Abstract
We describe a novel prokaryotic expression system for the production of cationic antimicrobial peptides (AMPs). The method relies on a translationally coupled two-cistron system, in which the termination codon for the first cistron (which encodes the anionic polypeptide mIFc2, a derivative of human gamma interferon) overlaps with the initiation codon for the second cistron (which encodes a cationic AMP) in the sequence of 5′-TAATG-3′. By forming an insoluble complex with the AMP upon translation, the mIFc2 protein efficiently neutralized the toxicity of the coexpressed cationic AMP and minimized the sensitivity of AMP to proteolytic degradation in a host. The AMPs were retrieved from the insoluble inclusion bodies without any chemical or enzymatic cleavage step by simple cation-exchange chromatography. With our system, ∼100 mg of various AMPs (buforin IIb, parasin I, and pexiganan) were obtained from 1 liter of Escherichia coli culture. Our expression system may represent a universal cost-effective solution for the mass production of intact AMPs in their natural forms.
Of worldwide concern is the increasing development of bacterial and fungal strains that are resistant to currently available antimicrobial drugs. This worsening situation has spurred Herculean efforts to develop new classes of antibiotics with novel targets and modes of action (19). Cationic antimicrobial peptides (AMPs) play a key role in the primary host defense of living organisms against infections by pathogenic microorganisms. Because their mechanisms of antimicrobial action differ from those of conventional antibiotics, AMPs have received increasing attention as a potential new class of therapeutic substances (22, 30).
In contrast to bacterial growth in the presence of commonly prescribed antibiotics, the growth of bacteria in the presence of AMPs does not easily give rise to the selection of pathogenic drug-resistant mutant strains. This is because AMPs rapidly kill microbes by a variety of mechanisms, including (i) fatal depolarization of the normally energized bacterial membrane, (ii) creation of physical holes that cause cellular contents to leak out, (iii) degradation of the cell wall, (iv) disturbance of membrane functions, and/or (v) damaging of critical intracellular targets after internalization of the AMPs (7, 11, 19, 22, 30). Moreover, AMPs activate the host's innate (nonspecific) immune response without acting as a foreign antigen target of the host's adaptive immune system (23, 30). Despite the fact that AMPs show great potential as a novel class of antibiotics, the lack of a cost-effective means of mass production has limited the development of these peptides as human therapeutics (8).
Numerous biological expression systems have been introduced for the cost-effective production of AMPs in Escherichia coli (9). To decrease their natural destructive behavior toward microorganisms and sensitivity to proteolytic degradation, AMPs are often produced as fusion proteins in heterologous hosts (12, 16). These studies show that certain fusion partner proteins neutralize the toxicity of AMPs and improve their stability against proteolysis in an expression host. In another series of experiments, recombinant AMP-containing fusion proteins are expressed in tandem repeats in an attempt to increase AMP production. As expected, multimeric expression further enhanced the yield of AMP fusion proteins (9, 12, 16). However, all of these methods require that the AMP be separated from its fusion partner, and recombinant fusion proteins, including multimeric ones, are usually cleaved with enzymes such as furin or chemicals such as CNBr (12, 16). This additional process results in inefficient cleavage and thus poor recovery of AMPs from fusion partners. Moreover, unwanted amino acid residue(s) are often included in the AMPs after the cleavage reaction and can decrease antimicrobial activity and cause problematic side effects (18). Therefore, a new approach for producing an intact and biologically active AMP without the inclusion of an enzymatic or chemical cleavage step is needed.
We have developed here a novel translationally coupled, two-cistron expression system for the production of recombinant AMPs in their natural forms. Using this system, we were able to produce, from 1 liter of E. coli culture, ∼100 mg of a potent AMP, buforin IIb (BIIb) (15), without a cleavage step, and other cationic AMPs (parasin I [24] and pexiganan [6]) were also successfully produced.
MATERIALS AND METHODS
Bacterial strains, plasmids, and enzymes.
E. coli XL1-Blue (Stratagene, La Jolla, CA) was used as a host for subcloning, and E. coli BL21(DE3) (Invitrogen, Carlsbad, CA) was used for gene expression. E. coli cells were grown in Luria-Bertani (LB) medium at 37°C, and ampicillin (50 μg/ml) was added for the growth of plasmid-containing cells. The pET21c vector (Novagen, Madison, WI) was used for the expression of the AMP fusion proteins. Restriction enzymes were purchased from New England Biolabs (Beverly, MA), Taq polymerase was purchased from Takara (Otsu, Japan), and all enzymes were used according to the recommendations of the suppliers. All recombinant DNA techniques were performed as described by Sambrook and Russell (26).
Construction of translationally coupled mIFn-AMP cistrons.
The mIF gene (the modified human gamma interferon [hIFN-γ] gene was termed mIF), which was used as the first cistron of our two-cistron system, was synthesized by assembling eight synthetic oligonucleotides (oligos) in an asymmetric PCR (2). The hIFN-γ gene was mutated so as to produce the mIF gene (450 bp) that encoded a negatively charged protein with six amino acid (aa) substitutions relative to hIFN-γ (mIF; Table 1); these changes were made to facilitate an ionic interaction between the anionic mIF and the cationic AMPs. In addition, a region composed of 12 bp (5′-GAGGAGGTGGAA-3′, which encodes amino acid sequence EEVE) including a Shine-Dalgarno (SD) sequence (underlined), was introduced into the C terminus of the mIF gene in order to improve translational initiation of the product of the second cistron.
TABLE 1.
Characteristics of anionic coexpression partners and cationic AMPs
| Peptide | Mass (Da) | pI | Amino acid sequence (N to C terminus)a |
|---|---|---|---|
| Coexpression partners | |||
| mIF | 17,575 | 4.70 | CYCQDPYVKEAENLKKYFNAGHSDVADNGTLFLGILKNWKEESDEDIMQSQIVSFYFKLFKNFKDDQSIQKSMETIKEEMNVKFFNSNKKKRDDFEKLTNYSVTDLNVQRKAIHELIQVMAELSDAAKTGEDEDSQMLFRGRRASQEEVE |
| mIFc1 | 10,881 | 4.35 | CYCQDPYVKEAENLKKYFNAGHSDVADNGTLFLGILKNWKEESDEDIMQSQIVSFYFKLFKNFKDDQSIQKSMETIKEEMNVKFFNSNEEVE |
| mIFc2 | 7,371 | 4.12 | CYCQDPYVKEAENLKKYFNAGHSDVADNGTLFLGILKNWKEESDEDIMQSQIVSFYFKLEEVE |
| mIFc3 | 5,541 | 4.14 | CYCQDPYVKEAENLKKYFNAGHSDVADNGTLFLGILKNWKEESDEEVE |
| AMPs | |||
| BIIb | 2,560 | 13.27 | RAGLQFPVGRLLRRLLRRLLR |
| Parasin I | 2,000 | 12.84 | KGRGKQGGKVRAKAKTRSS |
| Pexiganan | 2,478 | 11.71 | GIGKFLKKAKKFGKAFVKILKK |
Substitution of amino acids is indicated by underlining. The substitutions are as follows: R45E, K46D, K131E, R132D, K133E, and R134D. The region containing the SD sequence, EEVE, is indicated in boldface.
Briefly, the 450-bp mIF gene was divided into eight oligos (see Table S1 in the supplemental material) that varied in length from 32 to 100 nucleotides (nt). The eight oligos (PR1 to PR8), which each overlaps with its respective neighboring sequences by an average of 20 nt, were mixed, annealed, and amplified by PCR, generating the mIF gene. For the gene encoding buforin IIb (BIIb), two complementary oligos (PR9 and PR16 [see Table S1 in the supplemental material]) that contain an NdeI and a BamHI site on each end, were annealed, digested with NdeI and BamHI, and ligated into pET21c that had been digested with the same restriction enzymes, generating pBIIb.
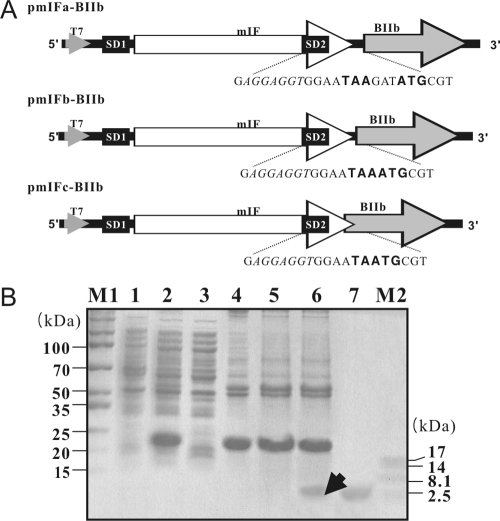
The plasmids that express both BIIb and one of each of the coexpression partner in a translationally coupled two-cistron system were constructed by recombinant PCR as follows. The genes encoding derivatives of mIF (the coexpression partners), followed by different intercistronic region at their 3′ ends, were placed in the first cistron, and the BIIb gene was placed in the second cistron. To identify the optimal distance between the two cistrons that resulted in the highest yield of the expressed proteins, we constructed three two-cistron plasmids (pmIFa-BIIb, pmIFb-BIIb, and pmIFc-BIIb) that contained a variety of intercistronic regions between the derivative of mIF and BIIb genes (Fig. 1A). To construct pmIFa-BIIb, pmIFb-BIIb, and pmIFc-BIIb, we amplified the mIFa, mIFb, and mIFc genes from the mIF template by using the primer pairs PR0/PR10, PR0/PR11, and PR0/PR12, respectively, and the BIIb genes from pBIIb by using the primer pairs PR13/PR16, PR14/16, and PR15/PR16, respectively. The resulting mIFa BIIb-, mIFb BIIb-, and mIFc BIIb-containing DNA fragments were then combined by using the primer pair PR0/PR16 (see Table S1 in the supplemental material), digested with NdeI and BamHI, and ligated into pET21c that had been digested with the same restriction enzymes, generating pmIFa-BIIb, pmIFb-BIIb, and pmIFc-BIIb, respectively (Fig. 1A). In pmIFa-BIIb and pmIFb-BIIb, the termination codon (TAA) of the mIF gene and the initiation codon (ATG) of the BIIb gene are 3 (5′-TAAGATATG-3′) and 0 nt (5′-TAAATG-3′) apart, respectively. In pmIFc-BIIb, the termination codon of mIF gene overlaps with the initiation codon of BIIb gene by 1 nt (5′-TAATG-3′).
FIG. 1.
Schematic representation of the translationally coupled two-cistron systems and their products. (A) Schematic representation of the translationally coupled two cistrons encoding mIF and BIIb. The mIF and BIIb genes are represented as white and gray arrows, respectively. The two-cistron expression plasmids have two SD sequences: one (SD1) upstream of the 5′ end and the other (SD2) at the 3′ end of the first cistron. The stop codon (TAA) and start codon (ATG) are indicated in boldface, and the SD sequences are shown in italics. The T7 promoter is from bacteriophage T7. (B) SDS-PAGE analysis of the products of several two-cistron expression systems. Lanes 1 and 2 show total cell protein fractions before and after IPTG induction, respectively. IPTG (0.5 mM) was added when the OD600 of the E. coli culture reached 0.6. Lane 3 represents the soluble fraction obtained from the total cell protein fraction shown in lane 2. Lanes 4, 5, and 6 represent proteins from solubilized inclusion bodies (in 3 M urea, pH 10) expressed by pmIFa-BIIb, pmIFb-BIIb, and pmIFc-BIIb, respectively. Lane 7 indicates synthetic BIIb. The arrow indicates recombinant BIIb. Lanes M1 and M2 show molecular size markers, with the actual molecular sizes (in kDa) given by the numbers that flank the gel photograph.
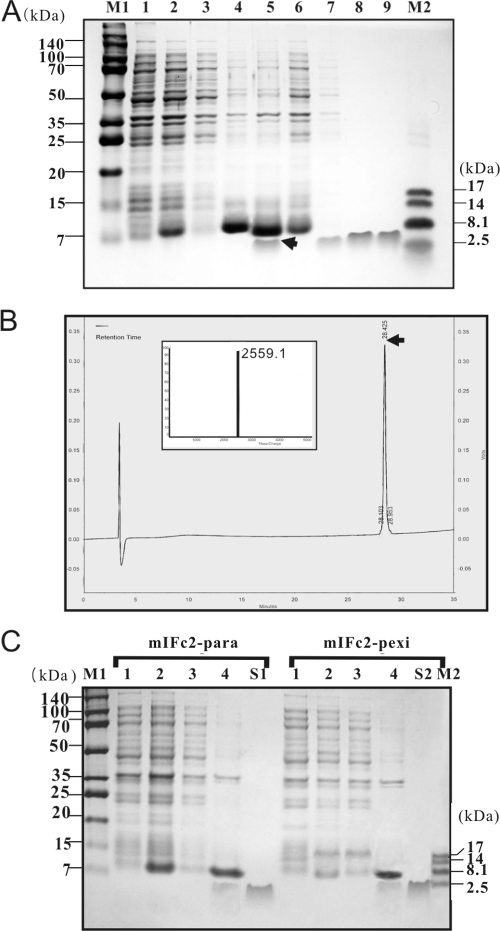
To identify the minimal length of the coexpression partner, we constructed expression plasmids that contain truncated versions of the mIFc gene (mIFc1, mIFc2, and mIFc3) as follows. The mIFc1, mIFc2, and mIFc3 genes were amplified from mIFc by PCR using the primer pairs PR0/PR17, PR0/PR18, and PR0/PR19, respectively, and BIIb was amplified from pBIIb by PCR by using PR15/PR16. The resulting fragments (mIFc1, mIFc2, or mIFc3 and BIIb) were combined by PCR using PR0/PR16 as described above, producing pmIFc1-BIIb, pmIFc2-BIIb, and pmIFc3-BIIb. In these three vectors, the termination codons of mIFc1, mIFc2, and mIFc3 genes overlap with the initiation codon of BIIb gene as in pmIFc-BIIb. The parasin I and pexiganan genes were also synthesized by PCR using the complementary oligos PR20/PR21 and PR22/PR23, respectively. The parasin I and pexiganan expression vectors, pmIFc2-para and pmIFc2-pexi, were constructed by the method described above for pmIFc2-BIIb.
Expression of the translationally coupled mIFn-AMP genes in E. coli.
E. coli BL21(DE3) cells were transformed with the various expression vectors described above (pmIFc-BIIb, pmIFc1-BIIb, pmIFc2-BIIb, and pmIFc3-BIIb). E. coli transformants that harbored either pmIFc-BIIb, pmIFc1-BIIb, pmIFc2-BIIb, or pmIFc3-BIIb were inoculated into 3 ml of LB medium supplemented with ampicillin and grown at 37°C for 12 h. Each culture was then diluted 1:100 into fresh medium and grown at 37°C. At optical density at 600 nm (OD600) of 0.6, we induced expression of the mIFn (n = c, c1, c2, and c3) and BIIb genes by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 0.5 mM. The cells were harvested 4 h after induction by centrifugation at 6,000 × g for 10 min at 4°C and lysed by sonication (6 × 30 s) at 4°C (B. Braun instruments, Allentown, PA). We determined the amount of fusion proteins in whole-cell lysates obtained from the induced cultures (which were all at an equivalent OD600) by quantifying the protein bands in each lane of sodium dodecyl sulfate (SDS)-polyacrylamide gels by densitometry at 600 nm (Bio/Profile image analysis software; Bio-1D, Vilber Lourmat, France).
Production and purification of the recombinant AMPs.
For mass production of BIIb, mIFc2 was selected as the coexpression partner because it was the smallest version of the mIFc protein that displayed a neutralizing effect that was as robust as that of the larger version of mIFc (Fig. 2B). E. coli cells harboring the AMPs expression vectors pmIFc2-BIIb, pmIFc2-para, or pmIFc2-pexi were cultivated in 1 liter of LB medium in shaking flasks. At an OD600 of 0.6, IPTG was added to a final concentration of 0.5 mM to induce fusion gene expression, and the cells were harvested 4 h after induction (OD600 of 2.4). After lysis of the cells by sonication as described above, inclusion bodies were recovered by centrifugation at 10,000 × g for 30 min at 4°C and washed with 50 mM Tris-HCl buffer (pH 8.0).
FIG. 2.
Effect of the truncation of coexpression partners (derivatives of mIF) on AMP expression. (A) Schematic representation of the various translationally coupled, two-cistron vectors that expressed truncated mIF and BIIb. (B) SDS-PAGE analysis of the mIF- and BIIb-containing complexes expressed. Lanes 1, 4, 7, and 10 show total cell proteins from E. coli cells that harbored pmIFc-BIIb, pmIFc1-BIIb, pmIFc2-BIIb, or pmIFc3-BIIb, respectively, before IPTG induction. Lanes 2, 5, 8, and 11 show total cell proteins from E. coli cells that contained pmIFc-BIIb, pmIFc1-BIIb, pmIFc2-BIIb, or pmIFc3-BIIb, respectively, after IPTG induction. IPTG (0.5 mM) was added when the OD600 of the E. coli culture reached 0.6. Lanes 3, 6, 9, and 12 show proteins from solubilized inclusion bodies (in 3 M urea, pH 10) isolated from E. coli cells that contained pmIFc-BIIb, pmIFc1-BIIb, pmIFc2-BIIb, or pmIFc3-BIIb, respectively. Lane 13 shows synthetic BIIb, and the arrow indicates recombinant BIIb. Lanes M1 and M2 show molecular size markers, with the actual molecular sizes (in kDa) given by the numbers that flank the gel photograph. (C) Relative amounts of BIIb to mIFn (n = c, c1, or c2) in the mIFn-BIIb complexes isolated from mIFn- and BIIb-expressing E. coli.
The inclusion bodies were then solubilized in 3 M urea (in 10 mM glycine-NaOH buffer at pH 10.0), and the solubilized coexpression mixture was applied to a Resource 15S cation-exchange column (Pharmacia LKB Biotechnology, Inc., Uppsala, Sweden). The bound peptides were eluted by applying a linear 0 to 0.5 M NaCl gradient in elution buffer [20 mM 2-(N-morpholino)ethanesulfonic acid (pH 6.0) with 0.5 M NaCl] and concentrated by lyophilization. The lyophilized peptides were further purified by reversed-phase high-pressure liquid chromatography (HPLC) on a Delta-Pak C18 column (3.9 mm by 300 mm [inner diameter], 15 μm, 300 Å pore size; Waters, Milford, MA) by using a linear elution gradient of 0 to 50% acetonitrile in 0.1% (vol/vol) trifluoroacetic acid at 1 ml/min for 1 h. The amounts of total proteins in the crude extracts and inclusion bodies (resuspended in Tris-HCl buffer) were determined by the bicinchoninic acid protein assay using bovine serum albumin as a standard, and the proteins collected after Resource 15S cation-exchange chromatography and reversed-phase HPLC were quantified by using a LavaPep-peptide quantification kit (Fluorotechnics, Sidney, Australia). In addition, mIF-BIIb protein complexes in the crude extracts and inclusion bodies and BIIb after Resource 15S cation-exchange chromatography and reversed-phase HPLC were also quantified by measuring the protein bands of SDS-polyacrylamide gels by densitometry at 600 nm according to the manufacturer's protocol (Bio/Profile Image Analysis Software; Bio-1D). In brief, the intensity of each protein band was measured as the integrated volume of pixels (with linear dimensions x and y in millimeters and the z axis as the relative absorbance) associated with each Coomassie blue-stained band, and the amount of mIF-BIIb complex or BIIb was calculated by multiplying the ratio of the intensity of the mIF-BIIb complex or BIIb band over the sum of the intensities of all protein bands in the same lane of a SDS gel by the amount of the total proteins determined above.
Characterization of recombinant BIIb.
The molecular weight and homogeneity of the recombinant BIIb preparation were analyzed by mass spectrometry (MS) on a matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer (Kartos Kompact MALDI, Manchester, England). The amino acid sequence of recombinant BIIb was determined by Edman degradation performed on a gas-phase sequencer (model 470A; Applied Biosystems, Foster City, CA). The antimicrobial activity of recombinant BIIb was determined against nine representative microorganisms, including gram-positive and gram-negative bacteria and fungi, using the broth microdilution assay as described by Park et al. (25).
RESULTS
Construction of translationally coupled two-cistron plasmids and expression of the encoded AMPs.
To express a potent antimicrobial peptide BIIb (15) in its naturally occurring form, translationally coupled, two-cistron plasmids were constructed by aligning a coexpression partner gene as the first cistron and a BIIb gene as the second cistron. For the successful expression of large amounts of BIIb, the coexpression partner protein must be able to shield the host-lethal effects of BIIb by (i) forming a complex with the AMP upon expression (20) and (ii) reinforcing the formation of inclusion bodies to protect the expressed peptides from proteolytic degradation by host proteases. We chose hIFN-γ as a coexpression partner because high-level expression of hIFN-γ in E. coli results in the distribution of >90% of the accumulated gene product into inclusion bodies (32).
For the first cistron, we made mutations in the hIFN-γ gene that resulted in the replacement of six basic amino acids (arginines and lysines) with six acidic amino acids (aspartates and glutamates) (the specific mutations were R45E, K46D, K131E, R132D, K133E, and R134D). These changes lowered the isoelectric point (pI) of hIFN-γ so as to efficiently facilitate complex formation (20) with cationic BIIb (the resulting modified hIFN-γ was mIF). In addition, we introduced an SD sequence (termed SD2) into C terminus of the first cistron for the efficient translation of the gene product of the second cistron (Fig. 1A).
It has been reported that efficient translational coupling between two cistrons occurs when the cistrons are in close proximity to each other (13, 20). Therefore, in order to analyze the effect of the distance between the coexpression partner and the AMP genes on the expression of AMPs, we made three different constructs (pmIFa-BIIb, pmIFb-BIIb, and pmIFc-BIIb) in which we varied the distance between the termination codon of the first cistron (mIF) and the initiation codon of the second cistron (BIIb) (Fig. 1A). The termination codon of the first cistron and the subsequent initiation codon of the second cistron were separated by 3 nt in the pmIFa-BIIb (5′-TAAGATATG-3′) and 0 nt in the pmIFb-BIIb (5′-TAAATG-3′); finally, in the pmIFc-BIIb, the termination codon of the first cistron overlapped with the subsequent initiation codon by 1 nt (5′-TAATG-3′) (Fig. 1A).
When the pmIFc-BIIb was expressed in E. coli, mIF efficiently neutralized the toxicity of the coexpressed BIIb by forming insoluble complexes. The expression of BIIb was confirmed after solubilization of the mIF- and BIIb-containing inclusion bodies in 3 M urea (Fig. 1B, lane 6). On the other hand, BIIb was barely expressed either when the mIF- and BIIb-encoding cistrons were placed either 3 or 0 nt apart (Fig. 1B, lanes 4 and 5, respectively) or in the absence of the mIF-encoding cistron (data not shown). These results clearly indicate that coexpressed anionic mIF and cationic BIIb form insoluble complexes through electrostatic interactions in our translationally coupled, two-cistron system when the termination codon of the first cistron overlaps with the subsequent initiation codon by 1 nt. Therefore, we selected the mIFc-BIIb vector for further expression experiments.
Selection of a minimal mIF derivative for the BIIb.
Although BIIb could be successfully expressed by translational coupling with mIF, the yield of BIIb from the mIF-BIIb complex is only ∼12%; this is because mIF (17.6 kDa) is a much larger protein than BIIb (2.6 kDa). To increase the relative yield of BIIb in the insoluble complex, we made translationally coupled, two-cistron constructs that encoded C-terminally truncated variants of mIF. The resulting derivatives—mIFc1, mIFc2, and mIFc3—were shorter than the intact mIF by 58, 87, and 102 aa residues, respectively, but still maintaining acidic pIs of 4.35, 4.12, and 4.14, respectively. Furthermore, the C-terminal region, including SD2, was identical to that of the mIFc-BIIb vector (Table 1 and Fig. 2A).
When these vectors were expressed in E. coli, the anionic truncated mIF derivatives interacted effectively with cationic BIIb (pI 13.27), and the production yield of BIIb increased significantly with the decreasing size of the coexpression partners (Fig. 2B and C). A nearly twofold increase in the yield of BIIb was achieved when the length of mIF was reduced from 152 to 63 aa (mIFc2 [see mIFc2-BIIb in Fig. 2C]), but further reduction in the length of mIF to 48 aa (mIFc3) dramatically reduced the yield of BIIb (Fig. 2B). We could reduce the size of the coexpression partner mIF down to 63 aa without compromising the yield of BIIb. Therefore, the pmIFc2-BIIb was selected for the remainder of the experiments.
Purification and characterization of recombinant BIIb, parasin I, and pexiganan.
After solubilization of the mIFc2- and BIIb-containing complex in 3 M urea (pH 10), the mixture was subjected to Resource 15S cation-exchange chromatography (Fig. 3A, lane 7), followed by reversed-phase HPLC (Fig. 3A, lane 8). A total of ∼100 mg of pure BIIb was obtained from 1 liter of E. coli culture after reversed-phase HPLC, and the purity of the BIIb preparation was more than 95%, which was estimated on the basis of MALDI-TOF-MS results (Table 2). The molecular mass (2,559.1 Da; Fig. 3B) and amino acid sequence of the recombinant BIIb were identical to those of natural BIIb. This recombinant BIIb displayed antimicrobial activity identical to that of chemically synthesized BIIb (Table 3).
FIG. 3.
Expression and purification of recombinant AMPs in E. coli BL21(DE3) that harbor pmIFc2-BIIb, pmIFc2-para, or pmIFc2-pexi expression vectors. (A) SDS-PAGE analysis of mIFc2- and BIIb-containing complexes expressed in E. coli by the mIFc2-BIIb vector. Lanes 1 and 2 show total cell protein fraction before and after IPTG induction, respectively. Lane 3 represents the soluble fraction of the total cell proteins shown in lane 2. Lanes 4 and 5 show the insoluble fraction (inclusion bodies) of the total cell proteins shown in lane 2 and the inclusion bodies solubilized in 3 M urea (pH 10), respectively. Lanes 6 and 7 show flowthrough and the bound fraction that passed through Resource 15S cation-exchange chromatography, respectively. Lanes 8 and 9 show the recombinant BIIb purified by reversed-phase HPLC and synthetic BIIb, respectively. IPTG (0.5 mM) was added when the OD600 of the E. coli culture reached 0.6. Lanes M1 and M2 show molecular size markers, with the actual molecular sizes (in kDa) given by the numbers that flank the gel photograph. (B) Purification of the recombinant BIIb by reverse-phase HPLC of the bound fractions from Resource 15S cation-exchange chromatography and determination of purity by MALDI-TOF-MS (inset). The arrow indicates the reversed-phase HPLC fraction that was collected and analyzed by MALDI-TOF-MS. (C) SDS-PAGE analysis of mIFc2- and para- or mIFc2- and pexi-containing complexes expressed in E. coli by the pmIFc2-para (lanes 1 to 4) and pmIFc2-pexi (lanes 1 to 4). Lanes 1 and 2 show total cell proteins before and after IPTG induction, respectively. Lane 3 represents the soluble fraction of the total cell proteins shown in lane 2. Lane 4 shows the insoluble fraction (inclusion bodies) of the total cell proteins fraction shown in lane 2 after it was solubilized in 3 M urea (pH 10). Lanes S1 and S2 show synthetic parasin I and pexiganan, respectively. IPTG (0.5 mM) was added when the OD600 of the E. coli culture reached 0.6. Lanes M1 and M2 show molecular size markers, with the actual molecular sizes (in kDa) given by the numbers that flank the gel photograph.
TABLE 2.
Purification of recombinant BIIb expressed from pmIFc2-BIIb from E. colia
| Purification step | Total protein (mg)b | Amt of protein of interest (mg)c | Yield (%)d |
|---|---|---|---|
| Crude extractse | 1,280 | 665 | 100 |
| Inclusion bodies | 950 | 632 | 95 |
| Cation-exchange chromatography | 126 | 117 | 18 |
| HPLC | 100 | 100 | 15 |
All of the values were the average of five independent experiments.
The amounts of total proteins in the crude extracts and inclusion bodies (resuspended in Tris-HCl buffer) were determined with the BCA protein assay using bovine serum albumin as a standard. The proteins after Resource 15S cation-exchange chromatography and reversed-phase HPLC were quantified with the LavaPep peptide quantification kit.
The amounts of mIF2-BIIb protein complexes in the crude extracts and inclusion bodies, and the amounts of BIIb after Resource 15S cation-exchange chromatography and reversed-phase HPLC were also determined by quantifying the protein bands of SDS-polyacrylamide gels by densitometry at 600 nm.
The yields were calculated based on the amount of the protein of interest.
The starting material for purification was a crude extract obtained by cell lysis of 1 liter of IPTG-induced E. coli culture as described in Materials and Methods.
TABLE 3.
Antimicrobial activities of recombinant and synthetic BIIb
| Microorganism | MIC (μg/ml)a
|
|
|---|---|---|
| Recombinant BIIb | Synthetic BIIb | |
| Gram-positive bacteria | ||
| Bacillus subtilis | 2 | 2 |
| Staphylococcus aureus | 0.5 | 0.5 |
| Streptococcus mutans | 2 | 2 |
| Gram-negative bacteria | ||
| Escherichia coli | 2 | 2 |
| Pseudomonas putida | 2 | 2 |
| Salmonella enterica serovar Enteritidis | 1 | 1 |
| Fungi | ||
| Cryptococcus neoformans | 2 | 2 |
| Saccharomyces cerevisiae | 2 | 2 |
| Candida albicans | 1 | 1 |
The MIC represents the amount of AMP required to inhibit growth of the microorganism. Each MIC was determined from two independent experiments performed in triplicate.
To verify the applicability of our two-cistron system to other AMPs, we expressed two other cationic AMPs, parasin I and pexiganan, from their corresponding mIFc2-AMP expression vectors. Parasin I is a potent 19-residue peptide isolated from the skin mucus of wounded catfish (24). Pexiganan (MSI-78) is a frog magainin derivative consisting of 22 aa residues that is being developed to treat foot ulcers in diabetic patients (6). The pIs of parasin I and pexiganan are 12.84 and 11.72, respectively. As shown in Fig. 3C, parasin I and pexiganan were successfully produced through translational coupling with mIFc2 in our two-cistron expression system, although their production yields were slightly lower than the BIIb yield.
DISCUSSION
Cationic AMPs have been regarded as a potential solution to the worldwide emergence and rapid horizontal spread of antibiotic-resistant traits in bacteria of human and veterinary clinical significance (9). Numerous biological expression systems have been introduced for the cost-effective production of AMPs (9). However, because AMPs display a natural destructive behavior toward microorganisms and a relative sensitivity to proteolytic degradation, most of the expression systems have been based on the production of AMP-partner protein fusions in heterologous hosts (9, 12, 16). Neutralization of AMP toxicity and increased expression of recombinant AMPs can be achieved with such systems, but they also require complicated, yield-compromising cleavage steps to release AMPs from their partner proteins after purification. Therefore, a cost-effective method for the production of intact and biologically active AMPs is sorely needed. In the present study, we described a procedure that permits the production of large amounts of potent AMPs in their naturally occurring forms without any cleavage step. Our method is based on translational coupling of the mIFc2 gene, which encodes an anionic polypeptide, with an AMP gene, which encodes a cationic polypeptide, in a two-cistron expression system that reduces the inherent toxicity of AMPs toward host bacterial cells.
It is well known that, in bacterial cells and some retroviruses or long terminal repeat retrotransposons, several protein genes can be coexpressed from a single polycistronic mRNA and ribosomes can reinitiate efficiently for the coupling of many closely juxtaposed genes (1, 10, 14, 31). A typical polycistronic mRNA is composed of several cistronic and intercistronic regions. In most polycistronic mRNAs, each cistron has a ribosome-binding site in the intercistronic region upstream of the initiation codon of the next cistron, which helps the translation of the gene products to proceed sequentially through the various linked cistrons. When translational coupling occurs, efficient translation of a downstream coding region in a polycistronic mRNA is partially or completely dependent on the prior translation of the adjacent upstream coding region (4). When the termination codon of the first cistron is positioned near the initiation codon of the next cistron, translation between the adjacent cistrons is directly linked; this is because the ribosome bound to the first cistron can easily gain access to the initiation codon of the second cistron (17). In the reinitiation mode, the 70S ribosome dissociates when it encounters the termination codon of the first cistron, and the 50S particle gets released, while the 30S particle remains attached to the mRNA and reinitiates translation when it finds the second SD (SD2) (17, 21).
We have adapted this two-cistron expression system for the expression of AMPs. Even though translational coupling was found to occur where the termination codon of the first cistron is positioned near the initiation codon of the second cistron (27-29), efficient translational coupling of forming an insoluble complex between AMP and a coexpression partner was achieved only when the sequence of the termination codon of the first cistron (which encodes the mIFc2 gene) overlapped with the initiation codon of the second cistron (which encodes the BIIb gene) in the sequence 5′-TAATG-3′. In our translational coupling system, it seems that the product of the first cistron (mIFcn) interacts with the product of the second cistron (AMP) immediately after translation before the product of the second cistron (AMP) exerts its lethal effect to the host.
In our translational coupling system, mIFc2 and its derivatives were chosen as anionic and insoluble coexpression partners, because (i) through translational coupling, mIFc2 protein reinforces the formation of inclusion bodies and hence prevents the host-lethal effect and proteolytic degradation of the expressed AMP; (ii) the mIFc2 protein has acidic pI and thus can be easily separated from cationic AMPs; and (iii) mIFc2 protein is nontoxic to our host bacterium, E. coli. This translational coupling of the BIIb and mIFc2 polypeptides led to the accumulation of large amounts of an insoluble complex of BIIb and mIFc2 in the E. coli host. The translational coupling of BIIb and mIFc2 polypeptides seems to be absolutely required for the process of forming a joint insoluble complex, because E. coli cells did not grow well and thus the joint insoluble complex was not formed when BIIb and mIFc2 polypeptide were expressed separately from two different plasmids (data not shown).
In addition, we found that AMP yield is enhanced by decreasing the length of the mIFc coexpression partner. The expression level of BIIb was significantly improved in accordance with the truncation of mIF (Fig. 2B and C). However, the mIF derivative that was <63 aa did not form an insoluble complex with or enhance the expression of BIIb.
Purification of recombinant BIIb produced by our two-cistron expression system was very simple and quite economical, in part because no cleavage step was required. In addition, we used 3 M urea (at pH 10.0) to dissolve the insoluble mIFc2-AMP complex, rather than 6 to 8 M urea, which is typically used to dissolve inclusion bodies. We used these milder conditions because our purpose was only to dissociate the recombinant BIIb from the insoluble complex and not to fully denature the peptide. BIIb that had been dissociated from the complex could then be retrieved from the dissolved inclusion bodies simply by using cation-exchange chromatography.
Until now, the only method that allows to produce AMPs from recombinant fusion protein without the addition of chemicals or enzymes was the use of a self-cleavage system, such as an intein-mediated method (3). However, the AMP yield from such a system has been very low (<2 mg/liter), because the intein tag is much larger than the AMPs and thus uses the majority of the translation resources provided by the host cell (3, 5). In contrast to the intein system, our translationally coupled system generated ∼100 mg of cationic BIIb (15) from 1 liter of E. coli culture. Moreover, other cationic AMPs (parasin I [24] and pexiganan [6]) were also successfully expressed through our system. These results reveal that our expression system could be a universal mass production method for cationic and small AMPs comprised of natural amino acids and may constitute a cost-effective solution for the mass production of AMPs in their intact and natural forms.
Supplementary Material
Acknowledgments
This study was supported in part by grants from the Research Program of New Drug Target Discovery (M10748000314-08N4800-31410) of the Ministry of Education, Science, and Technology and a Korea Science and Engineering Foundation grant (R01-2008-000-20559-0) and by the 21C Frontier Program of Microbial Genomics and Applications (MG08-0204-1-0).
Footnotes
Published ahead of print on 10 April 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Baughman, G., and M. Nomura. 1983. Localization of the target site for translational regulation of the L11 operon and direct evidence for translational coupling in Escherichia coli. Cell 34:979-988. [DOI] [PubMed] [Google Scholar]
- 2.Cello, J., A. V. Paul, and E. Wimmer. 2002. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science 297:1016-1018. [DOI] [PubMed] [Google Scholar]
- 3.Chong, S., F. B. Mersha, D. G. Comb, M. E. Scott, D. Landry, L. M. Vence, F. B. Perler, J. Benner, R. B. Kucera, C. A. Hirvonen, J. J. Pelletier, H. Paulus, and M. Q. Xu. 1997. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene 192:271-281. [DOI] [PubMed] [Google Scholar]
- 4.Das, A., and C. Yanofsky. 1984. A ribosome binding site sequence is necessary for efficient expression of the distal gene of a translationally coupled gene pair. Nucleic Acids Res. 11:4757-4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diao, H., C. Guo, D. Lin, and Y. Zhang. 2007. Intein-mediated expression is an effective approach in the study of beta-defensins. Biochem. Biophys. Res. Commun. 357:840-846. [DOI] [PubMed] [Google Scholar]
- 6.Ge, Y., D. L. MacDonald, K. J. Holroyd, C. Thornsberry, H. Wexler, and M. Zasloff. 1999. In vitro antibacterial properties of pexiganan, an analog of magainin. Antimicrob. Agents Chemother. 43:782-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hancock, R. E., and H. G. Sahl. 2006. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24:1551-1557. [DOI] [PubMed] [Google Scholar]
- 8.Hilpert, K., R. Volkmer-Engert, T. Walter, and R. E. Hancock. 2005. High-throughput generation of small antibacterial peptides with improved activity. Nat. Biotechnol. 23:1008-1012. [DOI] [PubMed] [Google Scholar]
- 9.Ingham, A. B., and R. J. Moore. 2007. Recombinant production of antimicrobial peptides in heterologous microbial systems. Biotechnol. Appl. Biochem. 47:1-9. [DOI] [PubMed] [Google Scholar]
- 10.Inokuchi, Y., A. Hirashima, Y. Sekine, L. Janosi, and A. Kaji. 2000. Role of ribosome recycling factor (RRF) in translational coupling. EMBO J. 19:3788-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenssen, H., P. Hamill, and R. E. Hancock. 2006. Peptide antimicrobial agents. Clin. Microbiol. Rev. 19:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, J. M., S. A. Jang, B. J. Yu, B. H. Sung, J. H. Cho, and S. C. Kim. 2008. High-level expression of an antimicrobial peptide histonin as a natural form by multimerization and furin-mediated cleavage. Appl. Microbiol. Biotechnol. 78:123-130. [DOI] [PubMed] [Google Scholar]
- 13.Kimura, S., T. Umemura, and T. Iyanagi. 2005. Two-cistronic expression plasmids for high-level gene expression in Escherichia coli preventing translational initiation inhibition caused by the intramolecular local secondary structure of mRNA. J. Biochem. 137:523-533. [DOI] [PubMed] [Google Scholar]
- 14.Kojima, K. K., T. Matsumoto, and H. Fujiwara. 2005. Eukaryotic translational coupling in UAAUG stop-start codons for the bicistronic RNA translation of the non-long terminal repeat retrotransposon SART1. Mol. Cell. Biol. 25:7675-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, H. S., C. B. Park, J. M. Kim, S. A. Jang, I. Y. Park, M. S. Kim, J. H. Cho, and S. C. Kim. 2008. Mechanism of anticancer activity of buforin IIb, a histone H2A-derived peptide. Cancer Lett. 271:47-55. [DOI] [PubMed] [Google Scholar]
- 16.Lee, J. H., M. S. Kim, J. H. Cho, and S. C. Kim. 2002. Enhanced expression of tandem multimers of the antimicrobial peptide buforin II in Escherichia coli by the DEAD-box protein and trxB mutant. Appl. Microbiol. Biotechnol. 56:790-796. [DOI] [PubMed] [Google Scholar]
- 17.Lewin, B. 2004. Genes VIII, p. 142-144. Pearson Education, Inc., Upper Saddle River, NJ.
- 18.Li, L., J. X. Wang, X. F. Zhao, C. J. Kang, N. Liu, J. H. Xiang, F. H. Li, S. Sueda, and H. Kondo. 2005. High level expression, purification, and characterization of the shrimp antimicrobial peptide, Ch-penaeidin, in Pichia pastoris. Protein Expr. Purif. 39:144-151. [DOI] [PubMed] [Google Scholar]
- 19.Makovitzki, A., D. Avrahami, and Y. Shai. 2006. Ultrashort antibacterial and antifungal lipopeptides. Proc. Natl. Acad. Sci. USA 103:15997-16002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mashko, S. V., V. P. Veiko, A. L. Lapidus, M. I. Lebedeva, A. V. Mochulsky, I. I. Shechter, M. E. Trukhan, K. I. Ratmanova, B. A. Rebentish, V. E. Kaluzhsky, and V. G. Debabov. 1990. TGATG vector: a new expression system for cloned foreign genes in Escherichia coli cells. Gene 88:121-126. [DOI] [PubMed] [Google Scholar]
- 21.Moll, I., G. Hirokawa, M. C. Kiel, A. Kaji, and U. Bläsi. 2004. Translation initiation with 70S ribosomes: an alternative pathway for leaderless mRNAs. Nucleic Acids Res. 32:3354-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mookherjee, N., and R. E. Hancock. 2007. Cationic host defense peptides: innate immune regulatory peptides as a novel approach for treating infections. Cell. Mol. Life Sci. 64:922-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mygind, P. H., R. L. Fischer, K. M. Schnorr, M. T. Hansen, C. P. Sönksen, S. Ludvigsen, D. Raventós, S. Buskov, B. Christensen, L. De Maria, O. Taboureau, D. Yaver, S. G. Elvig-Jørgensen, M. V. Sørensen, B. E. Christensen, S. Kjaerulff, N. Frimodt-Moller, R. I. Lehrer, M. Zasloff, and H. H. Kristensen. 2005. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature 437:975-980. [DOI] [PubMed] [Google Scholar]
- 24.Park, I. Y., C. B. Park, M. S. Kim, and S. C. Kim. 1998. Parasin I, an antimicrobial peptide derived from histone H2A in the catfish, Parasilurus asotus. FEBS Lett. 23:258-262. [DOI] [PubMed] [Google Scholar]
- 25.Park, I. Y., J. H. Cho, K. S. Kim, Y. B. Kim, M. S. Kim, and S. C. Kim. 2004. Helix stability confers salt resistance upon helical antimicrobial peptides. J. Biol. Chem. 279:13896-13901. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Schoner, B. E., R. M. Belagaje, and R. G. Schoner. 1986. Translation of a synthetic two-cistron mRNA in Escherichia coli. Proc. Natl. Acad. Sci. USA 83:8506-8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schoner, B. E., R. M. Belagaje, and R. G. Schoner. 1987. Expression of eukaryotic genes in Escherichia coli with a synthetic two-cistron system. Methods Enzymol. 183:401-416. [DOI] [PubMed] [Google Scholar]
- 29.Schoner, B. E., R. M. Belagaje, and R. G. Schoner. 1990. Enhanced translational efficiency with two-cistron expression system. Methods Enzymol. 185:94-103. [DOI] [PubMed] [Google Scholar]
- 30.Scott, M. G., E. Dullaghan, N. Mookherjee, N. Glavas, M. Waldbrook, A. Thompson, A. Wang, K. Lee, S. Doria, P. Hamill, J. J. Yu, Y. Li, O. Donini, M. M. Guarna, B. B. Finlay, J. R. North, and R. E. Hancock. 2007. An anti-infective peptide that selectively modulates the innate immune response. Nat. Biotechnol. 25:465-472. [DOI] [PubMed] [Google Scholar]
- 31.Sprengart, M. L., E. Fuchs, and A. G. Porter. 1996. The downstream box: an efficient and independent translation initiation signal in Escherichia coli. EMBO J. 15:665-674. [PMC free article] [PubMed] [Google Scholar]
- 32.Wetzel, R., P. L. Jeanne, and C. Veilleux. 1991. Mutations in human interferon gamma affecting inclusion body formation identified by a general immunochemical screen. Bio/Technology 9:731-737. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.