Abstract
Summary: Bacteria live in environments that are subject to rapid changes in the availability of the nutrients that are necessary to provide energy and biosynthetic intermediates for the synthesis of macromolecules. Consequently, bacterial survival depends on the ability of bacteria to regulate the expression of genes coding for enzymes required for growth in the altered environment. In pathogenic bacteria, adaptation to an altered environment often includes activating the transcription of virulence genes; hence, many virulence genes are regulated by environmental and nutritional signals. Consistent with this observation, the regulation of most, if not all, virulence determinants in staphylococci is mediated by environmental and nutritional signals. Some of these external signals can be directly transduced into a regulatory response by two-component regulators such as SrrAB; however, other external signals require transduction into intracellular signals. Many of the external environmental and nutritional signals that regulate virulence determinant expression can also alter bacterial metabolic status (e.g., iron limitation). Altering the metabolic status results in the transduction of external signals into intracellular metabolic signals that can be “sensed” by regulatory proteins (e.g., CodY, Rex, and GlnR). This review uses information derived primarily using Bacillus subtilis and Escherichia coli to articulate how gram-positive pathogens, with emphasis on Staphylococcus aureus and Staphylococcus epidermidis, regulate virulence determinant expression in response to a changing environment.
INTRODUCTION
All bacteria require carbon and energy for growth and replication. These are usually derived from the enzymatic breakdown of more-complex organic molecules such as carbohydrates, lipids, and proteins, a process collectively known as catabolism. Nonpathogenic bacteria can derive carbon and energy from the environment, either free-living or symbiotically from a host. However, pathogenic bacteria, at least transiently, derive their carbon and energy parasitically or destructively from a host organism. This is accomplished in part by the synthesis of a wide assortment of virulence determinants that are capable of killing host cells and catabolizing macromolecules. Therefore, it is not surprising that the regulation of many virulence determinants is controlled by nutrient availability. This connection between virulence determinant (erepsin/protease) synthesis and nutrient availability is one of the earliest physiological observations to have been reported and studied by numerous researchers (see, e.g., references 16 and 114). Similarly, from the examination of Staphylococcus aureus cells growing on blood agar plates to the earliest quantitative measurements of toxin accumulation, it has been observed that a connection between growth conditions and toxin biosynthesis existed (27, 39, 57, 93, 188, 233). While the connection between growth conditions and nutrient availability was first made a century ago, the study of bacterial physiology and studies of virulence have progressed independently. In this review, we will examine the more-recent data that demonstrate how metabolism has profound effects on the biosynthesis of virulence determinants and antibiotic resistance. We will use staphylococci as the major genus of focus, but parallels to other gram-positive organisms as well as gram-negative bacteria will be drawn.
OVERVIEW OF STAPHYLOCOCCAL VIRULENCE FACTOR REGULATION
In order to understand the relationship between virulence factor expression and metabolism, a basic understanding of staphylococcal virulence determinant regulation is required. The first S. aureus regulator of toxin biosynthesis identified was the accessory gene regulator (Agr) (181). The agr system consists of two divergently transcribed loci (∼3 kb) controlled by two promoters, P2 and P3, that regulate the transcription of a cell-density-sensing two-component regulator and a regulatory RNA known as RNAIII (166). There are four genes controlled by P2: agrA, agrC, agrD, and agrB. AgrC is the transmembrane histidine kinase component of the agr two-component regulatory system, with AgrA being the response regulator. AgrD is a small peptide that is processed by AgrB into a cyclic thiolactone peptide also known as the autoinducing peptide (108, 150). When the extracellular concentration of the autoinducing peptide reaches a threshold level, the probability that it will complex with AgrC is increased, leading to the activation of the kinase domain (136). Upon the transfer of a phosphate from AgrC to AgrA, AgrA is activated and increases transcription from the P2 and P3 promoters (122). Transcription from P2 is a type of autocrine regulation that increases the biosynthesis of the agr cell-density-sensing system. Transcription from P3 produces the predominantly untranslated riboregulator RNAIII (107, 167). RNAIII enhances the synthesis of secreted virulence determinants (e.g., alpha-toxin and serine protease) and represses the synthesis of surface-associated proteins (e.g., protein A) (181). The regulatory effect of RNAIII is elicited primarily through its action as an antisense RNA that anneals to agr-regulated mRNAs such as hla (alpha-toxin) and spa (protein A) (13, 21, 100, 155). In addition, RNAIII facilitates the degradation of RNAIII-mRNA complexes by directing the activity of endoribonuclease III (RNase III) (100). Although cell density sensing by the agr system is of critical importance for the regulation of virulence determinant synthesis, a postexponential growth phase signal independent of agr is also required (227).
The second major regulatory locus to be identified in S. aureus was found during Tn917 transposon mutagenesis, and it contained the staphylococcal accessory regulator (sar) (33). Encoded within the sar locus is SarA, a 124-amino-acid (14.7-kDa) DNA binding protein (35, 138). The sar locus contains three promoters (P1, P2, and P3), each capable of producing a transcript encoding SarA. The transcription of sarA from the P1 and P2 promoters is dependent upon σ70, while transcription from the P3 promoter requires the alternative sigma factor σB (18; see below). The inactivation of sarA increases the transcription of spa (protein A) and represses alpha-toxin (hla) biosynthesis (31, 32). SarA functions, in part, by regulating the transcription of agrACDB and RNAIII (30, 156). While there is no question that SarA is required for virulence determinant regulation, the mechanism by which SarA manifests its regulatory effects remains to be settled (36, 64, 138, 186, 217). Compounding the difficulty in defining SarA's mechanism of action is the fact that SarA represents one member of the SarA family of proteins, which consists of at least 11 members (34).
RNA polymerase is a multisubunit enzyme that binds to dissociable initiation factor sigma (σ) to start the process of transcription (78). When bound to RNA polymerase, the σ factor provides DNA sequence specificity and the ability to form an open promoter complex. S. aureus and Staphylococcus epidermidis have three known sigma factors: σ70, which is responsible for the transcription initiation of most genes; σH, which initiates transcription at a limited set of competence related genes (157); and σB, which is activated under stress conditions, during growth phase transitions, and during morphological changes (17, 204, 237). The transcription and synthesis of staphylococcal virulence determinants are influenced by environmental and nutritional stresses; hence, the stress-dependent activation of σB has been a focal point of staphylococcal research into the environmental regulation of virulence factor biosynthesis (72, 121, 126, 172). The activation of σB resembles that found in the closely related organism Bacillus subtilis and involves an anti-sigma factor (RsbW) and an anti-anti-sigma factor (RsbV) (121, 204). In the absence of environmental stimuli, σB is bound in a complex with RsbW. Stress-inducing stimuli are hypothesized to activate the phosphatase RsbU to dephosphorylate (activate) the anti-anti-sigma factor RsbV, which then binds RsbW in a competitive manner to increase the concentration of free σB (72, 120, 121, 172). Increasing the concentration of free σB allows for its association with core RNA polymerase and subsequent binding to σB-dependent promoters, resulting in alterations of virulence, stress, and metabolic gene transcription (17, 95, 127). In addition to these regulatory elements, the S. aureus virulon is temporally regulated by the overall energy metabolism of the cell (165).
OVERVIEW OF STAPHYLOCOCCAL METABOLISM
All macromolecules in a bacterium can be synthesized from 13 biosynthetic intermediates that are derived from the Embden-Meyerhof-Parnas (glycolytic), pentose phosphate, and tricarboxylic acid (TCA) cycle pathways. S. aureus and S. epidermidis have complete metabolic pathways for all three pathways; however, the TCA cycle lacks a glyoxylate shunt in both species. Carbohydrates are catabolized primarily through the glycolytic and pentose phosphate pathways, but TCA cycle activity is largely repressed when nutrients are abundant in the culture medium (40, 219). Glycolysis produces two molecules of pyruvate for every molecule of glucose consumed and in the process reduces two molecules of NAD+ to NADH. The catabolic fate of pyruvate is determined by the growth conditions, specifically, the availability of oxygen. During anaerobic growth, pyruvate is reduced primarily to lactic acid (115, 125), with the concomitant reoxidation of NADH allowing for the continuation of glycolysis. In aerobically grown staphylococci, pyruvate is enzymatically oxidized to acetyl coenzyme A (acetyl-CoA) and CO2 by the pyruvate dehydrogenase complex (71). Acetyl-CoA can be further oxidized by the TCA cycle when grown in the presence of certain citric acid cycle intermediates (77); however, the amount of acetyl-CoA that enters into the TCA cycle is low during nutrient-rich growth (40, 214, 219). In the exponential growth phase, acetyl-CoA is used to generate the small phosphodonor acetyl-phosphate, which serves as a substrate for acetate kinase in substrate-level phosphorylation to generate ATP and acetic acid (acetogenesis). Exit from the exponential phase of growth occurs when the concentration of an essential nutrient (e.g., glucose) decreases to a level where it can no longer sustain rapid growth or by the accumulation of growth-inhibitory molecules (e.g., lactic acid and acetic acid). If conditions are favorable, then entry into postexponential growth corresponds with the catabolism of nonpreferred carbon sources such as acetate (213). The postexponential catabolism of nonpreferred carbon sources requires TCA cycle activity and coincides with a large increase in TCA cycle enzymatic activity (215). Because the TCA cycle provides biosynthetic intermediates, ATP, and reducing potential, the transition to TCA cycle-driven metabolism dramatically alters the staphylococcal metabolome.
The TCA cycle-mediated alteration of the staphylococcal metabolome increases the availability of biosynthetic precursors for use in a complete set of amino acid and nucleic acid biosynthetic pathways (4, 5, 50, 74, 92, 129). Because the biosynthesis of many amino acids and nucleic acids requires oxidized dinucleotide cofactors (i.e., NAD+, FAD+, and NADP+) and TCA cycle activity generates reduced dinucleotides, the postexponential growth phase also coincides with increased oxidative phosphorylation activity. Therefore, the transition from the exponential phase to the postexponential phase of growth coincides with a dramatic change in the redox status of the bacteria. The oxidation of large amounts of NADH via oxidative phosphorylation requires the electron transfer chain.
S. aureus grows well aerobically or anaerobically and can use oxygen, nitrate, or nitrite as electron acceptors in the electron transfer chain. Despite this versatility, S. aureus, like most gram-positive bacteria, synthesizes only one quinone, menaquinone, which is used for growth at all levels of oxygen (133). During aerobic growth, electrons enter into the electron transport chain from NADH and are transferred to menaquinone by the NADH dehydrogenase complex (10). Menaquinone transfers the electrons to oxidized cytochrome c, generating the reduced form of cytochrome c. The electrons are then transferred to oxygen by cytochrome c oxidase (this process can also generate reactive oxygen species [ROS]), generating water and driving protons across the membrane to produce a pH and electrochemical gradient. Protons return to the cytoplasm by way of the Fo subunit of the FoF1 ATP synthase complex and drive the dissociation of newly formed ATP. The inactivation of the electron transfer chain, either by genetic inactivation (63, 124) or by extreme iron restriction (62), results in a growth phenotype similar to that of anaerobic/fermentative growth. The inactivation of the electron transfer chain produces a clinically relevant phenotype in staphylococci known as the small-colony variant phenotype.
METABOLIC AND NUTRIENT-RESPONSIVE REGULATORS
Catabolite-Responsive Regulators
CcpA.
One of the first types of regulation to be defined was glucose-mediated regulation, later termed catabolite repression (49, 145, 175). The purpose of metabolic regulation is to avoid wasting carbon- and energy-synthesizing macromolecules when the synthesis of these macromolecules would be deleterious to the organism. As an example, Escherichia coli cells growing on glucose as a carbon source do not require β-galactosidase; hence, the transcription of the lac operon is repressed to prevent the diversion of carbon and energy from growth to transcription and translation of the lac operon. This repression of the lac operon requires the presence of a regulatory protein, the Lac repressor (LacI) (73). The derepression of the lac operon occurs during basal-level lactose catabolism when a secondary metabolite (1,6-allolactose) is generated. When LacI binds 1,6-allolactose, the affinity of LacI for the operator site diminishes; hence, transcription is derepressed. Like the lac operon, many staphylococcal virulence genes are regulated by catabolite repression.
In gram-positive bacteria, carbon catabolite repression is mediated primarily by catabolite control protein A (CcpA) (89), a member of the GalR-LacI repressor family (231). CcpA regulatory activity is controlled by interactions with the phosphorylated corepressor HPr (histidine-containing protein) or Crh (catabolite repression HPr) (46, 47, 70, 236). Importantly, it is not glucose or some other rapidly catabolized carbohydrate that directs the repression of catabolite-repressed genes but the glycolytic intermediates derived from these carbohydrates such as glucose-6-phosphate and fructose-1,6-bisphosphate (139). The glycolytic intermediates increase the ATP-dependent phosphorylation of the corepressor HPr by enhancing the activity of HPr kinase (48). Similarly, the phosphorylation of Crh is also enhanced by glycolytic intermediates in an HPr kinase-dependent fashion (70). In addition to enhancing the phosphorylation of the corepressors HPr and Crh, glucose-6-phosphate and fructose-1,6-bisphosphate are bound by CcpA when it is complexed with the phosphorylated corepressors (199). When CcpA is in its DNA binding-competent form, it regulates transcription through interactions with catabolite-responsive elements (cre) located in or near carbon catabolite-repressed promoters (164). This repression appears to involve interacting with RNA polymerase to block transcription initiation but not DNA binding (117). Overall, this intricate control of CcpA activation serves to link the bacterial metabolic status to transcriptional regulation.
Staphylococcus aureus cells grown under carbohydrate-rich conditions repress the synthesis of capsular polysaccharides (202) and toxigenic exoproteins (42, 52, 183, 201, 211). In contrast, the biosynthesis of surface proteins including microbial surface components recognizing adhesive matrix molecules is enhanced under carbohydrate-rich conditions (169, 183). Because the biosynthesis of exoproteins and surface proteins is inversely correlated and regulated by RNAIII, it was hypothesized that the carbohydrate-dependent repression of exoprotein synthesis indirectly involves the agr system (183). Consistent with this hypothesis, Seidl and coworkers found that RNAIII was synthesized only when glucose was depleted from the culture medium (202). Despite these similar observations, the glucose-mediated repression of exoprotein synthesis appears to be independent of the agr system (182, 201). The effort to identify the mechanism by which staphylococci regulate virulence determinant synthesis was greatly aided by the discovery of a functional CcpA homolog in Staphylococcus xylosus (55), the subsequent identification of HPr kinase (101), and the demonstration that CcpA-dependent regulation required HPr kinase (106). With these data, it seemed reasonable to speculate that carbon catabolite repression of virulence determinants in the closely related S. aureus might involve CcpA. However, as with most things relating to staphylococcal virulence regulation, it is more complex than advertised.
The inactivation of ccpA in S. aureus results in a lower level of RNAIII (202), suggesting that the synthesis of RNAIII positively regulated virulence determinants, like alpha-toxin and capsule, would decrease. In contrast to this expectation, ccpA inactivation increased the transcription, or mRNA stability, of capsular polysaccharide biosynthetic genes and capsule biosynthesis (202); however, the effect on alpha-toxin biosynthesis was less clear. The level of transcription of hla (alpha-toxin) was slightly decreased in a ccpA mutant strain relative to that of the isogenic parental strain, suggesting that CcpA has only a minor function in the carbon catabolite repression of alpha-toxin synthesis (202). Because protein A (spa) synthesis is repressed by RNAIII, the lower level of RNAIII in a ccpA mutant strain would be predicted to increase the biosynthesis of protein A, and this was observed (100). Adding to the confusion about CcpA-mediated catabolite repression in S. aureus is the fact that cre sites were found upstream of hla and spa; however, no cre sites were identified near agr or the capsule genes (202). Perhaps the strongest case for the direct CcpA-mediated repression of a staphylococcal virulence factor is that of toxic shock syndrome toxin 1 (TSST-1). TSST-1 synthesis is repressed by glucose (194) and has a cre site located within the promoter region (201), and ccpA mutants derepress tst transcription (201). Taken together, these observations suggest that CcpA has a complex regulatory function in S. aureus and that another regulatory protein(s) and RNAs are involved in carbon catabolite repression of virulence factors.
CodY.
As stated above in this review, some of the earliest physiological observations relating to gram-positive virulence factors were that carbohydrates suppress ammonia generation in protein-based culture media (9, 15, 16). Specifically, carbohydrates were repressing three processes: (i) the proteolytic breakdown of proteins into peptides and amino acids, (ii) the transport of peptides and amino acids, and (iii) amino acid catabolism. It is the latter process that was the cause of the suppression of ammonia generation in the culture medium, as an essential step in amino acid catabolism is the deamination of amino acids. Although unknown at the time, the first process demonstrates a linkage between virulence determinant expression (proteases) and bacterial nutrient status. On the surface, the carbohydrate-dependent suppression of ammonia generation would appear to be CcpA mediated; however, transcriptional profiling studies suggest that CcpA has only a limited role in regulating gram-positive proteolysis and amino acid catabolism (119, 228). However, CcpA can act additively or antagonistically with another regulator, CodY, to regulate carbon and nitrogen metabolism (206, 207), raising the possibility that these early observations were the first descriptions of the regulatory affects of CodY.
CodY is a highly conserved gram-positive repressor that responds to intracellular concentrations of branched-chain amino acids (BCAA) and GTP (79, 207, 210, 216, 222). By responding to BCAA, CodY is sensing carbon and nitrogen availability, and by responding to GTP, CodY is linked with the stringent response (103). CodY binds to a 15-bp A/T-rich region of DNA (45) as a dimer through a winged helix-turn-helix domain (19, 134). The interaction of CodY with BCAA is mediated through a cyclic GMP, adenylyl cyclase, FhlA (GAF) domain; however, the mechanism of interaction with GTP remains speculative (134). CodY's affinity for its DNA binding site is increased by binding, but not hydrolyzing, GTP, and this affinity is increased further by a BCAA interacting with the GAF domain (82). These observations highlight the central role of CodY in gram-positive metabolic regulation.
The vital role of CodY in gram-positive metabolic regulation, and the fact that many virulence determinants display metabolic regulation, led to the hypothesis that CodY regulates virulence factor synthesis (149, 216). This hypothesis has proven to be correct for Bacillus cereus, Clostridium difficile, Listeria monocytogenes, S. aureus, Streptococcus pneumoniae, and Streptococcus pyogenes (14, 51, 87, 132, 147, 149). Similar to CodY in B. subtilis, CodY in pathogenic bacteria represses genes involved in amino acid transport, catabolism, and biosynthesis. In S. pyogenes, CodY influences the transcription of the global regulators Mga and RopB and the two-component regulatory system CovRS, which in turn affect the expression of numerous virulence determinants including cysteine protease (speB), M protein, capsule, and the surface-bound C5a peptidase (scpA) (148, 149). B. cereus CodY coordinates the regulation of protease activity and peptide transport via the dipeptide permease (99). The inactivation of codY in S. pneumoniae dramatically decreases adherence to nasopharyngeal cells and derepresses the synthesis of oligopeptide permeases, suggesting that the decreased adherence to nasopharyngeal cells may be linked to increased proteolytic activity (87). In C. difficile, CodY binds to the promoter region of the tcdR gene, which codes for an alternative sigma factor that is required for the synthesis of the large clostridial cytotoxins toxin A and toxin B (51). CodY in L. monocytogenes affects amino acid transport, catabolism, and biosynthesis; however, its function in pathogenesis is less clear (14). The inactivation of codY in L. monocytogenes affects motility and chemotaxis but has only a slight effect on virulence in a murine listeriosis model (14). The genetic inactivation of codY in two clinical isolates of S. aureus derepresses the synthesis of the agr loci, polysaccharide intercellular adhesin (PIA), and alpha-toxin (hla) (147). In addition, S. aureus CodY coordinately represses the expression of cysteine protease, serine protease, V8 protease, peptide and amino acid transporters, and amino acid catabolic genes (C. Majerczyk et al., unpublished observations). One interesting effect of CodY is its differential effect on bacterial growth in a biofilm. Streptococcus mutans and B. cereus codY mutant strains have reduced capacities to form biofilms (99, 132); however, S. aureus codY mutants have an increased capacity to form biofilms (147). Although further research is needed to understand these differences in biofilm growth, one possible explanation may be due to differences in the relative importance of exopolysaccharides versus proteins for facilitating adherence to the substratum.
Nitrogen-Dependent Regulators
Although less well studied in staphylococci than catabolite repression, amino-acid-dependent regulation of virulence determinants and antibiotic resistance has been demonstrated (80, 218, 220). However, most studies involving staphylococci and amino acids have focused on the amino acid requirements for growth. Phenotypic studies of S. aureus that have attempted to determine the absolute amino acid requirements necessary for growth have met with diverse results (56, 61, 146, 168, 170, 220). Those studies concluded that S. aureus frequently possessed multiple amino acid auxotrophies because growth often required between 3 and 12 amino acids, with proline, arginine, valine, and cysteine being most frequently required. Interestingly, it was also observed that these auxotrophies would revert to a prototrophic state at a high frequency, suggesting that the auxotrophies were not due to the absence, or genetic inactivation, of biosynthetic pathways (56, 75). Whole-genome sequencing of S. aureus confirmed this suggestion when it was determined that biosynthetic pathways exist for all amino acids (4, 5, 50, 74, 92, 129).
GlnR.
Glutamine is synthesized by glutamine synthetase (glnA) from glutamate and ammonia, a process requiring ATP. This is a critical reaction because glutamine and glutamate are the major intracellular nitrogen donors, and glutamine is also involved in the osmotic protection of S. aureus (2). In order to maintain an optimal intracellular concentration of glutamine, bacteria strictly regulate ammonia assimilation and the catabolism of amino acids. In gram-positive bacteria, nitrogen-dependent transcriptional regulation is mediated primarily by GlnR and TnrA (196, 235). Although staphylococci do not appear to have TnrA, they do have a glutamine synthetase repressor (GlnR) (197, 218). GlnR is a member of the MerR family of regulators (162, 196) that regulates glnRA expression in B. subtilis (198). In S. aureus, glutamine and the regulation of glnRA have been demonstrated to be important for maintaining methicillin resistance in methicillin-resistant S. aureus strains (80, 218). Although GlnR in S. pneumoniae is not required for virulence, GlnR-regulated targets (i.e., glnA and glnP) are important for colonization and survival during an infection (88).
NreC.
Nitrogen has important functions in bacteria beyond ammonia assimilation. As stated previously, S. aureus can use oxygen, nitrate, or nitrite as electron acceptors in the electron transfer chain; thus, S. aureus must have a means to “sense” the presence or absence of these electron acceptors. S. aureus uses a two-component regulatory system encoded by NreABC that consists of a histidine sensor kinase (NreB) and a response regulator (NreC) (112, 192); the function of NreA has yet to be determined. NreB is a cytoplasmic protein that contains a cysteine-coordinated, oxygen-sensitive, iron-sulfur cluster similar to the fumarate nitrate reductase regulator (112). Aerobic conditions cause the reversible loss of the iron-sulfur and dramatically reduce the autophosphorylation of NreB. Under anaerobic conditions with Fe2+, the iron-sulfur cluster is intact, facilitating the autophosphorylation of NreB, which transfers the phosphate to NreC. In Staphylococcus carnosus, the transfer of the NreB phosphate to NreC activates the transcription of the nitrite reductase (annotated as nasDE or nirBD) and respiratory nitrate reductase (narGHJI) operons and the nitrite and/or nitrate transporter (narT) (59). Similarly, S. aureus NreC activates the transcription of the nitrate and nitrite reductase operons and the nitrite extrusion protein (NarK) and differentially regulates several nitrogen metabolism genes (e.g., arcABDC, ureBCEF, and glnRA) (192). Under anaerobic conditions and in the absence of nitrate, NreC derepresses the transcription of genes involved in fermentative metabolism, such as the dissimilatory lactate dehydrogenase (lctE) and the 2,3-butanediol pathway (alsS) (192). Interestingly, several of the genes repressed by NreC in S. aureus have been identified as being important for biofilm growth and/or maturation (e.g., lctE and alsS) (11, 184). NreC-mediated repression of biofilm-associated genes may explain the observation that nitrite represses PIA and biofilm formation (193).
CodY.
The biosynthesis of valine and leucine requires the α-keto acid pyruvate and the amino group from glutamate, meaning that the biosyntheses of valine and leucine are the products of two major metabolic pathways, glycolysis and the TCA cycle. Isoleucine is synthesized from α-ketobutyrate, which is required not only for isoleucine synthesis but also for sulfur metabolism. By responding to BCAA, bacteria are sensing the overall carbon, nitrogen, and sulfur metabolic status. For more information on CodY, see above.
REDOX-RESPONSIVE REGULATION
The function of many catabolic pathways requires oxidized dinucleotides such NAD+, NADP+, and FAD+, with the TCA cycle creating the greatest demand for NAD+. Thus, catabolism generates reducing equivalents (i.e., NADH, NADPH, and FADH2) that can channel electrons into the respiratory chain (see above). Importantly, the reduction and oxidation of dinucleotides create an internal signal to which certain regulatory proteins can respond.
Rex (YdiH)
Several gram-positive bacteria (i.e., Streptomyces coelicolor, Thermus aquaticus, B. subtilis, L. monocytogenes, and S. aureus) have a redox-dependent transcriptional repressor (Rex) that plays a key role in regulating anaerobic metabolism (23). Rex proteins have predicted molecular masses of between 22.8 and 26.8 kDa and sense the bacterial redox status through changes in the NADH-NAD+ poise (NADH/NAD+ ratio) of the bacterium (23, 208). Responding to the NADH/NAD+ ratio meets several important needs. First, the NADH/NAD+ ratio permits the redox status to be monitored under aerobic and anaerobic conditions (i.e., independent of oxygen). Second, the redox poise can change dramatically under different metabolic states without a change in oxygen availability; thus, it monitors metabolism and oxygen. Third, other stresses in the cell (e.g., membrane disruption or oxidative challenge) will alter the NADH/NAD+ ratio, thereby allowing increased production of NADH when needed.
Rex is a winged-helix DNA binding protein with a typical Rossmann fold dinucleotide binding site (69, 208). X-ray crystallographic data from T. aquaticus Rex suggest that Rex exists as a homodimeric protein, with each monomer containing winged helix and NAD(H) binding domains (208). The binding of NADH to Rex generates an allosteric change that prevents the association of the dinucleotide-protein complex with Rex-regulated operators, derepressing transcription (69, 189, 208). In other words, when intracellular NADH concentrations increase relative to NAD+ concentrations, Rex disassociates from the operator, allowing gene activation. In S. aureus, the Rex binding site is TGTGAa/t6TCACA (M. Pagels et al., unpublished data), with similar sequences in L. monocytogenes, B. subtilis, and S. coelicolor (23, 81, 130, 191). Computer modeling suggests that Rex interacts with both the major and minor DNA grooves (69).
The intracellular concentration of NADH transiently increases during glycolysis; however, a more sustained increase occurs with the activation of the TCA cycle or when organisms encounter anaerobic conditions. Based on these observations, it is not surprising that Rex regulates the transcription of genes involved in respiration and metabolism that decrease NADH concentrations and help maintain the optimal redox status in the cell (Pagels et al., unpublished). Specifically, Rex-NADH dissociates from the repressor sites, allowing the transcription of genes required for (i) the synthesis of components of the electron transport chains (e.g., hemA, cydBD, and nuoA to nuoN), (ii) regulators of nitrogen and anaerobic metabolism (e.g., nirR, nirC, ywcJ, and srrA), (iii) fermentative enzymes (adhE, adh1, and alsS), and (iv) enzymes involved in pyruvate/lactate metabolism (lctE and lctP) and anaerobic metabolism (e.g., pflB and arcA) (23, 81, 130; Pagels et al., unpublished). In addition to regulating genes involved in maintaining the redox status, Rex also appears to regulate some S. aureus toxins, such as the leukocidin gene lukM (Pagels et al., unpublished). As mentioned above, Rex-NAD+ is a repressor of the oxygen-dependent two-component regulator SrrAB (see below), which positively regulates S. aureus toxin expression, suggesting that the Rex repression of srrAB transcription may be one of the reasons that toxins are repressed during anaerobic growth in S. aureus.
OXYGEN-DEPENDENT REGULATORS
During in vivo and in vitro growth, bacteria encounter oxygen in several forms, from the beneficial triplet oxygen (O2) and water (H2O) to the more harmful oxidants superoxide (O2−), hydrogen peroxide (H2O2), and hydroxyl radicals (OH·). The beneficial forms are necessary for numerous biochemical reactions (e.g., the hydration of cis-aconitate to form isocitrate and the oxidation of reduced flavin adenine dinucleotide). However, in the example of reduced flavin adenine dinucleotide, the addition of one electron to O2 generates O2−, a ROS that can damage iron-sulfur clusters, DNA, and sulfhydryl groups (102). Dismutation of the superoxide anion radical generates the more-stable H2O2. If oxidative stress has induced the release of iron from iron-sulfur clusters, this creates an intracellular environment permissive to Fenton chemistry. Fenton chemistry generates the highly reactive OH· from ferrous iron and H2O2. Pathogenic bacteria must respond not only to endogenous metabolism-generated ROS but also to host-generated exogenous ROS. These observations highlight the bacterial need for oxygen-responsive regulators.
SrrAB
In B. subtilis, the two-component regulatory system ResDE is required for anaerobic respiration (161). ResE is a membrane-associated sensor kinase that transfers a phosphate to the response regulator ResD. ResD activates the transcription of anaerobically induced genes such as fnr, hmp, and nasD (160, 240). During aerobic growth, the transcription of ResDE-regulated genes is induced in the postexponential and stationary phases of growth; however, transcription is induced to a greater extent during oxygen-limited growth. In addition to anaerobic growth, ResD-mediated transcriptional activation is enhanced by nitric oxide (NO) (159). In B. cereus, ResDE acts as an anaerobic redox regulator to regulate fermentative metabolism and enterotoxin expression (53). The inactivation of resD in Bacillus anthracis blocks sporulation; however, its function in regulating the expression of toxin biosynthesis is unsettled (229, 232). Two independent groups constructed B. anthracis ResD mutants (also known as BrrA) and produced conflicting results regarding the effects on toxin biosynthesis (229, 232). A study by Vetter and Schleivert demonstrated that the inactivation of BrrA dramatically reduced the synthesis of lethal factor, edema factor, and protective antigen; however, the group led by Perego failed to observe a decrease in protective antigen synthesis (229, 232). In staphylococci, the ResDE homolog is known as the staphylococcal respiratory response (SrrAB, also known as SrhSR) (221, 238).
SrrB is a membrane-associated protein, and SrrA is a cytoplasmic DNA binding protein whose DNA binding activity is influenced by phosphorylation (178, 226). In S. aureus, SrrAB influences the expression of RNAIII, TSST-1, protein A, and IcaR in response to oxygen availability (177, 178). In addition, SrrA binds to the promoter region of icaADBC and activates transcription under anaerobic conditions; thus, the inactivation of srrAB decreases PIA biosynthesis (226). Concomitant with a decrease in PIA biosynthesis, srrAB inactivation increases the expression of the TCA cycle enzymes aconitase, succinate dehydrogenase, and fumarase and NADH dehydrogenase (221). Our laboratory recently demonstrated that PIA is synthesized when TCA cycle activity is repressed (190, 230); thus, the effect of srrAB on PIA biosynthesis is twofold, with the direct regulation of icaADBC transcription and indirect regulation via the repression of TCA cycle activity.
MgrA
Two independently derived transposon mutants of S. aureus identified a multiple-gene regulator (MgrA) that positively affects the expression capsular polysaccharide and nuclease; represses the expression of alpha-toxin, coagulase, and protein A; and represses autolysis (104, 143). In addition to regulating virulence determinants, MgrA represses the expression of several efflux systems (i.e., NorA, NorB, NorC, and Tet38) that confer resistance to multiple antibiotics (111, 224, 225). MgrA is a member of the MarR family of regulatory proteins and, as such, has a dimerization domain and a conserved helix-turn-helix domain (28). Interestingly, MgrA contains a single cysteine (Cys12) in the dimerization domain (located in α-helix 1) that is accessible to oxidizing agents. Because cysteinyl sulfhydryls can be oxidized to sulfenic acid by H2O2, it was postulated that MrgA could act as a sensor of peroxide stress (28). The oxidation of Cys12 to sulfenic acid results in the dissociation of MrgA with the sarV promoter, while the presence of reductants restores DNA binding activity (28). Taken together, MrgA creates a direct linkage between oxidative stress and antibiotic resistance. Why? Different classes of bactericidal antibiotics (e.g., β-lactams, aminoglycosides, and fluoroquinolones) all have different mechanisms of action; however, recent evidence suggests that a common mechanism induces bacterial death (54, 123). It is postulated that bactericidal antibiotics stimulate the oxidation of NADH via the electron transport chain, generating O2− (123). As stated above, O2− can damage iron-sulfur clusters, causing the release of Fe2+. In addition, the dismutation of the superoxide anion will generate H2O2, which, with the free Fe2+, can facilitate Fenton chemistry. Fenton chemistry leads to hydroxyl radical formation, damaging DNA, proteins, and lipids that result in death. Support for this common killing mechanism comes from the observation that the inactivation of the TCA cycle, a primary source of NADH, reduces the killing effect of bactericidal antibiotics (123). These data explain why S. aureus has evolved a linkage between ROS-sensing and antibiotic efflux systems and also why TCA cycle-defective S. aureus strains are found in clinical-strain collections (163, 215).
PerR
All oxidizable moieties in bacteria are potential targets of endogenous (metabolic) and exogenous (host-associated) ROS; because of this, bacteria have evolved means to detect and eliminate the threat of ROS. Gram-positive bacteria have a metal-dependent peroxide sensor that belongs to the ferric-uptake repressor (Fur) family of proteins, specifically, PerR (96, 154). In B. subtilis, PerR contains both Zn and Fe; however, Mn can compete with Fe for binding to PerR (90). PerR uses a very interesting mechanism to detect H2O2, a metal-catalyzed oxidation of histidine residues that coordinate the binding of Fe (131). By this mechanism, H2O2 stress induces the oxidation of the iron-coordinating histidine(s), resulting in the release of Fe and derepressing PerR-regulated genes (e.g., catalase and alkyl-hydroperoxide reductase) (131). Interestingly, the competition between Fe and Mn for binding to PerR creates a scenario where PerR can exist in two forms: PerR:Fe and PerR:Mn (90). These two forms of PerR differ in their sensitivities to H2O2 stress and allow for a graded response to low and high H2O2 stress levels.
PerR homologs have been identified in several pathogenic bacteria and investigated in a few, specifically, S. pyogenes (118), L. monocytogenes (180), and S. aureus (96, 97). The inactivation of PerR in all of these bacteria results in an attenuation of virulence and increased resistance to peroxide-induced stress (96, 180, 185). In S. aureus, the PerR-mediated increase in resistance to peroxide-induced stress is due to the derepression of genes encoding components of the oxidative stress response, such as catalase (KatA), alkyl-hydroperoxide reductase (AhpCF), ferritin (Ftn), bacterioferritin comigratory protein (Bcp), thioredoxin reductase (TrxB), and the Dps (DNA binding protein from starved cells) homolog MrgA (96). Similarly, in S. pyogenes, PerR influences and/or regulates the expression of peroxide and superoxide stress responses, csp (cold shock protein), mrgA, and pmtA (PerR-regulated metal transporter A) (24, 118, 185). Importantly, these data emphasize the interrelatedness of the oxidative stress response and metal ion homeostasis.
METAL-DEPENDENT REGULATORS
In prokaryotes and eukaryotes, metals act as cofactors for many enzymatic reactions and can also function as structural components in proteins. However, the ability of transition metals to transfer electrons facilitates the generation of ROS through Fenton and Haber-Weiss reactions. For these reasons, the transport of metal ions is very tightly regulated to maintain an appropriate intracellular concentration and to avoid the accumulation of metals to toxic levels. To maintain metal ion homeostasis, bacteria use active transporters and efflux systems because the bacterial membrane does not permit metals to enter into the cytoplasm by simple diffusion. This implies that a regulatory network exists to control the import and efflux of metal ions and, as discussed above, to control the response to ROS.
Iron
Fe serves as a cofactor in many enzymatic reactions and as a catalyst in electron transport processes; as such, most bacteria require iron for growth. In the eukaryotic host, iron is usually in a complex with heme, ferritin, or lactoferrin, meaning that free iron is largely inaccessible to invading bacteria. To counter the sequestration of iron, bacteria have evolved several means to acquire iron from the host. S. aureus cells synthesize and secrete high-affinity iron chelators called siderophores (i.e., staphyloferrins A and B, staphylobactin, and aureochelin) that, when complexed with iron, are transported into the bacterial cytoplasm, and the iron is extracted. Transport of the iron-siderophore complexes is mediated by the ATP binding cassette (ABC) transporters encoded within sirABC, sitABC, fhuCBG, and sstABCD (38, 43, 158, 200). In addition, S. aureus has an elaborate system to transport and extract iron from heme encoded within the iron-regulated surface determinant (isd) locus and the hrtAB and htsABC loci (151, 209, 223). This multitude of iron acquisition systems demonstrates the importance of iron for staphylococcal survival. The regulation of most genes coding for the above-mentioned components of the iron acquisition systems is mediated by the iron binding repressor ferric uptake regulation (Fur) protein (1). In low-G+C gram-positive bacteria, Fur regulates the transcription of genes coding for the biosynthesis and transport of siderophores, heme transport systems, metabolism, and the oxidative stress response.
Fur.
The Fur gene was discovered in Salmonella enterica serovar Typhimurium, when it was observed that a mutant strain constitutively produced iron-regulated outer membrane proteins (58). The mutation was introduced into E. coli, and the fur gene region was cloned and determined to encode an ∼18-kDa protein that mediates the iron-dependent repression of genes involved in iron transport (84, 85). The Fur protein has an N-terminal DNA binding domain, a dimerization domain, possibly a structural Zn atom, and a high-affinity Fe binding site (131, 176). In B. subtilis, the Fur protein binds as a homodimer to a 19-bp inverted repeat sequence called the Fur box (GATAATGATAATCATTATC [underlining highlights the inverted repeat]) (7). Because the expression of many virulence determinants is negatively regulated by iron (26) and the virulence of fur mutant strains is attenuated relative to that of the isogenic parental strains (86, 97, 180), it was reasonable to speculate that Fur regulates the transcription of virulence determinants; however, the number of virulence genes regulated by iron far outweighs the number of virulence genes identified as being regulated by Fur (1, 62). Therefore, there must be other means for iron to influence gene expression that is independent of Fur or minimally dependent upon Fur. As stated previously, many enzymes require iron for enzymatic activity; thus, it is not surprising that metabolic genes are subjected to iron-dependent regulation (8, 62, 234). This iron-dependent regulation will alter the metabolic status of the bacteria, creating metabolic signals that can be “sensed” by regulators responding to carbon and nitrogen, etc. (e.g., CodY, CcpA, and GlnR).
DtxR and IdeR.
High-G+C-content gram-positive bacteria (e.g., Corynebacterium sp. and Mycobacterium sp.) regulate iron-dependent gene expression using proteins that are orthologous to the diphtheria toxin regulator (DtxR) (e.g., the iron-dependent regulator [IdeR]) (22, 195). Similar in function to Fur, these iron-dependent regulators control the biosynthesis and transport of siderophores, metabolism, and the oxidative stress response (25, 128, 187). In addition, when complexed with iron, DtxR binds as a dimer to a 19-bp palindromic operator sequence of the diphtheria toxin (tox) promoter in C. diphtheria, repressing the transcription of tox (94). Like Fur, the DtxR-mediated derepression of tox transcription occurs when the level of iron decreases. DtxR-like proteins typically contain two domains connected by a flexible linker: the N-terminal domain contains a helix-turn-helix DNA binding motif, and the C-terminal domain contains a dimerization region, two metal ion binding sites, and an SH3-like domain within a flexible region (37, 176, 179). Both metal ion binding sites are believed to bind iron and are important for transcriptional repression.
Manganese
Mn2+ readily cycles between the 2+ and 3+ oxidation states; however, relative to Fe2+, Mn2+ is less likely to donate an electron to another molecule and facilitate an undesirable reduction reaction. Specifically, Mn2+ is a very poor mediator of Fenton chemistry (113). The higher reduction potential of Mn2+ means that bacteria can tolerate higher intracellular concentrations of Mn in an oxidizing environment with minimal deleterious redox consequences. Thus, these redox properties make Mn an ideal cofactor for certain oxidative stress response enzymes such as catalase and superoxide dismutase. The bacterial demand for manganese is met by two major classes of Mn transporters: the Nramp H+-Mn2+ transporters (named after the eukaryotic natural resistance-associated macrophage protein) and the ABC Mn permeases (174).
MntR.
In S. aureus, the Nramp H+-Mn2+ transporter is encoded by mntH, and the ABC Mn permease is encoded by mntABC (98). Both mntH and mntABC are important for maintaining the growth rate and yield in metal ion-limited culture medium (98). The transcriptional regulation of mntH and mntABC is mediated by Mn and the DtxR homolog MntR (98), also known as SirR in S. epidermidis (91). In addition to regulating Mn transport, MntR is important for modulating the PerR-mediated oxidative stress response (98) and for virulence (3, 98). The structure of MntR is similar to that of DtxR and IdeR; however, MntR lacks the C-terminal SH3-like domain (76). Interestingly, MntR displays a greater metal ion specificity than do DtxR and IdeR (135). MntR represses the transcription of mntABC when the intracellular concentrations of Mn are elevated by binding as a homodimer to a 19-bp MntR-box consensus sequence (3). Mn-responsive DtxR homologs have been identified in several gram-positive bacteria including S. pneumoniae (PsaR), Streptococcus gordonii (ScaR), Enterococcus faecalis (EfaR), and Bacillus anthracis (AntR) (105, 109, 140, 203). Although the degree to which mutants of these dtxR homologs have been characterized varies, all were identified as regulating Mn transport, and the S. gordonii and S. pneumoniae homologs have been implicated as being important for virulence.
Zinc
Zn functions as a cofactor for enzymatic reactions such as those catalyzed by alcohol dehydrogenase, metalloproteases (e.g., aureolysin), and the [Cu,Zn]-superoxide dismutases of eukaryotes and some gram-negative bacteria. However, one of the most important functions of Zn is maintaining the structure of proteins (20). Like any transition metal, too much Zn can be toxic because it competes with other metals for binding to the active centers of enzymes; hence, the importation of Zn must be tightly controlled. Most bacteria possess high-affinity ABC zinc transporters orthologous to the gram-negative bacterial znuABC operon (83). The transcriptional regulation of Zn transport is mediated primarily by the Zn-responsive Fur homolog known as Zur (66).
Zur.
Like Fur, Zur has two conserved domains: an N-terminal DNA-binding motif and a C-terminal dimerization domain; however, Zur has three metal ion binding sites per monomer (141). Of the three metal ion binding sites, two are located in the C-terminal dimerization domain, while the third is in the hinge region. All three metal ion binding sites are believed to bind Zn; however, it is uncertain if the third site actually binds Zn in vivo. When complexed with Zn, Zur binds as a homodimer to a 19-bp inverted repeat, AAATCGTAATNATTACGATTT, known as the Zur box (68), and represses transcription. Functional Zur homologs have been identified in Mycobacterium tuberculosis (153), Streptococcus suis (153), L. monocytogenes (44), S. aureus (137), Streptomyces coelicolor (205), and B. subtilis (66). Not surprisingly, Zur controls Zn transport through ABC transporters by repressing transcription when Zn is abundant (60, 67, 144, 173). In addition, when the availability of Zn is low, Zur also derepresses the transcription of paralogous forms of ribosomal protein genes, which have disrupted Zn ribbon motifs (171, 173). These paralogous ribosomal proteins are believed to function in place of the Zn-containing ribosomal proteins when Zn becomes growth limiting. Although Zur does regulate potential virulence determinants such as metalloproteases in S. suis and proteins belonging to early secretory antigen target 6 (ESAT-6) cluster 3 in M. tuberculosis, the effect of zur inactivation on virulence appears to be nominal (60, 137).
CENTRAL ROLE OF CENTRAL METABOLISM
The TCA cycle supplies biosynthetic intermediates, reducing potential, and a small amount of ATP. Under nutrient-rich growth conditions, the bacterial demand for biosynthetic intermediates is supplied exogenously; hence, under these conditions, TCA cycle activity is very low (40, 214, 219). When environmental conditions change and nutrients become growth limiting, staphylococci increase TCA cycle activity and catabolize nonpreferred carbon sources such as acetate (213). The transition to TCA cycle-associated metabolism is preceded by the derepression of genes coding for TCA cycle enzymes. Repression is mediated primarily by CcpA, CodY, SrrA, and, in B. subtilis, CcpC (110, 116, 202, 221). In addition, the repressor Fur appears to positively affect the transcription of TCA cycle genes coding for iron-dependent enzymes (62, 65). As stated above, these regulators are responding to changes in the availability of carbon, nitrogen, oxygen, and iron; thus, if the DNA binding ability of these regulators is changing, it means that the bacterial metabolic status is changing. This changing metabolic status is exacerbated by the induction of the TCA cycle, creating another layer of metabolic changes for regulatory proteins to “sense” (190, 214).
Numerous studies have implicated the TCA cycle in regulating or affecting staphylococcal virulence and/or virulence determinant biosynthesis (6, 12, 41, 152, 190, 211, 213, 214). The two most common types of regulation are metabolic regulation and genetic regulation. Therefore, the two more likely explanations for TCA cycle-mediated effects on virulence determinant biosynthesis are the metabolic inhibition of biosynthetic enzyme activity or the repression and/or induction of virulence gene transcription. The former possibility is exemplified by the metabolic inhibition of delta-toxin biosynthesis due to glutamate auxotrophy created by the inactivation of the TCA cycle (214). In this example, the message for delta-toxin (RNAIII) is present but not translated due to the absence of glutamate. The supplementation of the culture medium with excess glutamate reverses this metabolic inhibition of delta-toxin translation (214). Similarly, TCA cycle inactivation prevents the biosynthesis of oxaloacetate for conversion to phosphoenolpyruvate and use in gluconeogenesis, resulting in an inability to synthesize the biosynthetic precursors of capsule (C. Y. Lee and G. A. Somerville, unpublished observations). These data confirm that some effects of TCA cycle inactivation are the result of metabolic regulation.
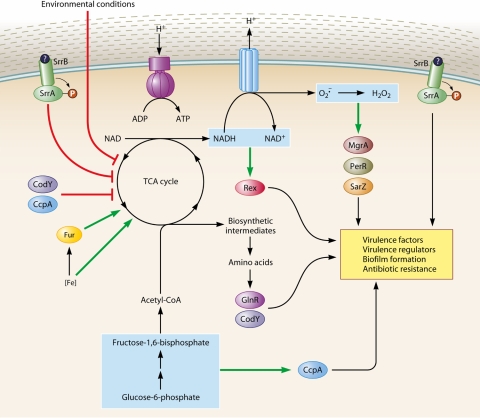
An example of TCA cycle-mediated genetic regulation is the TCA cycle-dependent repression of icaADBC transcription. Inhibiting TCA cycle activity dramatically increases levels of icaADBC transcription and PIA biosynthesis (190, 230); conversely, increasing TCA cycle activity decreases levels of icaADBC transcription and PIA biosynthesis (Y. Zhu and Y. Xiong, unpublished data). Although it is possible that an enzyme of the TCA cycle directly regulates PIA synthesis, a more likely mechanism is that regulatory proteins responding to TCA cycle-mediated changes in the metabolic status regulate icaADBC transcription. As stated above, the precedence for such a mechanism is well established: the transcription of the lac operon is derepressed when LacI binds the lactose metabolite 1,6-allolactose. Taken together, these observations lead us to propose that in staphylococci, the TCA cycle acts as a signal transduction pathway. In this model (Fig. 1), any environmental signal or TCA cycle regulator capable of altering TCA cycle activity (e.g., iron or oxygen limitation and CcpA, etc.) transforms the metabolic status of the bacterium, resulting in regulatory proteins responding to the new metabolic status to regulate the expression of genes coding for enzymes required for growth in the altered environment, such as the ica operon. Support for the TCA cycle being a central pathway for responding to external signals was recently reported by Kohanski et al. (123). In that study, the authors demonstrated that multiple classes of bactericidal antibiotics stimulate the production of hydroxyl radicals, which accelerated bacterial cell death in E. coli and S. aureus. Although different classes of antibiotics have unique mechanisms of action, that study suggested that they mediate killing through a common pathway involving hydroxyl radicals. Through an unknown mechanism, bactericidal antibiotics transiently increase the TCA cycle generation of NADH, resulting in an increased accumulation of superoxides by the electron transport chain. Superoxide damages iron-sulfur clusters, releasing iron, which can undergo Fenton chemistry to generate hydroxyl radicals (212). In support of their proposed mechanism, the genetic inactivation of the TCA cycle genes acnB (aconitase B) and icdA (isocitrate dehydrogenase) abrogated the killing effect of bactericidal antibiotics (123). Irrespective of whether or not the TCA cycle is a signal transduction pathway, these observations demonstrate that the TCA cycle is intimately integrated into regulating virulence factor synthesis, biofilm formation, and antibiotic resistance.
FIG. 1.
Model of TCA cycle-dependent regulation. (For simplicity, the well-characterized virulence regulators [i.e., RNAIII, Rot, SarA, and σB] have been omitted from this model.)
Regulators Responding to TCA Cycle-Associated Metabolomic Changes
MgrA.
The TCA cycle generates the reducing potential that drives oxidative phosphorylation, resulting in the endogenous generation of superoxide anions. Increased oxidative stress will oxidize the free sulfhydryl of MgrA to sulfenic acid, inhibiting MgrA DNA binding activity, derepressing the transcription of autolytic genes, inhibiting the transcription of antiautolytic factors, and resulting in autolysis (142). In the absence of TCA cycle activity, the generation of superoxide anions via oxidative phosphorylation is greatly reduced, MgrA retains DNA binding activity, and autolysis is prevented. Support for this proposition can be seen in the fact that S. aureus TCA cycle mutants fail to undergo autolytic cell death (213). Similarly, this persistence phenotype has been observed in oxidative phosphorylation-deficient small-colony-variant staphylococci. (During the preparation of the manuscript, a functional homolog of MgrA, SarZ, was also found to respond to oxidative stress and to have a role in autolysis [29].)
GlnR.
GlnR is responsive to the concentration of glutamate, glutamine, and ammonium in the growth medium (239). The carbon skeleton of glutamate and glutamine is the TCA cycle intermediate α-ketoglutarate. As stated previously, TCA cycle activity enhances killing by bactericidal antibiotics (123). The inactivation of glnR or the TCA cycle results in increased resistance to antibiotics (80, 123, 218), suggesting that the linkage of antibiotic resistance and a TCA cycle-responsive regulator was evolutionarily advantageous.
CodY.
The DNA binding affinity of CodY for its cognate promoters is increased in the presence of BCAA and GTP. The synthesis of isoleucine is complex, but it starts with the TCA cycle intermediate oxaloacetate. The synthesis of valine and leucine requires α-keto-isovalerate, which is derived from pyruvate. Interestingly, the inactivation of the TCA cycle in S. epidermidis increases the intracellular concentrations of valine, leucine, and isoleucine (190). This increased intracellular concentration of BCAAs is likely due to the redirection of pyruvate into mixed-acid fermentation and acetolactate biosynthesis, which would supply the biosynthetic precursors oxaloacetate and α-keto-isovalerate. These data suggest not only that CodY regulates TCA cycle activity but also that CodY may respond to changes in TCA cycle activity.
CcpA.
TCA cycle activity regulates icaADBC transcription (190). Recently, CcpA was shown to increase the level of transcription of icaADBC and the accumulation of PIA in S. aureus (202). As stated above, CcpA regulatory activity is controlled by interactions with phosphorylated HPr or Crh and fructose-1,6-bisphosphate or glucose-6-phosphate. TCA cycle inactivation increases the intracellular concentration of fructose-6-phosphate (190). Fructose-6-phosphate is the biosynthetic precursor of UDP-N-acetyl-glucosamine, the monomeric unit of PIA. In addition to its importance in PIA biosynthesis, fructose-6-phosphate can be reversibly isomerized to glucose-6-phosphate by glucose-6-phosphate isomerase, resulting in CcpA activation. Taken together, these observations lead to us hypothesize that TCA cycle activity influences the CcpA-mediated activation of icaADBC.
CONCLUSION
The interplay between metabolism and the production of virulence factors demonstrates a large array of influences. This was seen in the earliest studies of bacteria, wherein media constituents, growth conditions such as anaerobiosis, and stage of growth all influenced the accumulation of toxins. One hopes that a detailed understanding of these interactions may help in the design of more effective therapies for infections caused by staphylococci and other gram-positive pathogens.
Acknowledgments
We thank all of the members of our laboratories past and present that contributed intellectually and materially to this review.
The manuscript is a contribution of the University of Nebraska Agricultural Research Division, supported in part by funds provided through the Hatch Act and past support from the National Institute of General Medical Sciences and the American Heart Association to G.A.S. R.A.P. received funding from Kimberly Clark.
Biography
Greg Somerville's chief research focus is the elucidation of mechanisms by which bacteria control virulence factor expression in response to nutrient availability. Specifically, his interest is in the function of central metabolism in regulating pathogenesis. An assistant professor in the Department of Veterinary and Biomedical Sciences at the University of Nebraska-Lincoln (UNL), he is also affiliated with UNL's Department of Biochemistry, UNL's Redox Biology Center, and the Department of Pathology and Microbiology at the University of Nebraska Medical Center. Greg Somerville earned a Ph.D. degree in Biology from the University of Texas at Dallas under the supervision of Larry Reitzer. Prior to joining UNL, he was a postdoctoral research fellow in the Laboratory of Human Bacterial Pathogenesis at the Rocky Mountain Laboratories in Hamilton, MT.
Rich Proctor began research as an undergraduate student at the University of Michigan and became focused on bacterial infections when it was observed that antibiotic susceptibility was related to the inhibition of acid production in Robert Fekety's laboratory. After an Infectious Diseases Fellowship at the University of Wisconsin under the direction of Calvin Kunin and William Craig, Dr. Proctor's research interests turned to the pathogenesis of Staphylococcus aureus endocarditis. This led to the discovery of electron transport-defective bacteria that grew as small-colony variants (SCVs) and failed to damage endothelial cells due to lack of alpha-toxin production. This dramatic link between respiratory metabolism and virulence produced a long-term interest in the study of how metabolism influences virulence factor regulation. One of Dr. Proctor's first observations was that SCVs completely lacked RNA III. Dissecting out the interactions between metabolism and virulence factor production has provided many insights into these interactions, but we are left with many questions.
REFERENCES
- 1.Allard, M., H. Moisan, E. Brouillette, A. L. Gervais, M. Jacques, P. Lacasse, M. S. Diarra, and F. Malouin. 2006. Transcriptional modulation of some Staphylococcus aureus iron-regulated genes during growth in vitro and in a tissue cage model in vivo. Microbes Infect. 81679-1690. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, C. B., and L. D. Witter. 1982. Glutamine and proline accumulation by Staphylococcus aureus with reduction in water activity. Appl. Environ. Microbiol. 431501-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ando, M., Y. C. Manabe, P. J. Converse, E. Miyazaki, R. Harrison, J. R. Murphy, and W. R. Bishai. 2003. Characterization of the role of the divalent metal ion-dependent transcriptional repressor MntR in the virulence of Staphylococcus aureus. Infect. Immun. 712584-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba, T., T. Bae, O. Schneewind, F. Takeuchi, and K. Hiramatsu. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 190300-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 3591819-1827. [DOI] [PubMed] [Google Scholar]
- 6.Bae, T., A. K. Banger, A. Wallace, E. M. Glass, F. Aslund, O. Schneewind, and D. M. Missiakas. 2004. Staphylococcus aureus virulence genes identified by Bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci. USA 10112312-12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baichoo, N., and J. D. Helmann. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 1845826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 451613-1629. [DOI] [PubMed] [Google Scholar]
- 9.Bainbridge, F. A. 1911. The action of certain bacteria on proteins. J. Hyg. 11341-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayer, A. S., P. McNamara, M. R. Yeaman, N. Lucindo, T. Jones, A. L. Cheung, H. G. Sahl, and R. A. Proctor. 2006. Transposon disruption of the complex I NADH oxidoreductase gene (snoD) in Staphylococcus aureus is associated with reduced susceptibility to the microbicidal activity of thrombin-induced platelet microbicidal protein 1. J. Bacteriol. 188211-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beenken, K. E., P. M. Dunman, F. McAleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins, and M. S. Smeltzer. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 1864665-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Begun, J., C. D. Sifri, S. Goldman, S. B. Calderwood, and F. M. Ausubel. 2005. Staphylococcus aureus virulence factors identified by using a high-throughput Caenorhabditis elegans-killing model. Infect. Immun. 73872-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benito, Y., F. A. Kolb, P. Romby, G. Lina, J. Etienne, and F. Vandenesch. 2000. Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression RNA. 6668-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett, H. J., D. M. Pearce, S. Glenn, C. M. Taylor, M. Kuhn, A. L. Sonenshein, P. W. Andrew, and I. S. Roberts. 2007. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol. Microbiol. 631453-1467. [DOI] [PubMed] [Google Scholar]
- 15.Berman, N., and L. F. Rettger. 1916. Bacterial nutrition: a brief note on the production of erepsin by bacteria. J. Bacteriol. 1537-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berman, N., and L. F. Rettger. 1918. The influence of carbohydrate on the nitrogen metabolism of bacteria. J. Bacteriol. 3389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bächi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 1864085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bischoff, M., J. M. Entenza, and P. Giachino. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 1835171-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blagova, E. V., V. M. Levdikov, K. Tachikawa, A. L. Sonenshein, and A. J. Wilkinson. 2003. Crystallization of the GTP-dependent transcriptional regulator CodY from Bacillus subtilis. Acta Crystallogr. D Biol. Crystallogr. 59155-157. [DOI] [PubMed] [Google Scholar]
- 20.Blencowe, D. K., and A. P. Morby. 2003. Zn(II) metabolism in prokaryotes. FEMS Microbiol. Rev. 27291-311. [DOI] [PubMed] [Google Scholar]
- 21.Boisset, S., T. Geissmann, E. Huntzinger, P. Fechter, N. Bendridi, M. Possedko, C. Chevalier, A. C. Helfer, Y. Benito, A. Jacquier, C. Gaspin, F. Vandenesch, and P. Romby. 2007. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 211353-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyd, J., M. N. Oza, and J. R. Murphy. 1990. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc. Natl. Acad. Sci. USA 875968-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brekasis, D., and M. S. Paget. 2003. A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2). EMBO J. 224856-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brenot, A., B. F. Weston, and M. G. Caparon. 2007. A PerR-regulated metal transporter (PmtA) is an interface between oxidative stress and metal homeostasis in Streptococcus pyogenes. Mol. Microbiol. 631185-1196. [DOI] [PubMed] [Google Scholar]
- 25.Brune, I., H. Werner, A. T. Hüser, J. Kalinowski, A. Pühler, and A. Tauch. 2006. The DtxR protein acting as dual transcriptional regulator directs a global regulatory network involved in iron metabolism of Corynebacterium glutamicum. BMC Genomics 721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bullen, J., and E. Griffiths (ed.). 1999. Iron and infection, 2nd ed. John Wiley & Sons, Chichester, England.
- 27.Carpenter, D. F., and G. J. Silverman. 1974. Staphylococcal enterotoxin B and nuclease production under controlled dissolved oxygen conditions. Appl. Microbiol. 28628-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen, P. R., T. Bae, W. A. Williams, E. M. Duguid, P. A. Rice, O. Schneewind, and C. He. 2006. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat. Chem. Biol. 2591-595. [DOI] [PubMed] [Google Scholar]
- 29.Chen, P. R., S. Nishida, C. B. Poor, A. Cheng, T. Bae, L. Kuechenmeister, P. M. Dunman, D. Missiakas, and C. He. 2009. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol. Microbiol. 71198-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheung, A. L., M. G. Bayer, and J. H. Heinrichs. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J. Bacteriol. 1793963-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheung, A. L., Y. T. Chien, and A. S. Bayer. 1999. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect. Immun. 671331-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheung, A. L., K. Eberhardt, and J. H. Heinrichs. 1997. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. Infect. Immun. 652243-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 896462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheung, A. L., K. A. Nishina, M. P. Trotonda, and S. Tamber. 2008. The SarA protein family of Staphylococcus aureus. Int. J. Biochem. Cell Biol. 40355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung, A. L., and S. J. Projan. 1994. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 1764168-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chien, Y., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 27437169-37176. [DOI] [PubMed] [Google Scholar]
- 37.Chou, C. J., G. Wisedchaisri, R. R. Monfeli, D. M. Oram, R. K. Holmes, W. G. Hol, and C. Beeson. 2004. Functional studies of the Mycobacterium tuberculosis iron-dependent regulator. J. Biol. Chem. 27953554-53561. [DOI] [PubMed] [Google Scholar]
- 38.Cockayne, A., P. J. Hill, N. B. Powell, K. Bishop, C. Sims, and P. Williams. 1998. Molecular cloning of a 32-kilodalton lipoprotein component of a novel iron-regulated Staphylococcus epidermidis ABC transporter. Infect. Immun. 663767-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coleman, G. 1985. A comparison of the patterns of extracellular proteins produced by the high alpha-toxin-secreting organism Staphylococcus aureus (Wood 46) during aerobic and anaerobic growth. J. Gen. Microbiol. 131405-408. [DOI] [PubMed] [Google Scholar]
- 40.Collins, F. M., and J. Lascelles. 1962. The effect of growth conditions on oxidative and dehydrogenase activity in Staphylococcus aureus. J. Gen. Microbiol. 29531-535. [DOI] [PubMed] [Google Scholar]
- 41.Coulter, S. N., W. R. Schwan, E. Y. Ng, M. H. Langhorne, H. D. Ritchie, S. Westbrock-Wadman, W. O. Hufnagle, K. R. Folger, A. S. Bayer, and C. K. Stover. 1998. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol. Microbiol. 30393-404. [DOI] [PubMed] [Google Scholar]
- 42.Cripps, R. E., and E. Work. 1967. The accumulation of extracellular macromolecules by Staphylococcus aureus grown in the presence of sodium chloride and glucose. J. Gen. Microbiol. 49127-137. [DOI] [PubMed] [Google Scholar]
- 43.Dale, S. E., M. T. Sebulsky, and D. E. Heinrichs. 2004. Involvement of SirABC in iron-siderophore import in Staphylococcus aureus. J. Bacteriol. 1868356-8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dalet, K., E. Gouin, Y. Cenatiempo, P. Cossart, and Y. Héchard. 1999. Characterisation of a new operon encoding a Zur-like protein and an associated ABC zinc permease in Listeria monocytogenes. FEMS Microbiol. Lett. 174111-116. [DOI] [PubMed] [Google Scholar]
- 45.den Hengst, C. D., S. A. van Hijum, J. M. Geurts, A. Nauta, J. Kok, and O. P. Kuipers. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem. 28034332-34342. [DOI] [PubMed] [Google Scholar]
- 46.Deutscher, J., E. Küster, U. Bergstedt, V. Charrier, and W. Hillen. 1995. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram-positive bacteria. Mol. Microbiol. 151049-1053. [DOI] [PubMed] [Google Scholar]
- 47.Deutscher, J., J. Reizer, C. Fischer, A. Galinier, M. H. Saier, Jr., and M. Steinmetz. 1994. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J. Bacteriol. 176:3336-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deutscher, J., and M. H. Saier, Jr. 1983. ATP-dependent protein kinase-catalyzed phosphorylation of a seryl residue in HPr, a phosphate carrier protein of the phosphotransferase system in Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 806790-6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dienert, F. 1900. Sur la fermentation du galactose et sur l'accoutumance des levures à ce sucre. Ann. Inst. Pasteur 14139-189. [Google Scholar]
- 50.Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367731-739. [DOI] [PubMed] [Google Scholar]
- 51.Dineen, S. S., A. C. Villapakkam, J. T. Nordman, and A. L. Sonenshein. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 66206-219. [DOI] [PubMed] [Google Scholar]
- 52.Duncan, J. L., and G. J. Cho. 1972. Production of staphylococcal alpha toxin. II. Glucose repression of toxin formation. Infect. Immun. 6689-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duport, C., A. Zigha, E. Rosenfeld, and P. Schmitt. 2006. Control of enterotoxin gene expression in Bacillus cereus F4430/73 involves the redox-sensitive ResDE signal transduction system. J. Bacteriol. 1886640-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dwyer, D. J., M. A. Kohanski, B. Hayete, and J. J. Collins. 2007. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Egeter, O., and R. Brückner. 1996. Catabolite repression mediated by the catabolite control protein CcpA in Staphylococcus xylosus. Mol. Microbiol. 21739-749. [DOI] [PubMed] [Google Scholar]
- 56.Emmett, M., and W. E. Kloos. 1975. Amino acid requirements of staphylococci isolated from human skin. Can. J. Microbiol. 21729-733. [DOI] [PubMed] [Google Scholar]
- 57.Erickson, A., and R. H. Deibel. 1973. Production and heat stability of staphylococcal nuclease. Appl. Microbiol. 25332-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ernst, J. F., R. L. Bennett, and L. I. Rothfield. 1978. Constitutive expression of the iron-enterochelin and ferrichrome uptake systems in a mutant strain of Salmonella typhimurium. J. Bacteriol. 135928-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fedtke, I., A. Kamps, B. Krismer, and F. Götz. 2002. The nitrate reductase and nitrite reductase operons and the narT gene of Staphylococcus carnosus are positively controlled by the novel two-component system NreBC. J. Bacteriol. 1846624-6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feng, Y., M. Li, H. Zhang, B. Zheng, H. Han, C. Wang, J. Yan, J. Tang, and G. F. Gao. 2008. Functional definition and global regulation of Zur, a zinc uptake regulator in a Streptococcus suis serotype 2 strain causing streptococcal toxic shock syndrome. J. Bacteriol. 1907567-7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fildes, P., G. M. Richardson, B. C. J. G. Knight, and G. P. Gladstone. 1936. A nutrient mixture suitable for the growth of Staphylococcus aureus. Br. J. Exp. Pathol. 17481-484. [Google Scholar]
- 62.Friedman, D. B., D. L. Stauff, G. Pishchany, C. W. Whitwell, V. J. Torres, and E. P. Skaar. 2006. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fuchs, S., J. Pané-Farré, C. Kohler, M. Hecker, and S. Engelmann. 2007. Anaerobic gene expression in Staphylococcus aureus. J. Bacteriol. 1894275-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fujimoto, D. F., E. W. Brunskill, and K. W. Bayles. 2000. Analysis of genetic elements controlling Staphylococcus aureus lrgAB expression: potential role of DNA topology in SarA regulation. J. Bacteriol. 1824822-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gaballa, A., H. Antelmann, C. Aguilar, S. K. Khakh, K. B. Song, G. T. Smaldone, and J. D. Helmann. 2008. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc. Natl. Acad. Sci. USA 10511927-11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gaballa, A., and J. D. Helmann. 1998. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J. Bacteriol. 1805815-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gaballa, A., T. Wang, R. W. Ye, and J. D. Helmann. 2002. Functional analysis of the Bacillus subtilis Zur regulon. J. Bacteriol. 1846508-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gabriel, S. E., F. Miyagi, A. Gaballa, and J. D. Helmann. 2008. Regulation of the Bacillus subtilis yciC gene and insights into the DNA-binding specificity of the zinc-sensing metalloregulator Zur. J. Bacteriol. 1903482-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gajiwala, K. S., and S. K. Burley. 2000. Winged helix proteins. Curr. Opin. Struct. Biol. 10110-116. [DOI] [PubMed] [Google Scholar]
- 70.Galinier, A., J. Haiech, M. C. Kilhoffer, M. Jaquinod, J. Stulke, J. Deutscher, and I. Martin-Verstraete. 1997. The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc. Natl. Acad. Sci. USA 948439-8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gardner, J. F., and J. Lascelles. 1962. The requirement for acetate of a streptomycin-resistant strain of Staphylococcus aureus. J. Gen. Microbiol. 29157-164. [DOI] [PubMed] [Google Scholar]
- 72.Giachino, P., S. Engelmann, and M. Bischoff. 2001. σB activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 1831843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gilbert, W., and B. Müller-Hill. 1966. Isolation of the Lac repressor. Proc. Natl. Acad. Sci. USA 561891-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 1872426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gladstone, G. P. 1937. The nutrition of Staphylococcus aureus; nitrogen requirements. Br. J. Exp. Pathol. 18322-333. [Google Scholar]
- 76.Glasfeld, A., E. Guedon, J. D. Helmann, and R. G. Brennan. 2003. Structure of the manganese-bound manganese transport regulator of Bacillus subtilis. Nat. Struct. Biol. 10652-657. [DOI] [PubMed] [Google Scholar]
- 77.Goldschmidt, M. C., and D. M. Powelson. 1953. Effect of the culture medium on the oxidation of acetate by Micrococcus pyogenes var. aureus. Arch. Biochem. Biophys. 46154-163. [DOI] [PubMed] [Google Scholar]
- 78.Gruber, T. M., and C. A. Gross. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57441-466. [DOI] [PubMed] [Google Scholar]
- 79.Guedon, E., P. Serror, S. D. Ehrlich, P. Renault, and C. Delorme. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol. Microbiol. 401227-1239. [DOI] [PubMed] [Google Scholar]
- 80.Gustafson, J., A. Strässle, H. Hächler, F. H. Kayser, and B. Berger-Bächi. 1994. The femC locus of Staphylococcus aureus required for methicillin resistance includes the glutamine synthetase operon. J. Bacteriol. 1761460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gyan, S., Y. Shiohira, I. Sato, M. Takeuchi, and T. Sato. 2006. Regulatory loop between redox sensing of the NADH/NAD+ ratio by Rex (YdiH) and oxidation of NADH by NADH dehydrogenase Ndh in Bacillus subtilis. J. Bacteriol. 1887062-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Handke, L. D., R. P. Shivers, and A. L. Sonenshein. 2008. Interaction of Bacillus subtilis CodY with GTP. J. Bacteriol. 190798-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hantke, K. 2005. Bacterial zinc uptake and regulators. Curr. Opin. Microbiol. 8196-202. [DOI] [PubMed] [Google Scholar]
- 84.Hantke, K. 1984. Cloning of the repressor protein gene of iron-regulated systems in Escherichia coli K12. Mol. Gen. Genet. 197337-341. [DOI] [PubMed] [Google Scholar]
- 85.Hantke, K. 1981. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. Gen. Genet. 182288-292. [DOI] [PubMed] [Google Scholar]
- 86.Harvie, D. R., S. Vilchez, J. R. Steggles, and D. J. Ellar. 2005. Bacillus cereus Fur regulates iron metabolism and is required for full virulence. Microbiology 151569-577. [DOI] [PubMed] [Google Scholar]
- 87.Hendriksen, W. T., H. J. Bootsma, S. Estevão, T. Hoogenboezem, A. de Jong, R. de Groot, O. P. Kuipers, and P. W. Hermans. 2008. CodY of Streptococcus pneumoniae: link between nutritional gene regulation and colonization. J. Bacteriol. 190590-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hendriksen, W. T., T. G. Kloosterman, H. J. Bootsma, S. Estevão, R. de Groot, O. P. Kuipers, and P. W. Hermans. 2008. Site-specific contributions of glutamine-dependent regulator GlnR and GlnR-regulated genes to virulence of Streptococcus pneumoniae. Infect. Immun. 761230-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Henkin, T. M., F. J. Grundy, W. L. Nicholson, and G. H. Chambliss. 1991. Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacl and galR repressors. Mol. Microbiol. 5575-584. [DOI] [PubMed] [Google Scholar]
- 90.Herbig, A. F., and J. D. Helmann. 2001. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol. Microbiol. 41849-859. [DOI] [PubMed] [Google Scholar]
- 91.Hill, P. J., A. Cockayne, P. Landers, J. A. Morrissey, C. M. Sims, and P. Williams. 1998. SirR, a novel iron-dependent repressor in Staphylococcus epidermidis. Infect. Immun. 664123-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Holden, M. T., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 1019786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Holland, K., J. Wright, and D. Taylor. 1998. Does Staphylococcus aureus produce TSST-1 under anaerobic conditions? Int. Congr. Symp. Ser. R. Soc. Med. 229140-143. [Google Scholar]
- 94.Holmes, R. K. 2000. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J. Infect. Dis. 181(Suppl. 1)S156-S167. [DOI] [PubMed] [Google Scholar]
- 95.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 1845457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Horsburgh, M. J., M. O. Clements, H. Crossley, E. Ingham, and S. J. Foster. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 693744-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Horsburgh, M. J., E. Ingham, and S. J. Foster. 2001. In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Horsburgh, M. J., S. J. Wharton, A. G. Cox, E. Ingham, S. Peacock, and S. J. Foster. 2002. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol. Microbiol. 441269-1286. [DOI] [PubMed] [Google Scholar]
- 99.Hsueh, Y. H., E. B. Somers, and A. C. Wong. 2008. Characterization of the codY gene and its influence on biofilm formation in Bacillus cereus. Arch. Microbiol. 189557-568. [DOI] [PubMed] [Google Scholar]
- 100.Huntzinger, E., S. Boisset, C. Saveanu, Y. Benito, T. Geissmann, A. Namane, G. Lina, J. Etienne, B. Ehresmann, C. Ehresmann, A. Jacquier, F. Vandenesch, and P. Romby. 2005. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 24824-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huynh, P. L., I. Jankovic, N. F. Schnell, and R. Brückner. 2000. Characterization of an HPr kinase mutant of Staphylococcus xylosus. J. Bacteriol. 1821895-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Imlay, J. A. 2002. How oxygen damages microbes: oxygen tolerance and obligate anaerobiosis. Adv. Microb. Physiol. 46111-153. [DOI] [PubMed] [Google Scholar]
- 103.Inaoka, T., and K. Ochi. 2002. RelA protein is involved in induction of genetic competence in certain Bacillus subtilis strains by moderating the level of intracellular GTP. J. Bacteriol. 1843923-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ingavale, S. S., W. Van Wamel, and A. L. Cheung. 2003. Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol. Microbiol. 481451-1466. [DOI] [PubMed] [Google Scholar]
- 105.Jakubovics, N. S., A. W. Smith, and H. F. Jenkinson. 2000. Expression of the virulence-related Sca (Mn2+) permease in Streptococcus gordonii is regulated by a diphtheria toxin metallorepressor-like protein ScaR. Mol. Microbiol. 38140-153. [DOI] [PubMed] [Google Scholar]
- 106.Jankovic, I., O. Egeter, and R. Brückner. 2001. Analysis of catabolite control protein A-dependent repression in Staphylococcus xylosus by a genomic reporter gene system. J. Bacteriol. 183580-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Janzon, L., S. Löfdahl, and S. Arvidson. 1989. Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator (agr) of Staphylococcus aureus. Mol. Gen. Genet. 219480-485. [DOI] [PubMed] [Google Scholar]
- 108.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 2762027-2030. [DOI] [PubMed] [Google Scholar]
- 109.Johnston, J. W., D. E. Briles, L. E. Myers, and S. K. Hollingshead. 2006. Mn2+-dependent regulation of multiple genes in Streptococcus pneumoniae through PsaR and the resultant impact on virulence. Infect. Immun. 741171-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jourlin-Castelli, C., N. Mani, M. M. Nakano, and A. L. Sonenshein. 2000. CcpC, a novel regulator of the LysR family required for glucose repression of the citB gene in Bacillus subtilis. J. Mol. Biol. 295865-878. [DOI] [PubMed] [Google Scholar]
- 111.Kaatz, G. W., R. V. Thyagarajan, and S. M. Seo. 2005. Effect of promoter region mutations and mgrA overexpression on transcription of norA, which encodes a Staphylococcus aureus multidrug efflux transporter. Antimicrob. Agents Chemother. 49161-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kamps, A., S. Achebach, I. Fedtke, G. Unden, and F. Götz. 2004. Staphylococcal NreB: an O(2)-sensing histidine protein kinase with an O(2)-labile iron-sulphur cluster of the FNR type. Mol. Microbiol. 52713-723. [DOI] [PubMed] [Google Scholar]
- 113.Kehres, D. G., and M. E. Maguire. 2003. Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol. Rev. 27263-290. [DOI] [PubMed] [Google Scholar]
- 114.Kendall, A. I., A. A. Day, and A. W. Walker. 1913. Observations on the relative constancy of ammonia production by certain bacteria. J. Infect. Dis. 13425-428. [Google Scholar]
- 115.Kendall, A. I., T. E. Friedemann, and M. Ishikawa. 1930. Quantitative observations on the chemical activity of “resting” Staphylococcus aureus. J. Infect. Dis. 47223-228. [Google Scholar]
- 116.Kim, H. J., A. Roux, and A. L. Sonenshein. 2002. Direct and indirect roles of CcpA in regulation of Bacillus subtilis Krebs cycle genes. Mol. Microbiol. 45179-190. [DOI] [PubMed] [Google Scholar]
- 117.Kim, J. H., Y. K. Yang, and G. H. Chambliss. 2005. Evidence that Bacillus catabolite control protein CcpA interacts with RNA polymerase to inhibit transcription. Mol. Microbiol. 56155-162. [DOI] [PubMed] [Google Scholar]
- 118.King, K. Y., J. A. Horenstein, and M. G. Caparon. 2000. Aerotolerance and peroxide resistance in peroxidase and PerR mutants of Streptococcus pyogenes. J. Bacteriol. 1825290-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kinkel, T. L., and K. S. McIver. 2008. CcpA-mediated repression of streptolysin S expression and virulence in the group A streptococcus. Infect. Immun. 763451-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Knobloch, J. K., K. Bartscht, A. Sabottke, H. Rohde, H. H. Feucht, and D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 1832624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Knobloch, J. K., S. Jäger, M. A. Horstkotte, H. Rohde, and D. Mack. 2004. RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor σB by repression of the negative regulator gene icaR. Infect. Immun. 723838-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Koenig, R. L., J. L. Ray, S. J. Maleki, M. S. Smeltzer, and B. K. Hurlburt. 2004. Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J. Bacteriol. 1867549-7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kohanski, M. A., D. J. Dwyer, B. Hayete, C. A. Lawrence, and J. J. Collins. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130797-810. [DOI] [PubMed] [Google Scholar]
- 124.Kohler, C., C. von Eiff, G. Peters, R. A. Proctor, M. Hecker, and S. Engelmann. 2003. Physiological characterization of a heme-deficient mutant of Staphylococcus aureus by a proteomic approach. J. Bacteriol. 1856928-6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Krebs, H. A. 1937. Dismutation of pyruvic acid in Gonococcus and Staphylococcus. Biochem. J. 31661-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kullik, I., and P. Giachino. 1997. The alternative sigma factor sigmaB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch. Microbiol. 167151-159. [DOI] [PubMed] [Google Scholar]
- 127.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 1804814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kunkle, C. A., and M. P. Schmitt. 2003. Analysis of the Corynebacterium diphtheriae DtxR regulon: identification of a putative siderophore synthesis and transport system that is similar to the Yersinia high-pathogenicity island-encoded yersiniabactin synthesis and uptake system. J. Bacteriol. 1856826-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 3571225-1240. [DOI] [PubMed] [Google Scholar]
- 130.Larsson, J. T., A. Rogstam, and C. von Wachenfeldt. 2005. Coordinated patterns of cytochrome bd and lactate dehydrogenase expression in Bacillus subtilis. Microbiology 1513323-3335. [DOI] [PubMed] [Google Scholar]
- 131.Lee, J. W., and J. D. Helmann. 2006. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440363-367. [DOI] [PubMed] [Google Scholar]
- 132.Lemos, J. A., M. M. Nascimento, V. K. Lin, J. Abranches, and R. A. Burne. 2008. Global regulation by (p)ppGpp and CodY in Streptococcus mutans. J. Bacteriol. 1905291-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lester, R. L., and F. L. Crane. 1959. The natural occurrence of coenzyme Q and related compounds. J. Biol. Chem. 2342169-2175. [PubMed] [Google Scholar]
- 134.Levdikov, V. M., E. Blagova, P. Joseph, A. L. Sonenshein, and A. J. Wilkinson. 2006. The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in gram-positive bacteria. J. Biol. Chem. 28111366-11373. [DOI] [PubMed] [Google Scholar]
- 135.Lieser, S. A., T. C. Davis, J. D. Helmann, and S. M. Cohen. 2003. DNA-binding and oligomerization studies of the manganese(II) metalloregulatory protein MntR from Bacillus subtilis. Biochemistry 4212634-12642. [DOI] [PubMed] [Google Scholar]
- 136.Lina, G., S. Jarraud, G. Ji, T. Greenland, A. Pedraza, J. Etienne, R. P. Novick, and F. Vandenesch. 1998. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol. Microbiol. 28655-662. [DOI] [PubMed] [Google Scholar]
- 137.Lindsay, J. A., and S. J. Foster. 2001. zur: a Zn(2+)-responsive regulatory element of Staphylococcus aureus. Microbiology 1471259-1266. [DOI] [PubMed] [Google Scholar]
- 138.Liu, Y., A. C. Manna, C. H. Pan, I. A. Kriksunov, D. J. Thiel, A. L. Cheung, and G. Zhang. 2006. Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 1032392-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lopez, J. M., and B. Thoms. 1977. Role of sugar uptake and metabolic intermediates on catabolite repression in Bacillus subtilis. J. Bacteriol. 129217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Low, Y. L., N. S. Jakubovics, J. C. Flatman, H. F. Jenkinson, and A. W. Smith. 2003. Manganese-dependent regulation of the endocarditis-associated virulence factor EfaA of Enterococcus faecalis. J. Med. Microbiol. 52113-119. [DOI] [PubMed] [Google Scholar]
- 141.Lucarelli, D., S. Russo, E. Garman, A. Milano, W. Meyer-Klaucke, and E. Pohl. 2007. Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosis. J. Biol. Chem. 2829914-9922. [DOI] [PubMed] [Google Scholar]
- 142.Luong, T. T., P. M. Dunman, E. Murphy, S. J. Projan, and C. Y. Lee. 2006. Transcription profiling of the mgrA regulon in Staphylococcus aureus. J. Bacteriol. 1881899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Luong, T. T., S. W. Newell, and C. Y. Lee. 2003. Mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 1853703-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Maciag, A., E. Dainese, G. M. Rodriguez, A. Milano, R. Provvedi, M. R. Pasca, I. Smith, G. Palù, G. Riccardi, and R. Manganelli. 2007. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 189730-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Magasanik, B. 1961. Catabolite repression. Cold Spring Harb. Symp. Quant. Biol. 26249-256. [DOI] [PubMed] [Google Scholar]
- 146.Mah, R. A., D. Y. Fung, and S. A. Morse. 1967. Nutritional requirements of Staphylococcus aureus S-6. Appl. Microbiol. 15866-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Majerczyk, C. D., M. R. Sadykov, T. T. Luong, C. Lee, G. A. Somerville, and A. L. Sonenshein. 2008. Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 1902257-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Malke, H., and J. J. Ferretti. 2007. CodY-affected transcriptional gene expression of Streptococcus pyogenes during growth in human blood. J. Med. Microbiol. 56707-714. [DOI] [PubMed] [Google Scholar]
- 149.Malke, H., K. Steiner, W. M. McShan, and J. J. Ferretti. 2006. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: the action of CodY and RelA. Int. J. Med. Microbiol. 296259-275. [DOI] [PubMed] [Google Scholar]
- 150.Mayville, P., G. Ji, R. Beavis, H. Yang, M. Goger, R. P. Novick, and T. W. Muir. 1999. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. USA 961218-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mazmanian, S. K., E. P. Skaar, A. H. Gaspar, M. Humayun, P. Gornicki, J. Jelenska, A. Joachmiak, D. M. Missiakas, and O. Schneewind. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299906-909. [DOI] [PubMed] [Google Scholar]
- 152.Mei, J. M., F. Nourbakhsh, C. W. Ford, and D. W. Holden. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26399-407. [DOI] [PubMed] [Google Scholar]
- 153.Milano, A., M. Branzoni, F. Canneva, A. Profumo, and G. Riccardi. 2004. The Mycobacterium tuberculosis Rv2358-furB operon is induced by zinc. Res. Microbiol. 155192-200. [DOI] [PubMed] [Google Scholar]
- 154.Mongkolsuk, S., and J. D. Helmann. 2002. Regulation of inducible peroxide stress responses. Mol. Microbiol. 459-15. [DOI] [PubMed] [Google Scholar]
- 155.Morfeldt, E., D. Taylor, A. von Gabain, and S. Arvidson. 1995. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 14:4569-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Morfeldt, E., K. Tegmark, and S. Arvidson. 1996. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol. Microbiol. 211227-1237. [DOI] [PubMed] [Google Scholar]
- 157.Morikawa, K., Y. Inose, H. Okamura, A. Maruyama, H. Hayashi, K. Takeyasu, and T. Ohta. 2003. A new staphylococcal sigma factor in the conserved gene cassette: functional significance and implication for the evolutionary processes. Genes Cells 8699-712. [DOI] [PubMed] [Google Scholar]
- 158.Morrissey, J. A., A. Cockayne, P. J. Hill, and P. Williams. 2000. Molecular cloning and analysis of a putative siderophore ABC transporter from Staphylococcus aureus. Infect. Immun. 686281-6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nakano, M. M. 2002. Induction of ResDE-dependent gene expression in Bacillus subtilis in response to nitric oxide and nitrosative stress. J. Bacteriol. 1841783-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nakano, M. M., Y. Zhu, M. Lacelle, X. Zhang, and F. M. Hulett. 2000. Interaction of ResD with regulatory regions of anaerobically induced genes in Bacillus subtilis. Mol. Microbiol. 371198-1207. [DOI] [PubMed] [Google Scholar]
- 161.Nakano, M. M., P. Zuber, P. Glaser, A. Danchin, and F. M. Hulett. 1996. Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fnr upon oxygen limitation in Bacillus subtilis. J. Bacteriol. 1783796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nakano, Y., and K. Kimura. 1991. Purification and characterization of a repressor for the Bacillus cereus glnRA operon. J. Biochem. 109223-228. [PubMed] [Google Scholar]
- 163.Nelson, J. L., K. C. Rice, S. R. Slater, P. M. Fox, G. L. Archer, K. W. Bayles, P. D. Fey, B. N. Kreiswirth, and G. A. Somerville. 2007. Vancomycin-intermediate Staphylococcus aureus strains have impaired acetate catabolism: implications for polysaccharide intercellular adhesin synthesis and autolysis. Antimicrob. Agents Chemother. 51616-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nicholson, W. L., Y. K. Park, T. M. Henkin, M. Won, M. J. Weickert, J. A. Gaskell, and G. H. Chambliss. 1987. Catabolite repression-resistant mutations of the Bacillus subtilis alpha-amylase promoter affect transcription levels and are in an operator-like sequence. J. Mol. Biol. 198609-618. [DOI] [PubMed] [Google Scholar]
- 165.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 481429-1449. [DOI] [PubMed] [Google Scholar]
- 166.Novick, R. P., S. J. Projan, J. Kornblum, H. F. Ross, G. Ji, B. Kreiswirth, F. Vandenesch, and S. Moghazeh. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248446-458. [DOI] [PubMed] [Google Scholar]
- 167.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 123967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Nychas, G. J., H. S. Tranter, R. D. Brehm, and R. G. Board. 1991. Staphylococcus aureus S-6: factors affecting its growth, enterotoxin B production and exoprotein formation. J. Appl. Bacteriol. 70344-350. [DOI] [PubMed] [Google Scholar]
- 169.O'Neill, E., C. Pozzi, P. Houston, H. Humphreys, D. A. Robinson, A. Loughman, T. J. Foster, and J. P. O'Gara. 2008. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 1903835-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Onoue, Y., and M. Mori. 1997. Amino acid requirements for the growth and enterotoxin production by Staphylococcus aureus in chemically defined media. Int. J. Food Microbiol. 3677-82. [DOI] [PubMed] [Google Scholar]
- 171.Owen, G. A., B. Pascoe, D. Kallifidas, and M. S. Paget. 2007. Zinc-responsive regulation of alternative ribosomal protein genes in Streptomyces coelicolor involves zur and σR. J. Bacteriol. 1894078-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Palma, M., and A. L. Cheung. 2001. σB activity in Staphylococcus aureus is controlled by RsbU and an additional factor(s) during bacterial growth. Infect. Immun. 697858-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Panina, E. M., A. A. Mironov, and M. S. Gelfand. 2003. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl. Acad. Sci. USA 1009912-9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Papp-Wallace, K. M., and M. E. Maguire. 2006. Manganese transport and the role of manganese in virulence. Annu. Rev. Microbiol. 60187-209. [DOI] [PubMed] [Google Scholar]
- 175.Pardee, A. B., F. Jacob, and J. Monod. 1958. The role of the inducible alleles and the constrtutive alleles in the synthesis of beta-galactosidase in zygotes of Escherichia coli. C. R. Hebd. Seances Acad. Sci. 2463125-3128. (In French.) [PubMed] [Google Scholar]
- 176.Pohl, E., J. C. Haller, A. Mijovilovich, W. Meyer-Klaucke, E. Garman, and M. L. Vasil. 2003. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Microbiol. 47903-915. [DOI] [PubMed] [Google Scholar]
- 177.Pragman, A. A., Y. Ji, and P. M. Schlievert. 2007. Repression of Staphylococcus aureus SrrAB using inducible antisense srrA alters growth and virulence factor transcript levels. Biochemistry 46314-321. [DOI] [PubMed] [Google Scholar]
- 178.Pragman, A. A., J. M. Yarwood, T. J. Tripp, and P. M. Schlievert. 2004. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 1862430-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Qiu, X., C. L. Verlinde, S. Zhang, M. P. Schmitt, R. K. Holmes, and W. G. Hol. 1995. Three-dimensional structure of the diphtheria toxin repressor in complex with divalent cation co-repressors. Structure 387-100. [DOI] [PubMed] [Google Scholar]
- 180.Rea, R. B., C. G. Gahan, and C. Hill. 2004. Disruption of putative regulatory loci in Listeria monocytogenes demonstrates a significant role for Fur and PerR in virulence. Infect. Immun. 72717-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Recsei, P., B. Kreiswirth, M. O'Reilly, P. Schlievert, A. Gruss, and R. P. Novick. 1986. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol. Gen. Genet. 20258-61. [DOI] [PubMed] [Google Scholar]
- 182.Regassa, L. B., J. L. Couch, and M. J. Betley. 1991. Steady-state staphylococcal enterotoxin type C mRNA is affected by a product of the accessory gene regulator (agr) and by glucose. Infect. Immun. 59955-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Regassa, L. B., R. P. Novick, and M. J. Betley. 1992. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect. Immun. 603381-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Resch, A., R. Rosenstein, C. Nerz, and F. Götz. 2005. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol. 712663-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ricci, S., R. Janulczyk, and L. Björck. 2002. The regulator PerR is involved in oxidative stress response and iron homeostasis and is necessary for full virulence of Streptococcus pyogenes. Infect. Immun. 704968-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Roberts, C., K. L. Anderson, E. Murphy, S. J. Projan, W. Mounts, B. Hurlburt, M. Smeltzer, R. Overbeek, T. Disz, and P. M. Dunman. 2006. Characterizing the effect of the Staphylococcus aureus virulence factor regulator, SarA, on log-phase mRNA half-lives. J. Bacteriol. 1882593-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Rodriguez, G. M., M. I. Voskuil, B. Gold, G. K. Schoolnik, and I. Smith. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 703371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ross, R. A., and A. B. Onderdonk. 2000. Production of toxic shock syndrome toxin 1 by Staphylococcus aureus requires both oxygen and carbon dioxide. Infect. Immun. 685205-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Rossmann, M. G., M. J. Adams, M. Buehner, G. C. Ford, M. L. Hackert, A. Liljas, S. T. Rao, L. J. Banaszak, E. Hill, D. Tsernoglou, and L. Webb. 1973. Molecular symmetry axes and subunit interfaces in certain dehydrogenases. J. Mol. Biol. 76533-537. [DOI] [PubMed] [Google Scholar]
- 190.Sadykov, M. R., M. E. Olson, S. Halouska, Y. Zhu, P. D. Fey, R. Powers, and G. A. Somerville. 2008. Tricarboxylic acid cycle-dependent regulation of Staphylococcus epidermidis polysaccharide intercellular adhesin synthesis. J. Bacteriol. 1907621-7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Schau, M., Y. Chen, and F. M. Hulett. 2004. Bacillus subtilis YdiH is a direct negative regulator of the cydABCD operon. J. Bacteriol. 1864585-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Schlag, S., S. Fuchs, C. Nerz, R. Gaupp, S. Engelmann, M. Liebeke, M. Lalk, M. Hecker, and F. Götz. 2008. Characterization of the oxygen-responsive NreABC regulon of Staphylococcus aureus. J. Bacteriol. 1907847-7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Schlag, S., C. Nerz, T. A. Birkenstock, F. Altenberend, and F. Götz. 2007. Inhibition of staphylococcal biofilm formation by nitrite. J. Bacteriol. 1897911-7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Schlievert, P. M., and D. A. Blomster. 1983. Production of staphylococcal pyrogenic exotoxin type C: influence of physical and chemical factors. J. Infect. Dis. 147236-242. [DOI] [PubMed] [Google Scholar]
- 195.Schmitt, M. P., M. Predich, L. Doukhan, I. Smith, and R. K. Holmes. 1995. Characterization of an iron-dependent regulatory protein (IdeR) of Mycobacterium tuberculosis as a functional homolog of the diphtheria toxin repressor (DtxR) from Corynebacterium diphtheriae. Infect. Immun. 634284-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Schreier, H. J., S. W. Brown, K. D. Hirschi, J. F. Nomellini, and A. L. Sonenshein. 1989. Regulation of Bacillus subtilis glutamine synthetase gene expression by the product of the glnR gene. J. Mol. Biol. 21051-63. [DOI] [PubMed] [Google Scholar]
- 197.Schreier, H. J., S. M. Caruso, and K. C. Maier. 2000. Control of Bacillus subtilis glutamine synthetase expression by glnR from Staphylococcus aureus. Curr. Microbiol. 41425-429. [DOI] [PubMed] [Google Scholar]
- 198.Schreier, H. J., C. A. Rostkowski, J. F. Nomellini, and K. D. Hirschi. 1991. Identification of DNA sequences involved in regulating Bacillus subtilis glnRA expression by the nitrogen source. J. Mol. Biol. 220241-253. [DOI] [PubMed] [Google Scholar]
- 199.Schumacher, M. A., G. Seidel, W. Hillen, and R. G. Brennan. 2007. Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors glucose 6-phosphate and fructose 1,6-bisphosphate. J. Mol. Biol. 3681042-1050. [DOI] [PubMed] [Google Scholar]
- 200.Sebulsky, M. T., D. Hohnstein, M. D. Hunter, and D. E. Heinrichs. 2000. Identification and characterization of a membrane permease involved in iron-hydroxamate transport in Staphylococcus aureus. J. Bacteriol. 1824394-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Seidl, K., M. Bischoff, and B. Berger-Bächi. 2008. CcpA mediates the catabolite repression of tst in Staphylococcus aureus. Infect. Immun. 765093-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Seidl, K., C. Goerke, C. Wolz, D. Mack, B. Berger-Bächi, and M. Bischoff. 2008. Staphylococcus aureus CcpA affects biofilm formation. Infect. Immun. 762044-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sen, K. I., A. Sienkiewicz, J. F. Love, J. C. vanderSpek, P. G. Fajer, and T. M. Logan. 2006. Mn(II) binding by the anthracis repressor from Bacillus anthracis. Biochemistry 454295-4303. [DOI] [PubMed] [Google Scholar]
- 204.Senn, M. M., P. Giachino, D. Homerova, A. Steinhuber, J. Strassner, J. Kormanec, U. Flückiger, B. Berger-Bächi, and M. Bischoff. 2005. Molecular analysis and organization of the σB operon in Staphylococcus aureus. J. Bacteriol. 1878006-8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Shin, J. H., S. Y. Oh, S. J. Kim, and J. H. Roe. 2007. The zinc-responsive regulator Zur controls a zinc uptake system and some ribosomal proteins in Streptomyces coelicolor A3(2). J. Bacteriol. 1894070-4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Shivers, R. P., S. S. Dineen, and A. L. Sonenshein. 2006. Positive regulation of Bacillus subtilis ackA by CodY and CcpA: establishing a potential hierarchy in carbon flow. Mol. Microbiol. 62811-822. [DOI] [PubMed] [Google Scholar]
- 207.Shivers, R. P., and A. L. Sonenshein. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53599-611. [DOI] [PubMed] [Google Scholar]
- 208.Sickmier, E. A., D. Brekasis, S. Paranawithana, J. B. Bonanno, M. S. Paget, S. K. Burley, and C. L. Kielkopf. 2005. X-ray structure of a Rex-family repressor/NADH complex insights into the mechanism of redox sensing. Structure 1343-54. [DOI] [PubMed] [Google Scholar]
- 209.Skaar, E. P., M. Humayun, T. Bae, K. L. DeBord, and O. Schneewind. 2004. Iron-source preference of Staphylococcus aureus infections. Science 3051626-1628. [DOI] [PubMed] [Google Scholar]
- 210.Slack, F. J., P. Serror, E. Joyce, and A. L. Sonenshein. 1995. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol. Microbiol. 15689-702. [DOI] [PubMed] [Google Scholar]
- 211.Smith, J. L., M. M. Bencivengo, R. L. Buchanan, and C. A. Kunsch. 1986. Enterotoxin A production in Staphylococcus aureus: inhibition by glucose. Arch. Microbiol. 144131-136. [DOI] [PubMed] [Google Scholar]
- 212.Somerville, G. A. 2008. Oxidative stress and the host-pathogen interaction, p. 218-225. In R. Banerjee (ed.), Redox biochemistry. John Wiley & Sons, Hoboken, NJ.
- 213.Somerville, G. A., M. S. Chaussee, C. I. Morgan, J. R. Fitzgerald, D. W. Dorward, L. J. Reitzer, and J. M. Musser. 2002. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect. Immun. 706373-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Somerville, G. A., A. Cockayne, M. Dürr, A. Peschel, M. Otto, and J. M. Musser. 2003. Synthesis and deformylation of Staphylococcus aureus delta-toxin are linked to tricarboxylic acid cycle activity. J. Bacteriol. 1856686-6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Somerville, G. A., B. Saïd-Salim, J. M. Wickman, S. J. Raffel, B. N. Kreiswirth, and J. M. Musser. 2003. Correlation of acetate catabolism and growth yield in Staphylococcus aureus: implications for host-pathogen interactions. Infect. Immun. 714724-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Sonenshein, A. L. 2005. CodY, a global regulator of stationary phase and virulence in gram-positive bacteria. Curr. Opin. Microbiol. 8203-207. [DOI] [PubMed] [Google Scholar]
- 217.Sterba, K. M., S. G. Mackintosh, J. S. Blevins, B. K. Hurlburt, and M. S. Smeltzer. 2003. Characterization of Staphylococcus aureus SarA binding sites. J. Bacteriol. 1854410-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Strandén, A. M., M. Roos, and B. Berger-Bächi. 1996. Glutamine synthetase and heteroresistance in methicillin-resistant Staphylococcus aureus. Microb. Drug Resist. 2201-207. [DOI] [PubMed] [Google Scholar]
- 219.Strasters, K. C., and K. C. Winkler. 1963. Carbohydrate metabolism of Staphylococcus aureus. J. Gen. Microbiol. 33213-229. [DOI] [PubMed] [Google Scholar]
- 220.Taylor, D., and K. T. Holland. 1989. Amino acid requirements for the growth and production of some exocellular products of Staphylococcus aureus. J. Appl. Bacteriol. 66319-329. [DOI] [PubMed] [Google Scholar]
- 221.Throup, J. P., F. Zappacosta, R. D. Lunsford, R. S. Annan, S. A. Carr, J. T. Lonsdale, A. P. Bryant, D. McDevitt, M. Rosenberg, and M. K. Burnham. 2001. The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry 4010392-10401. [DOI] [PubMed] [Google Scholar]
- 222.Tojo, S., T. Satomura, K. Morisaki, J. Deutscher, K. Hirooka, and Y. Fujita. 2005. Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Mol. Microbiol. 561560-1573. [DOI] [PubMed] [Google Scholar]
- 223.Torres, V. J., D. L. Stauff, G. Pishchany, J. S. Bezbradica, L. E. Gordy, J. Iturregui, K. L. Anderson, P. M. Dunman, S. Joyce, and E. P. Skaar. 2007. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1109-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Truong-Bolduc, Q. C., P. M. Dunman, J. Strahilevitz, S. J. Projan, and D. C. Hooper. 2005. MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J. Bacteriol. 1872395-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Truong-Bolduc, Q. C., J. Strahilevitz, and D. C. Hooper. 2006. NorC, a new efflux pump regulated by MgrA of Staphylococcus aureus. Antimicrob. Agents Chemother. 501104-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Ulrich, M., M. Bastian, S. E. Cramton, K. Ziegler, A. A. Pragman, A. Bragonzi, G. Memmi, C. Wolz, P. M. Schlievert, A. Cheung, and G. Döring. 2007. The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol. Microbiol. 651276-1287. [DOI] [PubMed] [Google Scholar]
- 227.Vandenesch, F., J. Kornblum, and R. P. Novick. 1991. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J. Bacteriol. 1736313-6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.van der Voort, M., O. P. Kuipers, G. Buist, W. M. de Vos, and T. Abee. 2008. Assessment of CcpA-mediated catabolite control of gene expression in Bacillus cereus ATCC 14579. BMC Microbiol. 8:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Vetter, S. M., and P. M. Schlievert. 2007. The two-component system Bacillus respiratory response A and B (BrrA-BrrB) is a virulence factor regulator in Bacillus anthracis. Biochemistry 467343-7352. [DOI] [PubMed] [Google Scholar]
- 230.Vuong, C., J. B. Kidder, E. R. Jacobson, M. Otto, R. A. Proctor, and G. A. Somerville. 2005. Staphylococcus epidermidis polysaccharide intercellular adhesin production significantly increases during tricarboxylic acid cycle stress. J. Bacteriol. 1872967-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Weickert, M. J., and S. Adhya. 1992. A family of bacterial regulators homologous to Gal and Lac repressors. J. Biol. Chem. 26715869-15874. [PubMed] [Google Scholar]
- 232.Wilson, A. C., J. A. Hoch, and M. Perego. 2008. Virulence gene expression is independent of ResDE-regulated respiration control in Bacillus anthracis. J. Bacteriol. 1905522-5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Wong, A. C., and M. S. Bergdoll. 1990. Effect of environmental conditions on production of toxic shock syndrome toxin 1 by Staphylococcus aureus. Infect. Immun. 581026-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wong, D. K., B. Y. Lee, M. A. Horwitz, and B. W. Gibson. 1999. Identification of Fur, aconitase, and other proteins expressed by Mycobacterium tuberculosis under conditions of low and high concentrations of iron by combined two-dimensional gel electrophoresis and mass spectrometry. Infect. Immun. 67327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Wray, L. V., Jr., A. E. Ferson, K. Rohrer, and S. H. Fisher. 1996. TnrA, a transcription factor required for global nitrogen regulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 938841-8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Wray, L. V., Jr., F. K. Pettengill, and S. H. Fisher. 1994. Catabolite repression of the Bacillus subtilis hut operon requires a cis-acting site located downstream of the transcription initiation site. J. Bacteriol. 1761894-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Wu, S., H. de Lencastre, and A. Tomasz. 1996. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J. Bacteriol. 1786036-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Yarwood, J. M., J. K. McCormick, and P. M. Schlievert. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 1831113-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Zalieckas, J. M., L. V. Wray, Jr., and S. H. Fisher. 2006. Cross-regulation of the Bacillus subtilis glnRA and tnrA genes provides evidence for DNA binding site discrimination by GlnR and TnrA. J. Bacteriol. 1882578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zhang, X., and F. M. Hulett. 2000. ResD signal transduction regulator of aerobic respiration in Bacillus subtilis:ctaA promoter regulation. Mol. Microbiol. 371208-1219. [DOI] [PubMed] [Google Scholar]