Abstract
The biological functions of poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins (hnRNPs) are not well understood. However, it is known that hnRNPs are involved in the regulation of alternative splicing for many genes, including the Ddc gene in Drosophila. Therefore, we first confirmed that poly(ADP-ribose) (pADPr) interacts with two Drosophila hnRNPs, Squid/hrp40 and Hrb98DE/hrp38, and that this function is regulated by Poly(ADP-ribose) Polymerase 1 (PARP1) and Poly(ADP-ribose) Glycohydrolase (PARG) in vivo. These findings then provided a basis for analyzing the role of pADPr binding to these two hnRNPs in terms of alternative splicing regulation. Our results showed that Parg null mutation does cause poly(ADP-ribosyl)ation of Squid and hrp38 protein, as well as their dissociation from active chromatin. Our data also indicated that pADPr binding to hnRNPs inhibits the RNA-binding ability of hnRNPs. Following that, we demonstrated that poly(ADP-ribosyl)ation of Squid and hrp38 proteins inhibits splicing of the intron in the Hsrω-RC transcript, but enhances splicing of the intron in the Ddc pre-mRNA. Taken together, these findings suggest that poly(ADP-ribosyl)ation regulates the interaction between hnRNPs and RNA and thus modulates the splicing pathways.
INTRODUCTION
Covalent modification of proteins by Poly(ADP-ribose) polymerase (PARP1) has been found to be involved in a variety of biological processes, including DNA repair, apoptosis, and transcription regulation (1). In addition, PARP1-bound poly(ADP-ribose) (pADPr), or free pADPr, is able to bind to proteins through a conserved domain in a non-covalent manner (2). This very stable (3) non-covalent binding to pADPr is equally prevalent in cells and can also modulate protein functions (4–12).
The concentration and size of pADPr within a cell depend on the relative activity of PARP1 and poly(ADP-ribose) glycohydrolase (PARG), which degrades pADPr polymer (13,14). While PARP1 is potently up-regulated by environmental stress signals, such as DNA damage (1) and heat shock (15), PARP1 is also required for normal development (16,17). PARG plays a key role in recycling automodified PARP1 and maintaining homeostasis of pADPr levels in cells. Thus, PARG loss-of-function in both Drosophila and mice causes the accumulation of pADPr which, in turn, results in lethality (13,14,18).
The six major Drosophila heterogeneous nuclear ribonucleoproteins (hnRNPs) (Hrb87F/hrp36, Hrb98DE/hrp38, Squid/hrp40, Hrb27C/hrp48, Hrb57A/Bancal and hrp59) have been characterized. Among them, the following hnRNP proteins are similar to the human hnRNP A/B type, which has two RNA-binding domains and one glycine-rich and M9-like (nuclear shuttling signal) domain: Hrb87F/hrp36 (19); Hrb98DE/hrp38 (20); Squid/hrp40 (21) and Hrb27C/hrp48 (22). Hrb57A/Bancal encodes a homolog of the vertebrate hnRNP K protein (23,24), and Hrp59 encodes a homolog of the vertebrate protein hnRNP M containing three RNA-binding domains (25,26). Hrp36 (27), hrp38 (20), hrp40 (28), hrp48 (22,29) and hrp59 (25) have all been shown to be involved in pre-mRNA splicing, and hrp40 is required for proper RNA localization (21,30,31). Recently, we established that Hrb98DE/hrp38 is one of the major proteins associated with PARP1 in vivo (6). Previously, the human hnRNP A1 protein has been shown to have a pADPr-binding motif and can bind with pADPr by an in vitro binding assay (32). However, whether hnRNPs can interact with PARP1 and bind with pADPr in vivo is currently unknown. Furthermore, the biological functions of pADPr binding to hnRNPs have not yet been investigated.
Therefore, we use Drosophila as the model organism to study the regulation of hnRNPs by poly(ADP-ribosyl)ation. Our data demonstrate that pADPr binding to Squid/hrp40 and Hrb98DE/hrp38 does occur in vivo and is, furthermore, regulated by endogenous PARP1 and PARG. In addition, we find that pADPr binding to hnRNPs changes the RNA-binding ability of hnRNPs, further modulating splicing.
MATERIALS AND METHODS
Drosophila strains and breeding conditions
Flies were cultured on standard cornmeal-molasses-agar media at 22°C, unless otherwise indicated. Two Parp transgenic lines (UAST-PARPe-EGFP; 69B-GAL4/TM3 and G1-GAL4, UAST-PARP1-DsRed), both having ubiquitous expression of PARP1, were described in our previous reports (15,16). A Parg mutant (Parg27.1/FM7, Actin-GFP) was described previously (13,14). Hrb98DE/hrp38 GFP trap line (ZCL588) and Squid/hrp40 mutant (y1 w*; P{lacW}sqd j6E3, l(3)j6E3 j6E3/TM3, Sb1) were obtained from the Bloomington Drosophila Stock Center. One P-element insertion of the Hrb98DE/hrp38 gene (w*,P[XP]d05172/TM6B, Tb1), a Hrb98DE/hrp38 region deficiency line (w1118; Df(3R)Exel6209, P{XP-U}Exel6209/TM6B, Tb1), and a Parp mutant (C03256/TM6B, Tb1) were obtained from the Exelixis Collection at the Harvard Medical School. We replaced the balancer chromosomes in the Parp mutant, the Squid/hrp40 mutant, the P-element insertion of the Hrb98DE/hrp38 gene and the Hrb98DE/hrp38 region deficiency strain with the GFP-bearing balancer from w*; Sb1/TM3, P{ActGFP}JMR2, Ser1 for selecting the homozygous mutants. We also generated G1-GAL4, UAST-PARP1-DsRed; ZCL588 and Parg27.1/FM7, Actin-GFP; ZCL588 strains using the standard genetic methods.
Co-immunoprecipitation
The total proteins from about 25 third-instar wandering larvae of the appropriate genotypes grown at the normal culturing temperature, or heat shocked at 37°C for one hour using water bath, were extracted using RIPA buffer plus protease inhibitor tablets (Roche). The protein concentrations were measured using the RC DC protein assay kit (Bio-Rad). Equal amounts of total proteins (around 1 mg) were precleared with 50 μl protein A agarose (Invitrogen) and 5 μg rabbit IgG (Upstate) for 1 h. The precleared lysate was then precipitated using 5 μg rabbit IgG (the negative control), 5 μg anti-pADPr rabbit polyclonal antibody (Calbiochem) or anti-GFP rabbit polyclonal antibody (Torrey Pines Biolabs) with/without 10 μg RNase A (Sigma) and 100 units RNase T1 (Ambion) per mg protein overnight and further incubated with 30 μl protein A agarose (Invitrogen) for 2 h at 4°C. The IP complex was washed using RIPA buffer five times and eluted with 2× SDS loading buffer at 95°C.
To detect interaction between poly(ADP-ribose) and hrp38:GFP fusion protein, a Seize X protein A immunoprecipitation kit (Pierce) was used to eliminate the contamination of the 55 kDa heavy chain of the antibody, which has nearly the same molecular weight as the hrp38:GFP fusion protein. Basically, either the rabbit anti-pADPr polyclonal antibody (Calbiochem) or rabbit IgG (Upstate) was covalently crosslinked to immobilized Protein A-beaded agarose resin in a column, according to the protocol provided by the manufacturer (Pierce). Equal amounts of precleared lysates with 30 μl protein A agarose (Invitrogen) and 5 μg rabbit IgG (Upstate) were precipitated in the column containing the anti-pADPr rabbit polyclonal antibody (Calbiochem) or rabbit IgG (Upstate). The IP complex in the column was washed with RIPA buffer three times and twice with gentle binding buffer (Pierce), then finally eluted with gentle elution buffer (Pierce).
The IP complex obtained, using either the traditional IP method or Seize X protein A immunoprecipitation kit, was subjected to immuoblotting. The blot was incubated with mouse anti-Squid at dilution 1:2500 (8D2; a gift from Dr Dreyfuss) (33), anti-pADPr mouse monoclonal antibody at 1:500 dilution (10H; Calbiochem), anti-DsRed mouse monoclonal antibody at 1:500 dilution (Clontech), and anti-GFP rabbit polyclonal antibody at 1:1000 dilution (Torrey Pines Biolabs) as the primary antibody. After washing the blot with PBS-Tween 20, the blot was further incubated with horseradish peroxidase-conjugated secondary antiserum and detected using ECLTM reagents (GE Health). All IPs were repeated twice. The films were photographed with the white light transilluminator (Fisher) using the FoTo/analysis system (FOTODYNE), and the signals were measured using the Image J software (NIH).
RNA electrophoretic mobility shift assay (EMSA)
RNA EMSA was basically performed according to the protocol provided by the chemiluminescent EMSA kit (Pierce). RNA-binding reaction was done by incubating 100 ng GST (BioVision) or 100 ng GST-hnRNPA1 (Abnovo) with 25 fmol biotin-labeled human hnRNP A1 ‘winner’ sequence (UAUGAUAGGGACUUAGGGUG) (34) (Invitrogen) in a 25 µl volume, including 1× binding buffer (10 mM Tris–HCl, 50 mM KCl, 1 mM DTT, pH 7.5), 2.5% glycerol, 2 unit/µl RNAasin (Promega), 0.25 µg/µl purified BSA and 0.25 µg/µl yeast tRNA (Invitrogen) for 30 min at 30°C. For pADPr inhibition assay, 100 ng human GST-hnRNPA1 was preincubated with 70 ng or 140 ng pADPr (Biomol) in 1× binding buffer for 20 min at 25°C. Half of the reaction with 5× loading buffer (Pierce) was run on 6% DNA-retardation gel (Invitrogen) and transferred to the nylon membrane (Hybond-XL, Amersham Biosciences). The free RNA and RNA–protein complex were detected with stabilized streptavidin-horseradish peroxidase conjugate with chemiluminescent substrate (Pierce). EMSA was repeated three times, and the signals were measured using the Image J software (NIH).
RNA–protein co-immuoprecipitation
The RNA and protein complexes from about 75 third-instar wandering larvae of the appropriate genotypes were extracted using polysome lysis buffer (35). The precleared lysates were incubated with 20 μg anti-GFP rabbit polyclonal antibody (Torrey Pines Biolabs) or 20 μg rabbit IgG (Sigma) (the negative control) overnight at 4°C, followed by precipitating the antigen–antibody complexes with 120 μl protein A agarose (Invitrogen) for 2 h at 4°C. The IP complex was washed three times, using the lysis buffer, and eluted with 200 μl elution buffer [1% SDS, 50 mM NaCI, 50 mM Tris–HCI (pH 7.0), 5 mM EDTA and 100 units/ml RNAase inhibitor (Promega)] at 50°C for 30 min with rocking. RNA from elutes was further precipitated with 800 ml Trizol (Invitrogen) and treated with RQ1 RNAse-free DNAse (Promega) and cleaned with RNeasy mini kit (Qiagen) for the regular and real-time RT–PCR assay. The experiment was repeated twice.
Immunofluorescence
The polytene chromosomes prepared from the salivary glands of the intact nuclei of the wandering third-instar larvae grown at normal culturing temperature, or heat shocked at 37°C for one hour using the water bath, were stained for DNA with DRAQ5 dye (Biostatus). GFP was visualized using the Leica TCS-NT confocal microscope. The method for immunostaining of Squid and pADPr in the polytene chromosome from squashed nuclei was described previously by Tulin and Spradling (15). The primary antibodies were mouse anti-pADPr monoclonal antibody (10H; Calbiochem), used at 1:100 dilution against pADPr, and mouse anti-Squid monoclonal antibody, used at 1:100 dilution (8D2) against Squid. The secondary antibody was Alexa Fluor® 488 goat anti-mouse IgG (Invitrogen), used at 1:400 dilution, and propidium iodide (Sigma) was used to stain DNA. The confocal images were quantified using MetaMorph image analysis software (Molecular Devices).
Real-time RT–PCR
Either the wandering third-instar larvae of yw (wild-type) or non-GFP homozygous mutants were selected, or allowed to further develop into pharate adults, as indicated, for RNA extraction using RNeasy Mini Kit with on-column DNAse digestion (Qiagen). RNA concentration was quantified using the Agilent 2100 BioAnalyzer in combination with an RNA 6000 Nano LabChip (Agilent Technologies). RNA was reverse-transcribed using M-MLV reverse transcriptase (Ambion) and oligo(dT). Real-time PCR assays were performed using Power Sybr® Green PCR master mix and an ABI 7900 HT instrument (Applied Biosystems). The primer sequences were as follows: 5′ TGAAATAGGAAGCCAGTTGGG3′ (forward) and 5′ TATCGACTTTTCACGGATCGAT 3′ (reverse) for Hsrω-RA; 5′ CGAGCTCTTTGTTTGCCTTTC 3′ (forward) and 5′ CAAAGTCAGGCTGGCGAGA 3′ (reverse) for Hsrω-RC; 5′ CAGTTAACTAAAGTGCAACGATCGA 3′ (forward) and 5′CCTTTCGCGTATATTCTCCAGATATT 3′ (reverse) for Ddc-RB; 5′ AACACAATTCCAACAAAACAAACTGA 3′ (forward) and 5′ GCCTCCATGTCGATCGAAAC 3′ (reverse) for Ddc-RC. As an internal control, the expression level of the RpL32 gene was measured using the following primers: 5′ CCAAGGACTTCATCCGCCACC 3′ (forward) and 5′ GCGGGTGCGCTTGTTCGATCC 3′ (reverse). Cycling conditions were 95°C for 15 min, followed by 40 (2-step) cycles (95°C, 15 s; 60°C, 60 s). Ct (cycle threshold) values were converted to quantities (in arbitrary units) using a standard curve (4 points, 5-fold dilutions) established with a calibrator sample. For each sample per repeat, the value is an average of two PCR reactions performed with inputs of 200 (high input) and 40 ng (lower input) of total RNA. All the experiments were repeated twice, starting from the RNA extractions. The statistics analysis was done based on Student's t-test.
RESULTS
Drosophila hnRNPs have putative pADPr-binding sites
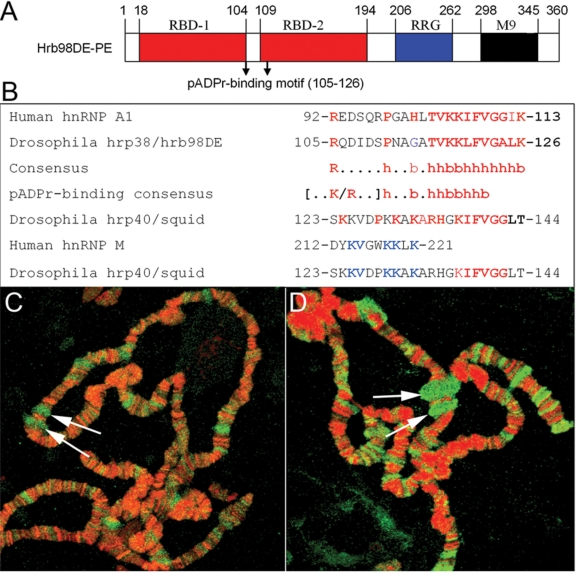
In an effort to identify the target proteins of PARP1, we found that Drosophila hrp38 is associated with PARP1 protein using mass spectrometry analysis (6). As previously described (19), hrp38 has 50% identity and conserved functional domains with human hnRNP A1 (Figure 1A). We found that hrp38 contains a pADPr-binding consensus (Figure 1B) previously identified in human hnRNP A1 (32). Squid also has the six residues ‘KIFVGG’ present in human hnRNP A1, which have been shown to be required for pADPr binding (32). In addition, Squid was also found to have a putative pADPr-binding site (Figure 1B), which is homologous to the pADPr-binding motif identified in human hnRNP M (32). Of added importance to the aims of the present study, we found that both pADPr and Squid accumulated in the ecdysone-inducible E74 and E75 puffs (Figure 1C and D), suggesting the association of pADPr with Squid. Based on these findings, we expected that pADPr could bind to hrp38 and Squid.
Figure 1.
The pADPr-binding motif in Drosophila Hrp38 and Squid protein. (A) Conserved functional domains of Drosophila hrp38 protein. RBD: RNA-binding domains. RGG: glycine-rich domain. M9: nuclear shuttling signal domain. (B) Alignment of putative pADPr-binding sequences of human and Drosophila hnRNPs with the consensus of pADPr-binding motif. The conserved residues in the pADPr-binding motif among human hnRNP A1, hrp38 and Squid are marked in red; among human hnRNP M and Drosophila Squid, they are marked in blue (b: basic, h: hydrophobic amino acid). (C) The accumulation of pADPr in the puffs. Drosophila polytene chromosomes of the wild-type line (y,w) were immunostained with the anti-pADPr antibody (10H). pADPr: green; DNA: red. (D) The localization of Squid in the puffs. Drosophila polytene chromosomes of the wild-type line (y,w) were immunostained with the anti-Squid antibody. Squid: green; DNA: red. Arrows in (C) and (D) indicate the E74 and E75 puffs.
PARP1 overexpression and PARG loss-of-function increases level of pADPr-binding Squid
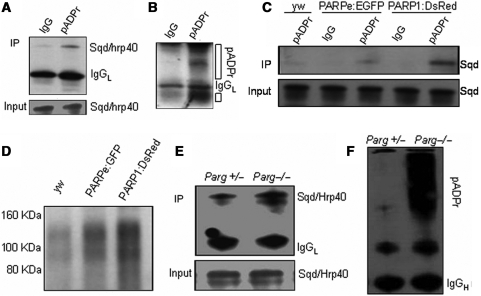
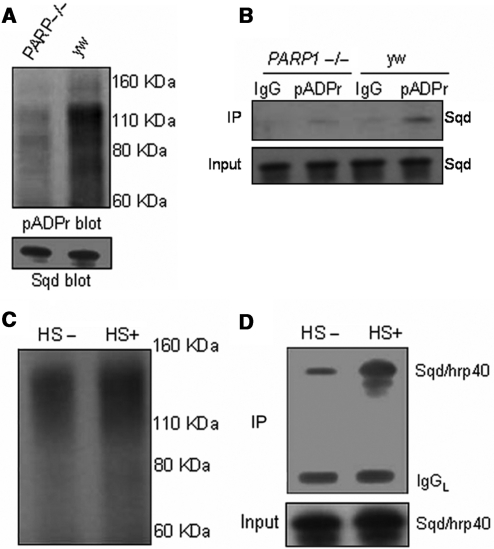
Using a co-immunoprecipitation strategy (9), we investigated if Squid binds to pADPr in vivo. First, we used anti-pADPr antibody to immunoprecipitate the poly(ADP-ribosyl)ated proteins from the wild-type line (yw) and immunoblotted the immunoprecipitated complex using the anti-Squid antibody (8D2) (33). The expression level of Squid was strongly reduced in the Squid mutant, further confirming the specificity of this Squid antibody (Supplementary Figure S1). The co-immunoprecipitation experiment showed that even the wild-type fly has a modest amount of Squid protein interacting with pADPr (Figure 2A). Since the co-immunoprecipitated Squid protein had the same molecular weight as the native Squid protein (Figure 2A), it appears that pADPr binds with Squid in a non-covalent way. To confirm immunoprecipitation efficiency, we reprobed the same blot with anti-pADPr antibody and observed that there were multiple bands, which are poly(ADP-ribosyl)ated proteins or free pADPr (Figure 2B). We further investigated if pADPr binding to Squid could be impacted by regulating the endogenous PARP1 and PARG. Increases of pADPr-binding Squid protein from two PARP overexpression transgenic lines (UAS-PARPe-EGFP; 69B-Gal4 and G1-Gal4, UAS-PARP1-DsRed) compared to the wild-type, by 2.3 times and 9.4 times, respectively, were observed (Figure 2C). In addition, overexpression of PARP1-DsRed, which represents full PARP activity (16), resulted in a 4-fold increase of pADPr-binding Squid protein over PARPe-EGFP overexpression, which does not have a catalytic domain, but is required for PARP1 transcription (16) and can be modified by PARP1 (6). Indeed, probing the same amount of immunoprecipitated complex from three different genotypes with anti-pADPr antibody showed an increasing order of pADPr level from the PARP overexpression lines (2.0-fold in UAS-PARPe-EGFP; 69B-Gal4 and 2.9-fold in G1-Gal4; UAS-PARP1-DsRed) when compared to the wild-type (yw) (Figure 2D). Co-immunoprecipitation experiments also showed that PARG loss-of-function resulted in a 2.7-fold increase of pADPr-binding Squid protein over that of the heterozygous genotype (Figure 2E). Reprobing the same blot with anti-pADPr antibody showed a greatly increased pADPr level in the Parg−/− mutant (Figure 2F), indicating that increased Squid bound to pADPr is positively correlated with pADPr accumulation by PARG loss-of-function. Taken together, both PARP overexpression and PARG loss-of-function result in increased amounts of Squid bound to pADPr. Therefore, we conclude that pADPr binding to Squid occurs in vivo and is regulated by PARP and PARG.
Figure 2.
Increased amounts of Squid bound to pADPr in vivo by PARP overexpression and Parg mutation. (A) The total protein from the wild-type line (yw) was immunoprecipitated using rabbit anti-pADPr antibody or preimmune rabbit IgG as a negative control. The immunoprecipitates and 1% input for immunoprecipitation were subjected to immunoblotting, using anti-Squid antibody. (B) The same blot was stripped and probed with rabbit anti-pADPr antibody. Open bar indicates pADPr. (C) Equal total proteins from the wild-type line (yw), PARPe-EGFP; 69B-Gal4 and G1-Gal4, PARP1-DsRed transgenic lines were immunoprecipitated using rabbit anti-pADPr antibody. The preimmune rabbit IgG was used as a negative control for immunoprecipitation. The immunoprecipitates and 1% input for immunoprecipitation were subjected to immunoblotting analysis using anti-Squid antibody. (D) The same amounts of the immunoprecipitates obtained with rabbit polyclonal anti-pADPr antibody from three different genotypes were immunoblotted with anti-pADPr monoclonal antibody (10H). (E) Equal amounts of the lysates from Parg+/− heterozygote and Parg−/− homozygotes were immunoprecipitated using rabbit anti-pADPr antibody. The immunoprecipitates and 1% input for immunoprecipitation were subjected to immunoblot analysis using anti-Squid antibody. (F) The same blot in E was stripped and probed with anti-pADPr monoclonal antibody (10H).
PARP1 overexpression and PARG loss-of-function increase level of pADPr-binding hrp38
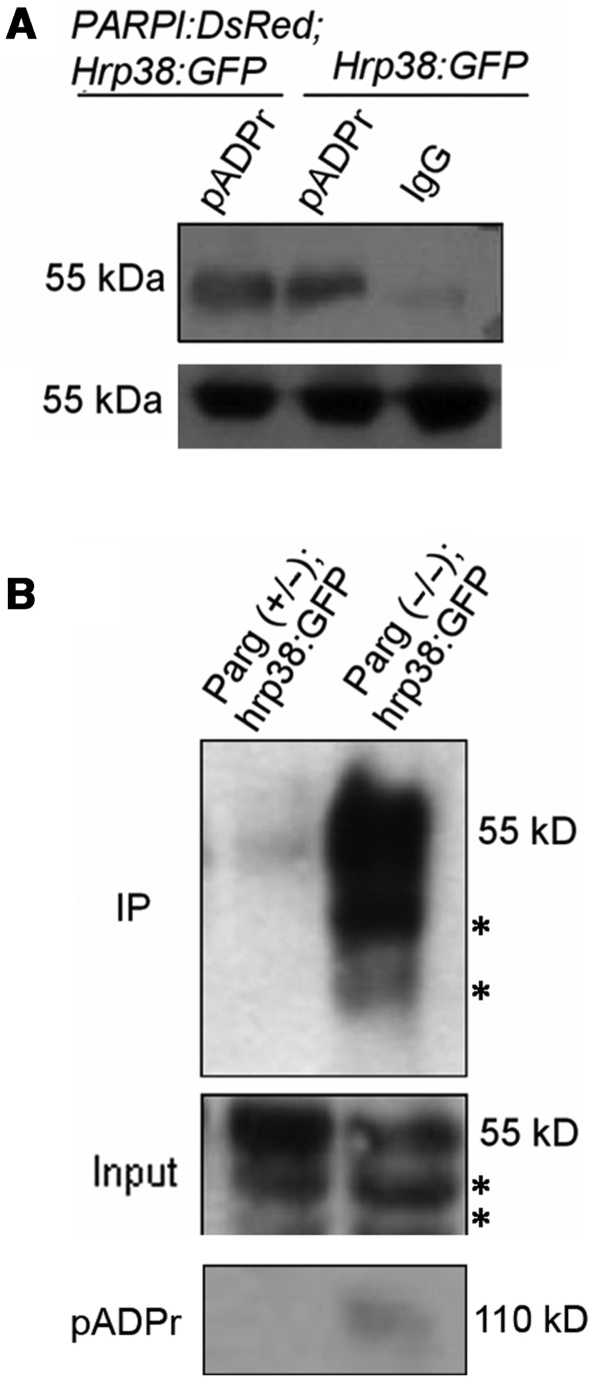
Next, to determine if hrp38 binds to pADPr in vivo, we generated G1-GAL4, UAST-PARP1-DsRed; ZCL588 (hrp38:GFP) and Parg27.1/FM7, actin-GFP; ZCL588 (hrp38:GFP) strains. ZCL588 is a protein trap line in which GFP was inserted into the first intron of the hrp38 (CG9983) gene to produce the hrp38:GFP fusion protein (36). Since the hrp38 (CG9983) gene has several alternative transcripts, we sequenced the RT–PCR products from the hrp38:GFP transcripts, and the results showed that GFP was spliced in the frame with hrp38-PE (CG9983-PE) (Supplementary Figures S2A and S2B). We used anti-pADPr antibody to immunoprecipitate the poly(ADP-ribosyl)ated proteins from the hrp38:GFP line and Parp1-DsRed; hrp38:GFP line and further immunoblotted the immunoprecipitated complex using anti-GFP antibody (Figure 3A, Supplementary Figures S2C and S2D). The result showed that pADPr interacts with the hrp38:GFP fusion protein and that there was an average 1.8-fold increase in the level of hrp38:GFP fusion protein bound to pADPr in the PARP1 overexpression transgenic line (Parp1-DsRed; hrp38:GFP) beyond what was observed in the hrp38:GFP line (Figure 3A). Consistent with the results obtained above, co-immunoprecipitation experiments also showed that Parg mutants expressing hrp38:GFP fusion protein (Parg−/−; hrp38:GFP) had a 7.4-fold higher level of fusion protein bound to pADPr than the heterozygous genotype (Figure 3B, upper panel). Probing the same immunoprecipitated complex with the anti-pADPr antibody confirmed immunoprecipitation efficiency (Figure 3B, bottom panel). Therefore, it appears that pADPr can bind with hrp38, also in a non-covalent way.
Figure 3.
Increased amounts of hrp38 bound to pADPr in vivo by PARP overexpression and Parg mutation. (A) Equal lysates of hrp38:GFP and PARP1-DsRed; hrp38:GFP transgenic lines were immunoprecipitated using rabbit anti-pADPr antibody or the preimmune IgG crosslinked to protein A agarose beads. The immunoprecipitates and 5% input for immunoprecipitation were subjected to immunoblot analysis using anti-GFP antibody. (B) Equal amounts of the lysates from Parg+/− heterozygote and Parg−/− homozygotes were immunoprecipitated using rabbit anti-pADPr antibody crosslinked to protein A agarose beads. The immunoprecipitates (upper panel) and 5% input for immunoprecipitation (middle panel) were subjected to immunoblot analysis using anti-GFP antibody. The same blot in the upper panel was stripped and probed with anti-pADPr monoclonal antibody (bottom panel). Asterisk in upper and middle panel indicates the degraded products from hrp38:GFP.
Automodified PARP1 is associated with hnRNPs in vivo
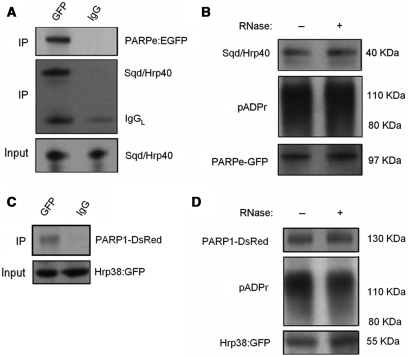
Since PARP1 itself is the predominant acceptor of pADPr, we further determined whether a PARP1-pADPr-Squid complex exists in vivo. To make this assessment, we first used anti-GFP antibody to immunoprecipitate the PARPe-EGFP fusion protein from the PARPe-EGFP transgenic line and further immunoblotted the immunoprecipitated complex using anti-Squid antibody. The 40 kDa Squid protein was observed from the immunoprecipitated complex with anti-GFP antibody, but not with preimmune rabbit IgG (Figure 4A), suggesting that an interaction between PARP1 and Squid does take place. The same blot was reprobed with anti-GFP antibody, and the 97 kDa PARPe-EGFP protein was detected, confirming immunoprecipitation efficiency (Figure 4A). Moreover, automodified PARPe-EGFP was also detected in the immunoprecipitated complex using anti-pADPr antibody (Figure 4B), suggesting that the interaction between PARP1 and Squid is mediated by pADPr. In addition, RNase treatment of the lysates did not disrupt the association between Squid and automodified PARPe-EGFP (Figure 4B), confirming that RNA did not mediate the interaction. Secondly, to determine whether an interaction between PARP1 and hrp38 exists in vivo, we used anti-GFP antibody to immunoprecipitate the hrp38:GFP fusion protein from the PARP1-DsRed; Hrp38:GFP fly strain, followed by immunoblotting the immunoprecipitated complex using anti-DsRed antibody. The PARP1-DsRed fusion protein (∼130 kDa) was observed from the immunoprecipitated complex with anti-GFP antibody, but not from the immunoprecipitated complex with preimmune rabbit IgG (Figure 4C). The automodified PARP1-DsRed was also detected in the immunoprecipitated complex using anti-pADPr antibody (Figure 4D). RNase treatment of the lysates did not disrupt the interaction between hrp38 and automodified PARP1-DsRed (Figure 4D), suggesting again that RNA did not mediate this association. Since the expression of hrp38:GFP fusion protein reflects the endogenous expression level of the hrp38 gene, we concluded that hrp38 is also associated with automodified PARP1 in vivo. Finding an interaction between automodified PARP1 and hrp38 or Squid, in turn, strongly suggests that pADPr binding to Squid or hrp38 is mediated by automodified PARP1 in vivo.
Figure 4.
The association of PARP with Squid and hrp38 in vivo. (A) An equal amount of lysate from PARPe-EGFP transgenic line was immunoprecipitated using rabbit anti-GFP antibody or preimmune rabbit IgG as a negative control, and the immunoprecipitated complex was subjected to immunoblotting analysis using anti-Squid antibody and reprobed with mouse anti-GFP antibody. One percent input was shown for the equal amount of protein for immunoprecipitation. (B) Equal amounts of lysates of PARPe-EGFP transgenic line with/without RNase treatment were immunoprecipitated using rabbit anti-GFP antibody. The immunoprecipitated complex was immunoblotted using anti-Squid antibody, anti-pADPr antibody (10H) and anti-GFP monoclonal antibody, respectively. (C) Equal amounts of lysates of the PARP1-DsRed; Hrp38:GFP transgenic line were immunoprecipitated using anti-GFP polyclonal antibody, or the normal rabbit IgG, as a negative control, followed by immunoblotting the immunoprecipitated complex using anti-DsRed antibody. Five percent input was immunoblotted using anti-GFP polyclonal antibody to show the equal input. (D) Equal amounts of lysates of the PARP1-DsRed; Hrp38:GFP transgenic line with/without RNase treatment were immunoprecipitated using anti-GFP polyclonal antibody. The immunoprecipitated complex was immunoblotted using anti-DsRed antibody and anti-pADPr antibody (10H). Five percent input was immunoblotted using anti-GFP polyclonal antibody to show that hrp38:GFP is present in the PARP1-DsRed; Hrp38:GFP transgenic line.
PARP1 controls pADPr binding to Squid during heat shock
Further testing was performed to determine whether PARP1 is required for pADPr binding to hnRNPs, using a partial loss-of-function PARP1 mutant (C03256), which was verified by northern blot (data not shown). This PARP1 mutation showed a 3.5-fold lower pADPr level than the wild-type (Figure 5A), further confirming it as a partial loss-of-function PARP1 mutant. We noticed that there was no obvious difference in the Squid expression level between this PARP1 mutant and the wild-type (Figure 5A). However, the co-immunoprecipitation experiment showed that this PARP1 mutant had 2.9-fold less Squid bound to pADPr compared to the wild-type (Figure 5B), further suggesting that PARP1 is required for pADPr binding to Squid. Our previous studies showed that heat shock results in pADPr accumulation at the heat shock-induced puffs (15). Therefore, we tested if heat-shock treatment alters pADPr binding to Squid. As expected, the pADPr level increased by 2.1 times after heat-shock treatment (Figure 5C). Subsequently, even greater amounts of Squid protein bound to pADPr (a 3.9-fold increase) were observed following the heat-shock treatment (Figure 5D), suggesting that the ability of pADPr to bind hnRNPs is up-regulated by the heat-shock treatment. This observation implies that pADPr binding to hnRNPs may play a role in regulating hnRNP upon environmental stresses such as heat shock.
Figure 5.
pADPr binding to Squid requires PARP1 and is potently induced by the heat-shock treatment: (A) Equal amounts of lysates from the wild-type line (yw) and a partial PARP1 homozygous mutant (PARP−/−) were immunoprecipitated with rabbit anti-pADPr antibody. The immunoprecipitates were subjected to immunoblotting with mouse anti-pADPr antibody (10H). One percent input was immunoblotted using anti-Squid antibody to compare Squid expression between wild-type and PARP1 mutant. (B) Equal amounts of lysates from the wild-type line (yw) and a partial PARP1 homozygous mutant (PARP−/−) were immunoprecipitated with rabbit anti-pADPr antibody or rabbit IgG (control). The immunoprecipitates and 1% input were subjected to immunoblotting with anti-Squid antibody. (C) Equal amounts of lysates from the wild-type line (yw) grown at 22°C or heat shocked at 37°C for 1 h at the third-instar larvae stage were immunoprecipitated with rabbit anti-pADPr antibody. The immunoprecipitates were subjected to immunoblot analysis using mouse anti-pADPr antibody (10H). (D) The same amount of immunoprecipitates as in Figure C and 1% input for immunoprecipitates were immunoblotted using anti-Squid antibody.
Poly(ADP-ribosyl)ation causes the relocalization of hnRNPs from chromatin to nucleoplasm
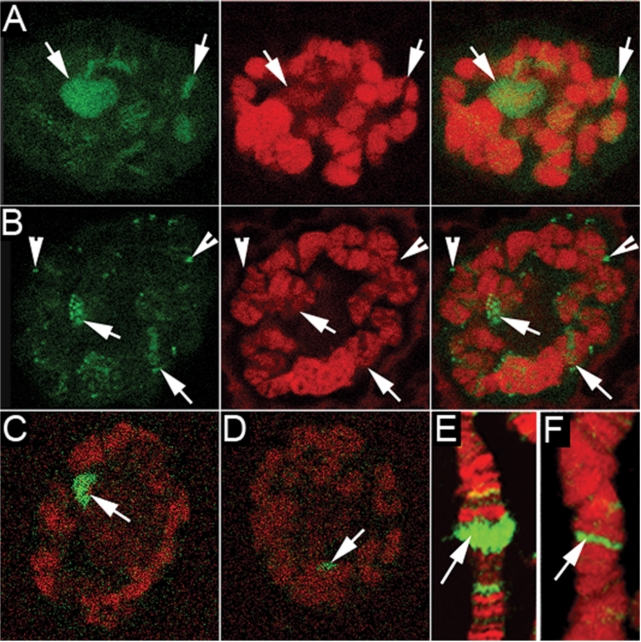
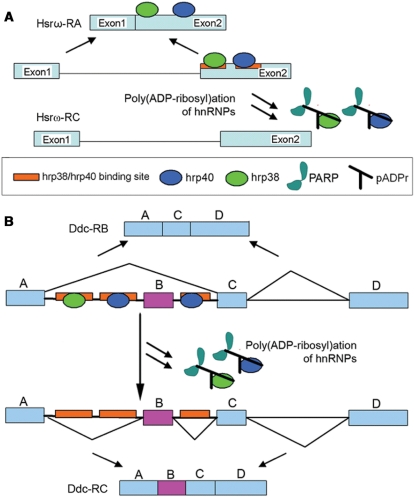
We further examined hrp38 expression patterns in polytene chromosomes prepared from the Parg; hrp38:GFP transgenic line. Hrp38 is mainly localized in the puffs and other transcription-activated loci in the hrp38:GFP line (Figure 6A). In the Parg mutant nuclei, hrp38:GFP dissociates from the puffs and accumulates in the nucleoplasmic particles (Figure 6B). In wild-type upon heat-shock treatment, hrp38:GFP dissociates from most of the loci and mainly binds to one heat shock-induced puff, Hsrω (93D) (Figure 6C and Supplementary Figure S3A). It is very likely that hrp38 is sequestered on the 14 kb non-coding RNA (Hsrω-n) at this locus as other hnRNP proteins (Squid and hrp36) were after heat shock (37). However, in Parg mutants, the accumulation of hnRNPs at the Hsrω (93D) locus is diminished by 3.3 times (Figure 6D). In addition, we found that Squid in the Parg mutant also had diminished accumulation by 4.2 times at the Hsrω (93D) locus upon heat shock (Figure 6F) compared with the wild-type (Figure 6E), suggesting that decreased amounts of Squid bind to the transcript in the Hsrω (93D) locus. As a control, immunoblotting showed that there is no significant difference in the expression levels of hrp38:GFP and Squid between hrp38:GFP and Parg; hrp38:GFP lines after heat-shock treatment (Supplementary Figure S3B). Therefore, we proposed that increased pADPr binding to hnRNPs caused by either heat shock or PARG loss-of function alters the RNA-binding ability of hnRNPs because of charge repulsion between pADPr and RNA, which, in turn, results in the dissociation of hnRNPs from most of the transcripts.
Figure 6.
The dissociation of hrp38 and Squid from the puffs in polytene chromosomes caused by heat shock and Parg null mutation: hrp38:GFP from the intact nuclei is shown as green in (A) through (D). Squid from the squashed nuclei is shown as green in (E) and (F). DNA is shown as red in (A) through (F). All images are from a single confocal section. (A) The localization of hrp38:GFP in the salivary glands polytene chromosomes of the hrp38:GFP transgenic line (ZCL588). Arrows indicate the major puffs. (B) The dissociation of hrp38:GFP from the puffs in polytene chromosomes of Parg27.1; hrp38:GFP transgenic lines. Arrowheads indicate extrachromosomal particles accumulating HRP proteins. (C) hrp38:GFP binding to the 93D puff in polytene chromosomes of the hrp38:GFP line after heat-shock treatment. Arrow indicates the 93D puff in (C) through (F). (D) Reduced amounts of hrp38:GFP binding to the 93D puff in the Parg27.1; hrp38:GFP line after heat-shock treatment. (E) Squid binding to the 93D puff in polytene chromosomes of the wild-type line (yw) after heat-shock treatment: Squid, green; DNA, red. (F) Reduced amounts of Squid binding to the 93D puff in the Parg27.1 line after heat-shock treatment: Squid, green; DNA, red.
Poly(ADP-ribose) reduces RNA-binding ability of hnRNPs in vitro and in vivo
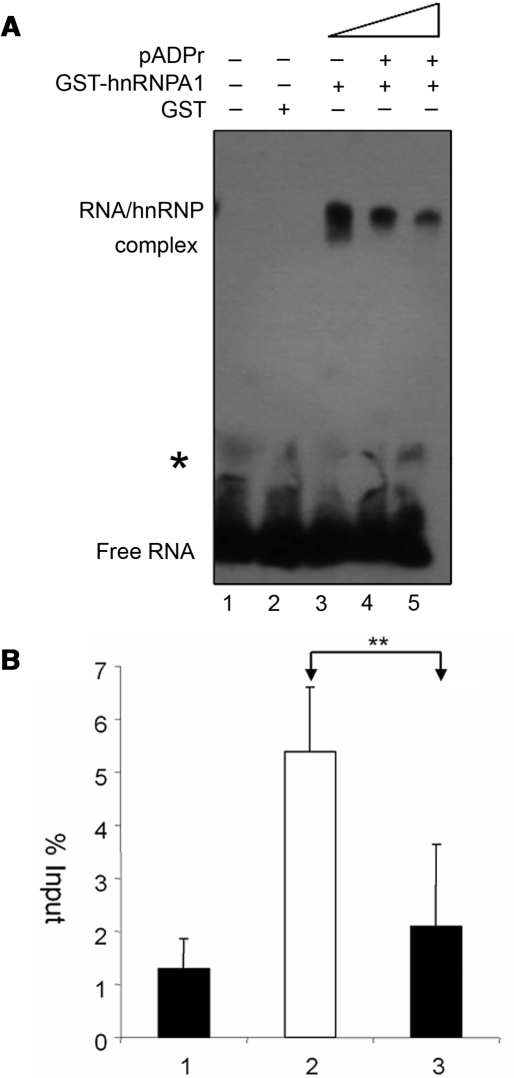
To further test our hypothesis that the interaction between pADPr and hnRNPs reduces the RNA-binding ability of hnRNPs, we used RNA EMSA to determine if pADPr can inhibit human GST-hnRNPA1 binding to its highest-affinity binding (‘winner’) sequence (34). At first, we confirmed that pADPr can bind with human GST-hnRNPA1 protein using dot-blot analysis (data not shown) as reported before (32). RNA EMSA showed that GST alone did not bind with RNA (Figure 7A, lane 2), while human GST-hnRNPA1 strongly bound with its ‘winner’ sequence to form the RNA–protein complex (Figure 7A, lane 3) as human hnRNP A1 shown before (38). However, after preincubation of 100 ng GST-hnRNPA1 with 70 ng or 140 ng pADPr (2-fold excess), the amounts of RNA bound by GST-hnRNPA1 were decreased by 1.8- and 2.7-fold, respectively (Figure 7A, lanes 4 and 5), suggesting that pADPr inhibits human hnRNPA1 binding to RNA in a dosage-dependent manner in vitro.
Figure 7.
Reduced RNA bound to hnRNPs by pADPr and PARG loss-of-function. (A) pADPr inhibits human GST-hnRNPA1 binding to RNA, as shown by EMSA. 25 fmol biotin-labeled hnRNPA1 binding sequence was incubated with the components as indicated: 1. Control; 2. 100 ng GST; 3. 100 ng human GST-hnRNP A1; 4. 100 ng human GST-hnRNP A1 preincubated with 70 ng pADPr; 5. 100 ng human GST-hnRNP A1 preincubated with 140 ng pADPr. *The trace of higher molecular weight RNA is present in all reactions, which likely comes from the synthetic RNA pool. (B) Reduced RNA bound to hrp38:GFP by PARG loss-of-function. The amount of hsrω-RA (spliced product) co-immunoprecipitated with hrp38:GFP as a percent of the input was measured by real-time RT–PCR: 1. hrp38:GFP strain immunoprecipitated with IgG as a control; 2. hrp38:GFP strain immunoprecipitated with anti-GFP; 3. Parg27.1; hrp38:GFP strain immunoprecipitated with anti-GFP. The error bar represents the standard deviation from two independent experiments (**P ≤ 0.01).
We also tested our hypothesis in vivo by co-immunoprecipitating the RNA-hrp38:GFP complex from the wild-type (hrp38:GFP) and the Parg mutants (Parg27.1; hrp38:GFP) using anti-GFP antibody. The co-immunoprecipitated RNAs were subjected to real-time RT–PCR to amplify a spliced transcript (Hsrω-RA) from the hsrω (93D) gene. We found that this transcript, hsrω-RA, was associated with hrp38:GFP and was significantly less co-immunoprecipitated from the Parg mutant (Parg27.1; hrp38:GFP) than the wild-type (hrp38:GFP) (Figure 7B). Since PARG loss-of function resulted in increased pADPr binding to hrp38 (Figure 3B), the reduction of the amount of hrp38:GFP fusion protein associated with the hsrω-RA transcript in the Parg mutant suggests that pADPr binding to hrp38:GFP abolishes the RNA-binding ability of hrp38:GFP. This result also agrees with the observation that hrp38:GFP in the Parg mutant was relocalized from the puffs at normal temperatures (Figure 6B). Taken together, both RNA EMSA and RNA–protein co-immunoprecipitation strongly suggest that pADPr binding to hnRNPs reduces the RNA-binding ability of hnRNPs.
PARP and PARG modulate splicing in vivo
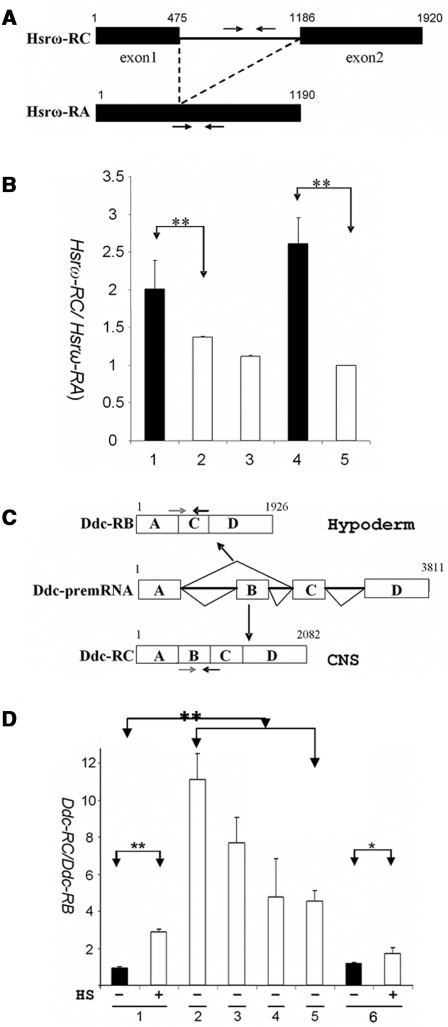
We further predicted that the dissociation of hnRNPs from RNA which results from pADPr binding to hnRNP would, as a consequence, influence pre-mRNA splicing, particularly since the hnRNP family of proteins has been shown to regulate pre-mRNA splicing. To confirm this, we tested the effect of PARG null mutation on the intron splicing in the hsrω (93D) gene, which encodes a 1.9 kb primary transcript (Hsrω-RC) and a 1.2 kb spliced transcript (Hsrω-RA) derived from Hsrω-RC (39). Using the qRT–PCR assay, the expression level of the Hsrω-RC transcript (unspliced) relative to the Hsrω-RA transcript (spliced) in the Parg mutant and the wild-type (yw) was quantified (Figure 8A). The result showed that the ratio of unspliced transcript (Hsrω-RC) to spliced transcript (Hsrω-RA) in the Parg mutant was 1.5 and 2.6 times greater than that of the wild-type counterpart when compared at the third-instar larvae stage and the pharate stage (Figure 8B, Supplementary Figures S4A and S4B), suggesting that PARG loss-of-function inhibited splicing of Hsrω-RC.
Figure 8.
Pre-mRNA splicing modulated by PARP and PARG in vivo. (A) The structures of Hsrω-RC (unspliced) and Hsrω-RA (spliced) transcripts in the hsrω (93D) locus. The primers specific for the intron sequence of Hsrω-RC (black arrow) were used for Hsrω-RC quantification. The forward primer spanning the exon 1 and exon 2 junctions, which are specific for Hsrω-RA, and the reverse primer in the exon 2 of Hsrω-RC and Hsrω-RA were used for Hsrω-RA quantification. (B) The ratios of Hsrω-RC/Hsrω-RA (unspliced to spliced transcript) in the indicated genotypes measured by real-time RT–PCR: 1. Parg (−/−), the third-instar larvae male; 2. yw, the third-instar larvae male (control); 3. Parg (+/−), the third-instar female; 4. Parg (−/−), pharate male, and 5. yw pharate male (control). (C) The structures of Ddc-RB (skipped isoform) and Ddc-RC (spliced isoform) transcripts in the Ddc gene. The positions of isoform-specific primers are indicated with arrows. CNS: central nervous system. (D) The ratios of Ddc-RC/Ddc-RB (spliced to skipped isoforms) in the indicated genotypes and treatment measured by real-time RT–PCR: 1. yw (the wild-type) as the control; 2. Squid−/− (sqd j6E3/sqd j6E3); 3. hrp38−/− (P[XP]d05172/Df(3R)Exel6209); 4. Parg−/− (Parg 27.1/Parg 27.1); 5. Parpe-EGFP(PARPe-EGFP; 69B-Gal4); 6. Parp−/− (C03256/C03256). All RNAs were extracted from the third-instar larvae. HS: 1-h heat shock of the third-instar larvae. The error bar represents the standard deviation from two independent experiments. *P ≤ 0.05; **P ≤ 0.01.
We also measured the expression level of the doublesex-male (dsx-M) transcript which is derived from alternative splicing of exon 5 in the dsx pre-mRNA (40) (Supplementary Figure S5). The results showed that the Parg mutant had only 47% and 21% of the dsx-M expression level in the control line at the third-instar larvae stage and the pharate stage, respectively, which suggests that PARG loss-of-function likely inhibits splicing exon 5 in the dsx pre-mRNA (Supplementary Figures S4A and S5).
In addition, we tested the effect of PARG and PARP on alternative splicing in the dopa decarboxylase (Ddc) gene. This gene encodes a 2.0 kb completely spliced isoform (Ddc-RC) expressed in the central nervous systems (CNS) and a 1.9 kb alternatively spliced isoform (Ddc-RB) after skipping the second exon, which is mainly expressed in the hypoderm (41) (Figure 8C). Consistent with previous findings (41), we found that the spliced transcript (Ddc-RC) relative to the skipped isoform (Ddc-RB), as expressed in the wild-type, increased about 2-fold after heat shock compared to that under the unstressed condition (Figure 8D). We therefore proposed that heat shock promotes splicing of exon B by displacing splicing repressors from the Ddc-pre-mRNA. To test whether hrp38 could be a splicing repressor, we characterized a hrp38 null mutation (P[XP]d05172) which has a P-element insertion in the encoding region of the hrp38 gene and fails to complement a hrp38 region deficiency (Df(3R)Exel6209). We observed that the expression level of the spliced isoform (Ddc-RC) relative to the skipped isoform (Ddc-RB) in both Squid homozygotes (Sqd−/−) and hrp38 hemizygote (Hrp38−/Df) increased around ten-fold compared to the wild-type under the unstressed condition, suggesting that hnRNPs are splicing repressors (Figure 8D and Supplementary Figure S6). Interestingly, both Parg mutant and Parp overexpression animals (PARPe-EGFP; 69B,GAL4/TM3) also had a very high expression level of the spliced isoform (Ddc-RC) relative to the skipped isoform (Ddc-RB) compared to the wild-type under the unstressed condition (Figure 8D and Supplementary Figure S6), suggesting that accumulation of pADPr enhances splicing of exon B, presumably by increased pADPr binding to the splicing repressors (Hrp38 and Squid). It was also observed that the enhanced splicing effect by heat shock was partially abolished in the PARP1 mutants (a 50% increase in the PARP1 mutant versus a two-fold increase in the wild-type) (Figure 8D and Supplementary Figure S6), suggesting that the PARP1 gene is required for the heat shock-induced splicing regulation. Taken together, we propose that pADPr binding to the splicing repressors (hrp38 and Squid) on the Ddc pre-mRNA results in the dissociation of the splicing repressors from the pre-mRNA, which thereby enhances exon B splicing.
DISCUSSION
Our results show that pADPr can bind with both Squid and hrp38 in a non-covalent way in vivo. We found that PARP1 is required for pADPr binding to Squid, and both PARP1 overexpression and PARG loss-of-function result in increased amounts of hnRNPs bound to pADPr. We also demonstrated that heat-shock treatment upregulates pADPr-bound Squid as a consequence of the increased pADPr level in the cells. Therefore, our findings demonstrate that the ability of pADPr binding to hnRNPs is subject to regulation by PARP and PARG at the genetic level, as well as physiological conditions in vivo, which ultimately determine pADPr synthesis and degradation rates in an organism. This is consistent with the observation that the binding affinity of pADPr to specific proteins depends on the pADPr chain length and branching complexity by in vitro studies (3,42).
Heat shock has a dramatic effect on the redistribution of hnRNPs in polytene chromosomes, including Squid (37) and Hrp38 (this report). Upon heat shock, hnRNPs dissociate from most of the transcripts and exclusively bind to one heat shock-induced transcript (hsrω-n) at the Hsrω locus, likely serving as the storage speckles (37). Here we propose a model which includes the following steps: (i) pADPr-PARP accumulated after heat shock binds with hnRNPs associated with the transcripts. (ii) This binding, in turn, results in the dissociation of hnRNPs from most of the transcripts as a result of either charge repulsion between pADPr and RNA or abolishment of RNA-binding ability of pADPr-bound hnRNPs. (iii) Then, a PARP-pADPr-hnRNPs complex moves into the nucleoplasm where PARG cleaves most of the pADPr. (iv) Finally, the free hnRNPs released from the PARP-pADPr complex bind with the non-coding RNA (hsrω-n) at the Hsrω locus. Consistent with our model, we observed that diminished amounts of Squid and hrp40:GFP in the Parg mutant are sequestered at the Hsrω (93D) locus upon heat shock. Thus, in the Parg mutant, it appears that PARG failed to cleave pADPr after heat shock so that only a minimum of free hnRNPs are available to bind to 93D.
It has been observed that hnRNPs can either promote or inhibit splicing by binding with exonic and intronic splicing enhancers (ESEs and ISEs) and silencers (ESSs and ISSs) (25,43,44). Since pADPr binding to hnRNPs causes the dissociation of hnRNPs from RNA, we have further proposed that splicing is either promoted or inhibited, depending on whether hnRNP binding takes place with splicing enhancers (ESEs and ISEs) or silencers (ESSs and ISSs) (Figure 9). In agreement with this hypothesis, we first observed that Parg null mutation partly inhibited splicing Hsrω-RC (Figure 8B) and splicing of the doublesex gene (Supplementary Figure S4). Our results also demonstrate that PARG loss-of-function and PARP overexpression can enhance splicing of the Ddc pre-mRNA, which is similar to the effect of heat shock on splicing of the Ddc pre-mRNA. It is well known that heat shock can inhibit splicing (45,46). However, the effect of heat shock on splicing of the Ddc pre-mRNA illustrates that heat shock also can also promote splicing (41). Our data strongly suggest that hrp38 and Squid are the splicing repressors for Ddc splicing since either hrp38 or Squid null mutation results in a very low expression of the skipped isoform which is produced by splicing inhibition. There are several G triplets or quartets in the intron 1 and intron 2 of the Ddc gene, which could be intronic splicing silencers (ISSs) for hrp38 and Squid binding (Supplementary Figure S8). We have observed that heat shock treatment results in increased amounts of hnRNPs bound to pADPr and the dissociation of hnRNPs from most of the transcripts. Based on this evidence, we proposed that heat shock induces the dissociation of the repressors (hrp38 or Squid) from the intronic splicing silencers in the Ddc pre-mRNA as a result of pADPr binding to hnRNPs and that this, in turn, enhances splicing of the Ddc pre-mRNA (Figure 9B). It has been reported that heat shock can induce dephosphorylation of a serine-arginine-rich splicing factor (SRp38) to inhibit splicing in human HeLa cells (47). Recently, it was also shown that pADPr can bind with human SR splicing factor (ASF/SF2) and further inhibit its phosphorylation (48). Therefore, pADPr binding to hnRNPs induced by heat shock could be an alternative to, or a mechanism combined with, the action of SRp38 dephosphorylation to regulate splicing upon heat shock.
Figure 9.
The model for the modulation of splicing by hnRNP poly(ADP-ribosyl)ation. (A) Splicing inhibition by poly(ADP-ribosyl)ation of hnRNPs. Hrp38 and Squid bind to the G triplets or quartets as exonic splicing enhancers (ESEs) in exon 2 of the Hsrω-RC transcript to promote splicing the intron (Supplementary Figure S7). However, the dissociation of hnRNPs from the transcript, such as Hsrω pre-RC, after poly(ADP-ribosyl)ation of hnRNPs, inhibits intron splicing. (B) Splicing enhancement by poly(ADP-ribosyl)ation of hnRNPs. In hypoderm tissues, the binding of Hrp38 and Squid to the G triplets or quartets as intronic splicing silencers (ISSs) in introns 1 and 2 of the Ddc pre-mRNA inhibits splicing and results in exon skipping. The dissociation of hnRNPs from the transcript, such as the Ddc pre-mRNA, by poly(ADP-ribosyl)ation of hnRNPs induced by heat shock or elevated PARP1 activity in CNS (13), enhances intron splicing.
In this report, we did not observe that Parp mutation has any effect on splicing at normal physiological condition in the two splicing events analyzed, Hsrω and Ddc genes. However, it has been reported that mammalian Parp-1 knockout cells have an increased incidence of alternative splicing within a subset of inflammatory response genes (48). Therefore, we suggest that the proposed model can be extended to explain tissue- or developmental-specific splicing under normal physiological conditions as a consequence of the spatial- and temporal-specific activity of PARP and PARG in the cell. For example, the maximal accumulation of pADPr was observed at the prepupal stage in the wild-type fly (49), and we also observed that both pADPr and Sqd accumulated at E74 and E75 loci (Figure 1C and D) where both genes have several alternative spliced isoforms. Therefore, it is reasonable to infer a dynamic scenario in which pADPr produced by activated PARP1 could transiently bind to hnRNP in puffs for the regulation of splicing at these sites. Further elucidation to confirm the developmental roles of hnRNP poly(ADP-ribosyl)ation on splicing will be undertaken in future investigation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (R01 GM077452) and the Ellison Medical Foundation (GM27875) [to A.V.T]. Funding for open access charge: National Institutes of Health (R01 GM077452)
Conflict of interest statement. None declared
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Dreyfuss for providing materials. Drs K. Zaret, R. Meyer and Mr D. Martin contributed valuable comments on the manuscript.
REFERENCES
- 1.D’Amours D, Desnoyers S, Silva D, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 2.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 3.Panzeter PL, Realini CA, Althaus FR. Noncovalent interactions of poly(adenosine diphosphate ribose) with histones. Biochemistry. 1992;31:1379–1385. doi: 10.1021/bi00120a014. [DOI] [PubMed] [Google Scholar]
- 4.Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl Acad. Sci. USA. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulikas T. Poly(ADP-ribosylated) histones in chromatin replication. J. Biol. Chem. 1990;265:14638–14647. [PubMed] [Google Scholar]
- 6.Pinnola A, Naumova N, Shah M, Tulin AV. Nucleosomal core histones mediate dynamic regulation of PARP1 protein binding to chromatin and induction of PARP1 enzymatic activity. J. Biol. Chem. 2007;282:32511–32519. doi: 10.1074/jbc.M705989200. [DOI] [PubMed] [Google Scholar]
- 7.Mendoza-Alvarez H, Alvarez-Gonzalez R. Regulation of p53 sequence-specific DNA-binding by covalent poly(ADP-ribosyl)ation. J. Biol. Chem. 2001;276:36425–36430. doi: 10.1074/jbc.M105215200. [DOI] [PubMed] [Google Scholar]
- 8.Malanga M, Pleschke JM, Kleczkowska HE, Althaus FR. Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions. J. Biol. Chem. 1998;273:11839–11843. doi: 10.1074/jbc.273.19.11839. [DOI] [PubMed] [Google Scholar]
- 9.Saxena A, Saffery R, Wong LH, Kalitsis P, Choo KH. Centromere proteins Cenpa, Cenpb, and Bub3 interact with poly(ADP-ribose) polymerase-1 protein and are poly(ADP-ribosyl)ated. J. Biol. Chem. 2002;277:26921–26926. doi: 10.1074/jbc.M200620200. [DOI] [PubMed] [Google Scholar]
- 10.Malanga M, Althaus FR. Poly(ADP-ribose) reactivates stalled DNA topoisomerase I and induces DNA strand break resealing. J. Biol. Chem. 2004;279:5244–5248. doi: 10.1074/jbc.C300437200. [DOI] [PubMed] [Google Scholar]
- 11.Reale A, Matteis GD, Galleazzi G, Zampieri M, Caiafa P. Modulation of DNMT1 activity by ADP-ribose polymers. Oncogene. 2005;24:13–19. doi: 10.1038/sj.onc.1208005. [DOI] [PubMed] [Google Scholar]
- 12.Haince JF, Kozlov S, Dawson VL, Dawson TM, Hendzel MJ, Lavin MF, Poirier GG. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J. Biol. Chem. 2007;282:16441–16453. doi: 10.1074/jbc.M608406200. [DOI] [PubMed] [Google Scholar]
- 13.Hanai S, Kanai M, Ohashi S, Okamoto K, Yamada M, Takahashi H, Miwa M. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2004;101:82–86. doi: 10.1073/pnas.2237114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tulin A, Naumova NM, Menon AK, Spradling AC. Drosophila poly(ADP-Ribose) glycohydrolase mediates chromatin structure and SIR2-dependent silencing. Genetics. 2006;172:363–371. doi: 10.1534/genetics.105.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tulin A, Spradling AC. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 16.Tulin A, Stewart D, Spradling AC. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 2002;16:2108–2119. doi: 10.1101/gad.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ménissier-de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Amé JC, Dierich A, LeMeur M, Sabatier L, Chambon P, de Murcia G. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, Nehls MC, Stöger T, Poirier GG, Dawson VL, Dawson TM. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl Acad. Sci. USA. 2004;101:17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haynes SR, Raychaudhuri G, Johnson D, Amero S, Beyer AL. The Drosophila Hrb loci: A family of hnRNA binding proteins. Mol. Biol. Rep. 1990;14:93–94. doi: 10.1007/BF00360430. [DOI] [PubMed] [Google Scholar]
- 20.Shen J, Zu K, Cass CL, Beyer AL, Hirsh J. Exon skipping by overexpression of a Drosophila heterogeneous nuclear ribonucleoprotein in vivo. Proc. Natl Acad. Sci. USA. 1995;92:1822–1825. doi: 10.1073/pnas.92.6.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norvell A, Kelley RL, Wehr K, Schupbach T. Specific isoforms of Squid, a Drosophila hnRNP, perform distinct roles in Gurken localization during oogenesis. Genes Dev. 1999;13:864–876. doi: 10.1101/gad.13.7.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammond L, Rudner DZ, Kanaar R, Rio RC. Mutations in the hrp48 gene, which encodes a Drosophila heterogeneous nuclear ribonucleoprotein particle protein, cause lethality and developmental defects and affect P-element third-intron splicing in vivo. Mol. Cell Biol. 1997;17:7260–7267. doi: 10.1128/mcb.17.12.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charroux B, Angelats C, Fasano L, Kerridge S, Vola C. The levels of the bancal product, a Drosophila homologue of vertebrate hnRNP K Protein, affect cell proliferation and apoptosis in imaginal disc cells. Mol. Cell Biol. 1999;19:7846–7856. doi: 10.1128/mcb.19.11.7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hovemann BT, Reim I, Werner S, Katz S, Saumweber H. The protein Hrb57A of Drosophila melanogaster closely related to hnRNP K from vertebrates is present at sites active in transcription and coprecipitates with four RNA-binding proteins. Gene. 2000;245:127–137. doi: 10.1016/s0378-1119(00)00027-5. [DOI] [PubMed] [Google Scholar]
- 25.Kiesler E, Hase ME, Brodin D, Visa N. Hrp59, an hnRNP M protein in Chironomus and Drosophila, binds to exonic splicing enhancers and is required for expression of a subset of mRNAs. J. Cell Biol. 2005;168:1013–1025. doi: 10.1083/jcb.200407173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hase ME, Yalamanchili P, Visa N. The Drosophila heterogeneous nuclear ribonucleoprotein M protein, HRP59, regulates alternative splicing and controls the production of its own mRNA. J. Biol. Chem. 2006;281:39135–39141. doi: 10.1074/jbc.M604235200. [DOI] [PubMed] [Google Scholar]
- 27.Zu K, Sikes ML, Haynes SR, Beyer AL. Altered levels of the Drosophila HRB87F/hrp36 hnRNP protein have limited effects on alternative splicing in vivo. Mol. Biol. Cell. 1996;7:1059–1073. doi: 10.1091/mbc.7.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc. Natl Acad. Sci. USA. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siebel CW, Kanaar R, Rio DC. Regulation of tissue-specific P-element pre-mRNA splicing requires the RNA-binding protein PSI. Genes Dev. 1994;8:1713–1725. doi: 10.1101/gad.8.14.1713. [DOI] [PubMed] [Google Scholar]
- 30.Goodrich JS, Clouse KN, Schupbach T. Hrb27C, Squid and Otu cooperatively regulate gurken RNA localization and mediate nurse cell chromosome dispersion in Drosophila oogenesis. Development. 2004;131:1949–1958. doi: 10.1242/dev.01078. [DOI] [PubMed] [Google Scholar]
- 31.Geng C, Macdonald PM. Imp associates with Squid and Hrp48 and contributes to localized expression of gurken in the oocyte. Mol. Cell Biol. 2006;26:9508–9516. doi: 10.1128/MCB.01136-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gagné JP, Hunter JM, Labrecque B, Chabot B, Poirier GG. A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose)-binding proteins. Biochem. J. 2003;371:331–340. doi: 10.1042/BJ20021675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matunis MJ, Matunis EL, Dreyfuss G. Isolation of hnRNP complexes from Drosophila melanogaster. J. Cell Biol. 1992;116:245–255. doi: 10.1083/jcb.116.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiríksdóttir E, Langel U, Eberwine J. Immunoprecipitation of mRNA-protein complexes. Nature Protocol. 2006;1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- 36.Morin X, Daneman R, Zavortink M, Chia M. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl Acad. Sci. USA. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prasanth K, Rajendra T, Lai AK, Lakhotia SC. Omega speckles – a novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J. Cell Sci. 2000;113:3485–3497. doi: 10.1242/jcs.113.19.3485. [DOI] [PubMed] [Google Scholar]
- 38.Expert-Bezançon A, Sureau A, Durosay P, Salesse R, Groeneveld H, Lecaer JP, Marie J. HnRNP A1 and the SR proteins ASF/SF2 and SC35 have antagonistic functions in splicing of {beta}-tropomyosin exon 6B. J. Biol. Chem. 2004;279:38249–38259. doi: 10.1074/jbc.M405377200. [DOI] [PubMed] [Google Scholar]
- 39.Garbe JC, Pardue ML. Heat shock locus 93D of Drosophila melanogaster: a spliced RNA most strongly conserved in the intron sequence. Proc. Natl Acad. Sci. USA. 1986;83:1812–1816. doi: 10.1073/pnas.83.6.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inoue K, Hoshijima K, Higuchi I, Sakamoto H, Shimura Y. Binding of the Drosophila transformer and transformer-2 proteins to the regulatory elements of doublesex primary transcript for sex-specific RNA processing. Proc. Natl Acad. Sci. USA. 1992;89:8092–8096. doi: 10.1073/pnas.89.17.8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen J, Beall CJ, Hirsh J. Tissue-specific alternative splicing of the Drosophila dopa decarboxylase gene is affected by heat shock. Mol. Cell Biol. 1993;13:4549–4555. doi: 10.1128/mcb.13.8.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fahrer J, Kranaster R, Altmeyer M, Marx A, Bürkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007;35:e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu J, Mayeda A, Krainer AR. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell. 2001;86:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]
- 44.Martinez-Contreras R, Fisette JF, Nasim FU, Madden R, Cordeau M, Chabot B. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biology. 2006;4:21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yost HJ, Lindquist S. RNA splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell. 1986;45:185–193. doi: 10.1016/0092-8674(86)90382-x. [DOI] [PubMed] [Google Scholar]
- 46.Bond U. Heat shock but not other stress inducers leads to the disruption of a subset of snRNPs and inhibition of in vitro splicing in HeLa cells. EMBO J. 1988;7:3509–3518. doi: 10.1002/j.1460-2075.1988.tb03227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin C, Feng Y, Manley JL. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature. 2004;427:553–558. doi: 10.1038/nature02288. [DOI] [PubMed] [Google Scholar]
- 48.Malanga M, Czubaty A, Girstun A, Staron K, Althaus FR. Poly(ADP-ribose) binds to the splicing factor ASF/SF2 and regulates its phosphorylation by DNA topoisomerase I. J. Biol. Chem. 2008;283:19991–19998. doi: 10.1074/jbc.M709495200. [DOI] [PubMed] [Google Scholar]
- 49.Kotova E, Jarnik M, Tulin AV. Poly (ADP-Ribose) polymerase 1 is required for protein localization to Cajal body. PLoS Genet. 2009;5:e1000387. doi: 10.1371/journal.pgen.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.