Abstract
Inactivation of the von Hippel-Lindau (VHL) tumor suppressor gene causes the familial cancer syndrome, VHL disease, characterized by a predisposition to renal cell carcinoma and other tumor types. Loss of VHL gene function also is found in a majority of sporadic renal carcinomas. A preponderance of the tumor-disposing inherited missense mutations detected in VHL disease are within the elongin-binding domain of VHL. This region mediates the formation of a multiprotein VHL complex containing elongin B, elongin C, cul-2, and Rbx1. This VHL complex is thought to function as an E3 ubiquitin ligase. Here, we report that VHL proteins harboring mutations which disrupt elongin binding are unstable and rapidly degraded by the proteasome. In contrast, wild-type VHL proteins are directly stabilized by associating with both elongins B and C. In addition, elongins B and C are stabilized through their interactions with each other and VHL. Thus, the entire VHL/elongin complex is resistant to proteasomal degradation. Because the elongin-binding domain of VHL is frequently mutated in cancers, these results suggest that loss of elongin binding causes tumorigenesis by compromising VHL protein stability and/or potential VHL ubiquitination functions.
Germline mutations in the von Hippel-Lindau (VHL) tumor suppressor gene cause VHL disease, a hereditary cancer syndrome characterized by a predisposition to various tumor types including renal cell carcinoma, pheochromocytoma, and hemangioblastomas of the central nervous system (1, 2). Inactivation of the VHL gene also is implicated in both sporadic renal cell carcinoma (reviewed in ref. 2) and sporadic central nervous system hemangioblastomas (2–4). Loss of VHL function appears to be an early event in renal cell carcinogenesis (5) and reintroduction of wild-type (WT) VHL expression restores VHL-null renal cells to a nontumorigenic state (6, 7). Thus, VHL inactivation can facilitate the development of renal cell carcinoma.
The VHL gene produces two native products, full-length VHLp24(MPR) (MPR = Met-Pro-Arg), and a more abundant, internally translated product, VHLp18(MEA) (MEA = Met-Glu-Ala) (7–9). In this paper, we will refer to both of these VHL proteins simply as pVHL. Biochemical studies have shown that pVHL forms a complex with elongins B and C (7, 8, 10, 11), cul-2 (12, 13), and Rbx1 (14). Because these VHL-binding proteins are similar to components of a yeast E3 ubiquitin ligase complex, a working hypothesis is that pVHL might function to target specific substrates for ubiquitin-mediated degradation (12–14). Although no such substrates have yet been molecularly identified, VHL-dependent degradation of hypoxia-inducible factor α subunits has been implicated (15). Furthermore, pVHL has been shown to be a component of a cellular complex with in vitro ubiquitin ligase activities (16, 17).
A majority of the inherited missense mutations associated with VHL disease are within the elongin-binding domain of pVHL (11, 13, 18). Many of these tumor-predisposing mutations have been demonstrated to disrupt elongin binding (10, 13, 19, 20), underscoring the biological importance of the pVHL/elongin complex. The crystal structure of this complex was solved (21) and showed that elongin C interacts directly with an α-helical domain of pVHL and forms a bridge to elongin B, as expected from previous studies (10, 22). Cul-2 also associates indirectly with the pVHL complex through elongins B and C (12, 13, 23). Rbx1 can interact independently with each member of the complex (14).
Although the pVHL/elongin/cul-2/Rbx1 complex is similar to a yeast ubiquitin ligase complex, the individual roles of each protein in the complex are not known. Elongins B and C also have been shown to bind to the SOCS (suppressor of cytokine signaling) family of proteins through the SOCS-box motif, which contains an elongin-binding domain (24). Interestingly, the binding of elongins B and C to a SOCS family member, SOCS-1, was shown to inhibit the degradation of the SOCS-1 protein. We hypothesized that elongin binding also might play a similar role for VHL proteins.
In this report, we demonstrate that VHL proteins that contain naturally occurring tumor-disposing mutations that disrupt the pVHL/elongin complex are unstable and degraded by a proteasome-dependent pathway. Moreover, we provide direct evidence that pVHL is stabilized by association with both elongins B and C. These data suggest that elongin binding plays two functions for pVHL: (i) to allow interaction with other molecules in the ubiquitin ligase complex, including cul-2, and (ii) to prevent proteasomal degradation of pVHL. Tumorigenesis resulting from VHL mutations that disrupt elongin binding may be caused by the loss of either or both of these functions.
Materials and Methods
Plasmids.
Flag-tagged VHLp24(MPR) plasmids were constructed by PCR amplification of the VHL cDNA vector, g7 (provided by Igor Kuzmin and Michael Lerman, National Cancer Institute, Frederick, MD), using a 5′-primer coding for the Flag epitope (DYKDDDDK) upstream of VHL sequences and 3′-primers containing stop codons following amino acids 154, 178, 197, or 213. PCR products were then directionally cloned into HindIII and XhoI sites in the vector, pCR3 (Invitrogen), as previously described (7). Internal deletion (amino acids 114–178) and point mutations of the VHL-coding region also were performed as previously described (7). HA-elongin B and Flag-elongin C expression constructs, both containing epitope tags at their respective N termini, were similarly created by PCR amplification (from plasmids provided by Arnim Pause, National Institutes of Health) and directional cloning into the pCR3 vector. Flag-tagged cul-2 was constructed by PCR amplification of a cul-2 cDNA vector (12) (kindly provided by Robert Stearman, National Institutes of Health) and directional cloning into BamHI and XhoI sites in the vector, pCR3 (Invitrogen), as previously described. All plasmids were confirmed by restriction enzyme digestion and DNA sequence analysis.
Cell Culture and Transfections.
786-O renal carcinoma cells were obtained from the American Type Culture Collection (Manassas, VA). 293T cells were a gift from Richard Pestell (Albert Einstein College of Medicine). All cells were grown in DMEM containing 10% FCS. Stable transfection of 786-O renal carcinoma cells was performed as described (7). 293T cells were transfected in 60-mm culture dishes with 8 μl of Lipofectamine Plus reagent, 12 μl of Lipofectamine (Life Technologies, Gaithersburg, MD), and a total of 6 μg of DNA per transfection (2 μg each of each plasmid tested and, when necessary, supplemented with empty pCR3 vector to a total of 6 μg of DNA). Forty-eight hours after transfection, cells from each transfection were divided equally into multiple 35-mm culture dishes, allowed to grow overnight, and then treated with either cycloheximide (100 μg/ml; Sigma) for various time points or lactacystin (10 μM final concentration; Calbiochem, La Jolla, CA) for 6 h.
Antibodies and Western Blotting.
mAb to the Flag epitope (M5) was purchased from Sigma and was used at an approximate concentration of 2 μg/ml for Western blotting. VHL mAb 11E12 supernatant (7) was used at a 1:20 dilution for Western blotting. Hybridoma (mAb) supernatant to HA-epitope (kindly provided by Liang Zhu, Albert Einstein College of Medicine) was used at a 1:4 dilution. For all Western blots, total cell lysates were prepared as described (7) and protein concentrations were determined by Bradford assay (Bio-Rad, Hercules, CA). Lysates were then normalized for equal protein loading (10 μg) in each lane.
Pulse/Chase Analysis.
Metabolic labeling was performed essentially as previously described (7), except cells were grown in 35-mm culture dishes and pulsed with 300 μl of medium containing 500 μCi/ml of [35S]methionine (EXPRE35S35S, New England Nuclear, Wilmington, DE, 1 μCi = 37 kBq) for 2 h. Cells were then washed with fresh medium and chased by incubating in unlabeled medium for 0, 2, 4, or 6 h. In some culture dishes, lactacystin (10 μM) was included during the 6-h chase. All subsequent steps were done at 4°C. Cells were lysed for 30 min in 250 μl of lysis buffer (50 mM Hepes, pH 7.6/250 mM NaCl/0.1% Nonidet P-40/5 mM EDTA/1 mM phenylmethylsulfonyl fluoride (PMSF) containing 1 μg/ml each of aprotinin, bestatin, and leupeptin), and lysates were clarified by microfuge for 15 min. Clarified lysate (50 μl) was then diluted to 300 μl with lysis buffer and incubated for 3 h with 50 μl of Flag-agarose (M2) beads (Sigma). Immunoprecipitated complexes were then washed four times with lysis buffer and were eluted, separated by SDS-PAGE, and visualized as previously described (7). Band intensities were quantified by using a Storm 860 PhosphorImager and imagequant software (Molecular Dynamics, Sunnyvale, CA).
Results
Elongin-Binding Mutants of pVHL Are Degraded by the Proteasome.
In the course of creating a series of cell lines stably transfected with various mutant VHL constructs, we were consistently unable to get high level expression of certain mutant VHL proteins. We hypothesized that these mutations in pVHL might render it susceptible to proteasomal degradation. To test this hypothesis, we compared the stability of WT and mutant VHL proteins, which were stably expressed in VHL-null 786-O renal carcinoma cells (Fig. 1).
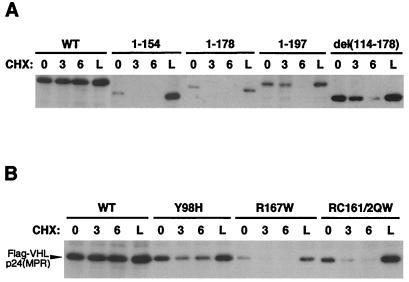
Figure 1.
Loss of elongin binding leads to increased proteasome-dependent degradation of pVHL. (A) 786-O cells stably expressing either WT or deletion mutant (containing amino acids indicated above blot) Flag-tagged pVHL were incubated with cycloheximide (CHX) for 0, 3, or 6 h, or with lactacystin (L) for 6 h, as indicated above blot. Whole cell lysates were normalized for equal protein loading (10 μg) in each lane. Western blotting was performed by using anti-Flag mAb. (B) 786-O cells stably expressing either WT or point mutant (containing amino acids substitutions indicated above blot) Flag-tagged pVHL were analyzed as in A. Position of WT and point mutant Flag-VHLp24(MPR) proteins are indicated by an arrowhead to the left of the blot.
Cells expressing WT or mutant VHL proteins (tagged with the Flag epitope) were treated with cycloheximide (to inhibit new protein synthesis) for various time points or lactacystin (to inhibit proteasomal degradation), and pVHL levels were determined by anti-Flag Western blotting. WT pVHL was extremely stable during cycloheximide treatment, and lactacystin did not increase the levels of WT pVHL (Fig. 1A, WT). In contrast, mutant VHL proteins demonstrated varying degrees of instability in this assay. A pVHL truncated at amino acid 154 (thereby removing the elongin-binding domain) was particularly unstable: steady–state levels of protein were lower than observed for WT pVHL and 3-h cycloheximide treatment led to undetectable protein levels (Fig. 1A, 1–154). Strikingly, lactacystin treatment restored pVHL () protein to levels seen with WT pVHL (Fig. 1A, 1–154, lane L), indicating that the mutant pVHL was undergoing proteasome-dependent degradation. A pVHL truncated at amino acid 178, which contains the elongin-binding domain, yet lacks adjacent residues necessary to sustain elongin binding (13), was similarly unstable (Fig. 1A, 1–178). A pVHL truncated at amino acid 197, which supports elongin binding (13), was more stable: the protein was still detected following 3-h cycloheximide treatment; however, it was undetectable after 6 h of cycloheximide (Fig. 1A, 1–197). These results suggested that elongin binding influences the stability of pVHL. Interestingly, an internal deletion, which alters the subcellular localization of pVHL [Fig. 1A, del(114–178) (A.R.S. and R.D.B., unpublished data)], was considerably more stable in this assay than other pVHL deletions that also lack elongin binding (e.g., 1–154 and 1–178). Thus, subcellular localization may play an important role in pVHL stability.
To assess the effect of elongin binding on pVHL stability, we also examined point mutations in pVHL using the cycloheximide/lactacystin assay (Fig. 1B). The elongin-binding mutations tested were the Arg-167 to Trp (R167W) mutation, a hotspot mutation in familial VHL disease shown to diminish elongin binding (19, 25), and an Arg-161 to Gln/Cys-162 to Trp double point mutation (RC161/2QW), which substitutes residues critical for contact with elongin C (13, 20). These were compared to WT pVHL and a pVHL with a Tyr-98 to His (Y98H) mutation in the β-domain of pVHL (21), which does not affect elongin binding (19). WT pVHL and the Y98H mutant were both stable in this assay (Fig. 1B, WT and Y98H). In contrast, pVHLs with point mutations in the elongin binding were degraded in a proteasome-dependent manner (Fig. 1B, R167W and RC161/2QW). Thus, mutations in the elongin-binding domain render pVHL unstable and susceptible to proteasomal degradation.
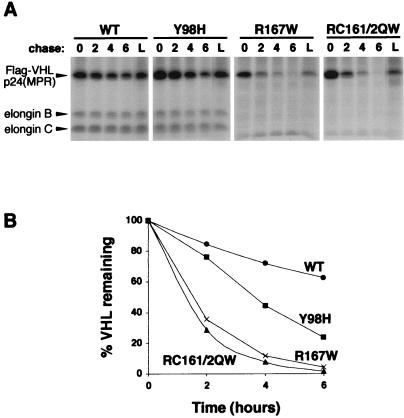
To more directly evaluate the degradation of WT and point mutant VHL proteins, we also performed pulse/chase analysis (Fig. 2). pVHL-expressing 786-O cell lines (used in Fig. 1B) were metabolically labeled with [35S]methionine, and VHL proteins were immunoprecipitated (Fig. 2A). Elongins B and C were confirmed to coimmunoprecipitate with WT pVHL and the Y98H mutant, but not with elongin-binding mutants R167W and RC161/2QW (Fig. 2A). As in the cycloheximide assay, WT and Y98H mutant pVHL were stable, whereas elongin-binding mutants were more rapidly degraded (Fig. 2 A and B). Again, proteasomal inhibition by lactacystin blocked degradation of mutant pVHL (Fig. 2A, L lanes). Protein half-lives were determined following quantitation of the band intensities (Fig. 2B). WT pVHL and the Y98H mutant had half-lives of 8.9 h and 3.3 h, respectively. Elongin mutants R167W and RC161/2QW had significantly diminished half-lives of 1.3 h and 1.1 h, respectively. Therefore, mutations in the elongin-binding domain led to more rapid proteasomal degradation of pVHL.
Figure 2.
Pulse/chase analysis of pVHL. (A) 786-O cells stably expressing either WT or point mutant (containing amino acids substitutions indicated above each panel) Flag-tagged pVHL were pulsed with [35S]methionine for 2 h. Cells were then chased with unlabeled media for 0, 2, 4, or 6 h, or for 6 h with lactacystin (L), as indicated above each autoradiograph panel. Flag-VHL proteins were immunoprecipitated with anti-Flag agarose beads (Sigma) and complexes were separated by SDS-PAGE and visualized by fluorography. Positions of Flag-VHLp24(MPR), elongin B, and elongin C proteins are indicated by arrowheads to the left. (B) Radioactive bands in A were quantified and graphically represented.
Elongin Binding Stabilizes pVHL.
We then sought to demonstrate that the instability of mutant pVHL directly results from loss of elongin binding and is not due to changes in protein conformation that may arise from amino acid substitutions in the elongin-binding domain. Toward this end, VHL expression vectors were transiently transfected into 293T cells. These cells have a high transfection efficiency and contain large T-antigen, allowing for amplification of plasmids containing an simian virus 40 origin of replication (such as those used in these experiments). Thus, high levels of VHL expression, probably in excess of elongin B and C levels, were obtained. In some transfections, elongin B and C expression vectors were cotransfected with VHL. Transfected cells were then treated with cycloheximide for various times and pVHL stability was determined by Western blotting (Fig. 3). In this assay, over-expressed WT pVHL was unstable: protein levels dropped below the level of detection after 24 h of cycloheximide treatment (Fig. 3A, Left panel). Strikingly, cotransfection of elongins B and C stabilized WT pVHL (Fig. 3A, Right panels). Elongin-mediated stabilization of pVHL did not occur when the RC161/2QW elongin-binding mutant was used (Fig. 3B). Thus, pVHL stabilization was a direct effect of elongin binding.
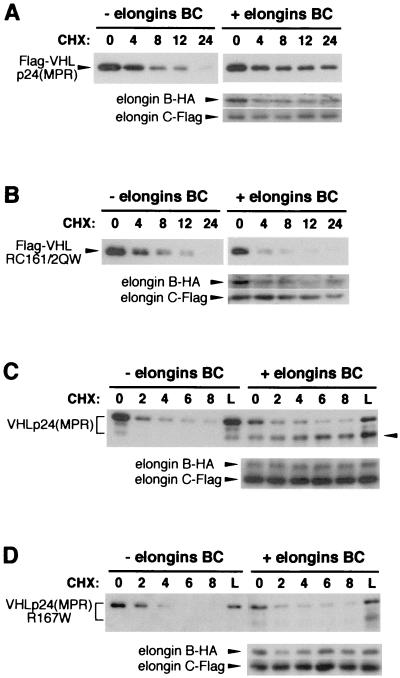
Figure 3.
Elongin binding stabilizes VHL products. VHL expression vectors were transiently transfected into 293T cells, either with no cotransfection (left panels, − elongins BC) or with cotransfection of both elongin B and elongin C vectors (right panels, + elongins BC). Transfected cells were treated with cycloheximide (CHX) for various time points (indicated above each blot) or with lactacystin (L). Whole cell lysates were normalized for equal protein loading (10 μg) in each lane. VHL Western blotting was performed by using mAbs to either Flag (A and B) or VHL (11E12, C and D). Positions of Flag-pVHL (A and B, Upper panels) are indicated by arrowheads to the left of each blot. Positions of untagged VHLp24(MPR) proteins (C and D, Upper panels) are indicated by a bracket to the left of each blot. Position of a faster-migrating VHLp24(MPR) polypeptide in C is indicated by an arrowhead to the right of the blot. Elongin B-HA and elongin C-Flag (A–D, Lower panels), detected with antibodies to the appropriate epitope-tag, also are indicated by arrowheads. VHL constructs used: Flag-VHLp24(MPR) (A); Flag-VHLp24(RC161/2QW) (B); VHLp24(MPR) (C); and VHLp24(R167W) (D).
To rule out possible effects of the Flag epitope (used in the previous experiments) on pVHL degradation, we also performed experiments with an untagged VHLp24(MPR) expression vector (7). Transiently expressed VHLp24(MPR) was detected as a heterogeneous population of at least two major polypeptides (Fig. 3C, denoted by bracket), in a pattern consistent with endogenous VHLp24(MPR) proteins in most cell lines (7). All forms of VHLp24(MPR) were degraded in the cycloheximide treatment assay (Fig. 3C, − elongins BC). Interestingly, cotransfection of elongins B and C had a minimal effect on the degradation of the slower-migrating (higher apparent molecular mass) form of VHLp24(MPR), but did stabilize the faster-migrating VHLp24(MPR) protein (Fig. 3C, + elongins BC, lower band denoted by arrowhead). Again, elongin-mediated stabilization of VHLp24(MPR) did not occur when the R167W elongin-binding mutant was used (Fig. 3D), demonstrating that elongin binding was directly responsible for pVHL stabilization.
Elongin cotransfection also increased the relative abundance of the WT faster-migrating (lower apparent molecular mass) form of VHLp24(MPR) in the absence of cycloheximide (Fig. 3C, + elongins BC, 0 h; compare to − elongins BC, 0 h) and levels of this polypeptide increased during cycloheximide treatment (Fig. 3C, + elongins BC, 2, 4, 6, and 8 h). This suggests that the slower-migrating VHLp24(MPR) may be converted to the faster-migrating form under these conditions. Elongin binding appears to be necessary for this conversion. Although the nature of these differently-migrating VHLp24(MPR) products in not known, the faster-migrating protein migrates closer to its expected molecular mass of 24 kDa. Thus, the slower-migrating (higher apparent molecular mass) forms of VHLp24(MPR) might result from posttranslational modification(s). The present data suggests that elongin binding might influence these potential VHL modifications, which may have consequences on pVHL stabilization.
Elongin B and C Binding Are Both Needed for Maximal pVHL Stabilization.
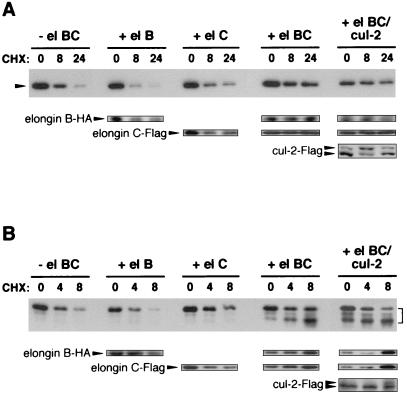
We next sought to determine which members of the pVHL/elongin/cul-2 complex are necessary for stabilization of pVHL. VHL expression vectors and various combinations of elongin B, elongin C, and cul-2 expression vectors were transiently transfected into 293T cells, and stability of pVHL (both Flag-tagged and untagged forms) was monitored by Western blotting after cycloheximide treatment (Fig. 4 A and B, top panels). Expression of elongins and cul-2 was also confirmed by Western blotting (Fig. 4 A and B, Lower). Cotransfection of elongin B and VHL had no effect on pVHL stability (Fig. 4 A and B). Cotransfection of VHL and elongin C led to a slight increase in pVHL stability (+ el B, + el C). However, cotransfection of both elongins B and C dramatically stabilized pVHL (Fig. 4 A and B, + el BC). Cul-2 (which was detected as two polypeptide bands, perhaps due to covalent attachment of NEDD8; ref. 26) did not affect the elongin-mediated stabilization of pVHL (Fig. 4 A and B, + el BC/cul-2). Thus, both elongins B and C, but not cul-2, are needed for maximal pVHL stabilization.
Figure 4.
Both elongins B and C are needed for pVHL stabilization. VHL expression vectors were transiently transfected into 293T cells, either with no cotransfection (− el BC) or with cotransfection of elongin B (+ el B), elongin C (+ el C), elongin B and elongin C (+ el BC), or elongin B, elongin C, and cul-2 vectors (+ el BC/cul-2). Transfected cells were treated with cycloheximide (CHX) for various time points (indicated above each blot). Whole cell lysates were normalized for equal protein loading (10 μg) in each lane. VHL Western blotting (Upper panels) was performed using either an anti-Flag mAb (A) or mAb 11E12 (B). Elongin B-HA, elongin C-Flag, and cul-2-Flag, detected with antibodies to the appropriate epitope-tag, are indicated by arrowheads (Lower panels). (A) Position of Flag-VHLp24(MPR) is indicated by an arrowhead to the left of the blot (Upper). (B) Positions of VHLp24(MPR) products are indicated by a bracket to the right of the blot (Upper).
Interestingly, cotransfection of VHL and both elongins B and C also led to stabilization of elongins B and C (Fig. 4 A and B, Lower, compare + el BC to + el B and + el C). Although the specific contribution of pVHL toward elongin stability was not assessed, one distinct possibility is that elongin C (which directly binds to pVHL) is responsible for pVHL stabilization and elongin B (which directly binds to elongin C, but not pVHL) stabilizes elongin C, leading to further stabilization of pVHL. Thus, the net result is that all components of the pVHL/elongin BC ternary complex are stabilized by these interactions. The formation of a complex that is resistant to proteasomal degradation may be important for VHL ubiquitination functions and may be necessary for VHL-mediated tumor suppression.
Discussion
In this report, we show that VHL proteins containing mutations that disrupt elongin binding are unstable and degraded in a proteasome-dependent manner. Furthermore, we demonstrate that pVHL is directly stabilized by complexing with elongins B and C. Substitution of the amino acids constituting the elongin-binding domain of pVHL occurs frequently in families with VHL disease (25, 27–29). Because many of these mutations have been proven to disrupt elongin binding (10, 13, 19, 20), it is likely that the majority of these mutant VHL proteins are unstable. The instability of these elongin-mutant VHL proteins suggests an additional or alternative mechanism for tumorigenesis in VHL disease: loss of VHL function as a result of proteasomal degradation of VHL proteins.
To function in a ubiquitin ligase complex, pVHL itself might need to be protected from ubiquitin-mediated degradation that may occur as a result of contact with other components of the ubiquitination machinery. Toward this end, it is interesting that cul-2 binds indirectly to VHL through elongin C. Thus, the formation of a proteasome-resistant pVHL/elongin BC complex is a prerequisite for cul-2 binding, ensuring a protected pVHL to participate in ubiquitination. The binding of elongins may therefore be primarily structural, serving to stabilize pVHL and provide a bridge to cul-2, which may play a more direct role in ubiquitination by interacting (with help from Rbx1) with an E2 ubiquitin conjugating enzyme. Elongin binding may represent a general mechanism whereby proteins, especially those involved in cellular ubiquitination, are themselves protected against (ubiquitin-mediated) proteasomal degradation. This notion is further supported by the observation that the SOCS-1 protein is inhibited from degradation by complexing with elongins B and C (24). It is likely that other SOCS-box proteins are similarly resistant to degradation when bound to elongins B and C.
Formation of the pVHL complex also may provide protection for the other members of the complex. This protection may result from interactions between these proteins. Yeast elongin C has been previously shown to be stabilized by interaction with either VHL or elongin A (30). Reciprocally, we observed that elongin C binding partially protected pVHL (Fig. 4). Contacts between α-helices of elongin C and pVHL (21) are likely to be responsible for these phenomena. Elongin B, initially noted for its role as an elongin C chaperone (22), binds to elongin C through interactions between β-sheet regions (21). The binding of elongin B greatly enhanced the stabilization of both pVHL and elongin C (Fig. 4), demonstrating that interactions between these proteins serve to strengthen the entire pVHL/elongin BC complex. It is interesting to speculate that cul-2 also may be protected from self-ubiquitination and degradation (in a manner similar to SUMO-1 protection of IκB-α ref. 31) through the covalent attachment of NEDD8 (26). The pVHL complex has been shown to promote this modification (26, 32). Thus, resistance to proteasomal degradation may be important for all members of the pVHL complex to function properly.
The subcellular localization of pVHL also might necessitate the protection of the pVHL complex. We have determined that pVHL is likely to be (A.R.S. and R.D.B., unpublished data) localized to the cytosolic face of the endoplasmic reticulum, a known cellular ubiquitination site (33). Proteins localized to this site may need to be protected from spurious ubiquitination. In fact, we also have observed that VHL fusion proteins which localize to this cellular region undergo ubiquitination (not self-ubiquitination, but performed by other ubiquitination molecules, presumably found at this subcellular locale), whereas VHL mutants that are unable to localize to the cytosol/endoplasmic reticulum do not get ubiquitinated. Accordingly, one such VHL deletion mutant with aberrant subcellular localization, del(114–178) (Fig. 1A), was considerably more stable than would be predicted from the fact that this mutant lacks the elongin-binding domain. Thus, subcellular localization may be an important factor in pVHL degradation.
Additionally, the chaperonin, TriC, also may play a role in pVHL degradation. Formation of the VHL/elongin complex requires TriC-mediated folding and assembly (34). After elongin binding, TriC is released from the pVHL/elongin complex. However, an elongin-binding point mutation of pVHL resulted in increased levels of bound TriC and was correlated with enhanced protease sensitivity (34). These results may suggest that TriC-associated pVHL remains in a partially unfolded, proteasome-sensitive conformation. However, a proteasome-resistant pVHL/elongin complex may be formed upon proper folding of pVHL by TriC. Thus, the proteasomal degradation of pVHL (not bound to elongins B and C) detected in our assays may be a result of TriC-mediated pVHL unfolding. Along these lines, it is interesting that a VHL deletion mutant, del(114–178), that does not bind to the TriC complex (ref. 33 and E.J.D., unpublished observations) demonstrated increased stability (relative to other mutant pVHL) despite lacking an elongin-binding domain (Fig. 1A).
The mechanisms whereby loss of VHL function leads to tumor formation have not been fully elucidated. Elongin-binding domain mutations have been speculated to cause tumorigenesis through the loss of elongin-dependent VHL cellular functions. For example, elongin-binding mutations have been shown to block VHL-mediated regulation of hypoxia-inducible mRNAs (including vascular endothelial growth factor) (13) and to abrogate VHL-associated ubiquitin ligase activities (16, 17). Although these VHL cellular functions may be important, they have not been causally linked to VHL tumor suppression. Thus, it is possible that VHL tumor suppression relies on activities that are performed by other regions of the VHL protein. This notion could account for the many tumor-promoting missense mutations which are not within the elongin-binding domain of VHL. Thus, it is possible that elongin-binding site mutations cause tumor formation solely because VHL proteins carrying these mutations are rapidly degraded, resulting in a loss of VHL function. Alternatively, tumor formation resulting from elongin-binding site mutations may occur through the synergistic effects of pVHL degradation and loss of crucial elongin/cul-2-dependent VHL functions such as ubiquitin ligase activity. The use of elongin B and C mutants, which are capable of stabilizing pVHL but unable to associate with cul-2, might help to resolve the specific contribution of pVHL stability toward VHL-mediated tumor suppression.
Acknowledgments
This work was supported in part by a grant from the VHL Family Alliance. A.R.S. was supported by a National Institutes of Health training grant (DK 07110). E.J.D. was supported by a National Institutes of Health training grant (DK 07218). DNA oligonucleotides were provided by the oligonucleotide facility of the Cancer Center of the Albert Einstein College of Medicine (partially supported by grant CA 13330). VHL mAb 11E12 was produced at the Hybridoma Facility of the Cancer Center of the Albert Einstein College of Medicine (partially supported by grant CA 13330).
Abbreviations
- VHL
von Hippel-Lindau
- pVHL
von Hippel-Lindau tumor suppressor gene products
- MPR
Met-Pro-Arg
- SOCS
suppressor of cytokine signaling
- WT
wild-type
- R167W
Arg-167 to Trp
- RC161/2QW
Arg-161 to Gln/Cys-162 to Trp
- Y98H
Tyr-98 to His
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Latif F, Tory K, Gnarra J, Yao M, Duh F M, Orcutt M L, Stackhouse T, Kuzmin I, Modi W, Geil L, et al. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 2.Linehan W M, Lerman M I, Zbar B. J Am Med Assoc. 1995;273:564–570. [PubMed] [Google Scholar]
- 3.Kanno H, Kondo K, Ito S, Yamamoto I, Fujii S, Torigoe S, Sakai N, Hosaka M, Shuin T, Yao M. Cancer Res. 1994;54:4845–4847. [PubMed] [Google Scholar]
- 4.Lee J Y, Dong S M, Park W S, Yoo N J, Kim C S, Jang J J, Chi J G, Zbar B, Lubensky I A, Linehan W M, et al. Cancer Res. 1998;58:504–508. [PubMed] [Google Scholar]
- 5.Lubensky I A, Gnarra J R, Bertheau P, Walther M M, Linehan W M, Zhuang Z. Am J Pathol. 1996;149:2089–2094. [PMC free article] [PubMed] [Google Scholar]
- 6.Iliopoulos O, Kibel A, Gray S, Kaelin W G. Nat Med. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 7.Schoenfeld A, Davidowitz E J, Burk R D. Proc Natl Acad Sci USA. 1998;95:8817–8822. doi: 10.1073/pnas.95.15.8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iliopoulos O, Ohh M, Kaelin W G., Jr Proc Natl Acad Sci USA. 1998;95:11661–11666. doi: 10.1073/pnas.95.20.11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blankenship C, Naglich J G, Whaley J M, Seizinger B, Kley N. Oncogene. 1999;18:1529–1535. doi: 10.1038/sj.onc.1202473. [DOI] [PubMed] [Google Scholar]
- 10.Duan D R, Pause A, Burgess W H, Aso T, Chen D Y T, Garrett K P, Conaway R C, Conaway J W, Linehan W M, Klausner R D. Science. 1995;269:1402–1406. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- 11.Kibel A, Iliopoulos O, DeCaprio J A, Kaelin W G. Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 12.Pause A, Lee S, Worrell R A, Chen D Y, Burgess W H, Linehan W M, Klausner R D. Proc Natl Acad Sci USA. 1997;94:2156–2161. doi: 10.1073/pnas.94.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lonergan K M, Iliopoulos O, Ohh M, Kamura T, Conaway R C, Conaway J W, Kaelin W G., Jr Mol Cell Biol. 1998;18:732–741. doi: 10.1128/mcb.18.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamura T, Koepp D M, Conrad M N, Skowyra D, Moreland R J, Iliopoulos O, Lane W S, Kaelin W G, Jr, Elledge S J, Conaway R C, et al. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 15.Maxwell P H, Wiesener M S, Chang G W, Clifford S C, Vaux E C, Cockman M E, Wykoff C C, Pugh C W, Maher E R, Ratcliffe P J. Nature (London) 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 16.Lisztwan J, Imbert G, Wirbelauer C, Gstaiger M, Krek W. Genes Dev. 1999;13:1822–1833. doi: 10.1101/gad.13.14.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwai K, Yamanaka K, Kamura T, Minato N, Conaway R C, Conaway J W, Klausner R D, Pause A. Proc Natl Acad Sci USA. 1999;96:12436–12441. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kishida T, Stackhouse T M, Chen F, Lerman M I, Zbar B. Cancer Res. 1995;55:4544–4548. [PubMed] [Google Scholar]
- 19.Duan D R, Humphrey J S, Chen D Y, Weng Y, Sukegawa J, Lee S, Gnarra J R, Linehan W M, Klausner R D. Proc Natl Acad Sci USA. 1995;92:6459–6463. doi: 10.1073/pnas.92.14.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohh M, Takagi Y, Aso T, Stebbins C E, Pavletich N P, Zbar B, Conaway R C, Conaway J W, Kaelin W G., Jr J Clin Invest. 1999;104:1583–1591. doi: 10.1172/JCI8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stebbins C E, Kaelin W G, Jr, Pavletich N P. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- 22.Aso T, Lane W S, Conaway J W, Conaway R C. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- 23.Pause A, Peterson B, Schaffar G, Stearman R, Klausner R D. Proc Natl Acad Sci USA. 1999;96:9533–9538. doi: 10.1073/pnas.96.17.9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamura T, Sato S, Haque D, Liu L, Kaelin W G, Jr, Conaway R C, Conaway J W. Genes Dev. 1998;12:3872–3881. doi: 10.1101/gad.12.24.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen F, Kishida T, Yao M, Hustad T, Glavac D, Dean M, Gnarra J R, Orcutt M L, Duh F M, Glenn G, et al. Hum Mutat. 1995;5:66–75. doi: 10.1002/humu.1380050109. [DOI] [PubMed] [Google Scholar]
- 26.Liakopoulos D, Busgen T, Brychzy A, Jentsch S, Pause A. Proc Natl Acad Sci USA. 1999;96:5510–5515. doi: 10.1073/pnas.96.10.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whaley J M, Naglich J, Gelbert L, Hsia Y E, Lamiell J M, Green J S, Collins D, Neumann H P, Laidlaw J, Li F P, et al. Am J Hum Genet. 1994;55:1092–1102. [PMC free article] [PubMed] [Google Scholar]
- 28.Crossey P A, Richards F M, Foster K, Green J S, Prowse A, Latif F, Lerman M I, Zbar B, Affara N A, Ferguson-Smith M A, et al. Hum Mol Genet. 1994;3:1303–1308. doi: 10.1093/hmg/3.8.1303. [DOI] [PubMed] [Google Scholar]
- 29.Zbar B, Kishida T, Chen F, Schmidt L, Maher E R, Richards F M, Crossey P A, Webster A R, Affara N A, Ferguson-Smith M A, et al. Hum Mutat. 1996;8:348–357. doi: 10.1002/(SICI)1098-1004(1996)8:4<348::AID-HUMU8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 30.Botuyan M V, Koth C M, Mer G, Chakrabartty A, Conaway J W, Conaway R C, Edwards A M, Arrowsmith C H, Chazin W J. Proc Natl Acad Sci USA. 1999;96:9033–9038. doi: 10.1073/pnas.96.16.9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desterro J M, Rodriguez M S, Hay R T. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 32.Wada H, Yeh E T, Kamitani T. J Biol Chem. 1999;274:36025–36029. doi: 10.1074/jbc.274.50.36025. [DOI] [PubMed] [Google Scholar]
- 33.Kopito R R. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- 34.Feldman D E, Thulasiraman V, Ferreyra R G, Frydman J. Mol Cell. 1999;4:1051–1061. doi: 10.1016/s1097-2765(00)80233-6. [DOI] [PubMed] [Google Scholar]