Abstract
Bluetongue virus (BTV), a nonenveloped insect-borne virus, is released from infected cells by multiple pathways. Unlike other nonenveloped viruses, in addition to cell lysis the newly synthesized virus particles also appear to use a unique “budding” process. The nonstructural protein NS3, the only membrane protein encoded by BTV in infected cells, has been implicated in this process, since it appears to interact not only with the outermost viral capsid protein VP2 but also with a component of the cellular ESCRT pathway. However, to date it had not been possible to obtain direct evidence for the involvement of NS3 in BTV morphogenesis due to the lack of a genetic system that would allow introducing the targeted mutation in NS3 gene. In this study, we have used the recently developed T7 transcript-based reverse genetics system for BTV to introduce mutations in the sequence of NS3 into the viral genome and have investigated the effect of these mutations in the context of a replicating virus. While certain NS3 mutations exhibited drastic effects on newly synthesized virus release, others had less pronounced effects. In particular, mutations of two residues in the Tsg101 binding motif, the putative L domain of NS3, altered normal virus egress patterns and left nascent particles tethered to the cellular membrane, apparently arrested in the process of budding. In cells infected with a mutant virus that was incapable of an NS3-VP2 interaction, no budding particles were visualized. These data suggest that NS3 may act like the membrane protein of enveloped viruses and is responsible for intracellular trafficking and budding of virus particles. NS3 is thus a bridge between the maturing virion particles and cellular proteins during virus egress.
The establishment of viral infection in host cells requires a highly specific series of consecutive processes dictated by the virus. Among these, the virus mechanism of entry into host cells has probably received the most attention. In contrast, the mechanism of egress of a newly synthesized progeny virus from an infected cell, which is equally important in relation to successful viral infection, has been investigated much less. Much of our knowledge of this process has been gained from studies involving a limited number of viruses, in particular, viruses that acquire cellular membranes during their egress from host cells. The acquisition of cellular membranes by these viruses (known as enveloped viruses as opposed to nonenveloped viruses), facilitates their release via budding from the cell membrane upon completion of the infection cycle. However, a range of viruses of various sizes and complexities lack envelopes, and these viruses must utilize alternative means to exit the cells. The dogma for nonenveloped viruses is that their egress is achieved through cell lysis. However, there is growing evidence that this is not the only mechanism for virus release.
We have been particularly interested in understanding the virus exit mechanism for a nonenveloped insect-borne virus, bluetongue virus (BTV), which has a complex, multiprotein-layered capsid structure. BTV is a relatively large virus (∼85 nm in diameter) in comparison with some other nonenveloped viruses such as picornaviruses (about ∼30 nm). Although BTV lacks a glycosylated membrane protein in its mature particle, in infected cells it does synthesize a nonstructural protein which is a glycosylated membrane protein, NS3, making it a likely candidate protein for aiding in virus release from infected cells (17, 18). Indeed, virus-encoded nonstructural proteins synthesized in infected cells often play important roles in virus replication and morphogenesis, although they are not included in the mature virus particles. Whether BTV NS3 has a direct role in virus egress or is involved in virus morphogenesis has not been addressed directly to date. This has mainly been due to the lack of techniques which would allow the introduction of site-specific mutations into the BTV genome, which is a double-stranded, multisegmented RNA genome. Recently we have successfully developed a reverse genetics system that allows the introduction of mutations into the viral genome, facilitating the examination of their phenotype in BTV replication and morphogenesis.
BTV is a member of the Reoviridae family, but unlike other members of the family such as rotaviruses and reoviruses, BTV is vectored by insect Culicoides spp. to vertebrate hosts (sheep, cattle, goat, etc.) and has the ability to replicate in both insect and mammalian hosts. The BTV particle consists of seven structural proteins organized in two capsids, an outer capsid and an inner capsid (core). Two major proteins, VP2 and VP5, form the outer capsid, which surrounds the core particle, composed of two other major proteins of the virus, VP7 and VP3. The three other structural proteins within the core are the minor enzymatic proteins VP1, VP4, and VP6, which are closely associated with the 10 genomic double-stranded RNA (dsRNA) segments (26). Each RNA segment encodes a single viral protein except the smallest segment, segment 10, which encodes two proteins, NS3 (229 residues) and the related NS3A (216 residues), a shorter form of NS3 lacking the 13 residues from the N-terminal end of NS3. These NS3 proteins are synthesized not only in smaller amounts than the two other virus-coded nonstructural proteins (NS1 and NS2) but also in different amounts depending on the different host cells (13, 20).
One of the striking characteristics of BTV infection is that although the majority of mature virions are released by cell lysis late in infection in mammalian cells, some particles are released from infected cells by local extrusion and budding from the plasma membrane (16). It is rational to expect this early release to be responsible for the cell-to-cell spread of the virus. Very little is known about the intracellular trafficking of the newly generated BTV virions, although there are some indications of involvement of the cytoskeleton, intermediate filaments, and vimentin during virus morphogenesis (4).
In contrast, in insect cells, release of this nonenveloped virus is almost entirely nonlytic. High-level expression of NS3 in invertebrate cells infected with BTV has been reported to correlate with nonlytic virus release (14). Furthermore, when BTV virus-like particles were expressed together with NS3/NS3A proteins in insect cells by recombinant baculoviruses, virus-like particles were released from the cellular membrane analogously to native virion particles (18). In addition, in virus-infected cells NS3 appears to be closely associated with the newly synthesized progeny viruses (17). Further, by the yeast-two-hybrid system we have shown previously that NS3 interacts with the outer capsid protein VP2 (3). These studies indicate that the NS3 protein is involved in virus egress; however, the underlying mechanism is not clear. The structure of NS3 and NS3A proteins comprises long N-terminal and short C-terminal cytoplasmic tails connected by two transmembrane domains, consistent with a role in membrane perturbation, and a short extracellular domain with a single asparagine glycosylation site (1, 3, 30).
Another intriguing feature of NS3 is that it possesses late (L) domain motifs similar to those of certain proteins of enveloped viruses which commonly use the budding process for release (10, 12). Through these L domains some viruses, in particular enveloped viruses, are able to hijack host proteins involved in the vacuolar protein-sorting pathway, a cellular budding network that gives rise to multivesicular bodies (MVB). Interestingly, we have shown in a previous study that, like enveloped viruses, BTV NS3 also interacts with a cellular protein component of the ESCRT-I complex, namely, the tumor susceptibility gene 101 protein (Tsg101) (29), which is involved in the last stages of membrane fission (28). These data suggest that NS3 may form a bridge between the virus particle and the host cell budding machinery and that it might have some controlling role in mature virus release from infected cells. However, the exact role of NS3 in this process and the role of cellular proteins such as Tsg101 in the release of nonenveloped viruses, such as BTV, were not investigated further due the lack of a genetic system that would allow the direct introduction of mutations into the virus genome. However, recently we have successfully generated BTV by transfection of permissive cells with a complete set of in vitro-synthesized T7 transcripts without the use of a helper virus (6).
In this study we have exploited this system to introduce a series of site-specific mutations in the BTV genome, particularly focusing on the two regions of NS3, the L-domain motif and the C-terminal cytoplasmic tail, that appear to interact with virus outer capsid VP2. The detailed analyses of these mutant viruses clearly demonstrate that NS3 plays a key role in the interaction of viral particles with cellular proteins and is involved in regulating virus egress. The data also suggest that a nonenveloped virus utilizes the same cellular exocytic pathway as some enveloped viruses.
MATERIALS AND METHODS
Cell lines and virus.
BSR (derived from baby hamster kidney) cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 5% fetal calf serum (FCS). The stable BSR/NS3 cell line was grown in DMEM-5% FCS supplemented with 7.5 μg/ml of puromycin (Sigma). Cell lines derived from Spodoptera frugiperda Sf21 and Sf9 were growth in TC100 medium (Sigma) supplemented with 10% FCS and in InsectExpress serum-free medium, respectively. The C6/36 insect cell line was maintained in L-15 medium (Gibco) supplemented with 5% FCS.
BTV-1 and reassortant BTV1/10 virus stocks were obtained by infecting BSR cells at low multiplicity of infection (MOI) and harvested when 100% cytopathic effect was evident. Titers of viral stocks were obtained by plaque assay and expressed as PFU per ml as described before (7). Viral stocks were stored at 4°C.
Recombinant baculoviruses based on Autographa californica nuclear polyhedrosis virus were propagated in Sf21 cells, and virus stocks were stored at 4°C.
Cloning and site-directed mutagenesis.
To introduce specific mutations in the BTV-10 NS3 sequence, two mutagenesis methods were used. For CT2 and CT4 mutations, site-directed mutagenesis by asymmetric PCR (21) with modifications was used. This method is based on two steps: the generation of a single-stranded DNA product containing the designed mutation and the use of this product as a primer in a second step of amplification. For CT1 and CT3, the method of Weiner et al. (27) with modifications was used; in this case, two complementary primers bearing the specific mutations are used for amplification. As a template for mutagenesis, the pYM1-NS3 baculovirus transfer vector bearing the sequence for BTV-10 NS3 protein that has described before (13) was used. The CT2 and CT4 mutations were introduced using the primers NS3BamHI (5′GTTAAAAAGGATCCCTGCCATGCTA3′; underlining indicates the BamHI recognition site), CT2 (5′ATATGATTGTGCTGCCATCACTTC3′), and CT4 (5′CTCTGTAAATCACATCCTCACTGCGTC3′). For CT1 and CT3 mutants the following primers were used: CT1-1 (5′CAACAGATAGATATGATAGCGGCCGCAGTGATGAAAAAACAATC3′), CT1-2 (5′GATTGTTTTTTCATCACTGCGGCCGCTATCATATCTATCTGTTG3′), CT3-1 (5′CAACAGATAGATATGATAGCGGCCGCAGTGATGGCAGCACAATC3′), and CT3-2 (5′GATTGTGCTGCCATCACTGCGGCCGCTATCATATCTATCTGTTG3′). All mutated versions of NS3 were sequenced (Value Read service of MWG Biotech) to confirm that only the designed mutations and no others were introduced during the amplification.
To introduce these mutations into pNS3BsmBI (20), which contains the exact copy cDNA of BTV-10 segment S10, the parental plasmid was digested with BstBI and Bsu36I and the original fragment was replaced by the corresponding mutated fragment.
To produce ASAP and GAAP mutants, site-directed mutagenesis was performed according to the method described by Weiner et al. (27) using pNS3BsmBI as the template and the primers NS3ASAP-F (5′CCGCCGCGGTATGCTGCGAGTGCACCGATGCCATCATC3′), NS3ASAP-R (5′CATCGGTGCACTCGCAGCATACCGCGGCGGTTGAGAAATGG3′), NS3GAAP-F (5′CCGCCGCGGTATGCTGGGGCTGCACCGATGCCATCATC3′), and NS3GAAP-R (5′CATCGGTGCAGCCCCAGCATACCGCGGCGGTTGAGAAATGG3′).
NS3-VP2 interaction: pull-down studies.
To analyzed the interaction between wild-type NS3 or mutants and VP2, Sf9 cells were coinfected with a recombinant baculovirus expressing an amino-terminal S-peptide fragment of RNase A-tagged VP2 (S-VP2) (15) and baculoviruses expressing either the wild-type or mutant NS3 proteins. After 48 h, the infected cells were harvested and washed twice with ice-cold phosphate buffered saline (PBS). The cell precipitate was then resuspended in radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate) supplemented with a cocktail of protease inhibitors (Complete, EDTA-free; Roche Applied Science) and incubated in ice for 20 min. Cell lysate fractions were clarified and incubated with 0.2 μg of a rabbit anti S-tag antibody (AbCam) for 2 h at 4°C, and then 10% protein A-Sepharose CL4B (Sigma) was added to the samples and incubated for a further 1 h at 4°C. The beads were centrifuged and then washed three times with buffer containing 100 mM Tris-HCl (pH 8.0), 500 mM LiCl, 1.5% NP-40, and 0.1% bovine serum albumin. To monitor the VP2-NS3 interaction, sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting were performed using a 1:5,000 dilution of a polyclonal mouse antibody against NS3 or the S-tag antibody (1:300) described previously (13).
Confocal microscopy.
The effect of the introduced mutations in NS3 on cellular localization of the protein and relative to VP2 protein was analyzed. Sf cells were infected with recombinant baculoviruses expressing the S-VP2 and either the wild-type or mutant NS3 proteins as before. At 24 h postinfection cells were fixed with 4% (wt/vol) paraformaldehyde in PBS for 20 min, permeabilized with 1% Triton X-100, and immunolabeled using the anti-NS3 and anti S-tag antibodies (1:100). As secondary antibodies, conjugated goat tetramethyl rhodamine isothiocyanate-anti-mouse and goat fluorescein isothiocyanate-anti-rabbit sera diluted 1:100 were used, and the nuclei were stained with 1:2,000 diluted Hoechst 33258 (Sigma). Samples were analyzed under a Zeiss LSM510 confocal microscope, and images were obtained using the LSM510 image browser software.
Transfection of mammalian cells with plasmid derived BTV transcripts: rescue of mutant BTVs.
The T7 BTV transcripts were prepared as described before (6) using a mMESSAGE mMACHINE T7 Ultra kit (Ambion). A total of 400 ng of each of the 10 BTV transcripts was incubated with 0.1 U/μl RNasin Plus (Promega) and mixed with Lipofectamine 2000 (Invitrogen). BSR cells were transfected with the mixture, and at 3 h posttransfection the transfected BSR cells were overlaid with 1.5% agarose type VII (Sigma) containing 2% FCS and further incubated at 35°C for 3 to 5 days posttransfection or until onset of cytopathic effect.
For rescue of the mutants BTVCT2 and BTVCT3, a stable cell line expressing the NS3 protein was constructed. The coding region for BTV-1 NS3 was amplified using the primers BTV1NS3PmeI (5′ggcccggggtttaaacgccatgctatccgggctgatcc3′) and BTV1NS3EcoNot (5′GGCCCGGGGCGGCCGCGAATTCTTATTAGGTTAATGGTAATTCGAAACC3′) and cloned into plasmid pCAGGS/MCS-PM1 (a generous gift from Y. Matsuura [24]) using PmeI and EcoRI restriction enzyme sites to obtain pCAGG/NS3. BSR cells were then transfected with pCAGG/NS3; at 48 h posttransfection the cells were trypsinized and 7.5 μg/ml of puromycin (Sigma) was added to the normal growth medium. Isolated resistant colonies were picked up, and the expression of the NS3 protein was tested using an appropriated antibody. The rescue of the CT2 and CT3 segments was performed in the BSR/NS3 stable cell line using the same procedure as described for normal BSR cells.
Genomic RNA purification and analysis.
Preparation of dsRNA from cells infected with control or mutant BTVs and analysis of those samples were performed as described before (6). cDNA copies of S10 from control or mutant BTVs were obtained by reverse transcription-PCR (RT-PCR) as describe before (6).
Virus growth kinetics and virus release.
For the growth curves of the mutant or control viruses, monolayers of BSR or BSR NS3 cells (as indicated) were infected at MOIs of 0.2 to 0.3. At 0, 16, 24, and 48 h postinfection (as indicated), cells and supernatant were harvested and disrupted by two freeze-thaw cycles and the total titer was determined by plaque assay.
For virus release studies, monolayers of BSR cells were infected at MOIs of 0.5 to 0.6 with BTVCT4, BTVASAP, BTVGAAP, or BTV1/10 (as a control) for about 1 h. Cells were washed with medium and incubated in DMEM supplemented with 2% FCS for 24 h. The supernatants were collected, and the cells were washed twice with DMEM before being harvested. Cell fractions were disrupted by freeze-thawing twice. The titer of each fraction was then determined by plaque assay.
Cell sectioning analysis.
BSR cells were infected and at 24 h postinfection were processed for cell sectioning. Briefly, after three washes with DMEM, monolayers were incubated in 2.5% glutaraldehyde-2% formaldehyde for 15 min followed by 2.5% paraformaldehyde-2.5% glutaraldehyde-0.1% sodium cacodylate (pH 7.4) and postfixed in 1% osmium tetroxide-0.1% sodium cacodylate. Cells were dehydrated in increasing concentrations of ethanol and embedded in epoxy resin (TAAB Laboratories Equipment Ltd., United Kingdom). Ultrathin sections were stained with Reynolds lead citrate (22).
For immunoelectron microscopy, infected cells were fixed with 4% paraformaldehyde followed by 2% formaldehyde-0.1% glutaraldehyde-0.2% HEPES. Ultrathin slices were cut and mounted onto nickel grids. The samples were incubated for 30 min in blocking buffer (PBS, 0.1% bovine serum albumin), followed by overnight incubation at 4°C with mouse anti-NS3 antibody (1:100). As secondary antibody, a 5-nm-gold-conjugated anti-mouse antibody (Sigma) was used.
RESULTS
Mapping the sequences in NS3 responsible for interaction with the outermost protein, VP2.
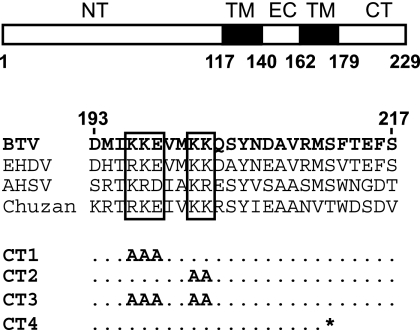
Our previous studies indicated that NS3 interacts with VP2, the outermost protein in the BTV capsid, and that the C-terminal cytoplasmic tail of NS3 mediates NS3-VP2 interactions (3). However, the exact nature of this interaction was not investigated. Therefore, we decided to create NS3 mutants with mutations in the C-terminal cytoplasmic tail in order to map the interacting region in more detail. To identify the putative target sequences, we compared the primary amino acid sequences of four different orbiviruses: BTV, epizootic hemorrhagic disease virus, African horse sickness virus, and Chuzan virus. A sequence alignment (based on ClustalW2) using the C-terminal cytoplasmic domains of NS3 from these viruses revealed certain conserved regions, suggesting that these residues are likely to be important for NS3 function (Fig. 1). Substitution mutations involving these conserved regions, in particular, the hydrophilic charged residues (such as KKE196-198 and KK201-202) likely to be more exposed and available for interactions with other proteins, were made. Each residue of KKE196-198 and KK201-202 was replaced with alanine to create two mutants, CT1 and CT2, and a third mutant, CT3, was created in which all five charged residues were replaced with alanine residue (Fig. 1). A fourth mutant, CT4, was also designed, in which a premature stop codon was inserted in the sequence of NS3. The resulting NS3 protein would lack the C-terminal 18 residues of the protein, but the charged residues highlighted would be left intact.
FIG. 1.
Site-directed mutagenesis based on NS3 sequence alignment. In a schematic representation of NS3 structure domains, the positions of the N-terminal (NT), C-terminal (CT), two transmembrane (TM), and extracellular (EC) domains are indicated. An alignment of a section of the primary amino acid sequences of C-terminal domains of NS3 from BTV (residues 193 to 217), epizootic hemorrhagic disease virus (EHDV), African horse sickness virus (AHSV), and Chuzan virus is shown. Conserved regions that were subjected to mutagenesis are in boxes. Specific changes in each mutant are indicated; *, stop codon.
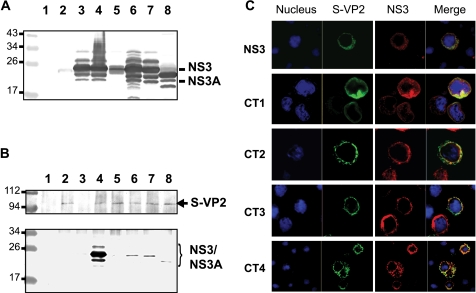
To determine if the changes introduced in the NS3 cytoplasmic domain have any effect on the interaction with VP2, each mutant NS3 was initially coexpressed with VP2 in the baculovirus system. Recombinant baculoviruses expressing NS3 and an S-tagged VP2 (S-VP2) had previously been generated (13, 15) and were used for protein-protein interaction study. Sf cells were coinfected with recombinant baculoviruses expressing the S-VP2 and expressing either the wild-type NS3 or one of the NS3 mutants. The expression of NS3 proteins in the cell lysate was detected by Western blotting using a monospecific NS3 antibody (Fig. 2A). Bands corresponding to NS3, NS3A, and derived glycosylated forms were clearly visible. Except for mutant CT1 protein, which was detected at a lower level in this fraction (Fig. 2A, lane 5), all NS3 mutants were detected at the same level as the unmodified NS3 when either expressed singly or coexpressed with VP2 (Fig. 2A, compare lane 3 with lanes 4, 6, 7, and 8). However, in a pull-down assay, while the wild-type NS3 interacted with VP2 (Fig. 2B, lower panel, lane 4) very strongly, none of the four mutants (Fig. 2B, lower panel, lanes 5 to 8) showed any strong interaction with VP2, indicating that each mutation introduced in the C-terminal cytoplasmic tail of NS3 disrupted the interaction between NS3 and VP2. The faint bands that were visible in each lane (lanes 5 to 8) and in the negative control lane (lane 3), where no VP2 was added, were most likely due to a nonspecific binding of NS3 to the beads.
FIG. 2.
Interaction of BTV NS3 with VP2 in the baculovirus system. Recombinant baculoviruses expressing wild-type (WT) NS3 or mutant CT1, CT2, CT3, or CT4 were obtained and used for coinfection of Sf cells with a recombinant baculovirus expressing an S-tagged VP2. (A) The expression of NS3 was assayed in cell lysates by Western blotting using a specific antibody: uninfected cells (lane 1); singly infected cells expressing only S-VP2 (lane 2) or WT NS3 (lane 3); or coinfected cells expressing S-VP2 and either WT NS3 (lane 4), CT1 (lane 5), CT2 (lane 6), CT3 (lane 7), or CT4 (lane 8). The unlabeled bands are likely to be the glycosylated forms of NS3 and NS3A. (B) Pull-down assay. A specific antibody against the S tag was used to coimmunoprecipitate S-VP2 and NS3 and used in Western blot analysis to detect S-VP2 in the pull-down fractions (upper panel). NS3 and NS3A proteins were detected in pull-down fractions by Western blotting using a specific antibody (lower panel). Noninfected Sf cells (lane 1); cells expressing only S-VP2 (lane 2) or WT NS3 (lane 3); or coinfected cells expressing S-VP2 and either WT NS3 (lane 4), CT1 (lane 5), CT2 (lane 6), CT3 (lane 7), or CT4 (lane 8) are shown. Positions of molecular mass standards are indicated in kDa. (C) Colocalization of NS3 and S-VP2 proteins. Sf cells were coinfected with baculoviruses expressing S-VP2 and either WT NS3 or CT1, CT2, CT3, or CT4 mutant protein, and at 24 h postinfection cells were fixed, permeabilized, and processed for confocal microscopy. S-VP2 (green) and NS3 (red) were detected.
In order to investigate whether these mutations had any effect on the distribution of NS3, Sf cells were coinfected with recombinant baculoviruses expressing VP2 and each of the mutant proteins as described above. The infected cells, at 48 h postinfection, were processed for intracellular staining for NS3 and VP2, and their colocalization was visualized by confocal microscopy. In contrast to the coimmunoprecipitation data, not only wild-type NS3 but also all NS3 mutant proteins were colocalized with VP2. There were no apparent differences in cellular distribution between the wild-type NS3 and NS3 mutants (Fig. 2C). These data suggest that the changes introduced in the C-terminal domain of NS3 did not affect the expression and localization of the protein in insect cells, although they disrupt the interaction between mutant NS3 and VP2 protein.
Generation of infectious BTV single-stranded RNA (ssRNA) with NS3 mutations and rescue of replicating viruses.
Although the protein expression studies indicated that the mutations in the NS3 cytoplasmic domain perturbed the interaction with VP2 in a heterologous expression system, it was necessary to demonstrate the importance of these residues in the context of BTV replication and morphogenesis. To investigate the importance of NS3 and VP2 interaction during virus infection, we have exploited our recently established reverse genetics system which allows manipulation of the viral genome (6). Whether infectious virus bearing an S10 of BTV-10 with either the CT1, CT2, CT3, or CT4 mutation together with nine RNA segments of BTV-1 could be rescued from ssRNA transfection of BSR cells was therefore assessed by the formation of plaques using light microscopy as established previously (6).
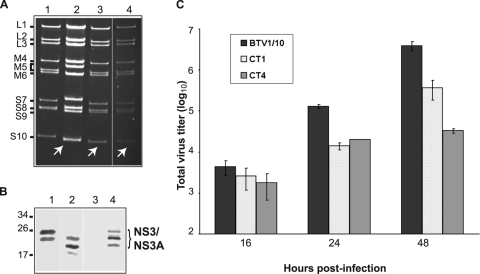
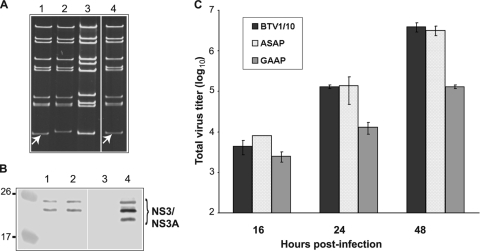
Typical BTV plaques were visible for CT1 (KKE196-198) and CT4 (deletion of last 18 residues) transfections, suggesting that those NS3 mutants were rescued successfully. It was not possible to recover infectious virus from two of the alanine mutations, CT2 (KK201-202) and CT3 (KKE196-198 plus KK201-202), both of which contained the alanine substitution of KK201-202, indicating that replacement of these two charged KK residues by uncharged hydrophobic residues had some effect on the virus growth. To confirm the replication of the two rescued viruses (CT1 and CT4), genomic dsRNAs from cells infected with independent plaques were extracted, purified, and analyzed on a nondenaturing 11% polyacrylamide gel. All nine dsRNA segments of both mutant viruses BTVCT1 and BTVCT4 exhibited the same migration profiles as BTV-1 (Fig. 3A, compare lanes 3 and 4 with lane 1), except for segment S10, which migrated at the same position as that of BTV-10 S10 (Fig. 3A, lane 2), indicating that both mutant viruses were successfully rescued as reassortant viruses.
FIG. 3.
Rescue of mutant BTVCT1 and BTVCT4 viruses. A T7 plasmid-based reverse genetics system was used to generate mutant BTVs (6). (A) Genomic dsRNA was purified from BSR cells infected with BTV-1 (lane 1), BTV-10 (lane 2), BTVCT4 (lane 3), or BTVCT1 (lane 4) and analyzed on an 11% nondenaturing polyacrylamide gel. The fast-migrating S10 from BTV-10 in control or reassortant viruses is indicated with white arrows. (B) Expression of NS3 in insect cells infected with BTVCT1 (lane 1), BTVCT4 (lane 2), or BTV1/10 (lane 4) was detected by Western blotting. A lysate of mock-infected cells (lane 3) is shown as control. (C) Total titers of mutant BTVCT1 and BTVCT4 viruses. BSR cells were infected at a low MOI and harvested at 16, 24, or 48 h postinfection. The total titer at each time point was determined by plaque assay, expressed as PFU/ml, and plotted on a logarithmic scale. Bars represent the averages and standard errors from three replicates.
The presence of the introduced mutations in the rescued BTVCT1 and BTVCT4 viruses was further confirmed by sequencing the RT-PCR products of the purified S10 dsRNA of each mutant virus. No other changes were detected in each S10 sequence (data not shown). The stability of the introduced mutations in BTVCT1 and BTVCT4 viruses was assessed by amplifying them an additional three times in BSR cells followed by sequencing the S10 segment as described previously. In each case, no extra changes in the sequence were detected and the introduced mutations were still present, indicating that those changes in the sequence of NS3 were stable in the context of replication of the viruses.
Virus replication and protein synthesis in cells infected by NS3 mutant viruses.
In order to characterize these mutant viruses, we investigated the protein expression profile during the replication in infected BSR cells. In addition, since mosquito-derived C6/36 cells have been demonstrated to support BTV replication and share properties with vector Culicoides cells, we used C6/36 cells in this experiment in order to compare them with BSR cells. A rescued reassortant virus consisting of nine segments of BTV-1 (L1 to S9) and one unmodified segment 10 from wild-type BTV-10 (BTV1/10) (6) was used as a positive control. When protein profiles of the rescued BTVCT1 and BTVCT4 viruses in C6/36 cells and mammalian BSR cells were analyzed by Western blotting using anti-BTV antibody, both structural and nonstructural proteins could be easily detected (data not shown). There was no apparent difference in the profiles between the mutant viruses and the control BTV1/10. To examine specifically the synthesis of NS3 by each virus, an additional Western blot analysis was performed using anti-NS3 antibody (Fig. 3B). Since NS3 is expressed at a high level in insect C6/36 cells (14), samples from insect C6/36 cells were included in this analysis. Major bands corresponding to NS3 and NS3A proteins were detected in the BTVCT1 cell lysate (Fig. 3B, lane 1). In the BTVCT4 cell lysate the bands were slightly smaller than the bands corresponding to wild-type NS3 protein, as this mutant has a premature stop codon (Fig. 3B, lanes 2 and 4, respectively). These results suggested that the specific point mutations introduced in the S10 sequence did not have any major effects on the normal protein expression profiles of these viruses in either insect or mammalian cells.
The total titer in mammalian cells of each of these viruses at 16, 24, or 48 h postinfection was also determined by plaque assay in comparison to the control reassortant virus (Fig. 3C). Although at 16 h postinfection the titers of mutants and control virus were not significantly different, at 24 h BTVCT1 and BTVCT4 showed a significant defect in replication in mammalian cells. Each of the mutant viruses exhibited about a 10-fold-lower titer (1.4 × 104 and 2 × 104 PFU/ml, respectively) than the control BTV1/10 virus (1.3 × 105 PFU/ml). At 48 h postinfection the difference between BTVCT1 and the control remained the same (3.7 × 105 versus 3.9 × 106 PFU/ml), but there was a more drastic effect on BTVCT4 replication, with a 100-fold difference in titer (3.3 × 104 PFU/ml). These results demonstrated that the replication characteristics of the mutant viruses differ from the kinetics of the control virus in mammalian cells.
Rescue of BTVCT2 and BTVCT3 viruses in a complementing cell line that expresses NS3 constitutively.
Since viruses with CT2 and CT3 mutations in NS3 could not be rescued in normal mammalian cells, we wanted to determine if they could be rescued if wild-type NS3 protein was supplemented in trans. In order to trans-complement the function of NS3 during the rescue experiment with CT2 and CT3 mutants, we decided to generate a stable cell line expressing BTV1 NS3 protein. A similar approach had been successfully used to rescue a mutant reovirus, another member of the Reoviridae family (23).
To create a cell line constitutively expressing NS3, BSR cells were transformed with the plasmid pCAGG/NS3 containing the coding region of NS3 under control of the cytomegalovirus promoter. The expression of NS3 in the BSR-NS3 cell line was analyzed by Western blotting (Fig. 4A) using an antibody against NS3. Two BSR/NS3 clones expressing NS3 protein at a level that could easily be detectable by Western blotting were obtained (Fig. 4A, lanes 1 and 2). One of these clones was chosen, and the expression of NS3 was further confirmed by immunofluorescence (Fig. 4B). To assess whether this stably NS3-expressing cell line could support the replication of BTV, the BSR/NS3 cells were infected with wild-type BTV, and after 48 h of infection, virus propagation in the NS3 stable cell line was monitored by measuring the titers of each infection using a plaque assay as described in Materials and Methods. Although the titer of BTV was slightly lower in the stably NS3-expressing cells than in normal BSR cells, the plaque morphology was normal (data not shown). Therefore, the BSR/NS3 stable cell line was then used for rescuing CT2 and CT3 mutant viruses. These cells were transfected with each mutant S10 segment together with the remaining nine segments of BTV-1 as described earlier. Plaques were evident at 4 days after transfection. Several plaques were isolated and amplified in the same complementing cell line. When the sequences of the S10 from BTVCT2 and BTVCT3 viruses were analyzed, no other changes were detected.
FIG. 4.
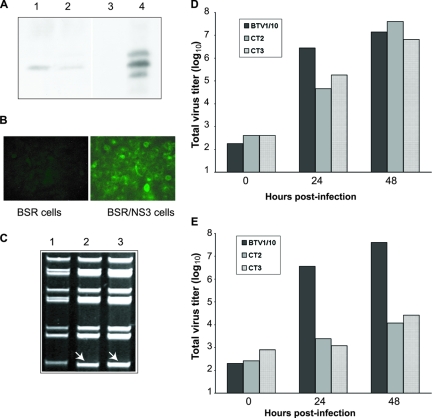
Rescue of mutant viruses BTVCT2 and BTVCT3 in the BSR/NS3 complementing cell line. (A) A BSR cell line stable transfected with NS3 protein was constructed as described in Materials and Methods, and the expression of NS3 was assayed by Western blotting. Two cell clones with detectable NS3 expression (lanes 1 and 2) were obtained. As controls, normal BSR (lane 3) and cells expressing NS3 (lane 4) were included. (B) Immunofluorescence staining of NS3 in the complementing cell line BSR/NS3 (right) or BSR as a control (left) using a specific antibody was performed. (C) Genomic dsRNA from BSR/NS3 cells infected with BTV-1 (lane 1), BTVCT2 (lane 2), or BTVCT3 (lane 3) was purified and run on a nondenaturing polyacrylamide gel. The fast-migrating S10 from mutant viruses is indicated with white arrows. (D and E) Titers of BTVCT2 and BTVCT3 in complementing BSR/NS3 cells (D) or BSR cells (E) at 0, 24, or 48 h postinfection were calculated, expressed as PFU/ml, and plotted on a logarithmic scale.
These viruses, which were amplified only once, were subsequently used to infect BSR/NS3 cells, and the genomic dsRNAs from each infection were purified. It was clear that both BTVCT2 and BTVCT3 could replicate in the BSR/NS3 stable cell line similarly to the control virus in the normal BSR cells (Fig. 4C, compare lanes 2 and 3 with lane 1), indicating that the defect in NS3 protein was successfully complemented by these cells.
To test whether these rescued mutant viruses from the stable cell line could be propagated as efficiently as the wild-type virus, BSR/NS3 cells were infected with either BTVCT2 or BTVCT3 virus. Virus titers were determined at 24 or 48 h postinfection (Fig. 4D). When the titers of these viruses were compared, it was evident that in the complementing BSR/NS3 cell line, the two mutant viruses behaved in a manner similar to the control virus, although at 24 h postinfection, mutant viruses had slightly lower titers than the control virus (Fig. 4D). Thus, the NS3 expressed constitutively by the stable cell line could compensate for the defect in the two NS3 mutant viruses. In parallel, the normal BSR cells were also infected with each of these viruses as a comparison, and virus titers were determined similarly (Fig. 4E). In contrast to the case for the BSR/NS3 cell line, the titers of the two mutants in the normal cells were considerably lower than that of the wild-type virus. At 48 h postinfection the titers of the mutants BTVCT2 and BTVCT3 were 1.2 × 104 and 2.6 × 104 PFU/ml, respectively, which were significantly lower than the control virus titer (4 × 107 PFU/ml). This result indicates that both of these NS3 mutations had a strong effect on virus growth in normal BSR cells.
Effect of mutations in the L domain of NS3 on virus replication.
The data obtained from the studies described above indicated the importance of the interaction between NS3 and VP2, the outermost protein of the virion particle, during virus replication. In addition to the interaction with VP2, NS3 also interacts with cellular proteins that are involved in exocytic pathways. To investigate whether NS3 does indeed act as a bridge between cellular exocytic pathways and the mature virion during virus egress, a second set of mutagenesis experiments was done. The role of the L-domain motif present in the sequence of NS3 during BTV release was first described by our group, and it was shown with the heterologous baculovirus expression system that NS3 interacts with Tsg101 through the L-domain motif, PSAP, present at the N-terminal region (residues 41 to 44) of NS3 (29). However, as a reverse genetics system was not available at the time, it was not possible to study the direct consequence of specific point mutations that disrupt the NS3-Tsg101 interaction in the context of a replicating BTV. To obtain direct evidence of the role of the NS3-Tsg101 interaction in virus egress, the PSAP sequence of the L domain was mutated to either ASAP or GAAP. In vitro, these mutations have been shown to have a drastic effect on Tsg101-NS3 interaction (29).
To examine whether the mutations in the L domain have an effect during virus replication, BSR cells were transfected with nine (L1 to S9) T7-derived ssRNAs of BTV-1 and either BTV-10 S10 ASAP or GAAP mutant segments. In both cases, typical BTV plaques were visualized. Each plaque was then isolated and amplified once in BSR cells, and dsRNA was purified from infected cells. When analyzed by polyacrylamide gel electrophoresis, the migration patterns of S10 of the rescued BTVASAP and BTVGAAP, as expected, were similar to that of wild-type BTV-10 (Fig. 5A, lanes 1, 4, and 3, respectively), while the remaining nine segments corresponded to that of BTV-1 (Fig. 5A, lane 2). To confirm that the introduced mutations were present in each rescued virus, the RT-PCR products of S10 of each virus were sequenced. The sequences revealed that the mutations had not reverted during virus amplification. The synthesis of viral proteins was monitored by Western blot analysis of infected cell lysates using a polyclonal BTV antibody. Both structural and nonstructural proteins were detected in the cell lysates infected with either BTVASAP or BTVGAAP (data not shown), similar to the case for the control virus. The synthesis of NS3 proteins in cell lysates was also detected by a specific NS3 antibody for both BTVASAP and BTVGAAP mutant viruses (Fig. 5B, lanes 1 and 2, respectively) and was equivalent to the control virus NS3 and NS3A (Fig. 5B, lane 4), indicating that the NS3 protein synthesis in infected cells was not perturbed due to the mutations.
FIG. 5.
Recovery of mutant BTVASAP and BTVGAAP viruses. (A) Genomic dsRNA corresponding to BTVASAP (lane 1), BTV-1 (lane 2), BTV-10 (lane 3), or BTVGAAP (lane 4) was analyzed in 11% nondenaturing polyacrylamide gels as previously. The fast-migrating S10 in mutant viruses is indicated with white arrows. (B) NS3 protein was detected by Western blotting in cell lysate fractions from insect cells infected with BTVASAP (lane 1), BTVGAAP (lane 2), or BTV1/10 (lane 4). As a control, lysate from noninfected cells was included (lane 3). (C) Replication kinetics of BTVASAP and BTVGAAP were determined by plaque assay at the indicated time points after infection.
Subsequently, in order to determine the virus replication profile, BSR cells were infected at a low MOI (∼0.1 to 0.2). The replication titers of these mutant viruses were determined as described previously. No major differences were detected between BTVASAP and the control BTV1/10 virus (Fig. 5C). However, BTVGAAP had a reduced replication rate at both 24 and 48 h after infection to the titer of the control virus (Fig. 5C). These results indicated that the mutations of the first two residues of the L domain had an important effect on virus growth.
Mutations in NS3 affect virus egress during virus infection.
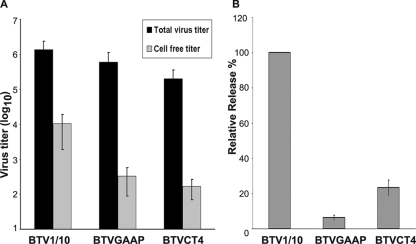
Although during BTV infection of mammalian cells the mature virion particles egress predominantly by cell lysis, a fraction of mature viruses could also be detected in the medium of the infected cultured cells prior to the onset of extensive cytopathic effect. Thus, in order to determine the importance of the NS3-VP2 and NS3-Tsg101 interactions in differential virus egress, we analyzed the virus release from infected mammalian cells. For this experiment, only BTVGAAP and BTVCT4 mutant viruses, as an example, were selected, as they have shown the most dramatic effect. To this end, BSR cells were infected with each mutant virus, both supernatant and cell fractions were harvested separately at 24 h postinfection, and the virus titer of each was determined. Total titer and cell-free titer were calculated and plotted (Fig. 6A). The cell-free fractions of both mutant viruses were lower than that of the control virus (Fig. 6A). In contrast the combined titer of both fractions of each mutant virus showed very little, if any, difference (Fig. 6A). The virus release was calculated as the proportion of cell-free titer to total titer and plotted as a relative percentage of the control virus release (Fig. 6B). A significant reduction in the relative release was observed in repeated experiments for both BTVGAAP and BTVCT4 viruses. On average, BTVGAAP exhibited about a 16 times reduction in the release of the virus relative to the control virus, and the reduction was about 4.3 times for BTVCT4. As a control, the same experiment was performed using BTVASAP virus, which showed no defect in virus growth in previous experiment (Fig. 5C). No defect in virus release was observed (data not shown), suggesting that the effect detected for BTVGAAP is specific. These data indicate that the interaction of NS3 with both VP2 and Tsg101 is essential for virus release.
FIG. 6.
Effect of NS3 mutations on virus release from mammalian cells infected with BTVGAAP or BTVCT4 virus. Monolayers of BSR cells were infected with mutant or control BTV1/10 virus, and at 24 h after infection, supernatant and cell fractions were harvested. The titer in each fraction was determined by plaque assay. (A) The total titer and cell-free titer were plotted on a logarithmic scale. (B) The relative release was calculated as the ratio of cell-free to total virus titer and normalized to 100% for the control virus release. Bars represent the averages and standard errors from three replicates of the experiment.
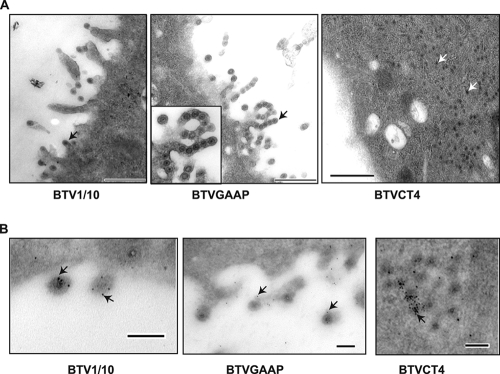
The defect in mutant virus egress was further confirmed by electron microscopy of ultrathin sections of the infected cell. Cells were infected with either BTVGAAP or BTVCT4, and at 24 h postinfection cells were processed for cell sectioning (Fig. 7A). Some particles could be detected budding from the surface of cells infected with the control virus (BTV1/10) (Fig. 7A, left panel), as reported by others (16) for BTV-infected cells. Although BTVGAAP virus has shown a drastic defect in virus release, many nascent particles could be seen on the cell surface, but these particles remained attached, tethered to the cell surface (Fig. 7A, middle panel). This result suggests that there was no apparent effect on the trafficking of the newly synthesized particles to the cell surface even though the final release of these particles seems to be strongly impaired. A completely different phenotype was detected in BTVCT4-infected cells (Fig. 7A, right panel). No membrane-associated particles could be detected, and the newly formed particles were detected only within the cytoplasm of the infected cells, suggesting that the trafficking of these particles to the cell membrane was perturbed.
FIG. 7.
Ultrastructural analysis of cells infected with mutant BTVGAAP or BTVCT4 virus. BSR cells were infected with the indicated mutant viruses or BTV1/10 as a control, fixed at 24 h postinfection, and processed for cell sectioning. (A) Virus particles in cells infected with BTV1/10 (left panel) or BTVGAAP (central panel) can be seen budding from the cell membrane (black arrows) or within the cytoplasm in cells infected with BTVCT4 (right panel, white arrows). Bar, 500 nm. (B) Immunogold staining of NS3 protein. Cell sections of cells infected with control BTV1/10 (left panel) or mutant BTVGAAP (central panel) or BTVCT4 (right panel) virus were incubated with an anti-NS3 antibody and an anti-mouse 5-nm gold-conjugated secondary antibody before staining. Gold particles are indicated by black arrows. Bar, 100 nm.
Finally, the thin sections were labeled with the specific NS3 antibody and 5-nm gold-conjugated anti-mouse antibody in order to localize NS3 protein within the cell sections. Gold particles were detectable in the nascent particles on the cell surface of BTVGAAP virus-infected cells, indicating the NS3 association (Fig. 7B, middle panel). Similarly, gold particle-labeled NS3 was also visualized in the cells infected with the control virus (Fig. 7B, left panel). In contrast, in the cells infected with mutant BTVCT4, NS3 was present predominantly within the cytoplasm along with virus particles but not on the cell surface (Fig. 7B, right panel). This suggests that although the NS3 CT4 mutant and VP2 were not interacting, they could still be trafficked to the same cell compartment during virus morphogenesis.
Thus, the accumulated data demonstrate that mutations in NS3 that affect its interaction either with VP2 or with Tsg101 have a drastic effect on virus release but at different stages during the egress process. While the GAAP mutation causes retention of the virion particles on the infected cell surface, the CT4 mutation, which failed to interact with the viral outermost protein VP2, hinders the trafficking of the virion particles to the surface and relocalizes the virions in the cytoplasm of infected cells.
DISCUSSION
Very little is known about the egress and release of nonenveloped viruses from infected cells. Although cell lysis seems to be the main mechanism used by nonenveloped viruses for release, some viruses also appear able to use nonlytic egress from the infected cells. However, the importance of this mechanism during virus replication is not yet understood. In cells infected with BTV, the onset of a drastic cytopathic effect appears after the virus is detected in the extracellular medium, suggesting that there is some mechanism of trafficking and release. It has already been shown that in addition to cell lysis, BTV is also released from infected cells by budding through the cellular membrane in the early stages of infection (16, 17). BTV is not the only nonenveloped virus that is released from infected cells before cell lysis. Jourdan et al. have demonstrated that rotavirus also is released from infected cells prior to cell lysis (19), and similar results have also been shown for simian virus 40, poliovirus, and parvovirus (2, 9, 25).
In this study, using the reverse genetics system developed for BTV, we have demonstrated that a viral nonstructural protein, NS3, plays a key role in the release and egress of progeny BTV particles from infected cells. Previously, by using yeast two-hybrid and recombinant baculovirus expression systems, we have shown that NS3 interacts with VP2, the outermost capsid protein of the virion particle, through its C-terminal cytoplasmic tail (3). In this study, we have identified domains within the C-terminal cytoplasmic domain of NS3 that are responsible for this interaction by introducing specific point mutations in critical sequences within this region of NS3. Two criteria were considered in designing three (mutations CT1 to CT3) of the four mutations. One is the charged residues, as we hypothesized that these residues are likely to be located on the surface of the protein, and the other is the conservative nature of the residues among different orbiviruses as an indication of constraint that may indicate their importance in interaction with other proteins. The fourth mutation was a drastic one in which a premature stop codon that removed the last 18 residues of the protein was introduced in the coding region of NS3. In the recombinant baculovirus expression system, all four mutant proteins failed to interact with VP2 (Fig. 2B), indicating that the C-terminal cytoplasmic domain is essential for VP2 binding in vitro. This effect was not due to a failure in the expression of the proteins in soluble forms or redistribution in infected cells, as all of them were detected in cell lysates and we were able to colocalize them with VP2 (Fig. 2A and C, respectively).
To understand the significance of VP2-NS3 interaction during the viral replication cycle, we took advantage of the recently developed reverse genetics system (6). This genetics system generates virus by the transfection of cells with transcripts derived from 10 plasmids containing exact cDNA copies of BTV RNA segments under the control of the T7 promoter without using a helper virus or poxvirus-vectored delivery of T7 RNA polymerase. Thus, this system allows the rapid recovery of virus and the effect of mutations to be analyzed in a completely defined genetic background. Each of these four mutants was created in this plasmid-based system. Mammalian cells were transfected with the in vitro-generated transcripts in order to generate the infectious viruses. S10 segments bearing the CT1 or CT4 mutation were successfully rescued, although the onset of the cytopathic effect was delayed, indicating that virus replication was significantly slower (5 to 7 days posttransfection) than that observed for the wild-type segment S10 (3 days posttransfection). The introduced mutations in the S10 correlate with the low rate of replication of BTVCT1 and BTVCT4 (Fig. 3C). However, analysis of the dsRNA and protein expression profiles indicated that both viruses were similar to the control reassortant (BTV1/10) virus and were able to replicate in both mammalian and insect cells (Fig. 3A and B). Thus, it is likely that the delay observed in the onset of the cytopathic effect was due to a defect of the spread of the virus and not due to a deficiency in the replication.
Initially, it was not possible to rescue viruses with segments carrying either the CT2 or CT3 mutation. We hypothesized that the function of NS3 may be complemented in trans by expression of the protein in the cells used for the rescue experiment. To test this hypothesis, we generated a mammalian cell line that constitutively and stably expresses NS3 protein (Fig. 4A and B). In this BSR/NS3 stable cell line both mutant viruses, BTVCT2 and BTVCT3, were successfully rescued, indicating that the function of NS3 protein can be trans-complemented and that the introduced changes in the S10 segment were not affecting packaging signals on BTV genomic RNA. Further, the success in the rescue of the mutant viruses BTVCT2 and BTVCT3 proved that the level of expression of NS3 in our complementing cell line is sufficient to improve the onset of visible plaques and virus growth. When BSR cells were used for BTVCT2 and BTVCT3 amplification, both viruses were able to replicate, although at very low rates. A growth kinetics curve has demonstrated that the replication of BTVCT2 and BTVCT3 was severely affected in mammalian (BSR) cells (Fig. 4E), supporting this hypothesis.
We note that CT2 and CT3 share a common mutation of KK201-202 to AA (Fig. 1). However, CT1 and CT4 have no modification of these two residues. It is also important to note that in particular the residue K201 is conserved for all orbiviruses (Fig. 1). It is therefore likely that these residues are critical to the function of the protein and therefore it was not possible to rescue these mutant viruses in normal BSR cells.
Our data strongly suggest that the interaction between NS3 and VP2 is not essential for virus replication, viral protein expression, or genomic dsRNA synthesis but that it is important for virus release.
Our laboratory has presented evidence of the interaction between NS3 and certain components of cellular transport machinery, in particular with ESCRT-I component Tsg101 through an L domain (PSAP) present in NS3 (29). However, it was not possible then to obtain direct evidence of the importance of this interaction in the context of a replicating virus. In that published report, a change in the first residue of the L-domain proline to alanine had little effect on the NS3-Tsg101 interaction, but a simultaneous change of the first two residues (PSAP to GAAP) had a drastic effect that abolished this interaction completely. Mutant BTV viruses bearing the mutation ASAP or GAAP in the L domain of NS3 were therefore generated, and their replication capabilities, in both insect and mammalian cells, were assessed. No major defects were detected in the genomic dsRNA synthesis or protein expression profiles, indicating that the replication of these viruses was not affected (Fig. 5A and B). However, when the replication kinetics were analyzed, it was clear that BTVGAAP virus had a strong delayed multiplication cycle in comparison to that of the control virus, suggesting that this mutation may have an impact on a late stage of virus morphogenesis, such as the virus release process.
The efficiency of virus release was therefore investigated further for BTVGAAP and BTVCT4 rescued viruses that had shown a strong defect in virus growth in normal mammalian cells, by measuring virus titers. Although both mutant viruses exhibited overall comparable titers in plaque assay compared to the control virus, the efficiency of virus egress was severely affected. When the ratio of cell-free fractions to total virus yield was calculated for each mutant virus and the relative percentage compared to the control virus was determined, a reduction in virus release was evident. For BTVCT4 virus a reduction of 4.3-fold was detected, and a much more stronger effect was observed for BTVGAAP virus, which had a 16-times reduction in virus release. These data strongly suggest the importance of the interaction of NS3 with both VP2 and Tsg101 during virus release. It is interesting to note that the reduction in the relative release of the mutant viruses BTVGAAP and BTVCT4 in mammalian cells at 24 h postinfection was mainly due to failure in the budding process, suggesting that BTV uses a highly controlled release pathway for spread of the newly generated progeny virus particles early in infection.
To substantiate these results, ultrathin sections of infected cells were prepared and virus distributions were visualized by electron microscopy. In cells infected with BTVCT4, virion particles remained predominantly within the cytoplasm, and no particles could be seen budding from the cell membrane. An immunogold analysis of BTVCT4-infected cells demonstrated that mutant NS3 is detected along with the particles in the cytoplasm of the cell. Our laboratory reported previously that NS3 colocalizes with the BTV structural protein VP2 (3) and with the other outer capsid protein, VP5 (5). Altogether, these results suggest that NS3 is the driving force that assists the newly synthesized virions for trafficking to the cell membrane.
In contrast to the case for BTVCT4, virion particles in cells infected with BTVGAAP were detected budding from the plasma membrane, similar to the case for wild-type virus. However, a defect in the final release was evident, as these particles were tethered in the cell membrane. There is ample evidence for the importance of the recruitment of components of the ESCRT-I to -III machinery through the L domains during the egress of enveloped viruses (10, 11). Recent studies have suggested that the ESCRT-III complex, recruited by ESCRT-I and Alix, an essential protein of the MVB pathway, is the membrane fission machinery that mediates the separation of nascent virus from cell membranes (8). Thus, it is tempting to hypothesize that the defect in the release of the virions in BTVGAAP-infected cells might be mainly due to a defect in the recruitment of Tsg101 and probably other components of the ESCRT complex that are essential for the late stages of virus release. The data presented here indicate that the maturation of BTVCT4 and BTVGAAP mutant viruses differs significantly due to the effect of mutation in two different functional sites of NS3 and that NS3 indeed is involved in virus trafficking and budding.
It is important to note that it has been reported previously that the cytoplasmic carboxyl-terminal end of NS3 is directly responsible for the interaction with VP2 (3). In a subsequent report (29) it was also shown that the L domain (PSAP motif) at the amino terminus of NS3 is solely responsible for Tsg101 interaction. The different phenotypes displayed by BTVCT4 and BTVGAAP also suggest that the NS3-VP2 interaction in BTVGAAP-infected cells was not affected, since the mature virus particles were able to reach the cell surface, in contrast to the phenotype displayed by BTVCT4 virus. These findings suggest that mutations in the L domain did not interfere with NS3-VP2 interaction. Thus, our data clearly show two different aspects of BTV morphogenesis. One is that the NS3-Tsg101 interaction is important for the budding of the virus from the plasma membrane. The second is that NS3 is involved in trafficking virus particles to the membrane via VP2 interaction.
These accumulating data support the model that we have proposed previously, that NS3 is a bridge between the newly virus particle and the cell components during virus egress (3). However, it is still not clear why a nonenveloped virus such as BTV hijacks components of the MVB machinery during virus release.
In this study, the effects of these mutations in virus egress have been examined only in mammalian cells and not in insect vector cells, where the virus release is mostly nonlytic. These mutations may have much more severe effect on BTV release from insect cells. Future studies using insect vector cells are planned.
Acknowledgments
We thank I. M. Jones (University of Reading, United Kingdom) and R. Noad for critical reading of the manuscript and Maria McCrossan (LSHTM) for technical help with electron microscopy experiments.
This work was funded by the BBSRC (United Kingdom).
Footnotes
Published ahead of print on 15 April 2009.
REFERENCES
- 1.Bansal, O. B., A. Stokes, A. Bansal, D. H. L. Bishop, and P. Roy. 1998. Membrane organization of bluetongue virus nonstructural glycoprotein NS3. J. Virol. 723362-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bär, S., D. L. Rommelaere, and J. P. Nüesch. 2008. Vesicular egress of non-enveloped lytic parvoviruses depends on gelsolin functioning. PLoS Pathog. 4e1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaton, A. R., J. Rodriguez, Y. K. Reddy, and P. Roy. 2002. The membrane trafficking protein calpactin forms a complex with bluetongue virus protein NS3 and mediates virus release. Proc. Natl. Acad. Sci. USA 9913154-13159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharya, B., R. J. Noad, and P. Roy. 2007. Interaction between bluetongue virus outer capsid protein VP2 and vimentin is necessary for virus egress. Virol. J. 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharya, B., and P. Roy. 2008. Bluetongue virus outer capsid protein VP5 interacts with membrane lipid rafts via a SNARE domain. J. Virol. 8210600-10612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyce, M., C. P. Celma, and P. Roy. 2008. Development of a reverse genetics system for bluetongue virus: recovery of infectious virus from synthetic RNA transcripts. J. Virol. 828339-8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyce, M., and P. Roy. 2007. Recovery of infectious bluetongue virus from RNA. J. Virol. 812179-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlton, J. G., and J. Martin-Serrano. 2007. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 3161908-1912. [DOI] [PubMed] [Google Scholar]
- 9.Clayson, E. T., L. V. Brando, and R. W. Compans. 1989. Release of simian virus 40 virions from epithelial cells is polarized and occurs without cell lysis. J. Virol. 632278-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demirov, D. G., and E. O. Freed. 2004. Retrovirus budding. Virus Res. 10687-102. [DOI] [PubMed] [Google Scholar]
- 11.Freed, E. O. 2004. Mechanisms of enveloped virus release. Virus Res. 10685-86. [DOI] [PubMed] [Google Scholar]
- 12.Freed, E. O. 2002. Viral late domains. J. Virol. 764679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.French, T. J., S. Inumaru, and P. Roy. 1989. Expression of two related nonstructural proteins of bluetongue virus (BTV) type 10 in insect cells by a recombinant baculovirus: production of polyclonal ascitic fluid and characterization of the gene product in BTV-infected BHK cells. J. Virol. 633270-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guirakhoo, F., J. A. Catalan, and T. P. Monath. 1995. Adaptation of bluetongue virus in mosquito cells results in overexpression of NS3 proteins and release of virus particles. Arch. Virol. 140967-974. [DOI] [PubMed] [Google Scholar]
- 15.Hassan, S. H., and P. Roy. 1999. Expression and functional characterization of bluetongue virus VP2 protein: role in cell entry. J. Virol. 739832-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyatt, A. D., B. T. Eaton, and S. M. Brookes. 1989. The release of bluetongue virus from infected cells and their superinfection by progeny virus. Virology 17321-34. [DOI] [PubMed] [Google Scholar]
- 17.Hyatt, A. D., A. R. Gould, B. Coupar, and B. T. Eaton. 1991. Localization of the non-structural protein NS3 in bluetongue virus-infected cells. J. Gen. Virol. 722263-2267. [DOI] [PubMed] [Google Scholar]
- 18.Hyatt, A. D., Y. Zhao, and P. Roy. 1993. Release of bluetongue virus-like particles from insect cells is mediated by BTV nonstructural protein NS3/NS3A. Virology 193592-603. [DOI] [PubMed] [Google Scholar]
- 19.Jourdan, N., M. Maurice, D. Delautier, A. M. Quero, A. L. Servin, and G. Trugnan. 1997. Rotavirus is released from the apical surface of cultured human intestinal cells through nonconventional vesicular transport that bypasses the Golgi apparatus. J. Virol. 718268-8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, J. W., and P. Roy. 1986. Nucleotide sequence of a cDNA clone of RNA segment 10 of bluetongue virus (serotype 10). J. Gen. Virol. 672833-2837. [DOI] [PubMed] [Google Scholar]
- 21.Perrin, S., and G. Gilliland. 1990. Site-specific mutagenesis using asymmetric polymerase chain reaction and a single mutant primer. Nucleic Acids Res. 187433-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reynolds, E. S. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17208-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roner, M. R., and W. K. Joklik. 2001. Reovirus reverse genetics: incorporation of the CAT gene into the reovirus genome. Proc. Natl. Acad. Sci. USA 988036-8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tani, H., Y. Komoda, E. Matsuo, K. Suzuki, I. Hamamoto, T. Yamashita, K. Moriishi, K. Fujiyama, T. Kanto, N. Hayashi, A. Owsianka, A. H. Patel, M. A. Whitt, and Y. Matsuura. 2007. Replication-competent recombinant vesicular stomatitis virus encoding hepatitis C virus envelope proteins. J. Virol. 818601-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tucker, S. P., C. L. Thornton, E. Wimmer, and R. W. Compans. 1993. Vectorial release of poliovirus from polarized human intestinal epithelial cells. J. Virol. 674274-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verwoerd, D. W., H. J. Els, E. M. De Villiers, and H. Huismans. 1972. Structure of the bluetongue virus capsid. J. Virol. 10783-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiner, M. P., G. L. Costa, Schoelttin, W., Cline, J., E. Mathur, and J. C. Bauer. 1994. Site directed mutagenesis of double stranded DNA by the polymerase chain reaction. Gene 151119-123. [DOI] [PubMed] [Google Scholar]
- 28.Williams, R. L., and S. Urbe. 2007. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 8355-368. [DOI] [PubMed] [Google Scholar]
- 29.Wirblich, C., B. Bhattacharya, and P. Roy. 2006. Nonstructural protein 3 of bluetongue virus assists virus release by recruiting ESCRT-I protein Tsg101. J. Virol. 80460-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu, X., S. Y. Chen, H. Iwata, R. W. Compans, and P. Roy. 1992. Multiple glycoproteins synthesized by the smallest RNA segment (S10) of bluetongue virus. J. Virol. 667104-7112. [DOI] [PMC free article] [PubMed] [Google Scholar]