Abstract
The function of the Staphylococcus aureus eukaryotic-like serine/threonine protein kinase PknB was investigated by performing transcriptome analysis using DNA microarray technology and biochemical assays. The transcriptional profile revealed a strong regulatory impact of PknB on the expression of genes encoding proteins which are involved in purine and pyrimidine biosynthesis, cell wall metabolism, autolysis, and glutamine synthesis. Functional activity of overexpressed and purified PknB kinase was demonstrated using the myelin basic protein as a surrogate substrate. Phosphorylation occurred in a time-dependent manner with Mn2+ as a preferred cofactor. Furthermore, biochemical characterization revealed regulation of adenylosuccinate synthase (PurA) activity by phosphorylation. Phosphorylated PurA showed a 1.8-fold decrease in enzymatic activity compared to unphosphorylated PurA. Loss of PknB led to formation of larger cell clusters, and a pknB deletion strain showed 32-fold-higher sensitivity to the cell wall-active antibiotic tunicamycin. The results of this study strongly indicate that PknB has a role in regulation of purine biosynthesis, autolysis, and central metabolic processes in S. aureus.
The phosphorylation of proteins is a key regulatory mechanism in the signal transduction pathways of both prokaryotes and eukaryotes. Typically, extracellular signals are translated into cellular responses. The phosphorylation of proteins is carried out by specific protein kinases and is coupled to dephosphorylation reactions catalyzed by protein phosphatases. In prokaryotes sensing of extracellular signals and transduction of information are usually mediated by two-component signal transduction systems consisting of histidine kinase sensors and their associated response regulators (42). In contrast, signal transduction in eukaryotes occurs via phosphorylation of serine, threonine, and tyrosine residues. Serine/threonine and tyrosine kinases and phosphatases control reversible phosphorylation of target proteins in eukaryotes and are essential for cell cycle control and differentiation (17, 19).
It has recently been shown in a number of studies that eukaryotic-type serine/threonine protein kinases (STPKs) and phosphatases are also expressed in many prokaryotes (2). Prokaryotic STPKs regulate various cellular functions, such as stress responses, biofilm formation, sporulation, and metabolic and developmental processes (20, 23, 30, 34, 37, 39, 46). STPKs also play a role in the virulence of many bacterial pathogens, such as streptococci, Mycobacterium tuberculosis, Yersinia pseudotuberculosis, and Pseudomonas aeruginosa (11, 16, 21, 36, 47). Although the functional roles of protein kinases have been described in previous studies, only a small number of target substrates have been identified so far. Moreover, the impact of phosphorylation and dephosphorylation of target protein functions has been investigated in only some cases (33, 38).
A single STPK has been found to be conserved in all sequenced strains of Staphylococcus aureus. Originally, this kinase was identified by using a transposon mutagenesis approach used to identify factors that modulate methicillin (meticillin) resistance in methicillin-resistant S. aureus (MRSA) (13). Recent work has demonstrated the functional kinase activity of PknB and has identified potential substrates. Most of the identified substrates of PknB are proteins which are involved in the central metabolism of bacteria, such as trigger factor, DnaK, enolase, pyruvate dehydrogenase, and the regulator MgrA (27, 44). These observations suggest a broad regulatory role for PknB in S. aureus. Interestingly, some of the MRSA strains in the database encode a second protein with a eukaryote-like tyrosine kinase domain located on the SCCmec element. In contrast to PknB, this putative protein kinase does not contain PASTA (penicillin-binding and Ser/Thr kinase-associated) domains. At present, it is not known if the second eukaryote-like tyrosine kinase is functionally active in S. aureus.
In this study, we functionally characterized PknB (SA1063) of S. aureus by constructing a pknB deletion mutant. To explore the role of PknB in gene expression, we studied expression of genes on a global scale by using comparative DNA microarray hybridization. We report here that pknB deletion affects the expression of genes belonging to specific regulons which are involved in central metabolic functions, including purine and pyrimidine biosynthesis, cell wall metabolism, and the citrate cycle. Furthermore, we show that purified PknB kinase phosphorylates myelin basic protein (MBP), which has been used as a surrogate substrate, in a time-dependent manner and has a preference for Mn2+ as a cofactor. In addition, we demonstrated that PknB specifically phosphorylates adenylosuccinate synthase PurA, a key enzyme in purine biosynthesis.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Escherichia coli and S. aureus were grown in Luria-Bertani (LB) medium. Liquid cultures were shaken at 220 rpm. The bacteria were grown at 37°C, unless indicated otherwise. Antibiotics were used at the following concentrations: 100 μg of ampicillin ml−1 and 10 μg of chloramphenicol ml−1 for E. coli and 10 μg of erythromycin ml−1 and 10 μg of chloramphenicol ml−1 for S. aureus.
TABLE 1.
Bacterial strains, plasmids, and PCR primers used in this study
| Strain, plasmid, or primer | Relevant genotype or phenotype or sequence | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F−endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1(A) Δ(argF-lac)U169 φ80dlacZΔM15 | BRL |
| BL21(DE3) | F−ompT hsdSB (rS− mS−) gal dcm (DE3) | Novagen |
| S. aureus strains | ||
| 8325 | NCTC 8325 (wild type, 11-bp deletion in rsbU) | Laboratory stock |
| RN4220 | NCTC 8325-4-r (restriction mutant, 11-bp deletion in rsbU) | Laboratory stock |
| 8325ΔpknB | pknB deletion in strain 8325 | This study |
| 8325ΔpknB/pRB473pknB | 8325 containing pRB473pknB for ΔpknB complementation | This study |
| COLΔpknB | pknB deletion strain COL | This study |
| Plasmids | ||
| pBT2 | Shuttle vector, Apr in E. coli, Cmr in S. aureus | 6 |
| pBT2ΔpknB | Deletion vector for pknB, ermB fragment flanked by fragments upstream and downstream of pknB in pBT2, Emr and Cmr in S. aureus | This study |
| pGEM-T | Apr | Promega |
| pEC1 | Apr EmrermB fragment in pUC18 | 6 |
| pRB473 | Shuttle vector, Apr Cmr | 6 |
| pRB473pknB | pRB473 containing pknB fragment for ΔpknB complementation | This study |
| pKG31 | Apr Emr EcoRI-PstI fragment of ermB in pGEM-T | This study |
| pET-28a(+) | His6 expression vector, Kanr | Novagen |
| pET-28aPknB | Encodes His6-PknB, cloned in pET28a(+), Kanr | This study |
| pET28aPurA | Encodes His6-PurA, cloned in pET28a(+), Kanr | This study |
| Primers | ||
| pknBEcoRI | 5′-CGGAATTCTATCACCTTCAATAGCCGCG-3′ | |
| pknBHindIII | 5′-CCCAAGCTTGTTTGGTGGTGTGAATGACC-3′ | |
| pknBBamHI | 5′-CGGGATCCAATAAATTCAGTCTCATAGCC-3′ | |
| pknBPstI | 5′-AACTGCAGAGTGACGATATTGATGAGGG-3′ | |
| ermBEcoRI | 5′-CGGAATTCGGTGACATCTCTCTATTGTG-3′ | |
| ermBPstI | 5′-AACTGCAGGGAAGCTGTCAGTAGTATACC-3′ | |
| SA1063-fw | 5′-AGCCATATGATAGGTAAAATAATAAATGAACG-3′ | |
| SA1063-rev | 5′-CCGCTCGAGTTATACATCATCATAGCTGACTTC-3′ | |
| SA0016-fw | 5′-GCTAGCTCATCAATCGTAGTAGTTGGG-3′ | |
| SA0016-rev | 5′-CGCCCTAGGCTACCACAATTCTTTTAATAGG-3′ |
Antibiotic susceptibility tests.
MICs were determined by microdilution according to the recommendations of the Clinical and Laboratory Standards Institute (9). The MICs were determined in 96-well microtiter plates using a final volume of 200 μl without agitation. The initial inoculum was 2 × 105 bacterial cells per well. The plates were incubated for 18 h at 37°C.
Construction of the S. aureus pknB deletion strain.
A ΔpknB mutant of S. aureus was constructed by replacing the coding sequence of the pknB gene with the coding sequence of the erythromycin resistance cassette (ermB) by a double-crossover event, as described by Brückner (6). Fragments upstream and downstream of the target gene were amplified by PCR, and restriction sites were added to the primers to facilitate cloning. All primer sequences are listed in Table 1. For the upstream fragment EcoRI and HindIII restriction sites were used (pknBEcoRI and pknBHindIII). The length of the fragment was 1,006 bp, and the fragment started 5 nucleotides before the start codon of pknB. For the downstream fragment BamHI and PstI restriction sites were used (pknBBamHI and pknBPstI). The length of the fragment was 1,018 bp, and the fragment contained 363 bp of the 3′ end of the open reading frame (ORF) of pknB. The erythromycin resistance cassette (ermB) was cloned between the EcoRI and PstI restriction sites. The ermB gene was amplified from the pEC1 vector (5), and EcoRI and PstI restriction sites were added to the primers (ermBEcoRI and ermBPstI). The length of the fragment was 1,364 bp. The fragments were first cloned into the pGEM-T vector (Promega, Mannheim, Germany) and then cut out with the corresponding restriction enzymes and cloned into the temperature-sensitive shuttle vector pBT2. Construction of this deletion vector was carried out using E. coli DH5α. The vector construct was introduced into S. aureus strain RN4220 by electroporation. Following propagation in RN4220, the vector was introduced into S. aureus strain 8325 by transduction with phage φ85. In this strain gene inactivation was carried out as described by Brückner (6). To rule out the possibility that the expression of downstream genes is affected by insertion of ermB and the possibility that the effects on the transcriptome are indeed caused by pknB and not by adjacent genes, we tested expression of downstream genes (SA1064, SA1065, and SA1066) by performing a reverse transcription (RT)-PCR analysis. We did not observe any difference in the expression rates of these genes between the wild type and the mutant. Likewise, in the microarray experiments these genes were not deregulated in the mutant compared to the wild type.
Expression and purification of recombinant proteins for overexpression in pET28a.
The gene fragments corresponding to the entire coding sequence of purA (SA0016) and pknB (SA1063) were synthesized by PCR amplification using genomic DNA of S. aureus 8325 as the template and primers listed in Table 1. Each DNA fragment synthesized was restricted with appropriate enzymes and ligated into the pET28a vector (Novagen, Madison, WI). The resulting plasmids were transformed into E. coli BL21(DE3) cells for protein expression. The resulting recombinant polyhistidine-tagged proteins were purified under native conditions by affinity chromatography on Protino Ni-TED columns by following the manufacturer's instructions (Macherey-Nagel, Düren, Germany) exactly. To exclude the possibility that His tagging resulted in nonspecific in vitro phosphorylation on serine and threonine residues, thrombin cleavage of the His tag of PknB and PurA was performed according to the manufacturer's instructions (Qiagen, Hilden, Germany). Protein purity was checked by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and protein concentrations were determined using the Roti-Nanoquant assay (Roth, Karslruhe, Germany).
RNA techniques.
Total RNA was isolated from S. aureus cultures in exponential growth phase (optical density at 600 nm [OD600], 1.0). Bacteria were harvested by addition of RNA Protect (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The cells were centrifuged for 10 min at 5,000 × g, and the bacterial pellet was resuspended in 1 ml RLT buffer and mechanically disrupted with glass beads in a Fast Prep shaker (Qbiogene, Heidelberg, Germany) two times for 45 s at a speed of 6.5. The cell lysate was centrifuged for 30 s at 13,000 × g, and the supernatant was used for RNA isolation. RNA was isolated using an RNeasy mini kit (Qiagen) according to the standard Qiagen RNeasy protocol. The isolated RNA was treated with RNase-free DNase I (Roche, Penzberg, Germany) to remove the DNA template. The integrity of RNA was monitored by analysis with a bioanalyzer (Agilent Technologies). Only probes with an RNA integrity number of >9 were used in subsequent experiments.
Real-time PCR.
Real-time PCR was performed by using the MyiQ device (Bio-Rad, Munich, Germany). RT was carried out using 2 μg RNA, 200 ng of a random hexamer primer mixture (Amersham, Freiburg, Germany), and Superscript III reverse transcriptase (Invitrogen, Karlsruhe, Germany) at 50°C for 1 h. The reaction was stopped by incubating the mixture at 70°C for 15 min. The cDNA was amplified in different PCRs (including negative controls) with primers specific for the corresponding genes. The iQ SYBR green supermixture (Bio-Rad) was used for the amplification reaction. Relative quantification was performed as described by Pfaffl (35).
DNA microarray analysis.
S. aureus N315 full-genome microarrays containing PCR products of 2,666 genes were used for microarray analysis (Scienion, Berlin, Germany). Each slide contained 6,912 features corresponding to duplicate copies of each ORF and several controls (32). The RNA was isolated from cultures in the exponential growth phase at an OD600 of 1.0 at 37°C. Ten micrograms of total RNA from S. aureus 8325 was used for RT with random primers and Superscript III reverse transcriptase (Invitrogen). The integrity of RNA was analyzed with a bioanalyzer (Agilent Technologies). Fluorescent labeling was performed during the RT reaction by incorporating the dyes Cy3 and Cy5 according to the manufacturer's instructions (Scienion, Berlin, Germany). Four different biological experiments were performed, and a reverse labeling (dye switch) experiment was performed to minimize bias due to differential dye bleaching or incorporation of Cy3 and Cy5 dyes during the RT reaction. Microarray hybridization and washing of slides were carried out as recommended by the manufacturer (Scienion, Berlin, Germany). The intensity of the fluorescence of the microarray was determined with a GenePix 4000B laser scanner (Axon Instruments Inc., Union City, CA), and individual signal intensities were analyzed using Acuity 4.0 software (Axon Instruments Inc.) according to the manufacturer's instructions. Normalization was performed by applying the LOWESS algorithm. Finally, significant changes in gene expression were identified with the SAM (significance analysis of microarrays) software using the one-class response type and a false discovery rate of <1.5% (45). The data were filtered for genes having at least a 1.5-fold change in expression.
In vitro phosphorylation assay.
In vitro phosphorylation of about 1 to 2 μg of purified PurA and PknB was performed for 20 min at 37°C in 20 μl of a reaction buffer containing 50 mM HEPES (pH 7.5), 1 mM dithiothreitol, 0.01% Brij 35, 3 mM MnCl2, 3 mM MgCl2, and 200 μCi [γ-32P]ATP/ml. MBP (Sigma, Deisenhofen, Germany) was used as a surrogate substrate to test the divalent cation dependence of the phosphorylation reaction. In this experiment, 50 μg/ml MBP and 25 μg/ml PknB were used along with the cations Mn2+, Mg2+, and Ca2+ at appropriate concentrations. In each case, the reaction was stopped by adding 4× SDS-protein buffer. One-dimensional gel electrophoresis was performed as previously described (25). Finally, radioactive proteins were visualized by autoradiography using direct-exposure film.
Adenylosuccinate synthase (PurA) activity assay.
The activity of PurA was assayed as described previously (22). The assay used measures the increase in absorbance at 280 nm that accompanies the conversion of IMP to adenylosuccinate in the presence of aspartate. As a control, reference reactions without aspartate were run simultaneously. Specific activity was calculated using a molar extinction coefficient of 11,700 M−1 cm−1 as described previously and was expressed in U/mg protein (22).
Triton X-100-induced autolysis assay.
The autolysis assay was performed as described by Mani et al. (31). Bacteria were grown in LB medium containing 1 M NaCl to an OD600 of ∼1.0 at 37°C with shaking at 220 rpm. The bacteria were washed once with phosphate-buffered saline, and the cells were resuspended in the same volume of 0.05 M Tris-HCl buffer (pH 7.5) containing 0.1% Triton X-100. The bacteria were incubated at 37°C with shaking, and the OD600 was measured at 30-min intervals.
Scanning electron microscopy.
The bacteria were grown overnight in tryptic soy broth on polystyrene chamber slides at 37°C. The slides were washed with 1× phosphate-buffered saline, mounted on aluminum stubs, and shadowed with gold. For visualization, a scanning electron microscope (Zeiss DSM962) was used at 15 kV.
Metabolome analysis.
S. aureus wild-type strain 8325 and the isogenic ΔpknB mutant were grown to an OD600 of 1 in 100 ml of LB medium. Samples used for intracellular metabolite analysis were obtained by fast filtration over a 0.22-μm sterility filter with a vacuum. Cells were washed immediately with a cooled isotonic NaCl solution and quenched with liquid nitrogen. Metabolite extraction was performed by cell disruption with glass beads and an ethanol-water solution. Samples were dried and prepared as described by Liebeke et al. (26). Detection of metabolites was performed by using ion pair liquid chromatography-mass spectrometry. The system consisted of an Agilent System 1100 liquid chromatograph (Agilent Technologies, Santa Clara, CA) coupled with a micrOTOF (Bruker, Rheinstetten, Germany) operating in electrospray negative-ionization mode. Samples were resolved with distilled water prior to injection, and chromatographic separation was performed at room temperature using an RP18 Waters SymmetryShield column (150 mm by 4.6 mm; 3.5 μm) connected to a Waters C18 precolumn. The mobile phase consisted of the following components: 5% methanol and 95% water containing 10 mM tributylamine as an ion-pairing reagent and acetic acid for adjustment of the pH to pH 4.8 (component A) and 100% methanol (component B). The elution gradient started with 100% component A for 2 min, which was followed by increases from 0 to 20% component B in 2 min, from 20 to 31% component B in 11 min, from 31 to 60% component B in 19 min, and from 60 to 100% component B in 5 min, 100% component B for 15 min, a decrease from 100 to 0% component B in 6 min, and then 3 min with 0% component B. The gradient flow rate was 0.3 ml/min. For quantitative analysis, standard solutions of metabolites were prepared and analyzed as described above. Accurate masses were extracted, and integration of designated peaks was performed by using QuantAnalysis (Bruker, Rheinstetten, Germany). Normalization of acquired data was performed by comparing the area of the added internal standard Br-ATP for each sample. The P values reported below are the results of a one-sided unpaired t test. Differences were considered significant if the P value was ≤0.05.
Microarray data accession number.
Additional information for the microarray platform, as well as the processed and raw microarray data from this study, have been deposited in the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under GEO series accession number GSE15346.
RESULTS AND DISCUSSION
In silico identification of an STPK-encoding gene in S. aureus.
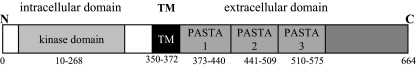
To identify S. aureus genes encoding an Ser/Thr kinase domain (S_TKc domain; SMART accession no. SM0020), domain library browse and search tools available at the SMART (http://smart.embl-heidelberg.de) and TIGR (www.tigr.org) databases were used. This search yielded a single gene (TIGR locus SA1063 in the S. aureus N315 genome), which we designated pknB due to a high level of sequence identity with pknB of M. tuberculosis (37% identity at the protein level) encoding a complete eukaryotic-type Ser/Thr kinase. Figure 1 shows the bipartite domain architecture of PknB, with the presumably cytoplasmic kinase domain separated by a putative transmembrane domain and three extracellular repeats of the PASTA domain (E values for PASTA domains, 6.37e-16, 2.38e-15, and 1.73e-17). The kinase domain of S. aureus PknB is predicted to belong to the STPK family with a high degree of statistical significance (E value, 7.12e-70) and contains all highly conserved residues characteristic of this family, including those required for catalytic activity. Thus, PknB of S. aureus possesses the typical domain architecture of the bacterial STPKs. Interestingly, a second protein with a eukaryote-like Ser/Tyr/Thr kinase domain has been found in the database for the prototype MRSA strain N315 (SA0077) and some other MRSA strains (JH1, JH9, MRSA252, and Mu50). The putative 502-amino-acid protein is encoded in the SCCmec element and contains a protein kinase domain (STYKc; E value, 5.55e-12) but no PASTA domains. However, the specificity of this class of kinases cannot be predicted, and they may possibly act as dual-specificity Ser/Thr/Tyr kinases. At present, it is not known if the kinase is functionally active.
FIG. 1.
Domain architecture for S. aureus PknB. TM, transmembrane domain. The domain library is available at SMART.
PknB is a manganese-dependent kinase.
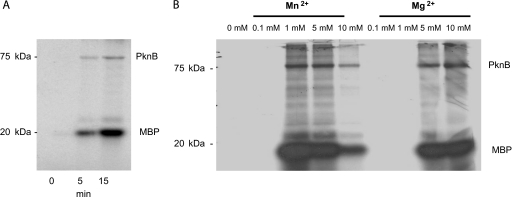
PknB contains all essential residues present in the kinase domain of STPKs (17). To study its phosphorylation activity, N-terminal His-tagged PknB was expressed under control of the T7 promoter in E. coli and purified as described in Materials and Methods. First, purified PknB was examined to determine its functional activity by performing phosphorylation assays using the surrogate substrate MBP, which is known to be phosphorylated by various STPKs, such as PknB from M. tuberculosis (1). As shown in Fig. 2A, purified PknB is able to phosphorylate MBP in a time-dependent manner. Next, we studied the phosphorylation reaction by varying the concentrations of bivalent cations. Maximal kinase activity was detected in the presence of up to 5 mM Mn2+, while concentrations between 5 mM and 10 mM were required for Mg2+; 10 mM Mn2+ inhibited phosphorylation of MBP as well as the absence of Mn2+ or Mg2+ (Fig. 2B). These experiments show that in vitro Mn2+ was more effective as a cofactor than Mg2+. However, the possibility that in vivo Mg2+ is the cofactor in phosphorylation reactions preferred by PknB cannot be ruled out as high concentrations of manganese may be toxic to bacterial cells. The presence of Ca2+ has no effect on the phosphorylation activity of PknB (data not shown).
FIG. 2.
Phosphorylation of MBP by PknB. Purified PknB was incubated with the surrogate kinase substrate MBP and [γ-32P]ATP in the presence or absence of Mn2+ or Mg2+. The reaction products were resolved on a 12% SDS-PAGE gel that was stained with Coomassie blue (not shown) and visualized by autoradiography. (A) Time-dependent phosphorylation of MBP. (B) Concentration-dependent phosphorylation with different Mn2+ or Mg2+ concentrations. The positions and masses (in kDa) of protein standards are indicated on the left. Besides phosphorylation of MBP autophosphorylation of PknB is visible.
Global transcription profile of ΔpknB mutant.
To investigate the functional role of PknB in S. aureus, DNA microarray experiments were performed. We compared exponentially growing (OD600, 1.0) mutant strain ΔpknB and parental strain 8325 using an S. aureus full-genome chip. The expression of 72 ORFs of the 2,666 genes evaluated was repressed in the ΔpknB strain, whereas the expression of 185 ORFs was increased in this mutant strain (Table 2 and 3). As an internal control, pknB transcripts were not detected for the ΔpknB mutant compared to the wild-type strain (Table 2). Importantly, the expression of genes involved in nucleotide biosynthesis, cell wall metabolism, and central metabolic pathways was affected by deletion of pknB (Table 2, 3).
TABLE 2.
Downregulated genes in the ΔpknB mutant strain
| N315 ORF | Gene | Description or predicted function | Change (fold)a | |||
|---|---|---|---|---|---|---|
| Purine and pyrimidine metabolism | ||||||
| SA0373 | xprT | Xanthine phosphoribosyltransferase | 3.7 | |||
| SA0917 | purK | Phosphoribosylaminoimidazole carboxylase carbon dioxide fixation chain PurK homolog | 1.4 | |||
| SA0920 | purQ | Phosphoribosylformylglycinamidine synthase I | 2.0 | |||
| SA0921 | purL | Phosphoribosylformylglycinamidine synthetase | 2.2 | |||
| SA0922 | purF | Phosphoribosylpyrophosphate amidotransferase | 3.0b | |||
| SA0923 | purM | Phosphoribosylformylglycinamidine cycloligase | 2.5b | |||
| SA0924 | purN | Phosphoribosylglycinamide formyltransferase | 2.2 | |||
| SA0925 | purH | Phosphoribosylaminoimidazole carboxamide formyltransferase | 1.8 | |||
| SA0926 | purD | Phosphoribosylamine glycine ligase | 6.2b | |||
| SA1041 | pyrR | Pyrimidine operon repressor chain A | 3.1 | |||
| SA1043 | pyrB | Aspartate transcarbamoylase chain A | 4.1 | |||
| SA1044 | pyrC | Dihydroorotase | 4.7 | |||
| SA1045 | pyrAA | Carbamoyl-phosphate synthase small chain | 5.6 | |||
| SA1046 | pyrAB | Carbamoyl-phosphate synthase large chain | 6.4 | |||
| SA1047 | pyrF | Orotidine-5-phosphate decarboxylase | 6.8 | |||
| SA1048 | pyrE | Orotate phosphoribosyltransferase | 5.6 | |||
| ABC transporter/transporter proteins | ||||||
| SA0255 | Hypothetical protein, similar to phosphotransferase system beta-glucoside-specific enzyme II | 2.2 | ||||
| SA0616 | vraF | ABC transporter ATP-binding protein | 1.8 | |||
| SA0617 | vraG | ABC transporter permease | 1.7 | |||
| SA0845 | oppB | Oligopeptide transport system permease protein | 1.8 | |||
| SA0847 | oppD | Oligopeptide transport system ATP-binding protein OppD homolog | 2.5 | |||
| SA0848 | oppF | Oligopeptide transport system ATP-binding protein OppF homolog | 2.0 | |||
| SA0849 | oppA | Hypothetical protein, similar to peptide-binding protein | 2.0 | |||
| SA1960 | mtlF | Phosphotransferase system, mannitol-specific IIBC component | 4.1 | |||
| SA2079 | fhuD2 | Hypothetical protein, similar to ferrichrome ABC transporter FhuD2 | 1.8 | |||
| SA2132 | Hypothetical protein, similar to ABC transporter | 2.2 | ||||
| SA2143 | Hypothetical protein, similar to ABC transporter | 2.6 | ||||
| SA2200 | Hypothetical protein, similar to ABC transporter, ATP-binding subunit | 1.6 | ||||
| SA2202 | Hypothetical protein, similar to ABC transporter periplasmic amino acid-binding protein | 1.9 | ||||
| SA2253 | Oligopeptide transporter putative membrane permease domain | 1.5 | ||||
| SA2302 | Hypothetical protein, similar to ABC transporter | 1.8 | ||||
| SA2434 | Fructose phosphotransferase system enzyme, FruA homolog | 2.5 | ||||
| Information pathways—DNA replication/RNA synthesis/protein synthesis | ||||||
| SA0442 | Probable DNA polymerase III, delta prime subunit | 1.7 | ||||
| SA1120 | Hypothetical protein, similar to transcription regulator GntR family | 1.9 | ||||
| SA1704 | map | Methionyl aminopeptidase | 1.6 | |||
| SA1717 | Glutamyl-tRNAGln amidotransferase subunit C | 1.7 | ||||
| SA1961 | Hypothetical protein, similar to transcription antiterminator BglG | 1.9 | ||||
| Metabolism of amino acids and related molecules | ||||||
| SA1121 | Hypothetical protein, similar to processing proteinase homolog | 1.8 | ||||
| SA1150 | glnA | Glutamine-ammonia ligase | 1.6 | |||
| Metabolism of carbohydrates and related molecules | ||||||
| SA1945 | Hypothetical protein, similar to mannnose-6 phospate isomerase Pmi | 2.5 | ||||
| Metabolism of nucleotides and nucleic acids | ||||||
| SA0468 | Hypoxanthine-guanine phosphoribosyltransferase homolog | 1.8 | ||||
| SA0687 | nrdF | Ribonucleoside-diphosphate reductase minor subunit | 1.7 | |||
| SA1921 | tdk | Thymidine kinase | 1.6 | |||
| SA2375 | Hypothetical protein, similar to dihydroorotate dehydrogenase | 2.1 | ||||
| SA2410 | Anaerobic ribonucleoside triphosphate reductase | 1.6 | ||||
| Metabolism of coenzymes and prosthetic groups | ||||||
| SA1442 | Hypothetical protein, similar to caffeoyl-CoA O-methyltransferase | 1.8 | ||||
| SA1734 | Pyrazinamidase/nicotinamidase homolog | 1.7 | ||||
| Miscellaneous | ||||||
| SA0231 | Hypothetical protein, similar to flavohemoprotein | 2.0 | ||||
| Cell envelope and cellular processes | ||||||
| SA0265 | lytM | Peptidoglycan hydrolase | 1.7 | |||
| SA0423 | Hypothetical protein, similar to autolysin | 2.1 | ||||
| SA0531 | proP | Proline/betaine transporter homolog ProP | 2.1 | |||
| SA0905 | atl | Autolysin (N-acetylmuramyl-l-alanine amidase and endo-β-N-acetylglucosaminidase) | 1.6 | |||
| SA1042 | pyrP | Uracil permease | 4.1 | |||
| SA1063 | pknB | STPK | 8.4 | |||
| SA1302 | gerCC | Heptaprenyl diphosphate syntase component II | 2.1 | |||
| SA2142 | Hypothetical protein, similar to multidrug resistance protein | 1.8 | ||||
| SA2203 | Hypothetical protein, similar to multidrug resistance protein | 1.7 | ||||
| Membrane bioenergetics (electron transport chain and ATP synthase) | ||||||
| SA0937 | Cytochrome d ubiquinol oxidase subunit 1 homolog | 1.6 | ||||
| Pathogenic factors | ||||||
| SA2097 | ssaA | Hypothetical protein, similar to secretory antigen precursor SsaA | 2.4 | |||
| SA2356 | isaA | Immunodominant antigen A | 1.6 | |||
| Detoxification | ||||||
| SA0781 | Hypothetical protein, similar to 2-nitropropane dioxygenase | 1.6 | ||||
| Unknown functions and hypothetical proteins | ||||||
| SA0269 | Hypothetical protein | 3.2 | ||||
| SA0289 | Conserved hypothetical protein | 1.5 | ||||
| SA0290 | Conserved hypothetical protein | 1.7 | ||||
| SA0291 | Hypothetical protein | 1.6 | ||||
| SA0467 | Hypothetical protein | 2.1 | ||||
| SA0725 | Conserved hypothetical protein | 1.8 | ||||
| SA0814 | Hypothetical protein | 2.5 | ||||
| SA0885 | Hypothetical protein | 2.6 | ||||
| SA1049 | Hypothetical protein | 5.6 | ||||
| SA1056 | Hypothetical protein | 1.6 | ||||
| SA1130 | Conserved hypothetical protein | 1.7 | ||||
| SA1890 | Conserved hypothetical protein | 1.8 | ||||
| SA2059 | Hypothetical protein | 1.5 | ||||
| SA2332 | Hypothetical protein | 2.6 | ||||
| SAS007 | Hypothetical protein | 1.6 | ||||
Data for genes with changes of ≥1.5-fold are shown. All genes had a q value that was <0.015, unless indicated otherwise.
Gene with a q value of <0.03.
TABLE 3.
Upregulated genes in the ΔpknB mutant strain
| N315 ORF | Gene | Description or predicted function | Change (fold)a | |||
|---|---|---|---|---|---|---|
| Citrate cycle | ||||||
| SA1244 | odhB | Dihydrolipoamide succinyltransferase | 2.4 | |||
| SA1245 | odhA | Oxoglutarate dehydrogenase E1 | 2.2 | |||
| Two-component system | ||||||
| SA0252 | lrgA | Holin-like protein LrgA | 2.8 | |||
| SA0253 | lrgB | Holin-like protein LrgB | 2.3 | |||
| SA1700 | vraR | Vancomycin resistance-associated two-component response regulator | 2.7 | |||
| SA1701 | vraS | Vancomycin resistance-associated two-component sensor histidine kinase | 2.5 | |||
| SA1843 | agrC | Accessory gene regulator C | 2.1 | |||
| SA1844 | agrA | Accessory gene regulator A | 2.3 | |||
| SA1882 | kdpD | Sensor protein | 2.8 | |||
| ABC transporter or transporter proteins | ||||||
| SA0209 | Maltose/maltodextrin transport permease homolog | 2.5 | ||||
| SA0599 | ATP-binding cassette transporter A | 2.5 | ||||
| SA1243 | ABC transporter homolog | 1.6 | ||||
| SA1744 | ABC-2-type transport system/permease protein | 3.9 | ||||
| SA1745 | Hypothetical protein, similar to ABC transporter, ATP-binding protein | 3.4 | ||||
| SA2227 | Truncated hypothetical protein, similar to d-serine/d-alanine/glycine transporter | 1.9 | ||||
| Phosphotransferase system | ||||||
| SA0186 | Phosphotransferase system, sucrose-specific IIBC component, putative | 3.1 | ||||
| SA0318 | Phosphotransferase system, unknown pentitol phosphotransferase enzyme IIC component | 2.9 | ||||
| SA0320 | Putative phosphotransferase system enzyme IIA component | 2.8 | ||||
| SA1255 | Phosphotransferase system, glucose-specific enzyme II, A component | 2.0 | ||||
| SA2114 | glvC | Phosphotransferase system, arbutin-like IIBC component | 1.7 | |||
| Membrane bioenergetics (electron transport chain and ATP synthase) | ||||||
| SA0210 | Hypothetical protein, similar to NADH-dependent dehydrogenase | 2.0 | ||||
| SA0411 | ndhF | NADH dehydrogenase subunit 5 | 2.5 | |||
| SA0799 | Hypothetical protein, similar to NADH dehydrogenase | 1.5 | ||||
| SA1904 | atpC | FoF1-ATP synthase epsilon subunit | 1.8 | |||
| SA1905 | atpD | ATP synthase beta chain | 1.6 | |||
| Cell envelope and cellular processes and cell wall | ||||||
| SA0115 | sbnD | Hypothetical protein, similar to multidrug resistance efflux pump | 2.3 | |||
| SA0172 | lmrP | Hypothetical protein, similar to integral membrane protein | 1.7 | |||
| SA0207 | Hypothetical protein, similar to maltose/maltodextrin-binding protein | 1.6 | ||||
| SA0248 | Hypothetical protein similar to beta-glycosyltransferase | 1.5 | ||||
| SA0259 | rbsD | Ribose permease | 1.9 | |||
| SA0260 | rbsU | Hypothetical protein, similar to ribose transporter | 1.7 | |||
| SA0303 | Hypothetical protein, similar to sodium-coupled permease | 2.2 | ||||
| SA0719 | trxB | Thioredoxin reductase | 1.5 | |||
| SA0758 | Hypothetical protein, similar to thioredoxin | 1.6 | ||||
| SA1283 | pbp2 | Penicillin-binding protein 2 | 1.5 | |||
| SA1691 | sgtB | Hypothetical protein, similar to penicillin-binding protein 1A/1B | 2.5 | |||
| SA1718 | putP | High-affinity proline permease | 2.1 | |||
| SA1926 | murZ | UDP-N-acetylglucosamine 1-carboxylvinyl transferase 2 | 1.5 | |||
| SA2437 | Hypothetical protein, similar to autolysin precursor | 1.9 | ||||
| SA2480 | drp35 | Drp35 | 4.1 | |||
| SAS023 | Hypothetical protein, similar to thioredoxin | 1.5 | ||||
| Information pathways: RNA synthesis, protein folding, and DNA modification and repair | ||||||
| SA0006 | gyrA | DNA gyrase subunit A | 1.5 | |||
| SA0009 | serS | Seryl-tRNA synthetase | 1.5 | |||
| SA0187 | Hypothetical protein, similar to transcription regulator | 3.9 | ||||
| SA0189 | hsdR | Probable type I restriction enzyme restriction chain | 1.9 | |||
| SA0321 | Hypothetical protein, similar to transcription antiterminator BglG family | 2.2 | ||||
| SA0480 | ctsR | Transcription repressor of class III stress gene homolog | 1.9 | |||
| SA0498 | rplL | 50S ribosomal protein L7/L12 | 1.5 | |||
| SA0652 | Hypothetical protein, similar to transcription regulation protein | 1.6 | ||||
| SA0653 | Hypothetical protein, similar to transcription repressor of fructose operon | 1.5 | ||||
| SA1257 | Peptide methionine sulfoxide reductase | 1.7 | ||||
| SA1287 | asnS | Asparaginyl-tRNA synthetase | 1.6 | |||
| SA1305 | hu | DNA-binding protein II (HB) | 1.6 | |||
| SA1360 | Xaa-Pro dipeptidase | 1.5 | ||||
| SA1586 | rot | Repressor of toxins | 1.8 | |||
| SA1659 | prsA | Peptidyl-prolyl cis/trans-isomerase homolog | 2.9 | |||
| SA1748 | Hypothetical protein, similar to transcription regulator, GntR family | 2.8 | ||||
| SA1836 | groEL | GroEL protein | 1.8 | |||
| SA2089 | sarR | Staphylococcal accessory regulator A homolog | 2.3 | |||
| SA2103 | lytR | Hypothetical protein, similar to Lyt divergon expression attenuator | 2.3 | |||
| SA2108 | Hypothetical protein, similar to transcription regulator, RpiR family | 2.2 | ||||
| SA2296 | Hypothetical protein, similar to transcriptional regulator, MerR family | 2.0 | ||||
| Metabolism of amino acids and related molecules | ||||||
| SA0008 | hutH | Histidine ammonia lyase | 1.9 | |||
| SA0818 | rocD | Ornithine aminotransferase | 2.5 | |||
| SA0819 | gudB | NAD-specific glutamate dehydrogenase | 2.0 | |||
| SA1348 | bfmBAA | Branched-chain alpha-keto acid dehydrogenase E1 | 1.5 | |||
| SA1365 | Glycine dehydrogenase (decarboxylating) subunit 2 homolog | 2.4 | ||||
| SA1366 | Glycine dehydrogenase (decarboxylating) subunit 1 | 2.2 | ||||
| SA1367 | Aminomethyltransferase | 1.5 | ||||
| SA1531 | ald | Alanine dehydrogenase | 2.3 | |||
| SA1585 | Proline dehydrogenase homolog | 3.3 | ||||
| SA1931 | blt | Hypothetical protein, similar to spermine/spermidine acetyltransferase | 1.5 | |||
| SA1968 | arg | Arginase | 2.4 | |||
| SA2084 | ureC | Urease alpha subunit | 1.7 | |||
| SA2085 | ureE | Urease accessory protein | 1.7 | |||
| SA2122 | hutU | Urocanate hydratase | 2.6 | |||
| SA2125 | Hypothetical protein, similar to formiminoglutamase | 1.9 | ||||
| SA2220 | Glycerate kinase | 2.1 | ||||
| SA2318 | Putative l-serine dehydratase | 2.4 | ||||
| SA2319 | Putative beta subunit of l-serine dehydratase | 2.0 | ||||
| SA2341 | rocA | 1-Pyrroline-5-carboxylate dehydrogenase | 3.5 | |||
| Metabolism of coenzymes and prosthetic groups | ||||||
| SA0181 | Hypothetical protein, similar to isochorismatase | 2.0 | ||||
| SA0915 | folD | FolD bifunctional protein | 1.7 | |||
| SA1587 | ribA | Riboflavin biosynthesis protein | 1.8 | |||
| SA1588 | ribB | Riboflavin synthase alpha chain | 1.6 | |||
| Metabolism of nucleotides and nucleic acids | ||||||
| SA0133 | dra | Deoxyribose phosphate aldolase | 1.7 | |||
| SA0511 | Hypothetical protein, similar to UDP-glucose 4-epimerase-related protein | 1.7 | ||||
| SA0545 | pta | Phosphotransacetylase | 1.8 | |||
| SA1308 | 30S ribosomal protein S1 | 1.8 | ||||
| SA1938 | pdp | Pyrimidine nucleoside phosphorylase | 1.7 | |||
| SA1939 | Deoxyribose phosphate aldolase | 1.8 | ||||
| Metabolism of lipids | ||||||
| SA0224 | Hypothetical protein, similar to 3-hydroxyacyl-CoA dehydrogenase | 2.6 | ||||
| SA0225 | Hypothetical protein, similar to glutaryl-CoA dehydrogenase | 3.1 | ||||
| SA1075 | hmrB | HmrB protein | 1.5 | |||
| SA2240 | Hypothetical protein, similar to para-nitrobenzyl esterase chain A | 1.5 | ||||
| SA2334 | mvaS | Hydroxymethylglutaryl-CoA synthase | 2.0 | |||
| Metabolism of carbohydrates and related molecules | ||||||
| SA0162 | aldA | Aldehyde dehydrogenase | 2.7 | |||
| SA0182 | Hypothetical protein, similar to indole-3-pyruvate decarboxylase | 1.8 | ||||
| SA0239 | Sorbitol dehydrogenase | 1.8 | ||||
| SA0258 | rbsK | Ribokinase | 2.1 | |||
| SA0304 | nanA | N-Acetylneuraminate lyase subunit | 2.0 | |||
| SA0342 | Acetyl-CoA C-acetyltransferase homolog | 1.9 | ||||
| SA0823 | pgi | Glucose-6-phosphate isomerase A | 1.5 | |||
| SA0945 | pdhC | Dihydrolipoamide S-acetyltransferase component of pyruvate dehydrogenase complex E2 | 1.5 | |||
| SA0946 | pdhD | Dihydrolipoamide dehydrogenase component of pyruvate dehydrogenase E3 | 1.5 | |||
| SA0958 | myo-Inositol-1 (or -4) monophosphatase homolog | 1.7 | ||||
| SA0995 | sdhA | Succinate dehydrogenase flavoprotein subunit | 1.7 | |||
| SA0996 | sdhB | Succinate dehydrogenase iron-sulfur protein subunit | 1.9 | |||
| SA1088 | sucC | Succinyl-CoA synthetase beta subunit | 1.9 | |||
| SA1089 | sucD | Succinyl-CoA synthetase alpha chain | 2.2 | |||
| SA1141 | glpK | Glycerol kinase | 1.8 | |||
| SA1142 | glpD | Aerobic glycerol-3-phosphate dehydrogenase | 1.8 | |||
| SA1184 | citB | Aconitate hydratase | 1.7 | |||
| SA1517 | citC | Isocitrate dehyrogenase | 1.6 | |||
| SA1553 | fhs | Formyltetrahydrofolate synthetase | 2.1 | |||
| SA1554 | acsA | Acetyl-CoA synthetase | 1.9 | |||
| SA1609 | pckA | Phosphoenolpyruvate carboxykinase | 2.6 | |||
| SA1669 | citG | Fumarate hydratase, class II | 1.7 | |||
| SA1679 | Hypothetical protein, similar to d-3-phosphoglycerate dehydrogenase | 1.5 | ||||
| SA1927 | fbaA | Fructose bisphosphate aldolase | 1.5 | |||
| SA2155 | Hypothetical protein, similar to malate:quinone oxidoreductase | 1.5 | ||||
| SA2304 | fbp | Fructose bisphosphatase | 1.7 | |||
| SAS020 | Hypothetical protein, similar to phosphoglycerate mutase | 1.6 | ||||
| Miscellaneous | ||||||
| SA0114 | sbnC | Siderophore biosynthesis protein | 1.8 | |||
| SA0117 | sbnF | Siderophore biosynthesis protein | 1.6 | |||
| SA0173 | Hypothetical protein, similar to surfactin synthetase | 1.8 | ||||
| SA0914 | Hypothetical protein, similar to chitinase B | 2.3 | ||||
| SA1238 | Hypothetical protein, similar to tellurite resistance protein | 1.6 | ||||
| SA1549 | htrA | Hypothetical protein, similar to serine proteinase Do, heat shock protein | 1.6 | |||
| SA1606 | Plant metabolite dehydrogenase homolog | 2.1 | ||||
| SA1617 | Hypothetical protein, similar to latent nuclear antigen | 1.7 | ||||
| SA1656 | hit | Hit-like protein involved in cell cycle regulation | 1.5 | |||
| Pathogenic factors | ||||||
| SA0091 | plc | 1-Phosphatidylinositol phosphodiesterase precurosr | 1.5 | |||
| SA0483 | clpC | Endopeptidase | 2.3 | |||
| SA0909 | fmtA | FmtA, autolysis and methicillin resistance-related protein | 2.3 | |||
| SA1007 | Alpha-hemolysin precursor | 3.1 | ||||
| SA1752 | hlb | Truncated beta-hemolysin | 23.1 | |||
| SA1811 | hlb | Truncated beta-hemolysin | 13.9 | |||
| SA2209 | hlgB | Gamma-hemolysin component B | 2.0 | |||
| SA2463 | lip | Triacylglycerol lipase precursor | 4.8 | |||
| Unknown functions and hypothetical proteins | ||||||
| SA0079 | Conserved hypothetical protein | 2.3 | ||||
| SA0129 | Hypothetical protein | 1.7 | ||||
| SA0174 | Conserved hypothetical protein | 2.4 | ||||
| SA0184 | Conserved hypothetical protein | 3.6 | ||||
| SA0185 | Conserved hypothetical protein | 4.5 | ||||
| SA0307 | Conserved hypothetical protein | 1.8 | ||||
| SA0372 | Hypothetical protein | 1.8 | ||||
| SA0403 | Hypothetical protein (pathogenicity island SaPIn2) lpl7 | 2.9 | ||||
| SA0412 | Conserved hypothetical protein | 2.7 | ||||
| SA0413 | Conserved hypothetical protein | 1.7 | ||||
| SA0546 | Conserved hypothetical protein | 1.9 | ||||
| SA0570 | Hypothetical protein | 1.8 | ||||
| SA0606 | Conserved hypothetical protein | 2.6 | ||||
| SA0607 | Conserved hypothetical protein | 2.7 | ||||
| SA0707 | Conserved hypothetical protein | 1.9 | ||||
| SA0752 | Hypothetical protein | 2.0 | ||||
| SA0861 | Conserved hypothetical protein | 1.6 | ||||
| SA0873 | Conserved hypothetical protein | 1.8 | ||||
| SA1001 | Hypothetical protein | 3.0 | ||||
| SA1019 | Conserved hypothetical protein | 2.0 | ||||
| SA1256 | Conserved hypothetical protein | 1.8 | ||||
| SA1281 | Conserved hypothetical protein | 1.9 | ||||
| SA1331 | Conserved hypothetical protein | 1.7 | ||||
| SA1432 | Conserved hypothetical protein | 1.6 | ||||
| SA1476 | Hypothetical protein | 2.8 | ||||
| SA1528 | Conserved hypothetical protein | 1.6 | ||||
| SA1529 | Conserved hypothetical protein | 1.7 | ||||
| SA1532 | Conserved hypothetical protein | 2.0 | ||||
| SA1578 | Conserved hypothetical protein | 1.6 | ||||
| SA1583 | Conserved hypothetical protein | 1.7 | ||||
| SA1618 | Conserved hypothetical protein | 1.7 | ||||
| SA1682 | Conserved hypothetical protein | 1.6 | ||||
| SA1702 | Conserved hypothetical protein | 2.4 | ||||
| SA1703 | Hypothetical protein | 2.0 | ||||
| SA1712 | Conserved hypothetical protein | 2.2 | ||||
| SA1726 | Hypothetical protein | 2.2 | ||||
| SA1746 | Hypothetical protein | 3.9 | ||||
| SA1867 | Conserved hypothetical protein | 1.5 | ||||
| SA1925 | Conserved hypothetical protein | 1.5 | ||||
| SA1937 | Conserved hypothetical protein | 1.7 | ||||
| SA1942 | Conserved hypothetical protein | 2.1 | ||||
| SA1976 | Conserved hypothetical protein | 2.3 | ||||
| SA2221 | Hypothetical protein | 2.4 | ||||
| SA2323 | Conserved hypothetical protein | 1.5 | ||||
| SA2331 | Hypothetical protein | 1.7 | ||||
| SA2366 | Conserved hypothetical protein | 1.5 | ||||
| SAS089 | Hypothetical protein | 1.6 | ||||
Data for genes with changes of ≥1.5-fold are shown. All genes had a q value that was <0.015.
Most strikingly, genes encoding proteins involved in de novo purine and pyrimidine biosynthesis were downregulated 2.0- to 6.8-fold in the ΔpknB mutant (Table 2). In particular, purK, purQ, purL, purF, purM, purN, purH, and purD of the purine biosynthesis pathway and pyrP, pyrR, pyrB, pyrC, pyrAA, pyrAB, pyrF, and pyrE involved in pyrimidine metabolism were significantly downregulated in the pknB-deficient strain compared to the wild-type strain. To confirm the decreased expression of genes involved in purine biosynthesis in the ΔpknB strain, we performed RT-PCR with selected target genes by using the same RNA samples that were used for microarray assays. As shown in Table 4, the purF and purH transcript levels were two- to threefold lower in the ΔpknB strain, confirming the results of the microarray analysis.
TABLE 4.
RT-PCR confirmation of microarray data
| Gene product | Gene | N315 ORF | DNA microarray expressiona | RT-PCR expressionb |
|---|---|---|---|---|
| Phosphoribosylpyrophosphate amidotransferase | purF | SA0922 | −3.0 | −3.0 |
| Phosphoribosylaminoimidazole carboxamide formyltransferase | purH | SA0925 | −1.8 | −2.0 |
| Autolysis and methicillin resistance-related protein | fmtA | SA0909 | 2.3 | 3.5 |
| Hypothetical protein, similar to penicillin-binding protein 1A/1B | sgtB | SA1691 | 2.5 | 4.0 |
| Holin-like protein | lrgA | SA0252 | 2.8 | 3.0 |
| Peptidyl-prolyl cis/trans-isomerase homolog | prsA | SA1659 | 2.9 | 3.4 |
| 1-Pyrroline-5-carboxylate dehydrogenase | rocA | SA2341 | 3.5 | 9.0 |
| Ornithine aminotransferase | rocD | SA0818 | 2.5 | 3.0 |
| Vancomycin resistance-associated two-component sensor histidine kinase | vraS | SA1701 | 2.5 | 10.0 |
Expression of genes was filtered by using a change of >1.5-fold and a false discovery rate of <1.5%. Negative values indicate downregulation in the ΔpknB mutant strain compared to isogenic wild-type strain S. aureus 8325. Positive values indicate upregulation in the ΔpknB mutant.
Expression levels were analyzed by RT-PCR. Negative values indicate downregulation in the ΔpknB mutant strain compared to isogenic wild-type strain S. aureus 8325. Positive values indicate upregulation in the ΔpknB mutant.
Effect of phosphorylation on PurA enzyme activity.
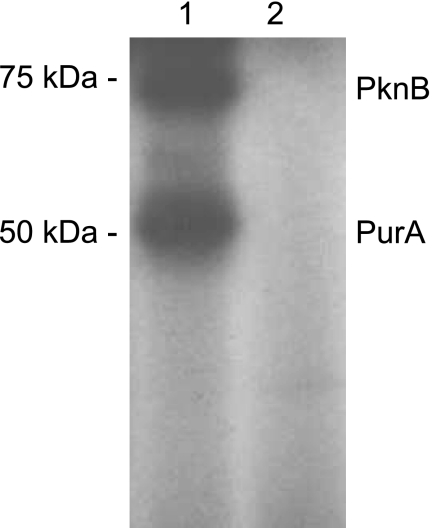
To further assess the role of PknB in purine biosynthesis, we investigated the effect of phosphorylation on the activity of PurA (adenylosuccinate synthase), an enzyme involved in synthesis of AMP. Recently, regulation of purine biosynthesis by phosphorylation and dephosphorylation of PurA has been described for Streptococcus agalactiae, in which the serine/threonine kinase Stk1 negatively affects PurA activity by phosphorylation (38). In our study, in vitro phosphorylation of PurA was performed in the presence of PknB and ATP. To exclude the possibility that His tagging results in nonspecific in vitro phosphorylation on serine and threonine residues as described by Boitel et al. (5), fusion proteins PknB-His6 and PurA-His6 were cleaved with thrombin to obtain recombinant PknB and PurA without a tag (Fig. 3). PurA assays were conducted as described in Materials and Methods. The results are summarized in Table 5. As controls we used reactions with unphosphorylated PurA lacking either PknB or ATP. Interestingly, we observed a 1.8-fold decrease in the activity of phosphorylated PurA (reactions performed after phosphorylation of PurA by PknB) compared to the activity of unphosphorylated PurA (control reactions without PknB or ATP). This indicates that phosphorylation of PurA by PknB decreases PurA enzyme activity. The results of these experiments support the idea that PknB phosphorylates and thereby inhibits PurA when intracellular A nucleotide pools (including the ATP concentration) increase. This would lead to suppression of further AMP and ATP synthesis. To prove this hypothesis, the intracellular concentrations of AMP, ADP, and ATP were measured by liquid chromatography-mass spectrometry. The AMP, ADP, and ATP concentrations were 1.2-fold to 1.4-fold higher in the pknB mutant than in the wild type (for AMP, 0.14 ± 0.02 μM/OD600 unit versus 0.11 ± 0.01 μM/OD600 unit; for ADP, 0.75 ± 0.02 μM/OD600 unit versus 0.54 ± 0.09 μM/OD600 unit; for ATP, 3.17 ± 0.29 μM/OD600 unit versus 2.53 ± 0.24 μM/OD600 unit). These results may have been due to the observed higher enzyme activity of PurA in its unphosphorylated form in cells lacking PknB, generating more AMP, ADP, and ATP.
FIG. 3.
Phosphorylation of PurA by PknB. The phosphorylation reaction was performed in the presence or absence of PknB as described in Materials and Methods. The His tag of both PknB and PurA was cleaved to exclude the possibility of a nonspecific in vitro phosphorylation effect. All reaction products were analyzed on a 12% SDS-PAGE gel, stained with Coomassie blue, and subjected to autoradiography. Lane 1 shows phosphorylation of PurA in the presence of PknB. Lane 2 contained the control without PknB. The positions and masses (in kDa) deduced from comparison with protein standards are indicated on the left.
TABLE 5.
Effect of phosphorylation on PurA activity
| Conditions | Sp act (U min−1 mg−1)a |
|---|---|
| PurA+ PknB + ATP | 1.42 ± 0.06 |
| PurA + PknB | 2.78 ± 0.27 |
| PurA+ ATP | 2.98 ± 0.09 |
| PurA | 2.68 ± 0.46 |
In vitro phosphorylation of PurA was performed in the presence of PknB and ATP as described in Materials and Methods. The controls included unphosphorylated PurA and reaction mixtures that did not contain either ATP or PknB. PurA enzyme assays were conducted in the presence or absence of aspartate. The average specific activities and standard deviations obtained in three independent experiments are shown. One unit was defined as the amount of enzyme required for the formation of 1 μmol of AMP per min.
This observation seems to be in contrast to the microarray expression data, where the pur operon was downregulated in the pknB mutant. However, in rich medium AMP, ADP, and ATP are preferentially generated via the salvage pathway that feeds the PurA reaction starting from IMP. Consequently, there is no need for de novo biosynthesis of either purines or pyrimidines in rich medium. The strong downregulation of purine biosynthesis genes observed in the microarray experiments may have resulted from a regulatory feedback loop in which elevated concentrations of the end products AMP and ATP inhibited transcription of the pur operon. Indeed, IMP concentrations were also increased in the ΔpknB strain (0.09 ± 0.02 μM/OD600 unit versus 0.05 ± 0.01 μM/OD600 unit). Some features of regulation of purine biosynthesis by PknB in S. aureus resemble the situation in S. agalactiae. In this pathogen, PurA activity is modulated by phosphorylation and dephosphorylation performed by the enzyme Stk1 homologous to PknB (38). In an stk1 mutant strain, AMP concentrations were also slightly increased, as shown for the pknB mutant of S. aureus in our study. In contrast to the findings for S. agalactiae, the guanine nucleotide concentrations were not altered in the PknB-deficient S. aureus strain, suggesting that there are differences in the regulation of A and G nucleotide metabolism in S. agalactiae and S. aureus (data not shown).
In gram-positive bacteria the organization of genes involved in purine metabolism and the regulation of these genes have been studied best in Bacillus subtilis. In contrast to E. coli, in which the purine biosynthesis genes are scattered throughout the genome, in B. subtilis and many other gram-positive organisms, such as S. aureus, almost all de novo purine biosynthesis genes are organized in an operon as a single transcriptional unit. The transcription of the operon is controlled by a repressor which is encoded by the purR gene and which at low 5-phosphoribosyl-1-pyrophosphate concentrations binds specifically to a DNA sequence in the promoter region (48). Thus, 5-phosphoribosyl-1-pyrophosphate acts as an indicator of purine availability. In general, regulation of purine biosynthesis is poorly characterized in S. aureus. Recently, it has been reported that the repressor of the pur operon PurR is functionally active in S. aureus, although binding of PurR to a consensus Pur box has not been shown (15). Fox et al. (15) observed variable PurM and PurH expression levels independent of PurR concentrations, and they concluded that in addition to PurR other regulators of purine biosynthesis in S. aureus are likely active in modulation of the expression levels of purine biosynthesis genes. We show here that PknB acts as an additional regulator of purine biosynthesis by regulating PurA activity via phosphorylation, thereby regulating A nucleotide synthesis. In our study, purR expression was not significantly changed in the microarray experiments.
Autolysis.
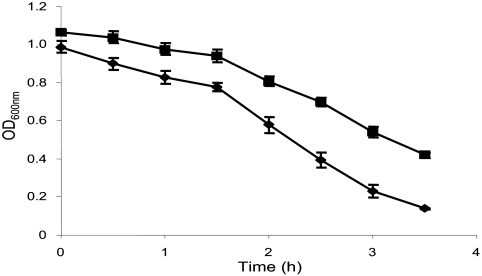
The microarray analysis showed that expression of two regulators of autolysis, fmtA and lytR, was induced ∼2.3-fold in the ΔpknB strain. Furthermore, expression of regulators of the murein hydrolase genes lrgA and lrgB was induced 2.3- to 2.8-fold in the ΔpknB strain (Table 3). In contrast, the expression of the autolysin gene atl was decreased 2.2-fold (Table 2). To determine the effect of pknB deletion on autolysis, an assay was performed by treating cells with Triton X-100. The ΔpknB strain showed decreased autolysis starting after 30 min of growth compared to the wild type, confirming the microarray data (Fig. 4). After 3.5 h of incubation ∼86% of the wild-type cells were lysed, whereas only ∼58% of the mutant cells were lysed at this time point.
FIG. 4.
Autolysis of whole cells of S. aureus wild-type strain 8325 (⧫) and the 8325 ΔpknB mutant (▪) during growth with Triton X-100. The results are expressed as decreases in the OD600 over time, which represented increases in autolysis (for details see Materials and Methods). The data are the means ± standard deviations of three independent experiments.
The LytR regulator of autolysis is a potential response regulator of a two-component system, which positively regulates lrgA and lrgB. Both the LrgA and LrgB proteins are involved in the regulation of peptidoglycan hydrolase activity (7, 8). LrgA and LrgB show similarities to a family of bacteriophage proteins known as holins (3). Holins are membrane-spanning proteins that allow phage cell wall hydrolases to access the cell wall murein by forming transmembrane holes. Deletion of the regulator lytR gene resulted in increased autolysis (8). The main autolysin Atl has been suggested to be involved in the separation of the daughter cells during development (4, 43). It is the predominant peptidoglycan hydrolase in staphylococci. An atlA mutant forms larger cell clusters and is defective in cell separation (4). Because our microarray data showed decreased atl expression together with increased transcription of fmtA and lytR, it is suggested that altered transcription of these factors contributes to the decreased autolysis detected in the mutant strain.
Role of pknB in cell wall metabolism.
Interestingly, the microarray data also showed that there was altered transcription of genes known to modulate the regulation of cell wall biosynthesis. We detected 2.4- to 2.7-fold upregulation of genes coding for the VraSR two-component system, which was confirmed by RT-PCR analysis (Table 4). VraSR is thought to be a positive regulator of cell wall peptidoglycan synthesis and is involved in the expression of β-lactam and glycopeptide resistance in S. aureus (24). Kuroda et al. (24) reported induction of VraSR by several cell wall synthesis inhibitors, suggesting that the sensor kinase VraS responds to damage to cell wall structure or inhibition of cell wall biosynthesis. Interestingly, 9 of 13 genes of the VraSR regulon (proP, SA2220, sgtB, murZ, prsA, SA1476, SA1702, SA1703, and SA2221) that were described by Kuroda et al. under non-antibiotic-induced conditions are upregulated in the pknB mutant.
In addition, several cell wall-active antibiotics have been tested, and the MICs of tunicamycin, methicillin, fosfomycin, d-cycloserine, vancomycin, bacitracin, cefepime, and ceftriaxone for the ΔpknB strain and the isogenic wild-type strain have been determined. Interestingly, tunicamycin, an inhibitor of MraY transferase, which catalyzes the formation of the first lipid intermediate of peptidoglycan synthesis, was 32-fold more active against the pknB mutant than against the wild type (Table 6). Other cell wall-active antibiotics only slightly affected MICs; e.g., the MICs of fosfomycin and methicillin observed for the wild type were twofold-higher than those for the ΔpknB strain. Interestingly, deletion of pknB in the methicillin-resistant strain COL led to a phenotype with reduced methicillin resistance (MIC for COL, 256 μg/ml; MIC for COLΔpknB, 16 μg/ml). This result supports the hypothesis that PknB has a role in cell wall metabolic pathways. In contrast, no differences in MICs between the wild type and the ΔpknB strain were detectable for d-cycloserine, bacitracin, vancomycin, cefepime, and ceftriaxone. In another study, Dèbarbouillè et al. found that the Δstk1 mutant expressed greater resistance to fosfomycin (2 logs) than wild-type strain 8325-4 expressed (12). This is in contrast to our results, and the difference may be due to the different strain backgrounds in the two studies.
TABLE 6.
MICs of antibiotics for S. aureus wild-type strain 8325 and the isogenic ΔpknB mutant
| Antibiotic | MIC(μg/ml)
|
|
|---|---|---|
| S. aureus wild-type strain 8325 | S. aureus 8325ΔpknB | |
| Fosfomycin | 16 | 8 |
| Tunicamycin | 64 | 2 |
| Methicillin | 0.25 | 0.125 |
| d-Cycloserine | 32 | 32 |
| Vancomycin | 1 | 1 |
| Bacitracin | 32 | 32 |
| Cefepime | 2 | 2 |
| Ceftriaxone | 2 | 2 |
| Kanamycin | 1 | 1 |
Most proteins that contain PASTA domains are directly involved in peptidoglycan metabolism (e.g., penicillin-binding proteins) by interacting with un-cross-linked peptidoglycan (49). Therefore, it has been postulated that STPKs such as PrkC of Enterococcus faecalis may monitor the integrity of the cell wall by detecting the accumulation of peptidoglycan precursors or un-cross-linked peptidoglycan polymers (23). In addition, it was shown recently by Shah and colleagues (40) that the STPK PrkC of B. subtilis is indeed capable of binding peptidoglycan. These workers tested various His-tagged domains of PrkC in a peptidoglycan binding assay and identified the extracellular PASTA domains as binding partners. A similar function may be played by PknB; however, there are no experimental data demonstrating that PASTA domains of PknB of S. aureus are involved in sensing cell wall precursors and thereby regulating parts of the cell wall synthesis machinery.
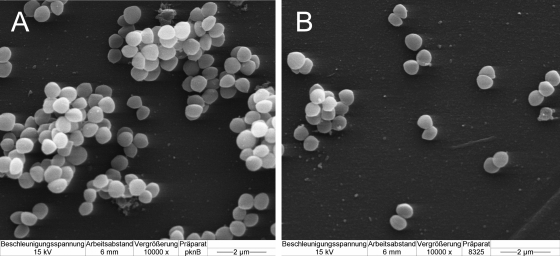
Furthermore, the morphology of ΔpknB and wild-type cells was investigated by scanning electron microscopy. As shown in Fig. 5A, mutant cells formed larger cell clusters than the wild-type cells formed (Fig. 5B). This phenotype of the mutant cells could be restored by complementation (not shown) and might be directly linked to decreased expression of atl, which results in the formation of larger cell clusters due to incomplete separation of cells during growth.
FIG. 5.
Scanning electron microscopy of the 8325 ΔpknB mutant (A) and wild-type strain 8325 (B) grown on polystyrene surfaces. Cells of the ΔpknB strain formed larger cell clusters than cells of the wild-type strain formed.
Collectively, the data from the microarray analysis, the formation of larger clusters by the mutant, and the increased sensitivity of the ΔpknB mutant to cell wall-active antibiotics indicate that PknB has a role in cell wall synthesis and metabolism. Our results support the hypothesis that the VraSR regulon is activated in part as a response to altered cell wall biosynthesis in the pknB mutant.
Transcription of genes involved in amino acid metabolism.
In addition to the altered expression of genes involved in nucleotide biosynthesis and cell wall metabolism, the microarray data revealed higher levels of expression of genes involved in amino acid metabolism. The rocA and rocD genes, encoding a 1-pyrroline-5-carboxylate dehydrogenase and an ornithine aminotransferase, are induced 3.5- and 2.5-fold, respectively, in the ΔpknB mutant. Both rocA and rocD encode proteins that are responsible for the degradation of l-ornithine to l-glutamate. Enhanced expression of these genes was also detected by RT-PCR (Table 4). Furthermore, the expression of gudB was increased ∼2-fold in the S. aureus ΔpknB strain. gudB encodes an NAD-specific glutamate dehydrogenase which catalyzes the conversion of l-glutamate to 2-oxoglutamate. To a large extent, genes involved in pyruvate metabolism are expressed at higher levels in the pknB mutant. For example, aldA (aldehyde dehydrogenase), pta (phosphotransacetylase), acsA (acetyl-coenzyme A [acetyl-CoA] synthetase), and pckA (phosphoenolpyruvate carboxykinase), as well as genes encoding proteins of the citrate cycle, such as odhB (dihydrolipoamide succinyltransferase) and odhA (oxoglutarate dehydrogenase E1), were induced in the ΔpknB mutant. These results suggest that in the ΔpknB mutant there is altered synthesis of central metabolic precursors compared to the wild type. Interestingly, phosphorylation of proteins involved in glycolysis, such as pyruvate dehydrogenase, enolase, phosphate acetyltransferase, and fructose biphosphate aldolase, has been demonstrated recently (27). Lomas-Lopez et al. (27) used purified PknB to study phosphorylation of these putative substrates. The results of our microarray analysis are consistent with the observation of Lomas-Lopez (27) that PknB is involved in regulation of central metabolic functions. PknB probably links substrate requirements of growing cells by sensing the pool of unlinked peptidoglycan precursors with synthesis of purines and pyrimidines and amino acids such as glutamine and lysine. Both glutamine and lysine are essential constituents of murein monomer precursors. To maintain the glutamine pool in the ΔpknB strain, activation of enzymes of the citrate cycle resulting in the production of 2-oxoglutarate from oxaloacetate might be important for the cells. Likewise, Cowley et al. described accumulation of glutamate and glutamine in an STPK deletion (ΔpknG) mutant of M. tuberculosis (10).
Conclusion.
Protein phosphorylation on serine and threonine residues seems to be a common theme in regulation of cellular functions in prokaryotes and eukaryotes. PknB probably is the only eukaryote-like STPK of S. aureus that is involved in the regulation of several central metabolic functions, such as purine and pyrimidine synthesis, cell wall synthesis, and glycolysis and the citrate cycle. Recently, the serine/threonine/tyrosine phosphoproteomes of the gram-positive model organism B. subtilis, the lactic acid bacterium Lactococcus lactis, and the gram-negative model bacterium E. coli have been determined (14, 28, 29, 30, 41). Comparison of phosphorylation sites in these three species revealed that the majority of phosphorylated proteins were species specific. Therefore, it has been concluded that phosphorylation by eukaryote-like STPKs probably coevolved with adaptation of the bacteria to specific ecological niches. Most strikingly, serine/threonine/tyrosine phosphorylation has been found in housekeeping pathways and in central carbon metabolism processes, such as glycolysis, in all three species. The results of these phosphoproteome studies are in agreement with our microarray data with respect to modulation of functions of central metabolism, such as glycolysis. Genes that are involved in central metabolic functions were clearly overrepresented in the deregulated genes in the pknB mutant compared to the wild-type strain. PurA and other enzymes involved in purine biosynthesis have not been found in recent phosphoproteome studies of B. subtilis and L. lactis, but this pathway has been shown to be regulated by an enzyme homologous to PknB in S. agalactiae. Obviously, phosphorylation on the functional level by eukaryote-like STPKs may be conserved among different bacterial species, and at least glycolysis seems to be a common theme of regulation by serine/threonine/tyrosine phosphorylation and dephosphorylation in many bacteria. However, specific Ser/Thr/Tyr phosphorylation has evolved more recently under the particular requirements of the niches in which these bacteria compete (e.g., B. subtilis in soil, L. lactis in dairy products, and S. aureus and S. agalactiae on mucosal surfaces).
The exact role of PknB in S. aureus remains to be determined; however, all the data available, such as the data for the regulation of PurA activity described in this report, suggest that there is fine regulation of enzymatic activity by PknB rather than an “on-off” mechanism due to phosphorylation and dephosphorylation of serine and threonine residues of target enzymes. Importantly, a lack of PknB resulted in decreased virulence in a murine model of kidney infection. The mutant strain showed significantly less ability to colonize kidneys after intravenous challenge (12). Based on our microarray data, it is more likely that the reduced virulence of the pknB mutant strain results from an impaired ability to survive in the host due to metabolic limitation rather than from reduced expression of virulence determinants.
Acknowledgments
This study was supported by grants from the Deutsche Forschungsgemeinschaft (TRR34, SFB630) and the EU (LSHM-CT-2006-019064-StaphDynamics).
We thank Ursula Wallner and Martin Eckart for technical assistance.
Footnotes
Published ahead of print on 17 April 2009.
REFERENCES
- 1.Av-Gay, Y., S. Jamil, and S. J. Drews. 1999. Expression and characterization of the Mycobacterium tuberculosis serine/threonine protein kinase PknB. Infect. Immun. 675676-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakal, C. J., and J. E. Davies. 2000. No longer an exclusive club: eukaryotic signalling domains in bacteria. Trends Cell Biol. 1032-38. [DOI] [PubMed] [Google Scholar]
- 3.Bayles, K. W. 2000. The bactericidal action of penicillin: new clues to an unsolved mystery. Trends Microbiol. 8274-278. [DOI] [PubMed] [Google Scholar]
- 4.Biswas, R., L. Voggu, U. K. Simon, P. Hentschel, G. Thumm, and F. Götz. 2006. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol. Lett. 259260-268. [DOI] [PubMed] [Google Scholar]
- 5.Boitel, B., M. Ortiz-Lombardia, R. Duran, F. Pompeo, S. T. Cole, C. Cervenansky, and P. M. Alzari. 2003. PknB kinase activity is regulated by phosphorylation in two Thr residues and dephosphorylation by PstP, the cognate phospho-Ser/Thr phosphatase, in Mycobacterium tuberculosis. Mol. Microbiol. 491493-1508. [DOI] [PubMed] [Google Scholar]
- 6.Brückner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 1511-8. [DOI] [PubMed] [Google Scholar]
- 7.Brunskill, E. W., and K. W. Bayles. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunskill, E. W., and K. W. Bayles. 1996. Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol. 1785810-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Document M7-A7. CLSI, Wayne, PA.
- 10.Cowley, S., M. Ko, N. Pick, R. Chow, K. J. Downing, B. G. Gordhan, J. C. Betts, V. Mizrahi, D. A. Smith, R. W. Stokes, and Y. Av-Gay. 2004. The Mycobacterium tuberculosis protein serine/threonine kinase PknG is linked to cellular glutamate/glutamine levels and is important for growth in vivo. Mol. Microbiol. 521691-1702. [DOI] [PubMed] [Google Scholar]
- 11.Curry, J. M., R. Whalan, D. M. Hunt, K. Gohil, M. Strom, L. Rickman, M. J. Colston, S. J. Smerdon, and R. S. Buxton. 2005. An ABC transporter containing a forkhead-associated domain interacts with a serine-threonine protein kinase and is required for growth of Mycobacterium tuberculosis in mice. Infect. Immun. 734471-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dèbarbouillè, M., S. Dramsi, O. Dussurget, M.-A. Nahori, E. Vaganay, G. Jouvion, A. Cozzone, T. Msadek, and B. Duclos. 24 April 2009. Characterization of a serine/threonine kinase involved in virulence of Staphylococcus aureus. J. Bacteriol. 1914070-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Lencastre, H., S. W. Wu, M. G. Pinho, A. M. Ludovice, S. Filipe, S. Gardete, R. Sobral, S. Gill, M. Chung, and A. Tomasz. 1999. Antibiotic resistance as a stress response: complete sequencing of a large number of chromosomal loci in Staphylococcus aureus strain COL that impact on the expression of resistance to methicillin. Microb. Drug Resist. 5163-175. [DOI] [PubMed] [Google Scholar]
- 14.Eymann, C., D. Becher, J. Bernhardt, K. Gronau, A. Klutzny, and M. Hecker. 2007. Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis. Proteomics 73509-3526. [DOI] [PubMed] [Google Scholar]
- 15.Fox, P. M., M. W. Climo, and G. L. Archer. 2007. Lack of relationship between purine biosynthesis and vancomycin resistance in Staphylococcus aureus: a cautionary tale for microarray interpretation. Antimicrob. Agents Chemother. 511274-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galyov, E. E., S. Hakansson, A. Forsberg, and H. Wolf-Watz. 1993. A secreted protein kinase of Yersinia pseudotuberculosis is an indispensable virulence determinant. Nature 361730-732. [DOI] [PubMed] [Google Scholar]
- 17.Hanks, S. K., A. M. Quinn, and T. Hunter. 1988. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 24142-52. [DOI] [PubMed] [Google Scholar]
- 18.Reference deleted.
- 19.Hunter, T. 1995. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80225-236. [DOI] [PubMed] [Google Scholar]
- 20.Hussain, H., P. Branny, and E. Allan. 2006. A eukaryotic-type serine/threonine protein kinase is required for biofilm formation, genetic competence, and acid resistance in Streptococcus mutans. J. Bacteriol. 1881628-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, H., and V. Pancholi. 2006. Identification and biochemical characterization of a eukaryotic-type serine/threonine kinase and its cognate phosphatase in Streptococcus pyogenes: their biological functions and substrate identification. J. Mol. Biol. 3571351-1372. [DOI] [PubMed] [Google Scholar]
- 22.Juang, R. H., K. F. McCue, and D. W. Ow. 1993. Two purine biosynthetic enzymes that are required for cadmium tolerance in Schizosaccharomyces pombe utilize cysteine sulfinate in vitro. Arch. Biochem. Biophys. 304392-401. [DOI] [PubMed] [Google Scholar]
- 23.Kristich, C. J., C. L. Wells, and G. M. Dunny. 2007. A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc. Natl. Acad. Sci. USA 1043508-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49807-821. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 26.Liebeke, M., D. C. Pother, N. van Duy, D. Albrecht, D. Becher, F. Hochgrafe, M. Lalk, M. Hecker, and H. Antelmann. 2008. Depletion of thiol-containing proteins in response to quinones in Bacillus subtilis. Mol. Microbiol. 691513-1529. [DOI] [PubMed] [Google Scholar]
- 27.Lomas-Lopez, R., P. Paracuellos, M. Riberty, A. J. Cozzone, and B. Duclos. 2007. Several enzymes of the central metabolism are phosphorylated in Staphylococcus aureus. FEMS Microbiol. Lett. 27235-42. [DOI] [PubMed] [Google Scholar]
- 28.Macek, B., F. Gnad, B. Soufi, C. Kumar, J. V. Olsen, I. Mijakovic, and M. Mann. 2008. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics 7299-307. [DOI] [PubMed] [Google Scholar]
- 29.Macek, B., I. Mijakovic, J. V. Olsen, F. Gnad, C. Kumar, P. R. Jensen, and M. Mann. 2007. The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6697-707. [DOI] [PubMed] [Google Scholar]
- 30.Madec, E., A. Laszkiewicz, A. Iwanicki, M. Obuchowski, and S. Seror. 2002. Characterization of a membrane-linked Ser/Thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol. Microbiol. 46571-586. [DOI] [PubMed] [Google Scholar]
- 31.Mani, N., P. Tobin, and R. K. Jayaswal. 1993. Isolation and characterization of autolysis-defective mutants of Staphylococcus aureus created by Tn917-lacZ mutagenesis. J. Bacteriol. 1751493-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michel, A., F. Agerer, C. R. Hauck, M. Herrmann, J. Ullrich, J. Hacker, and K. Ohlsen. 2006. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 1885783-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nariya, H., and S. Inouye. 2002. Activation of 6-phosphofructokinase via phosphorylation by Pkn4, a protein Ser/Thr kinase of Myxococcus xanthus. Mol. Microbiol. 461353-1366. [DOI] [PubMed] [Google Scholar]
- 34.Neu, J. M., S. V. MacMillan, J. R. Nodwell, and G. D. Wright. 2002. StoPK-1, a serine/threonine protein kinase from the glycopeptide antibiotic producer Streptomyces toyocaensis NRRL 15009, affects oxidative stress response. Mol. Microbiol. 44417-430. [DOI] [PubMed] [Google Scholar]
- 35.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajagopal, L., A. Clancy, and C. E. Rubens. 2003. A eukaryotic type serine/threonine kinase and phosphatase in Streptococcus agalactiae reversibly phosphorylate an inorganic pyrophosphatase and affect growth, cell segregation, and virulence. J. Biol. Chem. 27814429-14441. [DOI] [PubMed] [Google Scholar]
- 37.Rajagopal, L., A. Vo, A. Silvestroni, and C. E. Rubens. 2006. Regulation of cytotoxin expression by converging eukaryotic-type and two-component signalling mechanisms in Streptococcus agalactiae. Mol. Microbiol. 62941-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajagopal, L., A. Vo, A. Silvestroni, and C. E. Rubens. 2005. Regulation of purine biosynthesis by a eukaryotic-type kinase in Streptococcus agalactiae. Mol. Microbiol. 561329-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saskova, L., L. Novakova, M. Basler, and P. Branny. 2007. Eukaryotic-type serine/threonine protein kinase StkP is a global regulator of gene expression in Streptococcus pneumoniae. J. Bacteriol. 1894168-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah, I. M., M. H. Laaberki, D. L. Popham, and J. Dworkin. 2008. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135486-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soufi, B., F. Gnad, P. R. Jensen, D. Petranovic, M. Mann, I. Mijakovic, and B. Macek. 2008. The Ser/Thr/Tyr phosphoproteome of Lactococcus lactis IL1403 reveals multiply phosphorylated proteins. Proteomics 83486-3493. [DOI] [PubMed] [Google Scholar]
- 42.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69183-215. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi, J., H. Komatsuzawa, S. Yamada, T. Nishida, H. Labischinski, T. Fujiwara, M. Ohara, J. Yamagishi, and M. Sugai. 2002. Molecular characterization of an atl null mutant of Staphylococcus aureus. Microbiol. Immunol. 46601-612. [DOI] [PubMed] [Google Scholar]
- 44.Truong-Bolduc, Q. C., Y. Ding, and D. C. Hooper. 2008. Posttranslational modification influences the effects of MgrA on norA expression in Staphylococcus aureus. J. Bacteriol. 1907375-7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 985116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Umeyama, T., P. C. Lee, and S. Horinouchi. 2002. Protein serine/threonine kinases in signal transduction for secondary metabolism and morphogenesis in Streptomyces. Appl. Microbiol. Biotechnol. 59419-425. [DOI] [PubMed] [Google Scholar]
- 47.Wang, J., C. Li, H. Yang, A. Mushegian, and S. Jin. 1998. A novel serine/threonine protein kinase homologue of Pseudomonas aeruginosa is specifically inducible within the host infection site and is required for full virulence in neutropenic mice. J. Bacteriol. 1806764-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weng, M., P. L. Nagy, and H. Zalkin. 1995. Identification of the Bacillus subtilis pur operon repressor. Proc. Natl. Acad. Sci. USA 927455-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeats, C., R. D. Finn, and A. Bateman. 2002. The PASTA domain: a beta-lactam-binding domain. Trends Biochem. Sci. 27438. [DOI] [PubMed] [Google Scholar]