Abstract
Proper activation of transcriptional networks in complex organisms is central to the response to stimuli. We demonstrate that the selective activation of a subset of the estrogen receptor alpha (ERα) cistrome in MCF7 breast cancer cells provides specificity to the estradiol (E2) response. ERα-specific enhancers that are subject to E2-induced coactivator-associated arginine methyltransferase 1 (CARM1) action are critical to E2-stimulated gene expression. This is true for both FoxA1-dependent and independent enhancers. In contrast, a subset of E2-suppressed genes are controlled by FoxA1-independent ERα binding sites. Nonetheless, these are sites of E2-induced CARM1 activity. In addition, the MCF7 RNA polymerase II cistrome reveals preferential occupancy of E2-regulated promoters prior to stimulation. Interestingly, E2-suppressed genes tend to lie in otherwise silent genomic regions. Together, our results suggest that the transcriptional response to E2 in breast cancer cells is dependent on the interplay between polymerase II pre-occupied promoters and the subset of the ERα cistrome associated with coactivation.
The transcriptional response to estrogen in numerous tissues, including mammary gland, bone, and uterine tissues, and in diseases such as breast cancer is dependent on estrogen receptor alpha (ERα). Genome-wide positional analyses defining the set of cis-regulatory elements recruiting ERα, known as its cistrome, in breast cancer cells have revealed its predominant recruitment to enhancers as opposed to promoter regions (6, 7, 37, 39). As for many other transcription factors, genomic recruitment of ERα is restricted to a small proportion of its putative binding sites (<4.4%) offering a primary means of defining the response to estradiol (E2) (5, 7, 37). Similarly, the promoter predominant Pol II recruitment in breast cancer cells is restricted to a subset of promoters upon E2 stimulation (7, 32, 33, 35). Epigenetic modifications are central to the lineage-specific recruitment at enhancers and promoter regions. Indeed, promoters of activated genes harbor trimethylated histone H3 lysine 4 (H3K4me3) favoring the recruitment of chromatin remodeling enzymes and histone acetylases (1, 18, 42, 52, 56, 58). In contrast, promoters associated with transcriptional repression harbor trimethylated H3K27 (H3K27me3) (1, 3, 36, 42). Similarly, functional enhancers are associated with mono- and dimethylation of H3K4 (H3K4me1, me2) restricting the recruitment and the chromatin remodeling activity of the pioneer factor FoxA1, required for ERα binding, in a lineage-dependent manner, while levels of H3K9me2 are elevated on nonfunctional enhancers (15, 25, 40).
Despite these epigenetic constraints, RNA polymerase II (Pol II) and ERα together are recruited to more than 9,000 independent high-confidence (false discovery rate [FDR], 1%) sites across the genome of breast cancer cells upon E2 stimulation (7). Studies limited to a small number of ERα target sites have implicated coactivators, such as the coactivator associated arginine methyltransferase 1 (CARM1), in the E2 response (22). As they are recruited to ERα binding sites, coactivators allow for a series of posttranslational modifications on histones and other coactivator proteins in order to facilitate chromatin remodeling and cycling of the transcriptional unit essential for the E2 response (41, 57). In the case of CARM1, this involves dimethylation of arginine residues on histone H3 as well as on the coactivator AIB1 (8, 48). In addition, recent studies in Drosophila have revealed the dominant presence of poised Pol II at promoters of genes involved in the response to stimuli and developmental signals (47, 69). In the present study, we investigated the impact of CARM1 coactivator's activity on ERα binding sites and of Pol II at promoters in the transcriptional response to E2 through genome-wide positional analyses in human breast cancer cells.
MATERIALS AND METHODS
ChIP-microarray preparation.
Cells were hormone deprived for 3 days in phenol red-free medium (Invitrogen) supplemented with 10% charcoal dextran-treated fetal bovine serum. Cells were stimulated with the estrogen 17β-estradiol (10−8 M) for 45 min and cross-linked by using 1% formaldehyde. Samples were sonicated (Fisher Sonic Desmembrator, model 500) and immunoprecipitated, as previously described (40), using an antibody against histone H3 arginine 17 dimethylated (H3R17me2; Upstate Biotechnology, 07-214) and Pol II (Abcam, 4H8; Santa Cruz Biotechnology, H-224). Purified samples were labeled as previously described (6). The microarrays used were Affymetrix GeneChip Human Tiling 2.0R Array Sets. Genome-wide chromatin immunoprecipitation (ChIP)-on-ChIP analysis was conducted by using a model-based analysis of tiling-arrays program (MAT) (30). All ChIP-on-ChIP data used in the present study can be accessed at http://research.dfci.harvard.edu/brownlab/datasets/.
Cluster analysis.
We generated a set of genomic intervals derived from the union of all high-confidence sites associated with either ERα, FoxA1, or CARM1 activity or Pol II. Next, we assigned the score to each interval for each factor as the maximum MAT score falling within the interval for the given factor. For each factor MAT scores were trimmed at the 2.5 and 97.5 percentiles and scaled to lie between 0 and 1. Genomic regions were clustered by using k-means clustering.
Sequence analysis.
Genome-wide distribution as well as sequence conservation analysis of H3R17me2 chip-on-chip was determined by using cis-element annotation systems (28). Enriched motifs within clusters as well as the associations with gene expression were analyzed as described previously (40).
ChIP assays.
At 2 to 3 days before induction, MCF-7 cells were seeded in phenol red-free Dulbecco modified Eagle medium supplemented with 10% charcoal-dextran-treated fetal bovine serum (Omega Scientific, Inc., Tarzana, CA), 2 mM l-glutamine, and 100 U of penicillin-streptomycin/ml at a density of 5 × 106 cells per 150-mm plates. Cells were subsequently induced with 10−8 M E2 for 45 min. ChIP experiments were then performed as described previously (16). Antibodies to ERα (Lab Vision, Ab-10; Santa Cruz Biotechnology, sc-543), H3R17me2 (Upstate Biotechnology, 07-214), H3K18ac (Upstate Biotechnology, 07-354), H3K27ac (Upstate Biotechnology, 07-360), H4K12ac (Upstate Biotechnology, 07-595), H3 (Abcam, ab1791), p300 (Santa Cruz Biotechnology, sc-585), and SRC1 (Santa Cruz Biotechnology, sc-8995) were used for this assay. Purified DNA was used in quantitative PCR (qPCR) analysis. The primers used in this analysis are listed in Table S1 in the supplemental material. Immunoprecipitated DNA amounts were normalized to inputs and are expressed as the relative enrichment.
FAIRE analysis.
Formaldehyde-assisted isolation of regulatory elements (FAIRE) was performed as described in reference 21. The primers used in this analysis are listed in Table S1 in the supplemental material.
RESULTS
Distinct enhancer-rich clusters characterize genomic ERα recruitment.
In order to better characterize the impact of coactivator action on the ERα cistrome upon E2 stimulation, we have established the relative level of CARM1 activity across the genome of MCF7 breast cancer cells. This was achieved through ChIP studies combined with whole-genome tiling-path microarrays (ChIP-on-ChIP) using an antibody that recognizes exclusively sites of CARM1-dependent arginine methylation, including histone H3 dimethylated on arginine 17 (H3R17me2) and the CARM1-dependent arginine methylation of AIB1 (see Fig. S1 and S2 in the supplemental material) (9, 48, 67). More than 4,088 and 4,461 high-confidence sites were identified before and after E2 stimulation, respectively (FDR, 6%) (see Fig. S1A and B in the supplemental material). Interestingly, CARM1 activity was found predominantly (94.1%) at regions far from known promoters (see Fig. S1C in the supplemental material). The Pol II cistrome was also determined in the absence of E2 to address the role of promoter-associated factors in this system (see Fig. S3 in the supplemental material). As anticipated, of the 7,420 high-confidence sites (FDR, 5%) recruiting Pol II, 55.4% were recruited within 1 kb upstream of annotated transcription start sites (TSS) (see Fig. S3A to C in the supplemental material).
To establish the contribution of CARM1 activation and Pol II recruitment to E2 signaling, we combined our newly derived cistromes with previously published cistromes for FoxA1 in the presence or absence of E2, as well as ERα and Pol II in E2-treated MCF7 cells (7, 40). We first established the binding activity as determined by MAT score (29) for all factors across the 25,416 high-confidence regions recruiting at least one of these factors in MCF7 cells. The use of k-means clustering revealed five enhancer-rich clusters and two promoter-rich clusters (Fig. 1A to C). Interestingly, each cluster consisted of sites demonstrating high sequence conservation across vertebrate species (see Fig. S4 in the supplemental material). ERα was most significantly recruited after E2 stimulation to clusters Ec1 (23% of the 5782 ERα high-confidence sites) and Ec3 (47% of the 5782 ERα high-confidence sites), with <2.6% of the high-confidence sites found at promoters (Fig. 1A and C). The previously reported sites of FoxA1 recruitment favoring ERα binding were found predominantly in cluster Ec1 but not Ec3 (Fig. 1A and C). Correspondingly, both cluster Ec1 and Ec3 were highly enriched for the ERE half-site motif, while the Forkhead motif was only enriched in cluster Ec1 (Fig. 1D). In addition, both clusters demonstrated E2-induced CARM1 activity as measured by the MAT score (Fig. 1A and C and see Fig. S5 in the supplemental material). Cluster Ec2 consisted of sites found at <2.1% of promoters where FoxA1 was strongly recruited but where ERα had low binding activity (Fig. 1A to C and see Fig. S5 in the supplemental material). Accordingly, the Forkhead motif was enriched in this cluster, while the ERE half-site motif was not significantly enriched (Fig. 1D). In addition, CARM1 activity was not induced on sites from this cluster following E2 stimulation (Fig. 1A and C and see Fig. S5 in the supplemental material). Finally, sites from the enhancer-rich clusters Ec4 and Ec5, with <6.1% of sites at promoters, did not demonstrate strong ERα recruitment. However, sites from cluster Ec4 but not Ec5 associated with FoxA1 binding. In addition, ligand-independent CARM1 activity was associated with cluster Ec5 independently of E2 stimulation (Fig. 1A and C and see Fig. S5 in the supplemental material). Globally, these data reveal that various classes of regulatory elements are established under E2 stimulation, and those associated with ERα and FoxA1 recruitment, as well as CARM1 activity, are found predominantly in enhancer regions across the genome.
FIG. 1.
Establishing classes of enhancer-rich clusters under E2 treatment. (A) Cluster analysis according to the binding activity for the transcription factor ERα, the pioneer factor FoxA1, the mark of CARM1 activity (an antibody raised against dimethylation of arginine 17 on histone H3), and Pol II across the 25,416 high-confidence regions recruiting at least one factor from all analyzed cistromes established through unbiased genome-wide ChIP-on-ChIP in MCF7 breast cancer cells (E2, E2 treated for 45 min; O, vehicle treated). (B) Genomic distribution of binding sites found in each cluster with regard to the TSS of known genes using the cis-regulatory element annotation system (28). (C) Average MAT scores of ERα and FoxA1 and the difference in CARM1 activity between E2-treated and control MCF7 cells in each cluster. The average MAT score signal for ERα or FoxA1 for the various clusters significantly different from a 1.5 average MAT score is presented. Similarly, the average change in CARM1 activity MAT score significantly different from 1 between E2- and vehicle-treated cells is presented. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (D) Half-ERE and Forkhead (FKH) motif enrichment in sites from each cluster.
E2-induced CARM1 activity associates with coactivator recruitment, histone modifications, and chromatin opening.
A common feature of sites from clusters Ec1 and Ec3 predominantly involved in the E2-mediated regulation of gene expression is their association with the induction of CARM1 activity after E2 treatment (Fig. 1A and C). In order to better characterize the active state of these enhancer regions, we investigated the level of coactivator recruitment and histone modifications after E2 treatment. Sites recruiting ERα and associated with CARM1 activity significantly recruited other coactivators, such as p300 and SRC1, under E2 stimulation (Fig. 2A and see Fig. S6 in the supplemental material). Similarly, histone modifications, such as acetylation of lysine 18 or 27 on histone H3 (H3K18ac, H3K27ac), as well as on lysine 12 of histone H4 (H4K12ac), were significantly induced by E2 on these same sites (Fig. 2B). ERα binding sites not associated with the induction of CARM1 activity did not demonstrate any significant induction of coactivator recruitment or histone modification under E2 treatment (Fig. 2A and B). It is noteworthy that ERα binding sites undergoing coactivator recruitment and histone modifications after E2 treatment also associated with E2-induced chromatin opening measured by histone H3 density or extractability by FAIRE (21) (Fig. 2C). Considering that 30% of ERα binding sites are not associated with clusters Ec1 or Ec3 typified by E2-inducted CARM1 activity, our results reveal that the specific transcriptional response to E2 is in part dependent on the selective activation of a fraction of sites recruiting ERα.
FIG. 2.
E2-induced CARM1 activity at ERα sites associates with activating events. (A) Level of recruitment for the coactivators p300 and SRC1 under vehicle (O) or E2 treatment established by ChIP-qPCR on eight ERα sites associated and eight not associated with CARM1 activation in MCF7 breast cancer cells. (B) Levels of histone modifications, namely, H3K18ac, H3K27ac, and H4K12ac, were determined as in panel A. (C) Impact of E2 treatment on nucleosome density. The changes in occupancy of the core histone H3 were determined by ChIP-qPCR as in panel A. Alterations to the DNA accessibility were determined by using FAIRE (21). The results are derived from a minimum of two independent experiments. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
CARM1 activation on ERα binding sites drives the transcriptional response to E2.
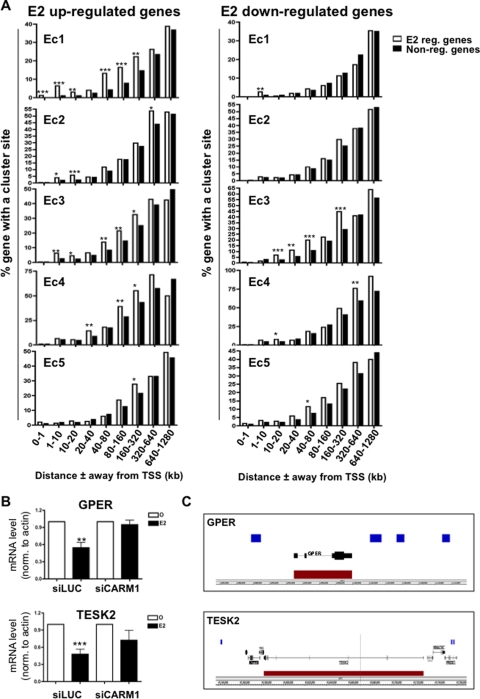
In order to address the role of the various enhancer-rich clusters in gene regulation, we established the proportion of genes regulated after 3 h of E2 treatment versus nonregulated genes with at least one binding site from a particular cluster within increasing window distances in kilobases from the TSS. This revealed a significant enrichment of E2 upregulated genes over nonregulated genes with regard to sites from clusters Ec1, Ec2, and Ec3 from various window distances from the TSS, as far as 160 to 320 kb for both Ec1 and Ec3 (Fig. 3A). Hence, our results suggest that the subset of the ERα cistrome subject to CARM1 activation upon E2 treatment, whether strongly or weakly associated with FoxA1 binding, is responsible for E2-mediated gene induction. Thus, the previously suggested role for CARM1 in mediating the E2 response (19, 67) is due to its activity at enhancer regions defined by a specific subset of the ERα cistrome. Interestingly, genes downregulated after E2 stimulation were significantly enriched over nonregulated genes near sites primarily from cluster Ec3 that could be as far away as 160 to 320 kb (Fig. 3A). Accordingly, silencing CARM1 (see Fig. S2A in the supplemental material) significantly prevented the E2-mediated repression of GPER and TESK2 (Fig. 3B and C). Hence, this finding suggests a predominant role for ERα sites associated weakly or not at all with FoxA1 and undergoing ligand-dependent CARM1 activity in E2-mediated gene downregulation.
FIG. 3.
Clusters associated with CARM1 activation drive the response under E2 treatment. (A) Proportion of E2 upregulated genes compared to nonregulated genes with at least one binding site from a specific cluster within increasing window distances from their TSS in MCF7 cells. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (B) GPER and TESK2 expression after CARM1 silencing in MCF7 was determined by reverse transcription-qPCR and revealed the requirement for CARM1 in the E2-induced repression of GPER and TESK2. siLUC was used as a control. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (C) Enrichment of Ec3 cluster sites (blue blocks) near GPER and TESK2 E2-downregulated target genes (red blocks).
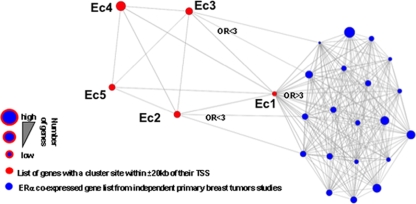
In order to address the physiological relevance of the different clusters, we compared the list of genes with a binding site from a particular cluster within 20 kb of their TSS to the top genes coexpressed with ERα in primary breast tumors from 20 independent studies (4, 10, 13, 20, 23, 26, 27, 43, 44, 50, 51, 54, 55, 60-62, 64-66, 68, 70). Remarkably, genes coexpressed with ERα defined in 19 out of the 20 independent studies were highly associated (odds ratio ≥ 3) with sites from Ec1 within 20 kb of their TSS (Fig. 4). Less significant association (odds ratio between 2 and 3) between ERα coexpressed genes and sites from Ec2 and Ec3 was also detected in 2 out of the 20 independent expression profiles from primary breast tumors (Fig. 4). Hence, these results further support the predominant regulatory role of sites from cluster Ec1 and less significantly from clusters Ec2 and Ec3 in the establishment of the phenotype of ERα-positive breast cancers.
FIG. 4.
ERα-positive primary breast tumor expression profile relates to clusters associated with CARM1 activation. Relationship between cluster-associated gene list (genes with a binding site from a given cluster within 20 kb of their TSS) and genes overexpressed in ERα-positive primary breast tumors (the top 1, 5, or 10% overexpressed genes from primary breast tumors were included in the analysis). Twenty independently defined ERα-positive primary breast tumor overexpressing gene signatures (blue) were compared using an Oncomine Concepts Map to the five enhancer clusters (red) derived gene lists. Odds ratios (OR) are presented when clusters are significantly associated with independent primary breast cancer overexpression gene signatures (P ≤ 6e−6).
Further evidence for the association between sites of ERα recruitment and their activation to mediate transcriptional program originates from the comparison of the MCF7 breast cancer and U2OS osteosarcoma cell lines. Indeed, E2 treatment in both cell lines allows for the recruitment of ERα to a number of common sites (34). Interestingly, the transcriptional program is cell line specific (34). For instance, PDK4 and FasL are two U2OS-specific E2-induced genes (Fig. 5A). Although ERα gets recruited to the PDK4 and FasL enhancers in both cell lines, they are coactivated only in U2OS cells (Fig. 5B). These results suggest that ERα recruitment associates with coactivation in order to mediate gene expression.
FIG. 5.
Cell-type specific coactivation of ERα binding sites associates with the transcriptional response. (A) Relative expression of PDK4 and FasL genes after E2 treatment for 3 h in MCF7 breast cancer and U2OS/ERα cells. (B) Relative enrichment of ERα and CARM1 activity established by ChIP in both MCF7 and U2OS/ERα cells after E2 treatment at the PDK4 and FasL enhancers.
Pol II occupies the promoter of E2 regulated genes prior to stimulation.
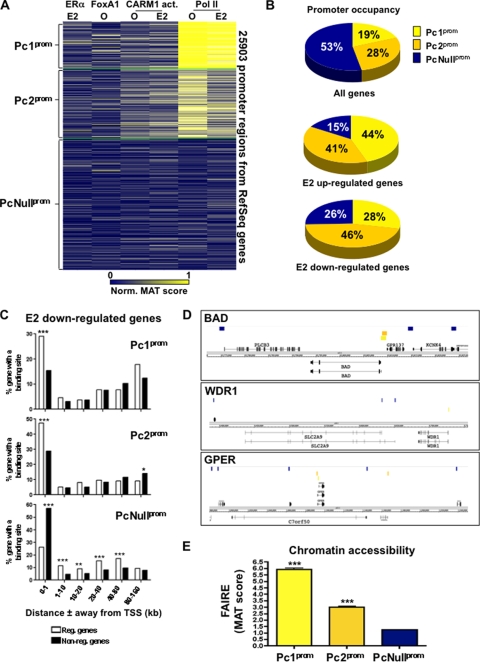
To address how different components of the transcriptional response to E2 signaling impact promoter activity in breast cancer cells, we performed k-means clustering on the 25,903 RefSeq gene promoters as described for Fig. 1A. Three distinct clusters could be defined (Fig. 6A). The first promoter cluster (Pc1prom) consisting of 4,846 sites revealed strong Pol II recruitment both prior to and after E2 stimulation and no significant recruitment of ERα, FoxA1 nor evidence of CARM1 activity (Fig. 6A). Similarly, the 7,316 promoters found in the second cluster (Pc2prom) were specifically enriched for Pol II both prior to and after E2 stimulation, albeit at lower levels than on sites from Pc1prom (Fig. 6A). Finally, 13,741 promoters (PcNullprom) in MCF7 cells were not significantly associated with the recruitment of Pol II, ERα, FoxA1, or CARM1 activity (Fig. 6A). Interestingly, more than 47% of promoters were associated with sites from either Pc1prom or Pc2prom; hence, with Pol II occupied promoters prior to E2 stimulation (Fig. 6A and B and see Fig. S7A in the supplemental material). Strikingly, more than 85% of the E2 upregulated and 74% of E2 downregulated genes had a promoter typical of either cluster Pc1prom or Pc2prom (Fig. 6B and see Fig. S7A in the supplemental material). Furthermore, downregulated genes were typically found in the regions of Pol II unoccupied promoters (Fig. 6C and D). In agreement with this, the promoters of genes surrounding downregulated genes were found in condensed chromatin measured by FAIRE (Fig. 6E) (17). This suggests that stimulus-dependent gene regulation is predominantly dependent on receptive promoters as defined by the presence of Pol II prior to stimulation and on the state of promoter occupancy in neighboring genes.
FIG. 6.
Pol II occupied promoter of E2 target genes prior to stimulation. (A) Cluster analysis performed as described for Fig. 1A across the 25,903 promoter regions associated with the RefSeq genes. (B) Proportions of all, E2-upregulated, and downregulated genes with a promoter typical of clusters Pc1prom, Pc2prom, or PcNullprom. (C) Enrichment of E2-downregulated versus nonregulated genes with at least one promoter of the Pc1prom, Pc2prom, or PcNullprom cluster within increasing window distances from the genes' TSS. (D) Specific examples of E2 downregulated genes surrounded by gene with Pol II deprived promoters. Sites from cluster Pc1prom (orange), Pc2prom (pink), or PcNullprom (dark blue) are presented with respect to E2-downregulated genes (red block). (E) Relative chromatin accessibilities of promoters from cluster Pc1prom, Pc2prom, or PcNullprom measured by FAIRE-ChIP. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
DISCUSSION
The selective utilization of enhancer regions and promoters is central to the establishment of lineage-specific transcriptional programs and stimuli specific responses. Exploiting the E2 signaling pathway, we have combined the cistromes from different components of a transcriptional response, namely, the pioneer factor FoxA1, the transcription factor ERα, a marker of the activity of the coactivator CARM1, and Pol II. Our results reveal that FoxA1-dependent and -independent ERα sites coactivated upon E2 treatment are predominantly driving the response to E2 in breast cancer cells (Fig. 7). Hence, the specific transcriptional program associated with E2 stimulation is not only dependent on the restricted genomic recruitment of ERα but also on the activation of a selected number of binding sites associated with coactivator recruitment and histone modifications. This is in agreement with the central role of coactivators in the response to E2 in both cell lines and mouse models (19, 46, 59, 67). Interestingly, our results also reveal an association between ERα binding sites displaying E2-induced CARM1 activity and gene repression. As we previously suggested, squelching and/or displacement of transcriptional units with greater regulatory capacities could account for this association (7). In addition, we demonstrate on a genome-wide scale that enhancers located as far as 160 to 320 kb from the TSS of the E2 target genes mediate the transcriptional response. This is in accordance with previous studies revealing the intrachromosomal interactions required for optimal transcriptional response upon E2 stimulation in MCF7 cells (6, 12). Considering the commonality of such long-range interactions between promoters and enhancers (38, 49, 63), defining how these are established on a genome-wide scale is of fundamental importance.
FIG. 7.
Model for the selection of functional and active enhancer sites in response to estrogen stimulation in breast cancer cells. A schematic representation of the transcriptional response to E2 stimulation in breast cancer cells is shown. The functional association between ERα recruiting sites undergoing coactivator (CoA) recruitment/activation and histone modifications with transcriptional regulation of the gene harboring Pol II at their promoters both prior to and after E2 stimulation is depicted. Sites of ERα recruitment not associated with these secondary events do not significantly impact E2-induced regulation of gene expression.
Furthermore, we show that Pol II promoter occupancy is typical of E2 responsive genes in MCF7 breast cancer cells that are both up- and downregulated. This is reminiscent of studies in Drosophila revealing the contribution of stalled or poised Pol II at the promoter of genes involved in the response to stimuli and developmental signals (47, 69). It also parallels previous work revealing Pol II at the promoter of unexpressed genes (2, 24, 31-33, 53). It is consistent with the concept of Pol II foci in the nucleus known as transcription factories that remain intact in the absence of transcription (45). In fact, postrecruitment regulation of Pol II was recently revealed to be central for the rapid signaling response to estrogen (33). Interestingly, we identified a difference between the genomic environments of E2 up- versus downregulated genes. Indeed, although Pol II typically occupies the promoter of E2 downregulated genes, the promoters of surrounding genes tend to be deprived of Pol II. Hence, the transcriptional response is dependent on the presence of a receptive promoter typified by Pol II occupancy prior to stimulation.
Globally, our results reveal that the specificity of the transcriptional response to E2 stimulation is dependent on the interplay between receptive promoters occupied by Pol II prior to stimulation and subclasses of ERα enhancers associated with E2-induced coactivator activity. Considering the unique expression profiles associated with ERα activation under distinct stimuli (11, 14), it remains to be established whether distinct subclasses of the ERα cistrome will be involved in these responses as well.
Supplementary Material
Acknowledgments
We thank Shannon T. Bailey for helpful discussion and review of the manuscript.
This study was supported by grants from the NIDDK (R01DK074967 to M.B.), the NCI (P01 CA8011105) and a DF/HCC Breast Cancer SPORE Grant (to M.B.), the DFCI Women's Cancers Program, and the U.S. Department of Defense Breast Cancer Research Program Awards (W81XWH-08-1-0214 to M.L.).
Footnotes
Published ahead of print on 13 April 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Azuara, V., P. Perry, S. Sauer, M. Spivakov, H. F. Jorgensen, R. M. John, M. Gouti, M. Casanova, G. Warnes, M. Merkenschlager, and A. G. Fisher. 2006. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8532-538. [DOI] [PubMed] [Google Scholar]
- 2.Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129823-837. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, B. E., T. S. Mikkelsen, X. Xie, M. Kamal, D. J. Huebert, J. Cuff, B. Fry, A. Meissner, M. Wernig, K. Plath, R. Jaenisch, A. Wagschal, R. Feil, S. L. Schreiber, and E. S. Lander. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125315-326. [DOI] [PubMed] [Google Scholar]
- 4.Bild, A. H., G. Yao, J. T. Chang, Q. Wang, A. Potti, D. Chasse, M. B. Joshi, D. Harpole, J. M. Lancaster, A. Berchuck, J. A. Olson, Jr., J. R. Marks, H. K. Dressman, M. West, and J. R. Nevins. 2006. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 439353-357. [DOI] [PubMed] [Google Scholar]
- 5.Bourdeau, V., J. Deschenes, R. Metivier, Y. Nagai, D. Nguyen, N. Bretschneider, F. Gannon, J. H. White, and S. Mader. 2004. Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol. Endocrinol. 181411-1427. [DOI] [PubMed] [Google Scholar]
- 6.Carroll, J. S., X. S. Liu, A. S. Brodsky, W. Li, C. A. Meyer, A. J. Szary, J. Eeckhoute, W. Shao, E. V. Hestermann, T. R. Geistlinger, E. A. Fox, P. A. Silver, and M. Brown. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the Forkhead protein FoxA1. Cell 12233-43. [DOI] [PubMed] [Google Scholar]
- 7.Carroll, J. S., C. A. Meyer, J. Song, W. Li, T. R. Geistlinger, J. Eeckhoute, A. S. Brodsky, E. K. Keeton, K. C. Fertuck, G. F. Hall, Q. Wang, S. Bekiranov, V. Sementchenko, E. A. Fox, P. A. Silver, T. R. Gingeras, X. S. Liu, and M. Brown. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 381289-1297. [DOI] [PubMed] [Google Scholar]
- 8.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 2842174-2177. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, D., J. Cote, S. Shaaban, and M. T. Bedford. 2007. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell 2571-83. [DOI] [PubMed] [Google Scholar]
- 10.Chin, K., S. DeVries, J. Fridlyand, P. T. Spellman, R. Roydasgupta, W. L. Kuo, A. Lapuk, R. M. Neve, Z. Qian, T. Ryder, F. Chen, H. Feiler, T. Tokuyasu, C. Kingsley, S. Dairkee, Z. Meng, K. Chew, D. Pinkel, A. Jain, B. M. Ljung, L. Esserman, D. G. Albertson, F. M. Waldman, and J. W. Gray. 2006. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 10529-541. [DOI] [PubMed] [Google Scholar]
- 11.Cunliffe, H. E., M. Ringner, S. Bilke, R. L. Walker, J. M. Cheung, Y. Chen, and P. S. Meltzer. 2003. The gene expression response of breast cancer to growth regulators: patterns and correlation with tumor expression profiles. Cancer Res. 637158-7166. [PubMed] [Google Scholar]
- 12.Deschenes, J., V. Bourdeau, J. H. White, and S. Mader. 2007. Regulation of GREB1 transcription by estrogen receptor alpha through a multipartite enhancer spread over 20 kb of upstream flanking sequences. J. Biol. Chem. 28217335-17339. [DOI] [PubMed] [Google Scholar]
- 13.Desmedt, C., F. Piette, S. Loi, Y. Wang, F. Lallemand, B. Haibe-Kains, G. Viale, M. Delorenzi, Y. Zhang, M. S. d'Assignies, J. Bergh, R. Lidereau, P. Ellis, A. L. Harris, J. G. Klijn, J. A. Foekens, F. Cardoso, M. J. Piccart, M. Buyse, and C. Sotiriou. 2007. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin. Cancer Res. 133207-3214. [DOI] [PubMed] [Google Scholar]
- 14.Dudek, P., and D. Picard. 2008. Genomics of signaling crosstalk of estrogen receptor alpha in breast cancer cells. PLoS ONE 3e1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eeckhoute, J., J. S. Carroll, T. R. Geistlinger, M. I. Torres-Arzayus, and M. Brown. 2006. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 202513-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eeckhoute, J., E. K. Keeton, M. Lupien, S. A. Krum, J. S. Carroll, and M. Brown. 2007. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 676477-6483. [DOI] [PubMed] [Google Scholar]
- 17.Eeckhoute, J., M. Lupien, C. A. Meyer, M. P. Verzi, R. A. Shivdasani, X. S. Liu, and M. Brown. 2008. Cell-type selective remodeling defines the active subset of FOXA1-bound enhancers. Genome Res. 19372-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flanagan, J. F., L. Z. Mi, M. Chruszcz, M. Cymborowski, K. L. Clines, Y. Kim, W. Minor, F. Rastinejad, and S. Khorasanizadeh. 2005. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 4381181-1185. [DOI] [PubMed] [Google Scholar]
- 19.Frietze, S., M. Lupien, P. A. Silver, and M. Brown. 2008. CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 68301-306. [DOI] [PubMed] [Google Scholar]
- 20.Ginestier, C., N. Cervera, P. Finetti, S. Esteyries, B. Esterni, J. Adelaide, L. Xerri, P. Viens, J. Jacquemier, E. Charafe-Jauffret, M. Chaffanet, D. Birnbaum, and F. Bertucci. 2006. Prognosis and gene expression profiling of 20q13-amplified breast cancers. Clin. Cancer Res. 124533-4544. [DOI] [PubMed] [Google Scholar]
- 21.Giresi, P. G., J. Kim, R. M. McDaniell, V. R. Iyer, and J. D. Lieb. 2007. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res. 17877-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green, K. A., and J. S. Carroll. 2007. Oestrogen-receptor-mediated transcription and the influence of cofactors and chromatin state. Nat. Rev. Cancer 7713-722. [DOI] [PubMed] [Google Scholar]
- 23.Gruvberger, S., M. Ringner, Y. Chen, S. Panavally, L. H. Saal, A. Borg, M. Ferno, C. Peterson, and P. S. Meltzer. 2001. Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res. 615979-5984. [PubMed] [Google Scholar]
- 24.Guenther, M. G., S. S. Levine, L. A. Boyer, R. Jaenisch, and R. A. Young. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 13077-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heintzman, N. D., R. K. Stuart, G. Hon, Y. Fu, C. W. Ching, R. D. Hawkins, L. O. Barrera, S. Van Calcar, C. Qu, K. A. Ching, W. Wang, Z. Weng, R. D. Green, G. E. Crawford, and B. Ren. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39311-318. [DOI] [PubMed] [Google Scholar]
- 26.Hess, K. R., K. Anderson, W. F. Symmans, V. Valero, N. Ibrahim, J. A. Mejia, D. Booser, R. L. Theriault, A. U. Buzdar, P. J. Dempsey, R. Rouzier, N. Sneige, J. S. Ross, T. Vidaurre, H. L. Gomez, G. N. Hortobagyi, and L. Pusztai. 2006. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J. Clin. Oncol. 244236-4244. [DOI] [PubMed] [Google Scholar]
- 27.Ivshina, A. V., J. George, O. Senko, B. Mow, T. C. Putti, J. Smeds, T. Lindahl, Y. Pawitan, P. Hall, H. Nordgren, J. E. Wong, E. T. Liu, J. Bergh, V. A. Kuznetsov, and L. D. Miller. 2006. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 6610292-10301. [DOI] [PubMed] [Google Scholar]
- 28.Ji, X., W. Li, J. Song, L. Wei, and X. S. Liu. 2006. CEAS: cis-regulatory element annotation system. Nucleic Acids Res. 34W551-W554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, D. S., W. Li, D. B. Gordon, A. Bhattacharjee, B. Curry, J. Ghosh, L. Brizuela, J. S. Carroll, M. Brown, P. Flicek, C. M. Koch, I. Dunham, M. Bieda, X. Xu, P. J. Farnham, P. Kapranov, D. A. Nix, T. R. Gingeras, X. Zhang, H. Holster, N. Jiang, R. Green, J. S. Song, S. A. McCuine, E. Anton, L. Nguyen, N. D. Trinklein, Z. Ye, K. Ching, D. Hawkins, B. Ren, P. C. Scacheri, J. Rozowsky, A. Karpikov, G. Euskirchen, S. Weissman, M. Gerstein, M. Snyder, A. Yang, Z. Moqtaderi, H. Hirsch, H. P. Shulha, Y. Fu, Z. Weng, K. Struhl, R. M. Myers, J. D. Lieb, and X. S. Liu. 2008. Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome Res. 18393-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, W. E., W. Li, C. A. Meyer, R. Gottardo, J. S. Carroll, M. Brown, and X. S. Liu. 2006. Model-based analysis of tiling-arrays for ChIP-chip. Proc. Natl. Acad. Sci. USA 10312457-12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, T. H., L. O. Barrera, M. Zheng, C. Qu, M. A. Singer, T. A. Richmond, Y. Wu, R. D. Green, and B. Ren. 2005. A high-resolution map of active promoters in the human genome. Nature 436876-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kininis, M., B. S. Chen, A. G. Diehl, G. D. Isaacs, T. Zhang, A. C. Siepel, A. G. Clark, and W. L. Kraus. 2007. Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol. Cell. Biol. 275090-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kininis, M., G. D. Isaacs, L. J. Core, N. Hah, and W. L. Kraus. 2009. Postrecruitment regulation of RNA polymerase II directs rapid signaling responses at the promoters of estrogen target genes. Mol. Cell. Biol. 29:1123-1133. [DOI] [PMC free article] [PubMed]
- 34.Krum, S. A., G. A. Miranda-Carboni, M. Lupien, J. Eeckhoute, J. S. Carroll, and M. Brown. 2008. Unique ERα cistromes control cell type-specific gene regulation. Mol. Endocrinol. 222393-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwon, Y. S., I. Garcia-Bassets, K. R. Hutt, C. S. Cheng, M. Jin, D. Liu, C. Benner, D. Wang, Z. Ye, M. Bibikova, J. B. Fan, L. Duan, C. K. Glass, M. G. Rosenfeld, and X. D. Fu. 2007. Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc. Natl. Acad. Sci. USA 1044852-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, T. I., R. G. Jenner, L. A. Boyer, M. G. Guenther, S. S. Levine, R. M. Kumar, B. Chevalier, S. E. Johnstone, M. F. Cole, K. Isono, H. Koseki, T. Fuchikami, K. Abe, H. L. Murray, J. P. Zucker, B. Yuan, G. W. Bell, E. Herbolsheimer, N. M. Hannett, K. Sun, D. T. Odom, A. P. Otte, T. L. Volkert, D. P. Bartel, D. A. Melton, D. K. Gifford, R. Jaenisch, and R. A. Young. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125301-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, C. Y., V. B. Vega, J. S. Thomsen, T. Zhang, S. L. Kong, M. Xie, K. P. Chiu, L. Lipovich, D. H. Barnett, F. Stossi, A. Yeo, J. George, V. A. Kuznetsov, Y. K. Lee, T. H. Charn, N. Palanisamy, L. D. Miller, E. Cheung, B. S. Katzenellenbogen, Y. Ruan, G. Bourque, C. L. Wei, and E. T. Liu. 2007. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 3e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ling, J. Q., T. Li, J. F. Hu, T. H. Vu, H. L. Chen, X. W. Qiu, A. M. Cherry, and A. R. Hoffman. 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312269-272. [DOI] [PubMed] [Google Scholar]
- 39.Liu, Y., H. Gao, T. T. Marstrand, A. Strom, E. Valen, A. Sandelin, J. A. Gustafsson, and K. Dahlman-Wright. 2008. The genome landscape of ERα- and ERβ-binding DNA regions. Proc. Natl. Acad. Sci. USA 1052604-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lupien, M., J. Eeckhoute, C. Meyer, Q. Wang, Y. Zhang, W. Li, J. Carroll, X. Liu, and M. Brown. 2008. FoxA1 translates epigenetic signatures into enhancer driven lineage-specific transcription. Cell 132958-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Metivier, R., G. Penot, M. R. Hubner, G. Reid, H. Brand, M. Kos, and F. Gannon. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115751-763. [DOI] [PubMed] [Google Scholar]
- 42.Mikkelsen, T. S., M. Ku, D. B. Jaffe, B. Issac, E. Lieberman, G. Giannoukos, P. Alvarez, W. Brockman, T. K. Kim, R. P. Koche, W. Lee, E. Mendenhall, A. O'Donovan, A. Presser, C. Russ, X. Xie, A. Meissner, M. Wernig, R. Jaenisch, C. Nusbaum, E. S. Lander, and B. E. Bernstein. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448553-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller, L. D., J. Smeds, J. George, V. B. Vega, L. Vergara, A. Ploner, Y. Pawitan, P. Hall, S. Klaar, E. T. Liu, and J. Bergh. 2005. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc. Natl. Acad. Sci. USA 10213550-13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minn, A. J., G. P. Gupta, P. M. Siegel, P. D. Bos, W. Shu, D. D. Giri, A. Viale, A. B. Olshen, W. L. Gerald, and J. Massague. 2005. Genes that mediate breast cancer metastasis to lung. Nature 436518-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell, J. A., and P. Fraser. 2008. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 2220-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Modder, U. I., A. Sanyal, J. Xu, B. W. O'Malley, T. C. Spelsberg, and S. Khosla. 2008. The skeletal response to estrogen is impaired in female but not in male steroid receptor coactivator (SRC)-1 knockout mice. Bone 42414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muse, G. W., D. A. Gilchrist, S. Nechaev, R. Shah, J. S. Parker, S. F. Grissom, J. Zeitlinger, and K. Adelman. 2007. RNA polymerase is poised for activation across the genome. Nat. Genet. 391507-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naeem, H., D. Cheng, Q. Zhao, C. Underhill, M. Tini, M. T. Bedford, and J. Torchia. 2007. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol. Cell. Biol. 27120-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osborne, C. S., L. Chakalova, K. E. Brown, D. Carter, A. Horton, E. Debrand, B. Goyenechea, J. A. Mitchell, S. Lopes, W. Reik, and P. Fraser. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 361065-1071. [DOI] [PubMed] [Google Scholar]
- 50.Perou, C. M., S. S. Jeffrey, M. van de Rijn, C. A. Rees, M. B. Eisen, D. T. Ross, A. Pergamenschikov, C. F. Williams, S. X. Zhu, J. C. Lee, D. Lashkari, D. Shalon, P. O. Brown, and D. Botstein. 1999. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc. Natl. Acad. Sci. USA 969212-9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pollack, J. R., T. Sorlie, C. M. Perou, C. A. Rees, S. S. Jeffrey, P. E. Lonning, R. Tibshirani, D. Botstein, A. L. Borresen-Dale, and P. O. Brown. 2002. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc. Natl. Acad. Sci. USA 9912963-12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pray-Grant, M. G., J. A. Daniel, D. Schieltz, J. R. Yates III, and P. A. Grant. 2005. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433434-438. [DOI] [PubMed] [Google Scholar]
- 53.Radonjic, M., J. C. Andrau, P. Lijnzaad, P. Kemmeren, T. T. Kockelkorn, D. van Leenen, N. L. van Berkum, and F. C. Holstege. 2005. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon Saccharomyces cerevisiae stationary phase exit. Mol. Cell 18171-183. [DOI] [PubMed] [Google Scholar]
- 54.Richardson, A. L., Z. C. Wang, A. De Nicolo, X. Lu, M. Brown, A. Miron, X. Liao, J. D. Iglehart, D. M. Livingston, and S. Ganesan. 2006. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 9121-132. [DOI] [PubMed] [Google Scholar]
- 55.Saal, L. H., P. Johansson, K. Holm, S. K. Gruvberger-Saal, Q. B. She, M. Maurer, S. Koujak, A. A. Ferrando, P. Malmstrom, L. Memeo, J. Isola, P. O. Bendahl, N. Rosen, H. Hibshoosh, M. Ringner, A. Borg, and R. Parsons. 2007. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc. Natl. Acad. Sci. USA 1047564-7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Santos-Rosa, H., R. Schneider, B. E. Bernstein, N. Karabetsou, A. Morillon, C. Weise, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2003. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell 121325-1332. [DOI] [PubMed] [Google Scholar]
- 57.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103843-852. [DOI] [PubMed] [Google Scholar]
- 58.Sims, R. J., III, C. F. Chen, H. Santos-Rosa, T. Kouzarides, S. S. Patel, and D. Reinberg. 2005. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J. Biol. Chem. 28041789-41792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith, C. L., D. G. DeVera, D. J. Lamb, Z. Nawaz, Y. H. Jiang, A. L. Beaudet, and B. W. O'Malley. 2002. Genetic ablation of the steroid receptor coactivator-ubiquitin ligase, E6-AP, results in tissue-selective steroid hormone resistance and defects in reproduction. Mol. Cell. Biol. 22525-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sorlie, T., C. M. Perou, R. Tibshirani, T. Aas, S. Geisler, H. Johnsen, T. Hastie, M. B. Eisen, M. van de Rijn, S. S. Jeffrey, T. Thorsen, H. Quist, J. C. Matese, P. O. Brown, D. Botstein, P. Eystein Lonning, and A. L. Borresen-Dale. 2001. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 9810869-10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sotiriou, C., S. Y. Neo, L. M. McShane, E. L. Korn, P. M. Long, A. Jazaeri, P. Martiat, S. B. Fox, A. L. Harris, and E. T. Liu. 2003. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc. Natl. Acad. Sci. USA 10010393-10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sotiriou, C., P. Wirapati, S. Loi, A. Harris, S. Fox, J. Smeds, H. Nordgren, P. Farmer, V. Praz, B. Haibe-Kains, C. Desmedt, D. Larsimont, F. Cardoso, H. Peterse, D. Nuyten, M. Buyse, M. J. Van de Vijver, J. Bergh, M. Piccart, and M. Delorenzi. 2006. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 98262-272. [DOI] [PubMed] [Google Scholar]
- 63.Spilianakis, C. G., M. D. Lalioti, T. Town, G. R. Lee, and R. A. Flavell. 2005. Interchromosomal associations between alternatively expressed loci. Nature 435637-645. [DOI] [PubMed] [Google Scholar]
- 64.van de Vijver, M. J., Y. D. He, L. J. van't Veer, H. Dai, A. A. Hart, D. W. Voskuil, G. J. Schreiber, J. L. Peterse, C. Roberts, M. J. Marton, M. Parrish, D. Atsma, A. Witteveen, A. Glas, L. Delahaye, T. van der Velde, H. Bartelink, S. Rodenhuis, E. T. Rutgers, S. H. Friend, and R. Bernards. 2002. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 3471999-2009. [DOI] [PubMed] [Google Scholar]
- 65.Wang, Y., J. G. Klijn, Y. Zhang, A. M. Sieuwerts, M. P. Look, F. Yang, D. Talantov, M. Timmermans, M. E. Meijer-van Gelder, J. Yu, T. Jatkoe, E. M. Berns, D. Atkins, and J. A. Foekens. 2005. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365671-679. [DOI] [PubMed] [Google Scholar]
- 66.West, M., C. Blanchette, H. Dressman, E. Huang, S. Ishida, R. Spang, H. Zuzan, J. A. Olson, Jr., J. R. Marks, and J. R. Nevins. 2001. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc. Natl. Acad. Sci. USA 9811462-11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yadav, N., J. Lee, J. Kim, J. Shen, M. C. Hu, C. M. Aldaz, and M. T. Bedford. 2003. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc. Natl. Acad. Sci. USA 1006464-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu, K., K. Ganesan, L. D. Miller, and P. Tan. 2006. A modular analysis of breast cancer reveals a novel low-grade molecular signature in estrogen receptor-positive tumors. Clin. Cancer Res. 123288-3296. [DOI] [PubMed] [Google Scholar]
- 69.Zeitlinger, J., A. Stark, M. Kellis, J. W. Hong, S. Nechaev, K. Adelman, M. Levine, and R. A. Young. 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 391512-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao, H., A. Langerod, Y. Ji, K. W. Nowels, J. M. Nesland, R. Tibshirani, I. K. Bukholm, R. Karesen, D. Botstein, A. L. Borresen-Dale, and S. S. Jeffrey. 2004. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol. Biol. Cell 152523-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.