Abstract
Receptor-linked protein-tyrosine phosphatases (RPTPs) are essential regulators of axon guidance and synaptogenesis in Drosophila, but the signaling pathways in which they function are poorly defined. We identified the cell surface receptor Tartan (Trn) as a candidate substrate for the neuronal RPTP Ptp52F by using a modified two-hybrid screen with a substrate-trapping mutant of Ptp52F as “bait.” Trn can bind to the Ptp52F substrate-trapping mutant in transfected Drosophila S2 cells if v-Src kinase, which phosphorylates Trn, is also expressed. Coexpression of wild-type Ptp52F causes dephosphorylation of v-Src-phosphorylated Trn. To examine the specificity of the interaction in vitro, we incubated Ptp52F-glutathione S-transferase (GST) fusion proteins with pervanadate-treated S2 cell lysates. Wild-type Ptp52F dephosphorylated Trn, as well as most other bands in the lysate. GST “pulldown” experiments demonstrated that the Ptp52F substrate-trapping mutant binds exclusively to phospho-Trn. Wild-type Ptp52F pulled down dephosphorylated Trn, suggesting that it forms a stable Ptp52F-Trn complex that persists after substrate dephosphorylation. To evaluate whether Trn and Ptp52F are part of the same pathway in vivo, we examined motor axon guidance in mutant embryos. trn and Ptp52F mutations produce identical phenotypes affecting the SNa motor nerve. The genes also display dosage-dependent interactions, suggesting that Ptp52F regulates Trn signaling in SNa motor neurons.
Receptor-linked protein-tyrosine phosphatases (RPTPs) are enzymes with extracellular (XC) domains, a single transmembrane domain, and one or two cytoplasmic protein tyrosine phosphatase (PTP) homology domains. Many RPTPs have XC sequences that resemble those of cell adhesion molecules (for a review, see reference 33). This sequence organization suggests that RPTPs can couple cell-cell recognition events to dephosphorylation of cytoplasmic substrates. Interestingly, while phosphotyrosine (PY) pathways involved in cell growth and differentiation typically involve receptor tyrosine kinases that bind to growth factors and are regulated by nontransmembrane PTPs, those that control axon guidance often use RPTPs and nontransmembrane TKs. This implies that the cues that affect PY signaling in axonal growth cones may interact with RPTPs rather than with receptor tyrosine kinases (reviewed in reference 14).
There are 17 active RPTPs encoded in the human genome, while Drosophila has six. Most of the mammalian RPTPs are expressed in nonneural tissues, but four of the six fly RPTPs are expressed only by central nervous system (CNS) neurons in late embryos. All published zygotic phenotypes produced by Rptp mutations are alterations in axon guidance or synaptogenesis. These results suggest that the major functions of the Drosophila RPTPs are in neural development (for a review, see reference 16). Analysis of axon guidance phenotypes in embryos bearing single or multiple Rptp mutations is consistent with the idea that RPTP interactions with ligands at growth cone choice points convey “information,” in the form of changes in substrate phosphorylation within growth cones, that is used to determine pathway decisions.
In the Drosophila neuromuscular system, 36 motor axons grow out within six nerve bundles in each abdominal hemisegment, and each axonal growth cone makes a series of genetically determined guidance decisions that direct it to the appropriate muscle fiber (for a review, see reference 27). Our work on Rptp mutant combinations suggests that each pathway decision uses a specific subset of the six RPTPs. RPTPs can exhibit functional redundancy, so that the loss of one does not produce a defect unless another RPTP is also absent, or competition, in which loss of one RPTP suppresses the phenotype produced by loss of another (5, 6, 31). Examination of RPTP expression patterns suggests that the RPTPs are expressed by most (or possibly all) CNS neurons, including motor neurons. If so, the requirements for individual RPTPs for execution of particular guidance decisions cannot be due to selective expression of these RPTPs on specific motor axons. These requirements might instead be determined by the expression patterns of RPTP ligands, so that only RPTPs whose ligands were localized to the vicinity of a growth cone choice point would participate in that pathway decision. Alternatively (or in addition), the necessity of a particular RPTP for a pathway decision might arise from selective expression of RPTP substrates, so that an RPTP would be important for guidance decisions made by a growth cone of a specific motor neuron only if that neuron expressed the relevant substrate(s).
Evaluation of such models requires identification of specific XC ligands and intracellular substrates for the Drosophila RPTPs. Only one set of ligands has been identified thus far. These are the heparan sulfate proteoglycans Syndecan (Sdc) and Dallylike (Dlp), which bind to the Lar RPTP with nanomolar affinity and contribute to its functions in axon guidance and synapse growth (9, 15). Similarly, little is known about substrate specificity in vivo. Lar can dephosphorylate the Enabled (Ena) protein, which regulates the growth cone cytoskeleton, and genetic interaction studies suggest that Ena may be an in vivo substrate for Lar (35). The transmembrane protein gp150 can be dephosphorylated by Ptp10D in cell culture and intact fly larvae, but genetics has not provided evidence that Ptp10D and gp150 are in the same signaling pathway in vivo (7).
The identification of in vivo substrates for RPTPs has been hampered by the fact that purified RPTP cytoplasmic domains often do not exhibit high selectivity in vitro when tested for dephosphorylation activity on peptides or proteins. The most fruitful method for finding substrates for both RPTPs and cytoplasmic PTPs has been the use of “substrate-trapping” mutants. The most effective substrate traps were devised by Tonks and coworkers, and are created by changing an invariant Asp (D) residue within the PTP active site to Ala (A) (8). The D residue has an abnormal pK and is thus able to donate a proton to the phosphorus-oxygen bond, facilitating displacement of the tyrosine (Y) OH by the invariant Cys (C) nucleophile of the enzyme. This creates a phosphoenzyme intermediate. The dephosphorylated substrate then dissociates, and water attacks the Cys-phosphate bond, releasing the phosphate and reconstituting the enzyme. In D→A mutants, the polarization of the phosphorus-oxygen bond by protonation cannot take place, and the PY substrate remains bound to the enzyme. Substrate-trapping mutants expressed in cells often bind to only a few phosphoproteins, suggesting that PTPs exhibit high specificity in vivo (see, for example, reference 11).
We conducted a modified yeast two-hybrid screen to find Drosophila phosphoproteins that bind selectively to RPTP substrate-trapping mutants. We identified the cell surface receptor Tartan (Trn) in this screen and showed that it is a substrate for the Ptp52F RPTP in Drosophila Schneider 2 (S2) cells. Axon guidance phenotypes in trn mutants are identical to those seen in Ptp52F mutants, and trn and Ptp52F exhibit dosage-dependent genetic interactions. These results suggest that Ptp52F is a regulator of Trn signaling in motor neurons in vivo.
MATERIALS AND METHODS
Yeast two-hybrid screening.
The cytoplasmic domains of the RPTPs were cloned in frame into a GAL4:DBD vector, pGBDUC2, a gift from Philip James; this bears the URA3 selectable marker. We made point mutations to change the invariant D to an A in these vectors. For Ptp52F, this is D1258 (numbered in the context of the entire 1,433-amino-acid [aa] preprotein). The activated c-Src construct in BTM116 (bearing a TRP1 marker) was a gift from Kathy Keegan. The library was a 0 to 24 h. cDNA library made in the −ACT (LEU2) vector and was a gift from Stephen Elledge. The insert size range is 0.5 to 6 kb, with an average insert size of 1.8 kb. The strains used for selection are described in reference 13 and were a gift from P. James. Yeast cells were transformed by using standard lithium acetate protocols. For the screen, “bait” cells, bearing the substrate-trap PTP vector and the c-Src vector, and “prey” cells, bearing the library, were grown up in selection medium, and equal numbers of bait and prey cells were mixed for mating and grown up for a few hours. The mated cells were plated onto −Ade plates and incubated at 30°C for 7 days. Positive colonies were patched onto −Ade plates and then tested for activation of the HIS3 and LacZ reporters. Colonies that activated all of the reporters were tested for bait dependence by using 5-fluoroorotic acid to remove the URA3 bait plasmid. They were tested for kinase dependence by replica plating onto +Trp and −Trp plates, followed by examination of whether the colonies lacking the c-Src vector were still ADE+. Bait-dependent plasmids were sequenced and then retransformed into yeast, together with each substrate trap or wild-type PTP bait, with or without c-Src (see Table 1 for results).
TABLE 1.
Summary of the modified yeast two-hybrid screen for RPTP substrates
| Probe (kinase plus bait) | Gene | Type of protein | No. of clones found | Additional interacting probe(s)a |
|---|---|---|---|---|
| src + 10D | Xmas-2 | RNA binding | 1 | src + 52F |
| CG12533 | Calponin homology (actin binding) | 9 | 10D | |
| src + 52F | trn | LRR cell surface receptor | 1 | None |
| CG15022 | Proline rich | 1 | None | |
| CG10283 | No defined domains | 1 | None | |
| src + 69D | CG9418 | DNA binding, HMG box | 4 | 69D |
| src + 99A | csp | Cysteine string protein (synaptic) | 1 | 99A, 10D, src + 10D |
| CG11110 | Leader peptidase | 1 | 99A |
That is, additional probe(s) with which the protein specified in columns 2 and 3 interacts.
Plasmid construction.
We subcloned the Ptp52F cytoplasmic domain into a modified version of the metallothionein (MT) promoter vector pRMHa3 (2) that includes an ATG and Src myristylation sequence, followed by restriction sites. We added a 9xMyc, His8 tag from the HTM53H plasmid (a gift from Ray Deshaies) to the C terminus. This produced the wild-type 52F plasmid used in Fig. 3. We introduced the D1258A mutation into this plasmid, generating the substrate-trapping mutant used in Fig. 3. The full-length untagged Trn cDNA sequence in the POT2 vector (GH10871 cDNA clone) was subcloned into pRMHa3 to produce the Trn-FL plasmid used in Fig. 3. The Trn-cyto-green fluorescent protein (GFP) plasmid used in Fig. 2 and 4 was made by subcloning the entire cytoplasmic domain of Trn into a derivative of pRMHa3 that contained both the myristylation sequence and the GFP sequence, flanking the cloning sites. The Trn-FL-GFP plasmid used in Fig. 3 was made by replacing the cytoplasmic domain of Trn in the Trn-FL plasmid with a cytoplasmic domain-GFP fused sequence from the Trn-cyto-GFP plasmid. The D-Src64B and D-Abl plasmids are described in reference 7 and were originally obtained from Alan Comer. The v-Src plasmid used in Fig. 2 and 3 was made by cloning the entire coding region of v-Src (from Tony Hunter) into the pRMHa3 vector. Details concerning cloning strategies (enzyme sites, primers, etc.) are available on request.
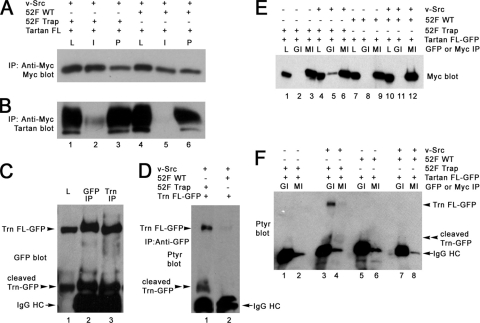
FIG. 3.
Phosphorylated Trn binds to the Ptp52F substrate-trapping mutant and is dephosphorylated by wild-type Ptp52F in S2 cells. (A) An anti-Myc immunoblot of lysates (L), anti-Myc IPs (I), and post-IP supernatants (P) from cells that were cotransfected with the Trn-FL and v-Src plasmids. As indicated by “+” and “−” signs above the blot, these cells were also transfected with the Ptp52F-trap-Myc plasmid (lanes 1 to 3) or the Ptp52F-wild-type-Myc plasmid (lanes 4 to 6). The region of the blot containing the ∼65-kDa Ptp52F-Myc band is shown. (B) Anti-Trn immunoblot of the same samples. Note that a strong Trn band was present in anti-Myc IPs of cells cotransfected with Ptp52F-trap-Myc (lane 2), but only a very faint signal was seen when Ptp52F-wild type-Myc was cotransfected (lane 5). This shows that the substrate-trapping mutant bound selectively to Trn-FL. (C) Anti-GFP immunoblot of lysate (L), an anti-GFP IP, and an anti-Trn IP from cells transfected with the Trn-FL-GFP plasmid. An ∼100-kDa band was present (labeled as Trn FL-GFP), as expected. An ∼60-kDa band that is likely to represent a cleavage product was also present (labeled as cleaved Trn-GFP). The cleavage site must be in the XC domain, close to the membrane, based on the size of this product. The IgG heavy chain (IgG HC) band is also labeled. (D) Anti-PY blot of anti-GFP IPs from S2 cells cotransfected with the Trn FL-GFP and v-Src plasmids, together with either the Ptp52F-trap-Myc (lane 1) or Ptp52F-wild type-Myc (lane 2) plasmids. Note that both the Trn-FL-GFP and cleaved Trn-GFP bands were tyrosine phosphorylated in lane 1; similar levels of phosphorylation were observed when the trap was not cotransfected. When Ptp52F-wild type-Myc was cotransfected, the PY signal was barely detectable for Trn-FL-GFP and undetectable for the cleaved product. This shows that Ptp52F caused dephosphorylation of v-Src-phosphorylated Trn-FL-GFP and the cleavage product. (E) Anti-Myc immunoblot of lysates (L), anti-GFP IPs (GI), and anti-Myc IPs (MI) from cells transfected with Trn-FL-GFP. Cells were also transfected with Ptp52F-trap-Myc (lanes 1 to 6) or Ptp52F-wild type-Myc (lanes 7 to 12) plasmids, with (lanes 4 to 6 and lanes 10 to 12) or without (lanes 1 to 3 and lanes 7 to 9) the v-Src plasmid. The region of the blot containing the ∼65-kDa Ptp52F-Myc band is shown. Note that this band was detected in anti-GFP IPs from cells cotransfected with Ptp52F-trap-Myc and v-Src (lane 5) but was not detectable in anti-GFP IPs from trap-transfected cells lacking the kinase (lane 2). A very faint signal was observed in anti-GFP IPs from cells transfected with Ptp52F-wild type-Myc and the kinase (lane 11). This shows that the substrate-trapping mutant bound to Trn-FL-GFP in a phosphorylation-dependent manner. (F) Anti-PY immunoblot of the same anti-GFP (GI) or anti-Myc (MI) IPs. Note that, as in panel D, the phosphorylated Trn-FL-GFP and cleavage product bands were present when Ptp52F-trap-Myc and v-Src were both transfected (lane 3) but were absent when Ptp52F-wild type-Myc and v-Src were transfected (lane 7). Faint signals were present for both the Trn-FL-GFP and cleavage product bands in lane 4, which is an anti-Myc IP from cells transfected with Ptp52F-trap-Myc and v-Src; this confirms that phosphorylated Trn-FL-GFP associated with the trap. The IgG HC band is also labeled.
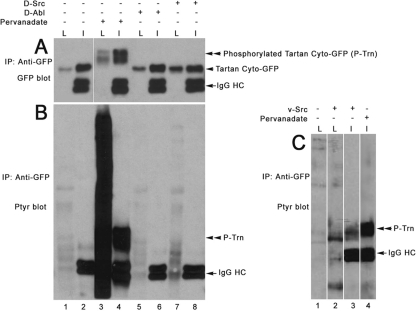
FIG. 2.
Tartan is tyrosine phosphorylated in v-Src-transfected S2 cells. (A) Anti-GFP immunoblot of lysates (L) and anti-GFP IPs (I) from cells transfected with the Trn cyto-GFP plasmid. As indicated by “+” and “−” signs above the blot, cells were left untreated (lanes 1 and 2), treated with pervanadate (lanes 3 and 4), or cotransfected with D-Abl (lanes 5 and 6) or D-Src64B plasmids (lanes 7 and 8). The immunoglobulin G heavy chain (IgG HC), unphosphorylated Trn-cyto-GFP, and tyrosine-phosphorylated Trn-cyto-GFP bands (P-Trn) are labeled. (B) Anti-PY immunoblot of the same samples. The IgG HC and P-Trn bands are labeled. (C) Anti-PY immunoblot of lysates and anti-GFP IPs from cells transfected with the Trn cyto-GFP plasmid. Cells were left untreated (lane 1), cotransfected with the v-Src plasmid (lanes 2 and 3), or treated with pervanadate (lane 4). The IgG HC and P-Trn bands are labeled. Note that the P-Trn band in lane 3 migrates more rapidly than that in lane 4, suggesting that Trn is less heavily tyrosine phosphorylated in v-Src-expressing cells than in pervanadate-treated cells.
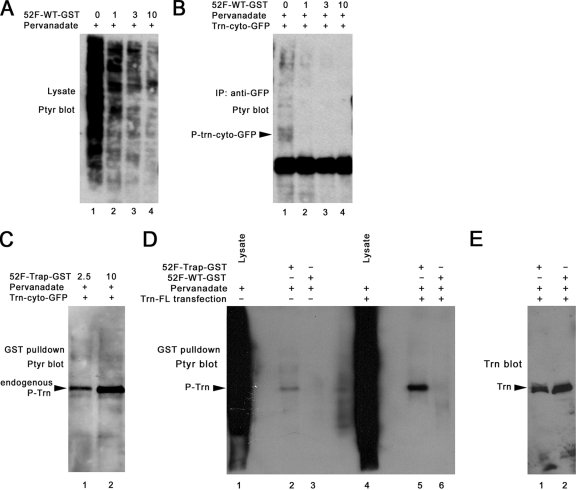
FIG. 4.
Phosphorylated Trn binds selectively to the Ptp52F substrate-trapping mutant in vitro. (A) Anti-PY immunoblot of a pervanadate-treated S2 cell lysate incubated with the indicated amounts (in μg) of Ptp52F-wild type-GST. (B) Anti-PY immunoblot of an anti-GFP IP from a lysate of pervanadate-treated S2 cells transfected with the Trn-cyto-GFP plasmid. The bead-bound protein was incubated with the indicated amounts (in μg) of Ptp52F-wild type-GST (100-μl reaction volume, 1 h, 20°C). The Trn-cyto-GFP band is indicated; other bands on the gel are presumably background. Note that the PY signal for Trn-cyto-GFP was eliminated by 1 μg of Ptp52F-wild type-GST (lane 2), showing that it dephosphorylated Trn, but many bands in Fig. 4A (lane 2) were also diminished in intensity by this same amount of fusion protein. (C) Anti-PY immunoblot of a GST “pulldown” experiment in which lysate from pervanadate-treated S2 cells transfected with the Trn-cyto-GFP plasmid was incubated with the indicated amounts of Ptp52F-trap-GST (in μg; 100-μl volume), followed by precipitation of the trap-bound proteins with glutathione-agarose beads. Only a single prominent band, of ∼85 kDa, was observed. The Trn-cyto-GFP band (∼60 kDa) was not detected. (D) Anti-PY immunoblot of a GST pulldown experiment in which lysate from pervanadate-treated untransfected or Trn-FL-transfected S2 cells was incubated with 10 μg of Ptp52F-trap-GST or Ptp52F-wild type-GST, followed by precipitation of the bound proteins as in panel C. Note that lysate from these cells produces a continuous smear but that only a single band is pulled down by Ptp52F-trap-GST, and the intensity of this band is increased when Trn-FL is overexpressed. (E) An anti-Trn immunoblot of transfected cell pulldowns from the same experiment. The Trn band is observed when either Ptp52F-trap-GST or Ptp52F-wild type-GST is used for pulldown, showing that wild-type Ptp52F dephosphorylates Trn but can then remain bound to it.
S2 cell transfection.
S2 cells were maintained in Schneider's medium supplemented with 100 U of penicillin-streptomycin/ml, 2.5 μg of amphotericin B/ml, and 10% heat-inactivated fetal bovine serum. Cells were grown at 25°C under standard conditions and were transiently transfected by calcium phosphate-mediated DNA transfer. Briefly, 3 × 106 cells per 60-mm plate were seeded in serum-only medium and expanded overnight till the cells reached 2 × 106 to 4 × 106 cells/ml. The cells were then transfected for 15 to 18 h by the addition of 600 μl of DNA-calcium phosphate coprecipitate mix (which included 3 μg of each plasmid DNA). For the findings shown in Fig. 3, experiments that examined the association of Trn with the Ptp52F trap, we cotransfected 3 μg each of Ptp52F-trap-Myc, the v-Src plasmid, and the Trn-FL, Trn cyto-GFP, or Trn FL-GFP plasmid. For the experiment to examine dephosphorylation of Trn FL-GFP by Ptp52F, the Ptp52F-wild-type-Myc plasmid was substituted for the trap plasmid. CuSO4 (500 μM final concentration) was added to the cells for 48 h prior to harvesting to induce expression from the MT promoter.
S2 transfection data for the other three candidate substrates.
We have not yet examined Xmas-2, which may be an RNA-binding protein, because the sequences of the proteins encoded by the Xmas genes are not well defined and full-length cDNA clones are not available. The clone identified in the yeast screen corresponds to part of Xmas-2. However, the Xmas-1 gene immediately downstream might be a part of the same functional unit, and published results do not clearly delineate the respective roles of these two coding sequences. CG15022 and CG10283 have no conserved domains, and they are not homologous to other proteins in nondrosophilid species. We obtained full-length cDNA clones for CG15022 and CG10283, attached their coding regions to N-terminal FLAG epitope tags, and cloned the tagged versions into MT promoter vectors. We transiently transfected these plasmids into S2 cells and induced expression from the MT promoter with copper. We observed that we could detect single bands on immunoblots of cell lysates that corresponded to the expected sizes of the tagged proteins, and both proteins were phosphorylated in pervanadate-treated cells. However, for CG15022, we discovered that phosphorylation was likely to be on the FLAG tag, which contains a Y, because a CG15022-GFP fusion was not phosphorylated under the same conditions. This may be of interest to other investigators who are examining the phosphorylation of FLAG-tagged proteins. We have not yet examined a CG10283-GFP fusion to determine whether CG10283-FLAG phosphorylation is also due to the FLAG tag.
Cell lysis and immunoprecipitation.
Cells were washed with cold phosphate-buffered saline (PBS) and lysed in ice-cold lysis buffer (125 mM NaCl, 100 mM Tris-Cl [pH 7.4], 0.2% Triton X-100, 1 mM EDTA, 600 mM phenylmethylsulfonyl fluoride, 1 μg of protease inhibitor cocktail/ml, 10% glycerol). Lysates were clarified by centrifugation at 35,000 × g for 20 min and subjected to immunoprecipitation (200-μl volume) using either 4 μl of a mouse anti-GFP antibody (Roche), 15 μl of a mouse anti-Myc antibody supernatant (9E10), or 1 μl of a rabbit anti-Trn antibody (a gift from Allen Laughon). Immune complexes were isolated by binding to protein G plus protein A-agarose beads (Oncogene Science, Inc.), followed by two cycles of lysis buffer washes. The washed immunoprecipitates were boiled in sodium dodecyl sulfate sample buffer, resolved on polyacrylamide gels, and transferred to polyvinylidene difluoride membranes. For Western blotting, we used a BM Chemiluminescence kit (Roche, catalog no. 1520709). Anti-Myc (9E10) supernatant (a gift from the Deshaies lab) was used at a 1:15 dilution for blotting, while anti-Trn, anti-GFP (Roche), and anti-PY (monoclonal antibody [MAb] 4G10; Millipore) were used at a 1:1,000 dilution.
GST fusion proteins, pulldowns, and enzymatic assays.
The Ptp52F-trap-glutathione S-transferase (GST) and Ptp52F-wild-type-GST fusion protein plasmids were created by PCR amplification of the Ptp52F cytoplasmic domain from the Ptp52F-Myc vectors described above. The Myc and His tags were included in the PCR product, but the myristylation sequence was not. The PCR products were subcloned in frame into the pGEX-2T vector to produce N-terminal GST fusions. These plasmids were transformed into the BL21 strain, and protein expression in liquid culture was induced with 50 μM IPTG (isopropyl-β-d-thiogalactopyranoside) at 30°C for 4 h. Fusion proteins, which bear the His8 tag, were purified by using Qiagen Ni-NTA Superflow resin according to the manufacturer's instructions. For enzymatic and pulldown assays, S2 cells (transfected or untransfected) were treated with pervanadate (2 mM Na3VO4, 3 mM H2O2) for 30 min at 25°C. Cells were washed with PBS and lysed in ice-cold lysis buffer with 5 mM iodoacetic acid. These lysates were incubated for 30 min with shaking in the cold, and then dithiothreitol was added to a final concentration of 10 mM. Lysates were centrifuged at high speed for 20 min in the cold. For dephosphorylation experiments such as those in Fig. 4A, lysate was mixed with various amounts of purified Ptp52F-wild type-GST and incubated for 1 h at 20°C. To assay dephosphorylation of Trn-cyto-GFP (Fig. 4B), lysate from transfected cells was first immunoprecipitated with mouse anti-GFP, and then Ptp52F-wild type-GST was added to the immunoprecipitate (IP) beads, and the mixture was incubated with rotation under the same conditions as for the lysate. For the GST pulldown experiment of Fig. 4C, the Ptp52F-trap-GST fusion protein was bound to glutathione-agarose beads for 2 h in the cold; pervanadate-treated lysate was then added, and the mixture was incubated with rotation overnight in the cold, after which the beads were spun down. The beads were washed with PBS and boiled in sodium dodecyl sulfate sample buffer. In some experiments, Ptp52F-trap-GST was incubated with lysate for 1 h at 20°C, after which glutathione-agarose beads were added, and the mixture was incubated with rotation overnight in the cold. This worked equally well. Samples from dephosphorylation and pulldown experiments were subjected to electrophoresis on polyacrylamide gels and analyzed by Western blotting with anti-PY or anti-Trn as described above.
Genetic analysis.
We combined the trn28.4 allele with the Ptp52F18.3 allele, crossing these stocks (over GFP balancers) to generate Ptp52F18.3/+, trn28.4/+ embryos. trn28.4 is a P element excision mutation that deletes coding sequence and is likely to be a null mutation. Our group has also examined other trn alleles. For example, Kurusu et al. (22) presented penetrance data for trns064117, which is a hypomorph; it generates the same phenotypes as trn28.4, but with a reduced penetrance. Ptp52F18.3 is a missense mutation that we defined as a possible null because it has the same phenotypic strength over itself or over a deficiency mutation (29). Two other Ptp52F alleles, Ptp52F7.8.1 and Ptp52F8.10.3, were also found (29) to produce the same phenotypes as Ptp52F18.3 but with a lower penetrance. In one experiment (see Fig. S1 in the supplemental material), a line with an “EP” (UAS-containing) P element upstream of the Ptp52F gene (obtained from Florenci Serras) was crossed to the Elav-GAL4 “driver line,” in backgrounds with or without the trn28.4 mutation. Thus, to remove trn function, both the driver and the EP lines were made heterozygous for the trn mutation over a GFP balancer, and embryos were sorted to find those which were EP52F × Elav-GAL4, trn/trn. Midline crossovers were scored in 1D4-stained embryos, and these data are shown in the bar graph in Fig. S1 in the supplemental material. Staining of fixed whole-mount embryos was done as described previously (25). Staining of live-dissected embryos was performed as described by Fox and Zinn (9). Two primary antibodies were used: mouse anti-FasII MAb 1D4 (Developmental Studies Hybridoma Bank), diluted 1:5, and rabbit anti-Trn (3), diluted 1:400. Cy3-conjugated or horseradish peroxidase (HRP)-conjugated secondary antibodies (Jackson Immunoresearch, West Grove, PA) were used at dilutions of 1:400 (Cy3) or 1:200 (HRP). Whole-mount embryos, stained using HRP immunohistochemistry, were dissected, and embryo “fillets” were photographed by using differential interference contrast optics on a Zeiss Axioplan microscope with a Magnafire camera. For quantitation of phenotypes, we scored segments A2 to A6, which have identical SNa morphologies.
RESULTS
Identification of candidate RPTP substrates using a modified yeast two-hybrid screen.
To search for substrates for the Drosophila RPTPs, we used methods similar to those devised by Keegan and Cooper (18) and by Noda and coworkers (10, 17). We cloned cytoplasmic domain sequences from four RPTPs into a yeast two-hybrid expression vector, inserting them in frame with the LexA DNA-binding domain (DBD). The four RPTPs are Ptp10D, Ptp52F, Ptp69D, and Ptp99A. We introduced substrate-trapping (D→A) mutations into each of these RPTP sequences. Ptp10D and Ptp52F have only one PTP homology domain, while Ptp69D and Ptp99A have two. However, the second PTP homology domain in Ptp99A has a D in place of the essential C residue, so it is unlikely to have enzymatic activity. For Ptp69D, it is unknown whether the second PTP homology domain is active, so we made a construct with D→A mutations in both domains.
To perform the screen, we introduced a plasmid encoding a version of chicken c-Src bearing three activating mutations (18), together with the RPTP-DBD plasmids and a cDNA-activation domain (AD) fusion library, and selected for ADE+ yeast bearing all three types of plasmids (13). (Constitutive expression of v-Src, which has additional activating mutations, is not tolerated well by yeast, while this version of c-Src can be expressed without adversely affecting growth.)
To screen these candidates, we first tested them for HIS+ and LacZ+, the other two selectable markers in the strain (13). We removed the URA3 (RPTP-DBD) “bait” plasmid and discarded candidates that were still ADE+ in the absence of the bait. The bait-dependent clones we obtained are listed in Table 1. For each of these, we determined whether the ADE+ phenotype required c-Src expression. Four clones that passed this test were identified, each encoding a different protein. Three were identified by screening with Ptp52F/c-Src, and one was identified by screening with Ptp10D/c-Src.
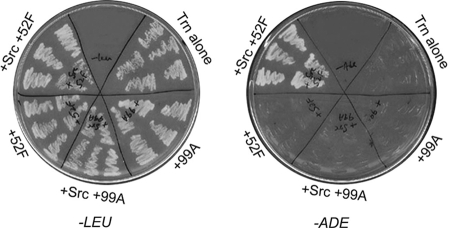
Each of these putative substrate clones was then reintroduced into yeast, together with each RPTP bait plasmid, with or without the c-Src plasmid. We also tested the four classes of non-Src-dependent clones in the same manner. Table 1 shows that the three clones identified with Ptp52F/c-Src are ADE+ only with this bait, while the Xmas-2 clone identified with Ptp10D/Src also interacts with Ptp52F/c-Src. Figure 1 shows plates with streaked yeast colonies harboring one Ptp52F/c-Src-selected clone encoding a fragment of Trn. Yeast expressing Trn-AD grew on plates lacking Ade only when they also contained the Ptp52F-trap-DBD and c-Src plasmids.
FIG. 1.
Selection of Trn in the yeast screen: growth on −Ade plates depends on both Ptp52F and c-Src. The left plate has streaks of yeast colonies (five independent colonies for each) transformed with Trn-AD (LEU+), with or without Ptp52F-trap-DBD or Ptp99A-trap-DBD, with or without c-Src, as indicated. All of these grew on plates lacking leucine (−LEU). The right plate has streaks of the same yeast colonies on plates lacking adenine (−ADE). Only the yeast with Ptp52F-trap-DBD and c-Src could express the ADE2 gene and grow on these plates.
Finally, we examined whether selection of these four putative Ptp52F substrate clones requires the substrate-trapping mutation by transforming them into yeast together with c-Src and a plasmid encoding the wild-type Ptp52F cytoplasmic domain fused to the DBD. None of the clones generated the ADE+ phenotype with wild-type Ptp52F. In summary, these data show that four Drosophila sequences, encoding portions of the Trn, Xmas-2, CG15022, and CG10283 proteins, selectively interact with the substrate-trapping mutant version of the RPTP in the presence of c-Src and therefore behave like substrates in this yeast assay.
Phosphorylation of Trn in cultured Drosophila cells.
We then wanted to determine whether the candidates identified in the yeast screen could exhibit phosphorylation-dependent interactions with RPTP substrate-trapping mutants in transfected Drosophila S2 cells. Such a demonstration would provide a rigorous test of whether interactions between the candidate substrates and the substrate-trapping mutant can occur when both proteins are present within cells from the species of origin. In a recent review of PTP-substrate interactions, substrate-trapping within cotransfected cells is defined as the most selective criterion for establishing that a candidate protein is a bona fide substrate (32).
We present here the S2 transfection results for Trn. The data for the other three candidates are in Materials and Methods. Trn is a cell surface protein with an XC domain composed primarily of leucine-rich repeats, which are ∼24-aa motifs that mediate protein-protein interactions. The Trn preprotein is 737 aa in length, and its cytoplasmic domain is 274 aa and contains 11 Y residues. The Trn coding fragment identified in the modified two-hybrid screen corresponds to cytoplasmic domain sequence from 38 aa located C-terminal to the transmembrane domain through to the stop codon.
Trn is normally expressed in S2 cells. Thus, in order to study phosphorylation of exogenous Trn and distinguish it from endogenous Trn, we needed to express “tagged” versions of truncated or full-length Trn by transfection. We initially expressed a fusion protein consisting of the myristylation sequence from Src, the complete cytoplasmic domain of Trn, and the complete GFP coding region. Myristylation of the protein should promote its association with the plasma membrane. We transfected MT promoter plasmids encoding this protein, Trn-cyto-GFP, into S2 cells and induced expression with copper. Tyrosine phosphorylation was induced by treating the transfected cells with H2O2 and Na3VO4 (pervanadate), which causes massive accumulation of PY in intact cells by inhibiting all PTP activity, for 30 min prior to lysis. This treatment efficiently induced phosphorylation of endogenous Trn (data not shown). We examined lysates and anti-GFP IPs from these cells by immunoblotting with anti-GFP and anti-PY. A single band of ∼60 kDa was seen, a finding consistent with the predicted molecular mass of Trn-cyto-GFP (Fig. 2A, lanes 1 and 2).
Immunoblotting of lysates from pervanadate-treated cells with anti-PY produced an intense signal at all molecular mass positions (Fig. 2B, lane 3). Pervanadate treatment of S2 cells generates so many phosphoprotein bands that they cannot be resolved on a gel, while discrete bands can sometimes be observed in lysates of pervanadate-treated mammalian cells (see, for example, reference 11). In anti-GFP IPs of these lysates, the 60-kDa Trn-cyto-GFP band (Fig. 2A, lane 2) disappeared and was replaced by two or three bands (Fig. 2A, lane 4) that migrated more slowly. These bands are likely to represent tyrosine-phosphorylated Trn-cyto-GFP species, since they were also detected by immunoblotting with anti-PY (Fig. 2B, lane 4, labeled as P-Trn).
We also attempted to induce Trn phosphorylation by expressing two Drosophila TKs, D-Src64B and D-Abl. We had previously shown that the TK expression plasmids we used for cotransfection can both increase phosphorylation of Ena in S2 cells, demonstrating that the kinases are functional (7). However, neither kinase was able to cause tyrosine phosphorylation of Trn-cyto-GFP (Fig. 2B, lanes 6 and 8), and the intensity of the tyrosine-phosphorylated bands seen in lysates was not greatly increased in cells expressing these kinases (Fig. 2B, compare lane 1 to lanes 5 and 7). Since neither of these Drosophila TKs could induce Trn phosphorylation, we next tested chicken v-Src, which is a highly active TK. Transfection of a v-Src expression plasmid produced more tyrosine phosphorylation in lysates than the D-Src or D-Abl plasmids (compare Fig. 2B, lanes 5 and 7, to Fig. 2C, lane 2). Trn-cyto-GFP was phosphorylated in cells expressing v-Src, although at lower levels than those attained with pervanadate treatment (Fig. 2C, lanes 3 and 4, P-Trn band).
Phosphorylated Trn binds to the Ptp52F substrate-trapping mutant in S2 cells.
Having demonstrated that the Trn cytoplasmic domain can be phosphorylated in S2 cells, we conducted cotransfection experiments to determine whether the Ptp52F substrate-trapping mutant could bind to v-Src-phosphorylated Trn. This experiment cannot be done in pervanadate-treated cells, because vanadate binds to the active sites of trap mutants and blocks their association with substrates.
Ptp52F is not endogenously expressed in S2 cells, so interactions between Trn and Ptp52F can only be studied in cells transfected with Ptp52F constructs. The predicted Ptp52F preprotein is 1,433 aa in length, and its cytoplasmic domain is 369 aa. Full-length Ptp52F cDNAs have never been isolated, and we had to use reverse transcription-PCR to define the complete sequence of its XC domain (29). Because of this, we decided to express the Ptp52F cytoplasmic domain with a myristylation sequence in S2 cells, so that it would be likely to associate with the plasma membrane and might therefore come into contact with Trn. We made wild-type and substrate-trapping mutant constructs with N-terminal Src myristylation sequences and 9xMyc tags at the C terminus. When these were transfected into S2 cells, a single band of ∼65 kDa was observed in lysates and anti-Myc IPs, which is consistent with the predicted molecular mass of the fusion protein (Fig. 3A).
Because we had demonstrated that Trn-cyto-GFP could be phosphorylated in S2 cells, we cotransfected Ptp52F-trap-Myc, Trn-cyto-GFP, and v-Src constructs in our initial experiments but did not find any evidence for association. This could have been due to inefficient localization of the Ptp52F and Trn proteins to the same compartment, to occlusion of trap binding to the cytoplasmic domain by GFP, or to insufficiently high expression of one of the proteins. To address these issues, we then made a construct (Trn-FL) with untagged full-length Trn. When transfected into S2 cells, this resulted in expression of wild-type Trn protein (∼85 kDa) at levels severalfold greater than those seen for endogenous Trn in untransfected cells (data not shown). When we cotransfected Ptp52F-trap-Myc and Trn-FL constructs into S2 cells together with v-Src, we were able to detect Trn in Myc IPs from these cells (Fig. 3B, lane 2). This Trn band was much weaker in Myc IPs from Ptp52F-wild type-Myc-cotransfected cells (Fig. 3B, lane 5). It can be clearly visualized upon longer exposure, however, showing that Ptp52F-wild type-Myc can also form a complex with Trn, albeit with a lower efficiency than the substrate-trapping mutant.
To further examine association between Ptp52F and Trn, we needed to make a tagged Trn fusion protein that could bind to the Ptp52F substrate-trap, so that we could distinguish this protein from endogenous Trn. We thus made a construct in which GFP was attached to the C terminus of full-length Trn. When a plasmid bearing this construct, Trn-FL-GFP, was transfected, we observed a band of ∼100 kDa that could be precipitated by either anti-GFP or anti-Trn antibodies (Fig. 3C). Interestingly, we also saw a second band of ∼60 kDa that reacted with both antibodies. This appears to be a cleavage product, and given its size and the fact that it must contain GFP, we infer that the XC domain can be cleaved near the plasma membrane.
To study dephosphorylation of Trn-FL-GFP, we cotransfected it together with Ptp52F-wild type-Myc or Ptp52F-trap-Myc and v-Src. Trn-FL-GFP and the cleavage product were both tyrosine phosphorylated when cotransfected with the Ptp52F trap and Src (Fig. 3D, lane 1). This was not due to protection from endogenous PTP activity by trap binding, because phosphorylation was detected at the same levels when Trn-FL-GFP and v-Src were cotransfected without the trap (data not shown). This is consistent with the observation that only a small fraction of Trn is bound by the trap (Fig. 3B). When wild-type Ptp52F was expressed together with Trn-FL-GFP and the kinase, tyrosine-phosphorylated Trn and its cleavage product were barely detectable (Fig. 3D, lane 2). These data show that Ptp52F can cause dephosphorylation of the Trn cytoplasmic domain.
To examine whether Ptp52F-trap-Myc could bind to Trn-FL-GFP, we immunoprecipitated the Trn protein with anti-GFP and then detected the Ptp52F protein by immunoblotting with anti-Myc. We observed a clear signal when Ptp52F-trap-Myc was cotransfected with both v-Src and Trn-FL-GFP (Fig. 3E, lane 5), but no association was detected when the kinase construct was omitted. These data show that the substrate-trapping mutant binds to Trn in a phosphorylation-dependent manner. With wild-type Ptp52F, a very faint band was observed in Myc immunoblots of anti-GFP IPs (Fig. 3E, lane 11), indicating that there was much less association between the wild-type RPTP and Trn. This band is clearly visible with longer exposure, however, demonstrating that the wild-type RPTP does form a complex with Trn-FL-GFP in cotransfected cells.
When lysates were immunoprecipitated with anti-Myc, we were able to observe the association of Trn-FL-GFP with Ptp52F-trap-Myc by immunoblotting with anti-PY (Fig. 3F, lane 4), although the signal was always weaker than that seen when anti-GFP IPs were probed with anti-Myc. Figure 3B, lane 2, shows that we can readily detect Trn-FL by blotting anti-Myc IPs with anti-Trn. However, for reasons we do not understand, we could not reproducibly detect association between Trn-FL-GFP and Ptp52F-trap-Myc by blotting anti-Myc IPs with anti-GFP.
Selective in vitro binding of Ptp52F and Trn.
The results described above demonstrate that the Ptp52F substrate-trapping mutant can bind to Trn in S2 cells and that wild-type Ptp52F can cause dephosphorylation of Trn. However, these data do not show that Trn is the only protein in S2 cells that binds to the Ptp52F trap. To address selectivity, it is necessary to examine binding to the entire spectrum of proteins that are tyrosine phosphorylated when cells are treated with pervanadate. This cannot be done by cotransfection, since vanadate occludes binding by PTP substrate-trapping mutants.
Many other investigators (see, for example, reference 11) have performed experiments in which they assessed binding of PTP substrate-trapping mutants purified as GST fusion proteins from Escherichia coli to proteins in a lysate from cells that were treated with pervanadate, followed by inactivation of the pervanadate with iodoacetate and dithiothreitol. We used this approach for Ptp52F, purifying substrate-trapping mutant and wild-type GST fusion proteins from E. coli lysates.
We first examined dephosphorylation by the wild-type Ptp52F fusion protein. To do this, we added various amounts of Ptp52F-wild type-GST to pervanadate-treated S2 cell lysates, followed by incubation for 1 h at 20°C before fractionating the samples on a gel and immunoblotting them with anti-PY. Figure 4A shows that all of the bands in the lysate were reduced in intensity as the amount of Ptp52F-wild type-GST was increased. It is not obvious that any bands were selectively dephosphorylated, although some bands appeared to be more resistant than others. The PTP-GST fusion protein appears to have a surprisingly low level of enzymatic activity, since 1 μg (∼150 nM) was required to achieve >75% dephosphorylation of most bands. However, other investigators have obtained similar results: for example, 70 nM GST-SHP1 was required to produce >75% dephosphorylation of the SHP-1 substrate Lck in vitro (4).
We also examined the dephosphorylation of Trn-cyto-GFP by analyzing anti-GFP IPs from pervanadate-treated lysates of cells transfected with the Trn-cyto-GFP plasmid. These data (Fig. 4B) show that phosphorylation of the Trn-cyto-GFP protein was eliminated by 1 μg of Ptp52F-GST fusion protein. This experiment suggests that the wild-type enzyme does not exhibit strong specificity for Trn-cyto-GFP as a substrate in vitro, since many bands in Fig. 4A were also reduced in intensity when 1 μg of Ptp52F-GST was used. In other experiments, we observed that ∼50% dephosphorylation of Trn-cyto-GFP can be achieved with 100 ng of Ptp52F-wild type-GST (∼15 nM). This amount of Ptp52F-wild type-GST produces no detectable effect when the entire lysate is analyzed. However, since there are so many PY bands in a pervanadate-treated S2 cell lysate that individual bands cannot be resolved (e.g., lane 1 of Fig. 4A, and lanes 1 and 4 of Fig. 4D), we would not be able to detect 50% dephosphorylation of other proteins (data not shown).
To address the selectivity of Ptp52F trap binding, we incubated various amounts of Ptp52F-trap-GST with pervanadate-treated S2 cell lysates and precipitated protein bound to the trap using glutathione-agarose beads. We then analyzed bead-bound proteins by blotting with anti-PY as in Fig. 4A. This produced a remarkable result: only a single prominent phosphoprotein band, of ∼85 kDa, was precipitated by the substrate-trapping mutant (Fig. 4C). The migration of this band was consistent with that of endogenous Trn, and it was recognized by anti-Trn antibody (Fig. 4E, lane 1).
Although the cells used for the experiment of Fig. 4C were transfected with Trn-cyto-GFP plasmid, no band corresponding to Trn-cyto-GFP was detectable on the blot. We also could not detect Trn-cyto-GFP or Trn-FL-GFP binding to Ptp52F-trap-GST when glutathione-agarose precipitates were analyzed by immunoblotting with anti-GFP. Perhaps the GFP and GST domains of the fusion proteins sterically interfere with each other to prevent binding.
To further evaluate the interaction between Ptp52F-GST and Trn, we then incubated 10 μg of Ptp52F-trap-GST or Ptp52F-wild type-GST with pervanadate-treated lysates from cells transfected with untagged Trn-FL plasmids (or left untransfected) and detected bound phosphoproteins by blotting the cells with anti-PY. Lysates from these cells are shown in lanes 1 and 4 of Fig. 4D. As in Fig. 4C, we observed that Ptp52F-trap-GST precipitated only a single ∼85-kDa band from the lysates and that the intensity of this band was increased when the cells had been transfected with Trn-FL, a finding consistent with the identification of this band as phospho-Trn (Fig. 4D, lanes 2 and 5).
When Ptp52F-wild type-GST was used for the pulldown, no band was detected on the anti-PY blot. Interestingly, however, when pulldowns from Trn-FL-transfected cells were blotted with anti-Trn, equal amounts of Trn were observed to be precipitated by the wild-type and substrate-trapping mutant proteins (Fig. 4E). These data show that the wild-type Ptp52F protein can also form a stable complex with Trn that persists after dephosphorylation. The stability of this complex, or the affinity of Trn and Ptp52F for each other, may be lower for the wild-type than for the substrate-trapping mutant, since much more Trn associated with the substrate-trapping mutant in cotransfected S2 cells (Fig. 3B, lane 2 versus lane 5; Fig. 3E, lane 5 versus lane 11). However, the vast excess of wild-type Ptp52F used in pulldown experiments is apparently capable of bringing down the same amount of Trn even if its affinity for dephosphorylated Trn is lower than that of the substrate-trapping mutant for phosphorylated Trn.
Genetic interactions between Ptp52F and tartan.
The results described above show that Trn is a preferred substrate for Ptp52F in S2 cells. It would be difficult to use biochemical techniques to determine whether this is also the case in vivo, because Trn is not an abundant protein, and it is unlikely to be heavily tyrosine phosphorylated during normal development. However, we can examine whether Ptp52F and Trn are involved in the same developmental processes by using genetics.
Ptp52F, like other Drosophila RPTPs, is selectively expressed in CNS neurons and is a regulator of axon guidance. When we identified the Ptp52F gene and isolated mutations in it, we discovered that Ptp52F mutant embryos have two strong axon guidance phenotypes. The longitudinal tracts in the CNS are disorganized and incomplete, and one branch of the motor axon network, the SNa nerve, exhibits guidance errors (29).
During embryonic development, the 36 motor axons in each abdominal hemisegment grow out from the CNS and segregate into six major branches, called SNa, SNc, ISN, ISNb, ISNd, and TN, which innervate the 30 body wall muscle fibers. Motor axons can be selectively visualized by staining dissected embryos with MAb 1D4 (34). Single mutations or combinations of mutations in the six Rptp genes affect the guidance of all motor axon branches (see reference 14 for a recent summary). Ptp52F is the only Rptp gene for which single mutations affect the SNa branch. In Ptp52F-null mutants, >40% of SNa nerves exhibit guidance errors (Fig. 5F), while 22 to 27% have phenotypes in hypomorphic (weaker) mutants. These are the only high-penetrance motor axon phenotypes seen in Ptp52F single mutants. There are also some ISN phenotypes (16%), but only rare ISNb defects (7%) (29).
FIG. 5.
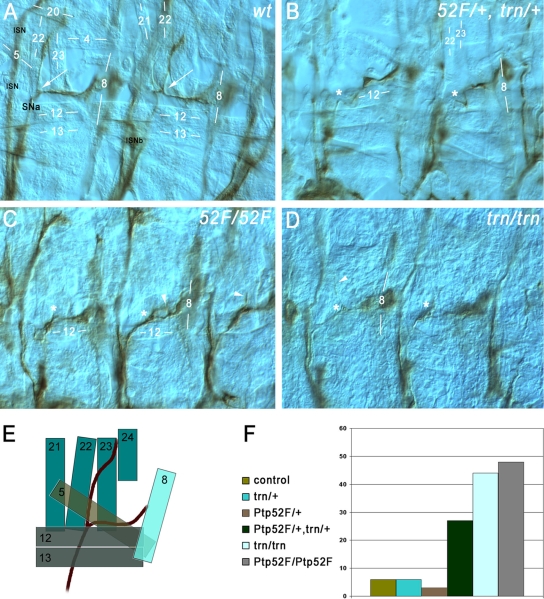
Ptp52F and tartan mutants have the same motor axon guidance phenotypes, and the genes display a dosage-dependent interaction. Motor axons (brown) in late-stage 16/early-stage 17 embryo “fillets” were stained with MAb 1D4, using HRP immunohistochemistry for visualization, and photographed using differential interference contrast optics. (A) Two hemisegments in a wild-type (wt) control embryo. SNa, the branch that exhibits phenotypes in both of these mutants, is labeled. Its bifurcation point is indicated by white arrows in both segments. The ISN and ISNb (out of focus) are also labeled. Muscle fibers are labeled by number (compare to diagram of panel E). The anterior SNa branch normally extends dorsally along muscle 22 and then across muscle 23 to reach muscle 24, and the posterior branch extends across muscle 5 to reach muscle 8. (B) Two hemisegments in a Ptp52F/+, trn/+ embryo (lacking one wild-type copy of each gene). The anterior branch of the SNa is missing in both segments. The approximate points at which bifurcation would have occurred if the anterior branches were present are indicated by asterisks. (C) Three hemisegments in a Ptp52F/Ptp52F embryo. The anterior branch of the SNa is missing, truncated, or misguided in all three. In the left-hand hemisegment the branch is missing, while in the middle hemisegment axons branch off near the normal bifurcation point (asterisks) but then grow posteriorly and rejoin the posterior SNa branch, forming a loop (arrowhead). In the right-hand hemisegment a thin and truncated anterior branch is observed (arrowhead). (D) Two hemisegments in a trn/trn embryo. In the right-hand segment, the anterior branch is missing, while in the left-hand hemisegment a single axon appears to have extended partway along the normal anterior branch pathway (arrowhead), leaving the SNa near the normal bifurcation point (asterisks). (E) Diagram of the SNa and adjacent muscle fibers in wild-type. The muscles are indicated as semitransparent to show their layering. The deepest (most external) muscles are 21 to 24, and they are overlaid by muscles 5, 12, 13, and 8. SNa extends underneath (external to) 12 and 13. (F) Bar graph of phenotypic penetrance for SNa guidance errors, in control (balancer/+); trn/+; Ptp52F/+; Ptp52F/+, trn/+; trn/trn; and Ptp52F/Ptp52F cells. The numbers of hemisegments examined and the distribution among phenotypic classes are shown in Table 2.
The SNa grows out from the CNS in the SN root and bifurcates dorsal to muscle 12. Its posterior (or ventral) branch innervates muscles 5 and 8, while its anterior (or dorsal) branch innervates muscles 22 to 24 (Fig. 5A and E). The most common SNa guidance error observed in Ptp52F mutants is a failure to bifurcate, so that one branch is missing. The anterior branch is lost more frequently than the posterior branch (Fig. 5C).
Trn is expressed in CNS neurons but also in a variety of other cell types (3). Motor axon phenotypes had not been previously analyzed in trn mutants. When we examined trn mutant embryos by staining with MAb 1D4, we made the remarkable discovery that they have SNa phenotypes that are identical to those of Ptp52F mutants (Fig. 5D and F and Table 2). trn-null embryos display SNa defects with >40% penetrance, and the most common phenotype is the absence of the anterior branch. They also have ISNb defects, which are not seen in Ptp52F mutants. Trn also has other functions in the neuromuscular system; a recent paper from our group identified Trn and several other leucine-rich repeat proteins as synaptic target selection cues on ventrolateral muscles. As part of this analysis, we quantified ISNb and SNa phenotypes in trn-null and trn-hypomorph embryos and also showed that these phenotypes are primarily due to loss of Trn from neurons (22).
TABLE 2.
Quantitation of SNa phenotypes
| Genotype | No. of hemisegments (A2 to A6) scored | Phenotypic percentage
|
|||
|---|---|---|---|---|---|
| Missing branches | Extra branches | Bypass | Total | ||
| balancer/+(control) | 339 | 5 | 1 | 0 | 6 |
| trn/+ | 178 | 4 | 2 | 0 | 6 |
| Ptp52F/+ | 131 | 3 | 0 | 0 | 3 |
| Ptp52F/+, trn/+ | 198 | 26 | 1 | 0 | 27 |
| trn/trn | 192 | 30 | 12 | 2 | 44 |
| Ptp52F/Ptp52F | 169 | 42 | 4 | 2 | 48 |
In the present study, our major purpose is to document the shared Ptp52F and trn SNa guidance phenotype and to determine whether the two proteins function within the same pathway or process. To do this, we examined dosage-sensitive genetic interactions between them. This is a standard method for establishing whether two gene products are functionally related. An example that is relevant to axon guidance is provided by the Roundabout (Robo) receptor and its ligand Slit; removal of 50% of gene function for both Slit and Robo (in slit/+, robo/+ embryos) pathway components produces defects that could otherwise only be generated by complete loss of Robo (20).
We analyzed Ptp52F and Trn in a similar manner and found that Ptp52F/+ and trn/+ embryos do not display SNa guidance errors (null mutations were used for both genes). However, Ptp52F/+, trn/+ embryos have SNa phenotypes like those seen in Ptp52F/Ptp52F or trn/trn embryos, with a penetrance of 27% (Fig. 5B and F and Table 2). The differences in penetrance between Ptp52F/+, trn/+ embryos and Ptp52F/+ or trn/+ embryos are statistically significant (P < 0.0001, chi-square test). Ptp52F/+, trn/+ embryos do not have CNS or ISNb phenotypes. These phenotypes are not shared between Ptp52F/Ptp52F and trn/trn and therefore presumably involve pathways that do not require both proteins. In summary, our results suggest that bifurcation of the SNa nerve is dependent on a pathway that includes both Ptp52F and Trn.
Another way to evaluate genetic interactions is through suppression of a gain-of-function phenotype for one gene by a loss of function for the other. Neuronal overexpression of Trn did not produce phenotypes, so the potential role of Ptp52F in altering Trn signaling through dephosphorylation could not be examined in this manner. Neuronal overexpression of Ptp52F, however, generates CNS axon guidance phenotypes (26), and these are suppressed by removal of trn function (see Fig. S1 in the supplemental material). This represents a requirement for Trn in mediating Ptp52F function, rather than control of Trn by Ptp52F. Perhaps the normal activity of Ptp52F in vivo requires formation of a complex with Trn.
DISCUSSION
In this study, we present evidence that the cell surface receptor Trn is a substrate for Ptp52F. Trn was identified in a yeast screen for phosphoproteins that bind selectively to a Ptp52F substrate-trapping mutant (Table 1 and Fig. 1). We showed that Trn can be phosphorylated on tyrosine in S2 cells (Fig. 2) and that phosphorylated Trn binds selectively to the substrate-trapping mutant of Ptp52F when it is coexpressed with Trn and the v-Src kinase (Fig. 3B, E, and F). Wild-type Ptp52F causes dephosphorylation of v-Src-phosphorylated Trn in S2 cells (Fig. 3D). A purified Ptp52F-wild-type-GST fusion protein can dephosphorylate Trn in a pervanadate-treated S2 cell lysate, but it also dephosphorylates many other proteins in the lysate. However, a Ptp52F-trap-GST fusion protein binds to only one phosphoprotein in pervanadate-treated S2 lysates, and we identified this protein as Trn (Fig. 4C to E). Ptp52F-wild type-GST forms a complex with Trn that persists after dephosphorylation (Fig. 4D and E). These data demonstrate that the Ptp52F substrate-trapping mutant has a strong specificity for Trn binding. Our results fulfill all three of the criteria listed by Tiganis and Bennett (32) as necessary for the rigorous definition of a protein as a PTP substrate: (i) direct interaction with the PTP substrate-trapping mutant in transfected cells, (ii) modulation of cellular substrate tyrosine phosphorylation by the PTP in transfected cells, and (iii) in vitro dephosphorylation of substrate by the PTP.
Our genetic results (Fig. 5) are consistent with a model in which Trn signaling in SNa motor neurons is necessary for correct axon pathfinding at the SNa bifurcation point, and Ptp52F regulates Trn via dephosphorylation. In trn and Ptp52F mutants, SNa axons destined for muscles 22 to 24 sometimes fail to separate from those destined for muscles 5 and 8. This results in a phenotype in which the anterior branch of the SNa is missing. In SNa growth cones, signaling through Trn and dephosphorylation of Trn by Ptp52F might be regulated by the interactions of these two receptors with unknown ligands on cells near the SNa bifurcation point. Activation of Ptp52F might involve a secreted protein called Folded gastrulation (Fog), which is expressed in this vicinity. Fog is a positive regulator of Ptp52F function in SNa neurons (26).
Trn is also important for ISNb axon guidance (22) and is involved in many other developmental processes, including tracheal development and the sorting of cells within imaginal discs and developing appendages (21, 23, 24, 28). Trn signaling in ISNb neurons and tracheae may be independent of Ptp52F, since trn and Ptp52F do not share ISNb or tracheal phenotypes.
How does Ptp52F regulate Trn signaling in neurons? One might have expected that Ptp52F would be a negative regulator of Trn, because dephosphorylation of Trn would prevent it from binding to SH2-domain downstream signaling proteins. Several Y residues in the Trn cytoplasmic domain are in sequence contexts that suggest that they could bind to SH2 domains if they were phosphorylated. In this model, however, mutation of Ptp52F should lead to an increase rather than a decrease in Trn signaling. We do not know what consequences might result from increasing Trn signaling, since overexpression of Trn in neurons does not produce phenotypes (data not shown). However, the fact that the trn and Ptp52F loss-of-function phenotypes are the same and that the two genes interact in a dosage-dependent manner suggests that Trn signaling is reduced in the absence of Ptp52F and thus that Ptp52F is actually a positive regulator of Trn.
Interestingly, these relationships between the Trn receptor, the unknown TK that phosphorylates Trn in vivo, and the RPTP resemble those described for the Robo receptor, which controls axon guidance across the CNS midline. Robo signaling is antagonized by the Abl TK (1), which can phosphorylate its cytoplasmic domain, and is facilitated by the Ptp10D and Ptp69D RPTPs (30). There are several models that could explain this apparent positive regulation by dephosphorylation. Tyrosine-phosphorylated Trn or Robo might be downregulated by endocytosis. It is known that Robo can be regulated in this manner: the transmembrane protein Commissureless, which downregulates Robo function in vivo, can remove it from the cell surface by diverting it into an endocytic pathway (12, 19). In another model, an SH2-domain protein might bind to a phosphorylated tyrosine residue in Trn or Robo and occlude binding of a positive regulator that normally binds to an adjacent site in a phosphorylation-independent manner.
Supplementary Material
Acknowledgments
We thank Allen Laughon for anti-Trn antibodies; Stephen Elledge for the cDNA library; Philip James for yeast vectors and strains; Kathy Keegan for the c-Src plasmid; Tony Hunter for a v-Src clone; Amy Cording (Zinn lab) for fixed embryos, discussions, and communication of results before publication; Ed Silverman (Zinn lab) for help with GST fusion protein purifications; and Girish Ratnaparkhi for S2 cells, vectors, protocols, and discussions. A.R. thanks L. S. Shashidhara and the Biology Division of IISER for the use of their facilities.
This study was supported by National Institutes of Health grant RO1 NS28182 to K.Z.
Footnotes
Published ahead of print on 30 March 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bashaw, G. J., T. Kidd, D. Murray, T. Pawson, and C. S. Goodman. 2000. Repulsive axon guidance: Abelson and Enabled play opposing roles downstream of the roundabout receptor. Cell 101703-715. [DOI] [PubMed] [Google Scholar]
- 2.Bunch, T. A., Y. Grinblat, and L. S. Goldstein. 1988. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res. 161043-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, Z., B. D. Price, S. Bockheim, M. J. Boedigheimer, R. Smith, and A. Laughon. 1993. Molecular and genetic characterization of the Drosophila tartan gene. Dev. Biol. 160315-332. [DOI] [PubMed] [Google Scholar]
- 4.Chiang, G. G., and B. M. Sefton. 2001. Specific dephosphorylation of the Lck tyrosine protein kinase at Tyr-394 by the SHP-1 protein-tyrosine phosphatase. J. Biol. Chem. 27623173-23178. [DOI] [PubMed] [Google Scholar]
- 5.Desai, C. J., J. G. Gindhart, Jr., L. S. Goldstein, and K. Zinn. 1996. Receptor tyrosine phosphatases are required for motor axon guidance in the Drosophila embryo. Cell 84599-609. [DOI] [PubMed] [Google Scholar]
- 6.Desai, C. J., N. X. Krueger, H. Saito, and K. Zinn. 1997. Competition and cooperation among receptor tyrosine phosphatases control motoneuron growth cone guidance in Drosophila. Development 1241941-1952. [DOI] [PubMed] [Google Scholar]
- 7.Fashena, S. J., and K. Zinn. 1997. Transmembrane glycoprotein gp150 is a substrate for receptor tyrosine phosphatase DPTP10D in Drosophila cells. Mol. Cell. Biol. 176859-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flint, A. J., T. Tiganis, D. Barford, and N. K. Tonks. 1997. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc. Natl. Acad. Sci. USA 941680-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox, A. N., and K. Zinn. 2005. The heparan sulfate proteoglycan syndecan is an in vivo ligand for the Drosophila LAR receptor tyrosine phosphatase. Curr. Biol. 151701-1711. [DOI] [PubMed] [Google Scholar]
- 10.Fukada, M., and M. Noda. 2007. Yeast substrate-trapping system for isolating substrates of protein tyrosine phosphatases. Methods Mol. Biol. 365371-382. [DOI] [PubMed] [Google Scholar]
- 11.Garton, A. J., A. J. Flint, and N. K. Tonks. 1996. Identification of p130Cas as a substrate for the cytosolic protein tyrosine phosphatase PTP-PEST. Mol. Cell. Biol. 166408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgiou, M., and G. Tear. 2003. The N-terminal and transmembrane domains of Commissureless are necessary for its function and trafficking within neurons. Mech. Dev. 1201009-1019. [DOI] [PubMed] [Google Scholar]
- 13.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 1441425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeon, M., H. Nguyen, S. Bahri, and K. Zinn. 2008. Redundancy and compensation in axon guidance: genetic analysis of the Drosophila Ptp10D/Ptp4E receptor tyrosine phosphatase subfamily. Neural Dev. 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, K. G., A. P. Tenney, A. Ghose, A. M. Duckworth, M. E. Higashi, K. Parfitt, O. Marcu, T. R. Heslip, J. L. Marsh, T. L. Schwarz, J. G. Flanagan, and D. Van Vactor. 2006. The HSPGs Syndecan and Dallylike bind the receptor phosphatase LAR and exert distinct effects on synaptic development. Neuron 49517-531. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, K. G., and D. Van Vactor. 2003. Receptor protein tyrosine phosphatases in nervous system development. Physiol. Rev. 831-24. [DOI] [PubMed] [Google Scholar]
- 17.Kawachi, H., A. Fujikawa, N. Maeda, and M. Noda. 2001. Identification of GIT1/Cat-1 as a substrate molecule of protein tyrosine phosphatase zeta/beta by the yeast substrate-trapping system. Proc. Natl. Acad. Sci. USA 986593-6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keegan, K., and J. A. Cooper. 1996. Use of the two hybrid system to detect the association of the protein-tyrosine-phosphatase, SHPTP2, with another SH2-containing protein, Grb7. Oncogene 121537-1544. [PubMed] [Google Scholar]
- 19.Keleman, K., S. Rajagopalan, D. Cleppien, D. Teis, K. Paiha, L. A. Huber, G. M. Technau, and B. J. Dickson. 2002. Comm sorts Robo to control axon guidance at the Drosophila midline. Cell 110415-427. [DOI] [PubMed] [Google Scholar]
- 20.Kidd, T., K. S. Bland, and C. S. Goodman. 1999. Slit is the midline repellent for the robo receptor in Drosophila. Cell 96785-794. [DOI] [PubMed] [Google Scholar]
- 21.Krause, C., C. Wolf, J. Hemphala, C. Samakovlis, and R. Schuh. 2006. Distinct functions of the leucine-rich repeat transmembrane proteins capricious and tartan in the Drosophila tracheal morphogenesis. Dev. Biol. 296253-264. [DOI] [PubMed] [Google Scholar]
- 22.Kurusu, M., A. Cording, M. Taniguchi, K. Menon, E. Suzuki, and K. Zinn. A screen of cell-surface molecules identifies leucine-rich repeat proteins as key mediators of synaptic target selection. Neuron, in press. [DOI] [PMC free article] [PubMed]
- 23.Milan, M., L. Perez, and S. M. Cohen. 2005. Boundary formation in the Drosophila wing: functional dissection of Capricious and Tartan. Dev. Dyn. 233804-810. [DOI] [PubMed] [Google Scholar]
- 24.Milan, M., U. Weihe, L. Perez, and S. M. Cohen. 2001. The LRR proteins capricious and Tartan mediate cell interactions during DV boundary formation in the Drosophila wing. Cell 106785-794. [DOI] [PubMed] [Google Scholar]
- 25.Patel, N. H. 1994. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Methods Cell Biol. 44445-487. [DOI] [PubMed] [Google Scholar]
- 26.Ratnaparkhi, A., and K. Zinn. 2007. The secreted cell signal Folded gastrulation regulates glial morphogenesis and axon guidance in Drosophila. Dev. Biol. 308158-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz-Canada, C., and V. Budnik. 2006. Introduction on the use of the Drosophila embryonic/larval neuromuscular junction as a model system to study synapse development and function, and a brief summary of pathfinding and target recognition. Int. Rev. Neurobiol. 751-31. [DOI] [PubMed] [Google Scholar]
- 28.Sakurai, K. T., T. Kojima, T. Aigaki, and S. Hayashi. 2007. Differential control of cell affinity required for progression and refinement of cell boundary during Drosophila leg segmentation. Dev. Biol. 309126-136. [DOI] [PubMed] [Google Scholar]
- 29.Schindelholz, B., M. Knirr, R. Warrior, and K. Zinn. 2001. Regulation of CNS and motor axon guidance in Drosophila by the receptor tyrosine phosphatase DPTP52F. Development 1284371-4382. [DOI] [PubMed] [Google Scholar]
- 30.Sun, Q., S. Bahri, A. Schmid, W. Chia, and K. Zinn. 2000. Receptor tyrosine phosphatases regulate axon guidance across the midline of the Drosophila embryo. Development 127801-812. [DOI] [PubMed] [Google Scholar]
- 31.Sun, Q., B. Schindelholz, M. Knirr, A. Schmid, and K. Zinn. 2001. Complex genetic interactions among four receptor tyrosine phosphatases regulate axon guidance in Drosophila. Mol. Cell Neurosci. 17274-291. [DOI] [PubMed] [Google Scholar]
- 32.Tiganis, T., and A. M. Bennett. 2007. Protein tyrosine phosphatase function: the substrate perspective. Biochem. J. 4021-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tonks, N. K. 2006. Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell. Biol. 7833-846. [DOI] [PubMed] [Google Scholar]
- 34.Vactor, D. V., H. Sink, D. Fambrough, R. Tsoo, and C. S. Goodman. 1993. Genes that control neuromuscular specificity in Drosophila. Cell 731137-1153. [DOI] [PubMed] [Google Scholar]
- 35.Wills, Z., J. Bateman, C. A. Korey, A. Comer, and D. Van Vactor. 1999. The tyrosine kinase Abl and its substrate enabled collaborate with the receptor phosphatase Dlar to control motor axon guidance. Neuron 22301-312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.