Abstract
The transcription factor Ikaros is essential for B cell development. However, its molecular functions in B cell fate specification and commitment have remained elusive. We showed that the transcription factor EBF rescued the generation of CD19+ pro-B cells from Ikzf1-/- hematopoietic progenitors. Intriguingly, these pro-B cells, in spite of expressing normal amounts of EBF and Pax5, were not committed to the B cell fate. They also failed to selectively undergo VH to DJH recombination at the immunoglobulin heavy-chain (Igh) locus. Ikaros induced VH gene rearrangements by activating Rag gene expression as well as by controlling VH gene accessibility and compaction of the Igh locus. Thus Ikaros is an obligate component of a network that regulates B cell fate commitment and Igh recombination.
Keywords: Ikaros, B cell fate commitment, Rag gene expression, Immunoglobulin heavy chain recombination
B lineage precursors are generated from hematopoietic stem cells (HSCs) through an ordered developmental pathway that is controlled by a hierarchical and dynamic regulatory network that includes the cytokine receptors Flt3 and interleukin 7R (IL-7R) and the transcription factors PU.1, Ikaros (A001169), Bcl11a, E2A, EBF (A000809) and Pax5 (A000403)1,2. PU.1 and E2A can be regarded as competence factors for this developmental pathway as these are required for B cell development in vivo but their functions can be bypassed using in vitro culture systems and single gene molecular complementation3-6. On the other hand, EBF and Pax5 are obligate factors as their functions cannot be bypassed by simple complementation assays. These two transcription factors directly control three distinct molecular processes in early B cell development that includes activation of B lineage specific genes, repression of alternate lineage genes and rearrangement of the immunoglobulin heavy chain (Igh) locus1,7,8. The molecular functions of Ikaros in the B lineage regulatory network have remained enigmatic although it has generally been assumed to function as a developmental competence factor.
Ikaros is the prototypical member of a Kruppel-like zinc finger transcription factor sub-family, which also includes Helios and Aiolos9,10. The canonical Ikaros protein contains two multi-zinc finger domains: the N-terminal domain mediates sequence-specific DNA binding, while the C-terminal domain is involved in homo-or hetero-dimerization11. The Ikaros gene (denoted Ikzf1) encodes multiple isoforms that are generated by alternative splicing and is widely expressed in the hematopoietic system including multipotent, self-renewing HSCs and their committed progenitors12. Ikaros expression increases in developing lymphocytes, is maintained in granulocytes but downregulated in the macrophage and erythrocyte lineages12,13. Consistent with its broad pattern of expression, Ikaros regulates various aspects of hematopoiesis. In mice, targeted disruption of Ikaros leads to a strong reduction of hematopoietic stem cell activity, a developmentally delayed and abnormal thymopoiesis as well as perturbed myelopoeisis14-16. The most striking phenotype associated with the loss of Ikaros is an early and complete block of B cell development14,15. In the absence of Ikaros, lymphoid-primed multipotent progenitors (LMPPs) are generated but are impaired in giving rise to common lymphoid progenitors (CLPs), the latter are currently regarded as primitive B lineage progenitors17,18. Due to the early nature of this block, the molecular functions of Ikaros in B lymphopoiesis remain to be delineated10. Moreover, no molecular complementation analysis using Ikzf1-/- progenitors has been reported.
The transcription factors EBF and Pax5 play major roles in B cell fate specification and commitment by activating B lineage genes and concomitantly repressing genes of alternate lineages8,19,20. EBF initiates restriction of alternate lineage options and B cell fate commitment independently of Pax5 (ref. 8). EBF induces the Pax5 gene4,21. Pax5 is required to maintain the committed state in part by functioning in a feedback loop to sustain the expression of the Ebf1 gene22,23. EBF and Pax5 appear to co-regulate (activate or repress) a number of genes and likely do so via molecular mechanisms involving both sequential and concerted action24. As is the case for EBF and Pax5, Ikaros also has the dual ability to activate or repress transcription9. Although Ikaros family proteins have been suggested to repress surrogate light chain genes (Igll1 and VpreB1) in pre-B cells25, the molecular functions of Ikaros in B cell fate specification and commitment remain to be explored.
The hallmark of B cell development is the sequential DNA rearrangement of the immunoglobulin heavy and light chain loci26. At the Igh locus, recombination is temporally ordered with rearrangement of diversity (D) and joining (J) segments preceding that of variable (V) gene segments. The latter recombination events at the Igh locus are specific to B lineage cells and coincide with cell fate commitment. V(D)J recombination is regulated at two major levels27. One level involves the regulated expression of the recombination-activating genes 1 and 2 (Rag1 and Rag2) that encode the V(D)J recombinase (A002009, A002010). The Rag1 and Rag2 genes are tightly linked in the genome and convergently transcribed. Distinct cis-acting elements and transcription factors regulate the expression of the Rag genes in developing B and T lymphocytes28-33. Ikaros proteins have been suggested to regulate Rag gene expression by virtue of binding sites in promoters or enhancers within the Rag locus. However, it remains to be determined if they do so, particularly in B lineage cells.
The second level of regulation of V(D)J rearrangement involves control of accessibility of immunoglobulin or T cell receptor gene segments to the recombinase26. Both large-scale and localized molecular changes have been suggested to control the sequential and selective accessibility of different regions of the Igh locus to the recombinase. Accessibility and recombination of the VH segments have been correlated with the repositioning of the Igh locus from the periphery to the center of the nucleus and its compaction to generate DNA loops22,34,35. Localized events regulating accessibility include changes in chromatin structure via histone modification and the activation of germline as well as antisense transcription36,37. The transcription factors STAT5 and the chromatin modifier Ezh2 regulate the accessibility of distal VH gene segments through distinct non-redundant molecular mechanisms38,39. Pax5 and more recently Yin Yang1 (YY1) have been shown to control the compaction or contraction of the Igh locus presumably by DNA looping22,40. No role for Ikaros proteins in regulating the accessibility and recombination of the Igh locus has been uncovered.
Here, we showed by complementation analysis that EBF can rescue the generation of B cell precursors from Ikzf1-/- hematopoietic progenitors thereby demonstrating that Ikaros initially functions as a B lineage competence factor. The EBF-rescued Ikzf1-/- pro-B cells provided us with a unique tool to analyze the molecular functions of Ikaros in B cell fate specification and commitment. Our analysis indicates that Ikaros was required for B cell fate commitment as Ikzf1-/- pro-B cells despite expressing normal amounts of EBF and Pax5 differentiated into macrophages. We also established that Ikaros plays a central role in the rearrangement of the immunoglobulin heavy chain locus at the pro-B stage by simultaneously regulating the expression of the Rag1 and Rag2 genes and controlling VH gene accessibility and compaction of the Igh locus. These results establish that Ikaros is also an obligate regulator of B cell development. Therefore, Ikaros promotes B cell identity by repressing alternate lineage myeloid genes and by concomitantly inducing lineage-specific VH segment recombination events at the Igh locus.
RESULTS
EBF rescues B cell development from Ikzf1-/- progenitors
We initially attempted to complement Ikzf1-/- Lin-Sca1hic-Kithi (LSK) cells with Ik-1, the largest Ikaros isoform11. Under B lymphoid culture conditions, both control and Ik-1 transduced Ikzf1-/- progenitors generated myeloid but not B lineage progeny (data not shown, see below). We next pursued functional bypass experiments to circumvent the block in B cell development caused by the loss of Ikaros. We have previously demonstrated that the transcription factor EBF is a primary B cell fate determinant8. It not only induces early B cell gene expression and Igh gene recombination but it also represses the myeloid developmental program. Wild-type and Ikzf1-/- multipotent progenitors (LSK cells) were transduced with a control retrovirus (MIGR1) or one encoding EBF (MIG-EBF). Retroviral transduction was monitored using green fluorescent protein (GFP) whose expression was driven by an internal ribosome entry site (IRES). Following infection, the cells were plated on OP9 stromal cells with the cytokines, stem cell factor (SCF), Flt3 ligand (FL) and interleukin 7 (IL-7). After 3 days, transduced cells were sorted based on GFP expression and plated on a fresh stromal cell layer in the presence of B lymphoid cytokines (see Methods). Under these conditions, wild-type progenitors infected with control virus proliferated and differentiated after 10 days into CD19+ B cell precursors (Fig. 1a). As expected, expression of EBF in these cells strongly enhanced the generation of B lineage progeny8 (Fig. 1a).
Figure 1. EBF expression restores B-cell development from Ikzf1-/-hematopoietic progenitors.
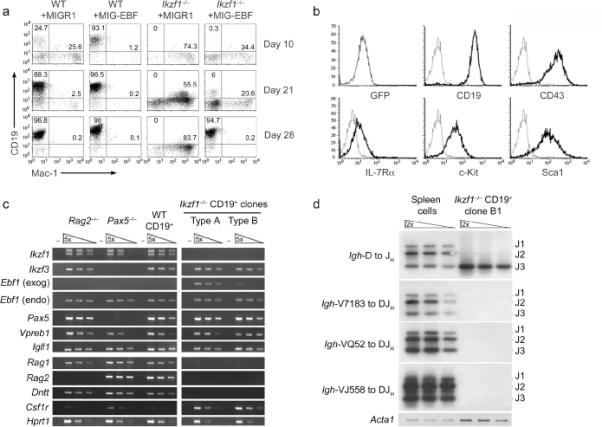
(a) Kinetic analysis of B cell development induced by the expression of EBF in Ikzf1-/- LSK progenitors. Wild-type or Ikzf1-/- LSK progenitors were transduced with MIGR1 or MIG-EBF viruses and cultured on OP9 stromal cells under B lymphoid conditions. Cultures were harvested at day 10, 21 and 28 and analyzed for generation of B lymphoid (CD19+) or myeloid (Mac-1+) precursors. (b) Flow cytometry of an Ikzf1-/- CD19+ clonal cell line rescued with EBF. (c) Gene expression analysis of Ikzf1-/- CD19+ clones. Type A clones express both exogenous (exog) and endogenous (endo) Ebf1 genes whereas type B clones only express the latter. Serial dilutions of cDNA were analyzed by RT-PCR after normalization to Hprt1. cDNAs from wild-type CD19+ cells generated in vitro (denoted WT CD19+) as well as Rag2-/- and Pax5-/- pro-B cell lines were used as controls. (d) PCR analysis of Igh rearrangements. Genomic DNA from Ikzf1-/- CD19+ clone B1 was analyzed using primers that detect DH to JH or VH7183-, VHQ52- and VHJ558-to-DJH rearrangements. PCR products were detected by Southern blotting. Genomic DNA from spleen cells was used as a control. DNA samples were normalized with Acta1 (encoding α-actin). All data are representative of at least two independent experiments.
Ikzf1-/- LSK cells transduced with the control vector failed to differentiate down the B cell pathway but in contrast gave rise to Mac-1+ (CD11b) myeloid progeny that continued to proliferate for an extended period of time in culture (Fig. 1a). The in vitro expansion of Mac-1+ precursors caused by the loss of Ikaros is reminiscent of the phenotype observed in the bone marrow of Ikzf1-/- mice15. Transduction of EBF in Ikzf1-/- LSK cells resulted in the accumulation of a double negative Mac-1- CD19- population and blocked the generation of Mac-1+ myeloid progeny (Fig. 1a). Interestingly, after 2-3 weeks of culture, we reproducibly detected a small fraction of CD19+ B lineage progeny (Fig. 1a). Once generated, these B lineage cells were highly responsive to IL-7 and rapidly became established as the dominant cell type in the cultures leading to the generation of homogenous cell lines. Morphological analysis by Wright staining revealed small round cells with large nuclei characteristic of lymphocyte progenitors (data not shown). Limiting dilution analysis revealed that approximately 1 in 1400 of the EBF-transduced LSK cells were able to give rise to CD19+ B lineage progeny (Supplementary Fig. 1 online). The generation of CD19+ B lineage cells from Ikzf1-/- LSK cells was specifically dependent on EBF as retroviral transduction of E2A or Pax5 did not yield such progeny (data not shown).
The EBF-rescued Ikzf1-/- CD19+ cell lines and clones isolated by limiting dilution were analyzed by flow cytometry. All clones displayed a homogeneous cell surface phenotype. In addition to CD19, they expressed CD43, CD24 (HSA), IL-7R (CD127), and c-Kit (CD117) but not Flt3 (CD135) or the myeloid markers Mac-1 and Gr-1 (Fig. 1b and not shown). We note that some clones no longer expressed GFP and retrovirally encoded EBF but nevertheless displayed an identical cell surface phenotype (Supplementary Fig. 2 online). Intriguingly Sca1, which is normally downregulated during B cell differentiation, remained highly expressed in a manner reminiscent of the phenotype of Pax5-/- pro-B cells19 (Fig. 1b and Supplementary Fig. 2). Notably, EBF-rescued Ikzf1-/- B lineage cells could be propagated in the presence of IL-7 and stromal cells without undergoing crisis or change of phenotype.
We next compared the gene expression profiles of the two types of EBF-rescued Ikzf1-/- clones; type A (GFP+) and type B (GFP-) with wild-type, Rag2-/- or Pax5-/- pro-B cells (Fig. 1c). Rag2-/- pro-B cells like their wild-type counterparts have undergone B cell fate specification and commitment although they retain their Ig loci in a germline configuration. Pax5-/- pro-B cells are specified but not committed to the B cell fate as they mis-express alternate lineage genes. Both types of Ikzf1-/- clones expressed the endogenous Ebf1 and Pax5 genes at comparable levels to wild-type cells, consistent with their CD19+ cell surface phenotype. Furthermore the two types of clonal derivatives expressed similar amounts of Cd79a (mb-1), Cd79b (B29), Igll1 (λ5) and Vpreb1 (VpreB) transcripts that encode components of the pre-B cell receptor (Fig. 1c and data not shown). Finally, they also expressed the Ikaros related family member Aiolos (encoded by Ikzf3) that is characteristically induced in pro-B cells41. Thus, in conjunction with the cell surface phenotype analysis, these molecular data demonstrate that EBF-rescued Ikzf1-/- CD19+ cells are pro-B cells. Hereafter, we refer to these cells as Ikzf1-/- pro-B cells and focus subsequent analysis on four independent type-B clones (denoted B1 to B4) as they express wild-type amounts of endogenous EBF.
Ikzf1-/- pro-B cells are defective in Igh recombination
In spite of the similarity to wild-type cells in the expression of genes encoding the pre-B cell receptor, Ikzf1-/- pro-B cells were strikingly defective in the expression of the recombination activating genes (Rag1 and Rag2) as well as the Dntt gene that encodes terminal deoxyribonucleotidyltransferase (TDT) (Fig. 1c). We next analyzed the cells for D-JH and V-DJH recombination events at the Igh locus. D-JH recombination events could be detected in all Ikzf1-/- clones (Fig. 1d). In contrast, no VH to DJH recombination events were detected by our analysis that encompassed both proximal (VH7183 and VHQ52) and distal (VHJ558) VH genes (Fig. 1d). Cloning and sequence analysis of Igh regions indicated differential usage of DH segments (Supplementary Fig.3 online). As appeared to be the case from the PCR analysis, a majority of the clones exhibited DJH recombination at only one allele while retaining the other in a germline configuration (Supplementary Fig.4 online). All of the DNA sequence analyses were consistent with the absence of TDT activity in the Ikzf1-/- clones as no N-region diversity could be detected in the recombined D-JH sequences (Supplementary Figs.3, 4). These data demonstrate that Ikaros is specifically required to promote Rag and Dntt gene expression in B lineage progenitors. In its absence, pro-B cells cannot recombine VH gene segments within the Igh locus and generate pre-B cells.
Ikzf1-/- pro-B cells retain latent myeloid potential
We also analyzed Ikzf1-/- pro-B cells for the expression of lineage-inappropriate genes. EBF and Pax5 repress these genes at the pro-B stage. Pax5 has been demonstrated to target the gene encoding the macrophage colony-stimulating factor receptor (Csf1r)42,43. As shown above, both the Ebf1 and Pax5 genes were expressed similar to wild-type cells in Ikzf1-/- pro-B cells (Fig. 1c). Despite normal Ebf1 and Pax5 expression, pro-B cells lacking Ikaros failed to properly repress the Csf1r gene (Fig. 1c). Interestingly, Csf1r expression was even higher in Ikzf1-/- pro-B clones than in Pax5-/- pro-B cells (3.3 fold +/- 0.5, n = 3). Consistent with this result, expression of CSF1R could be detected by flow cytometry on Ikzf1-/- clones but not on Pax5-/- cells (see below and data not shown). Mis-expression of Csf1r in pro-B cells in the absence of Ikaros is reminiscent of the phenotype of Pax5-/- pro-B cells. As is the case for their Pax5-/- counterparts42, Ikzf1-/- pro-B cells mis-expressed a number of myeloid, erythroid and T lymphoid genes (Supplementary Fig. 5 online).
We next examined whether Ikzf1-/- pro-B cells retained myeloid developmental potential. Ikzf1-/- pro-B cells were plated onto S17 stromal cells and cultured with M-CSF (8-14 days) instead of IL-7. In parallel, wild-type, Rag2-/- and Pax5-/- pro-B cells were cultured under the same conditions. As previously described42, Pax5-/- pro-B cells underwent a dramatic change in morphology and differentiated into macrophages that expressed Mac-1, F4/80 and high amounts of CSF1R (Fig. 2a,b). In contrast, wild-type or Rag2-/- pro-B cells underwent considerable cell death. The residual cells maintained their B cell identity and showed no signs of myeloid differentiation based on morphology and cell surface phenotype (Fig. 2a and data not shown). Under these conditions, a substantial number of Ikzf1-/- pro-B cells also underwent cell death but in contrast with Rag2-/- pro-B cells, these cells like their Pax5-/- counterparts trans-differentiated into macrophages. The differentiated cells displayed the characteristic morphology of large macrophages and expressed Mac-1, F4/80 and higher amounts of CSF1R (Fig. 2a,b). Although nearly all of the Ikzf1-/- pro-B cell clones tested (11 out of 12) generated macrophages, the frequency of trans-differentiation of Ikzf1-/- pro-B cells was substantially lower than that of Pax5-/- pro-B cells (ranging between 1/60 to 1/1000, Supplementary Fig. 6 online). We note that Ikzf1-/- pro-B cell clones displayed the ability to trans-differentiate into macrophages even after plating freshly sorted CD19+ cells (data not shown). Furthermore, upon exposure to M-CSF a large fraction of Ikzf1-/- pro-B cells downregulated CD19. Thus, it appears that trans-differentiation of Ikzf1-/- pro-B cells involves, initial loss of expression of B lineage genes followed by the acquisition of macrophage identity, the latter being an inefficient process. These results demonstrate that pro-B cells despite expressing wild-type amounts of EBF and Pax5 are not fully committed to the B cell fate in the absence of Ikaros.
Figure 2. Ikzf1-/- pro-B cells can trans-differentiate into macrophages.
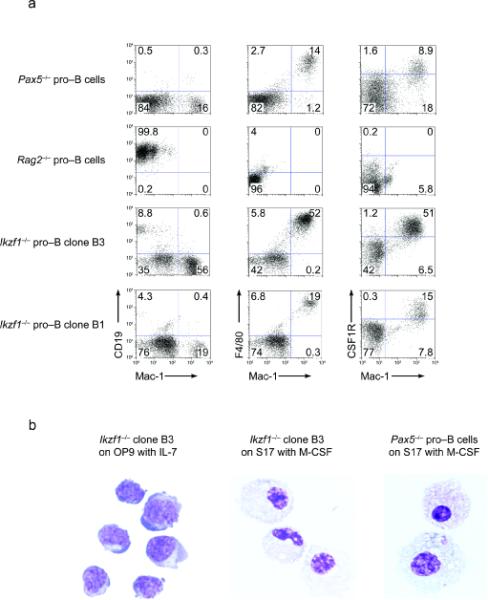
Pax5-/-, Rag2-/- and Ikzf1-/- pro-B cells were plated on S17 stromal cells under B lymphoid conditions in the presence of IL-7 for 2 days and then switched to myeloid culture conditions in the presence of M-CSF. (a) Flow cytometry of switched cultures (day 12) examining expression of CD19, Mac-1, F4/80 and CSF1R. (b) Cellular morphology of Mac-1+ cells generated under the switched culture conditions. Pax5-/- or Ikzf1-/- cells were sorted on the basis of Mac-1 expression and examined by Wright staining. Ikzf1-/- pro-B cells cultured on OP9 in the presence of IL-7 were used to compare morphology. All data are representative of three independent experiments.
Ik-1 promotes pro-B cell differentiation and commitment
To analyze the functions of Ikaros in B lineage cells we complemented Ikzf1-/- pro-B cells with Ik-1. Ikzf1-/- clones (type B) that did not sustain the expression of the EBF-IRES-GFP transgene were transduced with a retrovirus encoding Ik-1 (MIG-Ik-1) and monitored using GFP expression. Cells were analyzed 3 days after transduction. Ik-1 expressing cells were smaller and morphologically more similar to wild-type pro-B cells (data not shown). They maintained expression of CD43 and IL-7R but downregulated c-Kit and Sca1 in a manner that correlated with the level of Ik-1 expression (Fig. 3a). Importantly no cell surface phenotypic alterations were observed in the GFP- fraction of Ik-1 cultures or in cells transduced with the control retrovirus MIGR1 (Fig. 3a). Notably, Ik-1 expression resulted in downregulation of Csf1r transcripts and CSF1R expression on the cell surface (Fig. 3b). Furthermore, Ik-1 inhibited the ability of Ikzf1-/- pro-B cells to trans-differentiate into macrophages. In limiting dilution experiments, restoration of Ik-1 expression reduced the frequency of trans-differentiation to one-tenth that of cells transduced with the control vector (Supplementary Fig. 7a,b online). Thus Ikaros appears to repress multi-lineage as well as alternate lineage hematopoietic genes during B cell differentiation and promote B cell fate commitment.
Figure 3. Restoration of Ik-1 expression in Ikzf1-/- pro-B cells promotes B-lineage commitment.
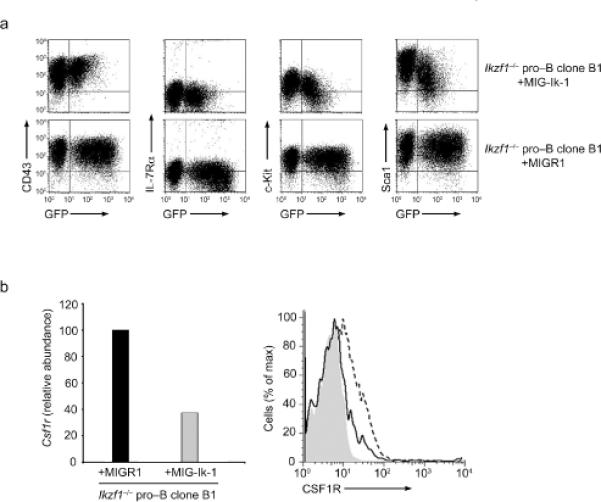
Ikzf1-/- CD19+ clones were transduced with a control vector (MIGR1) or a derivative encoding Ik-1 (MIG-Ik-1) and analyzed 3 days after transduction. (a) Flow cytometry examining expression of indicated cell surface markers in Ik-1 transduced (top) and control (bottom) cells. Transduced cells are distinguished on the basis of GFP expression. (b) Left, relative expression of Csfr1 transcripts in Ikzf1-/- CD19+ cells transduced with MIG-IK-1 or MIGR1 (set to 100). Right, flow cytometry of CSF1R protein on the surface of Ikzf1-/- CD19+ cells transduced with MIGR1 (dashed line) or MIG-Ik-1 (solid line). Isotype control is shown in grey. Analyses are gated on GFPhi cells. All data are representative of three independent experiments.
In contrast with repression of Csf1r transcription, Ik-1 substantially induced Rag1 and Rag2 gene expression (Fig. 4a). We therefore determined if Ik-1 stimulated VH DNA rearrangements at the Igh locus. As shown above, Ikzf1-/- cells transduced with the control vector harbored only DJH rearrangements (Fig. 4b). Expression of Ik-1 induced the rearrangement of both proximal (VH7183 and VHQ52) and to a lesser extent distal (VHJ558) VH gene segments (Fig. 4b). Thus Ikaros positively regulates VH segment recombination at the Igh locus in part by controlling activation of Rag gene transcription in pro-B cells (see below).
Figure 4. Restoration of Ik-1 expression in Ikzf1-/- pro-B cells induces Rag gene expression and VH to DJH rearrangements.
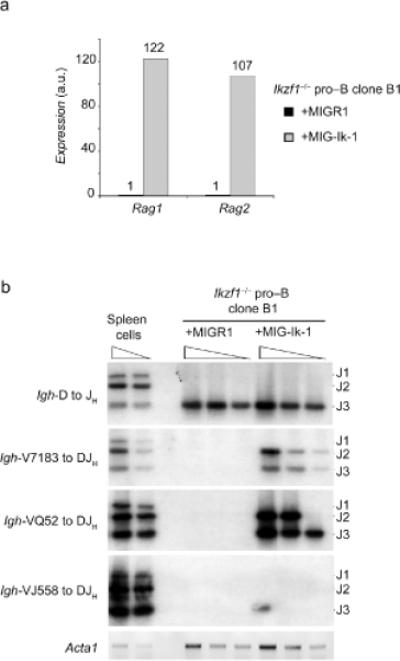
(a) Q-PCR analysis of Rag1 and Rag2 mRNA expression in Ikzf1-/- cells transduced with MIGR1 or MIG-Ik-1 vectors. Hprt1 transcripts were used for normalization. The relative expression of each gene is expressed as a fold difference in relation to that detected in MIGR1 transduced cells (set to 1). (b) PCR analysis of Igh rearrangements. Genomic DNA from Ikzf1-/- pro-B cells transduced with MIGR1 or MIG-Ik-1 was analyzed as described in Fig. 1d. Genomic DNA from the mouse spleen was used as a control. All data are representative of three independent experiments.
Ikaros directly activates expression of the Rag locus
Rag gene expression is regulated during the cell cycle44. Furthermore, we have recently shown that IL-7 signaling is a potent negative regulator of these genes in pre-B cells45. To test if Ikaros induced Rag expression via modulation of the cell cycle and/or IL-7 signaling, we cultured Ikzf1-/- clones in limiting concentrations of IL-7. Lowering the IL-7 concentration resulted in reduced cell proliferation (data not shown). Expression of Rag1 and Rag2 genes was assessed by QPCR. Irf4-/- Irf8-/- pre-B cells were used as controls46. As expected, Rag gene transcription was highly induced in Irf4-/-Irf8-/- pre-B cells upon attenuation of IL-7 signaling45. In striking contrast no induction of Rag gene expression was observed in the Ikzf1-/- pro-B cells (Supplementary Fig. 8 online). On the basis of these experiments, we ruled out the possibility that Ikaros regulates Rag gene transcription in pro-B cells by modulating the cell cycle and/or IL-7 signaling.
We next investigated whether the Rag locus is a direct transcriptional target of Ikaros. Several regulatory elements have been described in the Rag locus. In addition to the Rag1 and Rag2 promoters28,29, there are three distinct enhancers denoted proximal enhancer (Ep), distal enhancer (Ed) and Erag31-33 (Fig. 5a). Ed ensures lymphoid specificity while Ep and Erag are required for optimal expression of Rag1 and Rag2 genes specifically in developing B cells. Each of these elements contains several potential Ikaros binding sites. To determine whether Ikaros binds to these regulatory regions in vivo, we performed chromatin immunoprecipitation (ChIP) assays using Rag2-/- pro-B cells. We note that a portion of the Rag2 coding exon is deleted in these cells47 and they express wild-type amounts of Rag1 transcripts. The pericentromeric γ-satellite repeat sequences (γ-sat.) were used as a bona fide Ikaros target in these ChIP assays48 while chromatin from Ikzf1-/- mutant pro-B cells served as a negative control. Ikaros binding was observed at multiple regulatory elements of the Rag locus with the notable exception of the Rag2 promoter (Fig. 5b). Electrophoretic mobility-shift assays (EMSA) demonstrated the presence of Ikaros binding sites in the different regulatory elements analyzed by ChIP (Fig. 5c and Supplementary Fig. 9 online). To further analyze the molecular consequences of Ikaros binding to the Rag locus, we assessed the histone modification status of these regulatory regions in pro-B cells lacking or expressing Ikaros. The degree of histone-H3 acetylation was analyzed by ChIP assays in Ikzf1-/- pro-B cells and compared with that observed in Rag2-/- pro-B cells. Strikingly none of the Rag locus regulatory domains displayed this active chromatin modification in the absence of Ikaros (Fig. 5d). Thus Ikaros directly targets the Rag locus and appears to induce histone acetylation while activating Rag gene transcription.
Figure 5. Ikaros binds to the Rag locus and regulates histone acetylation.
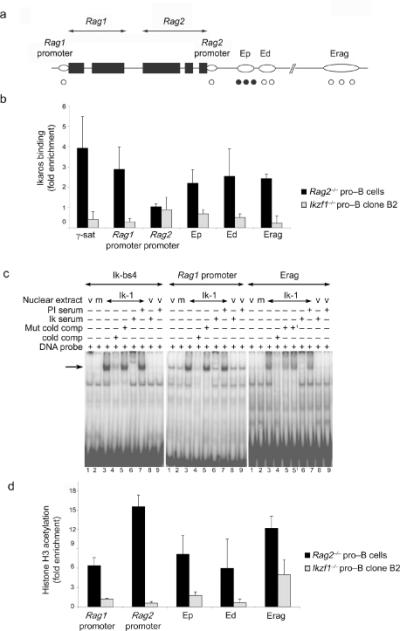
(a) Schematic representation of the Rag locus. Black boxes represent Rag1 and Rag2 exons while open ovals represent regulatory elements. Validated (•) and putative (Ο) Ikaros binding sites are indicated. (b) ChIP analysis of Ikaros binding to regulatory elements of the Rag locus. Binding was determined in Rag2-/- pro-B cells that express Ikaros and Rag1. Ikzf1-/- pro-B cells were used as negative controls. Relative enrichment of the bound DNA over input was determined by Q-PCR after normalization to Acta1. Pericentromeric g-satellite sequence (g-sat) was used as a positive control for binding of Ikaros. (c) EMSAs were performed using nuclear extracts of 293T cells transfected with the Ik-1 expression vector. Control extracts were generated from 293T cells transfected with an empty vector (lane 1) or with an Ikaros-159A DNA binding mutant (lane 2). Ik-bs4 probe contains a high affinity Ikaros binding site. Other probes spanned putative Ikaros binding sites in the Rag1 promoter and Erag enhancer. Where indicated binding reactions included competitor DNA in 50-fold molar excess or either pre-immune (PI) or anti-Ikaros antiserum (Ik serum). In lanes 5 and 51 two related mutant competitor DNAs were used. (d) ChIP analysis of histone H3 acetylation at regulatory elements of the Rag locus. Analysis was performed as described for Fig. 3b using Actb (encoding β-actin) for normalization. Error bars in panels (b) and (d) represent the standard deviation from the mean (n = 3). Data in (c) are representative of at least two independent experiments.
To reinforce the above conclusion, we tested if increased expression of Ikaros leads to upregulation of Rag1 and Rag2 expression in pro-B cells. Pax5-/- pro-B cells were transduced with MIG-Ik-1 retrovirus or the control vector. Transduced cells were sorted based upon GFP expression and Q-PCR was used to analyze Rag gene transcripts. Increased amounts of Ik-1 augmented Rag1 (2.9 fold) and Rag2 (5.6 fold) gene expression (Supplementary Fig. 10 online). Notably, the Ik-7 isoform that cannot bind DNA failed to upregulate Rag gene expression. Thus Ikaros regulates Rag gene expression in a concentration dependent manner in pro-B cells.
Ikaros controls accessibility and compaction of Igh locus
To determine if the molecular defect in VH gene recombination in Ikzf1-/- pro-B cells (Fig. 1d, 4b) was simply due to a failure to properly activate the Rag locus, we restored Rag gene expression in these cells. For these experiments we made use of retroviruses expressing RAG1 (RAG1-IRES-Dsred, denoted MIRR-Rag1) and RAG2 (RAG2-IRES-GFP, denoted MIGR-Rag2). Dsred, GFP doublepositive cells were FACS sorted after stepwise infection with the two retroviral vectors, expanded in culture in the presence of IL-7 and analyzed for DNA rearrangements at the Igh locus. As previously shown, cells transduced with control vectors did not recombine either proximal or distal VH segments. Expression of Rag1 along with Rag2 modestly stimulated VH to DJH rearrangements involving the proximal (VH7183 and VHQ52) VH segments (Fig. 6a, 4b). These data suggest that in the absence of Ikaros, RAG proteins cannot efficiently access the VH domain of the Igh locus. To further explore this possibility, we examined germline transcription across the Igh locus as an indicator of chromatin accessibility. Germline transcripts that initiate within the heavy-chain intronic enhancer, from the DQ52 segment promoter or from the proximal VH7183, VHQ52 promoters were unaffected by the loss of Ikaros (Fig. 6b). In contrast, germline transcripts from the distal VHJ558 gene segment promoters were substantially reduced in the Ikzf1-/- cells. Thus, Ikaros appears to promote the accessibility of VH gene segments to recombination, particularly that of the distal VHJ558 gene segments (see below).
Figure 6. Ikaros regulates accessibility of the VH distal segments.
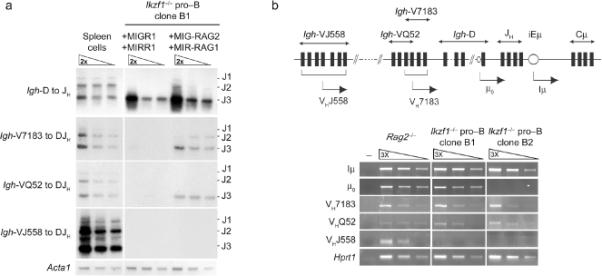
(a) Ikzf1-/- pro-B cells were successively transduced with MIG-Rag2 and MIRRag1 or with MIGR1 and MIR1 as controls. Genomic DNAs from indicated cells were analyzed as in Fig. 1d, 3b to determine Igh rearrangement status. (b) Analysis of Igh germline transcripts. Top, a schematic of the Igh locus with the different germline transcripts denoted VHJ558, VHQ52, VH7183, μ0 and Iμ. Bottom, RT-PCR analysis of Igh germline transcripts in Rag2-/- and Ikzf1-/- pro-B cells. Serial dilutions of cDNA were analyzed after normalization to Hprt1. All data are representative of at least two independent experiments.
Repositioning away from the nuclear lamina and large-scale compaction are two nuclear events that have been proposed to regulate the ability of the distal VH genes to undergo recombination22,34,35. Three-dimensional fluorescence in situ hybridization (3D Immuno-FISH) was used to analyze the nuclear disposition and compaction of Igh alleles in Ikzf1-/- pro-B cells. Rag2-/- pro-B and EL-4 T cells were used as controls. Unlike in EL-4 T cells, the Igh alleles in Ikzf1-/- pro-B cells were positioned away from the nuclear lamina similar to their disposition in Rag2-/- pro-B cells (Fig. 7a and Supplementary Fig. 11 online). We next analyzed compaction of the Igh locus by measuring the distances between distal VHJ558 genes and the constant CH regions on the same allele. As previously described39, Igh alleles in EL-4 nuclei demonstrated a wide range of intralocus distances while Rag2-/- pro-B cells displayed a more constrained distribution (Fig. 7a,b). The profile of Igh alleles in Ikzf1-/- pro-B cells resembled that of EL-4 T cells and not of Rag2-/- pro-B cells (Fig. 7a,b). The intralocus distance distributions of Ikzf1-/- and EL-4 cells were not significantly different from each other (P > 0.03) while both were statistically different from that of Rag2-/- cells (P < 0.0001 and P = 0.0003, respectively). Thus Ikaros is not required for repositioning of Igh alleles away from the nuclear lamina. Instead, it seems to regulate spatial constraints on the Igh locus that are manifested in its apparent compaction. Collectively, these results reveal Ikaros to be a regulator of both accessibility and compaction of the Igh locus.
Figure 7. Ikaros is required for compaction of the Igh locus.
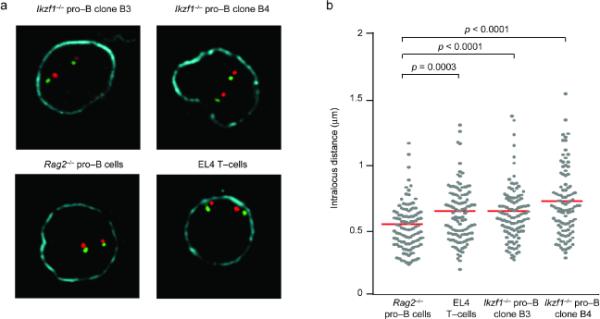
(a) Three-dimensional immuno-FISH analysis was performed on the Igh locus. Igh alleles detected by cohybridization with probes for the VHJ558 (red) and constant CH (green) regions with three-dimensionally preserved nuclei of EL-4 T cells, Rag2-/- pro-B cells and two independent Ikzf1-/- pro-B clones. Anti-lamin B1 was used to stain the nuclear periphery (blue). (b) Quantitative analysis of intralocus distances in the four cell types. Measurements are between VHJ558 (red in a) and CH (green in a) signals on the same allele. Distances represent 110 alleles per cell type. All data are representative of two independent experiments.
DISCUSSION
Recently we have proposed that the transcription factor EBF functions as the primary determinant of the B cell fate1,49. EBF is able to rescue B cell development in vitro from PU.1-, EBF- or E2A-deficient progenitors4,5,8. Herein, we demonstrate that EBF is also able to restore the B cell developmental potential of Ikzf1-/- progenitors. However, the generation of CD19+ cells was significantly delayed compared to the aforementioned experiments. Limiting dilution experiments demonstrate that the delayed kinetics reflect a low frequency of Ikzf1-/- LSK progenitors that can be rescued with EBF. We suggest that this is due to the fact that a large proportion of the LSK compartment in absence of Ikaros is poised for myeloid differentiation18. Consistent with this view, Ikzf1-/- LSK progenitors undergo rapid myeloid differentiation even when cultured in B lymphoid inducing conditions. Moreover the Ikzf1-/- LSK cellular compartment is characterized by upregulation of myeloid cell fate determinants and a failure to induce expression of early B cell genes18 (C. Spooner and H.S., unpublished data). EBF's ability to rescue Ikzf1-/- cells is reminiscent of its ability to redirect at low frequency myeloid progenitors toward the B lymphoid lineage8. When ectopically expressed in myeloid progenitors EBF not only induces the B cell program but also represses myeloid gene expression. It is notable that expression of EBF in Ikzf1-/- LSK cells promotes the proliferation of a progenitor population that is Mac-1- CD19-. These results are consistent with the ability of EBF to antagonize myeloid determinants such as PU.1 and C/EBPα8.
It has been proposed that Ikaros is essential for activating the Flt3 and Il7r genes that encode two key cytokine receptors for lymphoid differentiation1,16,50. The block to B cell development in Ikzf1-/- mice has been suggested to be due to deficiencies in these two signaling pathways. We tested this possibility by restoring expression of Flt3 or IL-7R in Ikzf1-/- LSK cells (data not shown). We were unable to functionally bypass the requirement for Ikaros in B cell development with either of these cytokine receptors. Although it remains possible, that co-expression of Flt3 and IL-7R may rescue B cell development from Ikzf1-/- LSK cells, our results are consistent with the demonstration that lymphoid-primed multipotent progenitors can be generated in absence of Ikaros18. Our analysis of Ikzf1-/- pro-B cells rescued with EBF indicates that Ikaros is not strictly necessary for the expression or functioning of IL-7R. Collectively, these results weaken the argument that Ikaros promotes B cell fate specification by controlling the expression of Flt3 and/or IL-7R. The successful rescue of Ikzf1-/- progenitors by EBF suggests instead that Ikaros is required for the developmental induction of the Ebf1 gene. In the future, it will be important to determine if the Ebf1 gene is an Ikaros target as is the case for the Rag genes (see below).
One of the primary functions of Ikaros in LMPPs may be to antagonize the myeloid program. Intriguingly, rescued Ikzf1-/- pro-B cells despite the normal expression of EBF and Pax5, are unable to properly repress alternate lineage genes including Csf1r. Thus we propose that Ikaros functions to attenuate the Csf1r gene in LMPPs but also to maintain its repression in pro-B cells. Ikzf1-/- pro-B cells are not committed to the B lineage as they are able to differentiate into macrophages upon culturing in M-CSF. This result predicts that conditional deletion of Ikzf1, the gene encoding Ikaros, at the pro-B cell stage should also result in loss of B cell commitment. We propose that Ikaros ensures B cell fate commitment by maintaining the repressed state of alternate lineage genes. Thus, B cell fate commitment is not simply a consequence of the action of a single transcription qfactor7 but instead dependent on an elaborate network of regulatory molecules that include EBF, Pax5 and now Ikaros8. These factors are likely to function in both a sequential and concerted manner during B cell development from LMPPs to restrict alternate cell fate options ultimately leading to commitment.
How might Ikaros function to repress myeloid genes? Several molecular mechanisms of repression have been described in the case of Ikaros. One is linked to its ability to associate with histone deacetylase (HDAC)-containing complexes NURD or Sin3 (ref. 9). In favor of this hypothesis, we note that the Csf1r locus exhibits a high degree of histone H3 acetylation in Ikzf1-/- pro-B cells unlike it's status in Ikaros expressing Rag2-/- pro-B cells (data not shown). Another possible mechanism involves a competition on overlapping binding sites as described with EBF in the Igll1 (γl5) locus or with Ets1 in the Dntt (TdT) locus25,51. A third mechanism involves the proposed ability of Ikaros to associate genes with pericentromeric hetrochromatin48,52. Our results demonstrate similarities as well as differences between Ikzf1-/- and Pax5-/- pro-B cells. Both express c-Kit and Sca1 and numerous alternate lineage genes19,42. Specifically, they fail to repress the Csf1r gene and are not committed to the B lineage and can differentiate into macrophages42. Notably, as recently described for EBF8, raising the concentration of Ikaros in Pax5-/- pro-B cells also leads to the repression of the Csf1r locus and restrains the ability to these cells to differentiate into macrophages (data not shown). Although EBF or Ikaros can bypass the requirement for Pax5 in repression of the Csfr1 gene in Pax5-/- pro-B cells, we suggest that in wild-type pro-B cells such repression is mediated by concerted action of the three transcription factors. Interestingly, Ikzf1-/- pro-B cells, in contrast to their Pax5-/- counterparts, failed to undergo T cell differentiation in vitro or in vivo (data not shown). Exposure of Ikzf1-/- pro-B cells to the Notch-ligand Delta1 induced expression of the Notch target genes Hes1 and Dtx1 and resulted in rapid apoptosis (data not shown). Thus Ikzf1-/- pro-B cells can differentiate into macrophages but not T lineage cells. We demonstrate that Ikaros specifically controls the activation of the Rag locus and subsequently the transition between the pro- and pre-B cell stages that requires the rearrangement of the Igh locus. These data are consistent with the analysis of a mouse strain expressing a hypomorphic allele of Ikaros (Ikzf1L) generated by insertion of lacZ53. These mice accumulate B220+CD43+IgM- cells, which comprise the pro-B and early pre-B compartment. The authors noted decreased expression of the Rag1, Rag2 and Dntt genes in B220+IgM- cells but it was unclear if this was due to a defect in pro-B cells. Our results suggest that a majority of the Ikzf1L/L B220+CD43+ cells are pro-B cells that have failed to recombine the Igh locus.
We show that Ikaros binds in vitro and in vivo to multiple sites within the Rag locus and regulates histone acetylation as well as transcription. As reported for Foxp154, Ikaros targets the most 5' distal enhancer designated Erag. Consistent with our findings, B lineage progenitors in Erag deficient mice are specifically impaired in VH-to-DJH rearrangements32. We note that loss of Ikaros leads to a more profound block in the expression of the Rag genes than deletion of Erag. This result is consistent with our data indicating that Ikaros binds other regulatory elements in the Rag locus such as the proximal and distal enhancers as well as the Rag1 promoter. How are Ikzf1-/- progenitors able to undergo DH-to-JH recombination? We suggest that a low level of Rag gene expression in lymphoid-primed multipotential progenitors that is independent of Ikaros may be sufficient to promote monoallelic DH-to-JH recombination. It is important to note that the Rag genes represent a rare example of targets that are activated by Ikaros. Previously Ikaros has been shown to bind and activate the Cd8a gene in T lineage cells55. As is the case for the Cd8a locus, the molecular mechanism by which Ikaros activates transcription of the Rag locus remains to be determined. Ikaros regulates histone acetylation at the Rag locus and may do so by recruiting chromatin-modifying complexes9. Our results indicate that Ikaros also positively regulates the expression of the Dntt gene in pro-B cells that is required for addition of non-templated nucleotides at coding joints within the assembled Ig segments. The Dntt gene promoter contains Ikaros binding sites that have been implicated in repression of the Dntt gene during differentiation of CD4+CD8+ thymocytes51. It is possible that Ikaros is able to activate the Dntt gene through these sites in the promoter at an earlier stage in T and B cell development. Finally, we note that Ikaros is not required for the expression of the Rag genes in T lineage cells. This dispensability could be due to replacement of Ikaros function by alternate members of the family or usage of other cis-regulatory elements in the Rag locus during T cell development.
Restoration of Rag gene expression in Ikzf1-/- pro-B cells modestly induces recombination of proximal VH gene segments. In contrast expression of Ik-1 in these cells promotes more efficient proximal VH gene recombination as well as limited recombination of distal VH genes. Although, it is possible that these differences in the efficiency and recombination bias of VH gene segments is due to differing concentrations of Rag protein expression, it is unlikely given that reduced Rag expression in Foxp1 or Erag mutant B lineage cells does not result in preferential usage of proximal VH genes. We suggest that the aforementioned differences are due to the fact that Ikaros functions in a dual capacity. It not only activates Rag gene expression but also promotes VH genes accessibility. Consistent with the latter possibility, germline transcription of the distal VH gene segments is compromised in Ikzf1-/- pro-B cells. Ikaros is unlikely to be regulating germline transcription of the distal VH segments by modulating IL-7 signaling and Stat5 activity as the Ikzf1-/- pro-B cells proliferate normally in response to IL-739. Although putative Ikaros binding sites have been described in the V regions of the Igh locus56 it remains to be determined if Ikaros regulates transcription and recombination accessibility via binding to these sites. At the level of nuclear compartmentalization, Ikaros is not required for repositioning of the Igh locus away from nuclear lamina. In contrast, Ikaros controls compaction of the Igh locus that has been proposed to promote the rearrangement of the distal VH genes. Compaction of Igh locus in Ik-/- pro-B cells is similar to that observed in Pax5-/- or Yy1-/- pro-B cells. However, since Ikzf1-/- pro-B cells express wild-type amounts of Pax5 and Yy1 transcripts, the reduced Igh locus compaction phenotype in these cells cannot be attributed to impaired expression of Pax5 or Yy1 (data not shown). Furthermore, the defect in germline transcription of distal VH genes observed in Ikzf1-/- pro-B cells argues for an independent mechanism. Notably, in contrast with our findings that demonstrate Ikaros as a positive regulator of Igh recombination, Ikaros appears to promote the association of the Igk locus with pericentromeric heterochromatin during allelic exclusion57,58. The molecular mechanisms underlying these distinct Ikaros functions during B cell differentiation remain to be explored.
Collectively, our results reveal key, non-redundant molecular functions of Ikaros during B cell development. Based on the findings that, Ikaros function in multipotent progenitors can be partially by-passed by the expression of EBF, we propose that Ikaros acts in LMPPs as a B lineage competence factor both by restraining the myeloid developmental program and by enabling the expression of EBF that is a primary B cell fate determinant. Moreover, we show that Ikaros is an obligate regulator of the early molecular program that is operative in pro-B cells. In this context Ikaros functions in a non-redundant manner to establish B cell identity by repressing alternate cell fate options. Our results suggest that commitment to the B lineage requires the sequential and concerted actions of Ikaros, EBF and Pax5. Finally, Ikaros is essential for positively regulating VH to DJH rearrangement at the Igh locus that is the defining hallmark of B cell fate commitment. Importantly Ikaros regulates this DNA recombination step by activating the expression of the Rag genes and by controlling the accessibility as well as compaction of the Igh locus.
METHODS
Retroviral vectors
The murine stem cell virus-internal ribosomal entry site-green fluorescent protein (MSCV-IRES-GFP, MIGR1) retroviral vector has been described previously3. The EBF, Ik-1 and Ik-7 vectors are derivatives of MIGR1 (refs. 4,59). Rag1 and Rag2 coding sequences were amplified by PCR from pEBB-Rag1 and pEBB-Rag2 plasmids (gifts from D.G. Schatz, Yale University, New Haven, CT) and inserted into XhoI and HpaI cloning sites of MIR1 (MSCV-IRES-Dsred) and MIGR1 respectively. Retroviruses were produced by transient transfection in Plat-E packaging cell line as described previously4.
Isolation, retroviral transduction and differentiation of Ikzf1-/- hematopoietic progenitors
Ikzf1-/- mice as well as wild-type control littermates were bred and maintained in specific pathogen-free conditions with oral antibiotics at the Animal Facilities of the University of Chicago. Mice used for this study were between 3 and 6 weeks old. Mouse handling involved protocols that were approved by the Institutional Animal Care and Use Committee. Hematopoietic progenitors were enriched from bone marrow cell suspensions by magnetic depletion (Miltenyi Biotec) using lineage-specific antibodies. The LSK population was isolated, by cell sorting, based on the expression of c-Kit and Sca1. LSK cells were suspended in retroviral supernatants with 12.5 mg/ml polybrene and centrifuged 1,400 g, at 30 °C, for 2 h. The cells were then washed and plated on OP9 stromal cells in Optimem medium (Invitrogen) supplemented with 5% fetal calf serum (FCS) in presence of SCF (10 ng/ml), FL (10 ng/ml), and IL-7 (5 ng/ml). After 72 h, transduced cells were sorted based on GFP expression and re-plated under the same culture conditions. Following sequential withdrawal of FL and SCF upon two-day intervals, cultures were maintained in IL-7. Medium was changed twice a week and cells were re-plated on fresh OP9 stroma once a week. Cellular differentiation was monitored on a weekly basis by flow cytometry.
Propagation and retroviral transduction of pro-B cells
Wild-type or Rag2-/- pro-B cells were propagated on OP9 stromal cells in Optimem medium supplemented with 5% FCS and IL-7 (5 ng/ml). Pax5-/- pro-B cells (a gift from M. Busslinger, Research Institute of Molecular Pathology, Vienna, Austria) were cultured according to a previously described protocol42.Cell lines were transduced with retroviral constructs as described above for primary cells. Transduced cells were sorted based on GFP expression and propagated in culture.
Assaying myeloid potential of pro-B cells
Wild-type or mutant (Ikzf1-/-, Rag2-/- or Pax5-/-) pro-B cells were transferred onto S17 stromal cells and initially cultured in α-MEM medium supplemented with 10% FCS and IL-7 (5 ng/ml). After two days, the cells were switched to myeloid culture conditions by replacing the above medium with α-MEM supplemented with 10% FCS and M-CSF (20 ng/ml).
Antibodies and Flow cytometry
Cell suspensions were stained using standard procedures. Cells were analyzed on a FACSCalibur using CellQuest or FlowJo software (BectonDickinson). Cells were sorted at the Flow cytometry facility of the University of Chicago using MoFlo-HTS (DakoCytomation) or FacsAria (BectonDickinson) cell sorters. The antibodies used in this study are listed in Supplementary Table 1 online.
Gene expression analysis
RNA was isolated using TRIzol LS reagent (Invitrogen) and reverse-transcribed with random hexamers and the SuperScript™ II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. For semi-quantitative analysis the reverse-transcription reaction products were serially diluted and specific transcripts were amplified by PCR. RT-PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. For quantitative analysis, real-time RT-PCR was performed using Brilliant SYBR Green and the Mx4000 system (Stratagene). Each RNA sample was normalized based on Hprt1 expression. Sequences of primers used for semi-quantitative and quantitative-PCR are provided in Supplementary Tables 2 and 3 online, respectively.
PCR analysis of Igh rearrangements
DNA recombination at the Igh locus was assessed as described previously39. Serial dilutions of genomic DNA were analyzed by PCR with primers specific for VHJ558, VH7183 or DH segments and a primer 3' of JH3 (Supplementary Table 4 online). PCR products were separated by electrophoresis on a 1% agarose gel and analyzed by Southern blotting with a DHFL16-JH4 probe.
Chromatin Immunoprecipitation assays (ChIP)
Chromatin crosslinking and Immunoprecipitation was performed as described previously39. The following antibodies were used; anti-acetylated Histone H3 (06-599; Upstate Biotechnology), anti-Ikaros-Ct (rabbit antiserum generated against a carboxy terminal fragment of Ikaros60) and control IgG (sc-2027; Santa Cruz). Immunoprecipitated DNA sequences were analyzed by quantitative real-time PCR as described above. Primers used in ChIP assays are listed in Supplementary Table 5 online.
Electrophoretic mobility-shift assays (EMSA)
Nuclear extracts were prepared from 293T cells transfected with the Ik-1 constructs cloned into the pcDNA3 expression vector as previously described48. Negative controls included nuclear extracts from cells transfected with vector alone or the Ikaros-159A mutant construct48. Ikaros expression was verified by Western blotting (data not shown). DNA binding probes were generated by annealing synthetic double-stranded oligonucleotides and end-labeling using polynucleotide kinase and 32P-labeled γ-ATP. Sequences of the probes are indicated in Supplementary Table 6 online. Ik-bs4 probe contains a high affinity consensus Ikaros binding site11,48. The anti-Ikaros-Ct serum described above for ChIP assays was used for supershifting Ikaros protein-DNA complexes. Pre-immune serum was used as negative control.
Three-dimensional fluorescencein situhybridization (3D Immuno-FISH)
3D Immuno-FISH experiments were performed as previously described39. DNA probes were made by nick translation of bacterial artificial chromosome clones corresponding to the VHJ558 and heavy-chain constant (CH) region of the Igh locus. Specific DNA sequences were detected with avidin-fluorescein isothiocyanate or mouse anti-digoxin-indocarbocyanine (Jackson ImmunoLabs). The nuclear lamina was demarcated by anti-Lamin B1 staining. Image analysis was performed using ImageJ software. At least 100 signal pairs for each cell type were assigned scores for analysis. The unpaired t-test was used for the statistical analysis of the intralocus distances.
Accession numbers
Ikaros; http://www.signaling-gateway.org/molecule/query?afcsid=A001169 EBF; http://www.signaling-gateway.org/molecule/query?afcsid=A000809 Pax5; http://www.signaling-gateway.org/molecule/query?afcsid=A000403 Rag1; http://www.signaling-gateway.org/molecule/query?afcsid=A002009 Rag2; http://www.signaling-gateway.org/molecule/query?afcsid=A002010
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Singh lab for helpful suggestions and critical comments. I.D. and H.S. are supported by the Irvington Institute Fellowship program of the Cancer Research Institute and the Research Council of Norway, respectively. Z.C. is sponsored by a fellowship from the China Scholarship Council. S.T.S. acknowledges support from NIH R01 DK43726. H. Singh is an Investigator with the Howard Hughes Medical Institute.
REFERENCES
- 1.Singh H, Medina KL, Pongubala JM. Contingent gene regulatory networks and B cell fate specification. Proc Natl Acad Sci USA. 2005;102:4949–4953. doi: 10.1073/pnas.0500480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 3.DeKoter RP, Lee HJ, Singh H. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity. 2002;16:297–309. doi: 10.1016/s1074-7613(02)00269-8. [DOI] [PubMed] [Google Scholar]
- 4.Medina KL, et al. Assembling a gene regulatory network for specification of the B cell fate. Dev Cell. 2004;7:607–617. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Seet CS, Brumbaugh RL, Kee BL. Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A. J Exp Med. 2004;199:1689–1700. doi: 10.1084/jem.20032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye M, Ermakova O, Graf T. PU.1 is not strictly required for B cell development and its absence induces a B-2 to B-1 cell switch. J Exp Med. 2005;202:1411–1422. doi: 10.1084/jem.20051089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 8.Pongubala JM, et al. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol. 2008;9:203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- 9.Georgopoulos K. Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat Rev Immunol. 2002;2:162–174. doi: 10.1038/nri747. [DOI] [PubMed] [Google Scholar]
- 10.Cobb BS, Smale ST. Ikaros-family proteins: in search of molecular functions during lymphocyte development. Curr Top Microbiol Immunol. 2005;290:29–47. doi: 10.1007/3-540-26363-2_3. [DOI] [PubMed] [Google Scholar]
- 11.Molnar A, Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol Cell Biol. 1994;14:8292–8303. doi: 10.1128/mcb.14.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelley CM, et al. Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors. Curr Biol. 1998;8:508–515. doi: 10.1016/s0960-9822(98)70202-7. [DOI] [PubMed] [Google Scholar]
- 13.Klug CA, et al. Hematopoietic stem cells and lymphoid progenitors express different Ikaros isoforms, and Ikaros is localized to heterochromatin in immature lymphocytes. Proc Natl Acad Sci USA. 1998;95:657–662. doi: 10.1073/pnas.95.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georgopoulos K, et al. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- 15.Wang JH, et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 16.Nichogiannopoulou A, Trevisan M, Neben S, Friedrich C, Georgopoulos K. Defects in hemopoietic stem cell activity in Ikaros mutant mice. J Exp Med. 1999;190:1201–1214. doi: 10.1084/jem.190.9.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allman D, et al. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol. 2006;7:382–391. doi: 10.1038/ni1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delogu A, et al. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 2006;24:269–281. doi: 10.1016/j.immuni.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 20.Schebesta A, et al. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity. 2007;27:49–63. doi: 10.1016/j.immuni.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 21.O'Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- 22.Fuxa M, et al. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roessler S, et al. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5. Mol Cell Biol. 2007;27:579–594. doi: 10.1128/MCB.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maier H, et al. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat Immunol. 2004;5:1069–1077. doi: 10.1038/ni1119. [DOI] [PubMed] [Google Scholar]
- 25.Thompson EC, et al. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity. 2007;26:335–344. doi: 10.1016/j.immuni.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 27.Oettinger MA, Jung D, Giallourakis C, Mostoslavsky R, Alt FW. How to keep V(D)J recombination under control: mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Immunol Rev. 2004;200:165–181. doi: 10.1111/j.0105-2896.2004.00172.x. [DOI] [PubMed] [Google Scholar]
- 28.Fuller K, Storb U. Identification and characterization of the murine Rag1 promoter. Mol Immunol. 1997;34:939–954. doi: 10.1016/s0161-5890(97)00000-x. [DOI] [PubMed] [Google Scholar]
- 29.Lauring J, Schlissel MS. Distinct factors regulate the murine RAG-2 promoter in B- and T-cell lines. Mol Cell Biol. 1999;19:2601–2612. doi: 10.1128/mcb.19.4.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu W, et al. Coordinate regulation of RAG1 and RAG2 by cell type-specific DNA elements 5' of RAG2. Science. 1999;285:1080–1084. doi: 10.1126/science.285.5430.1080. [DOI] [PubMed] [Google Scholar]
- 31.Wei XC, et al. Characterization of chromatin structure and enhancer elements for murine recombination activating gene-2. J Immunol. 2002;169:873–881. doi: 10.4049/jimmunol.169.2.873. [DOI] [PubMed] [Google Scholar]
- 32.Hsu LY, et al. A conserved transcriptional enhancer regulates RAG gene expression in developing B cells. Immunity. 2003;19:105–117. doi: 10.1016/s1074-7613(03)00181-x. [DOI] [PubMed] [Google Scholar]
- 33.Wei XC, et al. Characterization of the proximal enhancer element and transcriptional regulatory factors for murine recombination activating gene-2. Eur J Immunol. 2005;35:612–621. doi: 10.1002/eji.200425185. [DOI] [PubMed] [Google Scholar]
- 34.Kosak ST, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 35.Sayegh C, Jhunjhunwala S, Riblet R, Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 2005;19:322–327. doi: 10.1101/gad.1254305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chowdhury D, Sen R. Stepwise activation of the immunoglobulin mu heavy chain gene locus. EMBO J. 2001;20:6394–6403. doi: 10.1093/emboj/20.22.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolland DJ, et al. Antisense intergenic transcription in V(D)J recombination. Nat Immunol. 2004;5:630–637. doi: 10.1038/ni1068. [DOI] [PubMed] [Google Scholar]
- 38.Su IH, et al. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 39.Bertolino E, et al. Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangements by transcription factor STAT5. Nat Immunol. 2005;6:836–843. doi: 10.1038/ni1226. [DOI] [PubMed] [Google Scholar]
- 40.Liu H, et al. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morgan B, et al. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 1997;16:2004–2013. doi: 10.1093/emboj/16.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 43.Tagoh H, et al. The mechanism of repression of the myeloid-specific cfms gene by Pax5 during B lineage restriction. EMBO J. 2006;25:1070–1080. doi: 10.1038/sj.emboj.7600997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin WC, Desiderio S. V(D)J recombination and the cell cycle. Immunol Today. 1995;16:279–289. doi: 10.1016/0167-5699(95)80182-0. [DOI] [PubMed] [Google Scholar]
- 45.Johnson K, et al. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 2008;28:335–345. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 46.Lu R, Medina KL, Lancki DW, Singh H. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev. 2003;17:1703–1708. doi: 10.1101/gad.1104803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shinkai Y, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 48.Cobb BS, et al. Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes Dev. 2000;14:2146–2160. doi: 10.1101/gad.816400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Medina KL, Singh H. Genetic networks that regulate B lymphopoiesis. Curr Opin Hematol. 2005;12:203–209. doi: 10.1097/01.moh.0000160735.67596.a0. [DOI] [PubMed] [Google Scholar]
- 50.Sitnicka E, et al. Complementary signaling through flt3 and interleukin-7 receptor alpha is indispensable for fetal and adult B cell genesis. J Exp Med. 2003;198:1495–1506. doi: 10.1084/jem.20031152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trinh LA, et al. Down-regulation of TDT transcription in CD4(+)CD8(+) thymocytes by Ikaros proteins in direct competition with an Ets activator. Genes Dev. 2001;15:1817–1832. doi: 10.1101/gad.905601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown KE, et al. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 53.Kirstetter P, Thomas M, Dierich A, Kastner P, Chan S. Ikaros is critical for B cell differentiation and function. Eur J Immunol. 2002;32:720–730. doi: 10.1002/1521-4141(200203)32:3<720::AID-IMMU720>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 54.Hu H, et al. Foxp1 is an essential transcriptional regulator of B cell development. Nat Immunol. 2006;7:819–826. doi: 10.1038/ni1358. [DOI] [PubMed] [Google Scholar]
- 55.Harker N, et al. The CD8alpha gene locus is regulated by the Ikaros family of proteins. Mol Cell. 2002;10:1403–1415. doi: 10.1016/s1097-2765(02)00711-6. [DOI] [PubMed] [Google Scholar]
- 56.Johnston CM, Wood AL, Bolland DJ, Corcoran AE. Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J Immunol. 2006;176:4221–4234. doi: 10.4049/jimmunol.176.7.4221. [DOI] [PubMed] [Google Scholar]
- 57.Goldmit M, et al. Epigenetic ontogeny of the Igk locus during B cell development. Nat Immunol. 2005;6:198–203. doi: 10.1038/ni1154. [DOI] [PubMed] [Google Scholar]
- 58.Liu Z, et al. A recombination silencer that specifies heterochromatin positioning and ikaros association in the immunoglobulin kappa locus. Immunity. 2006;24:405–415. doi: 10.1016/j.immuni.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 59.Kathrein KL, Lorenz R, Innes AM, Griffiths E, Winandy S. Ikaros induces quiescence and T-cell differentiation in a leukemia cell line. Mol Cell Biol. 2005;25:1645–1654. doi: 10.1128/MCB.25.5.1645-1654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hahm K, et al. The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol Cell Biol. 1994;14:7111–7123. doi: 10.1128/mcb.14.11.7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


