Abstract
The majority of human microRNAs (miRNAs) are located in the introns of other genes (A. Rodriguez, S. Griffiths-Jones, J. L. Ashurst, and A. Bradley, Genome Res. 14:1902-1910, 2004). Based on the discovery that artificial insertion of pre-miRNAs in introns did not hamper mRNA production and that the miRNA-harboring introns were spliced more slowly than the adjacent introns, a model was previously proposed in which Drosha crops the pre-miRNA and the two cropped fragments from the pre-mRNA are subsequently trans spliced (Y. K. Kim and V. N. Kim, EMBO J. 26:775-783, 2007). However, the molecular basis for this model was not elucidated. To analyze the molecular mechanism of intronic miRNA processing, we developed an in vitro system in which both pre-miRNA processing and mRNA splicing are detected simultaneously. Our analysis using this system showed that pre-miRNA cropping from the pre-mRNA could occur kinetically faster than splicing. Glycerol gradient sedimentation experiments revealed that part of the pre-miRNA was cofractionated with the spliceosome. Furthermore, coimmunoprecipitation experiments with an anti-Drosha antibody demonstrated that Drosha was associated not only with the cropping products but also with a Y-shaped branch intron and a Y-shaped splicing intermediate. These results provide a molecular basis for the postulated existence of a pathway in which the Microprocessor complex becomes associated with the spliceosome, pre-miRNA cropping occurs prior to splicing, and trans splicing takes place between the cropped products.
MicroRNAs (miRNAs) are short, noncoding RNAs that mediate posttranscriptional gene silencing via base pairing with their target mRNAs (for reviews, see references 7, 23, and 24). The majority of miRNAs are transcribed by RNA polymerase II (31) and matured through several processing steps in both the nucleus and cytoplasm (6, 30, 31). Primary miRNA transcripts (pri-miRNAs) are cleaved by Drosha, an RNase III-type enzyme in the nucleus (29). Drosha functions in the context of a large protein complex, termed the Microprocessor complex, that also contains DGCR8/Pasha as well as several splicing factors (8, 11, 13, 28). Cleavage of pri-mRNAs by the Microprocessor complex results in the production of short, stem-loop-shaped RNAs called pre-miRNAs. These RNAs are subsequently exported to the cytoplasm by exportin 5 (5, 32, 52). In the cytoplasm, pre-miRNAs are cleaved by the cytoplasmic RNase III-type enzyme Dicer (4, 12, 16, 20, 26). The cleavage reaction, referred to as dicing, yields short RNA duplexes and, subsequently, incorporation of one strand of each duplex into the RNA-induced silencing complex (21, 46). The RNA-induced silencing complex binds to its target mRNAs through base pairing with the 3′ untranslated regions and induces translational repression, mRNA cleavage, or mRNA degradation (42, 49).
Most miRNAs were formerly thought to have their own transcriptional units in the intergenic regions. However, recent studies revealed that about 80% of miRNAs are encoded in the intronic regions of protein-encoding or noncoding genes in mammals (25, 43) and that this number is 75% in Xenopus tropicalis (48), implying that the phenomenon of the majority of miRNAs being encoded in the introns seems to be evolutionarily conserved in vertebrates. The expression of intronic miRNAs largely coincides with that of their host genes (2), suggesting that miRNAs are transcribed from their host gene promoters and processed posttranscriptionally from the host pre-mRNAs. In an analogy to intronic small nucleolar RNA (snoRNA), it was initially assumed that pre-miRNAs are produced from the excised introns after the completion of splicing, thereby requiring splicing for miRNA production (7, 10, 50). However, very recent studies showed that pre-miRNA could be produced by Drosha even by splicing-deficient pre-mRNA, and an artificial insertion of pre-miRNA sequences into the introns did not show a significant effect on host gene mRNA production in vivo (25). It was also demonstrated that the coding sequence of the miRNA-containing intron is spliced out more slowly than the adjacent introns (25). Based on these findings, a model was proposed in which Drosha cleaves the pre-miRNA before the intron is spliced out and the two RNA fragments, produced by the pre-miRNA cropping from the pre-mRNA, might subsequently be trans spliced (25). Yet the molecular basis for the model remained unclear. In the present study, we took advantage of an in vitro system with HeLa cell nuclear extracts in which pre-miRNA processing and mRNA splicing can be detected simultaneously, and our analysis using the in vitro system showed that pre-miRNA cropping could occur prior to mRNA splicing. Moreover, the presence of miRNA in the intron slowed splicing while increasing pre-miRNA production. The glycerol gradient sedimentation experiment demonstrated that part of the pre-miRNA and the two cropped fragments of pre-mRNA were cofractionated with the spliceosome. Immunoprecipitation with an anti-Drosha antibody precipitated not only Drosha-cleaved products derived from pre-mRNA but also a Y-shaped intron and a Y-shaped splicing intermediate, which is indicative of trans splicing. Furthermore, anti-Drosha antibody precipitated U2, U5, and U6 but not U1 or U4 spliceosomal U snRNP from HEK293T whole-cell lysates. Taken together, these results strongly suggest that the Microprocessor complex associates with the spliceosome and that the resultant large complex mediates both pre-miRNA cleavage and mRNA splicing, allowing production of two different functional RNAs from one pre-mRNA molecule.
MATERIALS AND METHODS
Plasmid construction.
Part of the human EML2 gene was PCR amplified from HeLa cell genomic DNA. The primers used for the EML2 pre-mRNA template vector were 5′-AAAGGATCCATGAGTAGCTTTGGAGCTGG-3′ (forward) and 5′-AAACTCGAGCTACCACACTGAAGATAACTTCTTTGG-3′ (reverse). The PCR product was inserted between the BamHI site and the XhoI site of myc-pCDNA3 vector (33, 36) and verified by sequencing. The resultant plasmid was named pEML2-330-WT. PCR-based site-directed mutagenesis was performed to generate a splicing-defective construct named pEML2-330 Δss. The primers used for 5′ splice site mutation were 5′-TTTAACCTCTACTTTCGCAGACCACCCC-3′ (forward) and 5′-GCTCCAAAGCTACTCATGGATCC-3′ (reverse), and the primers for 3′ splice site mutation were 5′-AAAAATGCCTCAAACCAAAGAAGTTATCTT-3′ (forward) and 5′-AAGGACAAAGAAAAAAAAAGGGTC-3′ (reverse). To generate miRNA lacking the EML2 gene construct (pEML2-330 ΔmiR), PCR-based deletion was carried out by using primers 5′-CTTACTCGGCCCCGTTTTCATCGG-3′ (forward) and 5′-ATCACACAGCACGAAGTGGCAGGG-3′ (reverse). To prepare pmyc-hnRNP K-7, part of the human hnRNP K gene was also PCR amplified from HeLa cell genomic DNA and cloned between the BamHI site and XhoI site of myc-pCDNA3 vector. The primers used for PCR were 5′-AAAGGATCCGAAGATCGGATCATTACCATTAC-3′ (forward) and 5′-AAACTCGAGTCTTGCATTAGAATCCTTCAACAT-3′ (reverse). PCR-based site-directed mutagenesis with a QuikChange kit (Stratagene) was performed to generate a splicing-defective construct, named pmyc-hnRNP K-7 Δss. The primers used for splice site mutations were 5′-CTATCTCAAAGCTAATCTTAATCTGTTCTGTTCTGCAGC-3′ (for 5′-splice-site mutation) and 5′-TCGTTCAACATGTGCATAGCGGTTCACAGGATAAAAGAA-3′ (for 3′-splice-site mutation). To generate miRNA lacking the hnRNP K gene construct (pmyc-hnRNP K-7 ΔmiR), PCR-based deletion was carried out by using primers 5′-TGAATCTTCTATATTTTCTACAGGCTCTGT-3′ (forward) and 5′-GACAAATGATTGGTGCTGTAAAATGCAGCA-3′ (reverse). In order to prepare pδ-Crystallin-miR330, the DNA fragment containing pre-miR330 was amplified by PCR, using pEML2-330-WT as a template. The primers used for this PCR were 5′-AAAGAGCTCTGATCTTTGGCGATCACTGC-3′ (forward) and 5′-AAAGAGCTCCGAGTAAGGGGCAGAGC-3′ (reverse). After digestion with SacI, the PCR fragment was inserted into the SacI site of pSP-14-15 (45).
The plasmid named pδ-Crystallin-miR126 was also prepared by the same procedure. The DNA fragment containing pre-miR126 was amplified by PCR, using a genomic fragment of the EGFL7 gene as a template. The primers used for this PCR were 5′-AAAGAGCTCGCCACGCCTCCGCTGGCGACGGGA-3′ (forward) and 5′-AAAGAGCTCGGCGTTTTCGATGCGGTGCCGTGG-3′ (reverse). After digestion with SacI, the fragment was also inserted into the SacI site of pSP-14-15 (45). The plasmid named pδ-Crystallin-miR126 Δss was generated by PCR-based site-directed mutagenesis with a QuikChange kit (Stratagene) by using primers 5′-GAGAAGCTGAAACCTTTTCTGGAGGGTAGAAATC-3′ (forward) and 5′-CTCCATGTTCTCCGTGTTGACGGGCAAAGAGGAGAGGC-3′ (reverse). To prepare pδ-Crystallin-miR126 ΔmiR, PCR-based deletion was carried out by using primers 5′-TGAATCTTCTATATTTTCTACAGGCTCTGT-3′ (forward) and 5′-GACAAATGATTGGTGCTGTAAAATGCAGCA-3′ (reverse). The Flag-hDBR1 plasmid was prepared by inserting human debranching enzyme 1 (hDBR1) cDNA between the BamHI and XhoI sites of Flag-pCDNA3 (17).
In vitro transcription and splicing assay.
As templates for in vitro transcription, pEML2-330 WT, pEML2-330 Δss, pEML2-330 ΔmiR, pmyc-hnRNP K-7 WT, pmyc-hnRNP K-7 Δss, and pmyc-hnRNP K-7 ΔmiR were linearized with XhoI. The pSP14-15 (45), pδ-Crystallin-miR330, pδ-Crystallin-miR126 WT, pδ-Crystallin-miR126 Δss, and pδ-Crystallin-miR126 ΔmiR plasmids were linearized with SmaI. In vitro transcription and purification of the transcribed RNAs were performed as described previously (19, 45). HeLa cell nuclear extracts for in vitro splicing were obtained from Cilbiotech (Belgium). In vitro splicing assays were performed with 10-μl mixtures as described elsewhere (19, 45), and the mixtures were incubated for 10 to 120 min at 30°C. RNA products were analyzed by electrophoresis on a 6% acrylamide-7 M urea gel followed by autoradiography. The RNA bands were analyzed and quantitated with BAS-2500 (Fuji Film, Japan) and Image Gauge, version 3.45 (Fuji Film, Japan). The averages were calculated from three independent experiments, and statistical analysis (Student's t test) was performed using Excel (Microsoft).
Glycerol gradient centrifugation.
Glycerol gradient centrifugation was performed as described previously, with slight modification (27, 45). The splicing reaction was carried out in a 200-μl mixture as described elsewhere (19), using 600,000 cpm of 32P-labeled pre-mRNA, followed by incubation with 0.65 mg/ml heparin sodium salt (Sigma) for 10 min on ice. The mixture obtained by the procedure described above was separated in a 10% to 30% glycerol gradient in a gradient buffer (20 mM HEPES-KOH, pH 7.9, 50 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol) at 40,000 rpm in an SW41 rotor at 4°C for 3 h. Fractions (600 μl each) were collected, and the RNAs in each fraction were recovered and analyzed by electrophoresis on a 6% acrylamide-7 M urea gel followed by autoradiography.
Immunoblotting and immunoprecipitation.
Whole-cell extracts of HEK293T cells were prepared as described previously (17). To perform immunoblotting of Drosha, 16 μl of a HeLa cell nuclear extract and 20 μl of HEK293T whole-cell extract were subjected to 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Immunoblotting was performed as described previously (47). A 1:500 dilution of anti-Drosha antibody (Abcam) was used as the primary antibody. The primary antibody was probed with a 1:5,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit antibody (Jackson Immunoresearch), followed by detection with enhanced chemiluminescence (GE Healthcare).
Immunoprecipitation of RNA was performed essentially as described elsewhere (15, 19). Briefly, 1 μg of anti-Drosha antibody (Abcam) or anti-mouse immunoglobulin G control antibody (Sigma) was immobilized on 20 μl of protein A Sepharose (GE Healthcare). Either 50 μl of in vitro splicing reaction mix or 200 to 250 μl of a glycerol gradient fraction was incubated with the protein A Sepharose beads as described previously (15, 19). After recovery, the precipitated RNAs were analyzed in a 6% denaturing polyacrylamide gel. For U snRNA coprecipitation experiments, whole-cell extracts from 293T cells were prepared as described previously (15, 19). Briefly, HEK293T cells from three 10-cm plates were recovered by pipetting and washed twice with 500 μl of ice-cold phosphate-buffered saline. Cells were then resuspended in 400 μl of ice-cold buffer E (20 mM HEPES-KOH, pH 7.9, 100 mM KCl, 0.2 mM EDTA, 10% glycerol, 1 mM dithiothreitol). Five micrograms of anti-Sm (Y12; Thermo Fisher Scientific, Cheshire, United Kingdom) antibody, anti-Drosha antibody (Abcam), or anti-mouse immunoglobulin G control antibody (Sigma) was immobilized on 20 μl of protein A Sepharose (GE Healthcare). The protein A Sepharose beads were then mixed with 120 μl of HEK293T whole-cell extract as described previously (15, 19). After recovery, the precipitated RNAs were separated in an 8% denaturing polyacrylamide gel and transferred to a Hybond N+ membrane (GE Healthcare). Northern blot analysis and specific probes for U snRNAs were described previously (53).
Cell culture and transfection.
HEK293T cells were maintained in Dulbecco's modified Eagle's medium (Nacalai-Tesque, Japan) supplemented with 10% fetal bovine serum (Equitech-Bio, TX) and 1% penicillin-streptomycin (Nacalai-Tesque, Japan) under a 5% CO2 atmosphere in an incubator maintained at a temperature of 37°C. Cultured cells were transfected by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations.
Flag-hDBR1 protein expression in HEK293T cells and purification from whole-cell lysates.
Wild-type (WT) hDBR1 protein with a Flag tag was transiently expressed in HEK293T cells by transfection with the Flag vector to provide a control. HEK293T cells were transfected, and whole-cell lysates were prepared from the transfected cells as previously described (17). Flag-tagged proteins were purified from the whole-cell lysates by using anti-Flag M2 resin (Sigma) as described previously (41).
In vitro debranching assays.
A lariat intron of chicken δ-crystallin pre-mRNA was prepared by an in vitro splicing reaction. Briefly, the splicing reaction mixture was applied to a 6% denaturing acrylamide gel and the band corresponding to lariat intron RNA was excised and recovered from the gel. Y-shaped RNAs (designated a and b, respectively, in Fig. 6 and 7) were precipitated from the in vitro splicing mixture with anti-Drosha antibody and were also recovered from the gel. Forty to eighty attomoles of the RNA was incubated for 30 min at 30°C with approximately 50 μg of purified Flag-tagged proteins in 10 μl of debranching reaction mixture (20 mM HEPES-KOH, pH 7.9, 40 mM KCl, 3 mM MgCl2, 2.5 mM EDTA, 4% glycerol) (37, 39). The RNA was then recovered by proteinase K digestion, phenol extraction, and ethanol precipitation, followed by analysis using 6% denaturing polyacrylamide gel electrophoresis.
FIG. 6.
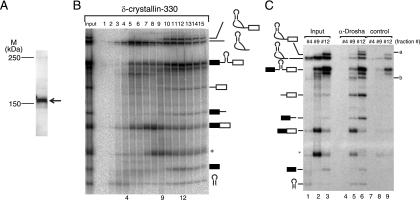
Identification of Drosha-associated RNAs. (A) Western blotting of HeLa cell nuclear extracts with anti-Drosha antibody. The band corresponding to Drosha is indicated by an arrow. The positions of size markers are indicated on the left. (B) Fractionation of in vitro splicing mixture by glycerol gradient centrifugation. In vitro splicing was performed with δ-crystallin-330 pre-mRNA and a 20-min incubation. The input lane contained 5% of the total splicing mixture. The structure of each RNA product is shown schematically at the right side of the panel. (C) Immunoprecipitation of RNAs from the fractions shown in panel B. The identities of RNA products are shown on the left side of the panel. The unidentified RNAs precipitated efficiently by anti-Drosha antibody are indicated on the right (a and b). The input lanes contained 5% of the fractions.
FIG. 7.
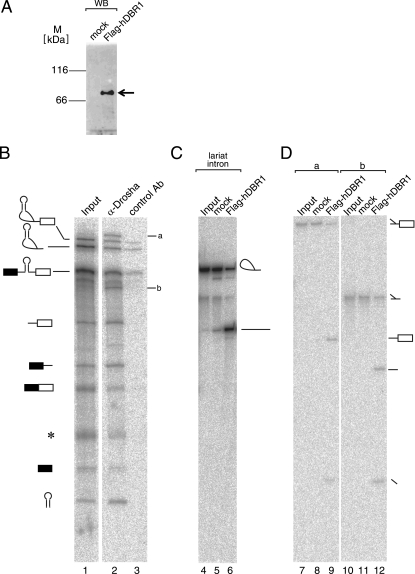
Drosha associates with intronic RNAs that contain a Y-shaped structure. (A) Purification of Flag-hDBR1 protein from HEK293T cells. The purified protein was detected by immunoblotting with anti-Flag M2 antibody (Sigma). An arrow indicates the band corresponding to Flag-hDBR1. The lane marked “mock” contained proteins purified from Flag vector-transfected HEK293T cells. The positions for size standards are shown on the left. (B) Immunoprecipitation of RNAs from in vitro splicing reaction mixture. 32P-labeled δ-crystallin-330 pre-mRNA was incubated with HeLa cell nuclear extracts under splicing conditions, followed by immunoprecipitation with the anti-Drosha and control antibodies (rabbit anti-mouse antibody). The precipitated RNAs were analyzed in a 6% denaturing acrylamide gel. The identities of the precipitated RNAs are shown on the left side of the panel. Two RNA products identified in Fig. 6C are indicated (a and b). The input lane contained 5% input RNA. (C) Debranching reaction of lariat intron RNA derived from δ-crystallin pre-mRNA. The lariat intron RNA was incubated with recombinant Flag-hDBR1 protein. After the reaction, RNAs were analyzed by 6% denaturing polyacrylamide gel electrophoresis. The structure of the RNA corresponding to each band is demonstrated schematically to the right side of the panel. (D) Debranching reactions of unidentified RNA products precipitated by anti-Drosha antibody. The a and b RNAs shown in panel B were incubated with recombinant Flag-hDBR1 protein. RNAs were analyzed and shown as in panel C.
RESULTS
In vitro splicing of a pre-mRNA harboring a pre-miRNA in its intron.
In order to analyze the relationship between miRNA processing and mRNA splicing, we first tried to devise an in vitro reaction system in which both pri-miRNA production and splicing of pre-mRNA could be detected simultaneously. We picked up several protein-encoding genes that harbor pre-miRNA sequences in their introns, including the genes for EGFL7, MYH6, hnRNP K, and EML2. Since expression of these miRNAs is correlated with that of the host genes (data not shown), they are most likely transcribed as a part of the host pre-mRNAs (43). We used PCR to amplify portions of these genes spanning the pre-miRNA-containing intron and the adjacent two exons and then prepared the corresponding pre-mRNAs by in vitro transcription and tested them for their in vitro splicing efficiency. Consequently, the pre-mRNA derived from the human EML2 gene was spliced most efficiently (data not shown) and therefore was chosen for further analyses. The intron of the pre-mRNA derived from human EML2 contains miR-330 (Fig. 1A, EML2-330 WT).
FIG. 1.
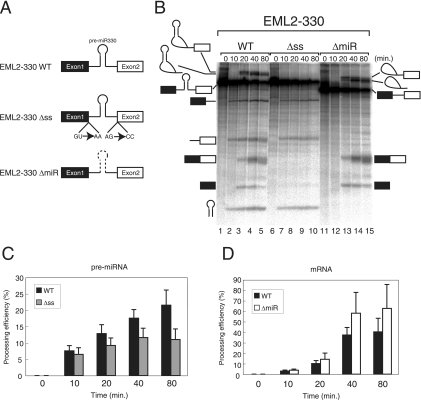
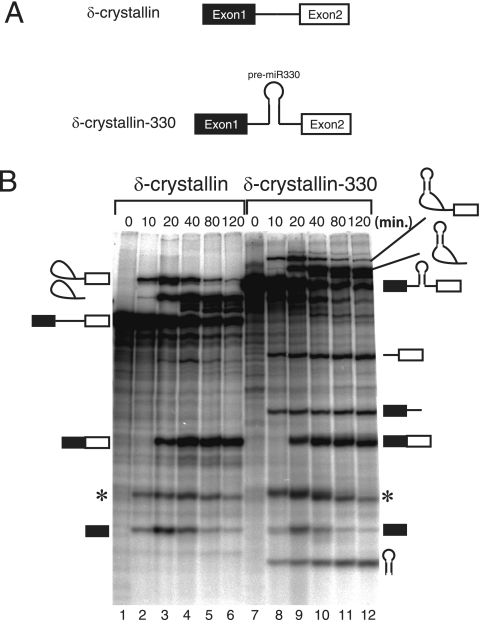
Intronic pre-miRNA can be processed from pre-mRNA in vitro. (A) Schematic representation of EML2 pre-mRNA and derivatives used for in vitro splicing reactions. Boxes and lines represent exons and introns, respectively. Mutations at both splice sites are shown for EML2-330 Δss pre-mRNA. A dashed line in EML2-330 ΔmiR indicates a deletion of the pre-miR330 stem-loop from pre-mRNA. (B) In vitro splicing reactions in HeLa cell nuclear extracts with EML2 WT and derivative pre-mRNAs shown in panel A. The identity of each RNA product is shown schematically at both sides. (C) Quantitation of mRNA production from either EML2-330 WT pre-mRNA (black bars) or EML2-330 Δss pre-mRNA (gray bars). Averages and standard deviations obtained from three independent experiments are shown. (D) Quantitation of pre-miRNA production from either EML2-330 WT pre-mRNA (black bars) or EML2-330 ΔmiR pre-mRNA (white bars). Averages and standard deviations from three independent experiments are shown.
As shown in Fig. 1B, the spliced products from the pre-mRNA were detected by 20 min and never were seen by 10 min (lanes 3 to 5). In addition to those of the splicing products, three additional major RNA bands were also detected (Fig. 1B, lanes 2 to 5). The smallest band corresponded to pre-miR330 RNA, while the others corresponded to the two RNA fragments that resulted from the cleavage of the pre-mRNA by Drosha. These RNA products appeared as early as 10 min after the start of analysis, before the splicing products were observed (Fig. 1B, lane 2). These results indicated that the system enabled detection of both pre-miRNA processing and mRNA splicing and that splicing was not required for pre-miRNA processing in vitro.
In order to confirm this finding, we used a mutant version of the pre-mRNA that had mutations at both the 5′ and 3′ splice sites (Fig. 1A, EML2-330 Δss). While, as expected, this pre-mRNA did not undergo mRNA splicing (Fig. 1B, lanes 6 to 10), the three RNA products produced by pre-miRNA processing could be detected. Moreover, depletion of ATP from the in vitro splicing of the WT pre-mRNA yielded the same results (data not shown) (also see Fig. 2 and 4). These results taken together indicate that excision of pre-miRNA from pre-mRNA by Drosha does not require an mRNA splicing reaction, as was the case in vivo (25), in strong contrast to snoRNA processing from the introns, which requires splicing (7, 10, 50). Careful comparison of the efficiency of pre-miRNA production from each pre-mRNA revealed that pre-miRNA production from EML2-330 WT pre-mRNA was reproducibly higher than that from EML2-330 Δss pre-mRNA (Fig. 1B, lanes 1 to 10, and Fig. 1C). The significance value P for EML2-330 was calculated to be 0.063. In addition, we could also observe reproducibly higher production of pre-miRNA from hnRNP K-7 Δss pre-mRNA than from hnRNP K-7 WT pre-mRNA (see Fig. S1A in the supplemental material) (P = 0.023). These results demonstrate that the presence of pre-miRNA in the spliceable intron stimulates the production of pre-miRNA from the intron, as reported for the in vivo situation (9, 25). This prompted us to test the effect of the presence of pre-miRNA in the intron on mRNA splicing. To this end, we prepared EML2-330 ΔmiR pre-mRNA that was lacking the pre-miR330 RNA sequence in the intron (Fig. 1A, EML2-330 ΔmiR). When in vitro splicing efficiencies were carefully compared between EML2-330 WT and EML2-330 ΔmiR, the splicing efficiency of EML2-330 WT was reproducibly lower than that of EML2-330 ΔmiR (Fig. 1B and D) (P = 0.18). A similar result was obtained when we compared in vitro splicing efficiencies of myc-hnRNP K-7 WT and ΔmiR pre-mRNAs (see Fig. S1B in the supplemental material) (P = 0.038). These results suggest that pre-miRNA excision from the intron caused a delay of mRNA splicing, as suggested in an in vivo system (25). Since the results obtained with our in vitro system were consistent with the results obtained with cultured cells, we concluded that this system was suitable for analyzing the relationship between splicing and pre-miRNA processing.
FIG. 2.
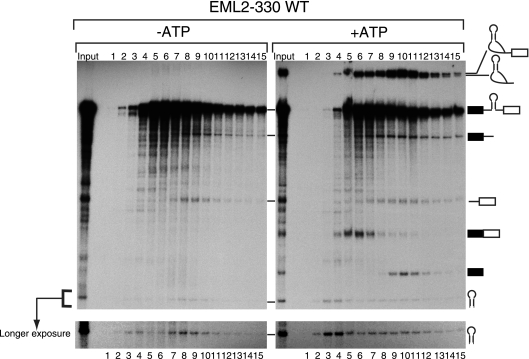
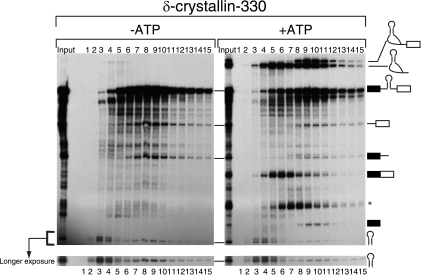
Pre-miRNA is cofractionated with the spliceosome in an ATP-dependent manner. In vitro splicing with EML2-330 WT pre-mRNA was carried out with or without exogenous ATP. The reaction mixture was separated by glycerol gradient centrifugation. The RNAs in each fraction were recovered and analyzed by 6% denaturing polyacrylamide gel electrophoresis. The identity of each RNA product is shown schematically at the right side. A longer-exposed image of the pre-miR330 area is shown at the bottom.
FIG. 4.
Comigration of pre-miRNA produced from heterologous pre-mRNA with the spliceosome. In vitro splicing reactions with δ-crystallin-330 pre-mRNA were carried out with HeLa cell nuclear extracts, and RNA products were analyzed as described in the legend to Fig. 2. The identity of each RNA product is shown schematically at the right side. The asterisk indicates an aberrant splicing product. Images with a longer exposure time to show pre-miR330 are presented at the bottom.
Pre-miRNA is cofractionated with the spliceosome in an ATP-dependent manner.
The results shown in Fig. 1 suggested a functional association between the miRNA processing machinery and the splicing machinery. In order to determine whether the miRNA processing machinery and the spliceosome are physically associated, we employed a glycerol gradient sedimentation method to separate complexes involved in pre-miRNA processing and mRNA splicing. When the reaction mixture was separated by this method, the spliceosome fraction corresponded to fraction 10 (Fig. 2, +ATP panel, lane 10), based on the peak of the splicing intermediates. Importantly, the two cropped fragments of pre-mRNA and the pre-miRNA also peaked in this fraction, suggesting a possibility that the pre-miRNA excision occurred in association with the spliceosome. The splicing reaction did not occur when we carried out the same sedimentation experiment in the absence of exogenous ATP (Fig. 2, −ATP panel), and although pre-miRNA processing was detected, the peak of pre-miR330 RNA and the two cropped fragments of pre-mRNA were found in fractions 4 and 8, not in fraction 10. Thus, the occurrence of the pre-miRNA excision in association with the spliceosome cannot be explained by the Microprocessor complex migrating by itself near the spliceosome fraction, as it is responsible for catalyzing pre-miRNA excision. The results obtained indicated that a part of pre-miRNA and the two cropped fragments of pre-mRNA were cofractionated with the spliceosome in a splicing-dependent manner, suggesting that the Microprocessor complex is physically associated with the spliceosome during the splicing of introns containing pre-miRNAs.
Pre-miR330 RNA is excised from the intron of a heterologous pre-mRNA prior to being spliced in vitro.
Pre-miRNA processing as a whole occurred kinetically faster than pre-mRNA splicing (Fig. 1B). However, the possibility remained that the EML2 pre-mRNA was simply not an efficient substrate for in vitro splicing and that the splicing reaction was slow enough for Drosha to crop pre-miRNA prior to splicing. In order to determine whether that was true, we artificially inserted the pre-miR330 sequence into the intron of a pre-mRNA derived from the chicken δ-crystallin gene (45), since chicken δ-crystallin pre-mRNA has been shown to be spliced very efficiently in vitro (19, 45). When the resultant pre-mRNA, termed δ-crystallin-330, and the parental δ-crystallin pre-mRNA (Fig. 3A) were subjected to in vitro splicing, both pre-mRNAs were spliced efficiently (Fig. 3B). The δ-crystallin-330 pre-mRNA also yielded the pre-miRNA and the cropped pre-mRNA products as early as 10 min into the reaction (Fig. 3B, lanes 7 to 12), when the splicing intermediates, but not the final spliced product, were observed (Fig. 3B, lane 8). These results indicated that pre-miRNA could be cropped even from a heterologous intron and that the cropping could be faster than mRNA splicing, even with an efficient splicing substrate. Based on these findings, we concluded that pre-miRNA cropping from pre-mRNA occurred prior to mRNA splicing, at latest by the end of the first reaction. We also prepared δ-crystallin-126 pre-mRNA, which has the miR-126 sequence in its intron. The pre-miR-126 RNA was also excised from the δ-crystallin intron (data not shown). When we compared the efficiencies of both mRNA splicing and pre-miRNA production between the WT and the Δss or Δmi mutant, the presence of miR-126 in the δ-crystallin intron slowed splicing, while pre-miRNA production was increased (see Fig. S1C and D in the supplemental material) (P = 0.024 and P = 0.038, respectively). These results are consistent with those shown in Fig. 1, suggesting that both stimulation of pre-miRNA cropping and inhibition of mRNA splicing by the presence of pre-miRNA in the intron are common phenomena, rather than specific to miR-330. We then analyzed spliceosome formation with δ-crystallin-330 pre-mRNA by glycerol gradient sedimentation in the presence or absence of exogenous ATP, as shown in Fig. 2 (Fig. 4). In the absence of ATP, no mRNA production was observed, and the pre-miRNA peaked in fractions 3 and 4 and in fractions 8 and 9 (Fig. 4, −ATP panel). In the presence of ATP, in contrast, pre-miRNA peaked in fractions 3 and 4 and in fraction 10, with the latter being the spliceosome fraction, judging from the peak of the splicing intermediates, i.e., the lariat intron with the 3′ exon and the 5′ exon (Fig. 4, +ATP panel). The cropped pre-mRNA products were also detected in fraction 10. These findings demonstrated that part of the pre-miRNA appeared in the same fraction as the spliceosome in a splicing-dependent manner, suggesting that the Microprocessor complex was physically associated with the spliceosome and that the resultant complex performed both pre-miRNA processing and mRNA splicing.
FIG. 3.
Pre-miRNA can be excised from heterologous pre-mRNA in vitro. (A) Schematic representation of both WT and pre-miR330-inserted chicken δ-crystallin pre-mRNAs. (B) In vitro splicing reactions in HeLa cell nuclear extracts, using pre-mRNAs shown in panel A. The identity of each RNA product is shown at both sides of the panel. The band marked by asterisks is a product of aberrant splicing.
The Microprocessor complex appears to assemble on the pre-mRNA in association with the spliceosome.
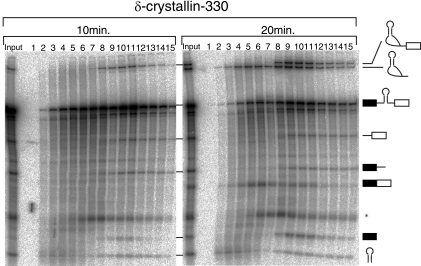
As shown in Fig. 2 and 4, the heaviest peak of pre-miRNA appeared in a splicing-dependent manner (fraction 10 of the +ATP panels in Fig. 2 and 4). Two other peaks (fractions 3 and 4 and fractions 8 and 9 of the −ATP panels in Fig. 2 and 4) were observed in the absence of ATP. For one of these peaks (fractions 8 and 9 of the −ATP panels in Fig. 2 and 4), the pre-miRNA and the cropped pre-mRNA products were also detected, indicating that the cropping of pre-miRNA is highly likely to have taken place in these fractions. Based on these results, it was possible that the pre-miRNA was cropped first by the Microprocessor complex alone in the lighter complex and that the spliceosome was assembled on the complex formed by the cropped RNAs and the Microprocessor complex, which resulted in the formation of the heavier complex. However, we were able to rule that possibility out. We performed glycerol gradient fractionation of an in vitro splicing reaction mixture, as shown in Fig. 5. Within 10 min, only the first step of splicing occurred, and the accumulation of splicing intermediates, such as the 5′ exon and the lariat intron with a 3′ exon, but not of the final spliced products, was observed (Fig. 5, 10 min; also see Fig. 3B, lane 8). In contrast, the second splicing step took place during the second 10 min of the incubation, and the final spliced products were detected within 20 min after the start of the incubation (Fig. 5, 20 min; also see Fig. 3B, lane 9). Under these conditions, part of the pre-miRNA had already peaked within 10 min, in fraction 11, in which the splicing intermediates, and hence the spliceosome, accumulated (Fig. 5, 10 min). In addition, the cropped pre-mRNA products were also seen in this spliceosome fraction. These results demonstrated that excision of pre-miRNA could take place by the end of the first step of splicing, at which point a pre-miRNA peak was detectable. Combined with the result that pre-miRNA cropping can occur prior to the splicing reaction, it is likely that the Microprocessor complex and the spliceosome assemble simultaneously on the pre-mRNA, rather than sequentially, and that pre-miRNA cropping occurs prior to the second step of splicing, in a large complex that contains both the spliceosome and the Microprocessor complex.
FIG. 5.
Pre-miRNA excision from pre-mRNA is kinetically faster than mRNA splicing. In vitro splicing reactions with δ-crystallin-330 pre-mRNA were performed with 10 min or 20 min of incubation. After the reaction, RNA products were analyzed as described in the legend to Fig. 2. The identity of each RNA product is shown schematically at the right. An asterisk shows an aberrant splicing product.
Identification of RNAs that associate with Drosha during cropping and splicing reactions in vitro.
To further confirm that the Microprocessor complex containing Drosha interacts with the spliceosome, we performed immunoprecipitation of RNAs from in vitro splicing reactions, using an anti-Drosha antibody. The antibody to Drosha was able to detect the endogenous Drosha protein in HeLa cell nuclear extracts (Fig. 6A). To obtain fractions for the immunoprecipitation experiments, we first performed a similar glycerol gradient experiment to that shown in Fig. 4 and 5 (Fig. 6B). Please note that the fraction numbers in the figure do not correspond to the fraction numbers in the previous figures. We chose two peak fractions for the pre-miRNA (Fig. 6, fractions 4 and 12). We also randomly chose a third fraction, fraction 9. Fraction 4 contained mostly the lariat intron and the pre-miRNA, whereas fraction 9 contained mainly the pre-mRNA, the lariat intron, and the mRNA (Fig. 6C, lanes 1 and 2). Fraction 12 contained the pre-mRNA, the lariat intron, the mRNA, and the splicing intermediates (Fig. 6C, lane 3), indicating that fraction 12 contained the spliceosome. The Drosha-cropped products, i.e., the pre-miRNA and two fragments of pre-mRNA, were efficiently precipitated from both fractions 9 and 12 (Fig. 6C, lanes 5, 6, 8, and 9), suggesting that Drosha, highly likely as a part of the Microprocessor complex, held these fragments together even after the cropping of the pre-miRNA from the pre-mRNA. The pre-mRNA, the mRNA, and the splicing intermediates were also precipitated (Fig. 6C, lanes 5, 6, 8, and 9), but not as efficiently as the Drosha-cleaved products. In addition to those RNAs, two novel RNA products, designated a and b, were highly enriched in the RNAs precipitated by anti-Drosha (Fig. 6C, lane 6). Almost no RNA precipitation was observed from fraction 4 (Fig. 6, lanes 4 and 7). These results strongly suggested that pre-miRNA cropping by Drosha took place in the large complex that consisted of the Microprocessor complex and the spliceosome, since Drosha-cropped fragments of pre-mRNA, the pre-miRNA, and the splicing intermediates were precipitated with Drosha.
Drosha associates with a Y-shaped intron, a Y-shaped splicing intermediate, and spliceosomal U snRNAs.
It was suspected from their unusual electrophoretic mobilities that the newly identified RNAs, a and b, might have a branch structure (Fig. 6C, lane 6). In order to test this possibility, we performed in vitro debranching assays. Large-scale immunoprecipitation like that performed for Fig. 6C was first performed (Fig. 7B). RNA bands a and b were recovered from the denaturing gel and subjected to debranching with recombinant hDBR1 (22). Flag-tagged hDBR1 proteins, which were detectable by anti-Flag antibody, were recovered from transfected HEK293T cells by use of anti-Flag M2 resin (Fig. 7A). If the RNA had a Y-shaped structure, the enzyme would debranch it, yielding two RNA fragments (18). As a control, we used the lariat intron derived from the chicken δ-crystallin pre-mRNA. The RNA was efficiently debranched by Flag-hDBR1 but not by a mock control, and the resultant linear intron showed faster migration in a denaturing gel (Fig. 7C, lanes 4 to 6). When bands a and b were incubated with Flag-hDBR1, they both yielded two bands after the reaction (Fig. 7D, lanes 9 and 12), indicating that these RNAs had a Y-shaped structure. These RNA products shared a common smaller fragment that corresponded to the portion of the pre-mRNA spanning from the 5′ splice site to the branch point (Fig. 7D, lanes 9 and 12). The other fragment corresponded to the 3′-half fragments of intron, with or without the 3′ exon (Fig. 7D, lanes 9 and 12). These results strongly suggest that the Drosha protein associates not only with pre-miRNA processing products but also with the Y-shaped lariat intron and the Y-shaped lariat intermediate, most likely through the interaction with the spliceosome.
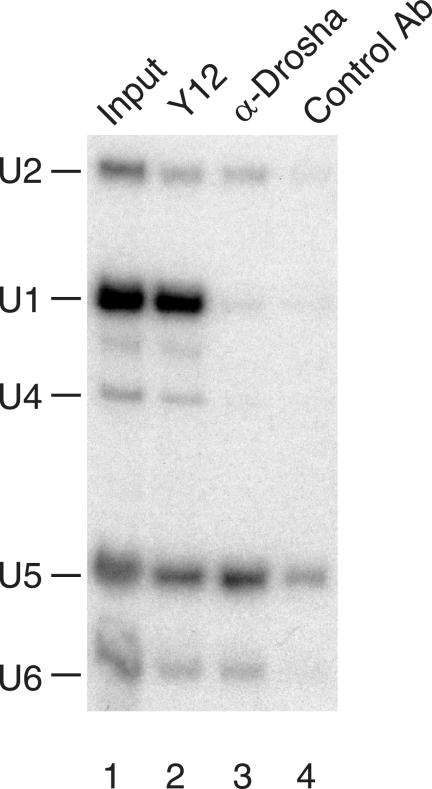
In order to further show the interaction between the Microprocessor complex and the spliceosome, we carried out immunoprecipitation experiments with HEK293T whole-cell extracts, followed by Northern blotting analysis using U snRNA-specific probes. As a positive control, anti-Sm antibody (Y12) was used, and it precipitated all spliceosomal U snRNAs (Fig. 8, lane 2). The anti-Drosha antibody precipitated U2, U5, and U6 but not U1 or U4 snRNA (lane 3), in contrast to the control antibody (lane 4). These results strongly suggested that the Microprocessor complex associates with the activated spliceosome in vivo. These results in sum provide molecular evidence that the Microprocessor is functionally associated with the spliceosome and that the two RNA fragments corresponding to the pre-miRNA-cropped pre-mRNA can be trans spliced (see Discussion for details).
FIG. 8.
Drosha associates with spliceosomal U snRNPs. Northern blotting analysis was performed to observe the spliceosomal U snRNAs in the immunoprecipitated fractions. Whole-cell extracts from HEK293T cells were subjected to immunoprecipitation with anti-Sm antibody (Y12) (lane 2), anti-Drosha (lane 3), or control antibody (lane 4). Coprecipitated RNAs were detected by Northern blot analysis probing for the five spliceosomal U snRNAs. The input lane contained 5% of the HEK293T whole-cell extract used for immunoprecipitation.
DISCUSSION
Model of coordination between pre-miRNA processing and pre-mRNA splicing.
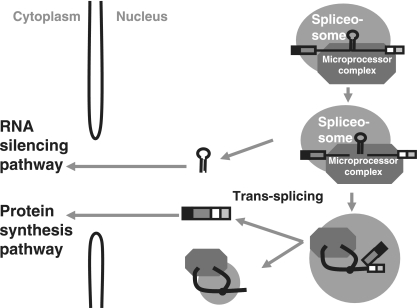
In this study, we analyzed intronic miRNA processing by using an in vitro system that enables detection of the production of both pre-miRNA and mRNA by incubating pre-mRNAs that harbor a pre-miRNA in the intron with HeLa cell nuclear extracts under splicing conditions (Fig. 1B). The production of the pre-miRNA was detected earlier than that of the mRNA, and part of the pre-miRNA was cofractionated with the spliceosome in the glycerol gradient sedimentation experiments. In addition, anti-Drosha antibody precipitated not only the pre-miRNA and the cropped pre-mRNA fragments but also the Y-shaped intron-containing products. These findings strongly suggest that the Drosha-containing complex, the Microprocessor complex, associates with the spliceosome prior to the splicing reaction and that the combined complex mediates both pre-miRNA cleavage and the following splicing reaction to produce two functional RNAs from one pre-mRNA (Fig. 9).
FIG. 9.
Model for the processing of miRNA from an intron. The model shows the association of the spliceosome and the Microprocessor complex during intronic miRNA processing. Details are described in the text.
At first, it was thought that the intronic miRNAs of mammals are processed from the excised introns by analogy to the biogenesis of the guide snoRNAs (7, 10, 50). For guide snoRNA processing in mammals, snoRNAs are transcribed as a part of the host gene introns. After being spliced, the excised introns are debranched by DBR1, and linearized introns are processed from both ends by exonucleases (22, 34). The snoRNA region of the intron is bound by several snoRNP proteins, which protect it from degradation (10, 50). Thus, splicing is required for guide snoRNA biogenesis in mammals. In contrast, for intronic pre-miRNA processing, both our own findings and those of others indicate that splicing is not essential for pre-miRNA production, since pre-miRNA was found to be produced from pre-mRNAs without splicing both in vivo (25) and in vitro (Fig. 1, 2, and 4).
Our immunoprecipitation experiments with an anti-Drosha antibody demonstrated that Drosha held the cleaved pre-mRNA molecules together even after cropping. It is likely that Drosha holds the cropped products through DGCR8, since DGCR8 but not Drosha was shown to interact with RNA directly (51). The anti-Drosha antibody also precipitated the Y-shaped intron and the Y-shaped splicing intermediate. Based on these results, it is reasonable to postulate that a pathway exists in which the pre-miRNA is cleaved first from the pre-mRNA and a splicing reaction takes place between the two cleaved fragments of pre-mRNA tethered by Drosha (Fig. 9). Of course, this model does not exclude the existence of another pathway in which the pre-miRNA is produced from the excised intron. In fact, it is possible that our in vitro system overlooked the latter pathway, since the intron turnover process appears to be inefficient under in vitro splicing conditions (data not shown). Similarly, we cannot completely exclude the possibility that Drosha cleaves the lariat splicing intermediate to produce the Y-shaped intermediate, although that seems unlikely. It is of interest to try to detect Y-shaped intron products in vivo. Depletion of both DBR1 and the exosome by RNA interference in mammalian cells is expected to help us to detect those otherwise unstable products.
Mechanisms of physical and functional interactions between the Microprocessor complex and the spliceosome.
Although our findings strongly suggest an association between Drosha, most likely in the context of the Microprocessor complex, and the spliceosome, how the Microprocessor complex is recruited to the spliceosome is unknown. Interestingly, several RNA binding proteins and RNA helicases that are involved in pre-mRNA splicing have been identified as components of the Microprocessor complex on the basis of their coprecipitation with Drosha and DGCR8/Pasha (8, 11, 13, 28). Most recently, it was demonstrated that Drosha became localized in SC35 speckles when pri-miRNA was overexpressed (35, 40). These results support an association of the Microprocessor complex with the splicing machinery in vivo. It would be interesting to know which splicing factor(s) can interact with Drosha and/or DGCR8/Pasha. Since cropping by Drosha seems to occur by the end of the first step of the reaction (Fig. 3 and 5), the Microprocessor complex is recruited to the early spliceosome. This should be related to results showing that the presence of an intron stimulates pre-miRNA production in vivo (9, 25) and in vitro (Fig. 1). It is still not known when and by which early spliceosomal complex, such as the H, E, A, or B form, Drosha cleaves pre-miRNA. We observed that cropping of myc-hnRNP K-7 pre-mRNA by Drosha occurred in the presence of 2.5 mM EDTA (N. Kataoka and M. Ohno, unpublished data). Under these conditions, the first reaction of splicing is inhibited and the spliceosome is accumulated (1). These results suggest that cropping by the Microprocessor complex takes place at latest by B complex formation. The coprecipitation of U2, U5, and U6 but not U1 or U4 snRNP with Drosha (Fig. 8) also supports this idea. On the other hand, the presence of miRNA in the intron slows splicing of the intron. The molecular mechanisms responsible for stimulation and suppression are currently unknown. The spliceosome may induce a conformational change in the pre-mRNA for the favor of Microprocessor complex recruitment. The recruited Microprocessor complex may inhibit splicing and crop the pre-miRNA. After cropping of pre-miRNA by Drosha, rearrangement of the complex may take place to allow completion of the splicing reaction. This may cause a delay of splicing of the pre-miRNA-harboring intron. RNA helicases that associate with Drosha and/or DGCR8/Pasha are strong candidates for such rearrangement.
Relationship with the mirtron pathway.
A novel class of miRNA-harboring introns was very recently reported for worms, flies, and mammals. This new type of intron is called a “mirtron” (3, 38, 44). Mirtrons are approximately 70 nucleotides in length, which is much smaller than the average size of canonical introns. The mirtrons are excised from pre-mRNAs in a lariat form, and the 5′ end of the intron forms a duplex with the 3′ end after debranching of the intron. The entire region of the mirtron then forms a stem-loop structure that corresponds to pre-miRNA. Thus, splicing and debranching are essential for mirtron processing, but Drosha activity is not required. Indeed, Drosha knockdown had almost no effect on the mirtron pathway (38). On the other hand, the results presented here strongly suggest that the Microprocessor complex interacts with the spliceosome. It is likely that the Microprocessor complex is incorporated into the spliceosome by recognizing the stem-loop structure of pre-miRNA in the context of pre-mRNA. These results suggest that the Microprocessor complex is not recruited to mirtrons. It was demonstrated that the Microprocessor complex requires three helical turns in its substrates for effective cleavage (14). Mirtrons are too short to form helical turns and to be cleaved effectively by Drosha.
Intronic pre-miRNAs that may contain their own promoters.
In this study, we analyzed an intronic miRNA whose expression correlated with its host gene's expression. This miRNA is expected to be transcribed as a part of the host gene introns under the control of host gene promoters. However, it is possible that there are some miRNAs that have their own promoters even though they are located in the intronic regions. In fact, we performed computational analysis using the ENCODE chromatin immunoprecipitation database with mammalian cells (http://genome.ucsc.edu/cgi-bin/hgTracks) and found that five miRNAs (miR-15b, 16-2, 335, 125b-2, and 199a-1) may have their own promoters in host gene introns (unpublished results). In those cases, pre-miRNAs are transcribed under the control of both the host promoters and their own promoters and therefore are likely to be regulated differently. Most recently, it was reported that pre-miRNA processing by the Microprocessor complex is coupled with transcription (35, 40). Further analysis of transcription units of these miRNAs should shed light on the global mechanisms of intronic miRNA expression in mammals.
Supplementary Material
Acknowledgments
We thank Makoto Kitabatake for suggestions and critical comments on the manuscript and Rei Yoshimoto for technical assistance.
This work was supported by CREST, JST, and by grants from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan. M.F. was supported by the 21st Century COE Program of the Ministry of Education, Culture, Sports, Science and Technology. N.K. was supported by the Program for Improvement of Research Environments for Young Researchers from Special Coordination Funds for Promoting Science and Technology (SCF), commissioned by MEXT, Japan.
Footnotes
Published ahead of print on 6 April 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abmayr, S. M., R. Reed, and T. Maniatis. 1988. Identification of a functional mammalian spliceosome containing unspliced pre-mRNA. Proc. Natl. Acad. Sci. USA 857216-7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baskerville, S., and D. P. Bartel. 2005. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berezikov, E., W. J. Chung, J. Willis, E. Cuppen, and E. C. Lai. 2007. Mammalian mirtron genes. Mol. Cell 28328-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409363-366. [DOI] [PubMed] [Google Scholar]
- 5.Bohnsack, M. T., K. Czaplinski, and D. Gorlich. 2004. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, X., C. H. Hagedorn, and B. R. Cullen. 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 101957-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullen, B. R. 2004. Transcription and processing of human microRNA precursors. Mol. Cell 16861-865. [DOI] [PubMed] [Google Scholar]
- 8.Denli, A. M., B. B. Tops, R. H. Plasterk, R. F. Ketting, and G. J. Hannon. 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432231-235. [DOI] [PubMed] [Google Scholar]
- 9.Du, G., J. Yonekubo, Y. Zeng, M. Osisami, and M. A. Frohman. 2006. Design of expression vectors for RNA interference based on miRNAs and RNA splicing. FEBS J. 2735421-5427. [DOI] [PubMed] [Google Scholar]
- 10.Filipowicz, W., and V. Pogacic. 2002. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 14319-327. [DOI] [PubMed] [Google Scholar]
- 11.Gregory, R. I., K. P. Yan, G. Amuthan, T. Chendrimada, B. Doratotaj, N. Cooch, and R. Shiekhattar. 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432235-240. [DOI] [PubMed] [Google Scholar]
- 12.Grishok, A., A. E. Pasquinelli, D. Conte, N. Li, S. Parrish, I. Ha, D. L. Baillie, A. Fire, G. Ruvkun, and C. C. Mello. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 10623-34. [DOI] [PubMed] [Google Scholar]
- 13.Han, J., Y. Lee, K. H. Yeom, Y. K. Kim, H. Jin, and V. N. Kim. 2004. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 183016-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han, J., Y. Lee, K. H. Yeom, J. W. Nam, I. Heo, J. K. Rhee, S. Y. Sohn, Y. Cho, B. T. Zhang, and V. N. Kim. 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125887-901. [DOI] [PubMed] [Google Scholar]
- 15.Hanamura, A., J. F. Caceres, A. Mayeda, B. R. Franza, Jr., and A. R. Krainer. 1998. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA 4430-444. [PMC free article] [PubMed] [Google Scholar]
- 16.Hutvagner, G., J. McLachlan, A. E. Pasquinelli, E. Balint, T. Tuschl, and P. D. Zamore. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293834-838. [DOI] [PubMed] [Google Scholar]
- 17.Kataoka, N., and G. Dreyfuss. 2004. A simple whole cell lysate system for in vitro splicing reveals a stepwise assembly of the exon-exon junction complex. J. Biol. Chem. 2797009-7013. [DOI] [PubMed] [Google Scholar]
- 18.Kataoka, N., S. Hashimoto, and Y. Shimura. 1993. Heat treatment of nuclear extract alters selection of the 3′ splice site in pre-mRNA splicing. Biochem. Biophys. Res. Commun. 190223-228. [DOI] [PubMed] [Google Scholar]
- 19.Kataoka, N., J. Yong, V. N. Kim, F. Velazquez, R. A. Perkinson, F. Wang, and G. Dreyfuss. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6673-682. [DOI] [PubMed] [Google Scholar]
- 20.Ketting, R. F., S. E. Fischer, E. Bernstein, T. Sijen, G. J. Hannon, and R. H. Plasterk. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 152654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khvorova, A., A. Reynolds, and S. D. Jayasena. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115209-216. [DOI] [PubMed] [Google Scholar]
- 22.Kim, J. W., H. C. Kim, G. M. Kim, J. M. Yang, J. D. Boeke, and K. Nam. 2000. Human RNA lariat debranching enzyme cDNA complements the phenotypes of Saccharomyces cerevisiae dbr1 and Schizosaccharomyces pombe dbr1 mutants. Nucleic Acids Res. 283666-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, V. N. 2005. Small RNAs: classification, biogenesis, and function. Mol. Cell 191-15. [PubMed] [Google Scholar]
- 24.Kim, V. N., and J. W. Nam. 2006. Genomics of microRNA. Trends Genet. 22165-173. [DOI] [PubMed] [Google Scholar]
- 25.Kim, Y. K., and V. N. Kim. 2007. Processing of intronic microRNAs. EMBO J. 26775-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knight, S. W., and B. L. Bass. 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 2932269-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konarska, M. M., and P. A. Sharp. 1987. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell 49763-774. [DOI] [PubMed] [Google Scholar]
- 28.Landthaler, M., A. Yalcin, and T. Tuschl. 2004. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 142162-2167. [DOI] [PubMed] [Google Scholar]
- 29.Lee, Y., C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Radmark, S. Kim, and V. N. Kim. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425415-419. [DOI] [PubMed] [Google Scholar]
- 30.Lee, Y., K. Jeon, J. T. Lee, S. Kim, and V. N. Kim. 2002. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 214663-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, Y., M. Kim, J. Han, K. H. Yeom, S. Lee, S. H. Baek, and V. N. Kim. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 234051-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lund, E., S. Guttinger, A. Calado, J. E. Dahlberg, and U. Kutay. 2004. Nuclear export of microRNA precursors. Science 30395-98. [DOI] [PubMed] [Google Scholar]
- 33.Michael, W. M., M. Choi, and G. Dreyfuss. 1995. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell 83415-422. [DOI] [PubMed] [Google Scholar]
- 34.Moore, M. J. 2002. Nuclear RNA turnover. Cell 108431-434. [DOI] [PubMed] [Google Scholar]
- 35.Morlando, M., M. Ballarino, N. Gromak, F. Pagano, I. Bozzoni, and N. J. Proudfoot. 2008. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 15902-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakielny, S., and G. Dreyfuss. 1996. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J. Cell Biol. 1341365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nam, K., R. H. Hudson, K. B. Chapman, K. Ganeshan, M. J. Damha, and J. D. Boeke. 1994. Yeast lariat debranching enzyme. Substrate and sequence specificity. J. Biol. Chem. 26920613-20621. [PubMed] [Google Scholar]
- 38.Okamura, K., J. W. Hagen, H. Duan, D. M. Tyler, and E. C. Lai. 2007. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 13089-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ooi, S. L., C. Dann III, K. Nam, D. J. Leahy, M. J. Damha, and J. D. Boeke. 2001. RNA lariat debranching enzyme. Methods Enzymol. 342233-248. [DOI] [PubMed] [Google Scholar]
- 40.Pawlicki, J. M., and J. A. Steitz. 2008. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J. Cell Biol. 18261-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pellizzoni, L., J. Baccon, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J. Biol. Chem. 2777540-7545. [DOI] [PubMed] [Google Scholar]
- 42.Pillai, R. S. 2005. MicroRNA function: multiple mechanisms for a tiny RNA? RNA 111753-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez, A., S. Griffiths-Jones, J. L. Ashurst, and A. Bradley. 2004. Identification of mammalian microRNA host genes and transcription units. Genome Res. 141902-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruby, J. G., C. H. Jan, and D. P. Bartel. 2007. Intronic microRNA precursors that bypass Drosha processing. Nature 44883-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawa, H., M. Ohno, H. Sakamoto, and Y. Shimura. 1988. Requirement of ATP in the second step of the pre-mRNA splicing reaction. Nucleic Acids Res. 163157-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwarz, D. S., G. Hutvagner, T. Du, Z. Xu, N. Aronin, and P. D. Zamore. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115199-208. [DOI] [PubMed] [Google Scholar]
- 47.Siomi, M. C., P. S. Eder, N. Kataoka, L. Wan, Q. Liu, and G. Dreyfuss. 1997. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J. Cell Biol. 1381181-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang, G. Q., and E. S. Maxwell. 2008. Xenopus microRNA genes are predominantly located within introns and are differentially expressed in adult frog tissues via post-transcriptional regulation. Genome Res. 18104-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valencia-Sanchez, M. A., J. Liu, G. J. Hannon, and R. Parker. 2006. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20515-524. [DOI] [PubMed] [Google Scholar]
- 50.Weinstein, L. B., and J. A. Steitz. 1999. Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol. 11378-384. [DOI] [PubMed] [Google Scholar]
- 51.Yeom, K. H., Y. Lee, J. Han, M. R. Suh, and V. N. Kim. 2006. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 344622-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yi, R., Y. Qin, I. G. Macara, and B. R. Cullen. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 173011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshimoto, R., N. Kataoka, K. Okawa, and M. Ohno. 2009. Isolation and characterization of post-splicing lariat-intron complexes. Nucleic Acids Res. 37891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.