Abstract
The elucidation of the human genome has had a major impact on histamine receptor research. The identification of the human H4 receptor by several groups has been instrumental for a new appreciation of the role of histamine in the modulation of immune function. In this review, we summarize the historical developments and the molecular and biochemical pharmacology of the H4 receptor.
Keywords: histamine H4 receptor, molecular pharmacology, biochemistry, signal transduction, species difference, isoforms, heterogeneity, selective agonists and antagonists
Introduction
Histamine is known since long to be an important mediator in different (patho)physiological conditions. In 1966, Ash and Schild suggested the presence of two distinct histamine receptor subtypes (Ash and Schild, 1966) based on the pharmacological effects of known histaminergic ligands on, for example, airway smooth muscle (H1) and the heart, uterus and stomach (H2). Sir James Black et al. at SK&F paved the way for the final acceptance of two distinct histamine receptor subtypes with the development of the first selective histamine H2 antagonists metiamide and burimamide (Black et al., 1972). Moreover, the recognition that different actions of histamine could be effectively inhibited by selective receptor antagonist eventually resulted in the development of various blockbuster H1 and H2 antagonists.
In 1983 a new histamine H3 receptor subtype was identified by Arrang et al. in the rat brain on the basis of classical pharmacology (Arrang et al., 1983). For a long time, this new histamine receptor subtype was not fully endorsed by the pharmaceutical industry because of the lack of molecular details of the receptor protein. While the gene for the histamine H1 (Yamashita et al., 1991) and H2 receptor (Gantz et al., 1991) were both cloned in 1991, for many years no definite proof of the molecular architecture of the H3 receptor was available. Finally, in 1999 a team of J&J researchers led by Tim Lovenberg cloned the complementary DNA (cDNA) of an α2 adrenergic-like EST sequence that, after characterization, turned out to encode for the human histamine H3 receptor (Lovenberg et al., 1999). This cloning of the H3 receptor gene resulted in a rapid increase in knowledge of its molecular pharmacology and biochemistry (e.g. expression, signal transduction, receptor isoforms) (Lovenberg et al., 1999; Drutel et al., 2001; Hancock, 2006; Bongers et al., 2007). Combined with the development of recombinant cell systems, these developments led to a huge interest from pharmaceutical companies (Leurs et al., 2000). Many new selective ligands have since then been reported in (patent) literature (Celanire et al., 2005; Wijtmans et al., 2007), and the H3 receptor is currently seen as a new target for the treatment of a variety of central nervous system disorders (e.g. attention deficit hyperactivity disorder, Alzheimer's disease and schizophrenia) (Leurs et al., 1995; Esbenshade et al., 2008).
The cloning of the H3 receptor gene also marked the entry of the histamine H4 receptor into the G protein-coupled receptor (GPCR) family of histamine receptors. Using the DNA sequence of the histamine H3 receptor, several research groups independently identified a previously unexplored GPCR sequence in the human genome as a new histamine H4 receptor (Nakamura et al., 2000; Oda et al., 2000; Liu et al., 2001a; Morse et al., 2001; Nguyen et al., 2001; Zhu et al., 2001). The new receptor showed a distinct pharmacology (see below) is highly expressed in immune cells (Nakamura et al., 2000; Oda et al., 2000; Liu et al., 2001a; Morse et al., 2001; Nguyen et al., 2001; Zhu et al., 2001) and is now regarded as an interesting target for, for example, inflammatory disorders and pruritis (de Esch et al., 2005; Thurmond et al., 2008). The first identification of the H4 receptor led to a further increase in the interest in histamine receptor pharmacology and a substantial rise in H4 receptor-related papers (112 in the period 2001–2009) and patents (36 in the period 2002–2008) in the last few years. In the present review we will highlight the molecular biology and pharmacology of the latest member of the histamine receptor family.
Cloning of the histamine H4 receptor gene
As reported above, the H4 receptor cDNA was finally identified in the human genome database on the basis of its overall homology (37%) to the H3 receptor sequence (Nakamura et al., 2000; Oda et al., 2000; Liu et al., 2001a; Morse et al., 2001; Nguyen et al., 2001; Zhu et al., 2001). The homology of the H4 receptor to the H1 and H2 receptor is only ∼19%, explaining why this subtype was only found after the long awaited identification of the H3 receptor gene in 1999 (Lovenberg et al., 1999). The size of the open reading frame of the H4 receptor cDNA is 1173 bp, encoding for a 390 amino acid protein (Figure 1), belonging to the large family of GPCRs. Besides a high sequence homology, the H4 receptor also shares its gene structure with the H3 receptor (Coge et al., 2001a; Coge et al., 2001b). The human H4 receptor gene is present in a single copy on chromosome 18q11.2. The gene spans 16.98 kb (position 20 294 591 to 20 311 567) and, like the H3 receptor (Drutel et al., 2001), is interrupted by two large introns (7867 bp and >17 500 bp). The introns divide the coding regions into three parts, encoding amino acid number 1–65, 66–119 and 120–390. Such an intron–exon distribution has led for the H3 receptor to the generation of a large number of alternatively spliced GPCR variants, sometimes with clearly altered functionalities (Drutel et al., 2001; Leurs et al., 2005). However, for the H4 receptor at present only two non-signalling, non-7TM H4 receptor isoforms have been identified (van Rijn et al., 2008) (Figure 1), designated as H4(67) and H4(302). The polypeptide of H4(67) is prematurely terminated at residue 67, while H4(302) lacks residues 68–155 (Figure 1, vide infra) (van Rijn et al., 2008).
Figure 1.
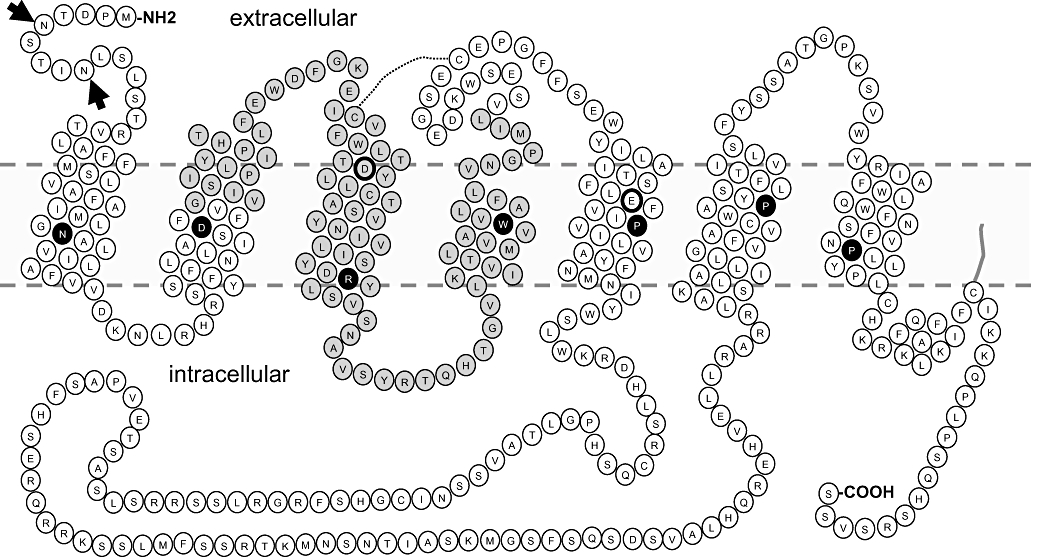
Snake plot of the human histamine H4 receptor. The full-length receptor consists of 390 amino acids, which form seven transmembrane helices, three extracellular loops and three intracellular loops, with an extracellular N-terminal and an intracellular C-terminal peptide; the dotted line represents the putative disulfide bridge that links the cysteines in the third transmembrane and second extracellular loop. Residues Asp3.32 and Glu5.46 that play important roles in histamine binding are marked with bold border. The H4R67 isoform only contains the first 67 N-terminal residues (marked in white), while the H4R302 lacks the residues marked in grey. The conserved residues in the family A of G protein-coupled receptors are depicted in black circles, while the putative glycosylation sites (Asn5 and Asn9) are indicated with arrows. A potential palmitoylation site (Cys374) at the C-terminal tail is suggested to be close to the membrane following membrane insertion of a putative attached palmitic acid.
The original H4 receptor amino acid sequence as reported by Oda et al. (2000) deviates at three positions from the sequences reported by four other research groups (Nakamura et al., 2000; Liu et al., 2001a; Morse et al., 2001; Zhu et al., 2001), that is, Val instead of Ala at position 138, Arg instead of His at position 206 and Arg instead of Gln at position 253. The Val138Ala and Arg206His polymorphisms have been confirmed by single nucleotide polymorphism (SNP) analysis involving samples from White, Chinese, Japanese, Afro-American and sub-Saharan African people (http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?locusId=59340). The occurrence of Ala138 or His206 alleles is higher than that of Val138 or Arg206 (91.1–100% vs. 0–9.2%). Two other SNPs have been found as well in the coding region, resulting in Cys284Ser mutation and a frame shift at residue L379. Besides those, 37 SNPs are found in intron 1, 45 in intron 2 and 21 in 3′-untranslated region (http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?locusId=59340). So far no functional comparisons of the various SNP variants or linkage studies have been reported.
As with the other histamine receptor family members, the H4 receptor protein possesses all consensus motifs identified for the Class A rhodopsin-like GPCRs, that is, N1.5033, D2.5061, R3.50112, W4.50140, P5.50186, P6.50318 and P7.50355[using the standardized GPCR nomenclature according to Ballesteros–Weinstein (Ballesteros and Weinstein, 1995) (Figure 1)]. With respect to histamine binding, a conserved aspartate residue in the third transmembrane (TM) aminergic GPCRs helix (D3.32, Figure 1) is also present in this newest member of the aminergic GPCR family. As for the other aminergic GPCRs, D3.32 is believed to interact with the protonated amine group of the agonist histamine (Shin et al., 2002). Moreover, in both H3 and H4 receptors a second negatively charged amino acid, E5.46, is present in the fifth TM helix (Figure 1). This amino acid has been shown to play an essential role in the binding of histamine as determined by site-directed mutagenesis analysis and computational analysis (Shin et al., 2002; Uveges et al., 2002). As is illustrated in Figure 2, E5.46 is a key anchor for the imidazole ring of histamine. The binding pocket for the imidazole ring is formed by E5.46, T5.42 and W6.48 (Jongejan et al., 2008). The latter residue adopts the ‘active’ rotamer conformation (Jongejan et al., 2005) by forming a hydrogen bond to the aforementioned E5.46 (box, Figure 2).
Figure 2.
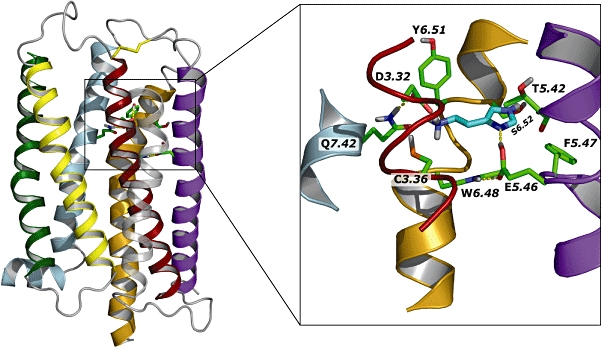
Schematic representation of a homology model of the human histamine H4 receptor. The figure shows the seven transmembrane domains and the position of the ligand binding site. In the box details of the histamine binding site, as determined using ab initio calculations is shown (reproduced with permission from Jongejan et al., 2008).
The H4R has two potential glycosylation sites (Asn5 and Asn9) (Figure 1, vide infra), whereas Cys374 might serve as a potential site for palmitoylation. The H4R also contains cysteine residues that potentially form a disulphide bridge, connecting the third TM with the second extracellular loop (EL2) (Figure 1). These cysteine residues are conserved among the GPCRs, and the disulphide bridge has been evidenced in the crystal structure of bovine rhodopsin and adrenergic β1 and β2 receptors (Palczewski et al., 2000; Cherezov et al., 2007; Warne et al., 2008). The disulfide bridge will bring some structural constraint to the extracellular loop two, which may be important in view of the suggested involvement of the EL2 in the binding of histamine (Lim et al., 2008).
Species differences
Following the identification of the human H4R, the cDNA sequence of mouse, rat, guinea pig, pig, dog and monkey H4 receptors have been reported and functionally expressed (Liu et al., 2001b; Oda et al., 2002; 2005). The monkey H4 receptor (Macaca fascicularis) shows a high sequence homology (92%) with the human orthologue (Oda et al., 2005). The H4 receptors of the other species are only moderately homologous to the human H4R, with sequence homology between 67% and 72% (Liu et al., 2001b; Oda et al., 2002; 2005; Jiang et al., 2008). Forty (partial) amino acid sequences of H4R orthologues from 34 other species have been extracted from various genomic ENSEMBL databases (http://www.ensembl.org/Homo_sapiens/Gene/Compara_Ortholog?db=core;g=ENSG00000134489;r=18:20294591-20313918;t=ENST00000256906), among others horse (Equus caballus), cow (Bos taurus), cat (Felis catus), rhesus monkey (Macaca mullata), chimpanzee (Pan troglodytes) and dolphin (Tursiops truncates) H4 receptor sequences are known (Figure 3). The H4R sequences from cynomogus (Macaca fascicularis) and pig (Sus scrofa), which are not listed as orthologues in the ENSEMBL database are also included in the phylogenetic tree in Figure 3. Because some of the sequences are still incomplete, changes in the phylogenetic tree are to be expected. Fish (zebra fish, fugu, tetraodon, medaka and stickleback) as well as frog (Xenopus tropicalis) have each two orthologues. However, these orthologues, together with the sequences from chicken (Gallus gallus) and opossum, show only 28–42% homology to the human H4 receptor, while all the others show a substantial higher homology to the human H4 receptor (>50%). In fact, these orthologues are more similar to the human H3R. Consequently, in the phylogenetic tree these H3/H4 orthologues cluster in a separate group (Figure 3). Within the extracted (partial) GPCR sequences, the typical aminergic GPCR features mentioned earlier (e.g. Asp3.32 in TM3 and Glu5.46 in TM5) can often be found. Detailed analysis of most of these species variants is for the moment however lacking, but may provide useful tools to dissect receptor–ligand binding. Previously, Lim et al. (2008), for example, showed that a sequence difference in the extracellular loop EL2 is responsible for the lower affinity of histamine for rat and mouse H4 receptors. Comparing the binding profiles of rat, mouse, guinea-pig, porcine, monkey and dog H4 receptors, one can identify for ligands like clozapine and clobenpropit (see also paragraph on H4 agonist) also substantial differences in binding affinity between the various species (Liu et al., 2001b;Oda et al., 2002; 2005). Differences in pharmacological activities of H4 receptor ligands between the different species might hamper preclinical development of future H4 receptor drugs.
Figure 3.
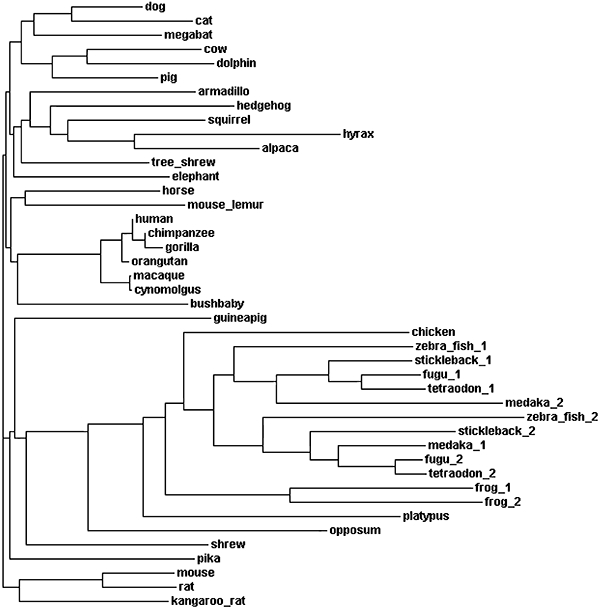
Phylogenetic tree of H4 receptor orthologues. The sequences are obtained from http://www.ensembl.org and the phylogram was created with ClustalW.
Signal transduction of the H4 receptor
The H4 receptor is coupled to pertussis toxin (PTX)-sensitive Gαi/o proteins, thereby inhibiting forskolin-induced cAMP and eventually the modulation of transcription of genes regulated by cAMP-responsive elements in cell lines recombinantly expressing the H4 receptor (Oda et al., 2000; Liu et al., 2001a; Zhu et al., 2001). In transfected HEK-293 cells H4 receptor stimulation also leads to a strong increase in mitogen-activated protein kinase phosphorylation via a PTX-sensitive pathway (Morse et al., 2001). Furthermore, the H4 receptor also signals to calcium via either PTX-sensitive Gαi/o proteins (Hofstra et al., 2003) or the promiscious Gα16 protein (Oda et al., 2000; Morse et al., 2001), which is selectively expressed in immune cells (Wilkie et al., 1991). In recombinant cell lines the H4 receptor constitutively inhibits forskolin-induced cAMP-responsive element-mediated reporter gene expression and activates [35S]GTPγS binding (Liu et al., 2001a; Morse et al., 2001; Lim et al., 2005). The agonist-independent activity can be inhibited by H4 inverse agonists, such as thioperamide.
Activation of endogenously expressed H4 receptors in for example eosinophils and mast cells leads to PTX-sensitive calcium mobilization (Buckland et al., 2003; Hofstra et al., 2003), via the activation of phospholipase C (Hofstra et al., 2003). The increased cytosolic calcium concentration is intimately linked to actin polymerization, cell shape change and eventually to the migration of mast cells, eosinophils and monocyte dendritic cells (Buckland et al., 2003; Hofstra et al., 2003; Ling et al., 2004; Gutzmer et al., 2005; Barnard et al., 2008).
Looking back in immunology literature with this new knowledge, early examples of H4 receptor-mediated signalling by histamine can be identified. Already in the 1975 Clark et al. observed histamine-induced eosinophil chemotaxis that could not be blocked by the known H1 and H2 receptor antagonists (Clark et al., 1975). Furthermore, in 1994 Raible et al. identified in eosinophils a PTX-sensitive cytosolic calcium increase induced by histamine, which could be inhibited by the presumed selective H3 receptor antagonist thioperamide. The potent H3 receptor reference agonist R-α-methylhistamine also induced a calcium increase, but at much lower potency than that of histamine itself. Based on these observations, the presence of a new histamine receptor subtype was suggested (Raible et al., 1994). Alas, this suggestion was largely ignored by the histamine research community and has only been fully exploited more than a decade after the discovery.
Anatomical Topography of the H4 receptor
The tissue distribution of the H4 receptor has been profiled at the mRNA level in a wide range of human (using reverse transcription polymerase chain reaction) and mouse (using in situ hybridization) tissues (Zhu et al., 2001). In human tissues, H4 receptor expression was most abundant in bone marrow, peripheral blood, spleen, thymus, small intestine, colon, heart and lung (Nakamura et al., 2000; Liu et al., 2001a; Morse et al., 2001; Nguyen et al., 2001; Zhu et al., 2001). Discrepancies have occurred in both human and rodent tissue based on these mRNA studies, which highlights the necessity for selective immunological anti-H4 receptor probes.
Anti-H4 receptor antibodies have been crucial tools to study the H4 receptor (van Rijn et al., 2006; 2008; Dijkstra et al., 2007; Baumer et al., 2008; Dijkstra et al., 2008; Morini et al., 2008). The majority of cells expressing H4 receptors are indeed hematopoietic in lineage, including neutrophils, mast cells, eosinophils, basophils, dendritic cells, monocytes and T cells (e.g. van Rijn et al., 2006; Dijkstra et al., 2007; Baumer et al., 2008; Dijkstra et al., 2008), but importantly, it has recently been shown that the H4 receptor is not exclusively expressed on hematopoietic cells. The H4 receptor has been identified in subsets of endocrine cells in the gastrointestinal tract (distinct from H3R containing cells) (Morini et al., 2008) and has been shown also to be expressed on dermal fibroblasts (Ikawa et al., 2008). Immunohistochemical studies have shown that H4 receptors are present on nerves from the human nasal mucosa (Nakaya et al., 2004) and in the enteric nervous system on the myenteric plexus of the rodent fundus (H3R absent) (PL Chazot, D Grandi, FC Shenton, G Morini, unpublished). Indeed, importantly, as with the other histamine receptor subtypes, the H4 receptor is expressed in the central nervous system (Connelly, et al., 2009). In agreement, the H4 receptor was detected on subpopulations of small and medium diameter cells within the lumbar sensory dorsal root ganglia and laminae I/II of the dorsal horn of the lumbar spinal cord (Strakhova et al., 2009; PL Chazot, NL Lethbridge, unpublished). These latter results indicate that the H4 receptor is present on synaptic terminals of primary sensory afferent neurons. Therefore, the H4 receptor is likely to subserve distinct roles in many parts of the body, and therefore may represent a target for an expanded array of therapeutic arenas, including neuropathic pain.
H4 receptor biochemistry
Immunological approaches have demonstrated that the H4 receptor comprises robust dimeric structures (van Rijn et al., 2006; 2008). It is clear from immunoblots that higher molecular weight species (consistent with dimers and/or higher oligomers) are present in both recombinant and native tissue (Figure 4). Immunoblotting identified three major protein species in human spleen lysate. Interestingly a low molecular weight species was clearly visible in the spleen lysate at 31 kDa, likely to be the receptor monomer. In contrast, monomeric species were not evident in either lymphocytes (polyhaemagglutinnin blasts) or in brain membranes (van Rijn et al., 2006; Connelly et al., 2009). The observed sizes detected in immunoblotting studies are not always completely consistent with H4R dimeric structures, with sizes ranging between 60 and 80 kD in HEK-293 H4R-transfected cells or in other native tissue preparations (Figure 4, van Rijn et al., 2006; Connelly et al., 2009). Several possibilities could account for these observations, including tissue-specific complements of H4R isoforms and differential tissue-specific post-translational modifications (e.g. palmitoylation, glycosylation).
Figure 4.
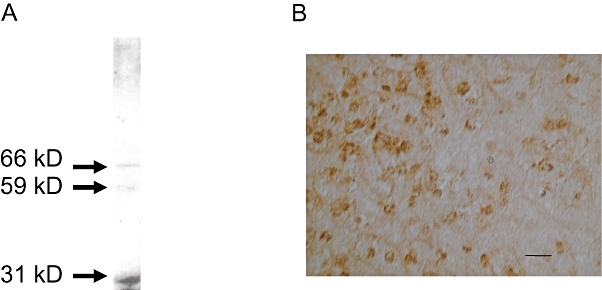
Anti-hH4(374–390) receptor antibody reacts with human H4 receptor protein in spleen tissue lysate and sections. (A) Immunoblot of human spleen lysate (10 µg). Anti-H4(374–390) antibodies were used at 2 µg·mL−1, incubated overnight at 4°C and developed as described in van Rijn et al. (2006). In addition to the putative monomer at Mw 31 kDa, two higher molecular weight species were detected at 59 and 66 kDa. (B) Immunostaining of human spleen slice (bar = 50 µm) using anti-H4(374–390) at 1 µg·mL−1 using an immunohistochemical protocol described in Chazot et al. (2001).
Anti-H4 receptor antibodies were also used to investigate the expression and potential role of the newly described human H4 receptor isoforms: H4(302) and H4(67) (van Rijn et al., 2008). Because mRNAs coding for all three isoforms are found in several white blood cell types, hetero-oligomerization may occur in native tissue. The two shorter isoforms are both non-functional as regards ligand binding and signalling. However they may play a regulatory role as they appear to reduce number of histamine binding to the H4(390) receptor by 55% [H4(302)] and 30% ]H4(67)] in heterologous expression studies. Based on surface biotinylation experiments, the three were expressed at the cell surface, although the shorter versions with greatly reduced efficiency. Furthermore co-expression of the H4(390) with either H4(302) or H4(67) dose-dependently reduced surface expression of the full length receptor (van Rijn et al., 2008). These results provide further evidence to support the role of splice isoforms as dominant negative regulatory elements, which maybe a common theme in GPCR-regulatory pathways (Bakker et al., 2006).
The majority of secreted and cell surface proteins that transit to the cell surface through the endoplasmic reticulum are N-glycosylated (95% of GPCRs) (Lanctot et al., 2005). In order for a GPCR to elicit intracellular signalling appropriately, the correct quantity of properly folded functional receptors must be available in the plasma membrane. It is equally essential that signalling can be terminated at the appropriate time and the receptors either recycled or permanently degraded. The regulatory mechanisms controlling export trafficking of GPCRs, including glycosylation, have recently been reviewed (Duvernay et al., 2005). The role of post-translational changes to receptors is complex and an area where there is growing interest. Several studies have shown that glycosylation is involved in GPCR transport from the endoplasmic reticulum through the Golgi to the cell surface (reviewed in Duvernay et al., 2005). Both the H3 and H4 receptors are indeed N-glycosylated (van Rijn et al., 2006; Shenton and Chazot, 2006). Interestingly, receptor dimerization in vitro does not appear to be dependent on N-glycosylation. However, in the case of endogenous H4 receptor expressed in human polyhaemagglutinnin blasts, chemical deglycosylation appears to destabilize the preformed dimeric species to individual monomers (van Rijn et al., 2006). The pharmacological influence of N-glycosylation of the H4 receptor is yet to be explored.
Pharmacological tools to study the H4 receptor
Already with the early description of histamine responsiveness of eosinophils in 1994, a clearly different pharmacology was described in comparison with the known receptor subtypes (Raible et al., 1994). The subsequent cloning of the H4 receptor cDNA and the analysis of the expressed protein also resulted in the rapid recognition of a completely distinct H4 receptor pharmacology (Hough, 2001). Since then, major progress has been made in the discovery of selective H4 receptor agonists and antagonists.
The early pharmacological and (patho)physiological characterization of the H4 receptor was performed using agonists that were originally developed for the homologous H3R receptor. Typically, these initial H3 and H4 receptor dual activity reference compounds all contain an imidazole heterocycle. The most important examples are the H3 and H4 receptor agonist, immepip (2) and the H3 antagonist and H4 agonist clobenpropit (3) (Lim et al., 2005) (Figure 5).
Figure 5.
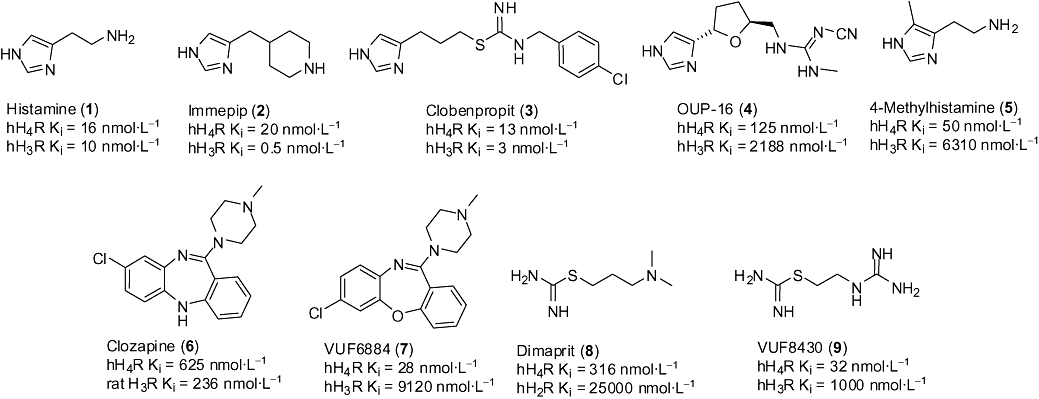
First and second generation H4 receptor agonists with different selectivity profiles.
The first ligand that was optimized for selective H4 receptor activation is OUP-16 (4) (Hashimoto et al., 2003) (Figure 5). This full agonist has a 40-fold selectivity for the H4 receptor over the H3 receptor. More recently, the potent H4 receptor agonist 4-methylhistamine (5) was described after evaluation of a large number of histaminergic compounds (Lim et al., 2005). Although originally developed in the H2 receptor research programme, 4-methylhistamine has a high affinity for the H4 receptor, while displaying at least a 100-fold selectivity over the other histamine receptor subtypes, including the H2 receptor. The first identified non-imidazole H4 receptor agonist was clozapine (6) (Oda et al., 2000). This anti-psychotic agent has affinity for a large number of GPCRs, but almost exclusively acts as antagonist. The dibenzodiazepine structure was optimized for H4 receptor affinity, leading to the dual activity H1 receptor antagonist and H4 receptor agonist VUF6884 (7) (Smits et al., 2006). Although promiscuous GPCR binders, this class of ligands provides detailed insight of the H4 receptor agonist binding site (Smits et al., 2006; Jongejan et al., 2008). Another non-imidazole H4 receptor agonist is dimaprit (8) and its analog VUF8430 (9) (Lim et al., 2005; Lim et al., 2006). The latter is a full agonist with high affinity for the H4 receptor and a 30-fold selectivity over the H3 receptor. In combination with 4-methylhistamine, VUF8430 forms a new useful pair of tools to study H4 receptor pharmacology (see Lim et al., 2009).
As is true for the first H4 receptor agonists, the first antagonists that were used to characterize the H4 receptor were imidazole-containing, dual activity H3 and H4 receptor ligands. Of particular use has been the H3 and H4 receptor inverse agonist thioperamide (10) (Figure 6; Buckland et al., 2003; Hofstra et al., 2003; Bell et al., 2004; Takeshita et al., 2004; Damaj et al., 2007). The H4 receptor research field has been greatly helped by the early discovery and disclosure of the selective non-imidazole neutral antagonist JNJ7777120 (11). The compound resulted from a high-throughput screening campaign and subsequent hit optimization efforts at Johnson and Johnson Pharmaceuticals (Carruthers et al., 2002). JNJ7777120 is equipotently active at human, mouse and rat H4Rs (Jablonowski et al., 2003; Thurmond et al., 2004). This H4R reference antagonist is more than a thousand fold selective over other histamine receptor subtypes and was reported to have no cross-reactivity in a panel of 50 other receptors, enzymes, transporters and ion-channels. JNJ7777120 has reasonable oral bioavailability (22%), but its use in disease models is hampered by a short in vivo half-life of about 0.8 h (Venable et al., 2005). Efforts to obtain compounds with improved properties by scaffold hopping led to VUF6002 (12) (Figure 6; Terzioglu et al., 2004; Venable et al., 2005). Although this benzimidazole derivative has an improved liver microsomes stability, the half-life remains limited (t1/2= 1.0 h).
Figure 6.
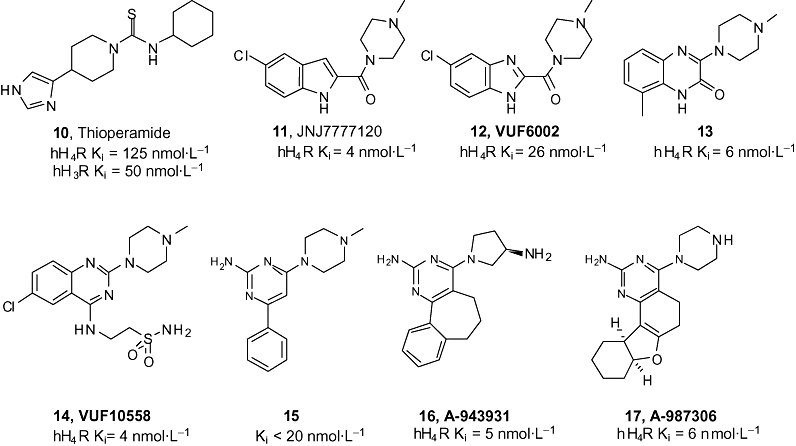
First and second generation H4 receptor antagonists. Except for thioperamide (10) the antagonists display at least a 100-fold selectivity for the histamine H4 receptor over the histamine H3 receptor.
Another series of H4R compounds that was recently claimed in a patent by Johnson and Johnson contain methylpiperazine substituted 2-quinoxalinones (Edwards and Venable, 2005), including compound 13. At VU University Amsterdam, the construction of a H4R pharmacophore model and rational design approaches resulted in the potent quinazoline compound 14 (Leurs et al., submitted). This compound was shown to have anti-inflammatory properties in vivo in the rat.
Bayer Healthcare AG has recently patented aminopyrimidines (e.g. 15) as H4 receptor antagonists (Sato et al., 2005a,b). Since this disclosure, several other companies have claimed other pyrimidines as H4 receptor ligands, including Abbott Laboratories who developed the rotationally restricted analogue A-943931 (16). This compound has been thoroughly characterized and displays a more than 640-fold selectivity over the H3 receptor, a 190-fold selectivity over α1 adrenergic receptors and a 470-fold selectivity over 5-HT1d receptors. A-943931 is reported to have excellent antagonistic activity both in vitro and in vivo across multiple species, while displaying excellent metabolic stability and good oral bioavailability (90%). This compound is a good anti-inflammatory agent in mice and also displays good efficacy in rat pain models (Cowart et al., 2008). Further exploration of the aminopyrimidine theme resulted in the recently described H4 receptor antagonist A-987306 (17), a potent and selective compound with an excellent pharmacokinetic profile, including a half-life of 3.7 h after p.o. dosing (Liu et al., 2008). A-987306 (17) has anti-inflammatory activity in a peritonitis model and is especially potent in a pain assay in rats, as was shown by the blockage of carrageenan induced thermal hyperalgesia.
In order to study the pharmacology of the H4 receptor, radioligand binding studies are instrumental. As with the H3 receptor, the H4 receptor binds the endogenous agonist histamine with high, nanomolar affinity (Nakamura et al., 2000; Oda et al., 2000; Liu et al., 2001a; Morse et al., 2001; Nguyen et al., 2001; Zhu et al., 2001). Consequently, [3H]histamine has been effectively used as radioligand since the discovery of the H4 receptor (Nakamura et al., 2000; Oda et al., 2000; Liu et al., 2001a; Morse et al., 2001; Nguyen et al., 2001; Zhu et al., 2001). In some studies also [3H]Nα-methylhistamine has been used as radiotracer (Liu et al., 2001a), but this has not become very popular. In 2004 J&J described the use of [3H]JNJ7777120 as antagonist radioligand for the H4 receptor (Thurmond et al., 2004). This new radioligand binds differently than histamine in the H4R ligand binding pocket (Jongejan et al., 2008), allowing the analysis of, for example, distinct H4 receptor mutants that have lost high affinity [3H]histamine binding (Jongejan et al., 2008).
Final remarks
With the cloning of the gene of the histamine H4 receptor in 1999/2000 the field of histamine research has been fueled with new excitement. Several laboratories have already identified a set of useful tools (agonists, antagonists, anti-H4 receptor antibodies), allowing the field to address the function of the H4 receptor in both the periphery and the brain. Moreover, in the last few years a large increase in knowledge on the biochemical pharmacology of the H4 receptor has been obtained. In view of the first preclinical in vivo data for H4 receptor ligands, this new histamine receptor family member is expected to become an important target in the area of inflammatory disorders and neuropathic pain. Moreover, also pruritis is associated with H4 receptor function as itching can be induced by H4 agonists such as clobenpropit and 4-methylhistamine and blocked by H4 antagonists such as JNJ7777120. It might nevertheless be an interesting challenge to develop H4 receptor drugs, as so far relatively high levels of H4 receptor antagonists are required to inhibit in vivo inflammatory effects despite the high potency and selectivity of the tool compounds. This may reflect limitations of the tool compounds with respect to for example pharmacokinetic properties, as well as for example high local histamine concentrations at the H4 receptor.
Acknowledgments
The authors acknowledge support from the EU-KP7 COST programme BM0806 (Histamine H4 receptor network).
Glossary
Abbreviations:
- cDNA
complementary DNA
- EL2
second extracellular loop
- GPCR
G protein-coupled receptor
- PTX
pertussis toxin
- SNP
single nucleotide polymorphism
- TM
transmembrane domain
Conflict of interest
None.
References
- Arrang JM, Garbarg M, Schwartz JC. Auto-inhibition of brain histamine release mediated by a novel class H3 of histamine receptor. Nature. 1983;302:832–837. doi: 10.1038/302832a0. [DOI] [PubMed] [Google Scholar]
- Ash AS, Schild HO. Receptors mediating some actions of histamine. Br J Pharmacol Chemother. 1966;27:427–439. doi: 10.1111/j.1476-5381.1966.tb01674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker RA, Lozada AF, van Marle A, Shenton FC, Drutel G, Karlstedt K, et al. Discovery of naturally occurring splice variants of the rat histamine H3 receptor that act as dominant-negative isoforms. Mol Pharmacol. 2006;69:1194–1206. doi: 10.1124/mol.105.019299. [DOI] [PubMed] [Google Scholar]
- Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- Barnard R, Barnard A, Salmon G, Liu W, Sreckovic S. Histamine-induced actin polymerization in human eosinophils: an imaging approach for histamine H4 receptor. Cytometry. 2008;73:299–304. doi: 10.1002/cyto.a.20514. [DOI] [PubMed] [Google Scholar]
- Baumer W, Wendorff S, Gutzmer R, Werfel T, Dijkstra D, Chazot P, et al. Histamine H4 receptors modulate dendritic cell migration through skin – immunomodulatory role of histamine. Allergy. 2008;63:1387–1394. doi: 10.1111/j.1398-9995.2008.01720.x. [DOI] [PubMed] [Google Scholar]
- Bell JK, McQueen DS, Rees JL. Involvement of histamine H4 and H1 receptors in scratching induced by histamine receptor agonists in Balb C mice. Br J Pharmacol. 2004;142:374–380. doi: 10.1038/sj.bjp.0705754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JW, Duncan WA, Durant CJ, Ganellin CR, Parsons EM. Definition and antagonism of histamine H2 receptors. Nature. 1972;236:385–390. doi: 10.1038/236385a0. [DOI] [PubMed] [Google Scholar]
- Bongers G, Bakker RA, Leurs R. Molecular aspects of the histamine H3 receptor. Biochem pharmacol. 2007;73:1195–1204. doi: 10.1016/j.bcp.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Buckland KF, Williams TJ, Conroy DM. Histamine induces cytoskeletal changes in human eosinophils via the H4 receptor. Br J Pharmacol. 2003;140:1117–1127. doi: 10.1038/sj.bjp.0705530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers NI, Chai W, Dvorak CA, Edwards JP, Grice CA, Jablonowski JA, et al. Heterocyclic compounds. Patent WO 02/072548.
- Celanire S, Wijtmans M, Talaga P, Leurs R, de Esch IJ. Keynote review: histamine H3 receptor antagonists reach out for the clinic. Drug Discov Today. 2005;10:1613–1627. doi: 10.1016/S1359-6446(05)03625-1. [DOI] [PubMed] [Google Scholar]
- Chazot PL, Hann V, Wilson C, Lees G, Thompson CL. Immunological identification of the mammalian H3 histamine receptor in the mouse brain. Neuroreport. 2001;12:259–262. doi: 10.1097/00001756-200102120-00016. [DOI] [PubMed] [Google Scholar]
- Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RA, Gallin JI, Kaplan AP. The selective eosinophil chemotactic activity of histamine. J Exp Med. 1975;142:1462–1476. doi: 10.1084/jem.142.6.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coge F, Guenin SP, Audinot V, Renouard-Try A, Beauverger P, Macia C, et al. Genomic organization and characterization of splice variants of the human histamine H3 receptor. Biochem J. 2001a;355:279–288. doi: 10.1042/0264-6021:3550279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coge F, Guenin SP, Rique H, Boutin JA, Galizzi JP. Structure and expression of the human histamine H4 receptor gene. Biochem Biophys Res Commun. 2001b;284:301–309. doi: 10.1006/bbrc.2001.4976. [DOI] [PubMed] [Google Scholar]
- Connelly WM, Shenton FC, Lethbridge N, Leurs R, Waldvogel HJ, Faull RLM, et al. The histamine H4 receptor is functionally expressed on neurons in the mammalian CNS. Br J Pharmacol. 2009;157:55–63. doi: 10.1111/j.1476-5381.2009.00227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowart MD, Altenbach RJ, Liu H, Hsieh GC, Drizin I, Milicic I, et al. Rotationally constrained 2,4-diamino-5,6-disubstituted pyrimidines: a new class of histamine H4 receptor antagonists with improved druglikeness and in vivo efficacy in pain and inflammation models. J Med Chem. 2008;51:6547–6557. doi: 10.1021/jm800670r. [DOI] [PubMed] [Google Scholar]
- Damaj BB, Becerra CB, Esber HJ, Wen Y, Maghazachi AA. Functional expression of H4 histamine receptor in human natural killer cells, monocytes, and dendritic cells. J Immunol. 2007;179:7907–7915. doi: 10.4049/jimmunol.179.11.7907. [DOI] [PubMed] [Google Scholar]
- Dijkstra D, Leurs R, Chazot P, Shenton FC, Stark H, Werfel T, et al. Histamine downregulates monocyte CCL2 production through the histamine H4 receptor. J Allergy Clin Immunol. 2007;120:300–307. doi: 10.1016/j.jaci.2007.03.024. [DOI] [PubMed] [Google Scholar]
- Dijkstra D, Stark H, Chazot PL, Shenton FC, Leurs R, Werfel T, et al. Human inflammatory dendritic epidermal cells express a functional histamine H4 receptor. J Invest Dermatol. 2008;128:1696–1703. doi: 10.1038/sj.jid.5701250. [DOI] [PubMed] [Google Scholar]
- Drutel G, Peitsaro N, Karlstedt K, Wieland K, Smit MJ, Timmerman H, et al. Identification of rat H3 receptor isoforms with different brain expression and signaling properties. Mol Pharmacol. 2001;59:1–8. [PubMed] [Google Scholar]
- Duvernay MT, Filipeanu CM, Wu G. The regulatory mechanisms of export trafficking of G protein-coupled receptors. Cell signal. 2005;17:1457–1465. doi: 10.1016/j.cellsig.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Edwards JP, Venable JD. Quinoxaline compounds. Patent US 2005/0070527.
- Esbenshade TA, Browman KE, Bitner RS, Strakhova M, Cowart MD, Brioni JD. The histamine H3 receptor: an attractive target for the treatment of cognitive disorders. Br J Pharmacol. 2008;154:1166–1181. doi: 10.1038/bjp.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Esch IJ, Thurmond RL, Jongejan A, Leurs R. The histamine H4 receptor as a new therapeutic target for inflammation. Trends Pharmacol Sci. 2005;26:462–469. doi: 10.1016/j.tips.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Gantz I, Munzert G, Tashiro T, Schaffer M, Wang L, DelValle J, et al. Molecular cloning of the human histamine H2 receptor. Biochem Biophys Res Commun. 1991;178:1386–1392. doi: 10.1016/0006-291x(91)91047-g. [DOI] [PubMed] [Google Scholar]
- Gutzmer R, Diestel C, Mommert S, Kother B, Stark H, Wittmann M, et al. Histamine H4 receptor stimulation suppresses IL-12p70 production and mediates chemotaxis in human monocyte-derived dendritic cells. J Immunol. 2005;174:5224–5232. doi: 10.4049/jimmunol.174.9.5224. [DOI] [PubMed] [Google Scholar]
- Hancock AA. The challenge of drug discovery of a GPCR target: analysis of preclinical pharmacology of histamine H3 antagonists/inverse agonists. Biochem Pharmacol. 2006;71:1103–1113. doi: 10.1016/j.bcp.2005.10.033. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Harusawa S, Araki L, Zuiderveld OP, Smit MJ, Imazu T, et al. A selective human H4 receptor agonist: (-)-2-cyano-1-methyl-3-[(2R,5R)-5-[1H-imidazol-4(5)-yl]tetrahydrofuran-2-y] methylguanidine. J Med Chem. 2003;46:3162–3165. doi: 10.1021/jm0300025. [DOI] [PubMed] [Google Scholar]
- Hofstra CL, Desai PJ, Thurmond RL, Fung-Leung WP. Histamine H4 receptor mediates chemotaxis and calcium mobilization of mast cells. J Pharmacol Exp Ther. 2003;305:1212–1221. doi: 10.1124/jpet.102.046581. [DOI] [PubMed] [Google Scholar]
- Hough LB. Genomics meets histamine receptors: new subtypes, new receptors. Mol Pharmacol. 2001;59:415–419. [PubMed] [Google Scholar]
- Ikawa Y, Shiba K, Ohki E, Mutoh N, Suzuki M, Sato H, et al. Comparative study of histamine H4 receptor expression in human dermal fibroblasts. J Toxicol Sci. 2008;33:503–508. doi: 10.2131/jts.33.503. [DOI] [PubMed] [Google Scholar]
- Jablonowski JA, Grice CA, Chai W, Dvorak CA, Venable JD, Kwok AK, et al. The first potent and selective non-imidazole human histamine H4 receptor antagonists. J Med Chem. 2003;46:3957–3960. doi: 10.1021/jm0341047. [DOI] [PubMed] [Google Scholar]
- Jiang W, Lim HD, Zhang M, Desai P, Dai H, Colling PM, et al. Cloning and pharmacological characterization of the dog histamine H4 receptor. Eur J Pharmacol. 2008;592:26–32. doi: 10.1016/j.ejphar.2008.06.095. [DOI] [PubMed] [Google Scholar]
- Jongejan A, Bruysters M, Ballesteros JA, Haaksma E, Bakker RA, Pardo L, et al. Linking agonist binding to histamine H1 receptor activation. Nat Chem Biol. 2005;1:98–103. doi: 10.1038/nchembio714. [DOI] [PubMed] [Google Scholar]
- Jongejan A, Lim HD, Smits RA, de Esch IJ, Haaksma E, Leurs R. Delineation of agonist binding to the human histamine H4 receptor using mutational analysis, homology modeling, and ab initio calculations. J Chem Inf Model. 2008;48:1455–1463. doi: 10.1021/ci700474a. [DOI] [PubMed] [Google Scholar]
- Lanctot PM, Leclerc PC, Clement M, Auger-Messier M, Escher E, Leduc R, et al. Importance of N-glycosylation positioning for cell-surface expression, targeting, affinity and quality control of the human AT1 receptor. Biochem J. 2005;390:367–376. doi: 10.1042/BJ20050189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leurs R, Vollinga RC, Timmerman H. The medicinal chemistry and therapeutic potentials of ligands of the histamine H3 receptor. Prog Drug Res. 1995;45:107–165. doi: 10.1007/978-3-0348-7164-8_4. [DOI] [PubMed] [Google Scholar]
- Leurs R, Hoffmann M, Wieland K, Timmerman H. H3 receptor gene is cloned at last. Trends Pharmacol Sci. 2000;21:11–12. doi: 10.1016/s0165-6147(99)01411-x. [DOI] [PubMed] [Google Scholar]
- Leurs R, Bakker RA, Timmerman H, de Esch IJ. The histamine H3 receptor: from gene cloning to H3 receptor drugs. Nat Rev Drug Discov. 2005;4:107–120. doi: 10.1038/nrd1631. [DOI] [PubMed] [Google Scholar]
- Lim HD, van Rijn RM, Ling P, Bakker RA, Thurmond RL, Leurs R. Evaluation of histamine H1-, H2-, and H3-receptor ligands at the human histamine H4 receptor: identification of 4-methylhistamine as the first potent and selective H4 receptor agonist. J Pharmacol Exp Ther. 2005;314:1310–1321. doi: 10.1124/jpet.105.087965. [DOI] [PubMed] [Google Scholar]
- Lim HD, Smits RA, Bakker RA, van Dam CM, de Esch IJ, Leurs R. Discovery of S-(2-guanidylethyl)-isothiourea (VUF 8430) as a potent nonimidazole histamine H4 receptor agonist. J Med Chem. 2006;49:6650–6651. doi: 10.1021/jm060880d. [DOI] [PubMed] [Google Scholar]
- Lim HD, Jongejan A, Bakker RA, Haaksma E, de Esch IJ, Leurs R. Phenylalanine169 in the second extracellular loop of the human histamine H4 receptor is responsible for the difference in agonist binding between human and mouse H4 receptors. J Pharmacol Exp Ther. 2008;327:88–96. doi: 10.1124/jpet.108.140343. [DOI] [PubMed] [Google Scholar]
- Lim HD, Adami M, Guaita E, Werfel T, Smits RA, de Esch IJP, et al. Pharmacological characterization of the new histamine H4 receptor agonist VUF 8430. Br J Pharmacol. 2009;157:34–43. doi: 10.1111/j.1476-5381.2009.00200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling P, Ngo K, Nguyen S, Thurmond RL, Edwards JP, Karlsson L, et al. Histamine H4 receptor mediates eosinophil chemotaxis with cell shape change and adhesion molecule upregulation. Br J Pharmacol. 2004;142:161–171. doi: 10.1038/sj.bjp.0705729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Ma X, Jiang X, Wilson SJ, Hofstra CL, Blevitt J, et al. Cloning and pharmacological characterization of a fourth histamine receptor (H4) expressed in bone marrow. Mol Pharmacol. 2001a;59:420–426. doi: 10.1124/mol.59.3.420. [DOI] [PubMed] [Google Scholar]
- Liu C, Wilson SJ, Kuei C, Lovenberg TW. Comparison of human, mouse, rat, and guinea pig histamine H4 receptors reveals substantial pharmacological species variation. J Pharmacol Exp Ther. 2001b;299:121–130. [PubMed] [Google Scholar]
- Liu H, Altenbach RJ, Carr TL, Chandran P, Hsieh GC, Lewis LG, et al. cis-4-(Piperazin-1-yl)-5,6,7a,8,9,10,11,11a-octahydrobenzofuro[2,3-h]quinazolin-2-amine (A-987306), a new histamine H4R antagonist that blocks pain responses against carrageenan-induced hyperalgesia. J Med Chem. 2008;51:7094–7098. doi: 10.1021/jm8007618. [DOI] [PubMed] [Google Scholar]
- Lovenberg TW, Roland BL, Wilson SJ, Jiang X, Pyati J, Huvar A, et al. Cloning and functional expression of the human histamine H3 receptor. Mol Pharmacol. 1999;55:1101–1107. [PubMed] [Google Scholar]
- Morini G, Becchi G, Shenton FC, Chazot PL, Grandi D. Histamine H3 and H4 receptors are expressed on distinct endocrine cell types in the rat fundic mucosa. Inflamm Res. 2008;57(Suppl.)(1):S57–S58. doi: 10.1007/s00011-007-0628-9. [DOI] [PubMed] [Google Scholar]
- Morse KL, Behan J, Laz TM, West RE, Jr, Greenfeder SA, Anthes JC, et al. Cloning and characterization of a novel human histamine receptor. J Pharmacol Exp Ther. 2001;296:1058–1066. [PubMed] [Google Scholar]
- Nakamura T, Itadani H, Hidaka Y, Ohta M, Tanaka K. Molecular cloning and characterization of a new human histamine receptor, HH4R. Biochem Biophys Res Commun. 2000;279:615–620. doi: 10.1006/bbrc.2000.4008. [DOI] [PubMed] [Google Scholar]
- Nakaya M, Takeuchi N, Kondo K. Immunohistochemical localization of histamine receptor subtypes in human inferior turbinates. Ann Otol Rhinol Laryngol. 2004;113:552–557. doi: 10.1177/000348940411300707. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Shapiro DA, George SR, Setola V, Lee DK, Cheng R, et al. Discovery of a novel member of the histamine receptor family. Mol Pharmacol. 2001;59:427–433. doi: 10.1124/mol.59.3.427. [DOI] [PubMed] [Google Scholar]
- Oda T, Morikawa N, Saito Y, Masuho Y, Matsumoto S. Molecular cloning and characterization of a novel type of histamine receptor preferentially expressed in leukocytes. J Biol Chem. 2000;275:36781–36786. doi: 10.1074/jbc.M006480200. [DOI] [PubMed] [Google Scholar]
- Oda T, Matsumoto S, Masuho Y, Takasaki J, Matsumoto M, Kamohara M, et al. cDNA cloning and characterization of porcine histamine H4 receptor. Biochim Biophys Acta. 2002;1575:135–138. doi: 10.1016/s0167-4781(02)00236-1. [DOI] [PubMed] [Google Scholar]
- Oda T, Matsumoto S, Matsumoto M, Takasaki J, Kamohara M, Soga T, et al. Molecular cloning of monkey histamine H4 receptor. J Pharmacol Sci. 2005;98:319–322. doi: 10.1254/jphs.sc0050033. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Raible DG, Lenahan T, Fayvilevich Y, Kosinski R, Schulman ES. Pharmacologic characterization of a novel histamine receptor on human eosinophils. Am J Respir Crit Care Med. 1994;149:1506–1511. doi: 10.1164/ajrccm.149.6.8004306. [DOI] [PubMed] [Google Scholar]
- van Rijn RM, Chazot PL, Shenton FC, Sansuk K, Bakker RA, Leurs R. Oligomerization of recombinant and endogenously expressed human histamine H4 receptors. Mol Pharmacol. 2006;70:604–615. doi: 10.1124/mol.105.020818. [DOI] [PubMed] [Google Scholar]
- van Rijn RM, van Marle A, Chazot PL, Langemeijer E, Qin Y, Shenton FC, et al. Cloning and characterization of dominant negative splice variants of the human histamine H4 receptor. Biochem J. 2008;414:121–131. doi: 10.1042/BJ20071583. [DOI] [PubMed] [Google Scholar]
- Sato H, Fukushima K, Shimazak M, Urbahns K, Sakai K, Ganter F, et al. 2-Aminopiperidine derivatives. Patent WO 2005/054239.
- Sato H, Tanaka K, Shimazak M, Urbahns K, Sakai K, Ganter F, et al. 2-Aminopyrimidine. Patent WO 2005/014556.
- Shenton FC, Chazot PL. Probing the importance of N-glycosylation for [3H] clobenpropit binding to human H3 receptors expressed in HEK 293 cells. 3. Molecular and chemical aspects of the histamine receptors. Inflamm Res. 2006;55(Suppl.)(1):S40–S41. doi: 10.1007/s00011-005-0033-1. [DOI] [PubMed] [Google Scholar]
- Shin N, Coates E, Murgolo NJ, Morse KL, Bayne M, Strader CD, et al. Molecular modeling and site-specific mutagenesis of the histamine-binding site of the histamine H4 receptor. Mol Pharmacol. 2002;62:38–47. doi: 10.1124/mol.62.1.38. [DOI] [PubMed] [Google Scholar]
- Smits RA, Lim HD, Stegink B, Bakker RA, de Esch IJ, Leurs R. Characterization of the histamine H4 receptor binding site. Part 1. Synthesis and pharmacological evaluation of dibenzodiazepine derivatives. J Med Chem. 2006;49:4512–4516. doi: 10.1021/jm051008s. [DOI] [PubMed] [Google Scholar]
- Strakhova MI, Nikkel AL, Manelli AM, Hsieh GC, Esbenshade TA, Brioni JD, et al. Localization of histamine H4 receptors in the central nervous system of human and rat. Brain Res. 2009;1250:41–48. doi: 10.1016/j.brainres.2008.11.018. [DOI] [PubMed] [Google Scholar]
- Takeshita K, Bacon KB, Gantner F. Critical role of L-selectin and histamine H4 receptor in zymosan-induced neutrophil recruitment from the bone marrow: comparison with carrageenan. J Pharmacol Exp Ther. 2004;310:272–280. doi: 10.1124/jpet.103.063776. [DOI] [PubMed] [Google Scholar]
- Terzioglu N, van Rijn RM, Bakker RA, De Esch IJ, Leurs R. Synthesis and structure-activity relationships of indole and benzimidazole piperazines as histamine H4 receptor antagonists. Bioorg Med Chem Lett. 2004;14:5251–5256. doi: 10.1016/j.bmcl.2004.08.035. [DOI] [PubMed] [Google Scholar]
- Thurmond RL, Desai PJ, Dunford PJ, Fung-Leung WP, Hofstra CL, Jiang W, et al. A potent and selective histamine H4 receptor antagonist with anti-inflammatory properties. J Pharmacol Exp Ther. 2004;309:404–413. doi: 10.1124/jpet.103.061754. [DOI] [PubMed] [Google Scholar]
- Thurmond RL, Gelfand EW, Dunford PJ. The role of histamine H1 and H4 receptors in allergic inflammation: the search for new antihistamines. Nat Rev Drug Discov. 2008;7:41–53. doi: 10.1038/nrd2465. [DOI] [PubMed] [Google Scholar]
- Uveges AJ, Kowal D, Zhang Y, Spangler TB, Dunlop J, Semus S, et al. The role of transmembrane helix 5 in agonist binding to the human H3 receptor. J Pharmacol Exp Ther. 2002;301:451–458. doi: 10.1124/jpet.301.2.451. [DOI] [PubMed] [Google Scholar]
- Venable JD, Cai H, Chai W, Dvorak CA, Grice CA, Jablonowski JA, et al. Preparation and biological evaluation of indole, benzimidazole, and thienopyrrole piperazine carboxamides: potent human histamine H4 antagonists. J Med Chem. 2005;48:8289–8298. doi: 10.1021/jm0502081. [DOI] [PubMed] [Google Scholar]
- Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, et al. Structure of a β1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijtmans M, Leurs R, de Esch I. Histamine H3 receptor ligands break ground in a remarkable plethora of therapeutic areas. Expert Opin Investig Drugs. 2007;16:967–985. doi: 10.1517/13543784.16.7.967. [DOI] [PubMed] [Google Scholar]
- Wilkie TM, Scherle PA, Strathmann MP, Slepak VZ, Simon MI. Characterization of G-protein alpha subunits in the Gq class: expression in murine tissues and in stromal and hematopoietic cell lines. Proc Natl Acad Sci USA. 1991;88:10049–10053. doi: 10.1073/pnas.88.22.10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Fukui H, Sugama K, Horio Y, Ito S, Mizuguchi H, et al. Expression cloning of a cDNA encoding the bovine histamine H1 receptor. Proc Natl Acad Sci USA. 1991;88:11515–11519. doi: 10.1073/pnas.88.24.11515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Michalovich D, Wu H, Tan KB, Dytko GM, Mannan IJ, et al. Cloning, expression, and pharmacological characterization of a novel human histamine receptor. Mol Pharmacol. 2001;59:434–441. doi: 10.1124/mol.59.3.434. [DOI] [PubMed] [Google Scholar]


