Abstract
The mammalian gastrointestinal microbiota exerts a strong influence on host lipid and cholesterol metabolism. In this study, we have characterized the interplay among diet, gut microbial ecology, and cholesterol metabolism in a hamster model of hypercholesterolemia. Previous work in this model had shown that grain sorghum lipid extract (GSL) included in the diet significantly improved the high-density lipoprotein (HDL)/non-HDL cholesterol equilibrium (T. P. Carr, C. L. Weller, V. L. Schlegel, S. L. Cuppett, D. M. Guderian, Jr., and K. R. Johnson, J. Nutr. 135:2236-2240, 2005). Molecular analysis of the hamsters' fecal bacterial populations by pyrosequencing of 16S rRNA tags, PCR-denaturing gradient gel electrophoresis, and Bifidobacterium-specific quantitative real-time PCR revealed that the improvements in cholesterol homeostasis induced through feeding the hamsters GSL were strongly associated with alterations of the gut microbiota. Bifidobacteria, which significantly increased in abundance in hamsters fed GSL, showed a strong positive association with HDL plasma cholesterol levels (r = 0.75; P = 0.001). The proportion of members of the family Coriobacteriaceae decreased when the hamsters were fed GSL and showed a high positive association with non-HDL plasma cholesterol levels (r = 0.84; P = 0.0002). These correlations were more significant than those between daily GSL intake and animal metabolic markers, implying that the dietary effects on host cholesterol metabolism are conferred, at least in part, through an effect on the gut microbiota. This study provides evidence that modulation of the gut microbiota-host metabolic interrelationship by dietary intervention has the potential to improve mammalian cholesterol homeostasis, which has relevance for cardiovascular health.
The mammalian gut microbiota interacts intimately with its host, affecting both host metabolic and immunological phenotypes with important consequences for health (18, 22, 32). Recent studies have revealed complex linkages between the gut microbiome and host metabolism, with the microbes exerting effects on the energy balance by influencing glucose and lipid metabolism (2, 7, 28). This intimate metabolic relationship is most likely the consequence of a long coevolutionary process that resulted in a mutualistic relationship between the host and its microbial partners (25). However, life in industrialized societies has introduced profound changes into the human environment (e.g., diet, antibiotics, hospital deliveries, hygiene, etc.) that are markedly different from the conditions to which humans have evolved and that are likely to have occurred too abruptly for the human microbiome to adjust. Consequently, aberrations of the gut microbiota induced through lifestyle factors could be relevant to the etiology of several complex human diseases whose occurrence has markedly increased in developed countries. Interestingly, imbalances in the gut microbiota have been reported for obesity, type 1 and 2 diabetes, some allergies, and inflammatory bowel diseases in humans and animal models (7, 24, 43, 45, 48). The connection between gut bacteria and disease suggests an intriguing paradigm on how to view and potentially treat complex diseases. Specific bacterial populations in the intestine could be pharmaceutical targets to maintain or restore metabolic functions (6, 17).
Coronary heart disease (CHD) continues to be a major cause of death in developed countries and is another example of a “western disease” that is less common in underdeveloped countries but increases in frequency with adoption of western customs (4). Most risk factors for CHD (obesity, high blood pressure, type 2 diabetes, heredity, high cholesterol, and diet) have been linked to the gut microbiota (7, 17, 20, 30, 45), and gut bacteria have been suggested to play a role in the etiology of cardiovascular disease (16, 33). Cholesterol metabolism is a key factor in susceptibility to CHD, and as early as 1959, it has been shown that germfree rats have higher serum cholesterol concentrations than their conventional counterparts do (12). Several mechanisms have been proposed by which gut bacteria could influence host cholesterol metabolism (13). Bacterial conversions of bile acids (such as the formation of secondary bile acids) are likely to play a role, as they affect enterohepatic circulation, de novo synthesis of bile acids, emulsification, and cholesterol absorption (10, 28, 30). A further mechanism by which gut bacteria might influence cholesterol metabolism is through Fiaf (fasting-induced adipocyte factor), which is selectively suppressed in the intestinal epithelium by the gut microbiota (1, 2). Fiaf is an important regulator of lipid metabolism (e.g., through its inhibition of lipoprotein lipase) and has been shown to increase total cholesterol and high-density lipoprotein (HDL) cholesterol levels when overexpressed in transgenic mice (26).
There are several reasons why hamsters are an excellent model for studying the metabolic relationships among diet, cholesterol metabolism, and gut microbiota in relation to health. First, hamsters are omnivorous, and their blood lipid profiles respond to diets in a predictive manner similar to humans (5). Second, unlike mice and rats which lack cholesterol ester transfer protein, hamsters exhibit all of the enzymatic pathways in lipoprotein and bile metabolism that are also present in humans. They exhibit limited hepatic synthesis of cholesterol and bile acids, resulting in more relevant data when extrapolating to humans (23). Third, hamsters develop atherosclerosis in a predictive manner in response to dietary manipulation (31).
Using the Golden Syrian hamster model, Carr and coworkers have shown that the hexane-extractable lipid fraction of grain sorghum whole kernels (GSL), when included in the hamsters' diet, leads to a significant reduction of plasma non-HDL and liver cholesterol levels while increasing HDL cholesterol levels (8). We extended this research and performed a comprehensive molecular characterization of the fecal microbiota of the hamsters by pyrosequencing of 16S rRNA tags, denaturing gradient gel electrophoresis (DGGE), and Bifidobacterium specific quantitative real-time PCR (qRT-PCR) in order to test whether metabolic effects of GSL were associated with specific modifications of the gut microbiota.
MATERIALS AND METHODS
Animal experiments.
The fecal samples analyzed here were obtained during a previous study that determined the effect of GSL included in the diet on the cholesterol metabolism of hamsters, and the handling of animals, feed composition, GSL composition, sample collection, and preparation have been described previously (8). Briefly, groups of seven or eight male F1B Syrian hamsters (Bio Breeders, Watertown, MA) were housed in cages (each hamster in an individual cage) and kept at 25°C with a 12-h light-12-h dark cycle. Hamsters were fed a modified AIN-93 M diet (37) supplemented with 0%, 1% and 5% grain sorghum lipid extract at the expense of cornstarch. Daily feed intake was determined to assess individual GSL ingestions. GSL was prepared from whole kernels obtained from a mixture of commercial red grain sorghum hybrids grown in Nebraska. Hamsters had free access to food and water throughout the study. After the hamsters were on their respective diets for 3 weeks, the complete fecal output for each hamster was collected over 7 days. The fecal samples were ground, weighed, and stored frozen at −80°C until the DNA extractions were performed.
DNA extraction.
The fecal samples were diluted in ice-cold phosphate-buffered saline (pH 7) in a 1:10 ratio and centrifuged at 8,000 × g for 5 min. This washing step was repeated two times. Bacterial cell pellets were resuspended in 750 μl lysis buffer (200 mM NaCl, 100 mM Tris [pH 8.0], 20 mM EDTA, 20 mg/ml lysozyme) and transferred to a microcentrifuge tube containing 300 mg of 0.1-mm zirconium beads (BioSpec Products). Samples were then incubated at 37°C for 20 min followed by the addition of 85 μl of 10% sodium dodecyl sulfate solution and 40 μl proteinase K (15 mg/ml). After incubation for 15 min at 60°C, 500 μl of phenol-chloroform-isoamyl alcohol (25:24:1) was added, and the samples were homogenized in a MiniBeadbeater-8 (BioSpec Products) at maximum speed for 2 min. Samples were cooled on ice before the layers were separated by centrifugation at 10,000 × g for 5 min. The top layer was extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1) and twice with chloroform-isoamyl alcohol; DNA was recovered by standard ethanol precipitation. The DNA pellets were dried for 30 min at room temperature and later resuspended in 100 μl of Tris-HCl buffer (10 mM, pH 8.0).
Analysis of the gut microbiota of hamsters by pyrosequencing of 16S rRNA tags.
The V1-V3 region of the 16S rRNA gene was amplified by PCR using bar-coded universal primers 8F and 518R containing the A and B sequencing adaptors (454 Life Sciences). The forward primer (A-8FM) was 5′-gcctccctcgcgccatcagAGAGTTTGATCMTGGCTCAG-3′ where the sequence of the A adaptor is shown in lowercase letters. The reverse primer (B-518) was 5′-gccttgccagcccgctcagNNNNNNNNATTACCGCGGCTGCTGG-3′ where the sequence of the B adaptor is shown in lowercase letters and N represents an eight-base bar code that is unique for each sample. Prior to sequencing, amplicons from the individual PCR samples were quantified using the Quant-iT PicoGreen double-stranded DNA assay (Invitrogen) and quality controlled on an Agilent 2100 bioanalyzer. The amplicons from each reaction mixture were mixed in equal amounts based on concentration and subjected to emulsion PCR, and amplicon libraries were generated as recommended by 454 Life Sciences. Sequencing was performed from the B end using the 454/Roche B sequencing primer kit using a Roche Genome Sequencer GS-FLX using the standard protocol. Samples were combined in a single region of the picotiter plate such that approximately 1,000 to 2,000 sequences were obtained from each animal. The data analysis pipeline removed low-quality sequences (i) that do not perfectly match the PCR primer at the beginning of a read, (ii) that are shorter than 200 bp in length, (iii) that contain more than two undetermined nucleotides (N), or (iv) that do not match a bar code. Sequences (1,000 to 2,000 per animal) were quality controlled and binned according to bar codes.
Taxonomy-based analyses were performed by assigning taxonomic status to each sequence using the CLASSIFIER program of the Ribosomal Database Project (47). To estimate species richness and diversity, taxonomy-independent methods were used. Sequences were aligned using Infernal Aligner; sequences from individual animals and then pooled sequences from all animals of a single treatment group were aligned. Cluster analysis was performed using the complete linkage clustering algorithm available through the Pyrosequencing pipeline of the Ribosomal Database Project (9). Clustering was done with a 97% cutoff for inclusion into an operational taxonomic unit (OTU) and was performed on alignments of sequences from individual animals. The number of species and species richness were estimated by further sampling-based (rarefaction) analyses of OTU data and of calculated Shannon diversity indices.
PCR-DGGE analysis.
PCR was performed using universal primers PRBA338fGC (5′CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGACTCCTACGGGAGGCAGCAG3′) and PRUN518r (5′-ATTACCGCGGCTGCTGG-3′ (34), which amplify the V3 region of the 16S rRNA gene. DGGE was performed by the method of Walter and coworkers (46) using a DCode universal mutation detection system (Bio-Rad, Hercules, CA). DNA bands in the DGGE gel were visualized by standard ethidium bromide staining and photographed using the InGenius gel documentation system (Syngene, Frederick, MD). DGGE images were analyzed using BioNumerics software version 5.0 (Applied Maths, Kortrijk, Belgium). Bands were manually assigned, and the normalized banding patterns were used to generate distance matrices by calculating the Pearson product moment correlation coefficients for all pair-wise combinations of patterns. This approach compares profiles in a pair-wise manner based on the entire densitometric curve, therefore accounting for both band position and intensity. DGGE fingerprints were transformed to peak profiles using the BioNumerics software, and the intensities of individual bands were determined as a percentage of the peak surface area relative to the surface area of the entire molecular fingerprint of the sample. To determine the effects of feeding hamsters GSL, normalized fragment intensities of all bands in DGGE fingerprints were determined and compared for the feeding groups.
In order to identify species represented by bands detected by DGGE, bands of fecal fingerprints from two or three animals were excised from the gel, purified, and reamplified by the method of ben Omar and Ampe (3), and cloned using the TOPO TA Cloning kit for sequencing (Invitrogen). Plasmids were isolated from three transformants per band using the QIAprep spin minprep kit (Qiagen), and inserts were sequenced by a commercial provider following the manual of the cloning kit. Closest relatives of the partial 16S rRNA sequences were determined using the nucleotide blast web tool at the NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and the Seqmatch web tool provided through the Ribosomal Database Project (http://rdp.cme.msu.edu/seqmatch/seqmatch_intro.jsp). A phylogenetic tree was generated from the consensus sequence of the F bands in three individual animals using the unweighted-pair group method using average linkages and neighbor-joining algorithms in the MEGA4 software package (42). There were a total of 178 positions in the final data set, and the evolutionary distances were computed by using the Kimura two-parameter method and are reported as the number of base substitutions per site.
Specific quantification of bifidobacteria by qRT-PCR.
Quantification of total bifidobacteria was performed by quantitative real-time PCR using primers BifFor (5′-TCGCGTCYGGTGTGAAAG-3′ and BifRev (5′-CCACATCCAGCRTCCAC-3′) (39). PCRs were performed using a Mastercycler Realplex2 (Eppendorf AG, Hamburg, Germany). Each PCR was done in a 25-μl volume. The reaction mixture comprised 11.25 μl of the 20× SYBR solution and 2.5× RealMasterMix (Eppendorf AG, Hamburg, Germany), 0.5 μM of each primer, and 1 μl of DNA template. The amplification program consisted of an initial denaturation step of 5 min at 95°C, followed by 35 cycles, where 1 cycle consisted of 15 s at 95°C (denaturation), 20 s at 58°C (annealing), and 30 s at 68°C (extension), and fluorescence at each step was measured. To control the specificity of the amplifications, a melting curve was done consisting of a denaturation step of 15 s at 95°C, an increase from 58°C to 95°C over a 20-min period, and a final step of 15 s at 95°C. Cultures of B. animalis ATCC 25527T and B. infantis ATCC 15697T were used to generate standard curves for absolute quantification of bifidobacteria in the fecal samples. Bacterial counts of overnight cultures (12 h) were determined by plate counting, and a 10-fold dilution series was performed in phosphate-buffered saline buffer for each strain. DNA was isolated from individual samples of the dilution series using the method for fecal samples. Standard curves were made by plotting the threshold cycle values obtained from DNA of the dilution series as a linear function of the base 10 logarithm of the number of bifidobacteria. Two individual qRT-PCR runs of all fecal DNA templates in duplicate were performed, and means of all four values were used for the analysis. Despite the use of two different strains of bifidobacteria to generate one standard curve, its correlation coefficient r2 was >0.96.
To quantify the Bifidobacterium animalis-like phylotype detected by DGGE, we used a specific primer (Bh1) based on a highly variable region of the sequence of fragment F in the DGGE gel (5′GGCAGGGGGTTTTCCTC3′). This primer was used in combination with primer BifRev (39) used for the Bifidobacterium genus-specific qRT-PCR. PCR was performed as described above. The specificity of the PCR was tested using DNA isolated from fecal samples from 10 human subjects and DNA from B. animalis ATCC 25527T and Bifidobacterium infantis ATCC 15697T. The PCRs all gave negative results with the primer combination Bh1 and BifRev and positive results with primers BifFor and BifRev (data not shown). As we had no cultural representative of the phylotype represented by band F, we used a standard curve generated as described above with B. animalis ATCC 25527T and B. infantis ATCC 15697T and primers BifFor and BifRev. Although the standard curve was generated with a different forward primer, it can be assumed that no significant bias is introduced, as the efficiencies of the two PCR systems were virtually identical (0.51 for primers BifFor and BifRev and 0.56 for primers Bh1 and BifRev).
Correlation analysis of gut microbiota-host metabolic functional relationships.
Correlation analysis between metabolic host parameters and bacterial populations at different taxonomic levels was performed by the method of Cani and coworkers (7). Metabolic parameters included in the correlation analysis were the levels of cholesterol absorption, fecal cholesterol, plasma total cholesterol, plasma HDL cholesterol, plasma non-HDL cholesterol, liver total cholesterol, liver-free cholesterol, liver triglycerides, liver phospholipids, and liver-esterified cholesterol. The determination of these metabolic phenotypes and the methods applied were reported previously (8).
Genome comparisons.
The web-based Integrated Genomics Platform of the Joint Genome Institute (JGI) was used to identify functions enriched in bifidobacteria (27). The Abundance Profile Search was used to identify clusters of orthologous groups of proteins (COGs) that were more abundant in individual bifidobacterial genomes (Bifidobacterium adolescentis ATCC 15703, Bifidobacterium adolescentis L2-32, Bifidobacterium animalis subsp. lactis HN019, Bifidobacterium dentium ATCC 27678, Bifidobacterium longum DJO10A, and Bifidobacterium longum NCC2705) compared to the genomes of a selection of bacteria commonly present in the mammalian gastrointestinal tract (Bacteroides caccae ATCC 43185, Bacteroides capillosus ATCC 29799, Bacteroides fragilis NCTC 9343, Bacteroides fragilis YCH46, Bacteroides ovatus ATCC 8483, Bacteroides stercoris ATCC 43183, Bacteroides thetaiotaomicron VPI-5482, Bacteroides uniformis ATCC 8492, Bacteroides vulgatus ATCC 8482, Clostridium acetobutylicum ATCC 824, Clostridium bartlettii DSM 16795, Clostridium bolteae ATCC BAA-613, Clostridium leptum DSM 753, Clostridium ramosum DSM 1402, Clostridium sp. strain L2-50, Clostridium sp. strain SS2/1, Clostridium thermocellum ATCC 27405, Collinsella aerofaciens ATCC 25986, Coprococcus eutactus ATCC 27759, Dorea formicigenerans ATCC 27755, Dorea longicatena DSM 13814, Enterobacter sp. strain 638, Enterococcus faecalis V583, Enterococcus faecium DO, Escherichia coli K-12, Eubacterium dolichum DSM 3991, Eubacterium siraeum DSM 15702, Eubacterium ventriosum ATCC 27560, Faecalibacterium prausnitzii M21/2, Lactobacillus reuteri 100-23, Lactobacillus reuteri F275, Lactobacillus salivarius subsp. salivarius UCC118, Methanobrevibacter smithii ATCC 35061, Parabacteroides distasonis ATCC 8503, Parabacteroides merdae ATCC 43184, Peptostreptococcus micros ATCC 33270, Providencia stuartii ATCC 25827, Ruminococcus gnavus ATCC 29149, Ruminococcus obeum ATCC 29174, Ruminococcus torques ATCC 27756, and Salmonella enterica serovar Typhimurium LT2). Functions associated with lipid metabolism were specifically selected from the enriched COGs and added to the function list. A function profile of these COGs was then generated for all of the gut species. Please refer to the IMG web page for details (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi).
Statistical analysis.
Results are presented as means ± standard deviations (SDs). Statistical tests for treatment effects of the GSL on the abundance of individual taxonomic ranks or DGGE band intensities were performed by one-way analysis of variance (ANOVA) analysis followed by Tukey's posthoc multiple comparison tests. The Mann-Whitney test was used to compare Shannon diversity indices of gut populations. Correlations between metabolic parameters and bacterial populations were assessed by Pearson's correlation test using GraphPad Prism version 5.00 (GraphPad Software, San Diego, CA).
RESULTS
Characterization of the hamster gut microbiota by pyrosequencing of 16S rRNA tags.
To determine whether proportional changes of the gut microbiota were associated with the effects of GSL on cholesterol metabolism in hamsters, we analyzed the fecal microbiota of hamsters fed 0% (n = 7), 1% (n = 7), and 5% (n = 7) GSL by pyrosequencing of the V3 region of the 16S rRNA gene. A total of 34,424 sequences were studied; the average sequence length was around 250 bp, and an average of 1,639 sequences per animal were studied. Taxonomy-based analysis showed that the composition of the hamster gut microbiota at the phylum level is similar to that of humans and mice, being dominated by Firmicutes and Bacteroidetes. An unusual feature of the hamster gut microbiota, however, was that Firmicutes comprised the vast majority of the taxa (94%) with Bacteroidetes making up only 4% of the population. The remaining bacteria belonged to the phyla Verrucomicrobia and Actinobacteria (each representing around 1% of the total sequence tags) and Proteobacteria and candidate division TM7 (0.07% and 0.024% of sequences, respectively).
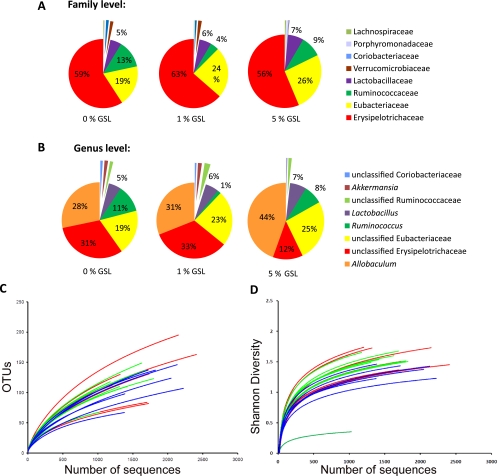
At the family level, the predominant groups in hamsters of the control group were the Erysipelotrichaceae, Eubacteriaceae, Ruminococcaceae, and Lactobacillaceae, represented by an average of 59%, 19%, 13% and 5% of the total fecal microbiota, respectively (Fig. 1A). Of the bacterial groups on the genus level, the most dominant were unclassified Erysipelotrichaceae, Allobaculum, unclassified Eubacteriaceae, Ruminococcus, and Lactobacillus, comprising 31%, 28%, 19%, 11%, and 5% of the total sequence pool on average in control animals, respectively (Fig. 1B). With the exception of Allobaculum, these genera are also shared with the gut microbiota reported for mice, humans, and primates (15, 24, 29, 44). As shown in Fig. S1 in the supplemental material, pyrosequencing revealed high animal-to-animal variability on both the family and genus levels.
FIG. 1.
Characterization of the gut microbiota composition of hamsters fed different amounts of GSL as determined by pyrosequencing of 16S rRNA tags (V3 region). Composition of the gut microbiota of hamsters fed 0%, 1%, and 5% GSL (n = 7 per group) at the family level (A) and the genus level (B). (C) Rarefaction curves of OTUs from sequences of fecal samples from individual hamsters fed 0% GSL (red), 1% GSL (green), and 5% GSL (blue). (D) Shannon diversity indices of the gut microbiota of individual hamsters fed 0% GSL (red), 1% GSL (green), and 5% GSL (blue). OTUs were identified using 97% cutoffs for rarefaction and Shannon diversity indices.
Effects of GSL on specific taxa of the hamster gut microbiota.
Sequence proportions determined by pyrosequencing were used to establish the effects of the GSL on the gut microbiota composition. To identify specific taxa that were affected by the feeding treatments, the proportions of taxa in each rank of each animal were tested for treatment effects. As shown in Table 1, ANOVA identified one family, the Coriobacteriaceae (P = 0.042), and two bacterial groups at the genus level, unclassified members of the family Erysipelotrichaceae (P = 0.0016) and genus Pseudoramibacter (P = 0.017), as being significantly affected by the inclusion of GSL to the hamsters' diet. Moreover, values for the genus Allobaculum (P = 0.096) and unclassified members of the family Coriobacteriaceae (P = 0.064) approached statistical significance.
TABLE 1.
Abundance of bacterial groups in the fecal microbiota of hamsters that changed by including GSL in the diet as determined by pyrosequencing of 16S rRNA tagsa
| Bacterial taxa | Abundance of bacterial groupb (% of total sequences obtained with sample [mean ± SD]) in hamsters fed:
|
||
|---|---|---|---|
| 0% GSL | 1% GSL | 5% GSL | |
| Family level Coriobacteriaceae | 1.22 ± 0.79 | 0.79 ± 0.63 | 0.31 ± 0.33c |
| Genus level | |||
| Allobaculum | 27.82 ± 15.9 | 30.63 ± 15.8 | 43.55 ± 7.0e |
| Pseudoramibacter | 0.11 ± 0.09 | 0.48 ± 0.3c | 0.35 ± 0.21c |
| Unclassified members of the following families | |||
| Coriobacteriaceae | 1.0 ± 0.7 | 0.69 ± 0.62 | 0.23 ± 0.31e |
| Erysipelotrichaceae | 31.0 ± 7.4 | 32.7 ± 14.0 | 12.39 ± 6.0d |
There were seven hamsters in each group.
Values that were significantly different or approaching statistical significance are shown in boldface type.
Statistically significantly different from the value for hamsters fed 0% GSL (P < 0.05) by ANOVA.
This value was statistically significantly different from the value for hamsters fed 0% GSL (P < 0.01) and from the value for hamsters fed 1% GSL (P < 0.01) by ANOVA.
Approaching statistical significance compared to the value for hamsters fed 0% GSL (P < 0.1) by ANOVA.
Taxonomy-independent analysis of the hamster gut microbiota from individual animals showed that with a conservative level of 97% identity as a cutoff for OTUs, nearly 200 OTUs were observed in the average of 1,600 sequences from each animal (Fig. 1C). Individual animals in the 5% GSL feeding group showed a trend toward fewer OTUs in the rarefaction curves. The Shannon diversity indices from individual animals also showed a trend of fewer OTUs in animals fed 5% GSL (blue lines) compared to animals fed 0% GSL (Fig. 1D). Grouping of the samples by GSL showed significant differences between 0% and 5% GSL (P < 0.0001, Mann-Whitney test). Thus, 5% GSL had the effect of reducing the diversity of the gut microbiota.
DGGE analysis of fecal microbiota of hamsters fed GSL.
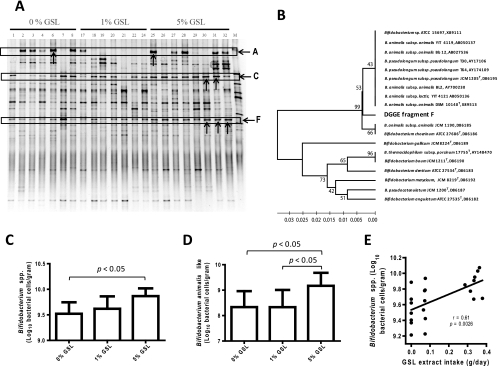
To validate the findings obtained with pyrosequencing, fecal bacterial populations of the hamsters were also analyzed by PCR-DGGE. The DGGE gel is shown in Fig. 2A, and the results of analysis of the gel are presented in Table 2. Feeding the hamsters 5% GSL significantly increased the staining intensity of band C (P = 0.037) and band F (P = 0.011). Band A showed a high animal-to-animal variation, and no consistent impact of GSL was detected. Sequence analysis of amplicons from these bands revealed that they represent bacteria related to Ruminococcus bromii (band A), Allobaculum stercoricanis (band C), and Bifidobacterium animalis (band F). Phylogenetic comparison of the sequence from band F with the closest hits in the RDP database revealed this sequence to be most similar to Bifidobacterium animalis, and it is referred to as the Bifidobacterium animalis-like phylotype in this article (a phylogenetic tree is shown in Fig. 2B).
FIG. 2.
Impact of GSL on the gut microbiota composition of hamsters fed 0% GSL (n = 7), 1% GSL (n = 7), and 5% GSL (n = 8) as determined by DGGE and qRT-PCR. (A) DGGE showing fingerprints of DNA isolated from the fecal samples of hamsters. Lanes 1 to 32 contain DNA from individual hamsters. Lane M contains markers from reference strains. Bands C and F showed significant increases in staining intensity in fecal fingerprints of hamsters fed 5% GSL. The bands A, C, and F marked by an arrow were excised, purified, and sequenced (Table 2). (B) Phylogenetic tree of DGGE band F with sequences that revealed highest similarities in GenBank. The tree was inferred using the unweighted-pair group method using average linkages, and the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. A neighbor-joining tree resulted in essentially the same phylogeny (data not shown). (C) Cell numbers of total bifidobacteria in hamster fecal samples as determined by qRT-PCR. (D) Quantification of the Bifidobacterium animalis-like phenotype detected by DGGE in hamster fecal samples by qRT-PCR. (E) Correlation of cell numbers of bifidobacteria with daily GSL intake.
TABLE 2.
Ratio of staining intensities of major bands as a proportion of total fingerprint intensity and results of sequence analysis of selected bands
| Band | Mean band intensitya (SD) in DNA from hamsters fed:
|
Closest GenBank hitd | Closest type straine | ||
|---|---|---|---|---|---|
| 0% GSL | 1% GSL | 5% GSL | |||
| A | 20.0 (17.5) | 0.06 (0.16)b | 14.2 (14.3) | AM265444, uncultured bacteria, clone ratBD050202C (99.4) | Ruminococcus bromii ATCC 27255T [L76600] (96.8) |
| B | 5.6 (4.5) | 7.8 (8.1) | 6.2 (6.9) | NDf | ND |
| C | 9.2 (7.9) | 7.9 (4.3) | 17.4 (8.2)c | EU777003, uncultured bacterium clone molerat_aai70g11 (92.4-93.0) | Allobaculum stercoricanis DSM 13633T [AJ417075] (92.0-92.6) |
| D | 11.4 (11.0) | 13.1 (7.1) | 10.3 (7.6) | ND | ND |
| E | 3.4 (3.2) | 7.5 (10.3) | 4.3 (5.1) | ND | ND |
| F | 5.7 (1.0) | 6.2 (3.4) | 11.1 (3.7)b,c | AB186296, Bifidobacterium animalis strain DBF 1307 (96.8) | Bifidobacterium animalis subsp. animalis DSM 10140T [X89513] (96.8) |
| G | 4.0 (1.6) | 3.2 (2.2) | 3.2 (1.3) | ND | ND |
| H | 3.0 (2.2) | 8.0 (7.6) | 1.7 (2.0) | ND | ND |
Ratio of staining intensities of major bands as a proportion of total fingerprint intensity (shown as a percentage). Values that were significantly different are shown in boldface type.
Statistically significantly different from the value for hamsters fed 0% GSL (P < 0.05) by ANOVA.
Statistically significantly different from the value for hamsters fed 1% GSL (P < 0.05) by ANOVA.
The GenBank accession number and species or clone is shown. The values in parentheses are the percentages of similarity.
The closest type strain is shown first. The GenBank accession number is shown in brackets. The values in parentheses are the percentages of similarity.
ND, not determined.
Although providing less depth, DGGE analysis showed good agreement with the results from pyrosequencing. Both methods detected the increase of bacteria related to Allobaculum and the high animal-to-animal variability of bacteria related to Ruminococcus. Relative species quantification obtained with pyrosequencing showed high correlations with staining intensities of DGGE bands representing the same bacterial groups (for Ruminococcus, r = 0.94; for Allobaculum, r = 0.81; both P < 0.0001) (see Fig. S2 in the supplemental material). This is remarkable, as both methods are only semiquantitative and entail multitemplate PCR susceptible to PCR bias. The main difference between the findings by pyrosequencing and DGGE was in the proportions of bifidobacteria, where a significant proportion could be detected only by DGGE, while only 0.03% of the total sequences obtained by pyrosequencing corresponded to bifidobacteria. The difference can be explained by the use of primer 8F in pyrosequencing, which shows three mismatches with the 16S rRNA genes from five Bifidobacterium species for which whole-genome sequences were available (data not shown). Accordingly, many studies employing direct analysis of 16S rRNA genes to study the human gut microbiota and using the primer 8F, which is one of the most commonly used primers for such approaches, resulted in a significant underrepresentation of Bifidobacterium species (15, 35, 41, 50).
Quantification of bifidobacteria using qRT-PCR.
Since primer 8F resulted in an underrepresentation of bifidobacteria in pyrosequencing and to confirm and quantify the bifidogenic effect of the GSL detected by DGGE analysis, a specific qRT-PCR procedure was used to determine the numbers of total bifidobacteria and the Bifidobacterium animalis-like phylotype. As shown in Fig. 2C and D, qRT-PCR analysis showed a significant increase in cell numbers of total bifidobacteria (P = 0.012) and the Bifidobacterium animalis-like phylotype (P = 0.019). As shown in Fig. 2E, the numbers of bifidobacteria from individual hamsters were highly variable, but a significant correlation between cell numbers and daily GSL intake was observed.
Bifidobacteria and Coriobacteriaceae showed high correlations with important markers of host cholesterol metabolism.
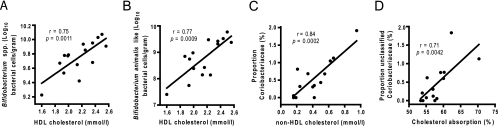
In a previous study using the animals studied here, dietary GSL reduced cholesterol absorption, plasma non-HDL cholesterol concentrations, and liver esterified cholesterol levels, while raising plasma HDL cholesterol levels (8). To determine whether alterations of the gut microbiota in hamsters fed GSL were associated with an improvement in cholesterol metabolism, a correlation analysis was used to determine correlations between all bacterial taxa at different taxonomic levels and host metabolic phenotypes. The analysis revealed highly positive correlations between HDL plasma concentrations and total bifidobacteria (r = 0.75; P = 0.0011), between HDL plasma concentrations and Bifidobacterium animalis-like phylotype (r = 0.77; P = 0.0009), among total Coriobacteriaceae and non-HDL plasma concentrations (r = 0.84; P = 0.0002), and between unclassified Coriobacteriaceae and both non-HDL plasma concentration (r = 0.82; P = 0.0004) and cholesterol absorption (r = 0.71; P = 0.0042). These high correlations were observed only in animals fed 1% and 5% GSL, and inclusion of the values from control animals significantly reduced correlations (Table 3). Graphs showing the highest correlations between bacterial taxa and metabolic phenotypes are shown in Fig. 3, and a metabolic network diagram linking GSL, bacterial phylotypes, and host cholesterol metabolism is shown in Fig. 4. Interestingly, the correlations between bifidobacteria and HDL cholesterol concentration and between Coriobacteriaceae and non-HDL concentration showed higher significance than correlations achieved between GSL intake and the respective host metabolic phenotypes.
TABLE 3.
Correlations between abundance of bacterial taxa and markers of cholesterol metabolisma
| Bacterial taxa | Correlationb (r value) between abundance of bacterial taxa and the following marker of cholesterol metabolism:
|
|||
|---|---|---|---|---|
| GSL intake | Non-HDL level | HDL level | Cholesterol absorption | |
| Family level Coriobacteriaceae | −0.54 (−0.50) | 0.84 (0.37) | −0.56 (−0.18) | 0.68 (0.50) |
| Genus level | ||||
| Bifidobacterium | 0.56 (0.61) | −0.61 (−0.33) | 0.75 (0.34) | −0.42 (−0.51) |
| Allobaculum | 0.50 (0.48) | −0.28 (−0.53) | 0.24 (0.50) | −0.45 (−0.37) |
| Pseudoramibacter | −0.24 (0.23) | 0.07 (−0.39) | −0.12 (0.19) | 0.08 (−0.40) |
| Unclassified members of the following families | ||||
| Coriobacteriaceae | −0.48 (−0.52) | 0.82 (0.40) | 0.52 (−0.19) | 0.71 (0.47) |
| Erysipelotrichaceae | −0.74 (−0.70) | 0.45 (0.34) | −0.48 (−0.32) | 0.68 (0.56) |
Values for animals fed 1% and 5% GSL are presented.
Values for all animals, including control animals, are presented in parentheses. Correlation coefficients with an r of >0.7 are shown in boldface type.
FIG. 3.
Specific bacterial populations in the guts of hamsters show high associations with both cholesterol metabolic phenotypes and GSL intake. (A and B) Correlations between cell numbers of total bifidobacteria (A) and the Bifidobacterium animalis-like phenotype (B) with HDL cholesterol. (C) Correlation between proportion of Coriobacteriaceae and non-HDL cholesterol. (D) Correlation between unclassified members of the family Coriobacteriaceae and cholesterol absorption. Data from control animals (0% GSL) were excluded from the analysis.
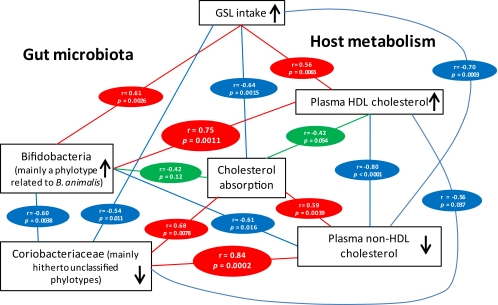
FIG. 4.
Metabolic network showing the associations between daily GSL intake, gut microbiota composition, and host cholesterol metabolism in hamsters fed 0%, 1%, and 5% GSL. Results of the correlations of cell numbers of bifidobacteria and proportions of Coriobacteriaceae and phenotypic markers were obtained with data from animals fed 1% and 5% GSL. Red connections indicate a positive correlation, while blue connections show correlations that are inverse. Green connections show associations with no statistical significance. Metabolic data were obtained by Carr and coworkers in a previous study (8).
Genome comparisons of bifidobacteria and other gut organisms.
By comparing the relative abundance of different functional categories in 47 genomes of gut bacteria, we observed that proteins belonging to the COG clusters COG0400 (predicted esterase), COG0657 (esterase/lipase), and COG2272 (carboxylesterase type B) are enriched in six Bifidobacterium genomes (see Table S1 in the supplemental material). Carboxylesterases represented by COG0400 and COG2272 belong to enzymes that hydrolyze a wide variety of substrates, ranging from methylcaplyrate to p-nitrobenzyl (21, 49).
DISCUSSION
In humans, CHD is associated with high levels of low-density lipoprotein and low levels of high-density lipoprotein. The characterization of the gut microbiota in a hamster model of hypercholesterolemia showed that dietary intervention with GSL had a major impact on the composition of the gut microbiota and that these modulations were highly associated with improvements in the HDL and non-HDL cholesterol equilibrium. With consumption of GSL, population levels of bifidobacteria increased and showed a strong positive association with increases in HDL cholesterol levels. In contrast, relative abundance of members of the family Coriobacteriaceae decreased with feeding the hamsters GSL, and a high positive correlation with non-HDL cholesterol and cholesterol absorption was discovered. The findings indicate that GSL intake influences the HDL/non-HDL equilibrium, at least in part, through an alteration of the gut microbiota. We infer this because correlation coefficients between bifidobacterial and Coriobacteriaceae populations and plasma cholesterol concentrations were higher than associations among GSL intake, host phenotypes, and cholesterol absorption (Fig. 4). In addition, if bacterial phylotype/host phenotype correlations were merely a result of GSL affecting both bacterial taxa and cholesterol metabolism independently, one would assume that all bacterial taxa whose abundance correlated with GSL intake would show an association with host phenotypes. However, much lower correlation coefficients with non-HDL and HDL plasma concentrations were observed between relative abundance of unclassified members of the family Erysipelotrichaceae and the genus Allobaculum, although these taxa showed significant associations with GSL intake (Table 3). However, it should be considered that bifidobacteria and Coriobactericeae are just two of hundreds of groups, and other bacteria, independent of GSL administration, are likely to interact with host cholesterol metabolism.
Changes in the composition of the hamsters' gut microbiota induced by GSL consumption were limited to a relatively small number of bacterial groups (Table 1 and 2). These compositional adjustments had the net effect of reducing the overall species richness (number of individual species per unit population). However, the overall composition of the microbiota at the phylum level was not affected by GSL. The reason for this finding was that increases of dominant bacterial taxa were “balanced” by a reduction of related bacteria, leaving the relative proportions of higher taxonomic taxa unaffected. Allobaculum belongs to the family Erysipelotrichaceae, and uncharacterized bacteria of this family declined as Allobaculum increased with feeding the hamsters GSL. Thus, the overall proportions within the family Erysipelotrichaceae and the phylum Firmicutes were not changed. Similar findings were obtained for the phylum Actinobacteria, where numbers of bifidobacteria increased while the abundance of members of the family Coriobacteriaceae declined. As shown in Fig. S3 in the supplemental material, significant inverse correlations were obtained between these related bacterial groups in individual animals. Similar diet-induced compositional adjustments of the gut microbiota that maintain the overall composition at higher taxonomic levels have also been observed in humans. For example, the decline of bacteria belonging to the Roseburia and Eubacterium rectale groups induced through reduced carbohydrate intake was balanced by an increase in the number of related members of the Clostridium coccoides cluster in human fecal samples (14). Furthermore, the diet of human infants appears to influence the Bifidobacterium/Coriobacteriaceae ratio, with higher numbers of bifidobacteria in breast-fed infants while there were higher numbers of coriobacteria when the infants were fed formula (19). Collectively, these findings indicate that homeostatic reactions that restore the overall equilibrium of the gut microbiota are often a natural consequence of compositional changes induced through diet. Nevertheless, as indicated by the correlation analysis in our study, an alteration of the gut microbiota at lower taxonomic levels is still likely to have important functional consequences for the host.
The mechanisms by which bifidobacteria and coriobacteria affect cholesterol metabolism remain an important field of future research. Including GSL in the diet reduced cholesterol absorption efficiency, which was directly correlated with non-HDL cholesterol concentration (8). The high correlations of unclassified members of the family Coriobacteriaceae with both non-HDL cholesterol and cholesterol absorption suggest that these bacteria could have a negative impact on cholesterol homeostasis through increasing cholesterol absorption. Bifidobacteria, on the other hand, showed high positive correlation with HDL cholesterol levels and no association with cholesterol absorption. Bifidobacteria have been shown to affect cholesterol and lipid metabolism in animal models when administered as probiotics or when stimulated by prebiotics (11, 13). The mechanism by which bifidobacteria achieve these effects remain speculative, but they might impact cholesterol metabolism indirectly by suppressing numbers of Coriobacteriaceae. For both bacterial groups, the capability to transform bile acids has been reported (38), and this phenotypic trait might influence host cholesterol metabolism through an impact on enterohepatic circulation. The strong correlations between bacterial taxa and cholesterol metabolism were observed only in animals fed GSL and not in control animals, suggesting that GSL influences the relative abundance of these organisms as well as metabolic characteristics.
The consumption of lipids has not yet been associated with increases in numbers of intestinal bifidobacteria. In contrast, Cani and coworkers (7) showed that a high-fat diet significantly lowered the number of bifidobacteria in mice. The composition of the GSL administered to the hamsters in our study contained not only mono-, di-, and triglycerides but also esters, alcohols, and other lipophilic compounds, such as waxes, sterols, and polycosanols (8). Carbohydrates or fiber are an unlikely explanation for the bifidogenic effect of GSL, as the amounts of fiber in hexane extracts of grains are negligible (8). Interestingly, genome comparisons revealed that bifidobacteria possess metabolic capacities that could allow them to utilize complex lipids, including lipids that may not be utilized by other members of the gut microbiota or the host. Schell and coworkers (40) detected four genes encoding long-chain fatty acyl-coenzyme A synthetases in the genome of Bifidobacterium longum, more than any other prokaryote genome available at that time, except for Streptomyces coelicolor and another gastrointestinal tract inhabitant, Bacteroides fragilis. These findings together with the enrichment of putative esterases in bifidobacterial genomes detected above indicate that bifidobacteria are likely to utilize specific components of GSL leading to the increase in numbers in the gut.
The correlation analysis identified the Bifidobacterium/Coriobacteriaceae equilibrium to be important for the plasma cholesterol levels in hamsters, with bifidobacteria being beneficial and coriobacteria being detrimental. While extrapolation of our observations to humans is still speculative, our findings suggest that bifidobacteria and coriobacteria could be potential targets for the prevention of metabolic aberrations that play a role in CHD. Clearly, it will be essential to first identify the exact bacterial taxa within the human gut microbiota that have strong correlations to cholesterol metabolism, which in itself is a challenging task. Unlike the inbred population of hamsters used in our study, human subjects have significant genetic diversity, and genetic factors that affect cholesterol metabolism play a more important role in humans than in the animal model. Furthermore, human subjects follow individual lifestyles and consume different diets, and they harbor variable and individual communities of the gut bacteria. All these factors will hamper the identification of bacterial contributors to human cholesterol metabolism. Nevertheless, it is tempting to speculate that the positive impact of breast-feeding on the Bifidobacterium/Coriobacteriaceae ratio in human infants (19) could be responsible for the higher HDL cholesterol levels observed in adults that were breast-fed in infancy (36).
This study provides new and important perspectives on dietary modulation of the mammalian gut microbiota and its effects on the host. The findings indicate that a complex mixture of lipids can exert a “prebiotic” effect that leads to improvements in host cholesterol metabolism. In conclusion, this study provided evidence that modulation of bacterial populations in the gut has the potential to improve mammalian cholesterol homeostasis, which has relevance in the prevention of CHD.
Supplementary Material
Acknowledgments
We thank the members of the University of Nebraska—Lincoln Nutraceutical Team and especially Curtis Weller, Vicki Schlegel, and Susan Cuppett for their contributions to the hamster feeding trial. We thank Ty Nguyen for programming the pyrosequencing data analysis pipeline.
Grant Wallace was supported by the UCARE Program of the University of Nebraska. This study was funded in part by the Nebraska Grain Sorghum Board.
Footnotes
Published ahead of print on 1 May 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Backhed, F., H. Ding, T. Wang, L. V. Hooper, G. Y. Koh, A. Nagy, C. F. Semenkovich, and J. I. Gordon. 2004. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 101:15718-15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backhed, F., J. K. Manchester, C. F. Semenkovich, and J. I. Gordon. 2007. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA 104:979-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ben Omar, N., and F. Ampe. 2000. Microbial community dynamics during production of the Mexican fermented maize dough pozol. Appl. Environ. Microbiol. 66:3664-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bickler, S. W., and A. DeMaio. 2008. Western diseases: current concepts and implications for pediatric surgery research and practice. Pediatr. Surg. Int. 24:251-255. [DOI] [PubMed] [Google Scholar]
- 5.Bravo, E., A. Cantafora, and G. Ortu. 1994. Why prefer the Golden Syrian hamster (Mesocritus auratus) to the Wistar rat in experimental studies on plasma lipoprotein metabolism? Comp. Biochem. Physiol. 107B:347-355. [Google Scholar]
- 6.Cani, P. D., and N. M. Delzenne. 2007. Gut microflora as a target for energy and metabolic homeostasis. Curr. Opin. Clin. Nutr. Metab. Care 10:729-734. [DOI] [PubMed] [Google Scholar]
- 7.Cani, P. D., A. M. Neyrinck, F. Fava, C. Knauf, R. G. Burcelin, K. M. Tuohy, G. R. Gibson, and N. M. Delzenne. 2007. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia 50:2374-2383. [DOI] [PubMed] [Google Scholar]
- 8.Carr, T. P., C. L. Weller, V. L. Schlegel, S. L. Cuppett, D. M. Guderian, Jr., and K. R. Johnson. 2005. Grain sorghum lipid extract reduces cholesterol absorption and plasma non-HDL cholesterol concentration in hamsters. J. Nutr. 135:2236-2240. [DOI] [PubMed] [Google Scholar]
- 9.Cole, J. R., Q. Wang, E. Cardenas, J. Fish, B. Chai, R. J. Farris, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, T. Marsh, G. M. Garrity, and J. M. Tiedje. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141-D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cowles, R. L., J. Y. Lee, D. D. Gallaher, C. L. Stuefer-Powell, and T. P. Carr. 2002. Dietary stearic acid alters gallbladder bile acid composition in hamsters fed cereal-based diets. J. Nutr. 132:3119-3122. [DOI] [PubMed] [Google Scholar]
- 11.Crittenden, R., and M. J. Playne. 2006. Modifying the human intestinal microbiota with prebiotics, p. 285-314. In A. C. Ouwehand and E. E. Vaughan (ed.), Gastrointestinal microbiology. Taylor & Francis, New York, NY.
- 12.Danielsson, H., and B. Gustafsson. 1959. On serum-cholesterol levels and neutral fecal sterols in germ-free rats: bile acids and steroids 59. Arch. Biochem. Biophys. 83:482-485. [DOI] [PubMed] [Google Scholar]
- 13.Delzenne, N. M., P. D. Cani, and A. M. Neyrinck. 2008. Prebiotics and lipid metabolism. In J. Versalovic and M. Wilson (ed.), Therapeutic microbiology: probiotics and related strategies. ASM Press, Washington, DC.
- 14.Duncan, S. H., G. E. Lobley, G. Holtrop, J. Ince, A. M. Johnstone, P. Louis, and H. J. Flint. 2008. Human colonic microbiota associated with diet, obesity and weight loss. Int. J. Obes. (London) 32:1720-1724. [DOI] [PubMed] [Google Scholar]
- 15.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fava, F., J. A. Lovegrove, R. Gitau, K. G. Jackson, and K. M. Tuohy. 2006. The gut microbiota and lipid metabolism: implications for human health and coronary heart disease. Curr. Med. Chem. 13:3005-3021. [DOI] [PubMed] [Google Scholar]
- 17.Flint, H. J., S. H. Duncan, K. P. Scott, and P. Louis. 2007. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ. Microbiol. 9:1101-1111. [DOI] [PubMed] [Google Scholar]
- 18.Gordon, H. A., and L. Pesti. 1971. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol. Rev. 35:390-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harmsen, H. J., A. C. Wildeboer-Veloo, J. Grijpstra, J. Knol, J. E. Degener, and G. W. Welling. 2000. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Appl. Environ. Microbiol. 66:4523-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes, E., R. L. Loo, J. Stamler, M. Bictash, I. K. Yap, Q. Chan, T. Ebbels, M. De Iorio, I. J. Brown, K. A. Veselkov, M. L. Daviglus, H. Kesteloot, H. Ueshima, L. Zhao, J. K. Nicholson, and P. Elliott. 2008. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 453:396-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong, K. H., W. H. Jang, K. D. Choi, and O. J. Yoo. 1991. Characterization of Pseudomonas fluorescens carboxylesterase: cloning and expression of the esterase gene in Escherichia coli. Agric. Biol. Chem. 55:2839-2845. [PubMed] [Google Scholar]
- 22.Hooper, L. V., and J. I. Gordon. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115-1118. [DOI] [PubMed] [Google Scholar]
- 23.Horton, J. D., J. A. Cuthbert, and D. K. Spady. 1995. Regulation of hepatic 7 alpha-hydroxylase expression and response to dietary cholesterol in the rat and hamster. J. Biol. Chem. 270:5381-5387. [DOI] [PubMed] [Google Scholar]
- 24.Ley, R. E., F. Backhed, P. Turnbaugh, C. A. Lozupone, R. D. Knight, and J. I. Gordon. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 102:11070-11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ley, R. E., D. A. Peterson, and J. I. Gordon. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837-848. [DOI] [PubMed] [Google Scholar]
- 26.Mandard, S., F. Zandbergen, E. van Straten, W. Wahli, F. Kuipers, M. Muller, and S. Kersten. 2006. The fasting-induced adipose factor/angiopoietin-like protein 4 is physically associated with lipoproteins and governs plasma lipid levels and adiposity. J. Biol. Chem. 281:934-944. [DOI] [PubMed] [Google Scholar]
- 27.Markowitz, V. M., E. Szeto, K. Palaniappan, Y. Grechkin, K. Chu, I. M. Chen, I. Dubchak, I. Anderson, A. Lykidis, K. Mavromatis, N. N. Ivanova, and N. C. Kyrpides. 2008. The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucleic Acids Res. 36:D528-D533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin, F. P., M. E. Dumas, Y. Wang, C. Legido-Quigley, I. K. Yap, H. Tang, S. Zirah, G. M. Murphy, O. Cloarec, J. C. Lindon, N. Sprenger, L. B. Fay, S. Kochhar, P. van Bladeren, E. Holmes, and J. K. Nicholson. 2007. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol. Syst. Biol. 3:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKenna, P., C. Hoffmann, N. Minkah, P. P. Aye, A. Lackner, Z. Liu, C. A. Lozupone, M. Hamady, R. Knight, and F. D. Bushman. 2008. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 4:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Midtvedt, T. 1999. Microbial functional activities, p. 79-96. In L. A. Hanson and R. H. Yolken (ed.), Probiotics, other nutritional factors, and intestinal microflora, vol. 42. Lippincott-Raven Publishers, Philadelphia, PA. [Google Scholar]
- 31.Mitchell, P. L., and R. S. McLeod. 2008. Conjugated linoleic acid and atherosclerosis: studies in animal models. Biochem. Cell Biol. 86:293-301. [DOI] [PubMed] [Google Scholar]
- 32.Nicholson, J. K., E. Holmes, and I. D. Wilson. 2005. Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Microbiol. 3:431-438. [DOI] [PubMed] [Google Scholar]
- 33.Ordovas, J. M., and V. Mooser. 2006. Metagenomics: the role of the microbiome in cardiovascular diseases. Curr. Opin. Lipidol. 17:157-161. [DOI] [PubMed] [Google Scholar]
- 34.Øvreås, L., L. Forney, F. L. Daae, and V. Torsvik. 1997. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63:3367-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmer, C., E. M. Bik, D. B. Digiulio, D. A. Relman, and P. O. Brown. 2007. Development of the human infant intestinal microbiota. PLoS Biol. 5:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parikh, N. I., S.-J. Hwang, E. Ingelsson, E. J. Benjamin, C. S. Fox, R. S. Vasan, and J. M. Murabito. 2007. Abstract 3498: the association of breastfeeding in infancy and adult cardiovascular disease risk factors: the Framingham Third Generation Cohort. Circulation 116(Suppl.):II_792. [Google Scholar]
- 37.Reeves, P. G., F. H. Nielsen, and G. C. Fahey, Jr. 1993. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr. 123:1939-1951. [DOI] [PubMed] [Google Scholar]
- 38.Ridlon, J. M., D. J. Kang, and P. B. Hylemon. 2006. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47:241-259. [DOI] [PubMed] [Google Scholar]
- 39.Rinttila, T., A. Kassinen, E. Malinen, L. Krogius, and A. Palva. 2004. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 97:1166-1177. [DOI] [PubMed] [Google Scholar]
- 40.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 43.Tannock, G. W. 2008. The search for disease-associated compositional shifts in bowel bacterial communities of humans. Trends Microbiol. 16:488-495. [DOI] [PubMed] [Google Scholar]
- 44.Turnbaugh, P. J., F. Backhed, L. Fulton, and J. I. Gordon. 2008. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3:213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turnbaugh, P. J., R. E. Ley, M. A. Mahowald, V. Magrini, E. R. Mardis, and J. I. Gordon. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027-1031. [DOI] [PubMed] [Google Scholar]
- 46.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munro, and T. Alatossava. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wen, L., R. E. Ley, P. Y. Volchkov, P. B. Stranges, L. Avanesyan, A. C. Stonebraker, C. Hu, F. S. Wong, G. L. Szot, J. A. Bluestone, J. I. Gordon, and A. V. Chervonsky. 2008. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455:1109-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zock, J., C. Cantwell, J. Swartling, R. Hodges, T. Pohl, K. Sutton, P. Rosteck, Jr., D. McGilvray, and S. Queener. 1994. The Bacillus subtilis pnbA gene encoding p-nitrobenzyl esterase: cloning, sequence and high-level expression in Escherichia coli. Gene 151:37-43. [DOI] [PubMed] [Google Scholar]
- 50.Zoetendal, E. G., M. Rajilic-Stojanovic, and W. M. de Vos. 2008. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57:1605-1615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.