Abstract
In streptococci, the secA2 locus includes genes encoding the following: (i) the accessory Sec components (SecA2, SecY2, and at least three accessory secretion proteins), (ii) two essential glycosyltranferases (GTs) (GtfA and GtfB), (iii) a variable number of dispensable additional GTs, and (iv) a secreted serine-rich LPXTG protein which is glycosylated in the cytoplasm and transported to the cell surface by this accessory Sec system. The secA2 locus of Streptococcus agalactiae strain NEM316 is structurally related to those found in other streptococci and encodes the serine-rich surface protein Srr1. We demonstrated that expression of Srr1 but not that of the SecA2 components and the associated GTs is regulated by the standalone transcriptional regulator Rga. Srr1 is synthesized as a glycosylated precursor, secreted by the SecA2 system, and anchored to the cell wall by the housekeeping sortase A. Srr1 was localized preferentially at the old poles. GtfA and/or GtfB, but not the six additional GTs, is essential for the production of Srr1. These GTs are involved in the attachment of GlcNac and sialic acid to Srr1. Full glycosylation of Srr1 is associated with the cell surface display of a protein that is more resistant to proteolytic attack. Srr1 contributes to bacterial adherence to human epithelial cell lines and virulence in a neonatal rat model. The extent of Srr1 glycosylation by GtfC to -H modulates bacterial adherence and virulence.
Adherence of bacteria to host tissues is a key event in the pathogenesis of many infectious diseases. It also plays an important role in bacterial colonization of medical devices coated with various cellular and extracellular host components. Gram-positive pathogens express numerous cell surface proteins which can mediate interactions with the components of the host extracellular matrix, adherence to and colonization and invasion of host cells and tissues, and evasion of host defenses (27, 29). The majority of secreted proteins in gram-positive bacteria utilize the SecA-dependent pathway for translocation across the cytoplasmic membrane irrespective of their subsequent targeting. Sec systems are composed of four main proteins: SecA, -Y, -E, and -G (34, 53). SecA is a cytoplasmic protein that targets the signal peptide-containing precursor polypeptide to the membrane translocase formed by SecYEG. Interestingly, some high- and low-GC gram-positive bacteria encode an extra SecA paralogue, designated SecA2, that is required for the export of proteins involved in virulence (4, 6, 28, 37).
In Mycobacterium tuberculosis and Listeria monocytogenes, the SecA2 pathway promotes export of a small subset of proteins devoid of signal peptide which are either well-described cytosolic enzymes like SodA (1, 7) or surface-located proteins like the multifunctional enzyme FbpA (14). Surprisingly, two lipoproteins encoded by Mycobacterium smegmatis that possess canonical signal peptides are also secreted through the SecA2 system (19).
In streptococci, the SecA2 pathway has been extensively studied in the species Streptococcus gordonii, where SecA2, SecY2, and three accessory secretory proteins (Asp1 to -3) (4, 49, 50) are dedicated to the export of the serine-rich glycoprotein GspB, which participates in the binding of bacteria to platelets (48). GspB is glycosylated in the cytoplasm and is transported to the cell surface via SecA2 (5). In Streptococcus parasanguinis (12) and Staphylococcus aureus (44), the SecA2 pathway is also involved in the secretion of unrelated serine-rich cell wall-anchored LPXTG adhesins, designated Fap1 and SraP, respectively. The secA2 locus of S. gordonii, S. parasanguis, Streptococcus pneumoniae, and S. aureus encodes the components of the SecA2 pathway (i.e., SecA2, SecY2, and Asp1 to -3), the substrate protein (i.e., GspB, Fap1, PsrP, or SraP, respectively), and associated glycosyltransferases (GTs) (see Fig. 1A).
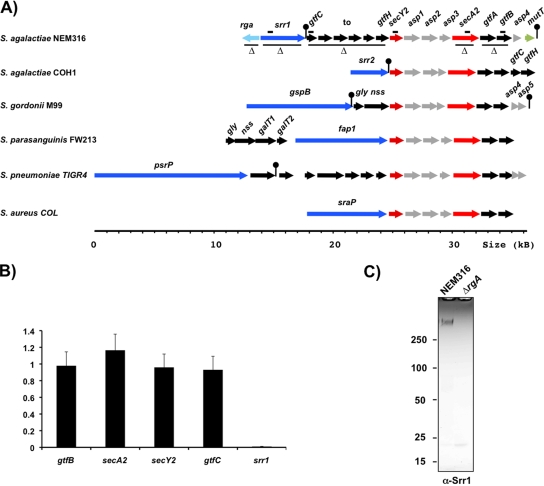
FIG. 1.
Characterization of the secA2 locus of S. agalactiae NEM316. (A) Comparison of the secA2 locus of NEM316 (http://genolist.pasteur.fr/SagaList/) with those of the ST17 GBS strain COH1 (accession number NZ_AAJR00000000), S. gordonii M99 (accession number AY028382), S. pneumoniae TIGR4 (GenBank accession number NC_003028), and S. aureus COL (accession number NC_002951). The NEM316 locus comprises a divergent regulatory gene designated rga (regulation of glycosylated adhesin) and 15 genes encoding a serine-rich cell wall-anchored LPXTG protein (srr1), 8 glycosyltransferases (gtfC to gtfH and gtfA and gtfB), and 6 proteins that are components of the SecA2 secretion apparatus (secA2, secY2, and asp1 to asp4). Seven orthologous genes (secA2, secY2, asp1 to asp3, gtfA, and gtfB) are present in all loci. Note that the NEM316 secA2 locus typified that present in most GBS strains with the remarkable exception of ST17 strains, which all possess the COH1 structure (see Fig. S1 in the supplemental material). The thin black lines below the NEM316 locus delineate the in-frame deletions carried out to inactivate rga, srr1, gtfCDEFGH, secA2, and gtfAB. The heavy black lines above the locus delineate the mRNA segments analyzed by qRT-PCR. The genes depicted in dark blue code the serine-rich LPXTG proteins that are secreted through the SecA2 system. (B) qRT-PCR analysis of the secA2 locus in NEM316 and the isogenic Δrga regulatory mutant. Total RNAs were extracted from bacteria cultivated in TH broth at 37°C and collected in late exponential phase (OD600 = 0.5). The expression of five genes scattered along the locus was measured in the two strains and normalized relative to the rpoB reference gene, and their expression ratio in the Δrga mutant relative to that in NEM316 was calculated. qRT-PCR experiments were performed in quadruplicate using two different RNA preparations. Similar results were obtained with the two preparations, and the mean values of one experiment are shown here. The standard deviation was calculated from the four experimental values of this analysis. (C) Western blot expression analysis of Srr1 in NEM316 and the isogenic Δrga regulatory mutant. Cell wall proteins were separated on 3-to-8%-gradient Tris-acetate Criterion XT SDS-PAGE gels, and Srr1 was detected by immunoblotting with rabbit anti-Srr1 pAb (α-Srr1). Ten μg of protein was loaded in each well.
Streptococcus agalactiae, also named group B streptococcus (GBS), is frequently found in the vaginal flora (20 to 40%) of pregnant women. It is the major cause of sepsis and meningitis in neonates (13). Colonization of the rectum and vagina of pregnant women by GBS, which causes infection of the amniotic cavity, is correlated with GBS sepsis in newborn infants with early-onset disease. In this case, newborns are colonized intrapartum by the aspiration of contaminated amniotic fluid. The lung is a likely portal entry for GBS into the bloodstream, since the bacterium can adhere to and invade alveolar epithelial (39) and endothelial (20) cells. The adherence of GBS to the mother's (intestinal and vaginal) and infant's (lung) epithelial cells is therefore considered essential for virulence. Genome analysis of S. agalactiae isolates (8) revealed the presence of two types of secA2 locus structurally related to those found in other streptococci. One locus is widespread among isolates from various serotypes and multilocus sequence types and encodes the serine-rich cell wall-anchored surface protein Srr1, whereas the other is apparently restricted to the hypervirulent sequence type 17 (ST17) lineage and encodes the serine-rich surface protein Srr2 (8, 43). It was recently reported that the Srr1 protein of the serotype III strain S. agalactiae 6313 was involved in adherence to epithelial HEp-2 cells by interacting with human keratin 4 (41).
In the present report, we carried out an extensive structural and functional analysis of the secA2 locus of S. agalactiae strain NEM316, which demonstrated the following: (i) expression of Srr1 but not that of the SecA2 components and associated GTs is regulated by the standalone transcriptional regulator Rga, (ii) Srr1 should be synthesized as a glycosylated precursor, and its secretion is SecA2 dependent, (iii) Srr1 contributes to bacterial adherence to human lung epithelial cell lines and virulence in a neonatal rat model, (iv) fully glycosylated Srr1 is more resistant to proteolysis, and (v) the extent of Srr1 glycosylation modulates bacterial adherence and virulence.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
S. agalactiae NEM316 was responsible for a fatal septicemia and belongs to the capsular serotype III, and its genome has been entirely sequenced (21). Escherichia coli DH5α (Gibco-BRL) was used for cloning experiments, and E. coli BL21(λDE3) (Novagen) was used for expression of His tag recombinant proteins. S. agalactiae was cultured in Todd-Hewitt (TH) broth or agar (Difco Laboratories, Detroit, MI) and E. coli in Luria-Bertani medium. Unless otherwise specified, antibiotics were used at the following concentrations: for E. coli, ampicillin, 100 μg/ml, and erythromycin, 150 μg/ml; for S. agalactiae, erythromycin, 10 μg/ml. S. agalactiae liquid cultures were grown at 37°C in standing filled flasks. The relevant characteristics of strains and plasmids used are shown in Table S1 in the supplemental material.
General DNA techniques.
Standard recombinant techniques were used for nucleic acid cloning and restriction analysis (40). Plasmid DNA from E. coli was prepared by rapid alkaline lysis using the QIAprep Spin Miniprep kit (Qiagen). Genomic DNA from S. agalactiae was prepared using the Boehringer-Roche High Purification kit. PCR was carried out with AmpliTaq Gold polymerase as described by the manufacturer (Applied Biosystems). Amplification products were purified on Sephadex S-400 columns (Pharmacia) and sequenced with an ABI 310 automated DNA sequencer, using the ABI PRISM dye terminator cycle sequencing kit (Applied Biosystems) to verify that no misincorporation of nucleotides had occurred during the PCR process.
Construction of strains.
The primers used for the construction of deletion alleles are listed in Table S2 in the supplemental material. In-frame deletion in gbs1530 (rga) (O1-O2; O3-O4), gbs1529 (srr1) (O5-O6; O7-O8), and gbs1518 (secA2) (O13-O14; O15-O16) and in the glycosyltransferase-encoding genes gbs1523 to gbs1528 (gtfC to gtfH) (O9-O10; O11-O12) and gbs1516-gbs1515 (gtfA and gtfB) (O17-O18; O19-O20) were constructed by using splicing-by-overlap extension PCR as described previously (15). To carry out chromosomal gene deletion, appropriate PCR fragments were cloned into the thermosensitive shuttle plasmid pG1 (see Table S1 in the supplemental material). Electroporation of GBS strains and allelic exchange were performed as described previously (15). The expected in-frame deletions were confirmed by PCR and sequence analysis.
The primers O23 and O24 were used to amplify the sequence encoding the NH2-terminal domain of Srr1 (residues 1 to 638). The PCR fragment was digested with appropriate enzymes and cloned into the shuttle vector pAT18P, downstream of the Streptococcus pyogenes gyrA promoter (33, 51). The resulting recombinant vector was introduced by electroporation into NEM316 to express the truncated Srr1 protein.
Real-time quantitative PCR.
Total RNAs were extracted from NEM316 and NEM316Δrga, harvested at an optical density at 600 nm (OD600) of 0.5 as previously described (26). cDNA synthesis was performed on DNase I-treated RNA (5 μg) with random hexamers (Roche) using Superscript II reverse transcriptase (RT) (Invitrogen) as recommended. Real-time quantitative PCR was performed as described previously (15) in a 25-μl reaction volume containing cDNA, 12.5 μl iQTM SYBR Supermix (Bio-Rad), and 0.5 μl gene-specific primer (10 μM) (see Table S2 in the supplemental material). Briefly, amplification, detection, and analysis were performed with the MyiQ single-color real-time iCycler PCR detection system and the MyiQ optical system software program (Bio-Rad). The specificity of the amplified products and the absence of primer dimer formation were verified by generating melting curves. The absence of contaminating genomic DNA was verified by testing each sample in control reactions without a prior reverse transcription step. The critical threshold cycle was defined for each sample. The expression levels of the tested genes were normalized using the NEM316 rpoB gene, whose transcription level did not vary under our experimental conditions. Each assay was performed in quadruplicate and repeated with two independent RNA samples. The change in the transcript level was calculated as described previously (17, 42).
Production of rabbit polyclonal antibodies.
The sequence encoding the NH2-terminal domain of Srr1 (residues 105 to 566) was cloned in pIVEX-2.4b-NdeI (Roche, Applied Science), and the corresponding six-His-tagged recombinant protein was produced in E. coli BL21(λDE3) and purified on nickel-nitrilotriacetic acid agarose. This purified truncated Srr1 protein, which does not contain the signal peptide or the serine-rich carboxylic half, was injected into rabbit to produce polyclonal anti-Srr1 antibodies as described previously (25). Rabbit antisera against the cell wall-anchored protein Gbs0791 (residues 34 to 361 with a carboxylic six-His tag) and the secreted protein Bsp/Gbs1420 (residues 40 to 374 with a carboxylic six-His tag), which were used as controls, were from our antibody collection. Rabbit polyclonal anti-Alp2 antibodies were kindly provided by G. Lindhal (University of Lund, Lund, Sweden).
Cell wall, protoplast, and secreted protein extracts.
To prepare cell wall and protoplast extracts, bacteria grown in 50 ml of TH medium were harvested in exponential phase (OD600 = 0.3). Cells were washed twice with ice-cold phosphate-buffered saline (PBS), resuspended at 100 U OD600 in 50 mM Tris-HCl (pH 7.5) containing sucrose (1 M), mutanolysin (500 U/ml) (Sigma), and a complete protease inhibitor cocktail used according to the manufacturer's recommendations (Roche Diagnostics), and incubated for 90 min at 37°C. The suspension was then centrifuged at 5,000 × g for 15 min at 4°C. The supernatant corresponding to the cell wall extract was collected. The pellets corresponding to protoplasts were lysed in 2 ml sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer.
Supernatant fractions were prepared from overnight cultures. After centrifugation (5,000 × g, 10 min), supernatants were filtrated through 0.22-μm membrane filters and concentrated 50-fold by ultrafiltration on Sartorius Vivaspin 20 devices (cutoff, 10 kDa) (Sartorius AG) in the presence of a complete protease inhibitor cocktail (Roche Diagnostics).
Proteins concentrations were measured with the bicinchoninic acid protein assay (Pierce).
Dot blots and in-gel detection of Srr1.
For dot blot analysis, bacteria harvested in exponential phase (OD600 = 0.3) were washed twice with cold PBS and resuspended at 1.5 U OD600, and 5 μl of bacterial suspension was spotted on nitrocellulose. Cell surface exposed proteins were detected using specific polyclonal antibody (pAb), horseradish peroxidase (HRP)-coupled antirabbit secondary antibodies (Zymed), and the Western Pico chemiluminescence kit (Pierce).
Cell wall extracts, protoplasts, and supernatant fractions prepared as described above were resolved on Criterion XT bis-Tris gels (4 to 12%) (Bio-Rad). Coomassie blue or silver-nitrate staining was used to verify that equivalent amounts of protein were loaded among similar samples. Gels were fixed in isopropanol-acetic acid (50:5) and probed for 1 h with rabbit anti-Srr1 pAb (1:1,000) or biotinylated lectin sWGA (Vector Laboratories) (1 μg/ml). After three PBS washes, the gels were probed for 1 h with DyLight 680-conjugated goat-antirabbit (1:10,000) or DyLight 800-conjugated streptavidin (1:10,000) (Pierce) and washed five times in PBS. Gels were scanned on a two-channel Odyssey infrared imaging system (Li-Cor Biosciences), and images were acquired with the Odyssey 2.0 software.
Immunofluorescence imaging.
Cells collected in exponential phase (OD600 = 0.3) were washed three times in PBS and resuspended at 1 U OD600 in PBS. Bacterial suspensions (50 μl) applied on glass coverslips were then incubated for 15 min at 25°C, washed twice with PBS, and fixed for 15 min in 0.5% paraformaldehyde. Cells were blocked for 30 min at 25°C with 5% fetal calf serum in PBS (FCS-PBS) and incubated for 1 h at 25°C with rabbit pAb directed against Srr1 (1:1,000), Alp2 (1:1,000), or glyceraldehyde-3-phosphate dehydrogenase (1/100) in FCS-PBS. After three PBS washes, the coverslips were incubated with Alexa Fluor A488-conjugated goat anti-rabbit immunoglobulin G (IgG) (1:10,000) in FCS-PBS for 1 h at 25°C in the dark. Slides were mounted after three PBS washes in mounting medium containing 4′,6′-diamidino-2-phenylindole (DAPI) to stain DNA. Cells were imaged with a Nikon Eclipse E600 epifluorescence microscope equipped with a mercury lamp and a Nikon DXM1200F charge-coupled device camera.
Trypsin sensitivity assay.
Bacteria harvested in exponential phase (OD600 = 0.3) were washed twice in PBS, resuspended in the same volume of PBS without or with trypsin (0.1 mg/ml), and incubated at 37°C for 5, 15, 30, and 60 min. After two washes in the same volume of PBS, 5-μl aliquots were dotted on nitrocellulose and cell surface proteins were detected using rabbit pAb directed against Srr1 or Gbs0791, used as a control LPXTG protein. After incubation with HRP-coupled antirabbit secondary antibodies (1:10,000) (Zymed), detection was achieved with the Western Pico chemiluminescence kit (Pierce). Images were acquired with the GeneGnome system (Syngen, Frederick, MD) and analyzed and integrated with the Genetools (Syngen) software program.
Lectin binding to bacterial cells.
Cells collected in exponential phase (OD600 = 0.3) were washed three times in PBS and resuspended at 1 U OD600 in PBS. One hundred μl of the bacterial suspension was then transferred in Maxisorp 96-well plates (Nunc; ThermoFisher Scientific) and incubated for 2 h at 25°C. Wells were then washed twice with 200 μl of PBS and incubated for 1 h at 25°C with 200 μl of biotinylated lectin (1 μg/ml in PBS containing 5% bovine serum albumin). After three 200-μl PBS washes, the plates were incubated for 1 h with streptavidin-conjugated horseradish peroxidase (1:10,000) in PBS-bovine serum albumin and washed three times with PBS, and 200 μl of OPD (o-phenylenediamine; 0.4 mg/ml in citrate buffer) was added. Fifteen minutes after OPD addition, 50 μl of 25% H2SO4 was added to stop the enzyme reaction, and the absorbance at 450 nm was measured. The absorbance at 450 nm of wells devoid of biotinylated lectins (control) was subtracted from all values. It is worth mentioning that all bacterial strains tested in this experiment displayed a similar attachment to plastic plates, as measured by crystal violet staining (see Materials and methods). Experiments were performed twice in triplicate, and similar results were obtained. Differences in lectin binding were compared by using an unpaired t test. Crystal violet staining of control wells revealed that all strains studied adhered similarly to the Maxisorp plates. The lectins sWGA (succinylated wheat germ agglutinin), WGA (wheat germ agglutinin), LEL (Lycopersicon esculentum lectin), MalII (Maackia amurensis lectin II), EBL (elderberry bark lectin), PSA (Pisum sativum agglutinin), and UEA (Ulex europaeus agglutinin) were from Vector Laboratories, Inc. (ABCYS, Paris, France).
Cell culture techniques and adherence assays.
The human cell lines A549 (ATCC CCL-185) from an alveolar epithelial carcinoma and TC7 (ATCC CCL-2), a subclone of the human intestinal cell line Caco-2 (11), were cultured in Quantum 286 medium (PAA laboratories). Cells were incubated in 10% CO2 at 37°C and were seeded at a density of 2 × 105 to 5 × 105 cells per well in 24-well tissue culture plates. Monolayers were used after 24 to 48 h of incubation.
Overnight cultures grown in TH broth (OD600 = 2; approximately 109 CFU/ml) were washed once in PBS and resuspended in Dulbecco's modified Eagle medium. Cells were infected at a MOI of 10 bacteria per cell for 1 h at 37°C in 10% CO2. The monolayers were then washed four times with PBS, and the cells were disrupted by the addition of 1 ml sterile deionized ice-cold water and repeated pipetting. Serial dilutions of the lysate were plated on TH agar for counting of viable bacteria. The percent adherence was calculated as follows: (CFU on plate count/CFU in original inoculum) × 100. Assays were performed in duplicate and were repeated at least three times.
Virulence assay.
Virulence of S. agalactiae NEM316 (wild-type [WT] strain), NEM316Δsrr1, and NEM316ΔgtfC-H was tested as described previously (56). Randomized groups of 10 2-day-old Sprague-Dawley rat pups (Janvier, Le Genest Saint Isle, France) were inoculated intraperitoneally with 100 μl of bacterial suspensions containing 5 × 106 CFU in 0.9% NaCl, and mortality was recorded over a 10-day period. The statistical significance (P < 0.05) between groups was evaluated using the chi-square test.
RESULTS
Sequence analysis of the srr1-secA2 locus of S. agalactiae NEM316.
The NEM316 srr1-secA2 locus includes at least 15 genes transcribed in the same direction (srr1 to gtfB) and one divergently transcribed regulator, designated rga (regulation of glycosylated adhesin) (Fig. 1A). The srr1 gene encodes a 130-kDa serine-rich protein and is followed by a palindromic sequence forming a possible stem-loop transcriptional terminator (ΔG = −16.6 kcal/mol). The downstream DNA segment encodes six putative GTs (GtfC to -H), five proteins participating in the SecA2 secretion system (SecA2, Asp1 to -3, and SecY2), two additional GTs (GtfA and -B), an orthologue of the accessory secretion protein Asp4 (Fig. 1A), and the MutT protein. It is worth noting that the genetic organization of the secA2-gtfB region is conserved in other gram-positive cocci, such as S. gordonii, S. pneumoniae, and S. aureus (Fig. 1A). In these bacteria, this region is located at varying distances downstream from a gene encoding a serine-rich LPXTG protein whose secretion is SecA2 dependent (Fig. 1A).
The structure of the srr1-secA2 locus of NEM316 typified that present in most GBS isolates belonging to different serotypes and multilocus sequence types, including the entirely (A909 and 2603V/R) and partially (CJB111, 18RS21, H36B, and 515) sequenced strains (see Fig. S1 in the supplemental material). In the completely sequenced strains, these loci differ by point mutations leading to synthesis of a truncated Rga regulator (A909 and 2603V/R) or by insertion of IS1381 within the glycosyltransferase-encoding gene gtfF (2603V/R) (see Fig. S1 in the supplemental material). The srr2-secA2 locus of strain COH1 (Fig. 1A) typified that present in all ST17 GBS strains (8). The level of sequence identity between the orthologous gene (secY2 to gtfB) of the secA2 locus of NEM316 and COH1 varies from 44% (secY2) to 53% (secA2). Surprisingly, the sequence identity between srr1 and srr2 is below 20%. The srr1-secA2 locus has the same location in the seven non-ST17 GBS genomes, whereas the srr2-secA2 locus is present at a different location in the ST17 strain COH1 (data not shown). This suggests the independent acquisition of either of these xenologue srr-secA2 loci by ancestral GBS clones.
Transcriptional analysis of the srr1-secA2 locus of S. agalactiae NEM316.
The divergently transcribed rga gene located upstream of srr1 in NEM316 encodes a 498-amino-acid (aa)-long polypeptide, Rga, that exhibits significant sequence homology with transcriptional regulators belonging to the Mga family (31), including the RofA regulators, which are restricted to streptococci (24). In particular, Rga contains in its NH2-terminal moiety the two putative helix-turn-helix DNA-binding regions (residues 7 to 65 and 71 to 157) that are invariably found in the corresponding region of members of this response regulator family (31).
Mga/RofA regulators frequently control the expression of adhesins in gram-positive pathogens. We deleted the rga gene in NEM316 and studied by quantitative RT-PCR (qRT-PCR) the expression of four genes (srr1, gtfC, secA2, and gtfB) of the srr1-secA2 locus in NEM316 and NEM316ΔrgA. As shown in Fig. 1B, a more than 100-fold decrease in transcription was measured for srr1 in the mutant, while the expression of gtfC, secA2, and gtfB was not affected. This result indicates that Rga is required for srr1 transcription and that srr1 and the downstream genes do not form an operon. We next examined Srr1 production in cell wall protein extracts by immunodetection using antibodies directed against a histidyl-tagged NH2-terminal Srr1 fragment (residues 105 to 566), i.e., a recombinant protein that does not contain either the signal peptide or the serine-rich carboxylic domain. A single high-molecular-mass protein of approximately 450 kDa was present in WT strain extracts that was not detected in NEM316ΔrgA extracts (Fig. 1C). This result confirms that the synthesis of Srr1 is dependent upon Rga. The apparent high molecular mass of Srr1 after SDS-PAGE, which greatly exceeds the theoretical value of 130 kDa, is probably due to the glycosylation of the protein (see below), as was observed for the protein GspB from S. gordonii (2, 3).
Secretion and cell wall anchoring of Srr1.
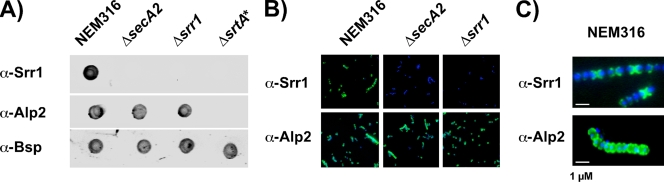
To further characterize the secA2 locus of NEM316, we deleted in-frame the genes coding for SecA2 and Srr1 (Fig. 1A). We also included in this analysis the NEM316ΔsrtA* mutant, which is unable to covalently anchor LPXTG proteins to the cell wall (16, 25) (S. Dramsi, unpublished results). The presence of Srr1 at the surface of NEM316 and mutant derivatives was first studied by immunoblotting whole-cell bacteria spotted on nitrocellulose membranes. This analysis revealed that Srr1 was displayed at the surface of NEM316, whereas no signal was detected with the ΔsecA2, Δsrr1, and ΔsrtA* mutants (Fig. 2A). Srr1 was found to accumulate in the culture supernatant of the ΔsrtA* mutant (see in Fig. 3C). In control experiments, we showed that the cell wall-anchored protein Alp2 was present at the surface of the WT and the ΔsecA2 and Δsrr1 mutants but not at the surface of the ΔsrtA* mutant. As an internal loading control, the surface-associated protein Bsp (Gbs1420) (36) was detected in all strains (Fig. 2A). These results indicate that secretion of Srr1 requires the SecA2 protein and that SrtA covalently attaches this surface protein to the cell wall.
FIG. 2.
Surface display of Srr1 in S. agalactiae NEM316 and isogenic mutant derivatives. (A) Five μl of bacteria cultivated in TH broth at 37°C and collected in stationary phase (OD600 = 1) were spotted onto a nylon membrane and analyzed with rabbit pAb directed against Srr1 (α-Srr1) (1/1,000), Alp2 (α-Alp2) (1/1,000), and Bsp (α-Bsp) (1/100). (B) Bacteria were fixed on cover slides and probed with α-Srr1 (1/1,000) and α-Alp2 (1/1,000). The presence of Srr1 and Alp2 was revealed by immunofluorescence following incubation with Alexafluor 488-conjugated goat anti-rabbit IgG (green), whereas bacterial DNA was stained with DAPI (blue). (C) Immunofluorescence localization of Srr1 and Alp2 at the surface of NEM316 with a magnified view revealing a different localization of the two cell wall-anchored LPXTG proteins.
FIG. 3.
Western blot analysis of the cell wall-anchored LPXTG protein Srr1 extracted from S. agalactiae NEM316 and isogenic mutant derivatives. Proteins were purified from the cell wall by mutanolysin treatment (A), bacterial protoplasts (B), or the culture supernatant (C). Extracts were separated on 3-to-8%-gradient Tris-acetate Criterion XT SDS-PAGE gels, and Srr1 was detected in-gel by immunoblotting with rabbit anti-Srr1 pAb (α-Srr1) or with the biotinylated lectin sWGA (succinylated wheat germ lectin). Ten μg of protein was loaded in each well.
The localization of Srr1 at the bacterial surface was studied by immunofluorescence. Wild-type bacteria probed with anti-Srr1 antiserum displayed a strong signal that was not observed with the ΔsecA2 or Δsrr1 mutant strain (Fig. 2B). As a control, we observed that the cell wall-anchored protein Alp2 was similarly displayed at the surface of the three strains. However, as opposed to Alp2, which was equally distributed along the bacterial surface, Srr1 presented a discrete localization with a preferential accumulation at the old poles of bacterial chains (Fig. 2C). This experiment confirms the surface display of Srr1 and indicates a specific localization pattern for the protein.
Glycosylation and secretion of Srr1.
To determine if glycosylation modulates the export of Srr1, we deleted the six genes gtfC to gtfH (in-frame fusion of the 5′ and 3′ extremities of gtfC and gtfH, respectively) and the genes gtfA and gtfB (in-frame fusion of the 5′ and 3′ extremities of gtfA and gtfB, respectively). Proteins from the cell wall, protoplasts, and supernatants of NEM316 and mutant derivatives were extracted, fractionated on gradient SDS-PAGE gels, and revealed by immunoblotting with anti-Srr1 pAb and with the lectin sWGA, which selectively binds N-acetylglucosamine (GlcNac binder) groups. An in-gel chemiluminescence detection assay was used in this analysis to circumvent problems inherent in the transfer and adsorption onto nitrocellulose of high-molecular-mass and potentially glycosylated proteins. A single high-molecular-mass protein was detected with anti-Srr1 serum in NEM316 and NEM316ΔgtfC-H cell wall extracts (Fig. 3A, lanes 1 and 4), whereas no reactivity was observed in those from the ΔgtfAB, ΔsecA2, and Δsrr1 mutants (Fig. 3A, lanes 2, 3, and 5). The apparent molecular masses of Srr1 in NEM316 (450 kDa) and NEM316ΔgtfC-H (350 to 400 kDa) are different but well above the calculated molecular mass (130 kDa) of this protein. The lectin sWGA, which selectively binds N-acetylglucosamine (GlcNac) groups, reacted with Srr1 bands in NEM316 and NEM316ΔgtfC-H cell wall extracts (Fig. 3A, lanes 1 and 1′ and lanes 4 and 4′). This confirmed that Srr1 was heavily glycosylated in NEM316 and NEM316ΔgtfC-H. The size difference between the lectin-reacting band in NEM316 and NEM316ΔgtfC-H suggests that the corresponding protein is more glycosylated in the WT than in the mutant strain.
Analysis of protoplast proteins revealed an accumulation of Srr1 in the ΔsecA2 mutant that reacted with both the anti-Srr1 pAb and the sWGA lectin (Fig. 3B, lanes 3 and 3′). The molecular mass of this sequestered form of Srr1 was similar to that detected in NEM316 cell wall extracts (Fig. 3A, lanes 1 and 1′). These results indicate that glycosylation takes place in the cytoplasm prior to protein export. Surprisingly, the protoplast extracts of the ΔgtfCDEFGH mutant contained the 350- to 400-kDa form of Srr1, which reacted strongly with the sWGA lectin but not with the anti-Srr1 serum (Fig. 3B, lanes 4 and 4′). Finally, we observed that Srr1 was released in the culture supernatant of the ΔsrtA* mutant but not in that of NEM316, confirming that the protein is a typical cell wall-anchored LPXTG protein (Fig. 3C, lane 6).
Taken together, these results demonstrate that Srr1 is glycosylated by the GtfA and/or -B and GtfCDEFGH enzymes and that its synthesis requires GtfA and/or -B but not GtfC to -H, which were therefore qualified as accessory GTs.
Protection against trypsin digestion.
Glycosylation of cell surface or secreted bacterial proteins can provide a protective mechanism against proteolytic digestion (52). To test this hypothesis, WT and mutant strains were incubated with trypsin and the sensitivity of Srr1 to proteolysis in its natural environment was investigated through quantitative dot blot and microscopy analysis. The protein possesses 40 predicted cleavage sites (100% probability) for trypsin, all located in the amino-terminal part (i.e., the segment produced in E. coli and purified to raise anti-Srr1 pAb). After 5 min of proteolytic treatment, Srr1 detection was more strongly affected in the ΔgtfCDEFGH mutant (65% decrease) than in the WT strain (45% decrease), but in both cases, no further decrease was observed up to 1 h of incubation (Fig. 4A). In contrast, the signal of the LPXTG protein Gbs0791 (33 cleavage sites scattered throughout the 512-aa sequence), which was used as a control, displayed a time-dependent decrease that was similar for the two strains. Microscopy analysis indicated that the amount and localization of Srr1 was the same in WT and mutant strains before treatment (Fig. 4B). Although Srr1 immunoreactivity was affected in both strains after 30 min of trypsin treatment, the protein level at the bacterial surface was more dramatically reduced with the mutant strain, where it was barely detectable, than with the WT strain (Fig. 4B). All together, these results demonstrate that GtfCDEFGH-mediated glycosylation protects Srr1 from proteolysis, an effect likely due to the modification of its NH2-terminal moiety.
FIG. 4.
Trypsin sensitivity assay. (A) Wild-type (▴ and ▵) or ΔgtfC-H mutant (▪ and □) strains cultivated in TH broth at 37°C and collected in exponential phase (OD600 = 0.3) were incubated in the presence of trypsin (0.1 mg/ml) for the indicated periods of time. Bacterial suspensions were spotted onto a nylon membrane and analyzed with rabbit pAb directed against Srr1 (α-Srr1) (1/1,000) (▴ and ▪) or Gbs0791 (▵ and □) used as a control protein (1/1,000). Chemiluminescent signals were acquired and integrated with the GeneGnome system and were normalized as 100% relative to the values measured at time zero. (B) Wild-type or ΔgtfC-H mutant bacteria were trypsinized for 30 min, fixed on cover slides, and probed with α-Srr1 (1/1,000). The surface protein Srr1 (red) was detected by immunofluorescence following incubation with Alexafluor 594-conjugated goat anti-rabbit IgG, whereas bacterial DNA (blue) was stained with DAPI (B).
Secretion and glycosylation of the NH2-terminal half of Srr1.
To study the glycosylation of the NH2-terminal part of Srr1, we investigated the expression of a truncated form of Srr1 devoid of its serine-rich carboxylic moiety in NEM316 and related mutants. The recombinant multicopy shuttle vector used in this analysis, which expressed a truncated protein corresponding to residues 1 to 638 of Srr1 (Fig. 5A), was introduced in NEM316 (WT) and in the ΔsecA2, ΔgtfAB, and ΔgtfCDEFGH mutants. The corresponding culture supernatants were analyzed by Western blotting with anti-Srr1 pAb and the sWGA lectin. In NEM316, two major bands (approximately 78 and 73 kDa) were detected with the anti-Srr1 serum (Fig. 5B, lane 1), whereas a single protein (approximately 71 kDa) was observed in the ΔgtfAB and ΔgtfCDEFGH mutants (Fig. 5B, lanes 2 and 4). No protein band was detected in the ΔsecA2 mutant and in the control strain NEM316 containing the corresponding empty shuttle vector (Fig. 5B, lanes 3 and 5). Interestingly, the sWGA lectin blot analysis revealed that only NEM316 expressed a glycosylated truncated Srr1 that corresponded to the 78-kDa protein (Fig. 5B, lane 1).
FIG. 5.
Secretion of a truncated recombinant Srr1 protein in NEM316 and isogenic mutant derivatives. (A) The structure of the cell wall-anchored LPXTG protein Srr1 is shown, and the relevant features encoded by the fragment cloned in pAT18P are indicated (Sp, signal peptide; Srr, serine-rich repeat). (B) Western blot analysis of the recombinant Srr1 protein in GBS strains. The 5′ half of the srr1 gene was amplified and cloned in pAT18P and the resulting plasmid introduced in NEM316 and the ΔgtfAB, ΔsecA2, and ΔgtfCDEFGH (ΔgtfC-H) mutant derivatives. NEM316 harboring pAT18P was used as a control strain. Culture supernatants of these strains were concentrated 50-fold and the proteins, separated on 3-to-8%-gradient Tris-acetate Criterion XT SDS-PAGE gels, were silver stained or revealed in-gel by immunoblotting with rabbit anti-Srr1 pAb (α-Srr1) or with the biotinylated lectin sWGA (succinylated wheat germ lectin). Ten μg of protein was loaded in each well.
These results indicated that the NH2 half of Srr1 was glycosylated and that this posttranslational modification, which required both GtfAB and GtfC to -H, was not essential for the SecA2-dependent secretion of this truncated protein. The ability of the NH2 half of Srr1 to be expressed and secreted by the ΔgtfAB mutant indicates that glycosylation of the missing segment of Srr1 (i.e., its SAST/M-rich carboxylic moiety) by at least one of the corresponding enzymes is absolutely required for synthesis/stability but not secretion of the full-length protein. This suggests that in the absence of GtfA/B, Srr1 remains unfolded in the cytoplasm and is degraded by intracellular proteases.
The secA2 locus controls GBS surface glycosylation.
Seven lectins were used in a whole-cell binding assay to determine if the pattern of carbohydrates exposed at the surface of the bacteria could be affected in secA2 locus mutants. This analysis revealed that the binding of the lectins WGA (GlcNac binder), sWGA (GlcNac and sialic acid binder), and EBL (Gal-2,6-linked sialic acid binder) to the four mutant strains tested was significantly decreased compared to that of the WT strain (Fig. 6). In contrast, the binding of MalII (α-2,3-linked sialic binder), LEL (N-acetylglucosamine oligomer binder), and lectins specific for mannose (PSA) or fucose (UEA) to the GBS mutants was not affected. These results suggest that GlcNac and sialic acid residues present on Srr1 are exposed at the bacterial surface. This hypothesis is consistent with the sWGA lectin reactivity of Srr1 in bacterial cell wall extracts (Fig. 3). Unfortunately, we were unable to detect EBL binding to Srr1 in our in-gel staining assay, presumably because of the lack of sensitivity of the method.
FIG. 6.
Lectin-binding patterns of S. agalactiae NEM316 and isogenic mutant derivatives. GBS strains were immobilized within the wells of 96-well plates, and the binding of seven biotinylated lectins was quantitated with streptavidin-conjugated HRP. sWGA, succinylated wheat germ lectin; WGA, wheat germ lectin; LEL, Lycopersicon esculentum lectin; MalII, Maackia amurensis lectin II; EBL, elderberry bark lectin; PSA, Pisum sativum lectin; UEA, Ulex europaeus lectin. The experiments were performed twice in triplicate, and similar results were obtained. The mean values of one representative experiment are shown here, and the standard deviation was calculated from the three experimental values of this analysis. Asterisks indicate significant (P < 0.05) binding differences between NEM316 and a mutant strain.
Taken together, these data demonstrate that the secA2 locus of NEM316 plays a role in the glycan decoration of the bacterial surface that might modulate the interactions with mammal host cells.
Srr1 biosynthesis and glycosylation modulate bacterial adherence and virulence.
Phenotypic comparison (morphology, growth rate, hemolysis, capsule biosynthesis, and antibiotic resistance) of NEM316 (WT), NEM316Δsrr1, NEM316ΔsecA2, NEM316ΔgtfAB, and NEM316ΔgtfC-H did not reveal significant differences between strains. Although we repeatedly observed that the four mutant strains present delayed sedimentation after overnight culture, microscopic analysis and biofilm formation were similar for all strains, indicating that Srr1 does not play a major role in association between bacterial cells.
Adherence of GBS to mucosal surfaces (intestine, vagina, and lung) constitutes an essential step in the infectious process. The properties of Srr1 suggest that it may act as an adhesin and therefore could contribute to bacterium-host cell interactions. Therefore, we compared adherence of NEM316 and various secA2 locus mutants to human lung (A549) and intestinal (TC7) epithelial cells at an MOI of 10 bacteria per cell (Table 1). The adherence of the Δsrr1, ΔsecA2, and ΔgtfAB strains, which do not express Srr1 at their surface, was significantly reduced (P < 0.05) compared to that of the parental strain (six-, three-, and fivefold reduction, respectively), and a similar defect was measured for the ΔsrtA* mutant used as an adhesion-deficient control strain (fourfold reduction) (Table 1). For the ΔgtfCDEFGH (ΔgtfC-H) mutant, a less severe but still significant twofold reduction (P < 0.05) in adhesion was measured. A similar adhesion default to intestinal TC7 epithelial cells was observed for the Δsrr1 mutant (Table 1). These results indicate that Srr1 exposure at the bacterial surface directly or indirectly favors adhesion of S. agalactiae to epithelial cells and that these adhesive properties are modulated by the GtfC-H-dependent glycosylation.
TABLE 1.
Capacities of S. agalactiae NEM316 (WT strain) and mutant derivatives to adhere to human lung (A549) and intestinal (TC7) epithelial cell linesa
| Strain | Relative % adherence to cell line:
|
|
|---|---|---|
| A549 | TC7 | |
| NEM316 | 100 ± 24 | 100 ± 22 |
| NEM316Δsrr1 | 17.7 ± 9 | 12.5 ± 7 |
| NEM316ΔsecA2 | 30.2 ± 14 | ND |
| NEM316ΔgtfAB | 21.7 ± 5 | ND |
| NEM316ΔgtfC-H | 51.5 ± 15 | 40.2 ± 12 |
| NEM316ΔsrtA* | 23.8 ± 4 | 20 ± 6 |
Cells were infected at a MOI of 10 bacteria per cell for 2 h at 37°C, and the adherence frequencies were calculated as the number of bacteria remaining attached to the cells after the incubation period with respect to the total number of inoculated bacteria. The level of adherence of the WT strain was set at 100, and the level of adherence of the derivative strains is given relative to that of the WT. Results are presented as mean values (± standard deviations) for at least three experiments performed in triplicate. In these experiments, NEM316ΔsrtA* was used as a control strain deficient for bacterial adherence. ND, not done.
The role of Srr1 in the virulence of NEM316 was evaluated in a neonatal rat sepsis model (18) by comparing the mortality of animals after intraperitoneal injection of 5 × 106 CFU of WT, ΔsecA2, Δsrr1, and ΔgtfCDEFGH strains. The mortality was followed for 10 days with 10 animals for each GBS strain in 2 independent experiments (Fig. 7). Nine out of the ten pups were killed within 2 days by the WT strain, whereas all the animals infected with the ΔsecA2 or Δsrr1 strain survived the infection. A strong reduction of virulence was observed for the ΔgtfCDEFGH mutant, which was more efficient in killing rat pups than the Δsrr1 mutant but produced significantly less mortality (30%) than the WT at the end of the experiment. Taken together, these results indicate that Srr1 acts as an important virulence factor of NEM316 in the neonatal sepsis model and that the activities of the accessory GTs GtfCDEFGH are involved in the bacterial aggressiveness.
FIG. 7.
Mortality curves of neonatal rats infected with S. agalactiae NEM316 (□), ΔsecA2 (•), Δsrr1 (▵), and ΔgtfCDEFGH (○) strains. Rat pups (10 per group) were injected intraperitoneally with 5 × 106 bacteria, and mortality was recorded during 10 days. Differences in mortality rates of animals infected with NEM316 and those infected with one of the mutant strains were statistically significant: NEM316 versus Δsrr1 mutant, P < 0.0001; NEM316 versus ΔgtfCDEFGH mutant, P = 0.0012.
DISCUSSION
In low-GC gram-positive cocci, the secA2 locus is apparently devoted to the secretion of a glycosylated serine-rich cell wall-anchored LPXTG protein encoded by the same locus. In these bacteria, the structure of this locus is conserved, and the simplest prototype version was found in S. aureus COL, where it includes the genes encoding the following: (i) the secreted glycosylated SecA2 substrate SraP, (ii) two GTs (GtfA and GtfB), and (iii) the accessory Sec components (SecA2, SecY2, and Asp1 to -3) (Fig. 1) (44). In streptococci, this basic structure was supplemented with varying numbers of additional glycosyltranferases and accessory secretion protein-encoding genes (Fig. 1), which suggests that substrate glycosylation and secretion may be different depending on the system considered. In particular, the secA2 locus of the GBS strain NEM316 encodes six additional GTs (GtfC to -H), whereas the GBS strain COH1 codes for only two additional GTs, GtfC and GtfH (Fig. 1; see also Fig. S1 in the supplemental material). GBS isolates contain a single secA2 locus type that directs synthesis of either Srr1 (NEM316) or Srr2 (COH1), two unrelated glycosylated surface proteins. These two loci were identified at different genome locations, suggesting their independent acquisition through horizontal transfer.
In S. agalactiae NEM316, the product of the upstream but divergently transcribed rga gene tightly controls expression of srr1 (Fig. 2). Surprisingly, Rga does not control the expression of the genes downstream of srr1, which are dedicated to Srr1 export and posttranslational modification. The presence of a rho-independent transcriptional terminator (ΔG = −16.6 kcal/mol) in the srr1-gtfC intergenic region is consistent with the observation that srr1 is not cotranscribed with the downstream genes. The regulation of Srr-encoding genes in other species has not been studied, but hairpin structures constituting putative transcriptional terminators could be found downstream of srr genes in S. gordonii (ΔG = −19.60 kcal/mol) and S. agalactiae COH1 (ΔG = −29.20 kcal/mol), which suggests that independent transcription of srr is not restricted to S. agalactiae NEM316. In S. pneumoniae, a putative terminator (ΔG = −15.60 kcal/mol) could be found downstream of the first glycosyltransferase-encoding gene located immediately downstream of srr (Fig. 1). Rga displays a domain organization similar to that found in Mga and RofA-like (RALP) transcriptional regulators of S. pyogenes. In group A streptococcus, the Mga and RALP regulatory networks control the infectious process through coordinated and growth-phase-dependent regulations of numerous genes involved in adhesion, immune evasion, bacterial persistence, and metabolic adaptations (23, 24). The genome of NEM316 contains three RALP-encoding genes (rogB [= gbs1479], rga [= gbs1530], and gbs1426) but no mga ortholog. RogB is a transcriptional activator that controls the expression of the P1-2A pilus operon (15, 22), whereas Gbs1426 has not yet been characterized. Interestingly, a recent transcriptome analysis showed that srr1 was upregulated during NEM316 growth in human blood (32), but it remains to be determined if Rga is involved in this activation.
Immunofluorescence, dot blot, and in-gel detection experiments revealed that secretion of Srr1 is SecA2 dependent and that this protein is anchored to the cell wall by the housekeeping sortase SrtA. Immunofluorescence and electronic microscopy analysis (data not shown) showed that Srr1 is preferentially localized at the old pole of cells, an observation reminiscent of the polar localization of the RofA-dependent group A streptococcus adhesin PrtF (10). The NH2-terminal signal sequence of PrtF was shown to be crucial in determining the site of export and hence the localization of the protein. Since Srr1 secretion is SecA2 dependent, this might suggest that this accessory secretion machinery is preferentially located at the old poles, and it would be of obvious interest to perform similar experiments with other species to determine if other SecA2-dependent Srr proteins share this property.
The discrepancy between the theoretical and apparent molecular mass of Srr1, as determined by SDS-PAGE, combined with its ability to bind the lectin sWGA indicate that it is a glycosylated protein. The carboxy-terminal domain of the protein (643 to 1262), made of more than 150 semiconserved repetitions of the motif SAST, is thought to be the main target of O-linked glycosylation (46). As mentioned previously, at least one of the two GT-encoding genes gtfA and gtfB, which are conserved among streptococci and staphylococci, is essential for the production and/or stability of Srr1. In the ΔgtfAB mutant, Srr1 was not detected in subcellular fractions (Fig. 2A and data not shown). A similar observation was made for S. gordonii, where the presence of GtfA and GtfB is required for the production of the mature form of GspB (49). In contrast, the deletion of the orthologous genes (gtf1 and gtf2, respectively) in S. parasanguis was associated with the production of a nonglycosylated form of the Srr protein Fap1 (55). These results indicate that the level of glycosylation of Srr proteins has a variable impact on their production depending on the species considered.
The NEM316 secA2 region displays six additional GTs or GT-associated encoding genes whose genetic organization is striking compared to that of other streptococcal loci. The three proteins GtfE, GtfF, and GtfG are similar to each other (32 to 38% of identity, with sizes varying between 394 and 430 aa residues), and they all belong to the family 8 of GT (GT-8), following the classification proposed by P. M. Coutinho in 2003 (http://www.cazy.org/). In S. gordonii, the Gly protein (682 aa), involved in GspB modification, possesses a GT-8 domain in its amino-terminal part, while its carboxy-terminal part is homologous to GtfC (55% identity), the first protein encoded by the GBS NEM316 SecA2 region that does not possess any known GT domain. We thus may hypothesize that GtfC is required for the proper functioning of one or several GTs. The two remaining GTs found in this region were GtfD (291 residues), which contains a GT-2 domain, and GtfH, which displays significant similarities with the galactofuranose transferase WefE of Streptococcus oralis (57). Deletion of the gtfCDEFGH region in NEM316 leads to a decrease of the apparent molecular mass of Srr1 after electrophoresis (Fig. 2A) but allows the export of Srr1 to the cell wall at the same polar location observed for the WT strain (Fig. 2A and 4B). These results indicate that although this set of GTs is involved in Srr1 glycosylation, it is not strictly necessary for the stability, export, and/or localization of the protein. Furthermore, the protein produced by the mutant NEM316ΔgtfC-H has retained its ability to bind the lectin sWGA (Fig. 2B), strongly suggesting that the remaining GTs, GtfA and/or GtfB, are involved in the attachment of GlcNac to Srr1. This hypothesis is consistent with the recent demonstration that transfer of the GlcNaC group to the Srr protein Fap1 of S. parasanguinis constitutes an early step in the glycosylation process (9). Interestingly, a significant fraction of Srr1 accumulates in the cytoplasm of the mutant NEM316ΔgtfC-H; this cytoplasmic form strongly reacts with the lectin sWGA, while it is not recognized by the Srr1 pAb (Fig. 2D). It is conceivable that in the absence of GtfC to -H, Srr1 is hyperglycosylated by GtfAB, which leads to a loss of epitopes and concomitantly interferes with its secretion. In S. gordonii, the deletion of the accessory GTs Gly and nss does not affect the migration of GspB but strongly decreases its export (49). Again, this indicates that notable differences exist in Srr biosynthesis among low-GC gram-positive cocci. Last, our results indicate that the Srr1 protein anchored in the cell wall of the ΔgtfC-H mutant is more prone to proteolytic digestion than its counterpart produced in the WT strain.
Analysis of the secretion and modification of the NH2-terminal domain of Srr1 shows that SecA2 is necessary for its export. We also observed that this segment is glycosylated when produced in the WT strain, and it is likely that the 70-aa serine-rich region is targeted by these modifications. In both the ΔgtfAB and ΔgtfCDEFGH strains, only one form of the protein was exported that was not responsive to the lectin. This result indicates that the two sets of GTs are required for the glycosylation of the NH2-terminal domain, suggesting that the corresponding enzymes could cooperate to elongate the glycan chain on this part of the protein.
A critical determinant of GBS virulence is the surface capsular polysaccharide, which invariably contains terminal sialic acids. The sialylation of capsular polysaccharide is considered an important factor enabling immune evasion through molecular mimicry (30, 54). A whole-cell lectin-binding assay shows that mutants deficient in the production, export, or full glycosylation of Srr1 display a similar default in the binding of GlcNaC-specific (sWGA and WGA) or sialic acid-specific (EBL) lectins. This result demonstrates that Srr1 is associated with sialic acid presentation at the bacterial surface and hence can contribute to evasion from host defense.
GspB and Fap1, produced by S. gordonii and S. parasanguis, respectively, and Srap, the Srr protein of S. aureus, are involved in the binding of the corresponding bacteria to platelets (4, 35, 45). GspB and Hsa are two synonymous Srr proteins encoded by the S. gordonii strains M99 and Challis, respectively, that directly bind the terminal sialic acid residues of GPIbα, the most abundant sialylated protein expressed at the surface of platelets (47, 48). In a recent study, Srr1 of S. agalactiae strain 6313 was shown to be involved in adhesion to the epithelial HEp-2 cell line originating from a larynx carcinoma. Human keratin 4 interacts with a recombinant Srr1 protein produced in E. coli and was therefore proposed to be a receptor mediating Srr1-dependent adhesion of GBS to HEp-2 (41). In S. pneumoniae TIGR4, the absence of PsrP was associated with a decrease in the bacterial adhesion to human pulmonary A549 and mouse bronchial LA-4 cell lines, respectively (38). We report here that adhesion of NEM316Δsrr1 to the human pulmonary A549 and intestinal TC7 epithelial cell lines is significantly decreased. The reduced adherence of the Δsrr1, ΔsecA2, and ΔgtfAB mutants, which do not display Srr1 at their surface, strongly suggests that this protein is an adhesin, but further biochemical evidences are needed to demonstrate this hypothesis. In the ΔgtfCDEFGH mutant, the adhesion defect was less pronounced but remained significant, indicating that glycosylation modulates the bacterium-host cell interactions either directly or indirectly by affecting the protein conformation or stability. Taken together, these studies indicate that unrelated Srr proteins contribute to bacterial adhesion to various eukaryotic cell lines. Their polymorphisms related to their primary structures and the nature or extent of glycosylation are likely important in determining their adhesive properties.
A major conclusion of this work is that Srr1 is important for the virulence of S. agalactiae NEM316, since the mortality rate of neonatal rats injected intraperitoneally with NEM316Δsrr1 was significantly lower than that for the WT strain. Interestingly, we observed that the ΔgtfCDEFGH mutant also displays a significant decrease in virulence, though less pronounced than that of the Δsrr1 mutant, and this observation highlights the role of these accessory GTs for GBS adaptation in vivo. The fact that the accessory glycosylation of Srr1 makes it more resistant to proteolysis could partially account for the virulence default of the ΔgtfCDEFGH mutant. In this animal model, the escape of the bacteria from the innate immune defenses in the peritoneal cavity is essential for the development of the infectious process. This suggests that Srr1, besides its cell adhesion capacity, could exert other functions in vivo. Further work is in progress to define more accurately the step in the infectious process where Srr1 is engaged.
Supplementary Material
Acknowledgments
M.-Y. Mistou is member of the scientific staff of INRA-Institut National de la Recherche Agronomique-France, and the support of Emmanuelle Maguin and Claude Gaillardin is acknowledged. We thank Christine Longin (INRA, Plate-forme de microscopie électronique, Jouy-en-Josas) for electron microscopy experiments and B. Kreikemeyer and A. Podbielski for the gift of pAT18P. We thank Mathieu Brochet and Sarah Dubrac for critical reading of the manuscript and helpful suggestions.
This work was supported by the Pasteur Institute (GPH no. 09) and by grants ANR-06-PATHO-001-01 from the Agence Nationale de la Recherche ERA-NET PathoGenoMics and LSHB-CT-2005-512061 from the Network of Excellence Europathogenomics.
Footnotes
Published ahead of print on 24 April 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Archambaud, C., M. A. Nahori, J. Pizarro-Cerda, P. Cossart, and O. Dussurget. 2006. Control of Listeria superoxide dismutase by phosphorylation. J. Biol. Chem. 28131812-31822. [DOI] [PubMed] [Google Scholar]
- 2.Bensing, B. A., B. W. Gibson, and P. M. Sullam. 2004. The Streptococcus gordonii platelet binding protein GspB undergoes glycosylation independently of export. J. Bacteriol. 186638-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bensing, B. A., J. A. Lopez, and P. M. Sullam. 2004. The Streptococcus gordonii surface proteins GspB and Hsa mediate binding to sialylated carbohydrate epitopes on the platelet membrane glycoprotein Ibα. Infect. Immun. 726528-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bensing, B. A., and P. M. Sullam. 2002. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol. Microbiol. 441081-1094. [DOI] [PubMed] [Google Scholar]
- 5.Bensing, B. A., D. Takamatsu, and P. M. Sullam. 2005. Determinants of the streptococcal surface glycoprotein GspB that facilitate export by the accessory Sec system. Mol. Microbiol. 581468-1481. [DOI] [PubMed] [Google Scholar]
- 6.Braunstein, M., A. M. Brown, S. Kurtz, and W. R. Jacobs, Jr. 2001. Two nonredundant SecA homologues function in mycobacteria. J. Bacteriol. 1836979-6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braunstein, M., B. J. Espinosa, J. Chan, J. T. Belisle, and W. R. Jacobs, Jr. 2003. SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 48453-464. [DOI] [PubMed] [Google Scholar]
- 8.Brochet, M., E. Couve, M. Zouine, T. Vallaeys, C. Rusniok, M. C. Lamy, C. Buchrieser, P. Trieu-Cuot, F. Kunst, C. Poyart, and P. Glaser. 2006. Genomic diversity and evolution within the species Streptococcus agalactiae. Microbes Infect. 81227-1243. [DOI] [PubMed] [Google Scholar]
- 9.Bu, S., Y. Li, M. Zhou, P. Azadin, M. Zeng, P. Fives-Taylor, and H. Wu. 2008. Interaction between two putative glycosyltransferases is required for glycosylation of a serine-rich streptococcal adhesin. J. Bacteriol. 1901256-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlsson, F., M. Stalhammar-Carlemalm, K. Flardh, C. Sandin, E. Carlemalm, and G. Lindahl. 2006. Signal sequence directs localized secretion of bacterial surface proteins. Nature 442943-946. [DOI] [PubMed] [Google Scholar]
- 11.Chantret, I., A. Rodolosse, A. Barbat, E. Dussaulx, E. Brot-Laroche, A. Zweibaum, and M. Rousset. 1994. Differential expression of sucrase-isomaltase in clones isolated from early and late passages of the cell line Caco-2: evidence for glucose-dependent negative regulation. J. Cell Sci. 107213-225. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Q., H. Wu, and P. M. Fives-Taylor. 2004. Investigating the role of secA2 in secretion and glycosylation of a fimbrial adhesin in Streptococcus parasanguis FW213. Mol. Microbiol. 53843-856. [DOI] [PubMed] [Google Scholar]
- 13.Doran, K. S., and V. Nizet. 2004. Molecular pathogenesis of neonatal group B streptococcal infection: no longer in its infancy. Mol. Microbiol. 5423-31. [DOI] [PubMed] [Google Scholar]
- 14.Dramsi, S., F. Bourdichon, D. Cabanes, M. Lecuit, H. Fsihi, and P. Cossart. 2004. FbpA, a novel multifunctional Listeria monocytogenes virulence factor. Mol. Microbiol. 53639-649. [DOI] [PubMed] [Google Scholar]
- 15.Dramsi, S., E. Caliot, I. Bonne, S. Guadagnini, M. C. Prevost, M. Kojadinovic, L. Lalioui, C. Poyart, and P. Trieu-Cuot. 2006. Assembly and role of pili in group B streptococci. Mol. Microbiol. 601401-1413. [DOI] [PubMed] [Google Scholar]
- 16.Dramsi, S., P. Trieu-Cuot, and H. Bierne. 2005. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res. Microbiol. 156289-297. [DOI] [PubMed] [Google Scholar]
- 17.Dubrac, S., I. G. Boneca, O. Poupel, and T. Msadek. 2007. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 1898257-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forquin, M. P., A. Tazi, M. Rosa-Fraile, C. Poyart, P. Trieu-Cuot, and S. Dramsi. 2007. The putative glycosyltransferase-encoding gene cylJ and the group B Streptococcus (GBS)-specific gene cylK modulate hemolysin production and virulence of GBS. Infect. Immun. 752063-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibbons, H. S., F. Wolschendorf, M. Abshire, M. Niederweis, and M. Braunstein. 2007. Identification of two Mycobacterium smegmatis lipoproteins exported by a SecA2-dependent pathway. J. Bacteriol. 1895090-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson, R. L., M. K. Lee, C. Soderland, E. Y. Chi, and C. E. Rubens. 1993. Group B streptococci invade endothelial cells: type III capsular polysaccharide attenuates invasion. Infect. Immun. 61478-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 451499-1513. [DOI] [PubMed] [Google Scholar]
- 22.Gutekunst, H., B. J. Eikmanns, and D. J. Reinscheid. 2003. Analysis of RogB-controlled virulence mechanisms and gene repression in Streptococcus agalactiae. Infect. Immun. 715056-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hondorp, E. R., and K. S. McIver. 2007. The Mga virulence regulon: infection where the grass is greener. Mol. Microbiol. 661056-1065. [DOI] [PubMed] [Google Scholar]
- 24.Kreikemeyer, B., K. S. McIver, and A. Podbielski. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11224-232. [DOI] [PubMed] [Google Scholar]
- 25.Lalioui, L., E. Pellegrini, S. Dramsi, M. Baptista, N. Bourgeois, F. Doucet-Populaire, C. Rusniok, M. Zouine, P. Glaser, F. Kunst, C. Poyart, and P. Trieu-Cuot. 2005. The SrtA sortase of Streptococcus agalactiae is required for cell wall anchoring of proteins containing the LPXTG motif, for adhesion to epithelial cells, and for colonization of the mouse intestine. Infect. Immun. 733342-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamy, M. C., M. Zouine, J. Fert, M. Vergassola, E. Couve, E. Pellegrini, P. Glaser, F. Kunst, T. Msadek, P. Trieu-Cuot, and C. Poyart. 2004. CovS/CovR of group B streptococcus: a two-component global regulatory system involved in virulence. Mol. Microbiol. 541250-1268. [DOI] [PubMed] [Google Scholar]
- 27.Lee, V. T., and O. Schneewind. 2001. Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 151725-1752. [DOI] [PubMed] [Google Scholar]
- 28.Lenz, L. L., and D. A. Portnoy. 2002. Identification of a second Listeria secA gene associated with protein secretion and the rough phenotype. Mol. Microbiol. 451043-1056. [DOI] [PubMed] [Google Scholar]
- 29.Lindahl, G., M. Stalhammar-Carlemalm, and T. Areschoug. 2005. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin. Microbiol. Rev. 18102-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marques, M. B., D. L. Kasper, M. K. Pangburn, and M. R. Wessels. 1992. Prevention of C3 deposition by capsular polysaccharide is a virulence mechanism of type III group B streptococci. Infect. Immun. 603986-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McIver, K. S., and R. L. Myles. 2002. Two DNA-binding domains of Mga are required for virulence gene activation in the group A streptococcus. Mol. Microbiol. 431591-1601. [DOI] [PubMed] [Google Scholar]
- 32.Mereghetti, L., I. Sitkiewicz, N. M. Green, and J. M. Musser. 2008. Extensive adaptive changes occur in the transcriptome of Streptococcus agalactiae (group B streptococcus) in response to incubation with human blood. PLoS ONE 3e3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakata, M., T. Köller, K. Moritz, D. Ribardo, L. Jonas, K. S. McIver, T. Sumitomo, Y. Terao, S. Kawabata, A. Podbielski, and B. Kreikemeyer. 2009. Mode of expression and functional characterization of FCT-3 pilus region encoded proteins in the Streptococcus pyogenes serotype M49 Masanobu. Infect. Immun. 7732-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papanikou, E., S. Karamanou, and A. Economou. 2007. Bacterial protein secretion through the translocase nanomachine. Nat. Rev. Microbiol. 5839-851. [DOI] [PubMed] [Google Scholar]
- 35.Plummer, C., H. Wu, S. W. Kerrigan, G. Meade, D. Cox, and C. W. Ian Douglas. 2005. A serine-rich glycoprotein of Streptococcus sanguis mediates adhesion to platelets via GPIb. Br. J. Haematol. 129101-109. [DOI] [PubMed] [Google Scholar]
- 36.Reinscheid, D. J., C. Stosser, K. Ehlert, R. W. Jack, K. Moller, B. J. Eikmanns, and G. S. Chhatwal. 2002. Influence of proteins Bsp and FemH on cell shape and peptidoglycan composition in group B streptococcus. Microbiology 1483245-3254. [DOI] [PubMed] [Google Scholar]
- 37.Rigel, N. W., and M. Braunstein. 2008. A new twist on an old pathway—accessory secretion systems. Mol. Microbiol. 69291-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rose, L., P. Shivshankar, E. Hinojosa, A. Rodriguez, C. J. Sanchez, and C. J. Orihuela. 2008. Antibodies against PsrP, a novel Streptococcus pneumoniae adhesin, block adhesion and protect mice against pneumococcal challenge. J. Infect. Dis. 198375-383. [DOI] [PubMed] [Google Scholar]
- 39.Rubens, C. E., S. Smith, M. Hulse, E. Y. Chi, and G. van Belle. 1992. Respiratory epithelial cell invasion by group B streptococci. Infect. Immun. 605157-5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Samen, U., B. J. Eikmanns, D. J. Reinscheid, and F. Borges. 2007. The surface protein Srr-1 of Streptococcus agalactiae binds human keratin 4 and promotes adherence to epithelial HEp-2 cells. Infect. Immun. 755405-5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmittgen, T. D. 2001. Real-time quantitative PCR. Methods 25383-385. [DOI] [PubMed] [Google Scholar]
- 43.Seifert, K. N., E. E. Adderson, A. A. Whiting, J. F. Bohnsack, P. J. Crowley, and L. J. Brady. 2006. A unique serine-rich repeat protein (Srr-2) and novel surface antigen (epsilon) associated with a virulent lineage of serotype III Streptococcus agalactiae. Microbiology 1521029-1040. [DOI] [PubMed] [Google Scholar]
- 44.Siboo, I. R., D. O. Chaffin, C. E. Rubens, and P. M. Sullam. 2008. Characterization of the accessory Sec system of Staphylococcus aureus. J. Bacteriol. 1906188-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siboo, I. R., H. F. Chambers, and P. M. Sullam. 2005. Role of SraP, a Serine-rich surface protein of Staphylococcus aureus, in binding to human platelets. Infect. Immun. 732273-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stephenson, A. E., H. Wu, J. Novak, M. Tomana, K. Mintz, and P. Fives-Taylor. 2002. The Fap1 fimbrial adhesin is a glycoprotein: antibodies specific for the glycan moiety block the adhesion of Streptococcus parasanguis in an in vitro tooth model. Mol. Microbiol. 43147-157. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi, Y., A. Yajima, J. O. Cisar, and K. Konishi. 2004. Functional analysis of the Streptococcus gordonii DL1 sialic acid-binding adhesin and its essential role in bacterial binding to platelets. Infect. Immun. 723876-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takamatsu, D., B. A. Bensing, H. Cheng, G. A. Jarvis, I. R. Siboo, J. A. Lopez, J. M. Griffiss, and P. M. Sullam. 2005. Binding of the Streptococcus gordonii surface glycoproteins GspB and Hsa to specific carbohydrate structures on platelet membrane glycoprotein Ibalpha. Mol. Microbiol. 58380-392. [DOI] [PubMed] [Google Scholar]
- 49.Takamatsu, D., B. A. Bensing, and P. M. Sullam. 2004. Genes in the accessory sec locus of Streptococcus gordonii have three functionally distinct effects on the expression of the platelet-binding protein GspB. Mol. Microbiol. 52189-203. [DOI] [PubMed] [Google Scholar]
- 50.Takamatsu, D., B. A. Bensing, and P. M. Sullam. 2005. Two additional components of the accessory Sec system mediating export of the Streptococcus gordonii platelet-binding protein GspB. J. Bacteriol. 1873878-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1991. Shuttle vectors containing a multiple cloning site and a lacZ alpha gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene 10299-104. [DOI] [PubMed] [Google Scholar]
- 52.Upreti, R. K., M. Kumar, and V. Shankar. 2003. Bacterial glycoproteins: functions, biosynthesis and applications. Proteomics 3363-379. [DOI] [PubMed] [Google Scholar]
- 53.van Wely, K. H., J. Swaving, R. Freudl, and A. J. Driessen. 2001. Translocation of proteins across the cell envelope of Gram-positive bacteria. FEMS Microbiol. Rev. 25437-454. [DOI] [PubMed] [Google Scholar]
- 54.Wessels, M. R., C. E. Rubens, V. J. Benedi, and D. L. Kasper. 1989. Definition of a bacterial virulence factor: sialylation of the group B streptococcal capsule. Proc. Natl. Acad. Sci. USA 868983-8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, H., S. Bu, P. Newell, Q. Chen, and P. Fives-Taylor. 2007. Two gene determinants are differentially involved in the biogenesis of Fap1 precursors in Streptococcus parasanguis. J. Bacteriol. 1891390-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamamoto, Y., C. Poyart, P. Trieu-Cuot, G. Lamberet, A. Gruss, and P. Gaudu. 2005. Respiration metabolism of group B Streptococcus is activated by environmental haem and quinone and contributes to virulence. Mol. Microbiol. 56525-534. [DOI] [PubMed] [Google Scholar]
- 57.Yoshida, Y., J. Yang, P. E. Peaker, H. Kato, C. A. Bush, and J. O. Cisar. 2008. Molecular and antigenic characterization of a Streptococcus oralis coaggregation receptor polysaccharide by carbohydrate engineering in Streptococcus gordonii. J. Biol. Chem. 28312654-12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.