Abstract
The gammaproteobacterium Xenorhabdus nematophila mutualistically colonizes an intestinal region of a soil-dwelling nematode and is a blood pathogen of insects. The X. nematophila CpxRA two-component regulatory system is necessary for both of these host interactions (E. Herbert et al., Appl. Environ. Microbiol. 73:7826-7836, 2007). Mutualistic association of X. nematophila with its nematode host consists of two stages: initiation, where a small number of bacterial cells establish themselves in the colonization site, and outgrowth, where these cells grow to fill the space. In this study, we show that the Cpx system is necessary for both of these stages. X. nematophila ΔcpxR1 colonized fewer nematodes than its wild-type parent and did not achieve as high a density as did the wild type within a portion of the colonized nematodes. To test whether the ΔcpxR1 host interaction phenotypes are due to its overexpression of mrxA, encoding the type I pilin subunit protein, we assessed the colonization phenotype of a ΔcpxR1 ΔmrxA1 double mutant. This mutant displayed the same colonization defect as ΔcpxR1, indicating that CpxR negative regulation of mrxA does not play a detectable role in X. nematophila-host interactions. CpxR positively regulates expression of nilA, nilB, and nilC genes necessary for nematode colonization. Here we show that the nematode colonization defect of the ΔcpxR1 mutant is rescued by elevating nil gene expression through mutation of nilR, a negative regulator of nilA, nilB, and nilC. These data suggest that the nematode colonization defect previously observed in ΔcpxR1 is caused, at least in part, by altered regulation of nilA, nilB, and nilC.
As part of its natural life cycle the gammaproteobacterium Xenorhabdus nematophila lives within two animal hosts: it is a mutualistic symbiont of the soil-dwelling nematode, Steinernema carpocapsae, and a pathogen toward a number of insect orders (14, 15, 18). X. nematophila colonizes a modified intestinal region of a free-living, nonfeeding stage of S. carpocapsae, which serves as its vector into the blood system (hemocoel) of susceptible insect hosts (15). X. nematophila is released from its nematode vector into the hemocoel and within 1 to 2 days kills the insect (15). The insect cadaver serves as a nutrient source for nematode and bacterium reproduction. Once nutrients inside the insect host are depleted, X. nematophila reassociates with the vector stage of the nematode, which migrates into the soil to seek a new insect host and repeat the cycle (16).
The S. carpocapsae vector-stage association with X. nematophila is species specific: of all Xenorhabdus species tested to date, only X. nematophila can colonize S. carpocapsae nematodes (1, 6). In a fully mature vector-stage nematode, the intestinal colonization site, known as a receptacle (48), or vesicle (2) can contain >100 CFU of X. nematophila (33). This bacterial population is founded by one or a few X. nematophila cells, and the process of colonization includes distinguishable initiation and outgrowth stages (33). Within the vesicle, X. nematophila cells adhere to a nematode-derived structure termed the intravesicular structure that is surrounded by a mucuslike material (32). These data have led to a model in which colonization initiation by X. nematophila cells includes binding to a specific sugar associated with the intravesicular structure (32). Newly colonized S. carpocapsae nematodes contain very few X. nematophila cells that increase in number as the nematodes age, reaching a fully colonized level after ∼6 days (33). X. nematophila mutants defective in both initiation and outgrowth have been isolated, lending insight into gene products and physiological processes necessary for nematode colonization by this bacterium (6, 23, 32, 49). For example, metabolic mutants defective in the biosynthesis of methionine or threonine can initiate colonization but do not grow within the vesicle (13, 34), while mutants lacking the sigma factor homolog RpoS cannot initiate colonization (23, 49).
Current evidence suggests that, although X. nematophila growth occurs in the nematode vesicle, it is a nutrient-limited and stressful environment (7, 19). When released from this environment into insect hemolymph, X. nematophila is expected to encounter a relatively nutrient rich environment (7, 17, 20, 38, 41) but also faces insect immune responses. We do not yet understand fully which characteristics of these environments are sensed by X. nematophila or how this information is transduced to the expression of symbiotic activities appropriate for each host. However, one regulator involved in this process is the CpxRA two-component signaling system, which is necessary for interactions with both hosts and is hypothesized to help regulate transitions between the nematode and insect environments (21). In other bacteria, the Cpx system senses cell surface-related parameters, such as proximity to a surface (39), changes in external conditions (27, 31, 36, 37, 50), or misfolded proteins in the periplasm (11). In response to sensor kinase (CpxA) activation by these signals, the response regulator, CpxR, regulates expression of genes that function in cell envelope integrity (43), motility and chemotaxis (10), biofilm formation (12), adherence and invasion of host cells (24, 31), pilus formation (25), type III secretion (35), and protein folding and degradation (8, 9, 42, 44).
When the cpxR gene is deleted in X. nematophila, the bacterium is less effective at killing a model insect host, Manduca sexta, and colonizes S. carpocapsae nematodes at lower levels than its wild-type parent (21). Phenotypic and gene expression studies of a cpxR deletion mutant (ΔcpxR1) led to the identification of multiple genes in the CpxR regulon, including genes known or thought to be involved in one or both host interactions. One of these genes is mrxA, encoding the X. nematophila type I pilin subunit (21) reported to be involved in X. nematophila-nematode mutualism (3) and virulence toward insects (3, 28-30). Previous studies have shown that the pilin subunit is toxic toward Helicoverpa armigera insects in a purified form (28-30) and that these pilin subunits are found in outer membrane vesicle blebs, which, perhaps not coincidentally, are also toxic toward H. armigera (28). In the ΔcpxR1 mutant, mrxA transcript is increased >3-fold over that of wild-type cells, indicating that the ΔcpxR1 mutant cell surface may contain an increased number of pili compared to wild-type cells. This has the potential to severely affect host interactions of a cell due to the potential of pili to stimulate immune systems (either of the nematode or insect) or attach to host ligands or other structures (26, 46).
Three other X. nematophila CpxR regulon members—nilA, nilB, and nilC (21)—are each necessary for nematode colonization and are thought to be involved in initiation of colonization, perhaps through signaling or attaching to a nematode surface (4-6). nilA, nilB, and nilC are encoded at the same locus, termed the symbiosis region 1 (23). nilA and nilB are cotranscribed, while nilC is divergently transcribed (23). NilC is a periplasmically oriented outer membrane lipoprotein (4), NilB is an outer membrane protein of unknown function (A. Bhasin and H. Goodrich-Blair, unpublished data), while NilA is predicted to be an inner membrane protein (23).
Both nil promoters are repressed by the synergistic activities of Lrp, the leucine-responsive regulatory protein that also plays a role in nematode mutualism and insect virulence, and NilR, a small helix-turn-helix containing protein (4, 5, 7). Maximal expression of the nil genes in liquid culture occurs in strains in which both nilR and lrp are disrupted (5). Derepression of the nil genes is thought to be necessary for successful colonization of nematodes, since a X. nematophila strain carrying an ectopic copy of NilR showed a 60-fold lower colonization level compared to the control strain (5).
The goal of the present study was to elucidate the mechanism(s) by which the Cpx signal transduction system contributes to X. nematophila mutualistic nematode colonization. Specifically, we tested whether CpxR regulation of mrxA and nil genes influences colonization and whether CpxR contributes to colonization initiation, outgrowth, or both.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains and plasmids used in the present study are listed in Table 1. All strains were grown in the dark in LB medium or LB agar plus 0.1% pyruvate at 30°C unless otherwise specified. Antibiotics were used at the following concentrations: ampicillin (150 μg ml−1), chloramphenicol (15 μg ml−1 for X. nematophila and 30 μg ml−1 for E. coli), gentamicin (25 μg ml−1), kanamycin (50 μg ml−1), and streptomycin (12.5 μg ml−1 for X. nematophila and 25 μg ml−1 for E. coli). Sucrose was used at a 5% final concentration.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| X. nematophila strains | ||
| HGB800 | X. nematophila ATCC 19061, wild type | American Type Culture Collection |
| HGB1227 | HGB800 Tn7 | 21 |
| HGB1230 | HGB800 ΔcpxR1::Str | 21 |
| HGB1231 | HGB800 ΔcpxR1::Str Tn7 | 21 |
| HGB1232 | HGB800 ΔcpxR1::Str Tn7-cpxRA | 21 |
| HGB1103 | HGB800 ΔnilR17::Kan | 5 |
| HGB1345 | HGB800 ΔnilR17::Kan ΔcpxR1::Str | This study |
| HGB1117 | HGB800 Tn7-nilA-lacZ | 5 |
| HGB1121 | HGB800 Tn7-nilB-lacZ | 5 |
| HGB1101 | HGB800 Tn7-nilC-lacZ | 4 |
| HGB1239 | HGB800 ΔcpxR1::Str Tn7-nilA-lacZ | 21 |
| HGB1240 | HGB800 ΔcpxR1::Str Tn7-nilB-lacZ | 21 |
| HGB1241 | HGB800 ΔcpxR1::Str Tn7-nilC-lacZ | 21 |
| HGB1119 | HGB800 ΔnilR17::Kan Tn7-nilA-lacZ | 5 |
| HGB1123 | HGB800 ΔnilR17::Kan Tn7-nilB-lacZ | 5 |
| HGB1346 | HGB800 ΔnilR17::Kan Tn7-nilC-lacZ | This study |
| HGB1347 | HGB800 ΔnilR17::Kan ΔcpxR1::Str Tn7-nilA-lacZ | This study |
| HGB1348 | HGB800 ΔnilR17::Kan ΔcpxR1::Str Tn7-nilB-lacZ | This study |
| HGB1349 | HGB800 ΔnilR17::Kan ΔcpxR1::Str Tn7-nilC-lacZ | This study |
| HGB1364 | HGB800 ΔmrxA1::Kan | This study |
| HGB1365 | HGB800 ΔcpxR1::Str ΔmrxA1::Kan | This study |
| E. coli S17-1(λpir) | Donor strain for conjugations | 47 |
| Plasmids | ||
| pPK13 | Allelic-exchange vector containing mrxA loci with aph cassette insertion | 3 |
| pTn7-nilA-lacZ | pEVS107+nilA 5′-flanking region fused to lacZ (Ermr Kanr) | 5 |
| pTn7-nilB-lacZ | pEVS107+nilB 5′-flanking region fused to lacZ (Ermr Kanr) | 5 |
| pTn7-nilC-lacZ | pEVS107+nilC 5′-flanking region fused to lacZ (Ermr Kanr) | 4 |
| pKRcpxRStr | Vector for allelic exchange of cpxR gene for aad, encoding streptomycin resistance | 21 |
Ermr, erythromycin resistance; Kanr, kanamycin resistance.
Molecular biological methods.
Standard biological methods were used in the present study (45). Plasmid and PCR purification kits (Zymo Research, Orange, CA) were used according to the manufacturer's recommendations. DNA amplification was performed by using ExTaq (Takara Otsu, Shiga, Japan) according to the manufacturer's directions.
β-Galactosidase assays.
β-Galactosidase assays were performed as previously described (4) on overnight cultures inoculated from LB medium plus ampicillin agar X. nematophila colonies. X. nematophila lacZ fusion strains were created by triparental mating of pTn7-nilA, pTn7-nilB, or pTn7-nilC (5) as previously described (21).
Bacterial mutant construction.
The cpxR gene was deleted in a ΔnilR17::Kan background (5) by allelic exchange using pKRcpxRStr (21). Candidate colonies resistant to ampicillin, kanamycin, and streptomycin, but sensitive to chloramphenicol were chosen. A ΔcpxR1/ΔmrxA1::ΩKan mutant was created by conjugating pPK13 (3) into the ΔcpxR1 mutant (21) background (HGB1230). Candidate colonies resistant to ampicillin, kanamycin, streptomycin, and sucrose but sensitive to chloramphenicol and gentamicin were chosen. The presence of an insertion within mrxA was confirmed by Southern analysis as described previously (3). Both mutants were tested for the presence of the appropriate insertions by PCR amplification of the loci. Primers EH72 (5′-GAATAGCGGAGTTTGTTATCG-3′) and EH15 (5′-CACTGGTAACAAGGAGTAAGC-3′) amplified the cpxR locus. EH193 (5′-GGATCCATGAAACTTAACACAATTGGC-3′) and EH194 (5′-CTCGAGGAGGAAATATGTCACCATCGG-3′) amplified the mrxA locus.
Nematode colonization.
Colonization assays were performed as previously described (4) using five replicates per bacterial strain per experiment. Day 1 of the assay corresponds to the day infective juvenile (IJ) nematodes were taken from water traps and surface sterilized. IJs were then kept in closed tissue culture flasks at room temperature in the dark for the remainder of the experiment. For all time points, 1,000 IJs were sonicated, dilution plated onto LB plates, and grown overnight at 30°C to determine the average bacterial CFU/IJ for the nematode population. For microscopic analysis, nematodes were paralyzed with levamisole (Sigma, St. Louis, MO) and viewed as previously described (33). Between 100 and 1,000 IJs were viewed per time point per bacterial strain.
Nematode survival and development.
Bacterial strains were grown in liquid culture overnight, plated on lipid agar plates, and grown overnight in the dark at 30°C. Nematode eggs were plated in equal volumes onto the bacterial lawns and, at various time points after hatching, the nematodes were washed off the plates with water, and a portion was counted and viewed microscopically to determine the total number of nematodes present, as well as the approximate stage of development.
RESULTS
cpxR contributes to initiation and outgrowth during nematode mutualism.
We previously reported that ΔcpxR1 mutant cells are recovered from a population of colonized nematodes at an average of ∼38% that of wild-type cells (21). To determine whether this nematode colonization defect is due to an inability of nematodes to survive or reproduce on lawns of ΔcpxR1 mutant cells, populations of nematodes growing on either wild-type X. nematophila, a ΔcpxR1 mutant, or the complemented ΔcpxR1 mutant (ΔcpxR1 Tn7-cpxRA) were monitored for 2 weeks to assess differences in nematode viability or development. At various time points after plating nematode eggs on bacterial lawns, the nematodes were rinsed off the lipid agar plates with water, counted microscopically to quantify total numbers, and classified by approximate stage of growth. No obvious differences were observed, either in number or growth stage of the nematodes present (data not shown).
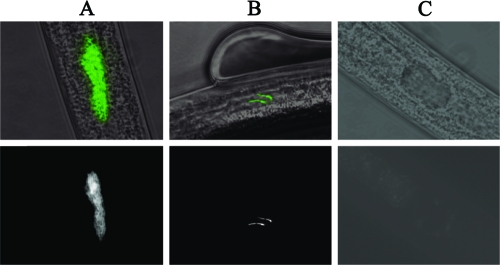
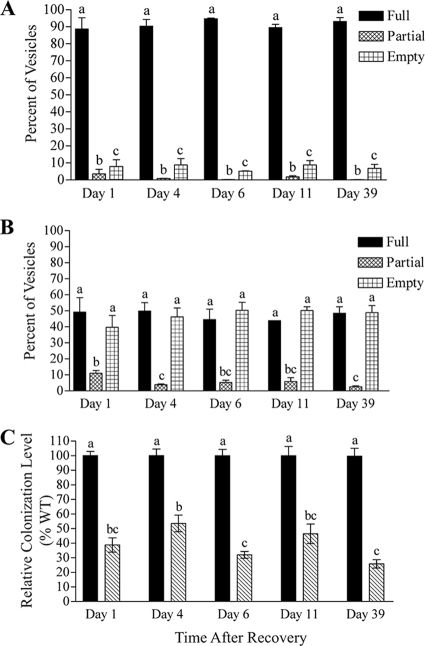
One possible explanation for the ΔcpxR1 mutant colonization defect is that, compared to the number of wild-type X. nematophila cells in colonized nematodes, ΔcpxR1 mutant-colonized nematodes each contain only 38% as many mutant cells, which would indicate an inability of the ΔcpxR1 mutant to maintain a fully colonized state or to grow past a certain limiting threshold within the nematode. A second possibility is that 38% of the nematodes exposed to the ΔcpxR1 mutant are fully colonized and that the remaining nematodes are uncolonized, which would indicate a defect in the ΔcpxR1 mutant's ability to initiate nematode colonization. To distinguish between these two possibilities, we used green fluorescent protein-labeled wild-type X. nematophila and ΔcpxR1 mutant cells to quantify by fluorescence microscopy the number of fully colonized (Fig. 1A), partially colonized (containing fewer than 10 bacterial cells within the vesicle) (Fig. 1B) and uncolonized (Fig. 1C) S. carpocapsae nematodes for each bacterial strain. In this experiment, wild-type X. nematophila cells fully colonized 89% of the nematodes counted, while the remaining 11% of nematodes counted were either uncolonized or partially colonized (Fig. 2A, day 1). However, of nematodes grown on the ΔcpxR1 mutant, only 49% were fully colonized, while 40% of nematodes were uncolonized and 11% were partially colonized (Fig. 2B, day 1).
FIG. 1.
A ΔcpxR1 mutant displays variable distribution within nematodes. Fluorescence microscope images of S. carpocapsae bacterial receptacles displaying various states of colonization by the ΔcpxR1 mutant are shown. The bottom row are fluorescence micrographs of nematode receptacles either fully colonized (A), partially colonized (containing 10 bacterial cells or fewer) (B), or uncolonized (C). The top-row images are phase-contrast micrographs of nematodes with false-colored fluorescence micrographs overlaid. All images were taken at a magnification of ×1,000. Bars, 10 μm.
FIG. 2.
A ΔcpxR1 mutant is defective in nematode colonization initiation and outgrowth. The proportion of fully colonized, partially colonized, and uncolonized nematode vesicles in nematodes colonized by wild-type X. nematophila (A) or the ΔcpxR1 mutant (B) over 39 days is shown. (C) The relative colonization level for the nematode population (as determined by dilution plating nematode sonicates) between the ΔcpxR1 mutant and the wild type was consistent over 39 days. Average values from three experiments are shown. Different letters indicate values that are statistically different from each other. Error bars indicate the standard error. WT, wild type.
Given the prolonged lag phase of ΔcpxR1 mutant growth in LB compared to wild-type cells (21), these results raised a third possible scenario, in which the ΔcpxR1 mutant grows more slowly in the vesicle but, given time, would eventually achieve full colonization levels in all nematodes in which colonization was initiated. Therefore, we continued our observation of nematode vesicles for 39 days past the initial surface sterilization to determine whether the ΔcpxR1 mutant showed delayed outgrowth. At days 4, 6, 11, and 39 after the initial surface sterilization, the nematodes were sonicated and plated to determine the average bacterial CFU per nematode, and the proportion of colonized versus uncolonized nematodes was assessed microscopically. Over 39 days, the average colonization of the ΔcpxR1 mutant (as determined by dilution plating the sonicated nematodes) consistently remained at ca. 26 to 54% that of nematodes colonized by wild-type X. nematophila cells (Fig. 2C). When fluorescent nematode vesicles were counted microscopically, wild-type X. nematophila cells fully colonized 89 to 95% of nematodes counted over the course of the experiment, as expected (Fig. 2A). The ΔcpxR1 mutant, however, fully colonized only 44 to 50% of the nematodes counted at all time points, and the number of fully colonized nematodes did not increase significantly over time. The percentage of partially colonized vesicles in both wild-type and ΔcpxR1-colonized nematodes decreased slightly during the course of the experiment (Fig. 2B).
Eliminating aberrant mrxA expression does not rescue ΔcpxR1 host interaction defects.
CpxR negatively influences the expression of mrxA (21), which encodes the X. nematophila type I pilin subunit protein. MrxA has been implicated in both mutualism (3) and the pathogenesis (3, 28-30) of X. nematophila, and we therefore hypothesized that abnormally high levels of the mrxA transcript in the ΔcpxR1 mutant (21) may contribute to ΔcpxR1 mutant-host interaction defects. For example, increased pili on the cell surface could mask lectins or receptors necessary for physical interactions within the nematode colonization site. Alternatively, within the insect or nematode host, an increase in surface pili on the ΔcpxR1 mutant may trigger an immune response (21). To test these ideas, we deleted mrxA in a ΔcpxR1 mutant background. If mrxA overexpression is responsible for the nematode colonization defect seen in the ΔcpxR1 mutant, then colonization may be restored to wild-type levels in the double mutant. One caveat of this approach is that mrxA itself was recently implicated as being necessary for nematode colonization (3). However, in contrast to that earlier study (3), we saw no decrease in nematode colonization of our ΔmrxA1 mutant, which colonized at 121% ± 6% that of the wild type (P < 0.05) (Fig. 3A). The absence of mrxA-encoded pilin subunit did not rescue the colonization defect of the ΔcpxR1 mutant (Fig. 3A). In this experiment, the ΔcpxR1 mutant colonized nematodes at 67% ± 4% that of the wild type (P < 0.001), while the ΔcpxR1 ΔmrxA1 mutant colonized at 73% ± 6%, a difference statistically different from the wild type (P < 0.01) but not different from the ΔcpxR1 mutant (Fig. 3A).
FIG. 3.
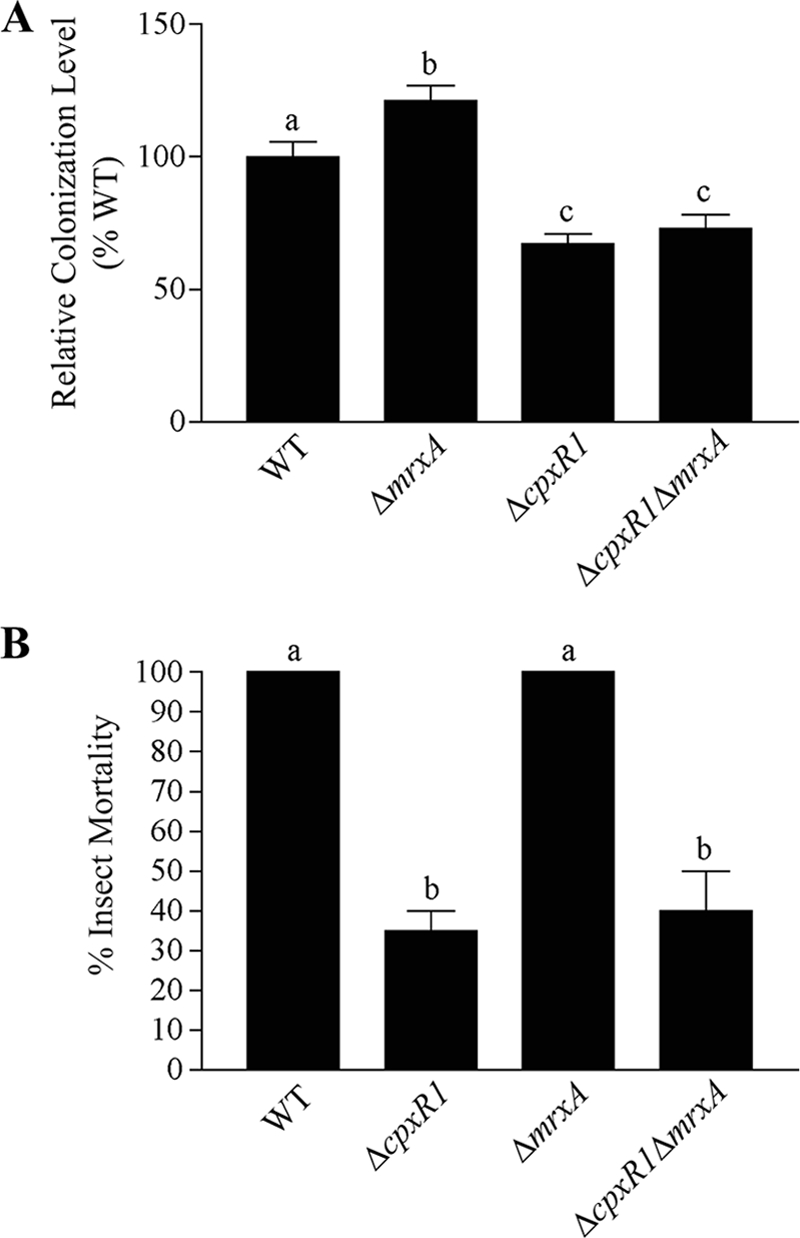
Eliminating aberrant mrxA expression in a ΔcpxR1 mutant does not rescue nematode colonization or virulence toward insects. For all graphs, error bars represent the standard error, and treatments labeled with different letters are significantly different from each other (P < 0.05). (A) The relative nematode colonization of ΔmrxA1, ΔcpxR1, and ΔcpxR1 ΔmrxA1 mutants is expressed as a percentage of the wild-type colonization. The average of two or more experiments is shown. (B) The percent mortalities of insects injected with wild type, ΔcpxR1, ΔmrxA1, and ΔcpxR1 ΔmrxA1 mutants are shown. The data represent the average of two or more experiments. WT, wild type.
Since mrxA has also been reported to play a role in insect virulence, we assessed whether the absence of mrxA rescues the virulence defect of the ΔcpxR1 mutant. However, virulence of the double mutant was not significantly different from that of the ΔcpxR1 mutant (Fig. 3B). Wild-type cells and the ΔmrxA1 mutant killed 100% ± 0% of M. sexta insects by 72 h after injection; however, the ΔcpxR1 and ΔcpxR1 ΔmrxA1 mutants each killed significantly fewer insects than did the wild type (P < 0.01) at 35% ± 5% and 40% ± 10% of the insects injected, respectively (Fig. 3B). This finding is also in contrast to previously reported data indicating that an mrxA mutant has reduced mortality toward H. armigera insects (3).
Elevated expression of nilA, nilB, and nilC, rescues the colonization defect of the ΔcpxR1 mutant.
CpxR positively regulates expression of three genes—nilA, nilB, and nilC (21)—that encode colonization factors (4, 23). We hypothesized that the colonization defect of the ΔcpxR1 mutant is caused by reduced expression of these genes. nilR encodes a repressor of nilA, nilB, and nilC expression (5), and an insertion mutation within nilR elevates nilABC expression above that of wild-type X. nematophila levels (5). To test whether elevation of nilABC expression in a ΔcpxR1 mutant would rescue the mutant's nematode colonization defect, we created a ΔnilR17 ΔcpxR1 double mutant that expresses greater than wild-type levels of nilA, nilB, and nilC (Fig. 4) and assessed its ability to colonize nematodes (Fig. 5). While a ΔcpxR1 mutant colonized nematodes at ca. 47% ± 5% that of the wild-type X. nematophila in this experiment (P < 0.001), the ΔnilR17 ΔcpxR1 double mutant colonized at 79% ± 7% that of the wild type (P > 0.05) (Fig. 5). As expected (5), the ΔnilR17 single mutant colonized nematodes at 117% ± 10%, a level not significantly different from that of the wild type (P > 0.05) (Fig. 5).
FIG. 4.
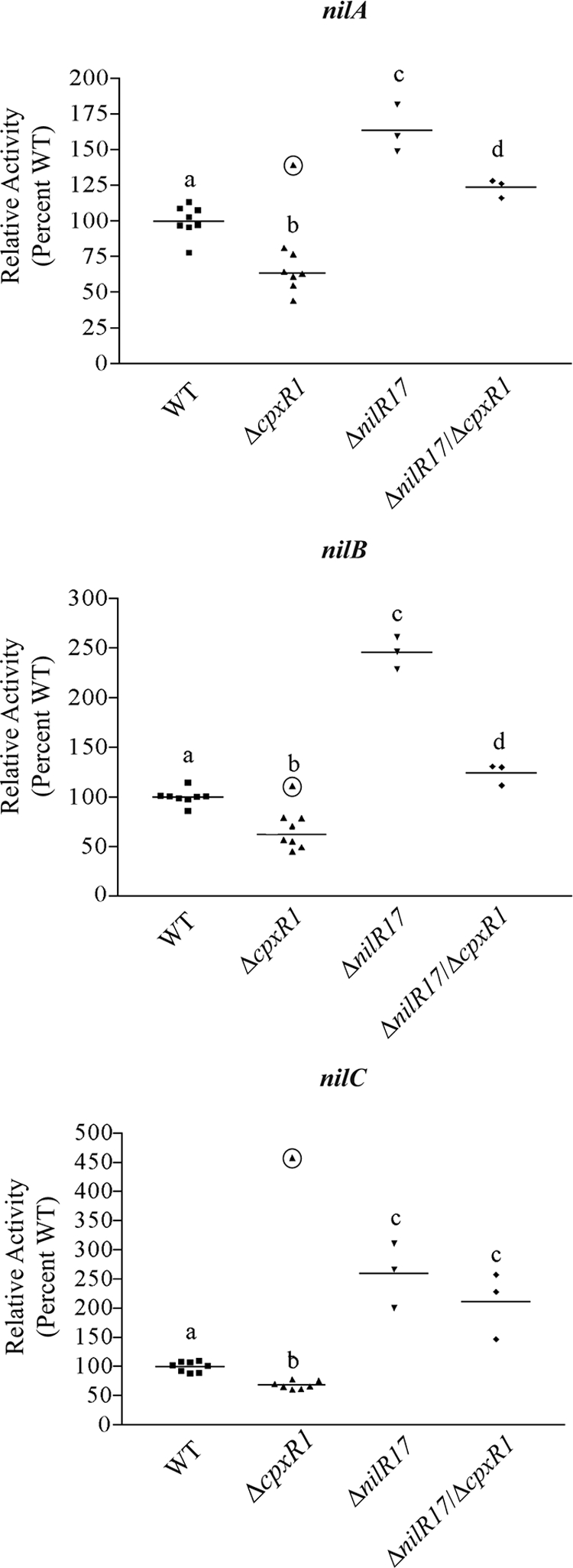
A mutation in nilR elevates nilABC expression in a ΔcpxR1 mutant. The relative expression of nilA, nilB, and nilC (expressed as a percentage of the wild-type activity) was measured by assaying the β-galactosidase activities of reporter fusions in wild-type, ΔcpxR1, ΔnilR17, and ΔnilR17 ΔcpxR1 strain backgrounds. The data points represent individual colonies tested, and bars represent strain averages. Different letters indicate values that are significantly different from the wild type (P < 0.01). Outlier ΔcpxR1 mutant colonies (circled) were left out of strain averages and statistical analysis. WT, wild type.
FIG. 5.
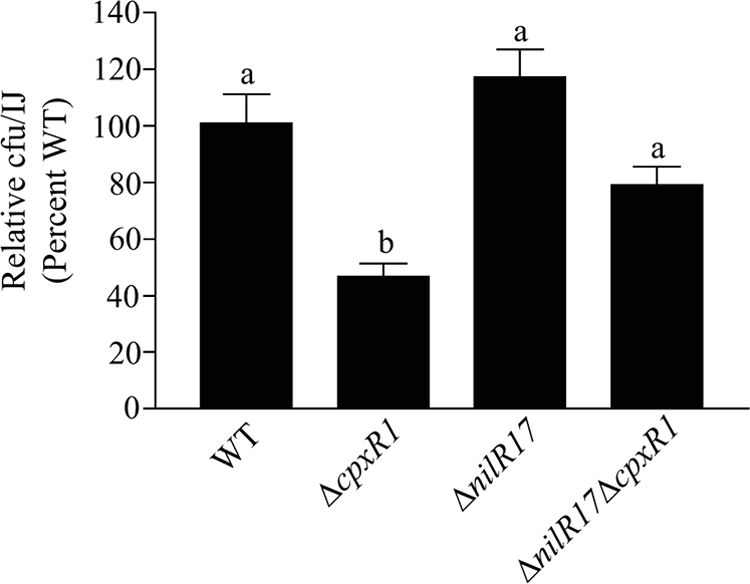
A mutation in nilR rescues the nematode colonization defect of a ΔcpxR1 mutant. The relative nematode colonization of ΔcpxR1, ΔnilR17, and ΔnilR17 ΔcpxR1 mutants is expressed as a percentage of the wild-type colonization. The average of two or more experiments is shown. Error bars represent the standard error. Different letters indicate values that are significantly different from wild-type colonization levels (P < 0.001). WT, wild type.
Interestingly, while a majority of the ΔcpxR1 single colony isolates tested demonstrated the previously reported reduction in nil gene expression, several ΔcpxR1 clones expressing lacZ fusions to one of the nil promoters exhibited abnormally high expression of nilA, nilB, or nilC (139, 111, and 458%, respectively, compared to that of the wild type) (Fig. 4). These clones, whose expression varied from the mean by more than twofold the standard deviation value, were not counted toward the average gene expression presented for a given strain. Potential causes of this phenomenon are discussed below.
DISCUSSION
In X. nematophila, the Cpx system is necessary for colonization of S. carpocapsae nematodes and virulence in M. sexta insects (21). In the present study we have shown that cpxR impacts two distinct stages of nematode colonization: initiation and outgrowth. Furthermore, we show that the likely cause of the colonization defect is that the ΔcpxR1 mutant does not express wild-type levels of nil genes encoding membrane-localized proteins necessary for nematode colonization.
A ΔcpxR1 mutant shows a defect in the colonization of S. carpocapsae nematodes, where, compared to wild-type cells, between 38 and 67% as many mutant cells are released from sonicated nematodes (21). Because these numbers represent an average number of bacterial cells recovered from a nematode population, we assessed whether the ΔcpxR1 mutant colonized only a portion of the nematode population or whether the entire population of nematodes was colonized to a lower level. We have shown, by fluorescence microscopy of nematodes cultivated with green fluorescent protein-labeled X. nematophila strains, that the ΔcpxR1 mutant failed to colonize 40% of the nematode population (Fig. 2). This indicates that the defect in this mutant is due in large part to an inability to initiate colonization of the nematode host.
The colonization and virulence defects of the ΔcpxR1 mutant were not rescued by deletion of mrxA (Fig. 3), a gene negatively regulated by CpxR (21). This indicates that elevated mrxA expression does not cause the host interaction defects of the ΔcpxR1 mutant. We found that an mrxA mutant does not have host interaction phenotypes, in contrast to a recent report stating that an mrxA mutant has decreased virulence toward H. armigera insects and reduced levels of nematode colonization (3). The authors of that study supplied the mrxA mutant construct that was used to create the strain used in the present study. Furthermore, quantitative PCR analysis confirmed that mrxA transcript is not expressed in the mrxA mutant made for the present study. Therefore, the discrepancies between the two reports cannot be explained by differences in the type of mutation. Both studies used X. nematophila ATCC 19061, although the strains were acquired at different times and may have diverged. The strain background used in the present study was the source of genomic DNA for sequencing (https://www.genoscope.cns.fr/agc/mage). The genome sequence contains another type I fimbrial subunit homologue, annotated as fimA (XN_978_8275/XN_1735). fimA is 48% identical in nucleotide sequence to mrxA and shares 36% amino acid identity. Therefore, our ΔmrxA1 mutant strain may not display defects in host interactions due to functional redundancy with fimA. Further experimentation will be required to determine whether this is the case and whether the loss of both type I fimbrial genes causes a decrease in host interactions in X. nematophila.
Our findings indicate that the overall colonization defects of the ΔcpxR1 mutant are due to insufficient levels of Nil colonization factors necessary for initiation. However, our data do not clarify whether CpxR transcriptional regulation of nil gene expression is necessary for colonization or whether overexpression of the nil genes simply is sufficient to overcome other defects of the ΔcpxR1 mutant. For example, the ΔcpxR1 mutant may have general envelope defects that destabilize Nil proteins in the membrane. Elevated expression of nil genes by removal of NilR-mediated repression might allow the expression of sufficient levels of Nil proteins to counteract such a general membrane defect.
Of the 60% of nematodes successfully colonized by the ΔcpxR1 mutant, 18% were only partially colonized (Fig. 2), suggesting that, in some nematodes, the mutant fails to grow. A similar combination of initiation and outgrowth defects has been posited for a nilA mutant that, unlike nilB and nilC mutants, is able to partially colonize a fraction (35%) of nematodes (6, 34). Therefore, the defects of the ΔcpxR1 mutant in both initiation and outgrowth potentially are due to insufficient expression of nil genes. Alternatively, CpxR may be necessary for acquisition of nutrients within the vesicle. Although the vesicle has sufficient levels of many amino acids and vitamins to support bacterial growth (34), it likely is nutrient limited. This idea is based on findings that X. nematophila grows slowly in the vesicle (∼10 h per doubling compared to 1 h per doubling in LB medium) (33) and that X. nematophila lrp mutants show delayed colony formation on LB plates when recovered from nematodes or minimal medium but not when recovered from insect hemolymph or LB medium (7). Two pieces of evidence argue against the idea that the outgrowth defects of the ΔcpxR1 mutant are due to nutrient limitation. First, the ΔcpxR1 mutant does not have a growth defect in defined medium containing leucine, glutamate, pyruvate, aspartic acid, and glucose (G. R. Richards, unpublished data), although growth of the ΔcpxR1 mutant in medium lacking sugars has not been tested. Second, metabolic mutants defective in the synthesis of methionine or threonine do not grow in the nematode vesicle. Such mutants exhibit spheroplast morphology and are eventually cleared from the population (34). Such spheroplast morphology has not been observed in colonizing ΔcpxR1 mutant cells (Fig. 1), and these mutant cells remain at stable levels over time (Fig. 2).
Unexpectedly, within the ΔcpxR1 population, several colonies displayed abnormally high levels of nil gene expression compared to the majority of other colonies in the population (21) (Fig. 4, circled data points). This phenomenon was seen in both previously stocked (21) and newly constructed nil fusion strains (data from both are shown in Fig. 4). There are several possible explanations to explain individual colonies with CpxR-independent expression of nil genes, including the presence of suppressor mutations, such as in nilR. Alternatively, nil gene expression may be affected by an as-yet-undescribed mechanism of phenotypic variation, in which a subpopulation of cells has altered regulation. For example, Lrp is necessary for the repression of nil gene expression and is known to control pleiotropic phenotypes in X. nematophila (7). Therefore, variation in Lrp activity or levels in subpopulations of the ΔcpxR1 mutant could cause variations in nil expression. In E. coli, Lrp and CpxR are known to compete for binding sites upstream of the operon encoding Pap pili (22). Binding of phosphorylated CpxR prevents the Lrp-mediated transition between phase OFF and phase ON states of pap gene expression (22). Although no CpxR consensus binding sites were identified upstream of the nil genes in X. nematophila (21), a similar regulatory mechanism acting directly or indirectly to influence nil gene expression may occur in X. nematophila. Consistent with this hypothesis, variant colonies with elevated levels of nil gene expression were not detected in the wild-type background, raising the possibility that the absence of cpxR increases the appearance of such variants.
X. nematophila was recently reported to undergo virulence modulation, in which a subpopulation of wild-type cells exhibited attenuated virulence and immune suppression in M. sexta insects but wild-type colonization levels in nematodes (40). Although the mechanisms controlling virulence modulation are not known, it is possible that nil gene expression is affected by this phenomenon. To address this possibility, it will be of interest to assess the colonization and virulence phenotypes of individual ΔcpxR1 clones exhibiting elevated nil gene expression and, conversely, to monitor nil gene expression in virulence-modulated strains.
Acknowledgments
This research was supported by an Investigators in Pathogenesis of Infectious Disease Award from the Burroughs-Wellcome Foundation and by National Institutes of Health (NIH) grant GM59776. E.E.H.T. was supported by an NIH National Research Service Award (T32 AI007414).
We thank Nirupama Banerjee for the gift of plasmid pPK13 and Elizabeth Hussa for experimental support.
Footnotes
Published ahead of print on 17 April 2009.
REFERENCES
- 1.Akhurst, R. J. 1983. Neoaplectana species: specificity of association with bacteria of the genus Xenorhabdus. Exp. Parasitol. 55:258-263. [DOI] [PubMed] [Google Scholar]
- 2.Bird, A. F., R. J. Akhurst. 1983. The nature of the intestinal vesicle in nematodes of the family Steinernematidae. Int. J. Parasitol. 13:599-606. [Google Scholar]
- 3.Chandra, H., P. Khandelwal, A. Khattri, and N. Banerjee. 2008. Type 1 fimbriae of insecticidal bacterium Xenorhabdus nematophila is necessary for growth and colonization of its symbiotic host nematode Steinernema carpocapsae. Environ. Microbiol. 10:1285-1295. [DOI] [PubMed] [Google Scholar]
- 4.Cowles, C. E., and H. Goodrich-Blair. 2004. Characterization of a lipoprotein, NilC, required by Xenorhabdus nematophila for mutualism with its nematode host. Mol. Microbiol. 54:464-477. [DOI] [PubMed] [Google Scholar]
- 5.Cowles, C. E., and H. Goodrich-Blair. 2006. nilR is necessary for co-ordinate repression of Xenorhabdus nematophila mutualism genes. Mol. Microbiol. 62:760-771. [DOI] [PubMed] [Google Scholar]
- 6.Cowles, C. E., and H. Goodrich-Blair. 2008. The Xenorhabdus nematophila nilABC genes confer the ability of Xenorhabdus spp. to colonize Steinernema carpocapsae nematodes. J. Bacteriol. 190:4121-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowles, K. N., C. E. Cowles, G. R. Richards, E. C. Martens, and H. Goodrich-Blair. 2007. The global regulator Lrp contributes to mutualism, pathogenesis and phenotypic variation in the bacterium Xenorhabdus nematophila. Cell. Microbiol. 9:1311-1323. [DOI] [PubMed] [Google Scholar]
- 8.Danese, P. N., and T. J. Silhavy. 1997. The sigma(E) and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 11:1183-1193. [DOI] [PubMed] [Google Scholar]
- 9.Dartigalongue, C., and S. Raina. 1998. A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J. 17:3968-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Wulf, P., O. Kwon, and E. C. Lin. 1999. The CpxRA signal transduction system of Escherichia coli: growth-related autoactivation and control of unanticipated target operons. J. Bacteriol. 181:6772-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiGiuseppe, P. A., and T. J. Silhavy. 2003. Signal detection and target gene induction by the CpxRA two-component system. J. Bacteriol. 185:2432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorel, C., O. Vidal, C. Prigent-Combaret, I. Vallet, and P. Lejeune. 1999. Involvement of the Cpx signal transduction pathway of Escherichia coli in biofilm formation. FEMS Microbiol. Lett. 178:169-175. [DOI] [PubMed] [Google Scholar]
- 13.Flores-Lara, Y., D. Renneckar, S. Forst, H. Goodrich-Blair, and P. Stock. 2007. Influence of nematode age and culture conditions on morphological and physiological parameters in the bacterial vesicle of Steinernema carpocapsae (Nematoda: Steinernematidae). J. Invertebr. Pathol. 95:110-118. [DOI] [PubMed] [Google Scholar]
- 14.Forst, S., and D. Clarke. 2002. Bacteria-nematode symbioses, p. 57-77. In R. Gaugler (ed.), Entomopathogenic nematology. CABI Publishing, Wallingford, United Kingdom.
- 15.Forst, S., B. Dowds, N. Boemare, and E. Stackebrandt. 1997. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu. Rev. Microbiol. 51:47-72. [DOI] [PubMed] [Google Scholar]
- 16.Forst, S., and K. Nealson. 1996. Molecular biology of the symbiotic-pathogenic bacteria Xenorhabdus spp. and Photorhabdus spp. Microbiol. Rev. 60:21-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furusawa, T., R. Rakwal, H. W. Nam, M. Hirano, J. Shibato, Y. S. Kim, Y. Ogawa, Y. Yoshida, K. J. Kramer, Y. Kouzuma, G. K. Agrawal, and M. Yonekura. 2008. Systematic investigation of the hemolymph proteome of Manduca sexta at the fifth instar larvae stage using one- and two-dimensional proteomics platforms. J. Proteome Res. 7:938-959. [DOI] [PubMed] [Google Scholar]
- 18.Georgis, R., A. M. Koppenhöfer, L. A. Lacey, G. Bélair, L. W. Duncan, P. S. Grewal, M. Samish, L. Tan, P. Torr, and R. W. H. M. van Tol. 2006. Successes and failures in the use of parasitic nematodes for pest control. Biol. Control 38:103-123. [Google Scholar]
- 19.Goodrich-Blair, H. 2007. They've got a ticket to ride: Xenorhabdus nematophila-Steinernema carpocapsae symbiosis. Curr. Opin. Microbiol. 10:225-230. [DOI] [PubMed] [Google Scholar]
- 20.Haunerland, N. H. 1996. Insect storage proteins: gene families and receptors. Insect Biochem. Mol. Biol. 26:755-765. [DOI] [PubMed] [Google Scholar]
- 21.Herbert, E. E., K. N. Cowles, and H. Goodrich-Blair. 2007. CpxRA regulates mutualism and pathogenesis in Xenorhabdus nematophila. Appl. Environ. Microbiol. 73:7826-7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernday, A. D., B. A. Braaten, G. Broitman-Maduro, P. Engelberts, and D. A. Low. 2004. Regulation of the pap epigenetic switch by CpxAR: phosphorylated CpxR inhibits transition to the phase ON state by competition with Lrp. Mol. Cell 16:537-547. [DOI] [PubMed] [Google Scholar]
- 23.Heungens, K., C. E. Cowles, and H. Goodrich-Blair. 2002. Identification of Xenorhabdus nematophila genes required for mutualistic colonization of Steinernema carpocapsae nematodes. Mol. Microbiol. 45:1337-1353. [DOI] [PubMed] [Google Scholar]
- 24.Humphreys, S., G. Rowley, A. Stevenson, M. F. Anjum, M. J. Woodward, S. Gilbert, J. Kormanec, and M. Roberts. 2004. Role of the two-component regulator CpxAR in the virulence of Salmonella enterica serotype Typhimurium. Infect. Immun. 72:4654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hung, D. L., T. L. Raivio, C. H. Jones, T. J. Silhavy, and S. J. Hultgren. 2001. Cpx signaling pathway monitors biogenesis and affects assembly and expression of P pili. EMBO J. 20:1508-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jonson, A. B., S. Normark, and M. Rhen. 2005. Fimbriae, pili, flagella, and bacterial virulence. Contrib. Microbiol. 12:67-89. [DOI] [PubMed] [Google Scholar]
- 27.Jubelin, G., A. Vianney, C. Beloin, J. M. Ghigo, J. C. Lazzaroni, P. Lejeune, and C. Dorel. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 187:2038-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khandelwal, P., and N. Banerjee-Bhatnagar. 2003. Insecticidal activity associated with the outer membrane vesicles of Xenorhabdus nematophilus. Appl. Environ. Microbiol. 69:2032-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khandelwal, P., R. Bhatnagar, D. Choudhury, and N. Banerjee. 2004. Characterization of a cytotoxic pilin subunit of Xenorhabdus nematophila. Biochem. Biophys. Res. Commun. 314:943-949. [DOI] [PubMed] [Google Scholar]
- 30.Khandelwal, P., D. Choudhury, A. Birah, M. K. Reddy, G. P. Gupta, and N. Banerjee. 2004. Insecticidal pilin subunit from the insect pathogen Xenorhabdus nematophila. J. Bacteriol. 186:6465-6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leclerc, G. J., C. Tartera, and E. S. Metcalf. 1998. Environmental regulation of Salmonella typhi invasion-defective mutants. Infect. Immun. 66:682-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martens, E. C., and H. Goodrich-Blair. 2005. The Steinernema carpocapsae intestinal vesicle contains a subcellular structure with which Xenorhabdus nematophila associates during colonization initiation. Cell. Microbiol. 7:1723-1735. [DOI] [PubMed] [Google Scholar]
- 33.Martens, E. C., K. Heungens, and H. Goodrich-Blair. 2003. Early colonization events in the mutualistic association between Steinernema carpocapsae nematodes and Xenorhabdus nematophila bacteria. J. Bacteriol. 185:3147-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martens, E. C., F. M. Russell, and H. Goodrich-Blair. 2005. Analysis of Xenorhabdus nematophila metabolic mutants yields insight into stages of Steinernema carpocapsae nematode intestinal colonization. Mol. Microbiol. 58:28-45. [DOI] [PubMed] [Google Scholar]
- 35.Mitobe, J., E. Arakawa, and H. Watanabe. 2005. A sensor of the two-component system CpxA affects expression of the type III secretion system through posttranscriptional processing of InvE. J. Bacteriol. 187:107-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakayama, S., A. Kushiro, T. Asahara, R. Tanaka, L. Hu, D. J. Kopecko, and H. Watanabe. 2003. Activation of hilA expression at low pH requires the signal sensor CpxA, but not the cognate response regulator CpxR, in Salmonella enterica serovar Typhimurium. Microbiology 149:2809-2817. [DOI] [PubMed] [Google Scholar]
- 37.Nakayama, S., and H. Watanabe. 1995. Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J. Bacteriol. 177:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orchard, S. S., and H. Goodrich-Blair. 2004. Identification and functional characterization of a Xenorhabdus nematophila oligopeptide permease. Appl. Environ. Microbiol. 70:5621-5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otto, K., and T. J. Silhavy. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA 99:2287-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, Y., E. E. Herbert, C. E. Cowles, K. N. Cowles, M. L. Menard, S. S. Orchard, and H. Goodrich-Blair. 2007. Clonal variation in Xenorhabdus nematophila virulence and suppression of Manduca sexta immunity. Cell. Microbiol. 9:645-656. [DOI] [PubMed] [Google Scholar]
- 41.Phalaraksh, C., E. M. Lenz, J. C. Lindon, J. K. Nicholson, R. D. Farrant, S. E. Reynolds, I. D. Wilson, D. Osborn, and J. M. Weeks. 1999. NMR spectroscopic studies on the haemolymph of the tobacco hornworm, Manduca sexta: assignment of 1H and 13C NMR spectra. Insect Biochem. Mol. Biol. 29:795-805. [Google Scholar]
- 42.Pogliano, J., A. S. Lynch, D. Belin, E. C. Lin, and J. Beckwith. 1997. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 11:1169-1182. [DOI] [PubMed] [Google Scholar]
- 43.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 44.Raivio, T. L., and T. J. Silhavy. 1997. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J. Bacteriol. 179:7724-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Sauer, F. G., M. A. Mulvey, J. D. Schilling, J. J. Martinez, and S. J. Hultgren. 2000. Bacterial pili: molecular mechanisms of pathogenesis. Curr. Opin. Microbiol. 3:65-72. [DOI] [PubMed] [Google Scholar]
- 47.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 48.Snyder, H., S. P. Stock, S. K. Kim, Y. Flores-Lara, and S. Forst. 2007. New insights into the colonization and release processes of Xenorhabdus nematophila and the morphology and ultrastructure of the bacterial receptacle of its nematode host, Steinernema carpocapsae. Appl. Environ. Microbiol. 73:5338-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vivas, E. I., and H. Goodrich-Blair. 2001. Xenorhabdus nematophilus as a model for host-bacterium interactions: rpoS is necessary for mutualism with nematodes. J. Bacteriol. 183:4687-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto, K., and A. Ishihama. 2006. Characterization of copper-inducible promoters regulated by CpxA/CpxR in Escherichia coli. Biosci. Biotechnol. Biochem. 70:1688-1695. [DOI] [PubMed] [Google Scholar]