attempts we were only able to obtain nine seals out of 129 attempts and could not achieve any stable whole-cell recordings. We therefore used the iodide efflux assay to evaluate native CaCCs in CFPAC-1 cells. Iodide efflux from CFPAC-1 cells increased 1 min after addition of 100 μm UTP from 1.2 ± 0.1 nmol min−1 to 10.2 ± 1.5 nmol min−1 (n= 8; P < 0.001) (Fig. 1A). Iodide efflux was also induced by ATP (100 μm) from 2.2 ± 0.2 nmol min−1 to 11. ± 2.4 nmol min−1 (n= 8; P < 0.001) and the calcium ionophore, ionomycin (2 μm) from 2.4 ± 0.4 nmol min−1 to 24.9 ± 2.5 nmol min−1 (n= 4; P < 0.001). In order to characterise the UTP-mediated stimulation of iodide efflux, CFPAC-1 cells were pretreated with the generic chloride channel blockers NA and DIDS before UTP stimulation was repeated. NA (200 μm) and DIDS (500 μm) reduced the UTP-mediated response from 9.1 ± 1.6 nmol min−1 (n= 8) to 1.8 ± 0.5 nmol min−1 (81% inhibition; n= 6; P < 0.001) and 0.9 ± 2.3 nmol min−1 (90% inhibition; n= 6; P < 0.001), respectively (Fig. 1B). To determine whether the UTP-stimulated iodide efflux from CFPAC-1 cells was Ca2+ dependent, the calcium chelator BAPTA-AM was used. BAPTA-AM (50 μm) completely blocked the UTP-stimulated iodide efflux from 11.9 ± 0.9 nmol min−1 to 1.9 ± 0.1 nmol min−1 (Fig. 1C, n= 6, P < 0.001). As our experiments were performed at room temperature, it was feasible that some CFTR protein might be delivered to the cell surface because F508del-CFTR is a temperature-sensitive mutation (Denning et al. 1992). To test whether CFTR was being functionally expressed in CFPAC-1 cells, 10 μm forskolin was used to stimulate the cells. Forskolin did not elicit any response from CFPAC-1 cells (1.7 ± 0.2 nmol min−1) and iodide efflux values were not significantly different from the vehicle control (1.5 ± 0.2 nmol min−1, Fig. 1D, n= 6, P > 0.05). Taken together these data confirm the presence of a native CaCC in CFPAC-1 cells.
Figure 1. Characterisation of native CaCCs in CFPAC-1 cells by iodide efflux.
A, mean data showing the increase in iodide efflux on addition of 100 μm UTP. B, summary data for the inhibition of either the ionomycin-induced (0.5 μm) or UTP-induced (100 μm) iodide effluxes with the chloride channel blockers niflumic acid (NA, 200 μm) and DIDS (500 μm). C, mean data showing the inhibition in the 100 μm UTP-induced iodide efflux by 50 μm BAPTA-AM. D, mean data showing the lack of stimulation of iodide efflux on addition of 10 μm forskolin. During the periods indicated by the filled bar either 100 μm UTP or 10 μm forskolin was added to the efflux buffer. The open bar indicates the pre-treatment of cells with 50 μm BAPTA-AM during their loading with iodide and the compound's presence in the efflux buffer. Control traces represent the addition to the efflux buffer of either water in the UTP experiments, or ethanol in the forskolin experiments. Symbols and error bars are means ±s.e.m. (n values indicated on figure) for each condition. Where not shown, error bars are smaller than symbol size. ***Significantly different from control (P < 0.001).
RNA expression of hBest1, 2, 3 and 4 in CFPAC-1 cells
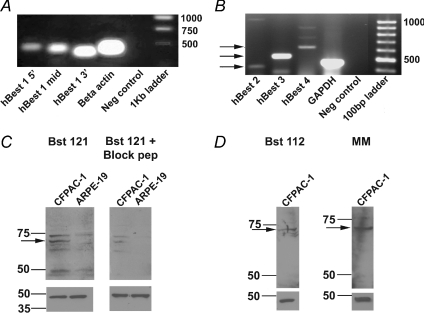
Using RT-PCR, we then investigated which bestrophin genes were present in CFPAC-1 cells. Primers for hBest1, 2, 3 and 4 were constructed (see Table 1) and used in RT-PCR reactions with RNA isolated from CFPAC-1 cells. Initially, RT-PCR was performed on CFPAC-1 RNA using three different primer pairs from the 5′, middle, and 3′ end of the hBest1 sequence (Fig. 2A, n= 4). A single band was visible for hBest1 5′ (Fig. 2A, first lane), middle (Fig. 2A, second lane) and 3′ (Fig. 2A, third lane) primers at 488, 460 and 407 base pairs, respectively. The size of these products was as predicted from the primer design. Sequence analysis of all three products demonstrated 100% sequence homology to the published hBest1 cDNA sequence. β-Actin primers (Fig. 2A, fourth lane) demonstrated a product at approximately 455 base pairs, as predicted by primer design. No product was visible in the negative control sample (Fig. 2A, fifth lane). RT-PCR reactions involving primers constructed to hBest2, 3 and 4 sequences also resulted in bands at 420, 515 and 609 base pairs, respectively (Fig. 2B, lanes 1–3 respectively, n= 3). Again these products matched the predicted size for the primer design and sequence analysis of all three products demonstrated 100% sequence homology to the published hBest2, 3 and 4 cDNA sequences. The positive control in this set of experiments was primers designed to GAPDH (Fig. 2B, lane 4). No product was visible in the negative control sample (Fig. 2B, lane 5). These results indicate that hBest1, 2, 3 and 4 RNA are present in CFPAC-1 cells.
Figure 2. RT-PCR analysis of bestrophin gene expression and Western blot detection of hBest1 in CFPAC-1 cells.
A, RT-PCR (n= 4) showing the presence of hBest1 in CFPAC-1 cells as indicated by predicted product sizes (see Table 1) for primers representing the 5′, middle and 3′ regions of hBest1 nucleotide sequence (Lanes 1–3). β-Actin primers were used as a positive control (Lane 4) and no template RNA was added to the negative control (Lane 5). A 1 kb ladder was used to estimate the products' size (Lane 6). B, RT-PCR (n= 3) showing the presence of hBest2, 3 and 4 in CFPAC-1 cells as indicated by predicted product sizes (shown by arrows and see Table 1) for primers representing hBest2, hBest3 and hBest4 (Lanes 1–3). GAPDH primers were used as a positive control (Lane 4) and no template RNA was added to the negative control (Lane 5). A 100 bp ladder was used to estimate the products' size (Lane 6). C, detection of hBest1 in CFPAC-1 and ARPE-19 cells using a rabbit polyclonal antibody, Bst121 (1 : 750 dilution, Fabgennix; n= 3) to the hBest1 amino acid sequence (see Table 2 for details). A band at 68 kDa (indicated by arrow on upper left panel) corresponds to the hBest1 protein. A blocking peptide (Fabgennix) was added to the Bst121 antibody from CFPAC-1 cells and from ARPE-19 cells (upper right panel). D, detection of hBest1 in CFPAC-1 cells using a different rabbit polyclonal antibody, Bst112 (1 : 1000 dilution, Fabgennix; n= 3). A band at 68 kDa (indicated by arrow on upper left panel) corresponds to the hBest1 protein. A band at 68 kDa (indicated by arrow on upper right panel) was also detected using a mouse monoclonal antibody, BstMM (1 : 500 dilution, Fabgennix; n= 2) to hBest1. A β-actin antibody (1 : 10000 dilution, Abcam) was used as a loading control for all blots in C and D and is shown in the lower panels.
Detection of hBest1 protein in CFPAC-1 cells
We then went on to determine whether hBest1 was expressed at the protein level in CFPAC-1 cells. Immunoblots were carried out on protein isolated from CFPAC-1 cells and retinal pigment epithelial (ARPE-19) cells. We used retinal pigmented epithelial cells as a control for these experiments as hBest1 was originally identified in these cells. Experiments were carried out using commercially available anti-hBest1 antibodies. Initially, Western blots were probed with the commercially available Bst121 antibody (FabGennix). This antibody has a blocking peptide available for purchase. Blots were incubated with either the Bst121 antibody (FabGennix) or Bst121 antibody pre-incubated with the blocking peptide. According to the company, this antibody should recognise a 68 kDa band. A 68 kDa band is evident in CFPAC-1 and ARPE-19 cell lysates (Fig. 2C upper left panel, indicated by the arrow, n= 3) although the band is much fainter in the ARPE-19 sample indicating that hBest1 expression in these cells is reduced in comparison to CFPAC-1 cells. Other bands were also observed with the use of Bst121, around 72, 66 and 63 kDa. Inclusion with the Bst121 antibody of the peptide against which the antibody was made reduced the intensity of the 68 kDa band on the immunoblot of the CFPAC-1 cells by 69% and completely blocked the immunoblot of the ARPE-19 cells (Fig. 2C, upper right panel). The lower bands may be degradation products of the protein, while the higher band may be a splice variant of hBest1 which has been reported in previous studies (Wistow et al. 2002); however, we have not investigated this further at the present time. We used two further commercially available antibodies. Blots were incubated with a rabbit polyclonal, Bst-112 (Fig. 2D, upper left panel, n= 3) or a mouse monoclonal to hBest1 (Fig. 2D, upper right panel, n= 2). Consistent with the expected specificity of these antibodies, a 68 kDa band was detect by both antibodies (Fig. 2D). The immunoblots were stripped and reprobed using anti-β-actin antibody as a loading control. These immunoblots demonstrated an intense band at the correct weight of 43 kDa, which was equally intense in all of the protein samples loaded (Fig. 2C and D, lower panels). Taken together these results indicate that hBest1 protein is expressed in CFPAC-1 cells.
Cellular localisation of hBest1 in CFPAC-1 cells
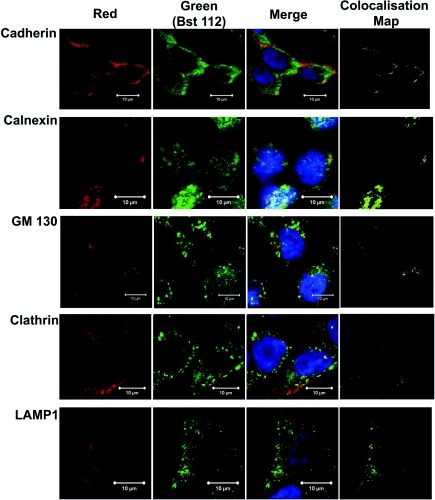
To further investigate the cellular expression of hBest1 in CFPAC-1 cells, colocalisation confocal imaging with the rabbit polyclonal antibody Bst112 (FabGennix) and a number of different cellular compartment marker antibodies was carried out (see Table 2 for details of antibodies used). As hBest1 is thought to represent a CaCC, it should be expressed on the cell surface. Initially, the expression of the cell membrane marker pan-cadherin (Fig. 3, 1st column, red, n= 3) and hBest1 (Fig. 3, 2nd column, green, n= 3) was studied (Fig. 3). The experiment demonstrated that hBest1 and cadherin colocalise on the cell surface (Fig. 3, 3rd column). To confirm that expression was colocalised, we performed a more detailed analysis of the merged image using the Volocity program and produced a colocalisation map (Fig. 3, 4th column). Analysis of the membrane area demonstrated that hBest1 colocalised with cadherin with a colocalisation coefficient of 0.97 ± 0.02 (n= 3). Further analysis also demonstrated that the majority of hBest1 was located in the cytoplasm with only 23.1 ± 2.6% hBest1 appearing on the cell surface. To begin to understand how hBest1 is processed through the cell and which intracellular compartments it may be associated with, hBest1 (Fig. 3, 2nd column, green) was also visualised alongside several intracellular markers including calnexin (n= 2), an ER marker; GM130 (n= 3), a Golgi marker; clathrin (n= 2), which labels clathrin coated vesicles; and LAMP-1 (n= 2), a lysosomal vesicle marker (all Fig. 3, 1st column, red). hBest1 was demonstrated in the ER, Golgi and lysosomal vesicles (Fig. 3, 3rd and 4th columns), but not clathrin-coated vesicles (Fig. 3, 3rd and 4th columns). There was no background fluorescence in any of the samples processed (data not shown). Taken together the data suggest that hBest1 is expressed in the cell membrane and specific cytoplasmic domains and during its biosynthesis follows the classical secretory pathway.
Figure 3. Confocal imaging of hBest1 expression and co-localisation with specific intracellular markers in CFPAC-1 cells.
The first column panels show expression of specific intracellular markers (red), cadherin (1 : 100 dilution; n= 3), calnexin (1 : 100 dilution; n= 2), GM130 (1 : 100 dilution; n= 3), Clathrin (1:100 dilution; n= 2), LAMP 1 (1:100 dilution; n= 2). The second column panels show expression of hBest1 (green) as determined using the Bst112 polyclonal antibody (1:200 dilution). The third column panels show the merged image of the first and second column panels. The fourth column panels are colocalisation maps created by the Volocity program. In all images hBest1 is fluorescently labelled with anti-rabbit Alexafluor 488 (green) and all specific membrane markers fluorescently labelled with anti-mouse Alexafluor 688 (red) and the nucleus of the cell labelled with DAPI (blue). Co-localisation is indicated by the yellow colour in the third column panels and the colocalisation maps of the fourth column panels. Scale bar given in each image corresponds to 10 μm. No background staining or autofluorescence was observed in CFPAC-1 cells when primary or secondary antibody was omitted (data not shown).
Further analysis of cell surface localisation of hBest1
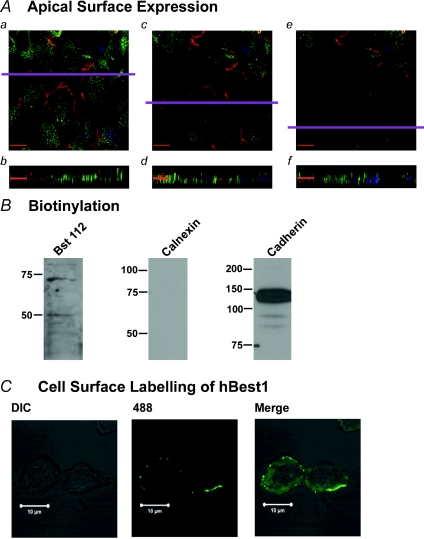
The above data suggest that hBest1 was expressed in the plasma membrane; however, they were obtained from CFPAC-1 cells that were not necessarily polarised as we did not grow them to full confluency. We therefore attempted to determine whether hBest1 is expressed on the apical or basolateral membranes of CFPAC-1 cells by growing the cells to confluency on glass coverslips (n= 2). Figure 4Aa, c and e shows sequential sections in the X–Y plane taken 0.43 μm apart through the cell at the apical region of the cell moving from the middle (1.29 μm; Fig. 4Aa) to the top of the cell (0.43 μm; Fig. 4Ae). A mouse monoclonal antibody to ZO-1 (BD Biosciences) was used to demonstrate that the cells formed tight junctions and that they were polarised. We also labelled hBest1 using the Bst112 antibody. ZO-1 labelling (red) is localised to the plasma membrane and showed a characteristic ‘chicken wire’ pattern at the apicolateral border of the cells, consistent with its expression at the tight junctions. The X–Y slices also show faint labelling of hBest1 (green) at the apical region of the plasma membrane of CFPAC-1 cells as evidenced by visualisation in the same plane as ZO-1. The expression of hBest1 is not uniform. Figure 4Ab, d and f shows X–Z profiles through several polarised cells (obtained in the plane indicated by the purple line on the X–Y sections), moving from the basolateral membrane at the bottom of the panel to the apical membrane at the top of the panel. These images show that although the majority of hBest1 appears to be located in the cytoplasm of the cells, a small fraction of hBest1 is located at the apical region of the plasma membrane. To confirm hBest1 expression in the plasma membrane we attempted to isolate the cell surface proteins from confluent CFPAC-1 cells by biotinylating these proteins and then isolating the cell surface fraction. This cell surface fraction was then immunoblotted for hBest1 using the rabbit polyclonal antibody, Bst112 (Fig. 4B, 1st column, n= 2). A 68 kDa band was demonstrated in the biotinylated protein isolates probed with Bst112, similar to previous results (see Fig. 2D) and representing hBest1. A lower band at approximately 50 kDa was also visible. In order to demonstrate that the biotinylated fraction contained only cell surface proteins we performed several immunoblot controls. The calnexin antibody was used as a negative control. This is an intracellular protein and should not be present in the cell surface fraction. The calnexin antibody did not detect any calnexin protein in the immunoblot (Fig. 4B, 2nd column, n= 2). The pan-cadherin antibody was used as a positive control. Cadherin is a cell surface protein and should be present in the cell surface fraction. Immunoblotting the cell surface fraction for cadherin detected an intense band at around 135 kDa (Fig. 4B, 3rd column, n= 2) the expected band size for this protein. We also performed some live cell labelling experiments. The mouse monoclonal antibody, BstMM, was incubated with CFPAC-1 cells and then viewed under confocal microscopy. hBest1 membrane expression was confirmed (Fig. 4C, n= 2), showing fluorescence on the outer surface of CFPAC-1 cell membrane. Taken together these data demonstrate that hBest1 is expressed predominantly in the cytoplasm of the CFPAC-1 cells but that there is also a small fraction that is expressed at the plasma membrane.
Figure 4. Cell surface expression of hBest1 in CFPAC-1 cells.
A, a series of confocal image sections taken 0.43 μm apart near the apical surface of the cell. In all images hBest1 is fluorescently labelled with anti-rabbit Alexafluor 488 (green) and ZO-1 (1 : 100 dilution; n= 2) is labelled with anti-mouse Alexafluor 688 (red) and the nucleus of the cell is labelled with DAPI (blue). a, c, e, X–Y sections moving from the tight junction level to top of cell, showing characteristic chicken wire pattern of ZO-1 labelling. b, d, f, X–Z sections obtained in the plane indicated by the purple line in the X–Y sections. Scale bar given in each image corresponds to 12 μm. B, CFPAC-1 cell surface protein was isolated by biotinylation. Bst112 (1 : 1000; n= 2) detected a single band at around 68 kDa demonstrating presence of hBest1 in cell surface fraction. Antibodies against the ER protein, calnexin (1 : 1000; n= 2), and the plasma membrane marker, cadherin (1 : 1000; n= 2), demonstrated the absence of intracellular protein in the isolate and confirmed that the isolate was the plasma membrane fraction. C, cell surface labelling of hBest1 using a mouse monoclonal antibody to hBest1 (BstMM). The antibody was used to live-stain CFPAC-1 cells, which were subsequently fixed and analysed using confocal microscopy (n= 2).
Knockdown of hBest1 demonstrates the CaCC activity of hBest1
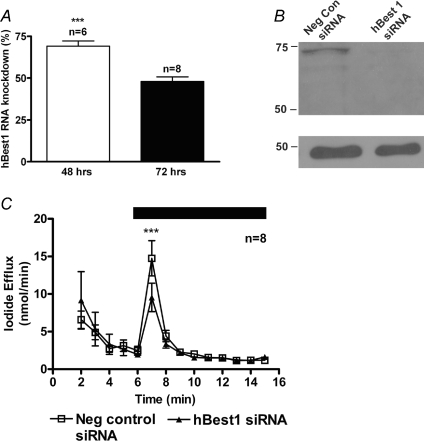
To investigate whether hBest1 is involved in the calcium-activated iodide efflux of CFPAC-1 cells, we used the small interfering RNA (siRNA) approach to knockdown hBest1 expression. Optimisation of hBest1 knockdown using single primer sets from Ambion (referred to as 03, 04 and 05) demonstrated an inconsistent level of knockdown for hBest1 in CFPAC-1 cells (data not shown). However, when two of the primer sets were pooled (referred to as 0305), a consistent level of knockdown was achieved. Real time PCR was utilised to demonstrate that the siRNAs did produce knockdown of hBest1 and the positive control, MAPK1. After transfection of the CFPAC-1 cells with siRNA for MAPK1, real time PCR showed that the MAPK1 gene expression was inhibited by 84 ± 2% and 78 ± 6% (n= 3) at 24 h and 48 h, respectively. After transfection of the siRNAs for hBest1, real time PCR demonstrated a gene knockdown of 69 ± 3% (n= 6) at 48 h, which was reduced to 48 ± 3% (n= 8) by 72 h (Fig. 5A). HBest1 gene expression was not affected by 24 h after transfection (data not shown). Western blot analysis of hBest1 expression using the Bst112 antibody in the hBest1 siRNA treated cells demonstrated a 79 ± 9% (n= 2) reduction in band intensity in the knockdown protein samples by 72 h after transfection (Fig. 5B). This was the optimal time frame for the knockdown of the hBest1 protein. Iodide efflux was carried out 72 h after transfection of pooled 0305 hBest1 siRNAs in CFPAC-1 cells (Fig. 5C). The iodide efflux response to 100μM UTP was reduced from 14.8 ± 2.3 nmol min−1 in the negative control siRNA transfected cells to 9.5 ± 1.9 nmol min−1 in hBest1 siRNA-treated cells, a reduction of almost 40% (n= 8, P < 0.001). Taken together these data suggest that hBest1 is involved in the calcium-activated iodide efflux from CFPAC-1 cells.
Figure 5. siRNA against hBest1 reduces calcium-activated chloride conductance in CFPAC-1 cells.
A, siRNA 03 and 05 (30 nm) were simultaneously transfected into CFPAC-1 cells using electroporation (Amaxa). Real time PCR analysis of isolated RNA demonstrated knockdown of 69 ± 3% (n= 6) at 48 h, which was reduced to 48 ± 3% (n= 8) by 72 h. B, Western blot analysis of extracted protein at 72 h after siRNA 03 and 05 simultaneous transfection showed that hBest1 protein had a considerably reduced band intensity of 79 ± 9% (n= 2). Detection of hBest1 in CFPAC-1 cells was performed using the rabbit polyclonal antibody, Bst112 antibody (1 : 750 dilution, Fabgennix). A band at 68 kDa corresponds to the hBest1 protein. A β-actin antibody (Abcam) was used as a loading control and is shown in the lower panels. C, iodide efflux analysis of CFPAC-1 cells after transfection with pooled 03 and 05 siRNA (30 nm; n= 8). Negative control siRNA was used to standardise the response (30 nm; n= 8). During the period indicated by the filled bar 100 μm UTP was added to the efflux buffer. Symbols and error bars are means ±s.e.m. (n values indicated on figure) for each condition. Where not shown, error bars are smaller than symbol size. ***Significantly different from control (P < 0.001).
Discussion
As far as we are aware, this is the first description of the expression of bestrophin genes in a human pancreatic duct cell line. The expression of hBest1 in CFPAC-1 cells can be drawn from three pieces of evidence. Firstly, three areas of hBest1 mRNA sequence were successfully amplified and fully sequenced. These areas correspond to the 5′ region (base pairs 289–777), middle region (base pairs 756–1216) and 3′ region (base pairs 1090–1499) of the hBest1 mRNA sequence. The sequence products shared 100% sequence identity to that published in GenBank. Western blot analysis and confocal microscopy have demonstrated the expression of hBest1 protein in CFPAC-1 cells, and further investigation has shown hBest1 expression on the cell surface. Finally, using gene silencing techniques, we have also provided evidence that hBest1 is involved in CaCC conductance in CFPAC-1 cells as evidenced by a reduction in UTP-stimulated iodide efflux from these cells.
The role of hBest1 in retinal pigment epithelial (RPE) cells has been well defined. It is generally thought that a light peak substance (possibly ATP) is released from photoreceptors in the light and diffuses to the RPE cells where it activates P2Y receptors that lead to generation of inositol-1,4,5-trisphosphate (IP3). This increase in IP3 leads to an increase in intracellular calcium and activation of hBest1 chloride channels on the basolateral membrane. The activation of these channels leads to membrane depolarisation, which is recorded as the light peak (Hartzell et al. 2008). A defining feature of Best macular dystrophy (BMD) is an abnormal electro-oculogram caused by a defective hBest1 chloride channel.
Until recently, bestrophin expression had only been shown to occur in RPE cells (Petrukhin et al. 1998). Duta et al. (2004) were the first to provide evidence that bestrophins may be expressed in epithelial tissues, when they demonstrated expression of hBest1 in Calu-3 cells (Duta et al. 2004). Bestrophin expression has now been reported in several epithelial tissues including mouse trachea, colon and kidney and human airway cells (Barro Soria et al. 2006). This latter study (Barro Soria et al. 2006) showed that the expression of Best1 and 4 correlated well with the functional expression of CaCCs in these tissues. Silencing of bestrophin genes in these tissues led to a diminished CaCC-mediated response, although the authors argued that this did not implicate fully the role of bestrophin as a channel but could have also indicated the role of bestrophin as a regulator of an endogenous channel. Our study provides further evidence for the notion that bestrophins represent CaCC. We have also used gene silencing as a technique for studying the relationship between bestrophin expression and CaCC activity in the pancreatic duct cell line, CFPAC-1 cells. Our data illustrate that knockdown of the hBest1 gene in CFPAC-1 cells led to a 40% decrease in iodide efflux stimulated by the addition of UTP. We did not see 100% inhibition of the iodide efflux. The lack of complete inhibition of iodide efflux may be due to incomplete knockdown of the hBest1 gene and as a result incomplete knockdown of the hBest1 protein (as indicated in Fig. 5). However, given that we have also shown that hBest2, 3 and 4 are present in CFPAC-1 cells and that others (Tsunenari et al. 2003, 2006) have shown that hBest2 and hBest4 when expressed in HEK293 cells generate chloride conductances, our data may be interpreted to suggest that the CaCC-mediated iodide efflux in CFPAC-1 cells is mediated by another bestrophin protein in addition to hBest1. This hypothesis is supported by very recent data which show that both mBest1 and mBest2 are involved in the CaCC from mouse tracheal epithelial cells (Barro-Soria et al. 2008). The data are, however, also consistent with hBest1 representing a regulatory component for the CaCC. As a regulator, the level of hBest1 gene knockdown may allow some CaCC function to remain. Given the earlier studies on mBest2 and chloride channel function, there is a strong body of evidence to suggest that bestrophins represent chloride channel proteins (Qu et al. 2004; Qu & Hartzell, 2004) and we suspect that the incomplete knockdown may be due to other hBests contributing to the CaCC.
However, the very recent reports (Caputo et al. 2008; Schroeder et al. 2008; Yang et al. 2008) of the identification of anoctamin 1 (ANO-1), also known as TMEM16A, protein as a calcium-activated chloride channel does complicate the interpretation of the role of hBest1 in pancreatic duct cells. The report by Caputo et al. 2008 used CFPAC-1 cells to knockdown the ANO-1 (TMEM16A) protein using interfering RNA (siRNA). ANO-1 (TMEM16A) is therefore expressed in CFPAC-1 cells. The data from the above groups (Caputo et al. 2008; Schroeder et al. 2008; Yang et al. 2008) suggesting that ANO-1 is a CaCC are very compelling. It is interesting to note that Caputo et al. (2008) report that cells transfected with siRNA against TMEM16A show a 60–70% reduction in Ca2+-dependent I− influx compared to control siRNA transfected cells. Our data showed an approximately 40% reduction in I− efflux from hBest1 siRNA transfected CFPAC-1 cells. This could suggest that there may be several different kinds of CaCC and that hANO-1 and hBest1 are different CaCCs which are both expressed in CFPAC-1 cells and both contribute to the calcium activated chloride conductance in these cells.
As bestrophins are thought to represent CaCCs, it is presumed that bestrophin protein should be expressed on the cell surface. In our present study we have investigated the cell surface expression of bestrophin by using a number of experimental techniques including confocal microscopy, biotinylation of surface facing plasma membrane proteins and a live cell surface-labelling protocol. The confocal microscopy of the CFPAC-1 cells in the X–Y and X–Z planes, labelled with ZO-1 and hBest1, shows that the CFPAC-1 cells do form polarised monolayers. A characteristic ‘chicken wire’ labelling pattern was seen which is in agreement with similar studies of cell polarisation in another pancreatic cell line, HPAF-II (Rajasekaran et al. 2004). Our data (see Figs 3 and 4) also show that a small fraction of hBest1 is expressed in the apical region of the cells. Further analysis showed that the majority of hBest 1 was expressed in an intracellular location in CFPAC-1 cells with only 23% appearing on the cell surface. The gross expression of bestrophin in the cytoplasm of the cell has also been documented in several other studies; however the majority of these studies used cells overexpressing the bestrophin protein (Tsunenari et al. 2003; Fischmeister & Hartzell, 2005). The ratio of cytoplasmic to membrane-localised hBest1 was quantified in one of these investigations: Fischmeister & Hartzell (2005) reported that ∼12% of hBest1 was found on the cell surface and the remainder in the intracellular compartment. By contrast, our data suggest that in CFPAC-1 cells almost twice as much hBest1 is on the cell surface compared with recombinant cells. Taken together these experiments show that bestrophin is expressed at the apical region of the plasma membrane, but that the expression is not continuous around the membrane perimeter and tends to be clustered.
Several studies have reported the intracellular localisation of bestrophins (Sun et al. 2002; Tsunenari et al. 2003; Qu et al. 2004; Kunzelmann et al. 2007), although few have attempted to identify which compartments bestrophin may be expressed in. In our present study we have used confocal imaging and dual labelling of hBest1 and several intracellular compartment markers to address this in CFPAC-1 cells. Our data show hBest1 is associated with the endoplasmic reticulum, Golgi apparatus and lysosomal vesicles. This is consistent with hBest1's involvement in a regulated secretory pathway that could be upregulated by stimulation of CFPAC-1 cells by secretagogues, leading to an increase in the CaCC conductance. However, other groups have presented evidence to suggest that this is not the case at least in hBest1 expressing HEK293 cells and mouse M1 kidney cells (Kunzelmann et al. 2007). Given the colocalisation of hBest1 and lysosomal vesicles in CFPAC-1 cells, it is also interesting to speculate that hBest1 may be important in lysosomal function in CFPAC-1 cells, however this is again in contrast to the only other group that has investigated the expression of hBest1 and LAMP-1 and reported a lack of co-localisation between LAMP-1 and hBest1 in their study carried out in hBest1-expressing M1 cells (Kunzelmann et al. 2007). We demonstrate little association of hBest1 with clathrin coated vesicles in CFPAC-1 cells. We have not yet studied the association of hBest1 with other species of vesicle in detail.
Pancreatic duct cells produce a bicarbonate rich fluid. At the centre of this bicarbonate production on the apical membrane is the CFTR chloride channel and an anion exchanger. This anion exchanger has been proposed to be a member of the SLC26 family of proteins (Mount & Romero, 2004). In CFPAC-1 cells, SLC26A3 (DRA; down-regulated in adenoma) and/or SLC26A6 (PAT1; putative anion transporter-1) has been suggested to represent the apical membrane Cl−/HCO3− exchanger (Greeley et al. 2001). In CF, defective CFTR leads to an uncoupling between CFTR and the chloride/bicarbonate exchanger, leading to perturbation of bicarbonate secretion. In CF mouse models the presence of functional CaCCs in the pancreas seems to prevent the development of pancreatic disease, whereas in the gut, where CaCCs do not seem to be expressed, pathology occurs that is associated with CF (Clarke et al. 1994). A physiological role for CaCCs in ductal secretion has been demonstrated as several calcium mobilizing agents stimulate ductal secretion (Ashton et al. 1993; Ishiguro et al. 1999). Studies on human pancreatic duct cells have demonstrated CaCC expression (Chan et al. 1996; Winpenny et al. 1998). Furthermore, it has been demonstrated that activation of CaCC can encourage bicarbonate secretion from pancreatic duct cells, providing a promising therapeutic avenue for CF (Zsembery et al. 2000). The present study provides evidence for the expression of one molecular candidate for CaCC, hBest1, in CFPAC-1 cells and also for the involvement of hBest1 in the CaCC conductance (as interpreted by a decrease in iodide efflux). Furthermore, Qu & Hartzell (2008) have recently shown that hBest1, 2 and 4 all have a high bicarbonate permeability (PHCO3/PCl in the range 0.44–0.7) when expressed in HEK293 cells (Qu & Hartzell, 2008). This raises the possibility that bestrophins may contribute to the bicarbonate transporting capability of human pancreatic duct cells.
Recently several studies have been published that contradict the role of bestrophin as a CaCC. Some have suggested that hBest1 is a volume regulated anion channel (Fischmeister & Hartzell, 2005; Chien & Hartzell, 2008). These studies suggest that expression of hBest1 and mBest2 in HEK293, HeLa and ARPE-19 cells produces a chloride current that is sensitive to both calcium and cell volume (Fischmeister & Hartzell, 2005). A more recent study by the same group has demonstrated that Drosophila Best1 (dBest1) is the pore-forming subunit of a cell volume-regulated anion channel (VRAC) in Drosophila S2 cells. The authors also carried out experiments to confirm that this was the case in mammalian cells by comparing VRACs from peritoneal macrophages from wild-type mice and mice with both mBest1 and mBest2 disrupted. VRACs were identical in macrophages from both mice, suggesting that bestrophins are probably not responsible for the classical VRAC seen in mammalian cells (Chien & Hartzell, 2008). Others have suggested that hBest1 is a channel regulator (Marmorstein et al. 2006; Rosenthal et al. 2006). These investigations suggest that in the retinal pigment epithelium, bestrophin does not conduct the light peak but actually antagonises it and also that bestrophin alters the activity of L-type Ca2+ channels. It has been demonstrated that knockout of the mBest1 gene in mice leads to increased influx of Ca2+ through L-type channels, which has been suggested as a mechanism of light peak antagonism (Marmorstein et al. 2006). These studies are in disagreement with those published previously demonstrating that bestrophin acts as a chloride channel; however, there are other notable examples (e.g. the CFTR chloride channel) where ion channels have been shown to have a number of different functions apart from acting as an ion channel (Kunzelmann, 2001). More research is now required to determine the role of bestrophin in different tissues and species to determine its physiological function.
In summary, we have demonstrated the presence of bestrophins in the pancreatic duct cell line, CFPAC-1. As hBest1 is a candidate for the CaCC, it is of therapeutic value to the disease CF. hBest1 seems to be involved in the CaCC in CFPAC-1 cells, although it is likely that hBest1 does not solely represent the CaCC conductance in these cells. The aim of future work will be to delineate the functional role of the other bestrophins expressed in CFPAC-1 cells and whether they also have a CaCC capacity.
Acknowledgments
We thank Mrs Patricia Lunness for technical assistance and Mrs Alba Warn and Dr Paul Thomas for confocal image training. L.L.M. was funded by a Cystic Fibrosis Trust PhD studentship (grant number RS25).
Glossary
Abbreviation
- ABC
ATP-binding cassette
- ANO-1
anoctamin 1
- ARPE-19
spontaneously arising human retinal pigmented epithelial cells
- BAPTA-AM
1,2-bis(O-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid, tetraacetoxymethyl ester
- BSA
bovine serum albumin
- CaCC
calcium activated chloride channel
- CF
cystic fibrosis
- CFPAC-1
human cystic fibrosis pancreatic adenocarcinoma cell line
- CFTR
cystic fibrosis transmembrane conductance regulator
- CLCA
chloride channel, calcium activated
- DABCO
1,4-diazabicyclo(2,2,2)octane
- DAPI
4′,6-diamidino-2-phenylindole
- ER
endoplasmic reticulum
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HeLa
human cervical adenocarcinoma cell line
- hBest1
human bestrophin 1
- IP3
inositol-1,4,5-trisphosphate
- MAPK1
mitogen-activated protein kinase 1
- NA
niflumic acid
- PDEC
pancreatic duct epithelial cell
- PKA
protein kinase A
- PVDF
polyvinylidene fluoride
- RPE cell
retinal pigment epithelial cell
- siRNA
small interfering RNA
- TMEM16A
transmembrane protein 16A
- UTP
uridine 5′-triphosphate
- VMD2
vitelliform macular dystrophy 2
- VRAC
volume-regulated anion channel
- ZO-1
zonula occludens 1
Author contributions
L.L.M. and J.P.W. contributed equally to the conception, design, analysis and interpretation of the data reported in this article. L.L.M. performed the experiments detailed in this article in the Biomedical Research Centre at the University of East Anglia. L.L.M. and J.P.W. contributed equally to the drafting of the article and to critically revising the article. L.L.M. and J.P.W. have approved the final version of the article to be published.
References
- Argent B, Case RM. Pancreatic ducts: cellular mechanisms and control of bicarbonate secretion. In: Johnston LR, editor. Physiology of the Gastrointestinal Tract. 3rd edn. New York: Raven Press; 1994. pp. 1473–1497. [Google Scholar]
- Ashton N, Evans RL, Elliott AC, Green R, Argent BE. Regulation of fluid secretion and intracellular messengers in isolated rat pancreatic ducts by acetylcholine. J Physiol. 1993;471:549–562. doi: 10.1113/jphysiol.1993.sp019915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakall B, Marmorstein LY, Hoppe G, Peachey NS, Wadelius C, Marmorstein AD. Expression and localization of bestrophin during normal mouse development. Invest Ophthalmol Vis Sci. 2003;44:3622–3628. doi: 10.1167/iovs.03-0030. [DOI] [PubMed] [Google Scholar]
- Barro-Soria R, Schreiber R, Kunzelmann K. Bestrophin 1 and 2 are components of the Ca2+ activated Cl− conductance in mouse airways. Biochim Biophys Acta. 2008;1783:1993–2000. doi: 10.1016/j.bbamcr.2008.06.016. [DOI] [PubMed] [Google Scholar]
- Barro Soria R, Spitzner M, Schreiber R, Kunzelmann K. Bestrophin 1 enables Ca2+ activated Cl− conductance in epithelia. J Biol Chem. 2006;281:17460–17467. doi: 10.1074/jbc.M605716200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo A, Caci E, Ferrera L, Pedemonte N, Barsanti C, Sondo E, Pfeffer U, Ravazzolo R, Zegarra-Moran O, Galietta LJ. TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science. 2008;322:590–594. doi: 10.1126/science.1163518. [DOI] [PubMed] [Google Scholar]
- Chan HC, Cheung WT, Leung PY, Wu LJ, Chew SB, Ko WH, Wong PY. Purinergic regulation of anion secretion by cystic fibrosis pancreatic duct cells. Am J Physiol Cell Physiol. 1996;271:C469–477. doi: 10.1152/ajpcell.1996.271.2.C469. [DOI] [PubMed] [Google Scholar]
- Chappe V, Hinkson DA, Zhu T, Chang XB, Riordan JR, Hanrahan JW. Phosphorylation of protein kinase C sites in NBD1 and the R domain control CFTR channel activation by PKA. J Physiol. 2003;548:39–52. doi: 10.1113/jphysiol.2002.035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien LT, Hartzell HC. Rescue of volume-regulated anion current by bestrophin mutants with altered charge selectivity. J Gen Physiol. 2008;132:537–546. doi: 10.1085/jgp.200810065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien LT, Zhang ZR, Hartzell HC. Single Cl− channels activated by Ca2+ in Drosophila S2 cells are mediated by bestrophins. J Gen Physiol. 2006;128:247–259. doi: 10.1085/jgp.200609581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke LL, Grubb BR, Yankaskas JR, Cotton CU, McKenzie A, Boucher RC. Relationship of a non-cystic fibrosis transmembrane conductance regulator-mediated chloride conductance to organ-level disease in Cftr−/– mice. Proc Natl Acad Sci U S A. 1994;91:479–483. doi: 10.1073/pnas.91.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham SA, Awayda MS, Bubien JK, Ismailov II, Arrate MP, Berdiev BK, Benos DJ, Fuller CM. Cloning of an epithelial chloride channel from bovine trachea. J Biol Chem. 1995;270:31016–31026. doi: 10.1074/jbc.270.52.31016. [DOI] [PubMed] [Google Scholar]
- Denning GM, Anderson MP, Amara JF, Marshall J, Smith AE, Welsh MJ. Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature. 1992;358:761–764. doi: 10.1038/358761a0. [DOI] [PubMed] [Google Scholar]
- Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res. 1996;62:155–169. doi: 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]
- Durie PR, Forstner GG. Pathophysiology of the exocrine pancreas in cystic fibrosis. J R Soc Med. 1989;82(Suppl 16):2–10. [PMC free article] [PubMed] [Google Scholar]
- Duta V, Szkotak AJ, Nahirney D, Duszyk M. The role of bestrophin in airway epithelial ion transport. FEBS Lett. 2004;577:551–554. doi: 10.1016/j.febslet.2004.10.068. [DOI] [PubMed] [Google Scholar]
- Elble RC, Walia V, Cheng HC, Connon CJ, Mundhenk L, Gruber AD, Pauli BU. The putative chloride channel hCLCA2 has a single C-terminal transmembrane segment. J Biol Chem. 2006;281:29448–29454. doi: 10.1074/jbc.M605919200. [DOI] [PubMed] [Google Scholar]
- Fischmeister R, Hartzell HC. Volume sensitivity of the bestrophin family of chloride channels. J Physiol. 2005;562:477–491. doi: 10.1113/jphysiol.2004.075622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong P, Argent BE, Guggino WB, Gray MA. Characterization of vectorial chloride transport pathways in the human pancreatic duct adenocarcinoma cell line HPAF. Am J Physiol Cell Physiol. 2003;285:C433–445. doi: 10.1152/ajpcell.00509.2002. [DOI] [PubMed] [Google Scholar]
- Gibson A, Lewis AP, Affleck K, Aitken AJ, Meldrum E, Thompson N. hCLCA1 and mCLCA3 are secreted non-integral membrane proteins and therefore are not ion channels. J Biol Chem. 2005;280:27205–27212. doi: 10.1074/jbc.M504654200. [DOI] [PubMed] [Google Scholar]
- Gray MA, Winpenny J, Verdon B, O'Reilly C, Argent B. Properties and role of calcium-activated chloride channels in pancreatic duct cells. In: Fuller CM, editor. Calcium Activated Chloride Channels. New York: Academic Press; 2002. pp. 231–256. [Google Scholar]
- Greeley T, Shumaker H, Wang Z, Schweinfest CW, Soleimani M. Downregulated in adenoma and putative anion transporter are regulated by CFTR in cultured pancreatic duct cells. Am J Physiol Gastrointest Liver Physiol. 2001;281:G1301–1308. doi: 10.1152/ajpgi.2001.281.5.G1301. [DOI] [PubMed] [Google Scholar]
- Hartzell HC, Qu Z, Yu K, Xiao Q, Chien LT. Molecular physiology of bestrophins: multifunctional membrane proteins linked to best disease and other retinopathies. Physiol Rev. 2008;88:639–672. doi: 10.1152/physrev.00022.2007. [DOI] [PubMed] [Google Scholar]
- Hughes LK, Li H, Sheppard DN. Use of an iodide-selective electrode to measure CFTR Cl– channels activity. The European Working Group on CFTR Expression http://pen2.igc.gulbenkian.pt/cftr/vr/physiology.html.
- Ishiguro H, Naruse S, Kitagawa M, Hayakawa T, Case RM, Steward MC. Luminal ATP stimulates fluid and HCO3− secretion in guinea-pig pancreatic duct. J Physiol. 1999;519:551–558. doi: 10.1111/j.1469-7793.1999.0551m.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzelmann K. CFTR: interacting with everything? News Physiol Sci. 2001;16:167–170. doi: 10.1152/physiologyonline.2001.16.4.167. [DOI] [PubMed] [Google Scholar]
- Kunzelmann K, Milenkovic VM, Spitzner M, Soria RB, Schreiber R. Calcium-dependent chloride conductance in epithelia: is there a contribution by bestrophin? Pflugers Arch. 2007;454:879–889. doi: 10.1007/s00424-007-0245-z. [DOI] [PubMed] [Google Scholar]
- Lansdell KA, Kidd JF, Delaney SJ, Wainwright BJ, Sheppard DN. Regulation of murine cystic fibrosis transmembrane conductance regulator Cl− channels expressed in Chinese hamster ovary cells. J Physiol. 1998;512:751–764. doi: 10.1111/j.1469-7793.1998.751bd.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen ME, Forsyth GW. Structure and function of CLCA proteins. Physiol Rev. 2005;85:1061–1092. doi: 10.1152/physrev.00016.2004. [DOI] [PubMed] [Google Scholar]
- Marino CR, Matovcik LM, Gorelick FS, Cohn JA. Localisation of the CFTR in the pancreas. J Clin Invest. 1991;88:712–716. doi: 10.1172/JCI115358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein LY, Wu J, McLaughlin P, Yocom J, Karl MO, Neussert R, Wimmers S, Stanton JB, Gregg RG, Strauss O, Peachey NS, Marmorstein AD. The light peak of the electroretinogram is dependent on voltage-gated calcium channels and antagonized by bestrophin (best-1) J Gen Physiol. 2006;127:577–589. doi: 10.1085/jgp.200509473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt A, Stohr H, Passmore LA, Kramer F, Rivera A, Weber BH. Mutations in a novel gene, VMD2, encoding a protein of unknown properties cause juvenile-onset vitelliform macular dystrophy (Best's disease) Hum Mol Genet. 1998;7:1517–1525. doi: 10.1093/hmg/7.9.1517. [DOI] [PubMed] [Google Scholar]
- Mount DB, Romero MF. The SLC26 gene family of multifunctional anion exchangers. Pflugers Arch. 2004;447:710–721. doi: 10.1007/s00424-003-1090-3. [DOI] [PubMed] [Google Scholar]
- Petrukhin K, Koisti MJ, Bakall B, Li W, Xie G, Marknell T, Sandgren O, Forsman K, Holmgren G, Andreasson S, Vujic M, Bergen AA, McGarty-Dugan V, Figueroa D, Austin CP, Metzker ML, Caskey CT, Wadelius C. Identification of the gene responsible for Best macular dystrophy. Nat Genet. 1998;19:241–247. doi: 10.1038/915. [DOI] [PubMed] [Google Scholar]
- Pifferi S, Pascarella G, Boccaccio A, Mazzatenta A, Gustincich S, Menini A, Zucchelli S. Bestrophin-2 is a candidate calcium-activated chloride channel involved in olfactory transduction. Proc Natl Acad Sci U S A. 2006;103:12929–12934. doi: 10.1073/pnas.0604505103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z, Fischmeister R, Hartzell C. Mouse bestrophin-2 is a bona fide Cl− channel: identification of a residue important in anion binding and conduction. J Gen Physiol. 2004;123:327–340. doi: 10.1085/jgp.200409031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z, Hartzell C. Determinants of anion permeation in the second transmembrane domain of the mouse bestrophin-2 chloride channel. J Gen Physiol. 2004;124:371–382. doi: 10.1085/jgp.200409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z, Hartzell HC. Bestrophin Cl− channels are highly permeable to HCO3−. Am J Physiol Cell Physiol. 2008;294:C1371–1377. doi: 10.1152/ajpcell.00398.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z, Wei RW, Mann W, Hartzell HC. Two bestrophins cloned from Xenopus laevis oocytes express Ca2+-activated Cl− currents. J Biol Chem. 2003;278:49563–49572. doi: 10.1074/jbc.M308414200. [DOI] [PubMed] [Google Scholar]
- Rajasekaran SA, Gopal J, Espineda C, Ryazantsev S, Schneeberger EE, Rajasekaran AK. HPAF-II, a cell culture model to study pancreatic epithelial cell structure and function. Pancreas. 2004;29:e77–83. doi: 10.1097/00006676-200410000-00016. [DOI] [PubMed] [Google Scholar]
- Rosenthal R, Bakall B, Kinnick T, Peachey N, Wimmers S, Wadelius C, Marmorstein A, Strauss O. Expression of bestrophin-1, the product of the VMD2 gene, modulates voltage-dependent Ca2+ channels in retinal pigment epithelial cells. FASEB J. 2006;20:178–180. doi: 10.1096/fj.05-4495fje. [DOI] [PubMed] [Google Scholar]
- Schoumacher RA, Ram J, Iannuzzi MC, Bradbury NA, Wallace RW, Hon CT, Kelly DR, Schmid SM, Gelder FB, Rado A, Frizzell RA. A cystic fibrosis pancreatic adenocarcinoma cell line. Proc Natl Acad Sci U S A. 1990;87:4012–4016. doi: 10.1073/pnas.87.10.4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder BC, Cheng T, Jan YN, Jan LY. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 2008;134:1019–1029. doi: 10.1016/j.cell.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard DN, Welsh MJ. Structure and function of the CFTR chloride channel. Physiol Rev. 1999;79:S23–45. doi: 10.1152/physrev.1999.79.1.S23. [DOI] [PubMed] [Google Scholar]
- Steward MC, Ishiguro H, Case RM. Mechanisms of bicarbonate secretion in the pancreatic duct. Annu Rev Physiol. 2005;67:377–409. doi: 10.1146/annurev.physiol.67.031103.153247. [DOI] [PubMed] [Google Scholar]
- Stohr H, Marquardt A, Nanda I, Schmid M, Weber BH. Three novel human VMD2-like genes are members of the evolutionary highly conserved RFP-TM family. Eur J Hum Genet. 2002;10:281–284. doi: 10.1038/sj.ejhg.5200796. [DOI] [PubMed] [Google Scholar]
- Sun H, Tsunenari T, Yau KW, Nathans J. The vitelliform macular dystrophy protein defines a new family of chloride channels. Proc Natl Acad Sci U S A. 2001;99:4008–4013. doi: 10.1073/pnas.052692999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Tsunenari T, Yau KW, Nathans J. The vitelliform macular dystrophy protein defines a new family of chloride channels. Proc Natl Acad Sci U S A. 2002;99:4008–4013. doi: 10.1073/pnas.052692999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavsanli BC, Pappu KS, Mehta SQ, Mardon G. Dbest1, a Drosophila homolog of human bestrophin, is not required for viability or photoreceptor integrity. Genesis. 2001;31:130–136. doi: 10.1002/gene.10013. [DOI] [PubMed] [Google Scholar]
- Trezise AE, Chambers JA, Wardle CJ, Gould S, Harris A. Expression of the cystic fibrosis gene in human foetal tissues. Hum Mol Genet. 1993;2:213–218. doi: 10.1093/hmg/2.3.213. [DOI] [PubMed] [Google Scholar]
- Tsunenari T, Nathans J, Yau KW. Ca2+-activated Cl− current from human bestrophin-4 in excised membrane patches. J Gen Physiol. 2006;127:749–754. doi: 10.1085/jgp.200609527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunenari T, Sun H, Williams J, Cahill H, Smallwood P, Yau KW, Nathans J. Structure-function analysis of the bestrophin family of anion channels. J Biol Chem. 2003;278:41114–41125. doi: 10.1074/jbc.M306150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winpenny JP, Harris A, Hollingsworth MA, Argent BE, Gray MA. Calcium-activated chloride conductance in a pancreatic adenocarcinoma cell line of ductal origin (HPAF) and in freshly isolated human pancreatic duct cells. Pflugers Arch. 1998;435:796–803. doi: 10.1007/s004240050586. [DOI] [PubMed] [Google Scholar]
- Winpenny JP, Verdon B, McAlroy HL, Colledge WH, Ratcliff R, Evans MJ, Gray MA, Argent BE. Calcium-activated chloride conductance is not increased in pancreatic duct cells of CF mice. Pflugers Arch. 1995;430:26–33. doi: 10.1007/BF00373836. [DOI] [PubMed] [Google Scholar]
- Wistow G, Bernstein SL, Wyatt MK, Fariss RN, Behal A, Touchman JW, Bouffard G, Smith D, Peterson K. Expressed sequence tag analysis of human RPE/choroid for the NEIBank Project: over 6000 non-redundant transcripts, novel genes and splice variants. Mol Vis. 2002;8:205–220. [PubMed] [Google Scholar]
- Yang YD, Cho H, Koo JY, Tak MH, Cho Y, Shim WS, Park SP, Lee J, Lee B, Kim BM, Raouf R, Shin YK, Oh U. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 2008;455:1210–1215. doi: 10.1038/nature07313. [DOI] [PubMed] [Google Scholar]
- Zsembery A, Strazzabosco M, Graf J. Ca2+-activated Cl− channels can substitute for CFTR in stimulation of pancreatic duct bicarbonate secretion. FASEB J. 2000;14:2345–2356. doi: 10.1096/fj.99-0509com. [DOI] [PubMed] [Google Scholar]