Abstract
Summary: Biofilms are communities of microorganisms that live attached to surfaces. Biofilm formation has received much attention in the last decade, as it has become clear that virtually all types of bacteria can form biofilms and that this may be the preferred mode of bacterial existence in nature. Our current understanding of biofilm formation is based on numerous studies of myriad bacterial species. Here, we review a portion of this large body of work including the environmental signals and signaling pathways that regulate biofilm formation, the components of the biofilm matrix, and the mechanisms and regulation of biofilm dispersal.
INTRODUCTION
What is the definition of biofilm? This question has been debated in frequent, lengthy, and sometimes heated discussions. Yet, a consensus has been elusive. For the purposes of this review, we will dissect the word and then unite the parts to create the definition. A “film” is a thin coating. “Bio” refers to the living nature of this film. In other words, a biofilm is a thin coating comprised of living material. In this review, we will focus on bacterial biofilms and, in particular, on gram-negative biofilms, which have been intensively studied. However, under many topics we also include examples of gram-positive organisms (For reviews of biofilm formation by gram-positive organisms, see references 191, 242, and 285).
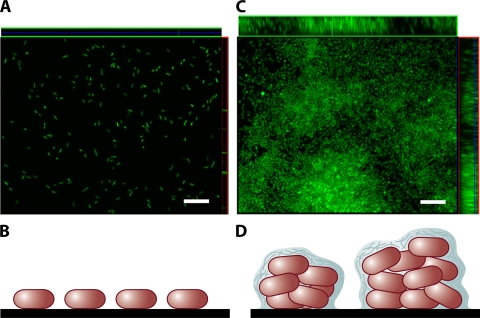
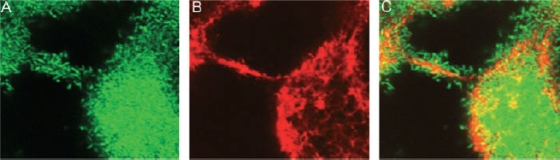
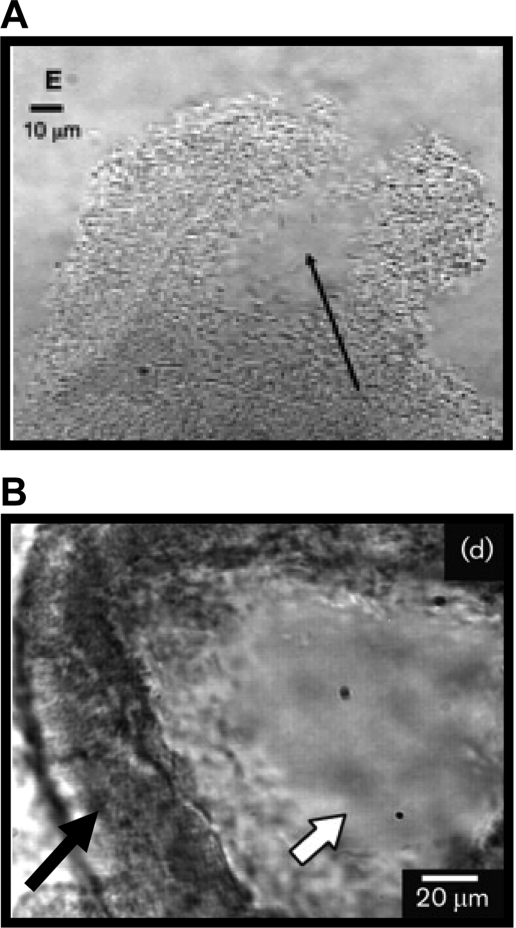
Biofilms can form on environmental abiotic surfaces such as minerals, the carapaces of dead organisms, or air-water interfaces. They can also form on biotic surfaces in the natural environment, such as plants, other microbes, and animals. In the human body, bacteria are present in biofilms in essentially every niche that they colonize. These include both pathogenic and nonpathogenic skin flora, pathogenic and nonpathogenic oropharyngeal and nose flora, commensal and pathogenic intestinal flora, and bacteria adherent to endovascular structures such as native and prosthetic heart valves, central venous catheters, and endovascular thromboses. In each of these environments, the bacteria are guided to or away from the biofilm by environmental signals. Once at the surface, the bacteria may attach either as single cells or as clusters of cells. If single cells form attachments to the surface, a monolayer biofilm is formed (Fig. 1A and B). We define a monolayer biofilm as one in which the bacterium is attached only to the surface. If the bacteria attach as clusters of cells or if a monolayer biofilm remodels to form clusters, a multilayer biofilm is formed (Fig. 1C to D). We define a multilayer biofilm as one in which the bacterium is attached both to the surface and to neighboring bacteria. The multilayer biofilm often forms in tandem with an extracellular matrix that may include exopolysaccharides, proteins, and DNA.
FIG. 1.
Monolayer and multilayer biofilms. (A and C) Transverse and vertical cross-sections through monolayer (A) and multilayer (C) biofilms of V. cholerae O139. (B and D) Schematic representations of side views of the monolayer (B) and multilayer (D) biofilms. In the monolayer, bacteria are distributed on the surface as a single layer. In the multilayer biofilm, pillars composed of multiple layers of bacteria encased in an extracellular matrix form. Biofilms were grown for 24 h in minimal medium without glucose and in LB broth for monolayer and multilayer biofilms, respectively, as described in reference 232. Biofilms were stained with Syto 9 and visualized using confocal scanning laser microscopy with an LSM 510 META confocal scanning system. Bars, ∼10 μm.
Biofilms are characterized by the environmental conditions and surfaces that favor their formation, the gene products that are required for their formation, the genes that are activated and required to maintain the biofilm, the architecture of the biofilm, and the types of extracellular products that are concentrated in the biofilm matrix. There are as many different types of biofilms as there are bacteria, and even one bacterium may make several different types of biofilms under different environmental conditions. Here we review the diverse array of environmental signals, gene products, extracellular matrices, and architectures, as well as dispersal mechanisms that have been uncovered as the biofilms of many different bacterial species have been defined. We focus mostly on research involving single-species biofilms studied under laboratory conditions. For biofilm studies of industrial and medical systems, the reader is referred to other reviews (21, 37, 42, 252, 334).
THE MONOLAYER BIOFILM
The monolayer biofilm is defined as a single layer of surface-adherent cells. This type of structure is favored when cell-surface interactions rather than cell-cell interactions predominate. Much attention has been given to the multilayer biofilm. However, because it affords every attached bacterium proximity to the surface, the monolayer biofilm may actually be the more pervasive surface-attached state in both the natural environment and the interaction of the bacterial pathogen with its host.
For bacteria with flagellar motility, formation of the monolayer biofilm is easily observed over time and has been described (232). For these bacteria, monolayer formation occurs in two steps. Bacteria that approach the surface closely become tethered to the surface. Most bacteria break the forces tethering them to the surface shortly after they are formed. This process is known as transient attachment. In a process that appears to be stochastic, a few bacteria remain attached to the surface for extended periods of time. In this case, the bacteria are said to have undergone the transition from transient to permanent attachment. We hypothesize that the bias toward permanent attachment is modulated by environmental signals, but to date, no such environmental signal has been elucidated. Recent evidence suggests, however, that changes in the membrane potential (ΔΨ) may alter the bias toward permanent attachment (328). In the sections below, we will outline what is known about the adhesive structures that mediate transient and permanent surface attachment, the transition to permanent attachment, and the monolayer transcriptome.
Types of Adhesive Structures Used To Form the Monolayer Biofilm
To date, three classes of adhesive structures have been defined in the formation of the monolayer biofilm. In the first class are preformed structures that increase transient attachments with the surface and thus accelerate formation of the monolayer biofilm. The synthesis of structures in the second class is coordinated with the transition to permanent attachment. The last class requires synthesis of specific adhesins and therefore may allow surface-specific adhesion.
Class 1: preformed adhesins. (i) The flagellum.
The phenotypes of aflagellate and paralyzed nonmotile bacterial mutants are difficult to reconcile. Bacterial mutants having a paralyzed flagellum are often completely defective for attachment, while aflagellate bacteria are able to progress through monolayer formation to the multilayer biofilm stage (187, 346). These apparently contradictory phenotypes can be reconciled by invoking a dual role for the flagellar structure. Motility itself is thought to enhance the initial interaction of the bacterium with the surface by enabling the bacterium to overcome long-range repulsive forces, thus increasing the likelihood of close approach (86). In fact, flagellar motility has been demonstrated to accelerate surface adhesion for many bacteria (169, 173, 192, 217, 224, 325, 344). However, under certain growth conditions, mutation of components of the flagellar structure leads to increased synthesis of the adhesive matrix that promotes interbacterial attachments and formation of a multilayer biofilm. Under these conditions, flagellar mutants are not deficient for surface attachment but rather are observed to form an increased multilayer biofilm (99, 187, 346). These observations suggest that later in the progression to multilayer biofilm development, sensing of flagellar arrest plays a role in priming the bacterium for formation of the multilayer biofilm.
Lastly, in Vibrio cholerae, the flagellar motor appears to play an essential role in monolayer formation that is independent of that played by either flagellar motility or the rotary portion of the flagellum. Mutants that are unable to synthesize a complete flagellum remain competent for both monolayer and multilayer biofilm formation. In contrast, a flagellar motor mutant is completely defective in formation of both monolayer and multilayer biofilms. Furthermore, in mutants lacking both the flagellum and the flagellar motor, the phenotype of the flagellar motor mutant is dominant (187, 328). This suggests that the flagellar motor plays a role in biofilm formation that is independent of that played either by flagellar motility or by the flagellar rotor. However, the mechanism underlying these observations has not yet been elucidated.
(ii) Pili.
Retractable pili are a common requirement for attachment of gram-negative bacteria to surfaces (20, 30, 80, 158, 220, 244, 254). Pili are long appendages found at the poles of some bacterial cells. Although not all types of pili have been demonstrated to be retractable, many types of pili are able to retract against great force (209, 225, 294). Thus, these structures are believed to pull bacteria either onto or along surfaces by attaching to the surface and retracting. As is hypothesized for the flagellum, therefore, these structures can also help the bacteria move through long-range repulsive forces to approach the surface more closely.
Class 2: conditionally synthesized adhesins.
In many bacteria, transient attachment is mediated by a retractable pilus. However, this attachment may be disrupted. In some bacteria, factors have been identified that stabilize this attachment, thus resulting in permanent attachment. It is likely that many more such factors remain to be identified. The transition of Pseudomonas fluorescens from transient to permanent attachment is mediated by LapA, a large secreted protein that associates with the surface of bacterial cells (134). Because secretion of LapA is inhibited by RapA, a phosphodiesterase that degrades the second messenger cyclic diguanylate monophosphate (c-di-GMP), it was hypothesized that c-di-GMP enables secretion of LapA (230). SadB, a protein that coordinates biofilm formation and swarming motility by an unknown mechanism, has also been implicated in the transition from transient to permanent attachment in Pseudomonas aeruginosa (43, 44). In Escherichia coli, the exopolysaccharide adhesin PGA has been postulated to mediate the transition from transient to permanent attachment (2).
The transition from transient to permanent attachment has perhaps been best defined for Caulobacter crescentus (13, 57, 110). In C. crescentus, a complex developmental program is associated with formation of the monolayer. The flagellum of cells destined for attachment is removed by a protease (3, 157). In its place, a protrusion called a holdfast composed of oligomers of N-acetylglucosamine appears (150, 296). The holdfast has a strong adhesive polysaccharide that ensures tight adhesion to the surface (197, 326). Monolayer formation is synchronized with cell division, and monolayer-associated cells give rise only to motile cells, known as swimmers, which move on to colonize new surfaces. Aspects of C. crescentus monolayer formation which have been most intensively studied, such as regulation of flagellar loss, timing of cell division, and differentiation into swimmer cells, may prove to be paradigms for the as-yet-unstudied monolayers formed by other bacteria.
Class 3: specific adhesins.
The attachment of bacterial pathogens to mammalian cells is a variation on the theme of monolayer formation. After transient attachment, which may involve the usual array of flagella and pili, these pathogens form specific, stable attachments to eukaryotic cells by adhering to cell surface receptors (123). In this case, the monolayer biofilm is a prelude to internalization rather than formation of a multilayer biofilm.
For instance, the enteric pathogens Yersinia pseudotuberculosis and Yersinia enterocolitica produce a bacterial cell surface protein known as invasin, which adheres to β1 integrin, a glycoprotein that is found on the surface of specialized intestinal epithelial cells known as M cells. The interaction of invasin with β1 integrin triggers internalization of Yersinia into M cells, providing an entry point to the underlying lymphoid tissue of Peyer's patches, where Yersinia can proliferate prior to dissemination (139-142). Other examples of specific adhesion of bacteria to mammalian cell surface proteins include the interaction of Listeria internalin with mammalian E-cadherin (188, 221) and the interaction of Neisseria meningitidis and Neisseria gonorrhoeae with carcinoembryonic antigen-related cell adhesion molecule, an immunoglobulin superfamily cell adhesion molecule (27, 45, 236).
A particularly fascinating example of a bacterium exploiting specific adhesion to form a monolayer on a cell surface is presented by the enterohemorrhagic and enteropathogenic strains of E. coli (46, 92). Finding no adequate preexisting receptor on the surface of mammalian cells, these bacteria use a type III secretion system (TTSS) to transfer their own, bacterially derived receptor into mammalian cells (72, 73, 167). Because it binds to the intimin protein on the bacterial cell surface, this receptor is termed Tir, for translocated intimin receptor. The interaction of Tir with intimin leads to the formation of an actin pedestal beneath the attached bacterium and activation of signaling cascades within the cell (36, 78, 91, 115, 178).
We propose, therefore, that the specific adhesion of bacterial cells to cells within their mammalian hosts is a special case of monolayer formation. Furthermore, it is likely that some of the principles of monolayer formation that have been elucidated under laboratory conditions will also apply to colonization of host tissues.
Transcriptional Program of the Monolayer Biofilm
There is much to be gained from understanding gene transcription and expression within the monolayer biofilm. Compared with the multilayer biofilm, the monolayer biofilm represents a more homogeneous collection of surface-attached cells. Furthermore, this biofilm affords us the opportunity to study the simple act of surface attachment in the absence of further modulation of the environment by elaboration of a matrix. However, studies of the monolayer biofilm also present additional challenges. Because surface-attached bacterial cells progress through the monolayer state to the multilayer state under most experimental growth conditions, it is difficult to isolate a pure monolayer biofilm in the laboratory. Furthermore, the monolayer biofilm is comprised of many fewer bacterial cells, presenting challenges in amassing enough RNA or protein for whole-genome or proteome studies. For V. cholerae, cultivation of the bacterium in the absence of sugars leads to arrest of surface attachment in the monolayer stage. This has paved the way for transcriptional studies of the monolayer biofilm (232, 233). These studies demonstrate that transcription of flagellar genes is repressed in the monolayer stage. This regulation is independent of activation of genes required for synthesis of the multilayer biofilm matrix. Furthermore, transcription of a large number of methyl-accepting chemotaxis genes is activated in the monolayer. Additional studies suggest that chemotaxis proteins influence monolayer formation. One possibility is that flagellar pausing, which plays a role in the response to chemoattractants, also enhances the transition to permanent attachment (183). Studies of the monolayer transcriptome have also allowed identification of genes that are differentially regulated in all studied surface-attached states. Interestingly, the formate-nitrate electron transport pathway is activated to the same degree in both monolayer and multilayer biofilms. This is an anaerobic respiration pathway that utilizes formate, a by-product of pyruvate metabolism, as an electron donor and nitrate as an electron acceptor. Because the cell densities are low, the monolayers are formed in the presence of agitation, and the air-fluid interface is within 5 mm of the bottom of the well, oxygen limitation is quite unlikely under the conditions of these monolayer experiments. Therefore, one possibility is that activation of genes involved in anaerobic respiration is a predetermined component of the transcriptional program of surface attachment rather than a response to the availability of oxygen in the environment.
Another interesting aspect of transcriptional control in the surface-attached state of V. cholerae is the repression of cholera toxin and a subset of its regulators. One possibility is that this regulation represents repression of virulence factors on a perceived “nonhost” surface. Another possibility is that V. cholerae inversely regulates surface attachment and cholera toxin because the expression of cholera toxin and the resulting massive diarrhea is likely to destabilize attachment of the bacterium to the intestinal epithelium.
Our understanding of transcription in the bacterial monolayer biofilm is limited by the paucity of studies both of V. cholerae and of other bacteria. General conclusions about the role and function of the monolayer biofilm must await additional studies of monolayer biofilms formed in other environments, on other surfaces, and by other bacteria.
THE MULTILAYER BIOFILM
Multilayer bacterial biofilms may form on the internal or external surfaces of another organism, an abiotic environmental surface, or an air-water interface. In fact, even suspended aggregates of cells display many of the characteristics that are associated with biofilms. A multilayer biofilm develops when bacteria are able to adhere to a surface and also to each other. Intercellular adhesions require an outer adhesive bacterial surface. In many environments, the surface characteristics of bacteria lead to repulsion. For instance, the chemical properties of the surfaces of gram-negative bacteria are generally determined by the O antigen, which is usually negatively charged. This negative charge may be neutralized by mutation of the O-antigen synthesis genes, addition of divalent cations to the medium, or synthesis of an adhesive matrix. The last strategy is, perhaps, the most easily regulated by the bacterium and the best studied by biofilm scientists. Components of the adhesive matrices synthesized by bacteria may include exopolysaccharide, protein, and DNA.
In this section, we will provide an overview of what is known about the environmental signals and regulatory networks that modulate formation of the biofilm matrix as well as the composition of the biofilm matrices and their role in biofilm structure and function.
Regulation
Signals.
The propensity to form a biofilm is guided by numerous environmental signals, some of which have been identified and many of which remain unstudied. Below we discuss a few of these signals that have been more extensively studied and are common to diverse bacteria.
(i) Mechanical signals.
Bacteria approaching a surface make a choice between the sessile and free-living lifestyle. This suggests that the surface itself must be sensed in order for biofilm formation to occur. Although definitive evidence has not been forthcoming, a variety of studies suggest that the flagellum may be the operative structure in surface sensing by motile bacteria. Transcriptional profiling studies of a variety of bacteria suggest that flagellar gene expression and biofilm matrix synthesis are inversely regulated (see, e.g., references 99, 232, 233, and 256). In some organisms the molecular mechanisms that underlie this inverse regulation has been elucidated. For example, in P. aeruginosa, the alternative sigma factor AlgT, which is a positive regulator of biofilm matrix synthesis, indirectly inhibits flagellar gene expression. AlgT promotes expression of the transcriptional regulator AmrZ, which then directly represses expression of FleQ, the master regulator of flagellar gene expression in this organism, thereby leading to loss of flagellar biosynthesis (316). Moreover, increases in both the synthesis of the biofilm matrix and the transcription of genes involved in the synthesis of the biofilm matrix are commonly observed in mutants lacking the flagellar structure, thus confirming the inverse relationship between motility and synthesis of the biofilm matrix. For example, for some strains of V. cholerae, mutants that lack a complete flagellar filament demonstrate exuberant synthesis of the biofilm matrix even in the absence of a surface. In contrast, flagellar motor mutants do not synthesize a biofilm matrix even in the presence of appropriate environmental signals (187, 346). One possibility is that the bacterium senses increased drag on the flagellar motor caused by its interaction with the surface. When the motor is not present, this signal is not transduced. While mutation of the flagellar structure bypasses the requirement for a surface in multilayer biofilm formation, the requirement for appropriate nutritional signals is not bypassed. This suggests that surface sensing and subsequent flagellar arrest is one of many checkpoints in the path toward multilayer biofilm formation.
An interesting twist in the inverse relationship between motility and surface attachment occurs in Bacillus subtilis. In this organism, EpsE acts as a molecular clutch that disengages the flagellum from its power source thereby immobilizing it (29). EpsE does this by binding flagellar switch protein FliG and presumably inhibiting this protein from interacting with the flagellar motor proteins. Interestingly, the epsE gene resides in an operon encoding biofilm matrix components. Thus, synthesis of the matrix components and flagellar immobilization are synchronized via the action of EpsE, thereby stabilizing the biofilm. If the conditions become unfavorable in the biofilm, the flagellar brake can be released, a process that is likely quicker and more energy efficient than de novo flagellar synthesis. It is proposed that this posttranslational mechanism would allow a quick and reversible transition between motile and sessile lifestyles.
(ii) Nutritional and metabolic cues.
Bacteria monitor and respond to the types and amounts of nutrients in their environment. Perhaps because of the energetic costs of joining and exiting the multilayer biofilm, the nutritional status of the environment has a great impact on the propensity of a bacterium to form a multilayer biofilm. Some bacteria, such as Salmonella enterica serovar Typhimurium, join a multilayer biofilm in response to nutrient limitation (104). In these organisms, the stationary-phase sigma factor, RpoS, participates in activation of many of the genes required for biofilm formation (103). In other bacteria, such as V. cholerae, nutrient-rich environments promote biofilm formation. In these organisms, RpoS participates in repression of genes required for biofilm formation (357). Therefore, we suggest that biofilm formation fulfills different needs depending on the environment which a bacterium inhabits. A number of the nutritional signals that affect biofilm formation are considered below.
(a) Glucose and catabolite repression. Glucose is a scarce and valuable commodity for many organisms living on Earth. For some bacteria, glucose and related sugars activate multilayer biofilm formation, while for others they serve as inhibitors of this type of surface attachment. Bacteria in the former group include Streptococcus mutans, Staphylococcus aureus, and Staphylococcus epidermidis (75, 203, 284, 286).
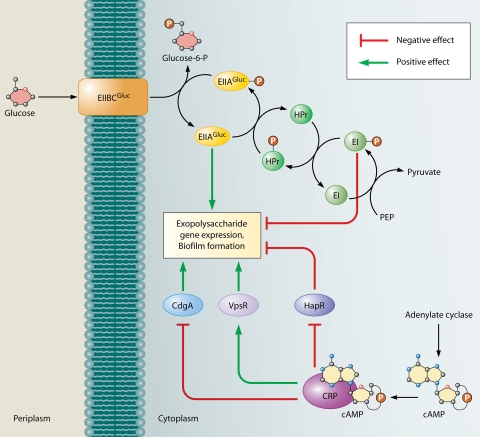
Glucose and other sugars are strong inducers of the V. cholerae biofilm matrix and multilayer biofilm formation (168). The sugars that induce synthesis of the multilayer biofilm matrix have in common their transport through the phosphoenoylpyruvate phosphotransferase system (PTS) (136). The PTS consists of a multiprotein phosphotransfer cascade that transfers a phosphate moiety from phosphoenoylpyruvate to incoming transported sugars (Fig. 2). General cytoplasmic proteins involved in this phosphotransfer cascade include enzyme I (EI) and histidine protein (Hpr). The final component of the cascade is a multisubunit, sugar-specific transport apparatus that consists of a cytoplasmic protein termed EIIA and a membrane-associated component termed EIIB, EIIC, or sometimes EIID. Because the level of phosphorylation of PTS components serves as a measure of the store of high-energy phosphate reserves within the cell and the level of favorable carbon sources in the environment, in E. coli, EI, Hpr, and EII regulate many functions within the cell, such as chemotaxis, glycogen synthesis, catabolite repression, and inducer exclusion (71). Recently, evidence has emerged that the PTS also regulates formation of the multilayer biofilm matrix in V. cholerae (Fig. 2) (136). When V. cholerae is grown in the presence of a PTS substrate, the PTS phosphotransfer cascade is depleted of phosphate due to transfer of phosphate to the incoming sugar. This leads to activation of exopolysaccharide gene transcription and biofilm formation. The PTS component most likely to be responsible for this regulation is EIIAGluc. While it is certain that this effect is not the result of catabolite repression, as catabolite repression leads to diminished biofilm formation, the complete signal transduction cascade responsible for this effect has not yet been fully delineated (89, 136). Under growth conditions in which a PTS substrate is not present or has been fully consumed, components of the PTS are fully phosphorylated. This leads to repression of the exopolysaccharide genes and decreased biofilm formation. Under growth conditions in which a PTS substrate is present, this is manifested as the entry of biofilm-associated cells into stationary phase. The entry of planktonic cells into stationary phase in such cultures does not show a similar dependence on the PTS. Deletion of the EI component of the PTS blocks this repression, leading to large increases in exopolysaccharide gene transcription and biofilm formation. In this case, we know that regulation by a PTS component rather than the act of transport is responsible for this phenomenon, because (i) the EI mutant biofilm phenotype can be rescued in a genetic background where sugar transport is not possible and (ii) supplementation of the growth medium with glucose-6-phosphate, which is not transported by the PTS, as the sole carbon source does not rescue the biofilm phenotype of an EI mutant. This supports the claim that for V. cholerae the nutritional status of the cell is an important consideration in the decision to form a multilayer biofilm.
FIG. 2.
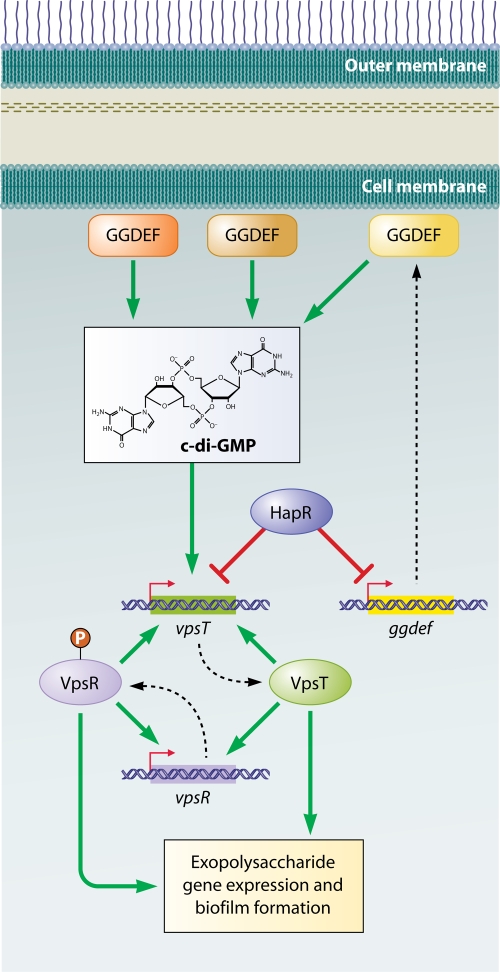
Effect of glucose transport and catabolite repression on V. cholerae biofilm formation. The PTS for glucose regulates biofilm formation in V. cholerae. Unphosphorylated EIIAGluc, which signals the presence of glucose in the environment, leads to activation of biofilm formation, whereas, phosphorylated EI, which signals the absence of glucose in the environment, leads to its repression. The cAMP-CRP complex has been shown to both activate and repress biofilm formation. The activation occurs as a result of negative regulation of the biofilm repressor HapR and positive regulation of the biofilm activator VpsR. The repression is due to negative regulation of the DGC CdgA, an activator of biofilm formation.
Unphosphorylated EIIA has been demonstrated to block transport of non-PTS sugars by direct interference with transport in a phenomenon known as inducer exclusion. In addition, in enteric bacteria, EIIA-P mediates catabolite repression or preferential utilization of glucose as a carbon source by enhancing the enzymatic activity of adenylate cyclase. When glucose is plentiful, uptake and utilization of alternative carbon sources are repressed. When glucose is scarce, high levels of EIIA-P activate adenylate cyclase, resulting in high levels of intracellular cyclic AMP (cAMP). cAMP interacts with the cAMP receptor protein (CRP) to relieve repression of genes controlling utilization of alternative carbon sources. In V. cholerae, EIIAGluc activates biofilm formation (136). Similarly, the mannose-specific PTS of S. mutans activates biofilm formation, as mutants lacking EIIABMan (a portion of the membrane-bound permease complex) have significantly impaired biofilm-forming capability (1).
Catabolite repression plays an important role in regulation of multilayer biofilm formation in many bacteria. In V. cholerae, the effect of catabolite repression on multilayer biofilm formation is complex (Fig. 2). In some V. cholerae studies, supplementation with cAMP was found to inhibit exopolysaccharide synthesis and multilayer biofilm formation, suggesting that catabolite repression decreases biofilm formation (136). Recent evidence has demonstrated that this is the result of repression of the diguanylate cyclase (DGC) CdgA by the cAMP-CRP complex (89). In contrast, in other studies, the cAMP-CRP complex was found to activate expression of the biofilm activator VpsR and the biofilm repressors HapR and CytR and also to repress expression of the biofilm activator VpsT (200). The net result was activation of exopolysaccharide synthesis by the cAMP-CRP complex. Taken together, these studies suggest that the effect of cAMP and catabolite repression on biofilm formation is likely to be the sum of multiple signal transduction cascades. Furthermore, the contributions of these various cascades may differ in different V. cholerae strains and under different environmental growth conditions, leading to different phenotypes for the cya (adenylate cyclase) and crp mutants in different studies.
Environmental glucose and catabolite repression inhibit multilayer biofilm formation in a variety of pathogenic and laboratory strains of E. coli, a number of clinical isolates of Enterobacteriaceae, and B. subtilis. In E. coli, the repressive effect of glucose is exerted through catabolite repression via the cAMP-CRP system (147). B. subtilis biofilm formation is activated when glucose is present in low concentrations but inhibited when glucose is present in high concentrations (302). When B. subtilis is grown in medium containing 0.1% glucose, a multilayer biofilm forms (302). The stimulatory effect of low glucose concentrations is, in part, due to the metabolism of glucose to acetoin, which stimulates Spo0A, a positive regulator of biofilm formation (260, 302). Other pathways for activation of multilayer biofilm formation by glucose may also exist. Catabolite repression of B. subtilis multilayer biofilm formation at high glucose concentrations is dependent on the catabolite control protein A (CcpA), a transcriptional regulator. When B. subtilis is grown in medium containing 1% glucose, deletion of ccpA leads to increased numbers of cells joining the multilayer biofilm.
In P. aeruginosa, catabolite repression enhances formation of the multilayer biofilm (243). Mutation of the gene encoding the catabolite repression protein, Crc, allows P. aeruginosa to catabolize sugars such as glucose even when tricarboxylic cycle intermediates, the preferred carbon source for this organism, are present in the environment. These mutants are also defective for biofilm formation. They attach to surfaces as single cells but fail to form microcolonies. The failure to form intercellular attachments has been linked to a defect in type IV pilus motility, which is required for multilayer biofilm formation in P. aeruginosa (243, 244).
(b) Indole. The amino acid tryptophan can be hydrolyzed by the enzyme tryptophanase to form indole and pyruvate, which are then used as a source of carbon and nitrogen under nutrient-depleted conditions (238). Indole has a stimulatory effect on biofilm formation in a variety of gram-negative bacteria. A study of the role of tryptophanase and indole in biofilm formation by a number of clinical isolates of E. coli, Klebsiella oxytoca, Providencia stuartii, Citrobacter koseri, Morganella morganii, and Haemophilus influenzae type b showed that the presence of a tryptophanase inhibitor in the culture medium inhibited biofilm formation but had no effect on growth (213). Interestingly, the stimulatory effect of indole on biofilms appears to be reversed by catabolite repression, at least in E. coli, where it has been shown to inhibit biofilms in the presence of glucose (76, 77). Another study showed that transposon insertions in the tryptophanase gene of V. cholerae led to a rugose-to-smooth shift in colony morphology, which was reversed by addition of exogenous indole (235). Because rugose-to-smooth shifts in colony morphology on a solid growth medium are usually accompanied by a decrease in biofilm formation and exopolysaccharide synthesis in broth, these results suggest that indole may also activate V. cholerae biofilm formation. Finally, biofilm formation by pseudomonads, which cannot synthesize indole, is increased when the growth medium is supplemented with indole (189). Thus, indole may be a commonly used intra- and interspecies biofilm signal that allows cells to detect and respond to nutritional depletion in the environment.
(c) Polyamines. Polyamines, such as putrescine, spermidine, and norspermidine, are linear organic molecules containing two or more amine groups that are positively charged at neutral pH (311). They are essential for cell growth, and their intracellular levels are tightly regulated by synthesis, import, export, and interconversion (311). Recently, several reports have suggested that polyamines may function as extracellular and/or metabolic signals that modulate biofilm formation. Norspermidine, a triamine, increases biofilm formation by V. cholerae (164). This effect is dependent on the presence of a periplasmic sensor protein, NspS, as well as the transmembrane protein MbaA, which is hypothesized to associate with NspS. Because NspS is a periplasmic protein, we hypothesize that norspermidine can exert its effect on NspS from the periplasm and therefore function as an extracellular signaling molecule (164). In Yersinia pestis, endogenous putrescine, a diamine, is required for biofilm development (249). Y. pestis mutants that are unable to synthesize putrescine are impaired in biofilm development. This defect can be rescued in a dose-dependent manner by supplementation of the growth medium with putrescine, suggesting that both exogenous and endogenous putrescine can activate biofilm formation (249). Furthermore, spermidine and putrescine transporters have been implicated in surface-associated growth of Agrobacterium tumefaciens and Pseudomonas putida (216, 280). Taken together, these findings suggest that polyamines may be regulators of surface-associated growth and biofilm formation in diverse bacteria.
(iii) Inorganic molecules.
(a) Iron. Iron is an essential and yet scarce nutrient for bacteria. Most of the iron in the environment of a microorganism either resides stably in ferric oxide hydrate complexes or is tightly bound either to specialized extracellular iron carrier proteins or to small molecules known as siderophores (227). Perhaps because iron is a rare commodity, it is also an activator of bacterial biofilm formation. The effect of iron limitation on biofilm formation depends on the bacterium under study. For example, several pieces of evidence suggest that iron limitation has an inhibitory effect on P. aeruginosa biofilm formation. First, subbacteriostatic amounts of the mammalian iron binding protein lactoferrin inhibit P. aeruginosa biofilm formation (292). Second, P. aeruginosa mutants that are unable to scavenge adequate amounts of iron from their environment are defective in biofilm formation (17). This effect is dependent on the ferric uptake repressor Fur, a global repressor of gene transcription in iron-rich environments found in P. aeruginosa and other members of the Proteobacteria (262, 330). Lastly, P. aeruginosa mutants that do not synthesize the siderophore pyoverdin are defective in biofilm formation (250). Similarly, V. cholerae biofilm formation is significantly reduced in iron-deficient medium (226).
In some cases, iron has an inhibitory effect on biofilms. For the oral bacterium Actinomyces naeslundii as well as for S. epidermidis, an opportunistic pathogen which is one of the most common causes of medical-device-related biofilm infections, iron limitation leads to increased biofilm formation (70, 228). In E. coli, CsgD, a positive regulator of biofilm formation, represses transcription of fecR, which encodes a transcriptional activator of genes involved in iron uptake. This suggests that iron uptake and biofilm formation are inversely regulated in this bacterium (41). Finally, even for P. aeruginosa, while some iron is required for biofilm formation, extracellular iron concentrations above 5 μM can lead to inhibition of biofilm formation (353). We propose that the effect of iron availability on biofilm formation by a particular organism reflects the nature of the surfaces available within its habitat.
(b) Phosphate. Levels of inorganic phosphate in a bacterium's environment are also important signals for biofilm formation. In Pseudomonas aurofaciens and P. fluorescens, phosphate limitation inhibits biofilm formation (230, 231). This signal is transduced by the Pho regulon, which is activated under phosphate starvation conditions. In P. fluorescens, this inhibition is a result of the activation of the phosphodiesterase RapA under phosphate limitation, which then decreases the levels of the secondary messenger c-di-GMP. Decreased c-di-GMP levels inhibit secretion of a surface adhesin, LapA, which is required for biofilm formation by this organism (230). Interestingly, phosphate limitation enhances biofilm formation by A. tumefaciens (64). The enhanced biofilm response of this bacterium is also mediated by the Pho two-component system (TCS) that is activated by phosphate limitation. The opposite effects of phosphate depletion on biofilm formation by these bacteria may be a reflection of the different environmental niches in which they live.
(iv) Osmolarity.
Osmolarity regulates biofilm formation in a number of bacterial species. In many cases, osmolarity inhibits biofilm formation although this effect may depend on the type of osmolyte in the environment. For example, P. fluorescens biofilm formation is inhibited in high-osmolarity environments produced by addition of NaCl and/or sucrose (245). In S. Typhimurium, growth in medium containing high concentrations of NaCl abolishes transcription of csgD, a central regulator of biofilm formation and curli production (272). Similarly, when E. coli is cultured in medium containing 100 mM NaCl, transcription of the curli genes is repressed by the transcription factor CpxR (155). In this case, addition of similar concentrations of sucrose does not produce the same effect, suggesting the possibility that the environmental signal is ionic strength rather than osmolarity. Interestingly, 200 mM NaCl activates transcription of the E. coli pga operon, which encodes the proteins required for synthesis of the biofilm-active polymer poly-N-acetylglucosamine (PNAG) (111). Strain-specific differences in regulation may be responsible for the seemingly contradictory effects of high-salt conditions on E. coli biofilms. Alternatively, different matrix components (PNAG versus curli) might be preferred under different environmental conditions. V. cholerae, a halophilic aquatic bacterium, will form a biofilm under high-salt conditions if cells are protected by the compatible solute glycine betaine (159). This response requires the ability to import the compatible solute glycine betaine into the cell. Therefore, the varied effects of osmolarity on bacterial biofilm formation most likely reflect differences in the physiology of these organisms.
(v) Host-derived signals.
Several pathogenic microorganisms respond to host-derived molecules by forming a biofilm. This, in turn, may increase survival within the host. For example, bile acids, which are detergents secreted into the small intestine through the bile duct, normally kill bacteria by solubilizing the bacterial cell membrane (22). V. cholerae, a diarrheal pathogen which is thought to colonize the small intestine, increases biofilm formation in response to bile acids (137). These results suggest that bile can actually have a protective effect on V. cholerae passing through the digestive system of the host by promoting biofilm formation. Another example is the response of P. aeruginosa to hydrogen peroxide, which is a product of the oxidative burst, a neutrophil-derived component of the host defense. Nonmucoid strains of P. aeruginosa become mucoid upon exposure to H2O2 (214). The mucoid colony morphology of P. aeruginosa reflects synthesis of the exopolysaccharide alginate. Alginate synthesis makes biofilm bacteria more resistant to antibiotics and further assault by the immune system. Therefore, the oxidative burst is a signal that causes bacteria to form biofilms that are more resistant to the action of the immune system.
(vi) Antimicrobials.
Antimicrobial compounds can also induce biofilm formation. Subinhibitory concentrations of the aminoglycoside antibiotic tobramycin has been shown to induce biofilm formation in P. aeruginosa (135). Another antimicrobial compound, triclosan, enhances transcription of cellulose synthesis genes in S. Typhimurium (310). Because cellulose is part of the biofilm matrix of S. Typhimurium, triclosan may activate biofilm formation in this organism.
(vii) Quorum signals.
Quorum-sensing circuits allow bacteria to coordinate their gene expression in a cell density-dependent manner. These circuits are activated by small molecules called autoinducers, which are secreted by bacteria and accumulate in the extracellular environment. The quorum-sensing circuit is activated when the autoinducer concentration exceeds a requisite threshold. The LuxI/LuxR system is a prototype of a quorum-sensing system used by many gram-negative bacteria (127). The details of this system were first elucidated in the luminescent marine bacterium Vibrio fischeri, in which quorum sensing regulates light production. LuxI-type proteins are enzymes that synthesize acylated homoserine lactone (AHL) autoinducers. AHLs then modulate the activity of LuxR-type transcriptional activators, which activate gene expression upon binding of the AHL.
Gram-positive bacteria such as S. aureus commonly use a more complex quorum-sensing system in which modified oligopeptides serve as autoinducers that are detected by two-component signal transduction pathways (127). Unlike AHLs, the oligopeptide does not enter the cell, but rather is detected extracellularly by a sensor kinase, which autophosphorylates and then transfers the phosphoryl group to its cognate response regulator. This regulator, in turn, activates the expression of target genes.
Some bacteria, such as Vibrio harveyi, use hybrid systems with components of both the gram-positive and gram-negative prototypical quorum-sensing systems. In these systems, AHL-type autoinducers are detected by a membrane-bound two-component hybrid sensor kinase. The phosphoryl group is transferred from the sensor kinase to a histidine phosphotransfer protein and then to a response regulator. In addition to the AHL-type autoinducer, which is species specific, an interspecies autoinducer called AI-2, a furonosyl borate ester, has been identified as a signal for hybrid quorum-sensing systems (127).
In the laboratory, cells cultured in tubes or on agar plates reach high densities, and threshold levels of autoinducer are easily achieved. In natural environments or in a eukaryotic host, where an abundance of nutrients is the exception rather than the rule, cell densities high enough to trigger quorum-sensing circuits are most likely achieved only in specific environmental niches. For example, in the V. fischeri/squid symbiosis, quorum sensing is activated only in the squid's light organ, which is colonized exclusively by V. fischeri (218, 274). At high cell densities, transcription of V. fischeri genes required for bioluminescence is activated. In the moonlight, the light emanating from the light organ protects the squid against predators by concealing its shadow. One might predict that the formation of a biofilm would favor cell densities high enough to activate the quorum-sensing circuit. In fact, genetic analysis of light organ colonization by V. fischeri suggests that the structure formed by V. fischeri within the light organ is a biofilm. In this case, quorum sensing positively regulates biofilm formation. However, this is not always the case. Quorum-sensing circuits can have positive or negative effects on biofilm formation. Below we discuss regulation of biofilm formation by quorum-sensing circuits in three different model organisms.
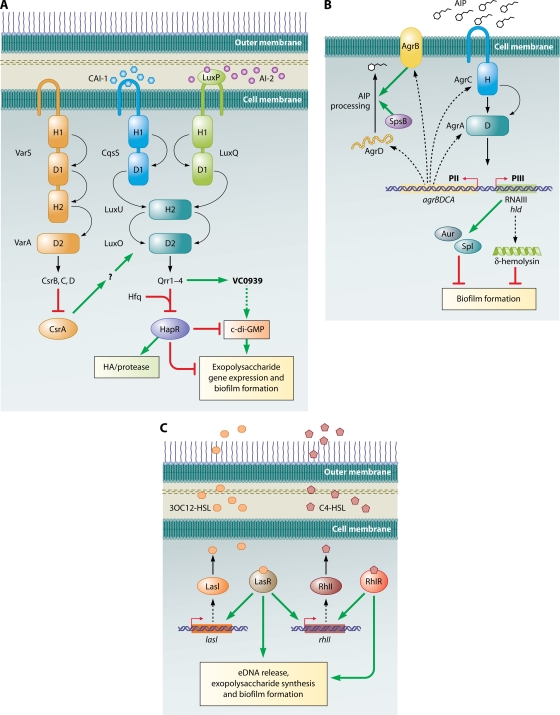
(a) Vibrio cholerae. In V. cholerae, increased cell density leads to inhibition of biofilm formation. The regulatory cascade leading to quorum sensing in V. cholerae is quite complex (Fig. 3A). Three different systems converge to regulate the expression of the transcriptional regulator HapR, which is at the bottom of the quorum-sensing regulatory cascade. In the first two systems, CqsS and LuxQ are membrane-bound sensor kinase-response regulator hybrid proteins which respond to autoinducers CAI-1 and AI-2, respectively. LuxQ detects AI-2 indirectly via the periplasmic binding protein LuxP. At low cell density, LuxQ and CsqS autophosphorylate. The phosphoryl group is transferred first to their receiver domains, then to the histidine phosphotransfer protein LuxU, and finally to the response regulator LuxO. Phospho-LuxO activates the transcription of the four small RNAs (sRNAs) Qrr1 to -4, which work with Hfq to block synthesis of HapR by destabilizing the mRNA encoding this protein. This relieves transcriptional repression of genes in the HapR regulon. At high cell density the flow of phosphate in the quorum-sensing signal transduction pathway is reversed, the half life of hapR mRNA increases, and more HapR is synthesized, leading to the transcriptional activation of genes favored under high-cell-density conditions and repression of those favored under low-cell-density conditions (119). The third quorum-sensing signal transduction cascade is composed of VarS/VarA, a two-component hybrid sensor kinase and response regulator pair, which activate transcription of the sRNAs CsrB, -C, and -D (193). These sRNAs bind and inhibit CsrA, which is an RNA binding protein involved in posttranscriptional regulation of a variety of processes. This system feeds into the LuxU-LuxO-Qrr-HapR relay at the level of LuxO, although the exact mechanism has not yet been elucidated. The third quorum-sensing signal, if any, for this signal transduction cascade has not yet been identified.
FIG. 3.
Quorum-sensing circuits and biofilm formation. (A) V. cholerae. Three quorum-sensing circuits converge on HapR to regulate biofilm formation. A HapR-independent quorum-sensing pathway involving Qrr1 to -4 and VC0939, which encodes a GGDEF family protein, has also been identified. This protein is likely to be a DGC that makes c-di-GMP, which is a positive activator of biofilm formation. HapR inhibits biofilm formation via multiple pathways, one of which is by indirectly decreasing c-di-GMP concentrations in the cell. Curved arrows denote the flow of phosphate under low-cell-density conditions. Phosphate flow is reversed at high density. H and D refer to the histidine and aspartate residues, respectively, which accept and shuttle the phosphoryl group. Dotted lines denote hypothesized effects. The question mark refers to a hypothesized intermediate effector in the pathway that has not been identified. (Adapted from reference 193 with permission of Blackwell Publishing Ltd.) (B) S. aureus: the Agr quorum-sensing pathway. A TCS composed of the histidine kinase AgrC and the response regulator AgrA responds to the presence of AIP. Phosphorylated AgrA activates transcription of the divergent PII and PIII operons. The PII operon encodes the machinery to synthesize, process, and detect AIP, while the PIII operon encodes RNAIII, the major effector of the quorum-sensing response. RNAIII regulates numerous downstream genes, two of which encode Aur and Spl proteases that are negative effectors of biofilm formation. The RNAIII transcript also encodes δ-hemolysin, which also inhibits biofilm formation. Curved arrows denote the flow of phosphate under high-cell-density conditions. H and D refer to the histidine and aspartate residues, respectively, which accept and shuttle the phosphoryl group. Broken lines connect the genes to their gene products. (C) P. aeruginosa. Las and Rhl pathways regulate quorum-sensing responses. The Rhl system is under the control of the Las system. LasR and RhlR, in the presence of their cognate autoinducers, activate a large number of genes, among which are those involved in exopolysaccharide production, eDNA, and biofilm formation. Broken lines connect the genes to their gene products.
In some strains of V. cholerae, HapR represses exopolysaccharide gene expression and biofilm formation in response to high cell density (119, 360). This repression is effected partially through degradation of the second messenger c-di-GMP (343). It has been proposed that these quorum-sensing cascades predominate in regulation of biofilm formation in the El Tor biotype of V. cholerae, whereas in the classical biotype, another phosphorelay consisting of VieS, VieA, and VieB predominates in regulation of biofilm formation. However, in many strains of V. cholerae of both biotypes, quorum-sensing circuits are inactivated by natural frameshift mutations or missense mutations in hapR. (119, 154, 361). Interestingly, the entire signal transduction cascade is preserved in some of these strains, as evidenced by the fact that repairing HapR restores quorum-sensing regulation of biofilm formation. These observations suggest that the role of quorum sensing in V. cholerae biofilm formation is not defined by biotype and raise the question of whether these mutations were acquired in the laboratory or in the wild.
HapR-independent quorum-sensing mechanisms have also been identified in V. cholerae. One such mechanism involves binding of Qrr1 to mRNA encoding the GGDEF domain-containing protein encoded at locus VCA0939. Expression of this mRNA is predicted to be inhibited by formation of a stem-loop structure that coincides with the binding site of Qrr1. Binding of Qrr1, therefore, is thought to inhibit stem-loop formation, leading to increased translation of VCA0939 mRNA (120). Although this regulation requires LuxO, Hfq, and Qrr1, it is independent of HapR.
HapR not only prevents biofilm formation; it may also promote detachment of cells from existing biofilms (360). One potential mechanism is by activation of the hapA gene. hapA codes for the hemagglutinin/protease, an enzyme that promotes detachment of V. cholerae from cultured epithelial cells (84, 153, 290). Furthermore, proteins also play a role in maintenance of the integrity of the V. cholerae VPS-dependent biofilm structure, and it is possible that HapA plays a role in detachment through degradation of these proteins. Thus, quorum-sensing circuits and HapR appear to provide V. cholerae with a strategy for exit from association with both biotic and abiotic surfaces. Interestingly, hapR mutants do not effectively colonize the mammalian intestine in an infant mouse model; however, this is not due to decreased expression of the virulence genes, as these mutants in fact have increased virulence gene expression (360, 361). Rather, this seemingly contradictory observation may be explained by the inability of hapR mutants to repress biofilm-coregulated genes inside the host. Biofilm formation therefore may interfere with colonization of the host epithelium or may promote clearance of V. cholerae by the innate immune system. Thus, quorum-sensing-regulated repression of biofilm formation appears to be necessary for efficient colonization of the host.
(b) Staphylococcus aureus. S. aureus biofilm formation is also negatively regulated by quorum sensing. In this organism, the autoinducer that activates the quorum-sensing cascade is a peptide (AIP) (Fig. 3B). AIP, which is encoded by the agrD gene, is synthesized as a linear peptide of approximately 46 residues. This peptide is then processed to yield a cyclic peptide containing a thiolactone ring. Depending on the particular staphylococcal strain, the final peptide is between seven and nine residues in length (239). The processing of the agrD gene product requires at least two proteins, AgrB and a type I signal peptidase, SpsB (165). Interestingly, the AIPs synthesized by various S. aureus strains fall into four specificity groups, which are defined by conserved amino acid residues. S. aureus strains producing AIPs in the same group are able to participate jointly to activate quorum sensing, whereas S. aureus strains producing AIPs of different groups may interfere with each other's quorum-sensing response. (239).
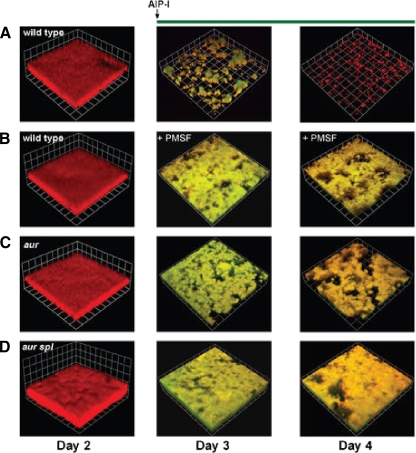
The detection system for AIPs is comprised of AgrC, which is a membrane-bound sensor-kinase, and AgrA, which is the response regulator. After phosphoryl group transfer from AgrC, AgrA activates transcription of the P2 and P3 operons (239, 240). The P2 operon includes the genes encoding AgrA, -B, -C, and -D. Thus, binding of AgrA to the P2 promoter leads to a rapid amplification of the quorum-sensing signal. The P3 promoter drives the expression of the RNAIII transcript, a 514-nucleotide regulatory RNA that is the primary effector of the quorum-sensing response. The RNAIII transcript also contains the hld gene which encodes the 26-amino-acid δ-hemolysin peptide, which inhibits biofilm formation, potentially due its surfactant-like properties (337). RNAIII positively regulates the transcription of genes encoding the metalloprotease aureolysin (Aur) and Spl serine proteases, which are extracellular proteases involved in dispersal of biofilms and are therefore negative effectors of biofilm formation (31).
AIP-deficient mutants have been shown to form more robust biofilms than the wild-type strain (337, 355), leading to the conclusion that the agr quorum-sensing system negatively regulates biofilm formation. Furthermore, the agr system is more active in cells that have detached from the biofilm, a finding consistent with the negative regulation of biofilm formation by quorum sensing (355). As has been observed in other organisms, biofilm formation in S. aureus is highly dependent on the culture medium, and the quorum-sensing response may play some role in this effect. For example, when S. aureus is cultured in the presence of glucose, agr gene expression is repressed (263). This may be partially responsible for the observation that glucose can promote S. aureus biofilm formation (31).
(c) Pseudomonas aeruginosa. Activation of quorum-sensing circuits in P. aeruginosa stimulates biofilm formation. P. aeruginosa possesses two LuxI/R-type quorum-sensing circuits, LasI/R and RhlI/R, which make and detect the autoinducers N-(3-oxododecanoyl)-l-homoserine lactone (3OC12-HSL) and N-butyryl-l-homoserine lactone (C4-HSL), respectively (Fig. 3C) (127). LasR bound to its cognate autoinducer activates a number of target genes, one of which is the rhlI gene; therefore, the Rhl system is under the control of the Las system (127).
P. aeruginosa quorum-sensing mutants make biofilms that have increased sensitivity to the detergent sodium dodecyl sulfate (SDS), suggesting that matrix synthesis is defective (69). In fact, production of matrix components is affected by the P. aeruginosa quorum-sensing systems. DNA is a major component of the P. aeruginosa biofilm matrix and is required for the integrity of biofilms formed by this bacterium (350). DNA release is controlled by quorum sensing, and the biofilm matrices of lasI rhlI double mutants contain less extracellular DNA (eDNA) than that of wild-type P. aeruginosa (5). Moreover, lasI and rhlR mutants are defective in matrix formation and activation of exopolysaccharide gene transcription (279). Quorum sensing may also provide P. aeruginosa with an advantage against other organisms in multicellular biofilms. In biofilms, wild-type P. aeruginosa has a growth advantage over A. tumefaciens. This advantage is decreased in the absence of the lasR and rhlR genes (8).
Chronic, intractable colonization and infection of the lungs with P. aeruginosa infections in the lungs of cystic fibrosis (CF) patients is a leading cause of morbidity and mortality in CF patients (112). P. aeruginosa is thought to exist as a biofilm in the CF lung. This hypothesis is based on (i) the presence of multicellular aggregates, (ii) production of the biofilm matrix polysaccharide alginate, and (iii) resistance to antibiotic treatment (248). Furthermore, the ratio of the quorum-sensing molecules 3OC12-HSL to C4-HSL measured in the sputa of CF patients colonized with P. aeruginosa was shown to be similar to the ratio that is found in biofilms and different from the ratio measured in planktonic cultures of P. aeruginosa, suggesting that the environment experienced by P. aeruginosa in the CF lung may bear some similarity to that experienced by P. aeruginosa in a biofilm (293). Involvement of quorum-sensing systems in biofilm formation in CF was underscored by a study that demonstrated the requirement for the Las quorum-sensing system for P. aeruginosa biofilm formation in an artificial medium designed to mimic conditions in the lungs of CF patients (301). Because of the role that quorum-sensing circuits play in regulation of biofilm formation in P. aeruginosa, they provide attractive targets for drugs to treat CF. Derivatives of furanone, compounds produced by the red seaweed Delisea pulchra, have shown promise as inhibitors of quorum sensing and biofilm formation (107, 212). For example, treatment of P. aeruginosa biofilms with a synthetic derivate of a natural furanone has lead to increased sensitivity of biofilm bacteria to antibiotics, H2O2, and phagocytosis by polymorphonuclear leukocytes as well as to increased rates of detachment from biofilms (128). It was proposed that administration of quorum-sensing inhibitors followed by antibiotic treatment may be a valid approach for treatment for lung infections. Moreover, the same drug led to increased clearance of P. aeruginosa in mouse models of both lung infections and foreign-body infections, further validating the promise held by these compounds (53, 130).
In addition to the many reports that have demonstrated an effect of quorum sensing on P. aeruginosa biofilm formation, contradictory results have also been reported (131, 257). Some of these discrepancies may be due to differences in experimental conditions such as the culture medium, flow conditions, and the specific P. aeruginosa strain used, all of which could have an influence on quorum-sensing regulation of biofilm formation (171, 172, 289). Thus, the relevance of quorum sensing in regulating P. aeruginosa biofilms is still debated. Of course, for all quorum-sensing bacteria, it is likely that this process will occur only in specific environmental niches where the appropriate environmental signals are present. The challenge of the researcher is to determine which of the conditions studied in the lab are relevant to environments experienced by the bacterium in the wild. Conditions such as the nutritional status of the environment and even the presence of conjugative plasmids can lead to quorum-sensing cues being bypassed or overridden (56, 105, 113, 264, 267). Presumably, all of these signals feed into intricate cellular signaling networks that ultimately results in the appropriate response to the prevailing conditions. Thus, the exact nature of regulation of biofilm formation by quorum sensing may require the elucidation of all of these networks.
Most of the signals described above have been studied only in the laboratory. A remaining challenge, therefore, is the correlation of the identified signals with the natural or host environments in which they are operative. A study of these signals in their natural setting is critical to an understanding of their role in adaptation of the bacterium to its environment.
Secondary messenger and protein networks.
In recent years, we have gained considerable knowledge about the numerous cellular networks that regulate biofilm formation. Even with this knowledge, we have only begun to understand how these networks function and, more importantly, how they work together to regulate biofilm formation. Below we review our current understanding of a number of these regulatory networks that have been more extensively studied.
(i) c-di-GMP.
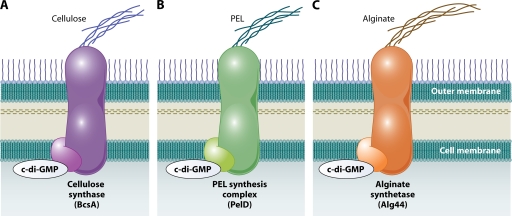
c-di-GMP, a ubiquitous second messenger widely used by bacteria, was discovered 2 decades ago as an allosteric activator of the cellulose synthase complex in Gluconacetobacter xylinus (273). More recently, we have come to appreciate the significant role that this molecule plays in adaptation of many different bacterial species to their environment. In particular, c-di-GMP has been firmly established as the central regulator of biofilm formation and the main switch between motile and sessile forms of existence in gram-negative bacteria (291). Surprisingly, gram-positive bacteria do not appear to use this molecule as extensively to regulate these phenotypes.
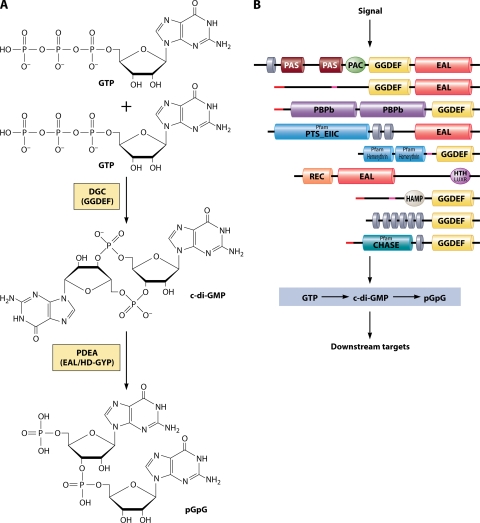
(a) GGDEF and EAL proteins. c-di-GMP is synthesized from two GTP molecules by DGCs, proteins that contain the ubiquitous GGDEF domain, which harbors their enzymatic activity (Fig. 4A). It is degraded to the linear dinucleotide pGpG by phosphodiesterase A's (PDEAs), proteins containing EAL or HD-GYP domains, which are responsible for the enzymatic activity (reviewed in reference 276). Many proteins belonging to the GGDEF/EAL superfamily contain both GGDEF and EAL domains; in this case the whole protein can act as either a DGC, a PDEA, or in some cases both, depending on the presence or absence of their interaction partners (52, 83, 170, 313).
FIG. 4.
c-di-GMP. (A) Synthesis and breakdown of c-di-GMP. c-di-GMP is synthesized by DGCs containing GGDEF domains from two GTP molecules and broken down to pGpG by PDEAs (containing either EAL or HD-GYP domains). (B) GGDEF and EAL/HD-GYP superfamily proteins are modular and diverse. Proteins that belong to this superfamily contain a variety of sensory domains that are likely to regulate the activity of the enzymatic domains based on the input signals. Shown here are a number of GGDEF/EAL proteins from V. cholerae that have different domain architectures (top to bottom, VC0072, VC0658, VC1067, VC1211, VC1216, VC1652, VC1370, VC1372, and VC1376). Sequences were obtained from http://cmr.jcvi.org. Domain architecture was analyzed using SMART (Simple Modular Architecture ResearchTool) (http://smart.embl-heidelberg.de) (283). Abbreviations: CHASE, cyclase/histidine kinase-associated sensory domain; GAF, domain present in phytochromes and cGMP-specific phosphodiesterases; HAMP, histidine kinases, adenylyl cyclases, methyl binding proteins, phosphatase domain; hemerythrin, hemerythrin HHE cation binding domain; HTH LUXR, helix-turn-helix, Lux regulon, PAS/PAC, Per (periodic clock protein), Arnt (aryl hydrocarbon receptor nuclear translocator protein), and Sim (single-minded protein) domain; PBPb, bacterial periplasmic substrate binding proteins; PTS_EIIC, PTS, EIIC; Rec, CheY-homologous receiver domain. Gray disks denote predicted transmembrane domains. Red lines denote predicted signal sequences. Pink lines denote segments of low compositional complexity. Pfam, Protein family database (http://pfam.sanger.ac.uk).
GGDEF and EAL domains and to a lesser extent HD-GYP domains appear in large numbers in bacterial genomes. A recent census identified over 4,200 GGDEF domains, over 2,500 EAL domains, and 200 HD-GYP domains in bacteria (275). Gram-negative bacteria generally have large numbers of genes encoding GGDEF and EAL family members; for example, V. cholerae and E. coli have 53 and 36 of these family members, respectively (97). Only a small number of genes encoding proteins belonging to the GGDEF/EAL superfamily are present in genomes of gram-positive bacteria, (e.g., seven in B. subtilis), which supports the conclusion that c-di-GMP may not play as fundamental a role in adaptation of these organisms to their environments (97). Most proteins that contain these domains are modular. In addition to their GGDEF, EAL, or HD-GYP domains, they have a variety of sensory domains (REC, PAS, GAF, etc.) that are likely to receive signals from the environment (Fig. 4B). These signals are thought to be transduced as an alteration of the enzymatic activity that would result in local or global fluctuations in c-di-GMP levels, which in turn would result in behavioral adjustments (152, 268, 275). Modulation of DGC activity as a result of phosphorylation of N-terminal REC domains has indeed been demonstrated for a number of GGDEF proteins (133, 251).
In most organisms and for most homologs, mutation of genes encoding DGCs decreases biofilm formation, while mutation of genes encoding PDEAs increases biofilm formation (135). Thus, DGCs usually promote biofilm formation whereas PDEAs inhibit it, indicating that c-di-GMP is a positive regulator of biofilm formation. Indeed, a number of studies in which the intracellular level of c-di-GMP was genetically manipulated have shown that intracellular levels of c-di-GMP are directly proportional to biofilm formation and transcription of exopolysaccharide genes (26, 133, 180, 201, 223, 291, 321). The possibility of pGpG, the degradation product of c-di-GMP, playing an active role in c-di-GMP signaling pathways has also been suggested; however, this possibility remains to be demonstrated (270). One of the curious characteristics of c-di-GMP signaling is that deletion of only one of the many EAL or GGDEF proteins encoded in a bacterial genome often leads to drastic phenotypic changes. Thus, the presence of other DGCs and PDEAs cannot compensate for the loss of one of these proteins. Although numerous GGDEF and EAL proteins have been identified as regulators of biofilm formation, a much smaller portion have been characterized in detail. Here we focus on a few systems that shed some light on various aspects of regulation of biofilm formation by GGDEF and EAL proteins.
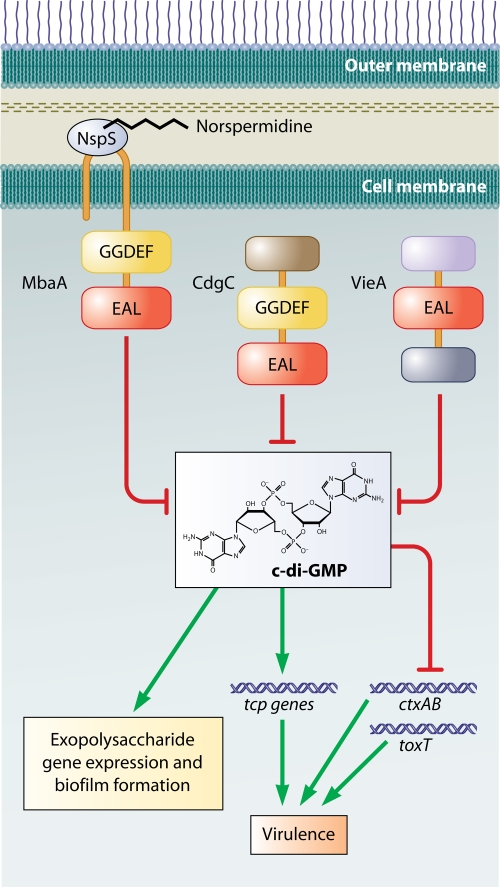
c-di-GMP inversely regulates biofilm formation and virulence in V. cholerae via VieA (Fig. 5). VieA is a two-component response regulator that is part of a three-component signal transduction system, VieS/A/B, that regulates cholera toxin expression. In addition to its phosphoryl group acceptor and DNA binding domains, VieA also has an EAL domain. This protein is a c-di-GMP phosphodiesterase that inhibits biofilm formation by decreasing cellular levels of c-di-GMP and also repressing vps gene expression (314, 321). Furthermore, the phosphodiesterase activity of VieA and the resultant low c-di-GMP levels are necessary for optimal transcription of the ctxAB genes, which encode cholera toxin, as well as toxT, a transcriptional activator of ctxAB (322). Also, a mutant strain with constitutively increased intracellular c-di-GMP as a result of a missense mutation in VieA is attenuated 10-fold in the infant mouse model of cholera (322). These results have suggested that in V. cholerae virulence gene expression and biofilm formation are inversely regulated by c-di-GMP. However, subsequent studies have reported increased expression of other virulence genes as a result of increased c-di-GMP levels or deletion of the phosphodiesterase CdgC, which is a negative regulator of biofilm formation (Fig. 5) (26, 202). Therefore, it is likely that there is a more complex relationship between biofilm formation and virulence gene expression in V. cholerae.
FIG. 5.
c-di-GMP regulation of virulence and biofilm formation in V. cholerae. MbaA is a predicted c-di-GMP phosphodiesterase. Association of MbaA with NspS is thought to inhibit its activity. Hypothesized binding of norspermidine to NspS is thought to increase this inhibition. CdgC is a phosphodiesterase that decreases c-di-GMP pools in the cell. Deletion of cdgC leads to an increase in intracellular c-di-GMP and an accompanying increase in the transcription of some of the genes in the tcp operon, which is required for virulence. VieA is also a phosphodiesterase which positively effects transcription of the virulence genes ctxAB and toxT via its negative effect on c-di-GMP.
Although many GGDEF/EAL proteins are involved in biofilm formation, the signals to which these proteins respond remain largely elusive. One signal that has been implicated in c-di-GMP regulation of biofilm formation is the polyamine norspermidine, which is transmitted via MbaA (Fig. 5). MbaA is a transmembrane protein which contains tandem GGDEF and EAL. Deletion of the mbaA gene leads to an increase in biofilm formation and exopolysaccharide gene transcription in V. cholerae (33, 164, 202). Because of its effect on biofilm formation, MbaA has been termed a repressor and is likely to have c-di-GMP phosphodiesterase activity, although this has not yet been shown experimentally. The mbaA gene is likely to be cotranscribed with the upstream gene, nspS, which encodes a protein with similarity to the periplasmic spermidine binding protein PotD of the ABC-type spermidine transport systems. Deletion of this gene results in a decrease in biofilm formation and exopolysaccharide gene transcription, suggesting that this protein is a positive regulator of biofilm formation that inhibits the proposed phosphodiesterase activity of MbaA (164). Furthermore, norspermidine increases biofilm formation in an NspS- and MbaA-dependent manner, suggesting that norspermidine is an extracellular signal detected and processed by the NspS-MbaA system. Because MbaA is a GGDEF-EAL domain protein, the norspermidine signal is likely to feed into the local or global c-di-GMP pools in the cell. As with most signaling systems that use c-di-GMP as a second messenger, the downstream effectors of this signaling pathway have not yet been identified.
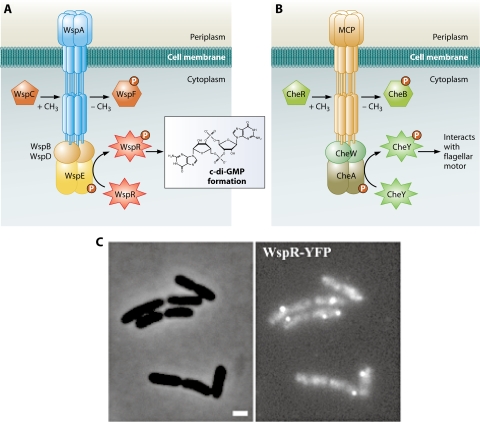
In P. aeruginosa, intracellular c-di-GMP levels can be regulated by a group of proteins similar to those controlling chemotactic responses in bacteria. For example, deletion of wspF, encoding a homolog of the methylesterase CheB, which is involved in adaptation to chemotactic stimuli, leads to increases in cellular c-di-GMP levels, transcription of exopolysaccharide synthesis genes, and biofilm formation (133). These phenotypes depend on the presence of wspR; wspR encodes a hybrid two-component response regulator/GGDEF protein whose DGC activity is enhanced by phosphorylation. wspF and wspR reside in an operon encoding homologs of all of the necessary components of a typical chemotaxis signaling system, including a methyl-accepting chemotaxis protein (wspA), a cheR methyltransferase (wspC), two cheW homologs (wspB and wspD), and a hybrid histidine kinase-response regulator (wspE), in addition to wspF and wspR (Fig. 6A). Regulation of c-di-GMP concentration by chemotaxis-like signaling networks is intriguing. It is hypothesized that because chemosensory networks mount rapid responses to chemical gradients, the Wsp signaling network may function to accelerate the transition between the planktonic and surface-associated states. Subsequent work on this system has demonstrated that in its phosphorylated form WspR forms clusters that are distributed in the cytoplasm (Fig. 6B) (117). While the exact function of these clusters has not been elucidated, it was speculated that clustering of WspR may result in localized synthesis of c-di-GMP, leading to areas of high and low c-di-GMP concentration within the cytoplasm. It is not difficult to imagine a scenario where activation of this chemotaxis-like signaling system leads to phosphorylation of WspR. This results in clustering of WspR as well as enhancement of its DGC activity, which in turn leads to microenvironments with increased c-di-GMP concentrations. The presence of c-di-GMP targets within these microenvironments could lead to spatial heterogeneity in the activation of c-di-GMP-responsive elements, while not significantly changing the average c-di-GMP concentration or requiring the activity of other diguanylate synthases within the cell. This work is particularly exciting because it describes a mechanism by which one c-di-GMP signaling cascade among many could potentially achieve independence in regulation of biofilm formation.
FIG. 6.
Regulation of intracellular c-di-GMP by a chemotaxis-like signaling system in P. aeruginosa. (A) Predicted organization of the Wsp signaling system. Wsp proteins regulate the c-di-GMP concentrations based on an as-yet-unidentified signal. The signal is predicted to be detected by the methyl-accepting chemotaxis protein homolog WspA; WspE autophosphorylates and transfers the signal to WspR. Phosphorylated WspR has increased DGC activity. WspA is coupled to WspE via WspB and -D. The methylation state of WspA is determined by opposing activities of the methyltransferase homolog WspC and the methylesterase homolog WspF. Demethylation of the receptor by WspF is thought to be involved in adaptation to the signal. (B) Predicted organization of the Che signaling system. The chemotaxis signaling pathway responds to external attractant and repellent molecules and results in swimming toward or away from these molecules, respectively. The output of the system is CheY, which in its phosphorylated form interacts with the flagellar switch proteins to determine the direction of flagellar rotation. (C) Clustering of WspR in the cell. The phase-contrast (left) and fluorescence (right) images of cells expressing a WspR translational fusion to yellow fluorescent protein (WspR-YFP) are shown. Notice clusters of fluorescence, indicating clustering of WspR-YFP. Bar, 1 μm. (Adapted from reference 117 with permission of Blackwell Publishing Ltd.)
In P. aeruginosa, SadC, a DGC, and BifA, a phosphodiesterase, control the surface attachment at the posttranscriptional level through modulation of the levels of c-di-GMP. SadC, an integral membrane DGC, stimulates biofilm formation and inhibits swarming motility due to synthesis of c-di-GMP (223). Furthermore, the action of SadC stimulates synthesis of exopolysaccharides, which is a characteristic of the process of biofilm formation. For instance, when SadC is expressed from a high-copy plasmid, P. aeruginosa forms a wrinkled colony with increased Congo red binding, both of which are phenotypes associated with increased exopolysaccharide synthesis. Both of these phenotypes are dependent on the presence of pelA and pelG genes, which are required for the synthesis of the matrix exopolysaccharide. However, transcription of these genes is not increased by the action of SadC, suggesting that SadC regulates exopolysaccharide synthesis at a posttranscriptional level (223). Conversely, BifA, a phosphodiesterase containing both GGDEF and EAL domains, enhances swarming and leads to reduced biofilm formation (180, 223). Deletion of the bifA gene increases exopolysaccharide production; this regulation also appears to occur at the posttranscriptional level (180, 223). Finally, either the deletion of bifA or overexpression of SadC results in increased cellular pools of c-di-GMP, confirming that these genes influence biofilm formation by affecting intracellular c-di-GMP pools. Thus, these two proteins work together to regulation biofilm formation at the posttranscriptional level.
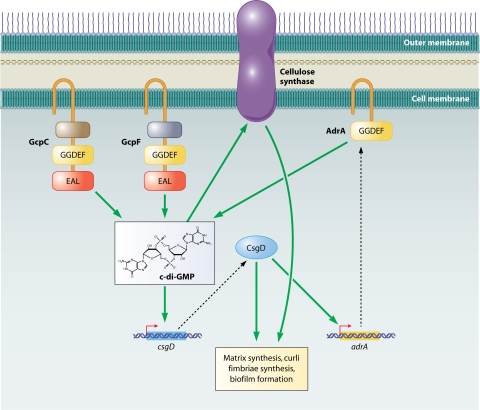
Several reports describing the interplay of multiple GGDEF proteins in the process of biofilm formation have revealed coordinated function (Fig. 7). In S. Typhimurium, increased c-di-GMP levels lead to increased curli expression, cellulose synthesis, and biofilm formation (156, 291). AdrA, one of the well-characterized GGDEF proteins in S. Typhimurium, is required for cellulose production and biofilm formation in LB broth, a rich growth medium, while GcpA, another GGDEF protein, is required for biofilm formation in the nutrient-deficient ATM medium (98). This observation lends credence to the idea that bacteria have multiple GGDEF proteins because different GGDEF proteins are active under different environmental conditions. Furthermore, different GGDEF proteins exert their effect on different steps of the regulatory network that controls biofilm formation in S. Typhimurium. For example, CsgD, the main transcriptional activator that regulates biofilm formation in this organism, activates transcription of the adrA gene, encoding the GGDEF protein AdrA; however, it has no effect on the transcription of the genes gcpA to -F, encoding GGDEF proteins GcpA to -F (98). Thus, unlike adrA, gcpA to -F are not downstream targets of CsgD. In fact, two of these proteins, GcpC and GcpF, are required for normal levels of CsgD expression, suggesting that they are upstream of CsgD in the signaling cascade (156). Moreover, measurements of CsgD levels over time in wild-type S. Typhimurium and gcpC and gcpF mutants grown on agar plates show that a gcpC mutant has lower levels of CsgD at 10 h than does the gcpF mutant. At 16 h, levels of CsgD are reduced in both mutants compared to the wild type. One explanation for this observation is that under the conditions of this experiment, first GcpC and then GcpF activates csgD expression during growth on agar plates (156). Thus, in S. Typhimurium, c-di-GMP affects biofilm formation by acting both upstream (via GcpC and GcpF) and downstream (via AdrA) of CsgD. Similar types of regulation using multiple GGDEF/EAL family proteins to regulate biofilm formation may exist in other bacteria.
FIG. 7.
Regulation of biofilm formation in S. Typhimurium by DGCs. Various diguanylated cyclases act both upstream and downstream in the signaling cascade that regulates biofilm formation. GcpC and -F positively affect CsgD, which is the main transcriptional activator of curli biosynthesis genes that are required for biofilm formation as well as adrA. AdrA is also a DGC. c-di-GMP then activates cellulose synthase leading cellulose production, which is part of the matrix of the biofilms made by S. Typhimurium. Broken lines connect the genes to their gene products.
(b) Downstream targets of c-di-GMP. One of the biggest puzzles in the regulation of biofilm formation by c-di-GMP is the mechanism by which this molecule brings about the various effects discussed above. Until recently, only two downstream targets of this molecule had been identified. The first is BcsA, the α subunit of the G. xylinus cellulose synthase enzyme for which c-di-GMP is an allosteric activator (7, 273, 349). The second downstream target is C. crescentus PleD, a DGC itself. c-di-GMP binds to the so-called I site of PleD and inhibits the activity of this enzyme by feedback inhibition (48, 50). Therefore, one mechanism by which c-di-GMP can affect its downstream targets is through modulation of enzyme activity. However, elucidation of c-di-GMP signaling will ultimately depend upon identification of all the downstream targets of this molecule and an understanding of its effects on each of these targets. In the last 2 years, important steps have been taken toward this goal as a result of identification of three other types of c-di-GMP targets, namely, PilZ domains, PelD, and riboswitches.
PilZ domains were named after the P. aeruginosa protein PilZ, a single-domain protein that is involved in fimbria biogenesis and twitching motility (6). Initially, these domains were proposed to bind c-di-GMP based on in silico analysis (7). A number of recent studies have experimentally shown that PilZ domains can indeed bind c-di-GMP (51, 222, 253, 278).
An analysis of five PilZ domain proteins in V. cholerae has demonstrated that c-di-GMP binds to two of these, PlzC and PlzD (253). PlzD does not appear to regulate biofilm formation, because the deletion of the plzD gene does not lead to any defects in biofilm formation. Deletion of plzC leads to a reduction of biofilm formation in a strain with artificially elevated intracellular concentrations of c-di-GMP but not in the wild-type strain. PlzB does not bind c-di-GMP in vitro; however, deletion of the plzB gene leads to a marked phenotype, including reduced ability to form biofilms, reduced motility, and a 10-fold attenuation in virulence in the infant mouse model of cholera. Furthermore, a point mutation in a conserved residue in the proposed c-di-GMP binding site leads to the same phenotypes as the deletion mutation, indicating that c-di-GMP binding is necessary for the function of PlzB. These data suggest that the function of PlzB does, in fact, depend on binding of c-di-GMP. The remaining two proteins, PlzA and PlzE, do not bind c-di-GMP in vitro; moreover, their deletion or overexpression does not result in any effect on biofilms, motility, or virulence. Thus, the presence of a PilZ domain may not be sufficient for binding c-di-GMP. Alternatively, these proteins may bind c-di-GMP only under particular conditions which have not been tested in these experiments. Therefore, although some PilZ domain proteins appear to be plausible downstream targets for c-di-GMP, the exact mechanism by which they regulate biofilm formation in V. cholerae remains to be elucidated.
Some PilZ domain proteins are involved in biofilm formation and/or synthesis of the matrix exopolysaccharide. P. aeruginosa protein Alg44, one of the eight PilZ domain proteins in this organism and a putative component of alginate synthetase, can bind c-di-GMP in vitro (222). Mutation of several conserved amino acids predicted to constitute the c-di-GMP binding site of Alg44 abolishes c-di-GMP binding. Strains expressing these mutants have significantly reduced alginate production, indicating that c-di-GMP binding to Alg44 is necessary for alginate synthesis (222). In this case, c-di-GMP regulates exopolysaccharide production by posttranslationally modulating the function of Alg44. Six of the remaining seven PilZ domain proteins have also been shown to bind c-di-GMP; however, it is not known whether these play a role in exopolysaccharide production or biofilm formation (222).
In P. aeruginosa, PelD, a transmembrane protein encoded by one of the genes in the pel operon that is required for pellicle production and PEL exopolysaccharide synthesis, is a downstream target of c-di-GMP (48, 190). Although the exact function of PelD is not known, it is likely to be part of the machinery that synthesizes the PEL exopolysaccharide. PelD has an RXXD motif which is found in the I sites of some DGCs such as PleD. Indeed, mutation of the arginine and glutamate residues in the RXXD sequence to alanines abolishes c-di-GMP binding to PelD, implicating this motif as part of the c-di-GMP binding site of PelD as well. Mutants unable to bind c-di-GMP are also unable to support pellicle formation, indicating that binding of c-di-GMP to PelD is necessary for synthesis of the PEL polysaccharide. The RXXD motif is also conserved in PelD orthologs from a number of bacterial species that contain pel operons, suggesting similar regulatory mechanisms involving c-di-GMP and PelD in these organisms (190). How binding of PelD to c-di-GMP affects PEL biosynthesis has not yet been elucidated. However, in addition to BcsA of cellulose synthase and Alg44 of alginate synthetase complexes, this protein is the third example of a c-di-GMP binding protein that is also a putative part of exopolysaccharide synthesis machinery. Thus, binding of c-di-GMP to a component of the exopolysaccharide synthesis machinery may be a common mechanism for regulation of exopolysaccharide production and biofilm development (Fig. 8) (190).
FIG. 8.
Regulation of exopolysaccharide synthesis machinery by c-di-GMP. Binding of c-di-GMP to both hypothesized and known components of the exopolysaccharide synthesis machinery has been shown for cellulose synthase (A), the PEL synthesis complex (B), and alginate synthetase (C). Activation of these enzymes by c-di-GMP could be a common mechanism of regulating exopolysaccharide synthesis by this second messenger.
In addition to the protein targets of c-di-GMP, riboswitches that bind this molecule with very high affinity (Kd of ∼1 nM) have been recently identified (307). Riboswitches are mRNA domains that bind a particular ligand and regulate expression of downstream genes in response to levels of this ligand. c-di-GMP binding riboswitches were identified upstream of genes encoding PDEs and DGCs, flagellar operons, and other genes whose expression is known to be regulated by c-di-GMP levels. Reporter fusions to several members of these c-di-GMP riboswitches showed that some were “on” switches that increased expression of their associated genes in response to high c-di-GMP levels and that others were “off” switches which decreased gene expression in response to high c-di-GMP levels. Of particular interest is the Cd1 riboswitch from Clostridium difficile that lies in the 5′ untranslated region of a flagellar operon. This riboswitch was shown to turn expression of its associated genes off in response to elevated c-di-GMP levels. Because high intracellular c-di-GMP levels are known to promote biofilm formation and decrease flagellar gene expression, this riboswitch provides a mechanism for inverse regulation of flagellar gene transcription and biofilm formation (307).
(ii) TCSs.
One of the most common mechanisms by which prokaryotes process environmental information is through phosphoryl group transfer. This is done by TCSs, which, in their simplest form, are composed of a sensor histidine kinase, which directly or indirectly senses a signal, and a response regulator, which receives the information from the histidine kinase and brings about the relevant response. The signal is relayed from the histidine kinase to the response regulator as a phosphoryl group transfer. In reality, very few of these systems are this simple; many of them are composed of multiple components and hybrid kinase-response regulators. As can be expected from their abundance in the prokaryotic world, TCSs are involved in regulating biofilm formation in a number of bacteria. The numbers of TCSs reported to be involved in biofilm formation are constantly increasing. We have already mentioned some of these in the context of signals affecting biofilm formation. Here we discuss a few more of the better-characterized TCSs that regulate biofilm formation.
One conserved TCS that has been shown to regulate exopolysaccharide production and biofilm formation in P. aeruginosa, E. coli, and V. cholerae is the GacS/GacA (BarA/UvrY) system. GacS is a membrane-bound sensor histidine kinase with tandem histidine kinase, phosphotransfer, and histidine kinase domains, and GacA is a typical response regulator with a receiver domain and a helix-turn-helix DNA binding domain (reviewed in reference 184). Upon phosphoryl group transfer from GacS, GacA activates the transcription of sRNAs (Rsm or Csr) that then bind the RNA binding protein CsrA (or RsmA). CsrA is a repressor of a multitude of genes; thus, binding of the sRNAs to this protein titrates CsrA away from its target mRNAs, thereby derepressing its target genes.
In P. aeruginosa, GacS/GacA/RsmZ inversely regulates biofilm formation and expression of genes encoding TTSS components. This regulation involves inputs from two other histidine kinases, RetS and LadS. RetS, a hybrid histidine kinase with two tandem response regulator domains, is a positive regulator of genes encoding the TTSS and a negative regulator of exopolysaccharide synthesis genes in P. aeruginosa (114). Deletion of retS was shown to result in an increase in biofilm formation, attachment to cultured mammalian cells, and exopolysaccharide gene expression (114). In addition, a retS mutant did not produce a cytotoxic response in host cells due to a lack of a functional TTSS, which is required for toxin delivery into host cells (114). As a result, this mutant was attenuated in virulence in a murine acute pneumonia model. Transposon mutagenesis screens to identify suppressors of the retS phenotype led to the isolation of multiple mutations in gacS, gacA, and rsmZ genes, indicating that these responses are coordinated through GacS/GacA/RsmZ signal transduction pathway. Furthermore, a gacA deletion introduced into the retS mutant background abrogated the hyperbiofilm response, confirming the transposon mutagenesis results. LadS, also a hybrid histidine kinase with a domain architecture similar to that of RetS, acts in a manner opposite to that of RetS (333). Deletion of ladS resulted in a decrease in biofilm formation and exopolysaccharide gene expression and an increase in TTSS gene expression and hypertoxicity. Furthermore, retS and ladS mutants had increased and decreased levels of the small regulatory RNA RsmZ, respectively. Thus, RetS and LadS appear to be a part of a regulatory network that converge on the GacS/GacA/RsmZ signal transduction pathway and inversely regulate type III secretion and biofilm formation. These studies confirm previously published work that showed that GacA is required for microcolony formation (247). Another signaling system, sadARS, a three-component system composed of two response regulators and one sensor histidine kinase, also regulates biofilm formation and TTSS gene expression in inverse manners (181). Mutations in these genes caused defects in maturation and macrocolony formation in flow cell biofilms and increased transcript levels of many genes encoding components of the TTSS (181). These regulatory inputs could potentially feed into the GacA/GacS system or work through an alternate route. It should be noted that newly colonized CF patients have P. aeruginosa strains that are capable of toxin delivery using the TTSS. These traits are lost from isolates from patients with chronic infection (114). Thus, elucidation of the signals detected by this multicomponent network that inversely regulates biofilm formation and TTSS gene expression should shed light on transition of P. aeruginosa from an organism that causes acute infections to one that causes chronic biofilm infections.
In E. coli, the BarA/UvrY TCS, an ortholog of the GacS/GacA TCS, is required for biofilm formation (309). This system modulates the transcription of the regulatory RNA CsrB (229). CsrB is an antagonist of the RNA binding protein CsrA, which is a repressor of biofilm formation in this organism (204). CsrA inhibits biofilms by repressing the translation of the pgaABCD mRNA, which is responsible for the synthesis of one type of matrix exopolysaccharide (341). Therefore, CsrB positively regulates biofilm formation in an indirect manner. Interestingly, the effect of BarA/UvrY TCS on biofilm formation is also seen in the absence of CsrB, indicating a CsrB-independent regulation of biofilm formation by this TCS (309). Many gram-negative bacteria have orthologs of the genes encoding the GacS/GacA TCS (125). The VarA/VarS system of V. cholerae mentioned above is another one of these systems that has already been shown to regulate biofilm formation, in this case, in response to quorum-sensing cues. Studies of similar systems in other bacteria will most likely show that these systems are widely used regulators of biofilm formation.
In V. cholerae, the two central regulators of vps (vibrio polysaccharide) gene expression and biofilm formation are VpsR and VpsT, both of which belong to the two-component response regulator family (Fig. 9). When cells are grown under static conditions, both VpsR and VpsT are required for biofilm formation, as vpsR and vpsT deletion mutants adhere to surfaces only as a single layer of cells and cannot form multilayer biofilms (47, 356). However, under flowthrough conditions, which reduce cell density effects such as accumulation of autoinducers, a vpsT mutant is able to form a well-developed biofilm, suggesting that vpsT regulation of biofilm formation is more sensitive to cell density effects (25). Under the same conditions, vpsR mutants still attach to the substratum as single cells, indicating that the requirement of vpsR for vps gene transcription and biofilm formation is absolute (25). Consistent with these observations, high cell density was recently shown to repress vpsT but not vpsR gene transcription (343). Furthermore, the quorum-sensing regulator HapR was shown to directly bind to the vpsT but not the vpsR promoter, suggesting that quorum sensing reduces vpsT gene transcription at least partially as a result of direct repression by HapR (343). VpsT and VpsR positively regulate their own expression as well as that of each other (25, 47). Various expression profiling studies have shown that increased c-di-GMP levels, overexpression of DGCs, and deletion of PDEAs increase vpsT gene transcription, suggesting that vpsT, either indirectly or directly, is a downstream target of c-di-GMP signaling pathways (25, 26, 47, 164, 202, 357). This regulation may also involve quorum-sensing cues, as HapR has been shown to bind promoters of several genes encoding GGDEF proteins, suggesting that it can reduce vpsT transcription indirectly via reduction of cellular c-di-GMP levels (343). Many of these same expression profiling studies do not show a change in vpsR gene transcript levels, suggesting that transcription of vpsR may not be as highly regulated as that of vpsT. It has been hypothesized that in cases where vpsR transcript levels do increase, it is a direct result of transcriptional activation of vpsR by VpsT (26). An expression profiling study designed to determine global effects of intracellular increases in c-di-GMP concentrations on gene transcription supports this hypothesis. In this study, an increase in vpsT transcript levels was detected at 15 min after induction of c-di-GMP synthesis whereas, an increase in vpsR transcript levels was detected only after 30 min (26). It is not known whether VpsR and VpsT activate vps gene expression directly or indirectly, although a VpsR binding motif has been identified in the in the promoter of one of the gene clusters (vpsL) encoding components of the matrix exopolysaccharide, implicating direct regulation. It is also not known whether environmental signals that increase biofilm formation by increasing vps gene transcription converge at one or both of these proteins. Mutational analysis of VpsR has suggested that phosphorylation of this response regulator is likely to be required for its positive effect on biofilm formation; however, its cognate histidine kinase has not yet been discovered (187). Identification of the cognate kinases of VpsT and VpsR and their environmental activators should shed more light on how environmental signals regulate V. cholerae biofilm formation via this TCS.
FIG. 9.
Two-component signaling pathways regulating biofilm formation in V. cholerae. The main regulator of exopolysaccharide gene expression, VpsR, is a two-component response regulator. It also activates its own transcription as well as that of vpsT, which encodes the second two-component response regulator involved in biofilm formation. VpsT also activates its own transcription as well as that of vpsR. vpsT gene transcription is activated by increased c-di-GMP levels and repressed by HapR. HapR also represses transcription of a number of genes encoding GGDEF proteins. Broken lines connect the genes to their gene products.
CsgD, a transcriptional activator belonging to the FixJ subfamily of two-component response regulators, is the central regulator for both curli and cellulose production in both E. coli and Salmonella (118, 269, 363). CsgD directly activates expression of the csgBAC operon, which encodes the structural genes for synthesis of the curli fimbriae, and indirectly activates cellulose biosynthesis via increased transcription of adrA, whose gene product activates cellulose biosynthesis posttranscriptionally (118, 271, 363). CsgD also regulates the expression of BapA, a protein component of the biofilm matrix in S. enterica serovar Enteritidis (186). Thus, CsgD is a master regulator of many of the genes encoding the matrix components in these organisms. The N-terminal receiver domain of this protein contains only two of the five conserved residues required for phosphorylation of response regulators. Therefore, although the putative phosphoacceptor aspartate is present, it is not known whether CsgD is phosphorylated, and a cognate kinase has not been identified for this protein (271). Expression of csgD itself is tightly regulated by a variety of environmental signals such as nutrient starvation, oxygen tension, temperature, osmolarity, and pH, as well as proteins, including integration host factor, H-NS, the two-component response regulators OmpR and CpxR, and the stationary-phase sigma factor RpoS (101, 155; reviewed in reference 102). The long csgD promoter region harbors binding sites for many of these proteins. Thus, the fine-tuning of csgD transcription in response to environmental cues is thought to be integrated at the csgD promoter by competition of these proteins for access to their binding sites (102).
In B. subtilis, Spo0A, the two-component response regulator which is responsible for initiation of the sporulation cascade, positively regulates biofilm formation (121). Spo0A achieves this regulation by inhibiting the two transcription factors AbrB and SinR. AbrB is a global regulator of functions associated with the transition from the exponential to the stationary phase of growth, and SinR is the master switch between the sessile and motile life styles (54, 166). Both AbrB and SinR bind to the promoters of the eps and the yqxM-sipW-tasA operons, which encode proteins required for the formation of the biofilm matrix (54, 55). The eps operon contains genes responsible for production of the matrix exopolysaccharide, and the yqxM-sipW-tasA operon regulates production and secretion of TasA, one of the protein components of the matrix (38, 54). The active form of Spo0A, which is phosphorylated, inhibits SinR indirectly by activating transcription of sinI, encoding SinI, a protein that antagonizes SinR (16, 166). Joint control of sporulation and biofilm formation by Spo0A is intriguing. Although both of these responses occur as a result of nutrient depletion, the outcomes, namely, formation of a spore and formation of a matrix-enclosed community, are very different and require very specific sets of genes to be activated. One study has shed light on the mechanism of this regulation by demonstrating that the activation of sinI required lower levels of Spo0A than activation of genes encoding components of later phases of sporulation (95). The authors hypothesized that lower levels of Spo0A than is necessary for activation of other sporulation genes might be sufficient to activate transcription of sinI; thus, the biofilm represents a preparative stage in the pathway to sporulation (95). Supporting this hypothesis is the observation that biofilms of B. subtilis indeed contain fruiting-body-like formations where sporulation takes place (39).
(iii) Solitary transcriptional regulators.
While many of the transcriptional regulators of biofilm formation are part of TCSs or c-di-GMP signaling cascades, a few that are not part of these networks have been identified. In V. cholerae, CytR is a transcriptional repressor of biofilm formation (124). In E. coli, CytR represses nucleoside uptake and catabolism in nucleoside-poor environments by decreasing expression of the udp gene, involved in nucleoside catabolism. Similar regulation of udp by CytR in response to cytidine levels was observed in V. cholerae (124). Deletion of cytR in V. cholerae leads to an increase in biofilm formation and vps gene transcription (124). This study has suggested that nucleoside concentrations in the environment regulate biofilm formation and vps gene expression; however, a direct effect of nucleosides acting through CytR on biofilm formation was not demonstrated. Interestingly, a HapR binding motif has been found in the cytR promoter, suggesting that HapR may negatively regulate biofilm formation partially by increasing cytR gene expression (357).
LeuO, a transcriptional activator of the leuABCD operon, was identified as a possible regulator of biofilm formation in V. cholerae in an expression profiling study aimed at identifying genes regulating synthesis of the biofilm matrix (233). In V. cholerae, genes required for the synthesis of the matrix exopolysaccharide reside in two clusters: vpsA to -K and vpsL to Q. vpsL gene expression is reduced in ΔvpsA strains, suggesting that cells can sense a block in synthesis of the extracellular matrix (232). Based on this observation, microarray analysis was used to identify genes whose transcription pattern was similar to that of the vps genes and that therefore might be involved in biofilm formation. One of the genes identified in this comparison was leuO. Deletion of leuO resulted in greatly diminished biofilm formation but did not affect vps gene expression. This is an unusual result given that most of the signals and regulators of biofilm formation also affect vps gene expression in a similar manner and points to the exciting possibility that LeuO is an activator of the genes encoding some of the non-VPS components of the biofilm matrix (233).
In E. coli, NhaR, a regulator that activates expression of the NhaA antiporter in response to sodium stress, also regulates biofilm formation (111). nhaR mutants are severely defective in biofilm formation as a result of their inability to produce the PNAG polysaccharide. NhaR was shown to bind the promoter of the pgaABCD operon and activate the expression of these genes in response to increasing amounts of NaCl, KCl, and LiCl, as well as increasing pH. The presence of orthologs of NhaR in other Enterobacteriaceae which have loci homologous to pga predicts conservation of this type of regulation in other species as well (111).
Composition of the Biofilm Matrix
Cells that reside in multilayer biofilms synthesize a variety of molecules that make up the matrix of the biofilm. The matrix can be likened to a sponge, which gives structural integrity to the biofilm and allows the flow of small molecules into and out of the biofilm. The biofilm matrix is believed to be highly hydrated, up to 97% water by some estimates (308). Polysaccharides, proteins, DNA, surfactants, lipids, glycolipids, membrane vesicles, and ions such as Ca2+ have been shown to be present in biofilm matrices made by various bacteria under various conditions. It is plausible that under different conditions and/or at different times during the maturation of a biofilm, different components of the biofilm matrix may be of more importance to the integrity and function of the biofilm. The view of the biofilm matrix as an inert structural casing has been changing to one that is dynamic and interactive and has been the subject of some excellent recent reviews (40, 85). We focus here on some of the matrix components.
Matrix components.
The most extensively studied components of the biofilm matrices are exopolysaccharides, followed by proteins and proteinaceous components such as fimbriae and pili and eDNA. Here we focus our discussion on these molecules in a variety of bacteria.
(i) Exopolysaccharides.
Exopolysaccharides are a major component of most biofilm matrices. In most cases, in the absence of exopolysaccharide synthesis and export, bacteria can adhere to surfaces but are unable to form multilayer biofilms; in some cases, synthesis of the polysaccharide is required for surface attachment as well. Bacteria capable of forming biofilms often have distinct genetic loci dedicated to synthesis and export of the matrix polysaccharides. While the composition of these polysaccharides usually varies among different bacteria, there are also some common polysaccharides produced by multiple species of bacteria. Moreover, some bacteria are capable of producing multiple kinds of polysaccharides.
One of the most common and most extensively studied matrix exopolysaccharides is a polymer of β-1,6-N-acetyl-d-glucosamine called PGA or PNAG. Diverse bacterial species, including E. coli, S. epidermidis, S. aureus, Yersinia pestis, Actinobacillus spp., Aggregatibacter actinomycetemcomitans, and Bordetella spp., utilize this exopolysaccharide to construct their biofilm matrices (58, 65, 126, 145, 146, 162, 246, 342). The synthesis and export of β-1,6-GlcNAc is carried out by genes in three different loci: icaADBC (in staphylococcal species), pgaABCD (in E. coli and other gram-negative bacteria), or hmsHFRS (in Yersinia species). pgaC and hmsR are orthologs of icaA which encodes a glycosyltransferase necessary for catalyzing the synthesis of the N-acetylglucosamine polymers (65, 100). pgaB and hmsF are orthologs of icaB, which is responsible for deacetylation of the N-acetylglucosamine polymer (65, 336). This step is necessary to anchor the PNAG in the cell envelope in staphylococci, as PNAG is released into the medium in icaB mutants (336). In E. coli, PNAG is also primarily associated with the cell under static growth conditions; however, in this case deacetylation of this polymer is necessary for its export to the cell surface (143). icaD and icaC, which are not similar to any of the genes in the pga and hmsF loci, are not well characterized, although they have been shown to be necessary for appropriate polymer length and transport of the polymer to the cell surface (143, 242). pgaD, a homolog of hmsS, is an inner membrane protein that is required for PNAG synthesis (143). pgaA is homologous to hmsH and is thought to encode a porin-like protein which forms a pore in the outer membrane of E. coli through which PNAG is secreted (143).
In E. coli, PNAG is required for both surface attachment and formation of multilayer biofilms (342). Mutations in this locus block attachment to surfaces even after prolonged incubation. Furthermore, treatment with metaperiodate, a chemical that disrupts this polymer, results in dispersal of biofilm-associated cells singly, suggesting that this polysaccharide mediates cell-cell adhesion in addition to cell-surface adhesion (342). In many S. aureus and S. epidermidis strains, the icaADBC locus is important for indwelling medical device-related biofilm infections (87, 198). Furthermore, in S. epidermidis, this locus was shown to be required for immune evasion and virulence, underscoring the importance of biofilms in the pathogenicity of this bacterium (336). In Y. pestis, the causative agent of bubonic plaque, the hmsHFRS locus is necessary for biofilm formation in digestive tracts of fleas (151). Transfer of biofilm bacteria as a result of a flea bite is thought to be the main mode of delivery of Y. pestis into its human host; therefore, the hmsHFRS locus is an important virulence factor for this organism (65, 151).
Another exopolysaccharide that is commonly found in biofilm matrices is cellulose, a linear polymer of (1-4)-β-linked glucose. Cellulose is a major component of the biofilm matrices of some E. coli strains and of some species of Salmonella, Citrobacter, Enterobacter, and Pseudomonas as well as Agrobacterium tumefaciens (66, 216, 297, 299, 327, 362, 363). In E. coli and S. Typhimurium, the synthesis of cellulose is carried out by the proteins encoded by the bacterial cellulose synthesis operons, bcsABZC-bcsEFG (297, 363). In some strains of E. coli the presence of cellulose in the matrix appears to be necessary for biofilm, formation whereas in others it is not. For example, deletion of several of the bcs genes in an E. coli commensal strain abolished the biofilm-forming ability of this strain (66). Furthermore, incubation of biofilms made by this strain with the enzyme cellulase led to dissolution of the biofilms, whereas this treatment did not affect biofilms made by E. coli K-12, which does not contain cellulose in its biofilm matrix (66).
In addition to PGA and cellulose, some E. coli strains, such as the laboratory derivatives of the K-12 strain, can make a third kind of exopolysaccharide called colanic acid. Colanic acid is a complex branched polymer whose synthesis requires 19 genes carried in the wca locus (303). Mutants that are unable to synthesize colanic acid can attach to surfaces as a one- to two-cell-thick compact layer but are unable to build more complex multilayer biofilms (63). Overexpression of curli fimbriae, adhesive proteinaceous appendages that are part of the biofilm matrix, may partially overcome this inability to produce multilayer biofilms (255). However, even under these conditions, the presence of colanic acid results in much thicker biofilms. Many of the E. coli strains whose chromosomes have been fully sequenced harbor the genetic information for producing all three of the above-mentioned exopolysaccharides. A particularly interesting question is whether all of these polysaccharides are produced concurrently in the biofilm matrix or whether synthesis of a particular exopolysaccharide is favored under certain conditions.
Another bacterium that is capable of synthesizing multiple types of matrix exopolysaccharide is P. aeruginosa. P. aeruginosa is one of the bacterial pathogens that colonize the lungs of patients with CF. The lungs are colonized initially by the nonmucoid forms of this bacterium, which then convert to a mucoid phenotype (116). Because this conversion occurs months to years after the initial colonization, biofilm formation by both nonmucoid and mucoid strains is considered to contribute to the progress of CF pathogenesis (116). Oxidative stress as a result of immune system attack is thought to induce nonmucoid strains to become mucoid (214). The mucoid phenotype is due to the overproduction of alginate (82), a polymer of β-1-4-linked mannuronic acid and guluronic acid (82). P. aeruginosa is believed to form alginate-based biofilms in the CF lung, and this is thought to contribute to the persistence of P. aeruginosa in the CF host (129, 182, 298). For instance, patients with CF undergo multiple rounds of antibiotic treatment during the course of the disease. In comparison with biofilms made by the nonmucoid strain PAO1, an isogenic strain that has been made mucoid as a result of deregulation of the alginate synthesis genes makes a biofilm that is 1,000 times more resistant to the antibiotic tobramycin (129).
The biofilm matrices of commonly used nonmucoid lab strains such as PAO1 and PA14 are devoid of alginate (351). Two different loci that contribute to the exopolysaccharide components of the matrix in the nonmucoid P. aeruginosa strains have been identified. The pel locus (referring to pellicle, a biofilm formed at the air-medium interface), containing the genes pelA to -G, is responsible for synthesis of the glucose-rich component of the matrix, whereas the psl locus (polysaccharide synthesis locus), containing the genes pslA to -O, is responsible for the mannose- and galactose-rich component (93, 94, 149, 208, 215). Both of these polysaccharides contribute to biofilm formation by mediating both cell-cell interactions (those that are likely to be present in the pellicle) and cell-surface interactions (94, 331). The psl locus can also mediate attachment to biotic surfaces such as mucin-coated surfaces and epithelial cells, pointing to an important role for this exopolysaccharide in establishment of P. aeruginosa in the human lung (207). Genomes of species as diverse as Nitrosospira multiformis, Geobacter metallireducens, Marinobacter aqueolei, and Burkholderia cenocepecia contain loci orthologous to pel, suggesting that this polysaccharide might be widely used to construct biofilm matrices (190).
Current data suggest that the Psl and Pel polysaccharides may be synthesized concurrently by P. aeruginosa. Several studies have shown upregulation of both psl and pel genes under conditions where cellular levels of c-di-GMP are increased (114, 133). Coregulation of these genes may be evidence that cells produce both kinds of polysaccharide simultaneously in biofilms. Furthermore, composition analysis of the exopolysaccharide isolated from biofilms of nonmucoid strains shows large amounts of glucose (a component of Pel) and mannose (a component of Psl) rather than one or the other. Conversely, there is evidence that alginate and Psl/Pel production may not take place simultaneously. Exopolysaccharide prepared from nonmucoid P. aeruginosa strain PAO1 is rich in glucose, mannose, and rhamnose and devoid of mannuronic acid, indicating the absence of alginate (351). PDO300, an isogenic mucA derivative of PAO1, shows the opposite carbohydrate profile. This strain is mucoid because a mutation in the anti-σ factor mucA, which inhibits the alternative sigma factor AlgT required for alginate biosynthesis, leads to overproduction of alginate (129). Exopolysaccharide isolated from this strain is rich in mannuronic acid and is devoid of any other sugars, suggesting that syntheses of alginate and Psl/Pel components may be mutually exclusive (351). More research will be necessary to explore the inverse and direct coregulation of the various exopolysaccharide synthesis loci in P. aeruginosa. (For more comprehensive reviews on alginate and Psl/Pel exopolysaccharides, see references 261 and 277).
For many different bacteria, only one type of biofilm matrix polysaccharide has been identified. Under the conditions studied thus far, only one set of genes has been associated with synthesis of the V. cholerae biofilm-associated exopolysaccharide. These genes reside in two operons, those of vpsA (vpsA to -K) and vpsL (vpsL to -Q), and encode proteins necessary for synthesis and export of the VPS exopolysaccharide (233, 358). These genes are also required for the distinctive rough or wrinkly colonies made by the so-called rugose variants of V. cholerae. Just as activation of vps gene transcription is correlated with biofilm formation, activation of vps gene transcription is also associated with the rugose colony morphology. This colony morphology is analogous to that of the wrinkly spreader (Wsp) phenotype of some Pseudomonas species or the Rdar (rough, dry, and red) phenotype of E. coli and Salmonella species which produce increased amounts of exopolysaccharide and make robust biofilms (291, 299). One study has investigated the composition of the polysaccharide component of the biofilm matrix, while two have investigated the composition of the polysaccharide associated with rugose colonies of V. cholerae. Each study has reported different exopolysaccharide compositions Carbohydrate analysis of the biofilm matrix of a nonrugose strain of V. cholerae O139, a capsulated pathogenic serotype, showed the presence of N-acetylglucosamine, glucose, galactose, and mannose (168). Carbohydrate analysis of the rugose polysaccharide isolated from V. cholerae El Tor (strain 92A1552) showed mostly glucose and galactose and to a lesser extent N-acetylglucosamine, mannose, and xylose. The presence of almost equal amounts of 4-linked galactose and glucose suggested that the backbone of the polysaccharide may be composed of these two subunits (358). A different group has reported the composition of the rugose polysaccharide from the TSI-4 strain of V. cholerae El Tor to contain mostly mannose and N-acetylglucosamine and to a lesser extent 6-deoxygalactose and galactose (338). Because the biofilm exopolysaccharide is so tightly associated with cells, it is possible that these polysaccharides are similar if not identical by composition and that the differences observed between the rugose polysaccharides and the biofilm polysaccharide reflect contamination from other exopolysaccharides present outside the cell, such as the O antigen and, when present, the O-antigen capsule. However, it is also possible that the observed differences reflect strain-to-strain variations in polysaccharide composition resulting from differences in the activities of the VPS synthesis proteins or the activity of an additional exopolysaccharide synthesis locus within the V. cholerae genome. Distinguishing between these possibilities will require careful purification and analysis of these polysaccharides under a variety of conditions and from a variety of V. cholerae strains.
Interestingly, while the vps genes are absolutely essential for V. cholerae biofilm formation in rich medium such as Luria-Bertani broth (LB) or in minimal medium supplemented with simple sugars such as mannose, this organism can also make VPS-independent biofilms. VPS-independent biofilms are more relevant in seawater and depend on the presence of Ca2+ (169). Removal of Ca2+ from the environment results in rapid dissolution of the VPS-independent biofilm (168). Thus, Ca2+ is proposed to be an important component of the VPS-independent biofilm that stabilizes the biofilm by bridging negatively charged moieties of the O-antigen polysaccharide.
(ii) Proteins.
(a) Pili and fimbriae. Curli fimbriae are proteinaceous appendages that confer adhesive properties to bacteria. E. coli and some Salmonella species produce curli fimbriae, which constitute part of the biofilm matrix (102). Two operons, csgBAC (encoding the structural subunits of curli) and csgDEFG (encoding CsgD, the transcriptional activator of the csgBAC operon and the curli-specific transport system), are involved in the production of curli fimbriae (118). In E. coli, curli contribute to biofilm formation by mediating both cell-substratum and cell-cell contacts and thus partially relieve the requirement for exopolysaccharides in biofilm formation (255). Fimbriae have also been implicated as a component of P. aeruginosa biofilm matrices. P. aeruginosa mutants that cannot synthesize CupA fimbriae make weak pellicles that are easily disrupted, suggesting that CupA fimbriae are likely to be a structural component of the pellicles (94). Furthermore, a number of transcriptional profiling studies have shown that fimbria/pilus gene expression is upregulated in biofilms compared to planktonic cultures, further lending support to the idea that these structures may be considered proteinaceous components of the matrix (24, 76).
(b) Bap family. A group of multidomain proteins that share structural similarities have been shown to promote biofilm formation in a number of bacterial species (reviewed in reference 185). They are referred to as Bap-related proteins due to their structural and functional similarity to the S. aureus Bap (biofilm-associated protein), which was shown to be required for biofilm formation in this bacterium. These proteins are generally large (greater than 1,800 amino acids and as large as 8,800 amino acids) and have a signal sequence at their N terminus followed by domains containing a number of tandem repeats that are thought to play a role in cellular adhesion. The repeats of the Bap family proteins Esp, BapA, and LapA exhibit 23 to 33% identity to that of Bap from Staphylococcus aureus (185). Most of these proteins are thought to be anchored to the surface of the cells, loosely associated with the surface of the cells, or secreted into the medium. Thus, they are thought to hold cells in the biofilm together possibly by interacting with similar proteins on the surface or in the vicinity of neighboring cells.
Disruption of bap in S. aureus strain V329 leads to inhibition of surface accumulation and intercellular adhesion. Furthermore, staphylococcal isolates from human clinical samples harboring this gene form thicker biofilms (60). Under some conditions, the presence of Bap and other related proteins appear to eliminate the requirement for exopolysaccharides in the biofilm matrix. For example, inactivation of the icaADBC operon in S. aureus strain V329 does not result in decreased biofilm formation if Bap is present (61). Also, many Staphylococcus species do not harbor the icaADBC operon and can still form biofilms, corroborating the above finding (324). A Bap-like protein, Esp, is required for biofilm formation by Enterococcus faecalis, although some strains can form biofilms in the absence of this protein (179, 323). In P. fluorescens and P. putida, a secreted Bap-like protein, LapA, is required for biofilm formation (81, 134). Mutations in lapA impair adhesion and biofilm formation of P. fluorescens and P. putida on various abiotic and biotic surfaces. In S. Enteritidis the Bap-like protein BapA is required for biofilm formation. Moreover, expression of bapA is coordinated with production of cellulose and curli fimbriae (186). When bapA-negative and -positive cells are mixed and allowed to form biofilms, the matrix contains only the bapA-positive cells, suggesting that BapA is tightly associated with the cell surface and can interact only with BapAs on the surfaces of other cells (186).
V. cholerae has two proteins, RbmC and Bap1, which are structurally similar to the Bap family of proteins, in that they are large, secreted proteins containing a number of tandem repeats. Bap1 and RbmC share 47% sequence similarity; however, they show no sequence similarity to proteins of the Bap family. Both of these proteins have been shown to be secreted and are likely to be protein components of the biofilm matrix (90). The tandem repeats show similarity to FG-GAP domains found in integrins, eukaryotic cell surface receptors which mediate cell-cell and cell-extracellular matrix interactions (138, 233). Expression of both rbmC and bap1 genes is coregulated with the vps genes, which is consistent with their proposed role as components of V. cholerae biofilm matrix (164, 233, 357). In V. cholerae El Tor A1552, deletion of neither rbmC nor bap1 leads to significant alterations in biofilm formation; however, deletion of both of these genes abolishes biofilm formation, suggesting a level of redundancy in their function (90). In contrast, deletion of bap1 from V. cholerae O139 leads to a significant reduction in biofilm formation (233). The differences in these results may reflect slight differences in regulation of biofilms in various serotypes of V. cholerae. Nevertheless, the involvement of RbmC and Bap1 proteins in biofilm formation marks cell-surface proteins or secreted proteins as important components of the biofilm matrix.
(c) Lectins and sugar binding proteins. Proteins that recognize carbohydrate moieties, known as lectins, can facilitate cell-matrix or cell-cell interactions by binding polysaccharide components of the matrix or sugar moieties on the surfaces of other cells, respectively. For example, in P. aeruginosa, the two lectins LecA and LecB have been implicated in biofilm formation. LecA, which is specific for d-galactose and its derivatives, was shown to be present in the biofilm matrix by immunoblot analysis and fluorescence microscopy of cells expressing translational fusions of lecA and the enhanced green fluorescent protein gene (74). Furthermore, incubation of preformed biofilms with isopropyl-β-d-thiogalactoside (IPTG), a galactoside with a strong affinity to LecA, led to dispersal of the biofilms. LecB, which is specific for l-fucose and its derivatives, was also shown to be required for biofilm formation (320). This protein, which is associated with the outer membrane, binds to the surface of biofilm cells as a result of its interaction with fucose-containing ligands, suggesting that it promotes cell-cell interactions (320). Consistent with these results, a recent expression profiling study showed that both lecA and lecB were induced in biofilms of P. aeruginosa (339). In the dental pathogen S. mutans, glucan (a glucose polymer) promotes cell aggregation and adhesion to dental surfaces. Four glucan binding proteins, GbpA to -D, which are either secreted or cell wall anchored, contribute to the biomass and architecture of biofilms formed by this bacterium (206). Of these four proteins, GbpC is the most important in mediating bacterium-polysaccharide interactions, since its loss leads to a significant reduction in biofilm biomass and cell aggregation (206). Biofilm formation by another dental pathogen, Eikenella corrodens, is also partially dependent on lectins, as strains that synthesize increased amounts of lectins demonstrate increased biofilm association (14). Biofilm formation in this bacterium is inhibited by the presence of N-acetyl-d-galactosamine in the growth medium, suggesting that the lectin responsible for biofilm formation binds this sugar moiety. In V. cholerae, RbmA, a secreted protein, is required for rugosity as well as robustness of V. cholerae El Tor A1552 biofilms. Biofilms formed by ΔrbmA mutants are quickly destroyed by SDS, whereas wild-type V. cholerae biofilms are able to withstand SDS treatment for longer periods of time. Structural analysis of this protein has suggested that it could potentially bind the polysaccharide component of the matrix, thus strengthening the biofilm (88). rbmA and rbmC (mentioned above) are part of a group of genes that reside between the two vps operons and that are coregulated with the vps genes, further supporting the possibility that these encode protein components of the biofilm matrix.
(d) Autotransporters. Autotransporter are proteins that are able to transport themselves to the cell surface without the need for other transport systems (106). The self-associating autotransporter subfamily of these proteins are capable of interacting with themselves or with other members of the family, thus mediating cell-cell interactions and leading to cell aggregation (177). Three glycoproteins in this family, Ag43, AIDA, and TibA, have been shown to promote biofilm formation in various toxigenic and nontoxigenic E. coli strains (62, 174, 287, 288). These proteins could potentially serve to maintain close-range interactions between some cells of the biofilm. Interestingly, the presence of fimbriae on the cell surface abolishes the intercellular interactions mediated by these proteins, possibly due to spatial constraints (122, 288). This finding suggests that bacterial adhesins may function in mutually exclusive manners.
(iii) DNA.
In addition to the exopolysaccharides and proteins, eDNA is also an important constituent of the biofilm matrix in a number of bacterial species. For example, the biofilm matrix in P. aeruginosa contains significant amounts of DNA, which are necessary for biofilm integrity (215, 350). Furthermore, addition of DNase to the culture medium inhibits biofilm formation by this organism and dissolves preformed biofilms (350). Mature biofilms formed by clinical isolates of P. aeruginosa are also dissolved by DNase treatment, corroborating the importance of eDNA in these biofilms (237). As shown by a recent study, DNA is present on the biofilm substratum in grid-like patterns (Fig. 10). It is also present on the surfaces of microcolonies of young (2-day-old) biofilms, on the stalks of the mushroom-like structures of 4-day-old biofilms, and throughout in 6-day-old biofilms. This pattern has lead to the speculation that DNA on the substratum could initially serve as a grid that allows bacteria to move using type IV pili, which in P. aeruginosa have been shown to bind DNA (329). The DNA on the stalks would then allow bacteria to “climb” on top and form the caps of the mushroom-like structures (5). The source of the eDNA, whose composition is similar to that of genomic DNA, is speculated to be a result of whole-cell lysis or secretion of outer membrane vesicles containing DNA into the biofilm matrix (5).
FIG. 10.
eDNA in the biofilm matrix. Horizontal optical sections in a 2-day-old biofilm formed by green fluorescent protein-tagged P. aeruginosa stained with the DNA binding dye DDAO [7-hydroxy-9H-(1,3-dichloro-9,9-dimethyl acridin-2-one)] are shown. The images show the green fluorescent bacteria (A), the red fluorescent eDNA (B), and an overlay of the two (C). (Reprinted from reference 5 with permission of Blackwell Publishing Ltd.)
Release of genomic DNA as a result of lysis of a population of cells appears to be the most likely source for eDNA. For example, in E. faecalis, chromosomal DNA is the source of biofilm matrix eDNA. This chromosomal DNA is released as a result of autolysis of a portion of cells in the biofilm (317). The extracellular protease GelE is responsible for this autolysis. Autolysis is inhibited by another extracellular protease, SprE. Thus, in this bacterium, release of eDNA is tightly regulated by the opposing effects of the two proteases GelE and SprE (317). In S. epidermidis, eDNA is released in to the biofilm matrix through cell lysis mediated by AtlE, the major autolysin involved in cell wall turnover, cell division, and cell lysis in this organism (28, 259). Likewise, eDNA is found in S. aureus biofilms and contributes to the strength of the biofilm matrix (266). The source of this DNA was shown to be chromosomal DNA by quantitative real-time PCR of four randomly selected genes found on the S. aureus chromosome. Furthermore, a cidA mutant, which exhibits decreased cell lysis as a result of loss of murein hydrolase activity, makes defective biofilms that also contain less eDNA. Therefore, cell lysis and subsequent release of genomic DNA may be a common mechanism for introduction of DNA into biofilm matrices.
The presence of multiple types of molecules such as polysaccharides, DNA, and proteins, in the biofilm matrix raises the question of the roles that these different matrices play in the health of the biofilm and whether these roles are conserved in different types of bacteria. This question has recently begun to be investigated. The results so far suggest that similar molecules may play different roles in different organisms. For example, both DNase I and dispersin B, an enzyme that degrades PNAG, inhibit biofilm formation in S. epidermidis and S. aureus; however, DNase I leads to dispersal of preformed S. aureus biofilms but not S. epidermidis biofilms. In contrast, dispersin B is able to disperse preformed S. epidermidis biofilms but not S. aureus biofilms (145). Consistent with these results, S. epidermidis biofilms are sensitized to killing by the cationic detergent cetylpyridinium chloride as a result of dispersin B treatment, whereas S. aureus biofilms are sensitized to killing by the same agent as a result of DNase I treatment. These results suggest that the PNAG polysaccharide and eDNA make different contributions to the integrity of the biofilm matrix in these two species.
Biofilm Architecture
The architecture of biofilms is influenced by both physical conditions, such as the flow rate of the medium that bathes the biofilm, and biological factors (257). Here we will focus our discussion on biological rather than hydrodynamic effects on biofilm architecture. In terms of architecture, biofilms can be divided into two main classes: (i) those that show an irregular topology characterized by mushroom-like structures separated by voids (which are most likely water channels) and low surface coverage and (ii) those that show a flat topology characterized by sheet-like compact layers and high surface coverage (Fig. 11). The main biological determinants of biofilm architecture are medium composition (particularly the carbon source in the medium), presence of surfactants, various types of motility (flagellar, twitching, and swarming), and quorum-sensing effects.
FIG. 11.
Flat versus irregular topology. Flow cell biofilms of green fluorescent protein-tagged P. aeruginosa grown with different carbon sources are shown. These images were acquired 48 h after inoculation of the system. Biofilms grown with succinate have a flat topology characterized by high surface coverage and low height; those grown with glucose have an irregular topology characterized by low surface coverage and dispersed pillars of bacteria. SV, side view (xz plane); TD, top-down view (xy plane). Gridlines are spaced 20.1 μm apart. (Reprinted from reference 289 with permission of Blackwell Publishing Ltd.)
For example, P. aeruginosa PAO1 makes flat and compact biofilms in flow chambers when grown on citrate, benzoate, and Casamino Acids as carbon sources, whereas it makes irregular biofilms with the typical mushroom-shaped structures when glucose is used as a carbon source (132, 175, 304). The effect of carbon sources on biofilm architecture can be different even for closely related species. Unlike P. aeruginosa, P. putida makes irregular biofilms in citrate medium (132).
Twitching motility using type IV pili is the most important factor in forming the flat biofilms in P. aeruginosa, since a type IV pilus mutant (ΔpilA) cannot not make flat biofilms (176). Thus, flat biofilms may result from bacteria spreading on the substratum using twitching motility. Flagellar motility is also implicated in formation of flat biofilms, as flagellar mutants of P. aeruginosa PAO1 form more irregular biofilms when grown on citrate-containing medium than wild-type P. aeruginosa PAO1 (176).
Quorum sensing also affects biofilm architecture. P. aeruginosa lasI mutants, which are defective in the synthesis of the autoinducer 3OC12-HSL, make thin and densely packed biofilms devoid of water channels and mushroom-like macrocolonies (69). This defect can be reversed by adding 3OC12-HSL to the culture medium. The second acyl-HSL signal does not appear to be involved in determining the architectural characteristics of the biofilm, because rhlI mutant biofilms are indistinguishable from those made by wild-type bacteria.
As discussed above, it has been well established that nutritional cues, motility, and quorum sensing all have significant effects on biofilm architecture. However, how these effects are linked is still not well understood. A recent study of P. aeruginosa demonstrated that changes in surface motility can be induced by different carbon sources and that these changes can account for differences in biofilm architecture. The same study also showed that carbon source dependence can be regulated by quorum-sensing cues (289). In that study, wild-type P. aeruginosa grown in medium with succinate as the carbon source made flat biofilms, whereas that grown in medium supplemented with glucose made biofilms that had irregular topology. Computer modeling predicted that increases and decreases in surface motility would lead to the flat topology and irregular topologies, respectively. Microscopic tracking and motility plates showed that succinate resulted in high swarming motility whereas glucose resulted in low motility, confirming the predictions of the computer model. In contrast, quorum-sensing mutants made irregular biofilms and exhibited poor swarming behavior in medium supplemented with either succinate or glucose. Thus, quorum sensing appears to regulate swarming motility in response to succinate in the growth medium; however, the mechanistic details of this effect are currently not elucidated (289). Interestingly, another carbon source, glutamate, led to high surface motility as well as formation of biofilms with flat topology even in the absence of active quorum-sensing circuits (289). Clearly, there are multiple mechanisms regulating surface motility and its impact on biofilm architecture.
Rhamnolipids, which are surfactants produced by P. aeruginosa at high cell density, affect biofilm architecture as well. Rhamnolipid production is regulated by quorum-sensing cues by the RhlI-RhlR system, which itself is under the control of LasI-LasR system (241). One study showed that an rhlA P. aeruginosa PAO1 mutant lacking the enzyme rhamnosyltransferase required for synthesis of rhamnolipids, has no defect in surface attachment and the early stages of biofilm formation (the first 4 to 5 days). In fact, rhamnolipids may have an inhibitory effect on early stages of biofilm formation, since addition of purified rhamnolipids can reduce biofilm formation by disrupting both cell-surface and cell-cell interactions (67).
In addition, transcription of rhlA is first detected in 2-day-old biofilms, suggesting that rhamnolipids are not utilized in early stages of biofilm formation. In agreement with this, more mature 6-day-old biofilms made by the rhlA mutant have flat topology (67). A later study showed that the expression of rhlA in P. aeruginosa PAO1 is confined to the stalks of the mushroom-like structures in more mature biofilms (3 to 5 days old) (194). Those authors suggested that the rhamnolipids produced by the cells forming the stalk “grease” the stalks to make it easier for other cells to climb up the stalks using type IV pili.
While many studies of the biological factors affecting biofilm architecture have been conducted with P. aeruginosa, a few experiments with other species have demonstrated that similar biological factors such as quorum sensing and medium composition affect biofilm architecture. For example, cell density-based signals have a significant effect on E. coli biofilm architecture. In this organism, the autoinducer AI-2 is actively imported into the cell via an ABC-type transporter whose components are encoded by the lsr operon. It is then phosphorylated by the LsrK kinase and in its phosphorylated form is thought to bind LsrR, the repressor of the lsr operon, leading to its derepression (312, 352). Therefore, lsrK mutants cannot import the autoinducer AI-2 because they cannot derepress the lsr operon, and lsrR mutants import large amounts of AI-2. lsrK and lsrR mutants both make biofilms that have flat topology, suggesting that functional quorum-sensing circuits are important for regulating biofilm architecture in E. coli (199). V. cholerae can form biofilms in a variety of aquatic environments, and the type of environment affects the architecture of the biofilms. For example, V. cholerae biofilms made in freshwater have an irregular topology, showing the typical mushroom-like macrocolonies and voids suggestive of water channels, whereas those formed in seawater are flatter, cover more surface area, and do not become as thick (169). These observations suggest once again that the environment is a significant factor in determining the structural features of biofilms.
DISPERSION OF BIOFILMS
It is widely accepted that the biofilm mode of life is advantageous for microorganisms under a variety of conditions. However, as biofilms grow in size, cells that reside in the innermost layers of the biofilm may not have access to nutrients or may suffer from accumulation of toxic waste products; therefore, their microenvironment can become unfavorable. Furthermore, if environmental conditions change, residence in a biofilm may become a liability. In either of these cases, bacteria must be able to detect and respond to the unfavorable environmental conditions by returning to the planktonic mode of existence. Thus, one would predict that biofilm dispersal should be a highly regulated process involving many sensory circuits. Passive dispersal of biofilms as a result of hydrodynamic parameters such as shear stress is a distinct process that will not be part of our discussion (49, 305, 306). In this section, we review signals, regulatory networks, and mechanisms that result in active dispersal of bacteria from biofilms.
Signals and Regulatory Networks
Nutritional cues.
The nutritional status of the environment most often dictates bacterial behavior, and the biofilm dispersal response is no exception. Indeed, both decreases and increases in environmental nutrients can lead to biofilm dispersal. For example, 4-day-old P. putida biofilms formed in flow chambers can dissolve within 15 min once the flow is turned off, suggesting that nutrient limitation rapidly leads to biofilm dissolution (108). The same phenomenon is observed if the carbon source citrate is removed from the flow medium, suggesting that carbon starvation induces dissolution of P. putida biofilms. A flagellar mutant also shows this rapid dissolution in response to carbon starvation, suggesting that this behavior does not depend on flagellar motility.
For some bacteria, an increase in environmental nutrients induces biofilm dispersal. An increase in the concentration of various carbon and/or nitrogen sources such as glutamate, succinate, citrate, glucose, and ammonium chloride in minimal medium leads to P. aeruginosa biofilm dissolution (282). Interestingly, a similar response is observed with P. putida, indicating that perhaps biofilm formation is advantageous only within a window of nutrient concentrations. The P. aeruginosa biofilm dispersion response is correlated with the loss of type IV pilus gene transcription and the onset of flagellar gene transcription. Thus, these bacteria repress twitching motility and activate flagellar motility to escape from a biofilm. Differences in the phosphoproteomes of the dispersed cells and biofilm cells have been noted. Moreover, inhibition of protein phosphorylation inhibits glutamate-induced dispersal, corroborating the results of the proteomic analysis (282). Because protein phosphorylation is utilized mainly by TCSs these results suggest that TCSs are important in biofilm dispersal.
Oxygen depletion and nitric oxide.
Studies using microelectrodes have shown that oxygen penetration into the biofilm core decreases with increasing biofilm thickness due to consumption of oxygen by the biofilm-based bacteria closest to the environmental interface (10, 340). Thus, bacteria residing at various layers of biofilms experience different in oxygen tensions. In fact, those bacteria in the deepest layers of the biofilm may require anaerobic metabolism for their survival. Both oxygen depletion and the by-products of anaerobic metabolism have been shown to induce biofilm dispersal. In Shewanella oneidensis, a sudden drop in molecular oxygen levels in the bulk medium leads to rapid detachment of cells from biofilms in a motility-independent manner. The dispersal response is reduced in mutants with mutations in genes encoding several transcriptional regulators, such as ArcA, CRP, and Etr, which are known to mediate responses to changing oxygen levels in other bacteria (319). While the identification of transcriptional regulators associated with biofilm dispersal in response to oxygen depletion suggests that oxygen-sensing circuits are at least partially responsible for the dispersal event, how these circuits mediate or coordinate the detachment response is currently not known.
Onset of anaerobic respiration in P. aeruginosa biofilms has also been demonstrated (18, 282, 359). Anaerobic respiration can result in production of reactive nitrogen intermediates, which in high doses can damage DNA, proteins, and lipids. This type of nitrosative stress can lead to dispersal of mature P. aeruginosa biofilms (18). In particular, nitric oxide (NO) or reactive species resulting from NO can cause dispersal events. The presence of ONOO− (peroxynitrite, produced from reaction of NO and O2) has been demonstrated inside macrocolonies of mature 7-day-old biofilms by the use of fluorescent dyes. Although NO was not detected inside macrocolonies, low doses of NO delivered to the biofilms using an NO donor (sodium nitroprusside) were shown to induce dispersal. Moreover, a ΔnirS mutant (a nitrite reductase mutant), which is unable to produce NO, forms biofilms that fail to disperse, whereas a ΔnorCB (NO reductase) mutant, which produces large amounts of NO, shows enhanced biofilm dispersal (18). In that study, increased dispersal was also correlated with increased cell lysis and the appearance of hollow voids inside macrocolonies.
c-di-GMP.
As we have seen in the previous section, c-di-GMP plays a significant role in regulating biofilm formation; therefore, it is not surprising that this molecule mediates dispersal events as well. For example, in P. putida, a genetic screen performed to determine the genetic basis of the starvation-induced dispersal response described above has identified a gene, PP0164, which contributes to the dispersal phenotype. Biofilms formed by a PP0164 deletion mutant do not disperse as a result of carbon starvation. PP0164, which encodes a putative periplasmic protein, is in an operon with another gene, PP0165, encoding a putative transmembrane protein with cytoplasmic GGDEF and EAL domains. In contrast, deletion of PP0165 blocks irreversible attachment to substrata (108). Thus, these two genes regulate P. putida biofilm formation and dissolution through c-di-GMP signaling; however, the exact mechanism of this regulation has not yet been elucidated. A subsequent study, again with P. putida, has shown that overexpression of an unrelated GGDEF domain protein (VCA0956 of V. cholerae) leads to biofilms that cannot dissolve in response to carbon-induced starvation, whereas overexpression of an EAL domain protein (YhjH of E. coli) leads to the rapid dissolution of established biofilms in this organism (109). A similar effect of E. coli YhjH has also been seen in S. oneidensis biofilms, where activation of transcription of the yhjH gene within established biofilms leads to rapid dispersal (318). Finally, nutrient-induced dispersion of P. aeruginosa biofilms is dependent on c-di-GMP signaling via a chemosensory regulator called BdlA (234). In a bldA mutant, dispersal is not observed in response to nutrient signals. Furthermore it has been noted that the intracellular c-di-GMP level of a bdlA mutant is five- to sixfold higher than that of wild-type P. aeruginosa (234). BdlA is a putative cytosolic methyl-accepting chemotaxis protein with two PAS domains, which are sensory domains that can detect light, oxygen, and redox potential. While it is unlikely that BldA directly degrades c-di-GMP, it is hypothesized that environmental changes detected by the PAS domains of BdlA could initiate a chemosensory signaling cascade which ultimately activates a phosphodiesterase. The components of this putative signaling cascade are not yet known, and the mechanism by which c-di-GMP levels lead to dispersal have not yet been elucidated. Given the abundance of proteins that regulate biofilm formation via c-di-GMP signaling, it seems likely that c-di-GMP will play a central role in dispersal of biofilms made by diverse bacteria. It will be interesting to see whether this will be the case as more studies of biofilm dispersal are conducted.
Quorum sensing.
Quorum-sensing systems play a role in dispersal of biofilms made by a number of bacterial species (31, 205). In S. aureus, the agr quorum-sensing system is activated in established biofilms and required for subsequent dispersal of the biofilm (31). Furthermore, addition of AIPs to an S. aureus biofilm results in dispersal (Fig. 12). Agr-mediated dispersal is partially dependent on production of extracellular proteases, including the agr-regulated Aur metalloprotease and the splABCDEF encoded serine proteases (31). In these strains, deletion of the ica locus does not have an effect on biofilm formation. Taken together, these results suggest that the biofilm matrix in these strains is composed primarily of proteinaceous material rather than exopolysaccharides. Agr-regulated detergent-like molecules called phenol-soluble modulins have also been implicated in detachment from biofilms made by S. epidermidis (354).
FIG. 12.
Quorum-sensing regulation of dispersal. Confocal scanning laser microscopy reconstructions of S. aureus biofilms are shown. The growth medium was supplemented on day 3 with AIP-1 (A) or AIP-1 and the serine protease inhibitor phenylmethylsulfonyl fluoride (PMSF) as indicated (B to D). aur, aureolysin. (Reprinted from reference 31 with permission.)
Mechanisms of Dispersal
While the studies described above shed light on the signals and signaling networks that lead to dispersal of biofilms, they do not provide a mechanism for this effect. Although the exact mechanistic details of dispersal have not been elucidated for any organism, biofilm researchers are beginning to gain insight into the events that take place during dispersal. These include synthesis of enzymes that degrade adhesins such as the biofilm matrix, the return of motility, surfactant production, and cell lysis. These events are discussed in detail below.
Degradation of the biofilm matrix.
Biofilm formation usually involves the production of an extracellular matrix which allows cells to adhere to each other and/or to a surface. One strategy for escape from a biofilm, therefore, is degradation of this matrix. A number of species have indeed been shown to secrete degradative enzymes with specificity for matrix components. For example, alginate, the biofilm exopolysaccharide of mucoid P. aeruginosa strains, can be degraded by alginate lyase, which is encoded by a gene in the alginate biosynthesis gene cluster. In model systems designed to measure detachment of P. aeruginosa cells from an agar surface, increased expression of alginate lyase was shown to accelerate detachment (34). S. mutans, an initiator of dental plaque, attaches to the surfaces of teeth via its cell surface adhesin P1 using a salivary receptor agglutinin (35). Degradation of P1 by exogenous addition of a surface protein-releasing enzyme (SPRE) leads to detachment of an S. mutants monolayer formed on saliva-conditioned epon-hydroxylapatite rods (332). The importance of SPRE in dispersal is underscored by the fact that SPRE-defective mutants of S. mutans are unable to detach from substrata. As mentioned above, extracellular proteases are involved in biofilm dispersal of some strains of S. aureus, suggesting that degradation of matrix proteins is important for dispersal (31). In Xanthomonas campestris pathovar campestris, biofilms can be dispersed by the enzyme ManA, an endo-β-(1,4)-mannanase, which is encoded in the X. campestris genome (79). Interestingly, a biofilm-specific substrate for this enzyme has not yet been discovered. Xanthan, the exopolysaccharide that is required for biofilm formation and cell aggregation in X. campestris, is not degraded by ManA (79). Production of ManA is regulated by the diffusible signal factor (DSF), a fatty acid signal synthesized and detected by proteins encoded by the rpf genes (59). The exact details of this regulation have not been elucidated; however, it is likely to involve c-di-GMP signaling, as RpfG, the response regulator of the Rpf sensory circuit, is a phosphodiesterase containing an HD-GYP domain (275). Recently, cis-2-decanoic acid, a DSF-like molecule produced by P. aeruginosa, was shown to induce dispersion not only of P. aeruginosa biofilms but also of those formed by a variety of gram-negative and gram-positive bacteria (68). Therefore, induction of biofilm dispersal by fatty acid signals may be a commonly used mechanism.
DspB (also called dispersin B), a β-hexosaminidase, can hydrolyze the glycosidic linkages of PNAG, the homopolymeric exopolysaccharide found in biofilm matrices of E. coli and a number of other bacteria, as discussed previously (144). Biofilm formation by several strains of E. coli, as well as S. epidermidis, S. aureus, P. fluorescens, various Bordetella species, and Y. pestis, can be inhibited completely in the presence of DspB (144, 161, 246). This enzyme was identified first in the human periodontopathogen Actinobacillus actinomycetemcomitans and later in Actinobacillus pleuropneumoniae, a porcine respiratory pathogen (160, 161). Both of these species contain PNAG as part of their biofilm matrix material, which can be degraded by DspB, leading to the dispersal of the biofilms made by these bacteria (161). Interestingly, DspB homologs are found only in Actinobacillus species (144). While mixed-species biofilms are likely to be the norm in nature and enzymes produced by one species can degrade matrix material produced by others, many of the bacteria that contain PNAG in their matrices are unlikely to be found in the same environmental niches together with bacteria that contain dispersin B. To explain this paradox, it has been hypothesized that another, as-yet-unidentified protein possessing PNAG hydrolase activity is encoded within the genomes of species that synthesize PNAG or cohabit with species that synthesize PNAG but whose genomes do not possess a dspB homolog (144).
Induction of motility.
Because biofilm formation by motile bacteria often coincides with cessation of flagellar motility, it is not surprising that onset of dispersal has been shown to coincide with the return of motility in a number of studies. For example, induction of CsrA in E. coli biofilms leads to biofilm dispersal (148). This protein is a positive regulator of flhDC, the master operon for flagellar biosynthesis. Therefore, synthesis of flagella and onset of motility are likely to be necessary for biofilm dispersal in this organism (348). Another study has shown that mature P. aeruginosa biofilms formed in flow cells go through architectural modifications which result in central hollowing of the mushroom-like pillars of the biofilm (Fig. 13A) (281). These central voids, which are open to the bulk fluid, are formed through evacuation of cells from the biofilm pillars by flagellar motility. The cells left behind in the outer shell remain nonmotile. In a subsequent study of P. aeruginosa biofilms, a similar gain in motility followed by hollowing of macrocolonies was observed (Fig. 13B) (258). This phenomenon was termed “seeding dispersal.” That study also showed that seeding dispersal is initiated once macrocolonies reach a certain size (∼80 μm in diameter). Thus, under the conditions of those experiments, biofilm residence became unfavorable when a certain macrocolony size was exceeded. Furthermore, quorum sensing was found to be important for seeding dispersal, as ΔlasI ΔrhlI mutants, which are unable to produce either of the acyl-HSL autoinducers, did not show this phenotype. In that study, rhamnolipids did not appear to contribute to the seeding dispersal; however, they were important in maintaining hollow pillars within biofilms, as these pillars collapsed into flat, homogeneous structures in ΔrhlA mutant biofilms. A clinical CF isolate, FRD1, did not show seeding dispersal, suggesting that a bacterial species could have multiple strategies for dispersal (or that pathogenesis depends on overriding dispersal mechanisms in CF) (258). Whether active swimming of cells from the biofilms in the absence of other dispersive processes is a common mechanism of dispersal remains to be seen. As the authors of the first study (281) point out, the ability of motile cells to swim away from the voids created inside macrocolonies indicates the absence of dense matrix material that would hinder flagellar movement. One could argue that the matrix material is degraded prior to the onset of motility; however, evidence for this has so far been lacking.
FIG. 13.
Induction of motility. (A) Cells have evacuated the pillars, leaving a hollow interior (arrow). (Reprinted from reference 281 with permission.) (B) Motile cells inside a wall of stationary cells. The white arrow points to the inside of the pillars where cells have gained motility. Motility is indicated by the blur in the image. The black arrow points to the “walls” of the pillars that are formed by cells that show no motility (see also the first movie in the supplemental material of reference 258). (Reprinted from reference 258 with permission of the publisher.)
Production of surfactants.
Rhamnolipids, surfactants produced by P. aeruginosa, can cause dispersal of biofilms formed by this organism (32). Biofilms formed by a strain with increased rhamnolipid production dispersed after 2 days, whereas wild-type biofilms formed under the same conditions did not disperse until day 10. Dispersal in this case was also preceded by hollowing of the centers of the macrocolonies as described above. Furthermore, exogenous addition of purified rhamnolipids to biofilms could induce central hollowing of the macrocolonies, suggesting that these molecules may act as external signals inducing biofilm dispersal, similar to the situation described above with DSF-type molecules. Another surfactant, SDS, also caused central hollowing, indicating that biofilm dispersal can be caused by many different types of surfactants. It was proposed that induction of dispersal could be related to changes in cell surface properties and decreasing adhesiveness within biofilms (32).
Cell death and cell lysis.
Several studies have shown that biofilm dispersal is preceded or accompanied by lysis of a subpopulation of cells within macrocolonies of mature biofilms. Viability staining of 10-day-old flow cell biofilms of P. aeruginosa showed extensive cell death as well as cell lysis in the centers of macrocolonies (347). Furthermore, the cause of cell lysis appeared to be infection with the Pf1 prophage, a filamentous lysogen of P. aeruginosa. Because lysogenic phages rarely cause lysis of their host, the lytic nature of this phage has prompted those authors to hypothesize that these phages arise as a result of increased mutation rates in the cells residing in the centers of macrocolonies in mature biofilms. In their model, nutrient limitation and accumulation of reactive oxygen species in the centers of macrocolonies would lead to the SOS response. This would result in adaptive mutations, which are means of creating genetic variability in times of stress, thereby maximizing chances of survival (219, 347). Adaptive mutations could potentially result in the lysogenic phage becoming lytic and hyperinfectious, leading to the cell lysis phenotype. Accumulation of reactive oxygen species in the center of the macrocolonies was indeed demonstrated in that study, supporting the authors' model. It is not clear how induction of lytic phages within biofilms can lead to dispersal of healthy and active cells from the biofilm. Nevertheless, the idea of phages contributing to cell lysis in biofilms is intriguing and could lead to development of novel therapies for use in biofilm-associated infections. In fact, the use of phages to combat biofilm infections has been the subject of a recent review (15).
In another study, extensive cell death could be observed in the center of macrocolonies after 48 h of biofilm development by the marine bacterium Pseudoalteromonas tunicata (210). Cell lysis was followed by detachment of the biofilm from the substrata (Fig. 14). Cell lysis was dependent on the presence of the protein AlpP, an extracellular protein that has antibacterial activity against a number of gram-negative and gram-positive bacteria, since biofilms formed by ΔalpP mutants do not exhibit this phenotype. AlpP appears to serve a dual role for P. tunicata as an inhibitor of other organisms and as an autotoxic protein which leads to lysis of the P. tunicata cells when necessary. The authors hypothesized that lysis of a subpopulation of cells precedes dispersal events to provide nutrients and energy to the remaining live cells, which would need this energy to disperse and colonize new surroundings. In a subsequent study by the same group, dispersal was indeed shown to occur from wild-type biofilms after 8 days; however, ΔalpP biofilms showed very little dispersal (211).
FIG. 14.
Cell death and cell lysis in biofilm dispersal, showing biofilm development and cell death of the P. tunicata wild-type strain. Biofilms were stained with the BacLight Live/Dead bacterial viability kit. Red propidium iodide-stained cells have a compromised cell membrane and are dead. Time points after inoculation are shown as follows: (C) 48 h; (D) 72 h; (E) 144 h; (F) 168 h. Cell death can be observed at 48 h, and cell lysis (arrow in panel D) and extensive cell death (arrow in panel E) are seen at 144 h, prior to complete dispersal of the biofilm at 168 h. Bars, 50 μm. (Reprinted from reference 210 with permission.)
While these experiments have established that biofilm dispersal is accompanied in at least some cases by cell death and cell lysis, the mechanism by which cell death and lysis occurs has been poorly understood. Recently, studies of cid/lrg systems have revealed a possible mechanism for these events (for extensive reviews on these systems and their potential role in biofilm development, see references 19 and 265). The cidABC and lrgAB operons regulate murein hydrolases, which cleave peptidoglycan and are therefore necessary for processes such as cell growth, cell division, and cell lysis. The cidA and lrgA genes encode proteins that are thought to function similarly to bacteriophage holins and antiholins, respectively. Holins and antiholins work together to control the timing of bacteriophage-induced host cell lysis by regulating access of murein hydrolases to peptidoglycan. Holins and CidA are positive regulators of murein hydrolase activity, whereas antiholins, as the name implies, and LrgA are negative regulators of murein hydrolase activity. A link between regulation of biofilm development and cid/lrg systems was described earlier in this review (266). While that study did not did not address dispersal events, it is tempting to speculate that cid/lrg systems may control the cell lysis events discussed in this section. These systems are conserved in a wide variety of bacteria, such as Pseudomonas spp., Vibrio spp., Staphylococcus spp., and E. coli, to name a few; therefore, cid/lrg systems could potentially be utilized by many different bacterial species to regulate cell lysis events accompanying biofilm dispersal (19). Furthermore, several regulatory inputs control transcription of the cidABC and lrgAB operons in S. aureus. For example, the LytSR TCS regulates the expression of the lrgAB operon in response to membrane potential, and the CidR protein positively regulates the transcription of both the cidABC and lrgAB operons in response to increased levels of acetic acid (19). It is plausible, therefore, that outputs of signals and signaling networks discussed above can converge at these operons to regulate cell lysis in response to a variety of environmental stimuli.
IS THERE A BIOFILM FINGERPRINT?
One of the interesting questions about biofilm development has been whether or not biofilms have distinct fingerprints, which we define here as a set of physiological and genetic parameters common to all biofilms. Numerous transcriptomic and proteomic studies of a variety of species have attempted to address this issue. Studies have also analyzed transcriptomes and proteomes of biofilm-associated cells over a period of time to trace the temporal changes in gene transcription and expression. These studies have shed some light on some common trends and highlighted important phenotypic characteristics of biofilms, as addressed by several recent reviews (9, 23). These trends include repression of flagellar gene expression, upregulation of matrix synthesis gene expression, and upregulation of genes involved in adaptation to stationary phase, environmental stress, and anaerobiosis (9, 23).
The stationary-phase character of biofilm cells is consistent with previous studies that have implicated both slow growth and high numbers of persister cells in the resistance of biofilm bacteria to some antibiotics (12, 300, 315; see references 11, 96, 195, and 295 for reviews on tolerance of biofilms to antimicrobials). Persisters are nondividing, multidrug-resistant cells that are genotypically identical to the rest of the bacterial population (reviewed in references 195 and 196). While persister cells are present in all bacterial cultures, their numbers are low during log phase and increase in stationary phase and in biofilms. Persisters are cells that are thought to have a different transcriptional program than other cells in the culture which protects them against the effects of antimicrobials. Stationary-phase cells have in fact been shown to be even more tolerant than biofilm cells to some antibiotics (300). Therefore, some researchers have questioned whether biofilms are merely immobilized stationary-phase cultures with few characteristics that are not found in stationary-phase cultures. This issue has been addressed in a number of recent studies. One study compared the proteome of Bacillus cereus grown under biofilm conditions with those of logarithmic- and stationary-phase planktonic cultures using principal-component analysis (335). The results of the analysis showed that the biofilm proteome was different from the stationary-phase proteome, arguing that biofilms are distinct from stationary-phase populations. Another study compared expression profiles of young and old P. aeruginosa biofilms with those of planktonic and stationary-phase cultures using cluster analysis (339). Similarly, this study also identified distinct expression signatures for biofilm-associated cells which were different from those for stationary-phase planktonic cells. A recent study has demonstrated that components of the V. cholerae PTS regulate the entry of biofilm cells but not planktonic cells into stationary phase, suggesting that the biofilm stationary phase is distinct from that of planktonic cells (136). Microarray and proteomic studies have also been able to identify stage-specific signatures for planktonic, monolayer, and multilayer biofilm cultures (4, 233).
However, these studies have not produced a unique biofilm fingerprint. First and foremost, we are realizing that the biofilms are highly heterogeneous communities. Global approaches to transcriptional and proteomic profiling, which measure the mean within a population, are not designed to detect these phenotypic heterogeneities. This is as true for single-species biofilms formed under highly regulated laboratory conditions as it is for multispecies biofilms found in nature. This is not surprising, since cells in the biofilm that are exposed to the bulk fluid experience an entirely different set of conditions than those that reside farther inside the biofilms in terms of amounts of available nutrients and oxygen, accumulation of toxic by-products or secondary metabolites, and cell density signals. It follows, then, that cells in the same microniche in the biofilm would respond to the local conditions similarly, while those in a different microniche would show a completely different phenotype. Therefore, we favor the hypothesis that while biofilm cells are distinct stationary-phase cells and certain traits are more common in biofilm-associated cells than in planktonic cells, there is no proteomic, transcriptomic, or matrix analysis that uniquely defines a fingerprint that can be used to characterize a bacterial assemblage as a biofilm.
CONCLUSION
In this review, we were able to include only a small fraction of the voluminous biofilm literature. As should be apparent from this discussion, much is known about the environmental signals, signal transduction pathways, and effectors that are required for formation of the bacterial biofilm. Furthermore, many components of the biofilm matrix have been delineated. However, many basic aspects of the process of biofilm formation remain to be elucidated. First, we are only beginning to understand the precise mechanisms by which second messengers such as cAMP and c-di-GMP control matrix synthesis and biofilm formation. Furthermore, the precise functions and molecular interactions of the various secreted biofilm matrix polymers, including proteins, polysaccharides, and DNA, have not been defined, and the contributions of these components to matrix integrity are poorly understood at the molecular level. Lastly and perhaps most importantly, while resistance to noxious chemicals, antibiotics, and harsh environments has been touted as a benefit of residence in a biofilm, very few studies have documented these benefits in natural biofilms where careful molecular analysis has confirmed a similarity to the laboratory biofilms that form the basis of our assertions. Molecular biological studies of biofilms found in natural environments will enable biofilm researchers to identify laboratory conditions that best model a particular environment. For instance, iron may activate biofilm formation under particular laboratory conditions but is unlikely to activate biofilm formation in iron-replete natural environments. High concentrations of monosaccharides are often required for the formation of thick multilayer biofilms having exopolysaccharide-based matrices. In which natural environments are sufficiently high levels of monosaccharides present to support the development of such a biofilm? How often and under which natural conditions does the thickness of a bacterial biofilm approach and exceed 100 μm? What similarity do the biofilms that we study in the laboratory bear to biofilms formed on the surfaces of ponds, on rocks in streams, on heart valves, on teeth, and in the intestine? To validate our approaches, we must develop new techniques or apply existing techniques to measure and spatially characterize gene transcription and protein expression in natural biofilms, where heterogeneous bacterial populations are the rule, numbers of any one species are low, and environmental factors that inhibit our molecular biological methods are present. We must define the signals present in natural environments, we must measure the composition of natural biofilm matrices, and we must observe natural biofilms over time to determine how they respond to environmental stresses. These may be the most challenging experiments yet performed on the bacterial biofilm, and yet perhaps this is where we now need to focus our efforts.
Acknowledgments
This work was supported, in part, by grant R01 AI050032 to P.W. and by a University Research Council grant to E.K.
E.K. acknowledges Mary Connell for providing chemicals for minimal media and Guichuan Hou for assistance with confocal scanning laser microscopy. The confocal scanning laser microscopy images were acquired at the William C. and Ruth Ann Dewel-College of Arts & Sciences Microscopy Facility at the Appalachian State University.
Biography
 Ece Karatan received her B.S. degree in biology at Boğaziçi University in Istanbul, Turkey. She obtained her Ph.D. in biochemistry at the University of Illinois at Urbana-Champaign, where she studied chemotactic signaling mechanisms in Bacillus subtilis. After graduation, she worked as a postdoctoral researcher in the Biosciences Division at Argonne National Laboratory on construction of antibody mimetic libraries using phage display. During this time, she realized that she wanted to go back to studying signal transduction in bacteria and became interested in biofilms. She completed her postdoctoral training at the Department of Geographical Medicine and Infectious Diseases at Tufts-New England Medical Center investigating the role of polyamines in Vibrio cholerae biofilm formation. She is currently an assistant professor in the Biology Department at Appalachian State University, where she teaches and continues her investigations of V. cholerae biofilms in her own laboratory.
Ece Karatan received her B.S. degree in biology at Boğaziçi University in Istanbul, Turkey. She obtained her Ph.D. in biochemistry at the University of Illinois at Urbana-Champaign, where she studied chemotactic signaling mechanisms in Bacillus subtilis. After graduation, she worked as a postdoctoral researcher in the Biosciences Division at Argonne National Laboratory on construction of antibody mimetic libraries using phage display. During this time, she realized that she wanted to go back to studying signal transduction in bacteria and became interested in biofilms. She completed her postdoctoral training at the Department of Geographical Medicine and Infectious Diseases at Tufts-New England Medical Center investigating the role of polyamines in Vibrio cholerae biofilm formation. She is currently an assistant professor in the Biology Department at Appalachian State University, where she teaches and continues her investigations of V. cholerae biofilms in her own laboratory.
 Paula I. Watnick is a physician-scientist who received her undergraduate degree in chemistry from Princeton University, her Ph.D. in biophysical chemistry from the California Institute of Technology and her medical degree from Yale Medical School. She has completed a residency program in Internal Medicine at The Beth Israel Hospital in Boston and an Infectious Diseases Fellowship at The Massachusetts General Hospital. She currently conducts research and practices medicine in the Division of Infectious Diseases at Children's Hospital, Boston, MA, and is an affiliate member of the Department of Microbiology and Molecular Biology at Harvard Medical School. Dr. Watnick became interested in the field of bacterial biofilms approximately 10 years ago as an outgrowth of the many problematic surface-associated bacterial infections she has encountered in clinical practice.
Paula I. Watnick is a physician-scientist who received her undergraduate degree in chemistry from Princeton University, her Ph.D. in biophysical chemistry from the California Institute of Technology and her medical degree from Yale Medical School. She has completed a residency program in Internal Medicine at The Beth Israel Hospital in Boston and an Infectious Diseases Fellowship at The Massachusetts General Hospital. She currently conducts research and practices medicine in the Division of Infectious Diseases at Children's Hospital, Boston, MA, and is an affiliate member of the Department of Microbiology and Molecular Biology at Harvard Medical School. Dr. Watnick became interested in the field of bacterial biofilms approximately 10 years ago as an outgrowth of the many problematic surface-associated bacterial infections she has encountered in clinical practice.
REFERENCES
- 1.Abranches, J., M. M. Candella, Z. T. Wen, H. V. Baker, and R. A. Burne. 2006. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J. Bacteriol. 1883748-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agladze, K., X. Wang, and T. Romeo. 2005. Spatial periodicity of Escherichia coli K-12 biofilm microstructure initiates during a reversible, polar attachment phase of development and requires the polysaccharide adhesin PGA. J. Bacteriol. 1878237-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldridge, P., and U. Jenal. 1999. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol. Microbiol. 32379-391. [DOI] [PubMed] [Google Scholar]
- 4.Allegrucci, M., F. Z. Hu, K. Shen, J. Hayes, G. D. Ehrlich, J. C. Post, and K. Sauer. 2006. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J. Bacteriol. 1882325-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allesen-Holm, M., K. B. Barken, L. Yang, M. Klausen, J. S. Webb, S. Kjelleberg, S. Molin, M. Givskov, and T. Tolker-Nielsen. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 591114-1128. [DOI] [PubMed] [Google Scholar]
- 6.Alm, R. A., A. J. Bodero, P. D. Free, and J. S. Mattick. 1996. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 17846-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amikam, D., and M. Y. Galperin. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 223-6. [DOI] [PubMed] [Google Scholar]
- 8.An, D., T. Danhorn, C. Fuqua, and M. R. Parsek. 2006. Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc. Natl. Acad. Sci. USA 1033828-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.An, D., and M. R. Parsek. 2007. The promise and peril of transcriptional profiling in biofilm communities. Curr. Opin. Microbiol. 10292-296. [DOI] [PubMed] [Google Scholar]
- 10.Anderl, J. N., J. Zahller, F. Roe, and P. S. Stewart. 2003. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 471251-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson, G. G., and G. A. O'Toole. 2008. Innate and induced resistance mechanisms of bacterial biofilms. Curr. Top. Microbiol. Immunol. 32285-105. [DOI] [PubMed] [Google Scholar]
- 12.Ashby, M. J., J. E. Neale, S. J. Knott, and I. A. Critchley. 1994. Effect of antibiotics on non-growing planktonic cells and biofilms of Escherichia coli. J. Antimicrob. Chemother. 33443-452. [DOI] [PubMed] [Google Scholar]
- 13.Ausmees, N., and C. Jacobs-Wagner. 2003. Spatial and temporal control of differentiation and cell cycle progression in Caulobacter crescentus. Annu. Rev. Microbiol. 57225-247. [DOI] [PubMed] [Google Scholar]
- 14.Azakami, H., H. Nakashima, H. Akimichi, Y. Noiri, S. Ebisu, and A. Kato. 2006. Involvement of N-acetyl-d-galactosamine-specific lectin in biofilm formation by the periodontopathogenic bacterium, Eikenella corrodens. Biosci. Biotechnol. Biochem. 70441-446. [DOI] [PubMed] [Google Scholar]
- 15.Azeredo, J., and I. W. Sutherland. 2008. The use of phages for the removal of infectious biofilms. Curr. Pharm. Biotechnol. 9261-266. [DOI] [PubMed] [Google Scholar]
- 16.Bai, U., I. Mandic-Mulec, and I. Smith. 1993. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev. 7139-148. [DOI] [PubMed] [Google Scholar]
- 17.Banin, E., M. L. Vasil, and E. P. Greenberg. 2005. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. USA 10211076-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barraud, N., D. J. Hassett, S. H. Hwang, S. A. Rice, S. Kjelleberg, and J. S. Webb. 2006. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 1887344-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bayles, K. W. 2007. The biological role of death and lysis in biofilm development. Nat. Rev. Microbiol. 5721-726. [DOI] [PubMed] [Google Scholar]
- 20.Bechet, M., and R. Blondeau. 2003. Factors associated with the adherence and biofilm formation by Aeromonas caviae on glass surfaces. J. Appl. Microbiol. 941072-1078. [DOI] [PubMed] [Google Scholar]
- 21.Beech, I. B., J. A. Sunner, C. R. Arciola, and P. Cristiani. 2006. Microbially-influenced corrosion: damage to prostheses, delight for bacteria. Int. J. Artif. Organs 29443-452. [DOI] [PubMed] [Google Scholar]
- 22.Begley, M., C. G. Gahan, and C. Hill. 2005. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29625-651. [DOI] [PubMed] [Google Scholar]
- 23.Beloin, C., and J. M. Ghigo. 2005. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 1316-19. [DOI] [PubMed] [Google Scholar]
- 24.Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J. M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51659-674. [DOI] [PubMed] [Google Scholar]
- 25.Beyhan, S., K. Bilecen, S. R. Salama, C. Casper-Lindley, and F. H. Yildiz. 2007. Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J. Bacteriol. 189388-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beyhan, S., A. D. Tischler, A. Camilli, and F. H. Yildiz. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 1883600-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Billker, O., A. Popp, S. D. Gray-Owen, and T. F. Meyer. 2000. The structural basis of CEACAM-receptor targeting by neisserial Opa proteins. Trends Microbiol. 8258-261. [DOI] [PubMed] [Google Scholar]
- 28.Biswas, R., L. Voggu, U. K. Simon, P. Hentschel, G. Thumm, and F. Gotz. 2006. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol. Lett. 259260-268. [DOI] [PubMed] [Google Scholar]
- 29.Blair, K. M., L. Turner, J. T. Winkelman, H. C. Berg, and D. B. Kearns. 2008. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 3201636-1638. [DOI] [PubMed] [Google Scholar]
- 30.Bodenmiller, D., E. Toh, and Y. V. Brun. 2004. Development of surface adhesion in Caulobacter crescentus. J. Bacteriol. 1861438-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boles, B. R., and A. R. Horswill. 2008. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4e1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boles, B. R., M. Thoendel, and P. K. Singh. 2005. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol. Microbiol. 571210-1223. [DOI] [PubMed] [Google Scholar]
- 33.Bomchil, N., P. Watnick, and R. Kolter. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 1851384-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyd, A., and A. M. Chakrabarty. 1994. Role of alginate lyase in cell detachment of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 602355-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brady, L. J., D. A. Piacentini, P. J. Crowley, P. C. Oyston, and A. S. Bleiweis. 1992. Differentiation of salivary agglutinin-mediated adherence and aggregation of mutans streptococci by use of monoclonal antibodies against the major surface adhesin P1. Infect. Immun. 601008-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brady, M. J., K. G. Campellone, M. Ghildiyal, and J. M. Leong. 2007. Enterohaemorrhagic and enteropathogenic Escherichia coli Tir proteins trigger a common Nck-independent actin assembly pathway. Cell. Microbiol. 92242-2253. [DOI] [PubMed] [Google Scholar]
- 37.Brady, R. A., J. G. Leid, J. H. Calhoun, J. W. Costerton, and M. E. Shirtliff. 2008. Osteomyelitis and the role of biofilms in chronic infection. FEMS Immunol. Med. Microbiol. 5213-22. [DOI] [PubMed] [Google Scholar]
- 38.Branda, S. S., F. Chu, D. B. Kearns, R. Losick, and R. Kolter. 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 591229-1238. [DOI] [PubMed] [Google Scholar]
- 39.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben-Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 9811621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Branda, S. S., S. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 1320-26. [DOI] [PubMed] [Google Scholar]
- 41.Brombacher, E., A. Baratto, C. Dorel, and P. Landini. 2006. Gene expression regulation by the curli activator CsgD protein: modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J. Bacteriol. 1882027-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bryers, J. D. 2008. Medical biofilms. Biotechnol. Bioeng. 1001-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caiazza, N. C., J. H. Merritt, K. M. Brothers, and G. A. O'Toole. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 1893603-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caiazza, N. C., and G. A. O'Toole. 2004. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J. Bacteriol. 1864476-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Callaghan, M. J., K. Rockett, C. Banner, E. Haralambous, H. Betts, S. Faust, M. C. Maiden, J. S. Kroll, M. Levin, D. P. Kwiatkowski, and A. J. Pollard. 2008. Haplotypic diversity in human CEACAM genes: effects on susceptibility to meningococcal disease. Genes Immun. 930-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campellone, K. G., and J. M. Leong. 2003. Tails of two Tirs: actin pedestal formation by enteropathogenic E. coli and enterohemorrhagic E. coli O157:H7. Curr. Opin. Microbiol. 682-90. [DOI] [PubMed] [Google Scholar]
- 47.Casper-Lindley, C., and F. H. Yildiz. 2004. VpsT is a transcriptional regulator required for expression of Vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 1861574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan, C., R. Paul, D. Samoray, N. C. Amiot, B. Giese, U. Jenal, and T. Schirmer. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. USA 10117084-17089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi, Y. C., and E. Morgenroth. 2003. Monitoring biofilm detachment under dynamic changes in shear stress using laser-based particle size analysis and mass fractionation. Water Sci. Technol. 4769-76. [PubMed] [Google Scholar]
- 50.Christen, B., M. Christen, R. Paul, F. Schmid, M. Folcher, P. Jenoe, M. Meuwly, and U. Jenal. 2006. Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 28132015-32024. [DOI] [PubMed] [Google Scholar]
- 51.Christen, M., B. Christen, M. G. Allan, M. Folcher, P. Jeno, S. Grzesiek, and U. Jenal. 2007. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 1044112-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Christen, M., B. Christen, M. Folcher, A. Schauerte, and U. Jenal. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 28030829-30837. [DOI] [PubMed] [Google Scholar]
- 53.Christensen, L. D., C. Moser, P. O. Jensen, T. B. Rasmussen, L. Christophersen, S. Kjelleberg, N. Kumar, N. Hoiby, M. Givskov, and T. Bjarnsholt. 2007. Impact of Pseudomonas aeruginosa quorum sensing on biofilm persistence in an in vivo intraperitoneal foreign-body infection model. Microbiology 1532312-2320. [DOI] [PubMed] [Google Scholar]
- 54.Chu, F., D. B. Kearns, S. S. Branda, R. Kolter, and R. Losick. 2006. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol. Microbiol. 591216-1228. [DOI] [PubMed] [Google Scholar]
- 55.Chu, F., D. B. Kearns, A. McLoon, Y. Chai, R. Kolter, and R. Losick. 2008. A novel regulatory protein governing biofilm formation in Bacillus subtilis. Mol. Microbiol. 681117-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collet, A., S. Vilain, P. Cosette, G. A. Junter, T. Jouenne, R. S. Phillips, and P. Di Martino. 2007. Protein expression in Escherichia coli S17-1 biofilms: impact of indole. Antonie van Leeuwenhoek 9171-85. [DOI] [PubMed] [Google Scholar]
- 57.Collier, J., and L. Shapiro. 2007. Spatial complexity and control of a bacterial cell cycle. Curr. Opin. Biotechnol. 18333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Gotz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 675427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crossman, L., and J. M. Dow. 2004. Biofilm formation and dispersal in Xanthomonas campestris. Microbes Infect. 6623-629. [DOI] [PubMed] [Google Scholar]
- 60.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 1832888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cucarella, C., M. A. Tormo, C. Ubeda, M. P. Trotonda, M. Monzon, C. Peris, B. Amorena, I. Lasa, and J. R. Penades. 2004. Role of biofilm-associated protein Bap in the pathogenesis of bovine Staphylococcus aureus. Infect. Immun. 722177-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Danese, P. N., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37424-432. [DOI] [PubMed] [Google Scholar]
- 63.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 1823593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Danhorn, T., M. Hentzer, M. Givskov, M. R. Parsek, and C. Fuqua. 2004. Phosphorus limitation enhances biofilm formation of the plant pathogen Agrobacterium tumefaciens through the PhoR-PhoB regulatory system. J. Bacteriol. 1864492-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Darby, C., J. W. Hsu, N. Ghori, and S. Falkow. 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417243-244. [DOI] [PubMed] [Google Scholar]
- 66.Da Re, S., and J. M. Ghigo. 2006. A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J. Bacteriol. 1883073-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 1851027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davies, D. G., and C. N. Marques. 2009. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 1911393-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280295-298. [DOI] [PubMed] [Google Scholar]
- 70.Deighton, M., and R. Borland. 1993. Regulation of slime production in Staphylococcus epidermidis by iron limitation. Infect. Immun. 614473-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deutscher, J., C. Francke, and P. W. Postma. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70939-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DeVinney, R., A. Gauthier, A. Abe, and B. B. Finlay. 1999. Enteropathogenic Escherichia coli: a pathogen that inserts its own receptor into host cells. Cell Mol. Life Sci. 55961-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.DeVinney, R., M. Stein, D. Reinscheid, A. Abe, S. Ruschkowski, and B. B. Finlay. 1999. Enterohemorrhagic Escherichia coli O157:H7 produces Tir, which is translocated to the host cell membrane but is not tyrosine phosphorylated. Infect. Immun. 672389-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Diggle, S. P., R. E. Stacey, C. Dodd, M. Camara, P. Williams, and K. Winzer. 2006. The galactophilic lectin, LecA, contributes to biofilm development in Pseudomonas aeruginosa. Environ. Microbiol. 81095-1104. [DOI] [PubMed] [Google Scholar]
- 75.Dobinsky, S., K. Kiel, H. Rohde, K. Bartscht, J. K. Knobloch, M. A. Horstkotte, and D. Mack. 2003. Glucose-related dissociation between icaADBC transcription and biofilm expression by Staphylococcus epidermidis: evidence for an additional factor required for polysaccharide intercellular adhesin synthesis. J. Bacteriol. 1852879-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Domka, J., J. Lee, T. Bansal, and T. K. Wood. 2007. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ. Microbiol. 9332-346. [DOI] [PubMed] [Google Scholar]
- 77.Domka, J., J. Lee, and T. K. Wood. 2006. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl. Environ. Microbiol. 722449-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Donnenberg, M. S. 1999. Interactions between enteropathogenic Escherichia coli and epithelial cells. Clin. Infect. Dis. 28451-455. [DOI] [PubMed] [Google Scholar]
- 79.Dow, J. M., L. Crossman, K. Findlay, Y. Q. He, J. X. Feng, and J. L. Tang. 2003. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. USA 10010995-11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Entcheva-Dimitrov, P., and A. M. Spormann. 2004. Dynamics and control of biofilms of the oligotrophic bacterium Caulobacter crescentus. J. Bacteriol. 1868254-8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Espinosa-Urgel, M., A. Salido, and J. L. Ramos. 2000. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J. Bacteriol. 1822363-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Evans, L. R., and A. Linker. 1973. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J. Bacteriol. 116915-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ferreira, R. B., L. C. Antunes, E. P. Greenberg, and L. L. McCarter. 2008. Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J. Bacteriol. 190851-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Finkelstein, R. A., M. Boesman-Finkelstein, Y. Chang, and C. C. Hase. 1992. Vibrio cholerae hemagglutinin/protease, colonial variation, virulence, and detachment. Infect. Immun. 60472-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Flemming, H. C., T. R. Neu, and D. J. Wozniak. 2007. The EPS matrix: the “house of biofilm cells.” J. Bacteriol. 1897945-7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fletcher, M. 1996. Bacterial attachment in aquatic environments: a diversity of surfaces and adhesion strategies, p. 1-24. In M. Fletcher (ed.), Bacterial adhesion: molecular and ecological diversity. Wiley-Liss, New York, NY.
- 87.Fluckiger, U., M. Ulrich, A. Steinhuber, G. Doring, D. Mack, R. Landmann, C. Goerke, and C. Wolz. 2005. Biofilm formation, icaADBC transcription, and polysaccharide intercellular adhesin synthesis by staphylococci in a device-related infection model. Infect. Immun. 731811-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fong, J. C., K. Karplus, G. K. Schoolnik, and F. H. Yildiz. 2006. Identification and characterization of RbmA, a novel protein required for the development of rugose colony morphology and biofilm structure in Vibrio cholerae. J. Bacteriol. 1881049-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fong, J. C., and F. H. Yildiz. 2008. Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J. Bacteriol. 1906646-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fong, J. C., and F. H. Yildiz. 2007. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J. Bacteriol. 1892319-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Francis, C. L., A. E. Jerse, J. B. Kaper, and S. Falkow. 1991. Characterization of interactions of enteropathogenic Escherichia coli O127:H6 with mammalian cells in vitro. J. Infect. Dis. 164693-703. [DOI] [PubMed] [Google Scholar]
- 92.Frankel, G., and A. D. Phillips. 2008. Attaching effacing Escherichia coli and paradigms of Tir-triggered actin polymerization: getting off the pedestal. Cell. Microbiol. 10549-556. [DOI] [PubMed] [Google Scholar]
- 93.Friedman, L., and R. Kolter. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51675-690. [DOI] [PubMed] [Google Scholar]
- 94.Friedman, L., and R. Kolter. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 1864457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fujita, M., J. E. Gonzalez-Pastor, and R. Losick. 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 1871357-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fux, C. A., J. W. Costerton, P. S. Stewart, and P. Stoodley. 2005. Survival strategies of infectious biofilms. Trends Microbiol. 1334-40. [DOI] [PubMed] [Google Scholar]
- 97.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 20311-21. [DOI] [PubMed] [Google Scholar]
- 98.Garcia, B., C. Latasa, C. Solano, F. Garcia-del Portillo, C. Gamazo, and I. Lasa. 2004. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol. Microbiol. 54264-277. [DOI] [PubMed] [Google Scholar]
- 99.Garrett, E. S., D. Perlegas, and D. J. Wozniak. 1999. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU). J. Bacteriol. 1817401-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gerke, C., A. Kraft, R. Sussmuth, O. Schweitzer, and F. Gotz. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 27318586-18593. [DOI] [PubMed] [Google Scholar]
- 101.Gerstel, U., A. Kolb, and U. Romling. 2006. Regulatory components at the csgD promoter—additional roles for OmpR and integration host factor and role of the 5′ untranslated region. FEMS Microbiol. Lett. 261109-117. [DOI] [PubMed] [Google Scholar]
- 102.Gerstel, U., C. Park, and U. Romling. 2003. Complex regulation of csgD promoter activity by global regulatory proteins. Mol. Microbiol. 49639-654. [DOI] [PubMed] [Google Scholar]
- 103.Gerstel, U., and U. Romling. 2003. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res. Microbiol. 154659-667. [DOI] [PubMed] [Google Scholar]
- 104.Gerstel, U., and U. Romling. 2001. Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environ. Microbiol. 3638-648. [DOI] [PubMed] [Google Scholar]
- 105.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412442-445. [DOI] [PubMed] [Google Scholar]
- 106.Girard, V., and M. Mourez. 2006. Adhesion mediated by autotransporters of Gram-negative bacteria: structural and functional features. Res. Microbiol. 157407-416. [DOI] [PubMed] [Google Scholar]
- 107.Givskov, M., R. de Nys, M. Manefield, L. Gram, R. Maximilien, L. Eberl, S. Molin, P. D. Steinberg, and S. Kjelleberg. 1996. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J. Bacteriol. 1786618-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gjermansen, M., P. Ragas, C. Sternberg, S. Molin, and T. Tolker-Nielsen. 2005. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ. Microbiol. 7894-906. [DOI] [PubMed] [Google Scholar]
- 109.Gjermansen, M., P. Ragas, and T. Tolker-Nielsen. 2006. Proteins with GGDEF and EAL domains regulate Pseudomonas putida biofilm formation and dispersal. FEMS Microbiol. Lett. 265215-224. [DOI] [PubMed] [Google Scholar]
- 110.Goley, E. D., A. A. Iniesta, and L. Shapiro. 2007. Cell cycle regulation in Caulobacter: location, location, location. J. Cell Sci. 1203501-3507. [DOI] [PubMed] [Google Scholar]
- 111.Goller, C., X. Wang, Y. Itoh, and T. Romeo. 2006. The cation-responsive protein NhaR of Escherichia coli activates pgaABCD transcription, required for production of the biofilm adhesin poly-beta-1,6-N-acetyl-d-glucosamine. J. Bacteriol. 1888022-8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gomez, M. I., and A. Prince. 2007. Opportunistic infections in lung disease: Pseudomonas infections in cystic fibrosis. Curr. Opin. Pharmacol. 7244-251. [DOI] [PubMed] [Google Scholar]
- 113.Gonzalez Barrios, A. F., R. Zuo, Y. Hashimoto, L. Yang, W. E. Bentley, and T. K. Wood. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188305-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Goodman, A. L., B. Kulasekara, A. Rietsch, D. Boyd, R. S. Smith, and S. Lory. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7745-754. [DOI] [PubMed] [Google Scholar]
- 115.Goosney, D. L., M. de Grado, and B. B. Finlay. 1999. Putting E. coli on a pedestal: a unique system to study signal transduction and the actin cytoskeleton. Trends Cell Biol. 911-14. [DOI] [PubMed] [Google Scholar]
- 116.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Guvener, Z. T., and C. S. Harwood. 2007. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol. Microbiol. 661459-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hammar, M., A. Arnqvist, Z. Bian, A. Olsen, and S. Normark. 1995. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18661-670. [DOI] [PubMed] [Google Scholar]
- 119.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50101-104. [DOI] [PubMed] [Google Scholar]
- 120.Hammer, B. K., and B. L. Bassler. 2007. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. USA 10411145-11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hamon, M. A., and B. A. Lazazzera. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 421199-1209. [DOI] [PubMed] [Google Scholar]
- 122.Hasman, H., T. Chakraborty, and P. Klemm. 1999. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J. Bacteriol. 1814834-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hauck, C. R. 2002. Cell adhesion receptors—signaling capacity and exploitation by bacterial pathogens. Med. Microbiol. Immunol. 19155-62. [DOI] [PubMed] [Google Scholar]
- 124.Haugo, A. J., and P. I. Watnick. 2002. Vibrio cholerae CytR is a repressor of biofilm development. Mol. Microbiol. 45471-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Heeb, S., and D. Haas. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant-Microbe Interact. 141351-1363. [DOI] [PubMed] [Google Scholar]
- 126.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 201083-1091. [DOI] [PubMed] [Google Scholar]
- 127.Henke, J. M., and B. L. Bassler. 2004. Bacterial social engagements. Trends Cell Biol. 14648-656. [DOI] [PubMed] [Google Scholar]
- 128.Hentzer, M., K. Riedel, T. B. Rasmussen, A. Heydorn, J. B. Andersen, M. R. Parsek, S. A. Rice, L. Eberl, S. Molin, N. Hoiby, S. Kjelleberg, and M. Givskov. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 14887-102. [DOI] [PubMed] [Google Scholar]
- 129.Hentzer, M., G. M. Teitzel, G. J. Balzer, A. Heydorn, S. Molin, M. Givskov, and M. R. Parsek. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 1835395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hentzer, M., H. Wu, J. B. Andersen, K. Riedel, T. B. Rasmussen, N. Bagge, N. Kumar, M. A. Schembri, Z. Song, P. Kristoffersen, M. Manefield, J. W. Costerton, S. Molin, L. Eberl, P. Steinberg, S. Kjelleberg, N. Hoiby, and M. Givskov. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 223803-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Heydorn, A., B. Ersboll, J. Kato, M. Hentzer, M. R. Parsek, T. Tolker-Nielsen, M. Givskov, and S. Molin. 2002. Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl. Environ. Microbiol. 682008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 1462395-2407. [DOI] [PubMed] [Google Scholar]
- 133.Hickman, J. W., D. F. Tifrea, and C. S. Harwood. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA 10214422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hinsa, S. M., M. Espinosa-Urgel, J. L. Ramos, and G. A. O'Toole. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49905-918. [DOI] [PubMed] [Google Scholar]
- 135.Hoffman, L. R., D. A. D'Argenio, M. J. MacCoss, Z. Zhang, R. A. Jones, and S. I. Miller. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 4361171-1175. [DOI] [PubMed] [Google Scholar]
- 136.Houot, L., and P. I. Watnick. 2008. A novel role for enzyme I of the Vibrio cholerae phosphoenolpyruvate phosphotransferase system in regulation of growth in a biofilm. J. Bacteriol. 190311-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hung, D. T., J. Zhu, D. Sturtevant, and J. J. Mekalanos. 2006. Bile acids stimulate biofilm formation in Vibrio cholerae. Mol. Microbiol. 59193-201. [DOI] [PubMed] [Google Scholar]
- 138.Hynes, R. O. 1987. Integrins: a family of cell surface receptors. Cell 48549-554. [DOI] [PubMed] [Google Scholar]
- 139.Isberg, R. R., and P. Barnes. 2001. Subversion of integrins by enteropathogenic Yersinia. J. Cell Sci. 11421-28. [DOI] [PubMed] [Google Scholar]
- 140.Isberg, R. R., and J. M. Leong. 1988. Cultured mammalian cells attach to the invasin protein of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 856682-6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Isberg, R. R., and G. T. Van Nhieu. 1995. The mechanism of phagocytic uptake promoted by invasin-integrin interaction. Trends Cell Biol. 5120-124. [DOI] [PubMed] [Google Scholar]
- 142.Isberg, R. R., D. L. Voorhis, and S. Falkow. 1987. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell 50769-778. [DOI] [PubMed] [Google Scholar]
- 143.Itoh, Y., J. D. Rice, C. Goller, A. Pannuri, J. Taylor, J. Meisner, T. J. Beveridge, J. F. Preston III, and T. Romeo. 2008. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-d-glucosamine. J. Bacteriol. 1903670-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Itoh, Y., X. Wang, B. J. Hinnebusch, J. F. Preston III, and T. Romeo. 2005. Depolymerization of beta-1,6-N-acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 187382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Izano, E. A., M. A. Amarante, W. B. Kher, and J. B. Kaplan. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Izano, E. A., I. Sadovskaya, E. Vinogradov, M. H. Mulks, K. Velliyagounder, C. Ragunath, W. B. Kher, N. Ramasubbu, S. Jabbouri, M. B. Perry, and J. B. Kaplan. 2007. Poly-N-acetylglucosamine mediates biofilm formation and antibiotic resistance in Actinobacillus pleuropneumoniae. Microb. Pathog. 431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jackson, D. W., J. W. Simecka, and T. Romeo. 2002. Catabolite repression of Escherichia coli biofilm formation. J. Bacteriol. 1843406-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jackson, K. D., M. Starkey, S. Kremer, M. R. Parsek, and D. J. Wozniak. 2004. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 1864466-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Janakiraman, R. S., and Y. V. Brun. 1999. Cell cycle control of a holdfast attachment gene in Caulobacter crescentus. J. Bacteriol. 1811118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jarrett, C. O., E. Deak, K. E. Isherwood, P. C. Oyston, E. R. Fischer, A. R. Whitney, S. D. Kobayashi, F. R. DeLeo, and B. J. Hinnebusch. 2004. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190783-792. [DOI] [PubMed] [Google Scholar]
- 152.Jenal, U., and J. Malone. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40385-407. [DOI] [PubMed] [Google Scholar]
- 153.Jobling, M. G., and R. K. Holmes. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 261023-1034. [DOI] [PubMed] [Google Scholar]
- 154.Joelsson, A., Z. Liu, and J. Zhu. 2006. Genetic and phenotypic diversity of quorum-sensing systems in clinical and environmental isolates of Vibrio cholerae. Infect. Immun. 741141-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jubelin, G., A. Vianney, C. Beloin, J. M. Ghigo, J. C. Lazzaroni, P. Lejeune, and C. Dorel. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 1872038-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kader, A., R. Simm, U. Gerstel, M. Morr, and U. Romling. 2006. Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 60602-616. [DOI] [PubMed] [Google Scholar]
- 157.Kanbe, M., S. Shibata, Y. Umino, U. Jenal, and S. I. Aizawa. 2005. Protease susceptibility of the Caulobacter crescentus flagellar hook-basal body: a possible mechanism of flagellar ejection during cell differentiation. Microbiology 151433-438. [DOI] [PubMed] [Google Scholar]
- 158.Kang, Y., H. Liu, S. Genin, M. A. Schell, and T. P. Denny. 2002. Ralstonia solanacearum requires type 4 pili to adhere to multiple surfaces and for natural transformation and virulence. Mol. Microbiol. 46427-437. [DOI] [PubMed] [Google Scholar]
- 159.Kapfhammer, D., E. Karatan, K. J. Pflughoeft, and P. I. Watnick. 2005. Role for glycine betaine transport in Vibrio cholerae osmoadaptation and biofilm formation within microbial communities. Appl. Environ. Microbiol. 713840-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kaplan, J. B., C. Ragunath, N. Ramasubbu, and D. H. Fine. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J. Bacteriol. 1854693-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kaplan, J. B., C. Ragunath, K. Velliyagounder, D. H. Fine, and N. Ramasubbu. 2004. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 482633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kaplan, J. B., K. Velliyagounder, C. Ragunath, H. Rohde, D. Mack, J. K. Knobloch, and N. Ramasubbu. 2004. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J. Bacteriol. 1868213-8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Reference deleted.
- 164.Karatan, E., T. R. Duncan, and P. I. Watnick. 2005. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J. Bacteriol. 1877434-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kavanaugh, J. S., M. Thoendel, and A. R. Horswill. 2007. A role for type I signal peptidase in Staphylococcus aureus quorum sensing. Mol. Microbiol. 65780-798. [DOI] [PubMed] [Google Scholar]
- 166.Kearns, D. B., F. Chu, S. S. Branda, R. Kolter, and R. Losick. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol. Microbiol. 55739-749. [DOI] [PubMed] [Google Scholar]
- 167.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91511-520. [DOI] [PubMed] [Google Scholar]
- 168.Kierek, K., and P. I. Watnick. 2003. Environmental determinants of Vibrio cholerae biofilm development. Appl. Environ. Microbiol. 695079-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kierek, K., and P. I. Watnick. 2003. The Vibrio cholerae O139 O-antigen polysaccharide is essential for Ca2+-dependent biofilm development in sea water. Proc. Natl. Acad. Sci. USA 10014357-14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kim, Y. K., and L. L. McCarter. 2007. ScrG, a GGDEF-EAL protein, participates in regulating swarming and sticking in Vibrio parahaemolyticus. J. Bacteriol. 1894094-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kirisits, M. J., J. J. Margolis, B. L. Purevdorj-Gage, B. Vaughan, D. L. Chopp, P. Stoodley, and M. R. Parsek. 2007. The influence of the hydrodynamic environment on quorum sensing in Pseudomonas aeruginosa biofilms. J. Bacteriol. 1898357-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kirisits, M. J., and M. R. Parsek. 2006. Does Pseudomonas aeruginosa use intercellular signalling to build biofilm communities? Cell. Microbiol. 81841-1849. [DOI] [PubMed] [Google Scholar]
- 173.Kirov, S. M., M. Castrisios, and J. G. Shaw. 2004. Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect. Immun. 721939-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kjaergaard, K., M. A. Schembri, C. Ramos, S. Molin, and P. Klemm. 2000. Antigen 43 facilitates formation of multispecies biofilms. Environ. Microbiol. 2695-702. [DOI] [PubMed] [Google Scholar]
- 175.Klausen, M., A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 5061-68. [DOI] [PubMed] [Google Scholar]
- 176.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 481511-1524. [DOI] [PubMed] [Google Scholar]
- 177.Klemm, P., R. M. Vejborg, and O. Sherlock. 2006. Self-associating autotransporters, SAATs: functional and structural similarities. Int. J. Med. Microbiol. 296187-195. [DOI] [PubMed] [Google Scholar]
- 178.Knutton, S., T. Baldwin, P. Williams, A. Manjarrez-Hernandez, and A. Aitken. 1993. The attaching and effacing virulence property of enteropathogenic Escherichia coli. Zentralbl. Bakteriol. 278209-217. [DOI] [PubMed] [Google Scholar]
- 179.Kristich, C. J., Y. H. Li, D. G. Cvitkovitch, and G. M. Dunny. 2004. Esp-independent biofilm formation by Enterococcus faecalis. J. Bacteriol. 186154-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kuchma, S. L., K. M. Brothers, J. H. Merritt, N. T. Liberati, F. M. Ausubel, and G. A. O'Toole. 2007. BifA, a c-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 1898165-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kuchma, S. L., J. P. Connolly, and G. A. O'Toole. 2005. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 1871441-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lam, J., R. Chan, K. Lam, and J. W. Costerton. 1980. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect. Immun. 28546-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lapidus, I. R., M. Welch, and M. Eisenbach. 1988. Pausing of flagellar rotation is a component of bacterial motility and chemotaxis. J. Bacteriol. 1703627-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lapouge, K., M. Schubert, F. H. Allain, and D. Haas. 2008. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 67241-253. [DOI] [PubMed] [Google Scholar]
- 185.Lasa, I., and J. R. Penades. 2006. Bap: a family of surface proteins involved in biofilm formation. Res. Microbiol. 15799-107. [DOI] [PubMed] [Google Scholar]
- 186.Latasa, C., A. Roux, A. Toledo-Arana, J. M. Ghigo, C. Gamazo, J. R. Penades, and I. Lasa. 2005. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol. Microbiol. 581322-1339. [DOI] [PubMed] [Google Scholar]
- 187.Lauriano, C. M., C. Ghosh, N. E. Correa, and K. E. Klose. 2004. The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J. Bacteriol. 1864864-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lecuit, M., H. Ohayon, L. Braun, J. Mengaud, and P. Cossart. 1997. Internalin of Listeria monocytogenes with an intact leucine-rich repeat region is sufficient to promote internalization. Infect. Immun. 655309-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lee, J., A. Jayaraman, and T. K. Wood. 2007. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lee, V. T., J. M. Matewish, J. L. Kessler, M. Hyodo, Y. Hayakawa, and S. Lory. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 651474-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lemon, K. P., A. M. Earl, H. C. Vlamakis, C. Aguilar, and R. Kolter. 2008. Biofilm development with an emphasis on Bacillus subtilis. Curr. Top. Microbiol. Immunol. 3221-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lemon, K. P., D. E. Higgins, and R. Kolter. 2007. Flagellar motility is critical for Listeria monocytogenes biofilm formation. J. Bacteriol. 1894418-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lenz, D. H., M. B. Miller, J. Zhu, R. V. Kulkarni, and B. L. Bassler. 2005. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol. 581186-1202. [DOI] [PubMed] [Google Scholar]
- 194.Lequette, Y., and E. P. Greenberg. 2005. Timing and localization of rhamnolipid synthesis gene expression in Pseudomonas aeruginosa biofilms. J. Bacteriol. 18737-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lewis, K. 2008. Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 322107-131. [DOI] [PubMed] [Google Scholar]
- 196.Lewis, K. 2004. Persister cells and the riddle of biofilm survival. Biochemistry (Moscow) 70267-274. [DOI] [PubMed] [Google Scholar]
- 197.Li, G., C. S. Smith, Y. V. Brun, and J. X. Tang. 2005. The elastic properties of the Caulobacter crescentus adhesive holdfast are dependent on oligomers of N-acetylglucosamine. J. Bacteriol. 187257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Li, H., L. Xu, J. Wang, Y. Wen, C. Vuong, M. Otto, and Q. Gao. 2005. Conversion of Staphylococcus epidermidis strains from commensal to invasive by expression of the ica locus encoding production of biofilm exopolysaccharide. Infect. Immun. 733188-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Li, J., C. Attila, L. Wang, T. K. Wood, J. J. Valdes, and W. E. Bentley. 2007. Quorum sensing in Escherichia coli is signaled by AI-2/LsrR: effects on small RNA and biofilm architecture. J. Bacteriol. 1896011-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Liang, W., A. J. Silva, and J. A. Benitez. 2007. The cyclic AMP receptor protein modulates colonial morphology in Vibrio cholerae. Appl. Environ. Microbiol. 737482-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lim, B., S. Beyhan, J. Meir, and F. H. Yildiz. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60331-348. [DOI] [PubMed] [Google Scholar]
- 202.Lim, B., S. Beyhan, and F. H. Yildiz. 2007. Regulation of Vibrio polysaccharide synthesis and virulence factor production by CdgC, a GGDEF-EAL domain protein, in Vibrio cholerae. J. Bacteriol. 189717-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lim, Y., M. Jana, T. T. Luong, and C. Y. Lee. 2004. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 186722-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Liu, M. Y., G. Gui, B. Wei, J. F. Preston III, L. Oakford, U. Yuksel, D. P. Giedroc, and T. Romeo. 1997. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 27217502-17510. [DOI] [PubMed] [Google Scholar]
- 205.Liu, Z., F. R. Stirling, and J. Zhu. 2007. Temporal quorum-sensing induction regulates Vibrio cholerae biofilm architecture. Infect. Immun. 75122-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lynch, D. J., T. L. Fountain, J. E. Mazurkiewicz, and J. A. Banas. 2007. Glucan-binding proteins are essential for shaping Streptococcus mutans biofilm architecture. FEMS Microbiol. Lett. 268158-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ma, L., K. D. Jackson, R. M. Landry, M. R. Parsek, and D. J. Wozniak. 2006. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 1888213-8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ma, L., H. Lu, A. Sprinkle, M. R. Parsek, and D. J. Wozniak. 2007. Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J. Bacteriol. 1898353-8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Maier, B., M. Koomey, and M. P. Sheetz. 2004. A force-dependent switch reverses type IV pilus retraction. Proc. Natl. Acad. Sci. USA 10110961-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mai-Prochnow, A., F. Evans, D. Dalisay-Saludes, S. Stelzer, S. Egan, S. James, J. S. Webb, and S. Kjelleberg. 2004. Biofilm development and cell death in the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 703232-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Mai-Prochnow, A., J. S. Webb, B. C. Ferrari, and S. Kjelleberg. 2006. Ecological advantages of autolysis during the development and dispersal of Pseudoalteromonas tunicata biofilms. Appl. Environ. Microbiol. 725414-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Manefield, M., R. de Nys, N. Kumar, R. Read, M. Givskov, P. Steinberg, and S. Kjelleberg. 1999. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 145283-291. [DOI] [PubMed] [Google Scholar]
- 213.Martino, P. D., R. Fursy, L. Bret, B. Sundararaju, and R. S. Phillips. 2003. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can. J. Microbiol. 49443-449. [DOI] [PubMed] [Google Scholar]
- 214.Mathee, K., O. Ciofu, C. Sternberg, P. W. Lindum, J. I. Campbell, P. Jensen, A. H. Johnsen, M. Givskov, D. E. Ohman, S. Molin, N. Hoiby, and A. Kharazmi. 1999. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 1451349-1357. [DOI] [PubMed] [Google Scholar]
- 215.Matsukawa, M., and E. P. Greenberg. 2004. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 1864449-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Matthysse, A. G., H. A. Yarnall, and N. Young. 1996. Requirement for genes with homology to ABC transport systems for attachment and virulence of Agrobacterium tumefaciens. J. Bacteriol. 1785302-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.McClaine, J. W., and R. M. Ford. 2002. Characterizing the adhesion of motile and nonmotile Escherichia coli to a glass surface using a parallel-plate flow chamber. Biotechnol. Bioeng. 78179-189. [DOI] [PubMed] [Google Scholar]
- 218.McFall-Ngai, M. 2008. Host-microbe symbiosis: the squid-Vibrio association—a naturally occurring, experimental model of animal/bacterial partnerships. Adv. Exp. Med. Biol. 635102-112. [DOI] [PubMed] [Google Scholar]
- 219.McKenzie, G. J., R. S. Harris, P. L. Lee, and S. M. Rosenberg. 2000. The SOS response regulates adaptive mutation. Proc. Natl. Acad. Sci. USA 976646-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Meibom, K. L., X. B. Li, A. T. Nielsen, C. Y. Wu, S. Roseman, and G. K. Schoolnik. 2004. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. USA 1012524-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Mengaud, J., H. Ohayon, P. Gounon, R. M. Mege, and P. Cossart. 1996. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84923-932. [DOI] [PubMed] [Google Scholar]
- 222.Merighi, M., V. T. Lee, M. Hyodo, Y. Hayakawa, and S. Lory. 2007. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65876-895. [DOI] [PubMed] [Google Scholar]
- 223.Merritt, J. H., K. M. Brothers, S. L. Kuchma, and A. O'Toole. 2007. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 1898154-8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Merritt, P. M., T. Danhorn, and C. Fuqua. 2007. Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J. Bacteriol. 1898005-8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 40798-102. [DOI] [PubMed] [Google Scholar]
- 226.Mey, A. R., S. A. Craig, and S. M. Payne. 2005. Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect. Immun. 735706-5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Miethke, M., and M. A. Marahiel. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71413-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Moelling, C., R. Oberschlacke, P. Ward, J. Karijolich, K. Borisova, N. Bjelos, and L. Bergeron. 2007. Metal-dependent repression of siderophore and biofilm formation in Actinomyces naeslundii. FEMS Microbiol. Lett. 275214-220. [DOI] [PubMed] [Google Scholar]
- 229.Mondragon, V., B. Franco, K. Jonas, K. Suzuki, T. Romeo, O. Melefors, and D. Georgellis. 2006. pH-dependent activation of the BarA-UvrY two-component system in Escherichia coli. J. Bacteriol. 1888303-8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Monds, R. D., P. D. Newell, R. H. Gross, and G. A. O'Toole. 2007. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 63656-679. [DOI] [PubMed] [Google Scholar]
- 231.Monds, R. D., M. W. Silby, and H. K. Mahanty. 2001. Expression of the Pho regulon negatively regulates biofilm formation by Pseudomonas aureofaciens PA147-2. Mol. Microbiol. 42415-426. [DOI] [PubMed] [Google Scholar]
- 232.Moorthy, S., and P. I. Watnick. 2004. Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol. Microbiol. 52573-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Moorthy, S., and P. I. Watnick. 2005. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol. Microbiol. 571623-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Morgan, R., S. Kohn, S. H. Hwang, D. J. Hassett, and K. Sauer. 2006. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J. Bacteriol. 1887335-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Mueller, R. S., D. McDougald, D. Cusumano, N. Sodhi, S. Kjelleberg, F. Azam, and D. H. Bartlett. 2007. Vibrio cholerae strains possess multiple strategies for abiotic and biotic surface colonization. J. Bacteriol. 1895348-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Muenzner, P., C. Dehio, T. Fujiwara, M. Achtman, T. F. Meyer, and S. D. Gray-Owen. 2000. Carcinoembryonic antigen family receptor specificity of Neisseria meningitidis Opa variants influences adherence to and invasion of proinflammatory cytokine-activated endothelial cells. Infect. Immun. 683601-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Nemoto, K., K. Hirota, K. Murakami, K. Taniguti, H. Murata, D. Viducic, and Y. Miyake. 2003. Effect of Varidase (streptodornase) on biofilm formed by Pseudomonas aeruginosa. Chemotherapy 49121-125. [DOI] [PubMed] [Google Scholar]
- 238.Newton, W. A., and E. E. Snell. 1964. Catalytic properties of tryptophanase, a multifunctional pyridoxal phosphate enzyme. Proc. Natl. Acad. Sci. USA 51382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 481429-1449. [DOI] [PubMed] [Google Scholar]
- 240.Novick, R. P., S. J. Projan, J. Kornblum, H. F. Ross, G. Ji, B. Kreiswirth, F. Vandenesch, and S. Moghazeh. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248446-458. [DOI] [PubMed] [Google Scholar]
- 241.Ochsner, U. A., and J. Reiser. 1995. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 926424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.O'Gara, J. P. 2007. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270179-188. [DOI] [PubMed] [Google Scholar]
- 243.O'Toole, G. A., K. A. Gibbs, P. W. Hager, P. V. Phibbs, Jr., and R. Kolter. 2000. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 182425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30295-304. [DOI] [PubMed] [Google Scholar]
- 245.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28449-461. [DOI] [PubMed] [Google Scholar]
- 246.Parise, G., M. Mishra, Y. Itoh, T. Romeo, and R. Deora. 2007. Role of a putative polysaccharide locus in Bordetella biofilm development. J. Bacteriol. 189750-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Parkins, M. D., H. Ceri, and D. G. Storey. 2001. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 401215-1226. [DOI] [PubMed] [Google Scholar]
- 248.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57677-701. [DOI] [PubMed] [Google Scholar]
- 249.Patel, C. N., B. W. Wortham, J. L. Lines, J. D. Fetherston, R. D. Perry, and M. A. Oliveira. 2006. Polyamines are essential for the formation of plague biofilm. J. Bacteriol. 1882355-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Patriquin, G. M., E. Banin, C. Gilmour, R. Tuchman, E. P. Greenberg, and K. Poole. 2008. Influence of quorum sensing and iron on twitching motility and biofilm formation in Pseudomonas aeruginosa. J. Bacteriol. 190662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Pavithra, D., and M. Doble. 2008. Biofilm formation, bacterial adhesion and host response on polymeric implants—issues and prevention. Biomed. Mater. 3034003. [DOI] [PubMed] [Google Scholar]
- 253.Pratt, J. T., R. Tamayo, A. D. Tischler, and A. Camilli. 2007. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 28212860-12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30285-293. [DOI] [PubMed] [Google Scholar]
- 255.Prigent-Combaret, C., G. Prensier, T. T. Le Thi, O. Vidal, P. Lejeune, and C. Dorel. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2450-464. [DOI] [PubMed] [Google Scholar]
- 256.Prigent-Combaret, C., O. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 1815993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Purevdorj, B., J. W. Costerton, and P. Stoodley. 2002. Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 684457-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Purevdorj-Gage, B., W. J. Costerton, and P. Stoodley. 2005. Phenotypic differentiation and seeding dispersal in non-mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology 1511569-1576. [DOI] [PubMed] [Google Scholar]
- 259.Qin, Z., Y. Ou, L. Yang, Y. Zhu, T. Tolker-Nielsen, S. Molin, and D. Qu. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 1532083-2092. [DOI] [PubMed] [Google Scholar]
- 260.Quisel, J. D., W. F. Burkholder, and A. D. Grossman. 2001. In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. J. Bacteriol. 1836573-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Ramsey, D. M., and D. J. Wozniak. 2005. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol. Microbiol. 56309-322. [DOI] [PubMed] [Google Scholar]
- 262.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54881-941. [DOI] [PubMed] [Google Scholar]
- 263.Regassa, L. B., R. P. Novick, and M. J. Betley. 1992. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect. Immun. 603381-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Reisner, A., B. M. Holler, S. Molin, and E. L. Zechner. 2006. Synergistic effects in mixed Escherichia coli biofilms: conjugative plasmid transfer drives biofilm expansion. J. Bacteriol. 1883582-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Rice, K. C., and K. W. Bayles. 2008. Molecular control of bacterial death and lysis. Microbiol. Mol. Biol. Rev. 7285-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Rice, K. C., E. E. Mann, J. L. Endres, E. C. Weiss, J. E. Cassat, M. S. Smeltzer, and K. W. Bayles. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 1048113-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Rice, S. A., K. S. Koh, S. Y. Queck, M. Labbate, K. W. Lam, and S. Kjelleberg. 2005. Biofilm formation and sloughing in Serratia marcescens are controlled by quorum sensing and nutrient cues. J. Bacteriol. 1873477-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Romling, U., and D. Amikam. 2006. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9218-228. [DOI] [PubMed] [Google Scholar]
- 269.Romling, U., Z. Bian, M. Hammar, W. D. Sierralta, and S. Normark. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Romling, U., M. Gomelsky, and M. Y. Galperin. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57629-639. [DOI] [PubMed] [Google Scholar]
- 271.Romling, U., M. Rohde, A. Olsen, S. Normark, and J. Reinkoster. 2000. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 3610-23. [DOI] [PubMed] [Google Scholar]
- 272.Romling, U., W. D. Sierralta, K. Eriksson, and S. Normark. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28249-264. [DOI] [PubMed] [Google Scholar]
- 273.Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinberger-Ohana, R. Mayer, S. Braun, E. de Vroom, G. A. van der Marel, J. H. van Boom, and H. Benziman. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid Nature 325279-281. [DOI] [PubMed] [Google Scholar]
- 274.Ruby, E. G., and M. J. McFall-Ngai. 1992. A squid that glows in the night: development of an animal-bacterial mutualism. J. Bacteriol. 1744865-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Ryan, R. P., Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y. W. He, L. H. Zhang, S. Heeb, M. Camara, P. Williams, and J. M. Dow. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 1036712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 276.Ryan, R. P., Y. Fouhy, J. F. Lucey, and J. M. Dow. 2006. Cyclic di-GMP signaling in bacteria: recent advances and new puzzles. J. Bacteriol. 1888327-8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ryder, C., M. Byrd, and D. J. Wozniak. 2007. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 10644-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Ryjenkov, D. A., R. Simm, U. Romling, and M. Gomelsky. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 28130310-30314. [DOI] [PubMed] [Google Scholar]
- 279.Sakuragi, Y., and R. Kolter. 2007. Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J. Bacteriol. 1895383-5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Sauer, K., and A. K. Camper. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 1836579-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 1841140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Sauer, K., M. C. Cullen, A. H. Rickard, L. A. Zeef, D. G. Davies, and P. Gilbert. 2004. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 1867312-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 955857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Seidl, K., C. Goerke, C. Wolz, D. Mack, B. Berger-Bachi, and M. Bischoff. 2008. Staphylococcus aureus CcpA affects biofilm formation. Infect. Immun. 762044-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Senadheera, D., and D. G. Cvitkovitch. 2008. Quorum sensing and biofilm formation by Streptococcus mutans. Adv. Exp. Med. Biol. 631178-188. [DOI] [PubMed] [Google Scholar]
- 286.Shemesh, M., A. Tam, and D. Steinberg. 2007. Expression of biofilm-associated genes of Streptococcus mutans in response to glucose and sucrose. J. Med. Microbiol. 561528-1535. [DOI] [PubMed] [Google Scholar]
- 287.Sherlock, O., M. A. Schembri, A. Reisner, and P. Klemm. 2004. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J. Bacteriol. 1868058-8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Sherlock, O., R. M. Vejborg, and P. Klemm. 2005. The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self recognizing and induces bacterial aggregation and biofilm formation. Infect. Immun. 731954-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Shrout, J. D., D. L. Chopp, C. L. Just, M. Hentzer, M. Givskov, and M. R. Parsek. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 621264-1277. [DOI] [PubMed] [Google Scholar]
- 290.Silva, A. J., K. Pham, and J. A. Benitez. 2003. Haemagglutinin/protease expression and mucin gel penetration in El Tor biotype Vibrio cholerae. Microbiology 1491883-1891. [DOI] [PubMed] [Google Scholar]
- 291.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Romling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 531123-1134. [DOI] [PubMed] [Google Scholar]
- 292.Singh, P. K., M. R. Parsek, E. P. Greenberg, and M. J. Welsh. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417552-555. [DOI] [PubMed] [Google Scholar]
- 293.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407762-764. [DOI] [PubMed] [Google Scholar]
- 294.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA 986901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Smith, A. W. 2005. Biofilms and antibiotic therapy: is there a role for combating bacterial resistance by the use of novel drug delivery systems? Adv. Drug Deliv. Rev. 571539-1550. [DOI] [PubMed] [Google Scholar]
- 296.Smith, C. S., A. Hinz, D. Bodenmiller, D. E. Larson, and Y. V. Brun. 2003. Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus. J. Bacteriol. 1851432-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Solano, C., B. Garcia, J. Valle, C. Berasain, J. M. Ghigo, C. Gamazo, and I. Lasa. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43793-808. [DOI] [PubMed] [Google Scholar]
- 298.Speert, D. P., J. E. Dimmick, G. B. Pier, J. M. Saunders, R. E. Hancock, and N. Kelly. 1987. An immunohistological evaluation of Pseudomonas aeruginosa pulmonary infection in two patients with cystic fibrosis. Pediatr. Res. 22743-747. [DOI] [PubMed] [Google Scholar]
- 299.Spiers, A. J., J. Bohannon, S. M. Gehrig, and P. B. Rainey. 2003. Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 5015-27. [DOI] [PubMed] [Google Scholar]
- 300.Spoering, A. L., and K. Lewis. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 1836746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Sriramulu, D. D., H. Lunsdorf, J. S. Lam, and U. Romling. 2005. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 54667-676. [DOI] [PubMed] [Google Scholar]
- 302.Stanley, N. R., R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2003. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J. Bacteriol. 1851951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Stevenson, G., K. Andrianopoulos, M. Hobbs, and P. R. Reeves. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 1784885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Stewart, P. S., B. M. Peyton, W. J. Drury, and R. Murga. 1993. Quantitative observations of heterogeneities in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 59327-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Stoodley, P., A. Jacobsen, B. C. Dunsmore, B. Purevdorj, S. Wilson, H. M. Lappin-Scott, and J. W. Costerton. 2001. The influence of fluid shear and AICI3 on the material properties of Pseudomonas aeruginosa PAO1 and Desulfovibrio sp. EX265 biofilms. Water Sci. Technol. 43113-120. [PubMed] [Google Scholar]
- 306.Stoodley, P., Z. Lewandowski, J. D. Boyle, and H. M. Lappin-Scott. 1999. Structural deformation of bacterial biofilms caused by short-term fluctuations in fluid shear: an in situ investigation of biofilm rheology. Biotechnol. Bioeng. 6583-92. [PubMed] [Google Scholar]
- 307.Sudarsan, N., E. R. Lee, Z. Weinberg, R. H. Moy, J. N. Kim, K. H. Link, and R. R. Breaker. 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321411-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Sutherland, I. W. 2001. The biofilm matrix—an immobilized but dynamic microbial environment. Trends Microbiol. 9222-227. [DOI] [PubMed] [Google Scholar]
- 309.Suzuki, K., X. Wang, T. Weilbacher, A. K. Pernestig, O. Melefors, D. Georgellis, P. Babitzke, and T. Romeo. 2002. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol. 1845130-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Tabak, M., K. Scher, E. Hartog, U. Romling, K. R. Matthews, M. L. Chikindas, and S. Yaron. 2007. Effect of triclosan on Salmonella typhimurium at different growth stages and in biofilms. FEMS Microbiol. Lett. 267200-206. [DOI] [PubMed] [Google Scholar]
- 311.Tabor, C. W., and H. Tabor. 1984. Polyamines. Annu. Rev. Biochem. 53749-790. [DOI] [PubMed] [Google Scholar]
- 312.Taga, M. E., S. T. Miller, and B. L. Bassler. 2003. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 501411-1427. [DOI] [PubMed] [Google Scholar]
- 313.Tal, R., H. C. Wong, R. Calhoon, D. Gelfand, A. L. Fear, G. Volman, R. Mayer, P. Ross, D. Amikam, H. Weinhouse, A. Cohen, S. Sapir, P. Ohana, and M. Benziman. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 1804416-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Tamayo, R., A. D. Tischler, and A. Camilli. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 28033324-33330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Tanaka, G., M. Shigeta, H. Komatsuzawa, M. Sugai, H. Suginaka, and T. Usui. 1999. Effect of the growth rate of Pseudomonas aeruginosa biofilms on the susceptibility to antimicrobial agents: beta-lactams and fluoroquinolones. Chemotherapy 4528-36. [DOI] [PubMed] [Google Scholar]
- 316.Tart, A. H., M. J. Blanks, and D. J. Wozniak. 2006. The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J. Bacteriol. 1886483-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Thomas, V. C., L. R. Thurlow, D. Boyle, and L. E. Hancock. 2008. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J. Bacteriol. 1905690-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Thormann, K. M., S. Duttler, R. M. Saville, M. Hyodo, S. Shukla, Y. Hayakawa, and A. M. Spormann. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 1882681-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Thormann, K. M., R. M. Saville, S. Shukla, and A. M. Spormann. 2005. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J. Bacteriol. 1871014-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Tielker, D., S. Hacker, R. Loris, M. Strathmann, J. Wingender, S. Wilhelm, F. Rosenau, and K. E. Jaeger. 2005. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology 1511313-1323. [DOI] [PubMed] [Google Scholar]
- 321.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Tischler, A. D., and A. Camilli. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 735873-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Toledo-Arana, A., J. Valle, C. Solano, M. J. Arrizubieta, C. Cucarella, M. Lamata, B. Amorena, J. Leiva, J. R. Penades, and I. Lasa. 2001. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 674538-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Tormo, M. A., E. Knecht, F. Gotz, I. Lasa, and J. R. Penades. 2005. Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology 1512465-2475. [DOI] [PubMed] [Google Scholar]
- 325.Toutain, C. M., N. C. Caizza, M. E. Zegans, and G. A. O'Toole. 2007. Roles for flagellar stators in biofilm formation by Pseudomonas aeruginosa. Res. Microbiol. 158471-477. [DOI] [PubMed] [Google Scholar]
- 326.Tsang, P. H., G. Li, Y. V. Brun, L. B. Freund, and J. X. Tang. 2006. Adhesion of single bacterial cells in the micronewton range. Proc. Natl. Acad. Sci. USA 1035764-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Ude, S., D. L. Arnold, C. D. Moon, T. Timms-Wilson, and A. J. Spiers. 2006. Biofilm formation and cellulose expression among diverse environmental Pseudomonas isolates. Environ. Microbiol. 81997-2011. [DOI] [PubMed] [Google Scholar]
- 328.Van Dellen, K. L., L. Houot, and P. I. Watnick. 2008. Genetic analysis of Vibrio cholerae monolayer formation reveals a key role for DeltaPsi in the transition to permanent attachment. J. Bacteriol. 1908185-8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.van Schaik, E. J., C. L. Giltner, G. F. Audette, D. W. Keizer, D. L. Bautista, C. M. Slupsky, B. D. Sykes, and R. T. Irvin. 2005. DNA binding: a novel function of Pseudomonas aeruginosa type IV pili. J. Bacteriol. 1871455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Vasil, M. L., and U. A. Ochsner. 1999. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol. Microbiol. 34399-413. [DOI] [PubMed] [Google Scholar]
- 331.Vasseur, P., I. Vallet-Gely, C. Soscia, S. Genin, and A. Filloux. 2005. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology 151985-997. [DOI] [PubMed] [Google Scholar]
- 332.Vats, N., and S. F. Lee. 2000. Active detachment of Streptococcus mutans cells adhered to epon-hydroxylapatite surfaces coated with salivary proteins in vitro. Arch. Oral Biol. 45305-314. [DOI] [PubMed] [Google Scholar]
- 333.Ventre, I., A. L. Goodman, I. Vallet-Gely, P. Vasseur, C. Soscia, S. Molin, S. Bleves, A. Lazdunski, S. Lory, and A. Filloux. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. USA 103171-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Videla, H. A., and L. K. Herrera. 2005. Microbiologically influenced corrosion: looking to the future. Int. Microbiol. 8169-180. [PubMed] [Google Scholar]
- 335.Vilain, S., and V. S. Brozel. 2006. Multivariate approach to comparing whole-cell proteomes of Bacillus cereus indicates a biofilm-specific proteome. J. Proteome Res. 51924-1930. [DOI] [PubMed] [Google Scholar]
- 336.Vuong, C., S. Kocianova, J. M. Voyich, Y. Yao, E. R. Fischer, F. R. DeLeo, and M. Otto. 2004. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 27954881-54886. [DOI] [PubMed] [Google Scholar]
- 337.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 1821688-1693. [DOI] [PubMed] [Google Scholar]
- 338.Wai, S. N., Y. Mizunoe, A. Takade, S. I. Kawabata, and S. I. Yoshida. 1998. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 643648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Waite, R. D., A. Paccanaro, A. Papakonstantinopoulou, J. M. Hurst, M. Saqi, E. Littler, and M. A. Curtis. 2006. Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Genomics 7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Walters, M. C., III, F. Roe, A. Bugnicourt, M. J. Franklin, and P. S. Stewart. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Wang, X., A. K. Dubey, K. Suzuki, C. S. Baker, P. Babitzke, and T. Romeo. 2005. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 561648-1663. [DOI] [PubMed] [Google Scholar]
- 342.Wang, X., J. F. Preston III, and T. Romeo. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 1862724-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Waters, C. M., W. Lu, J. D. Rabinowitz, and B. L. Bassler. 2008. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 1902527-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae biofilm. Mol. Microbiol. 34586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Reference deleted.
- 346.Watnick, P. I., C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter. 2001. Absence of a flagellum leads to altered colony morphology, biofilm development, and virulence in V. cholerae O139. Mol. Microbiol. 39223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 1854585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Wei, B. L., A. M. Brun-Zinkernagel, J. W. Simecka, B. M. Pruss, P. Babitzke, and T. Romeo. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40245-256. [DOI] [PubMed] [Google Scholar]
- 349.Weinhouse, H., S. Sapir, D. Amikam, Y. Shilo, G. Volman, P. Ohana, and M. Benziman. 1997. c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 416207-211. [DOI] [PubMed] [Google Scholar]
- 350.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 2951487. [DOI] [PubMed] [Google Scholar]
- 351.Wozniak, D. J., T. J. Wyckoff, M. Starkey, R. Keyser, P. Azadi, G. A. O'Toole, and M. R. Parsek. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. USA 1007907-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Xavier, K. B., and B. L. Bassler. 2005. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187238-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Yang, L., K. B. Barken, M. E. Skindersoe, A. B. Christensen, M. Givskov, and T. Tolker-Nielsen. 2007. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 1531318-1328. [DOI] [PubMed] [Google Scholar]
- 354.Yao, Y., D. E. Sturdevant, and M. Otto. 2005. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J. Infect. Dis. 191289-298. [DOI] [PubMed] [Google Scholar]
- 355.Yarwood, J. M., D. J. Bartels, E. M. Volper, and E. P. Greenberg. 2004. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 1861838-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 1831716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Yildiz, F. H., X. S. Liu, A. Heydorn, and G. K. Schoolnik. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53497-515. [DOI] [PubMed] [Google Scholar]
- 358.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 964028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Yoon, S. S., R. F. Hennigan, G. M. Hilliard, U. A. Ochsner, K. Parvatiyar, M. C. Kamani, H. L. Allen, T. R. DeKievit, P. R. Gardner, U. Schwab, J. J. Rowe, B. H. Iglewski, T. R. McDermott, R. P. Mason, D. J. Wozniak, R. E. Hancock, M. R. Parsek, T. L. Noah, R. C. Boucher, and D. J. Hassett. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3593-603. [DOI] [PubMed] [Google Scholar]
- 360.Zhu, J., and J. J. Mekalanos. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5647-656. [DOI] [PubMed] [Google Scholar]
- 361.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 993129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Zogaj, X., W. Bokranz, M. Nimtz, and U. Romling. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 714151-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Romling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 391452-1463. [DOI] [PubMed] [Google Scholar]