Abstract
Budding yeast telomerase is mainly activated by Tel1/Mec1 (yeast ATM/ATR) on Cdc13 from late S to G2 phase of the cell cycle. Here, we demonstrated that the telomerase-recruitment domain of Cdc13 is also phosphorylated by Cdk1 at the same cell cycle stage as the Tel1/Mec1-dependent regulation. Phosphor-specific gel analysis demonstrated that Cdk1 phosphorylates residues 308 and 336 of Cdc13. The residue T308 of Cdc13 is critical for efficient Mec1-mediated S306 phosphorylation in vitro. Phenotypic analysis in vivo revealed that the mutations in the Cdc13 S/TP motifs phosphorylated by Cdk1 caused cell cycle delay and telomere shortening and these phenotypes could be partially restored by the replacement with a negative charge residue. In the absence of Ku or Tel1, Cdk1-mediated phosphorylation of Cdc13 showed no effect on telomere length maintenance. Moreover, this Cdk1-mediated phosphorylation was required to promote the regular turnover of Cdc13. Together these results demonstrate that Cdk1 phosphorylates the telomerase recruitment domain of Cdc13, thereby preserves optimal function and expression level of Cdc13 for precise telomere replication and cell cycle progression.
INTRODUCTION
Telomeres are dynamic DNA–protein complexes that protect the ends of linear chromosomes, prevent detrimental chromosome rearrangements and defend against genomic instability and the associated risk of cancer (1–3). Telomeres, consisting of tandem repeats of short G-rich sequences, are synthesized by the enzyme telomerase (4,5). The catalytic core of telomerase is composed of a reverse transcriptase and an RNA subunit. The reverse transcriptase utilizes the RNA component as a template to add the G-rich repeats onto the 3′-ends of the chromosome (4–6). In most human somatic cells, telomerase activity is absent, and telomeres are gradually shortened with successive cell divisions due to incomplete replication, which eventually causes replicative senescence. Once telomeres become sufficiently short, they are thought to lose the ability to protect the ends of the chromosomes from being recognized as broken ends, and being subjected to nuclease digestion and active recombinational repair. Continuous telomere shortening in human fibroblasts leads to chromosome fusions, crisis and apoptosis (7). Very few human cells can bypass this crisis either through telomerase reactivation or through an alternative recombination pathway for telomere lengthening (8–10).
In budding yeast Saccharomyces cerevisiae, genes encoding components of telomerase have been identified and mutations in these genes cause a gradual loss of telomere length (11,12). EST2 and TLC1 encode the reverse transcriptase catalytic protein subunit and the templating RNA, respectively (12–14). In addition, the protein encoded by EST1 is associated with the RNA component of telomerase (15–18). Other accessory factors, such as Cdc13, are required for the in vivo action of telomerase. Cdc13 is a single strand telomere-binding protein (19,20). It forms a complex with Stn1 and Ten1. Cdc13, Stn1 and Ten1 show homology to Rfa1, Rfa2 and Rfa3, respectively. This replication protein A-like heterotrimeric complex specifically binds single strand telomeric sequences (21) and is required for both telomere protection and telomerase recruitment (19,20,22). It recruits telomerase to its site of action through an electrostatic interaction between Cdc13 and Est1 (22,23).
DNA replication must take place only once per cell cycle in eukaryotes. Cyclin-dependent kinases (CDKs), particularly the budding yeast Cdk1 (Cdc28), have been proven to phosphorylate and regulate a number of DNA replication factors, including components of the origin recognition complex, Orc1, Orc2 and Orc6, and MCM proteins (24,25). Previous studies also demonstrated that Cdk1 regulates telomere replication (26–28). cdk1 cells display defective telomere elongation and long single G strand tails (26,28). Furthermore, very recently, Cdk1 has been reported to activate Cdc13 (29).In eukaryotes, the maintenance of genome integrity relies on checkpoint to properly detect and repair DNA damage caused by environmental stresses or irregularities during DNA metabolisms. Damage and replication defects are recognized by the putative protein complex containing protein kinases such as Tel1 and Mec1. We previously reported that Cdc13 is a target of Tel1/Mec1-kinases (30). This regulation occurs from late S to G2 phase of the cell cycle and is required for telomerase recruitment (31–33). In addition, we also observed a TEL1/MEC1-independent phosphorylation of Cdc13 (30). We wondered how these phosphorylations mediate Cdc13 functions at telomere replication and which kinase contributes to the TEL1/MEC1-independent phosphorylation. In this work, we show that Cdc13 is phosphorylated in a cell cycle-dependent manner. Similar to the Tel1/Mec1-dependent regulation, Cdk1-mediated phosphorylation of Cdc13 occurs at the same cell cycle stage. We identified two phosphorylation sites. Inactivation of both Cdk1 phosphorylation sites in Cdc13 results in telomere shortening and delays the regular proteolysis of Cdc13 and cell cycle progression. Thus, we suggest that phosphorylation of Cdc13 by Cdk1 facilitates telomere replication.
MATERIALS AND METHODS
Strains, plasmids, yeast and telomere experiments
All the yeast operations were performed by standard methods (34). Yeast strains used in the study were derivatives of YPH501 (MATa/MATα ura3-52/ura3-52 lys2-801 amber/lys2-801 amber ade2-101 ochre/ade2-101 ochre trp1 Δ63/trp Δ63 his3 Δ200/his3 Δ200 leu2- Δ1/leu2- Δ1). The yeast strains carrying mec1 (all combined with sml1), tel1, cdc13, hdf1 and CDC13-Myc9 were described previously (15,35,36). cdc28-4 (37) and cdc28-as1 (38) strains were kindly provided by Steven Reed and David Morgan, respectively. pRS304cdc13-Myc9 was obtained from Virginia Zakian (36,39) and pRS306cdc13 was constructed by PCR containing the CDC13 open read frame and the downstream 200 nt. Point mutations were introduced into CDC13 using QuikChange site-directed mutagenesis (Stratagene). To generate chromosomal cdc13 mutants, pRS306cdc13 mutants were XhoI-digested and transformed into CDC13 strains, and the URA3 pop-out mutants were selected from the 5-FOA-resistant colonies using PCR analysis. pLD1(YIP, pGAL1-Sic1ΔN) (40) plasmid was kindly provided by John Diffley. pKB174(CEN CDC28-HA TRP1) and pKB173(CEN cdc28K40L-HA TRP1) (41) plasmids were generous gifts from Orna Cohen-Fix. All primer sequences for PCR and mutagenesis are available upon request. Telomere blot analysis was performed as previously described (42,43). Data shown are representatives of three or more experiments from independent colonies.
Immunoprecipitation, gel electrophoresis, western blot analysis and cell cycle analysis
To detect Cdc13, cells were grown in YPD broth. Whole cell proteins were extracted by TCA precipitation (44) and resolved by sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS–PAGE) or SDS–PAGE containing 10 μM of acrylamide-pendant Phos-tagTM (Phos-tag Consortium) (45) as previously described (46). Cdc13 was detected with a Myc antibody (Santa Cruz Biotechnology). A Rad53 antibody (gift of J. Diffley) was used to detect Rad53. Images were captured and quantified by a bioluminescence imaging system (UVP BioSpectrum AC Imagine System, UVP). Cells were stained with propidium iodide and the cell cycle analysis was performed by a flow cytometer (FACSCalibur 200, Becton Dickinson).
Inhibition of Cdk1 in cdc28-4 and cdc28-as1 strains
CDC13 in cdc28-4, W303 and cdc28-as1 strains was chromosomally tagged with Myc9. cdc28-4 arrests cells in G1, the stage that Cdc13 is usually under-phosphorylated, preventing us from distinguishing the lack of phosphorylation being directly due to the absence of Cdk1 from an indirect consequence of cell cycle arrest. To circumvent this problem, cdc28-4 cells were first arrested with nocodazole at 23°C and shifted to 37°C for 3 h. The cdc28-as1 cultures were split into halves for the 0.5 mM 4-amino-1-tert-butyl-3-(1′-naphthylmethyl) pyrazolo 3,4-d] pyrimidine (1-NMPP1) (47) and the mock treatments.
Phosphatase treatment of immune complexes of Cdc13
Agarose-bound Cdc13 immunocomplexes were washed twice with basic phosphatase (PPase) buffer (50 mM Tris–HCl, pH 7.9, 100 mM NaCl, 1 mM dithiothreitol, 10 mM MgCl2). The beads were then incubated with 100 U of calf intestinal PPase (New England Biolabs) for 30 min at 30°C. The reaction was stopped by addition of ice-cold basic kinase buffer containing 10 mM Na3VO4 and 10 mM NaF.
Recombinant protein purification
pGEX-4TCDC13(1–252) was constructed by ligating the PCR product containing amino acids 1–252 of Cdc13 from pRS314CDC13 into the NdeI- and EcoRI-treated pGEX-4T (GE). pGEX-4TCDC13(252–491) was constructed by ligating the EcoRI- and ClaI/Klenow-treated fragment, which contains amino acids 252–491 of Cdc13 from pRS314CDC13 into the EcoRI- and XhoI/Klenow-treated pGEX-4T. pGEX-4TCDC13(601–781) was constructed by ligating the BglII- and SacI/Klenow-treated fragment, which contains amino acids 601–781 of Cdc13 from pRS314CDC13 into the BamHI- and XhoI/Klenow-treated pGEX-4T. pGEX-4TCDC13(276–332) was constructed as previously described (30). Recombinant proteins were overexpressed in E. coli and purified as previously described (30). The bound GST-fusion proteins were detected by Coomassie blue staining.
Immunoprecipitation-kinase analysis
Log phase cells were lyzed and immunoprecipitated by incubation with an HA antibody and protein A beads. The immunoprecipitation (IP)-kinase assay was performed as described (48). Cdc28-HA or the kinase-dead Cdc28K40L-HA were purified from a protease-defected strain, BJ2168, carrying plasmid pKB174 (CDC28-HA) or pKB173 (cdc28K40L-HA). KSC1333 (MEC1-HA sml1::HIS3) (48) was kindly provided by Katsunori Sugimoto.
De novo telomere addition assay
De novo telomere addition assay was performed as previously described (49). UCC5706 strain was kindly provided by Daniel Gottschling. CDC13, cdc13-T308A, cdc13-T308D and cdc13-T308E were generated by one-step chromosomal insertion using XhoI-digested pRS304CDC13, pRS304cdc13-T308A, pRS304cdc13-T308D and pRS304cdc13-T308E, respectively.
RESULTS
Cdk1-dependent Cdc13 phosphorylation in vivo
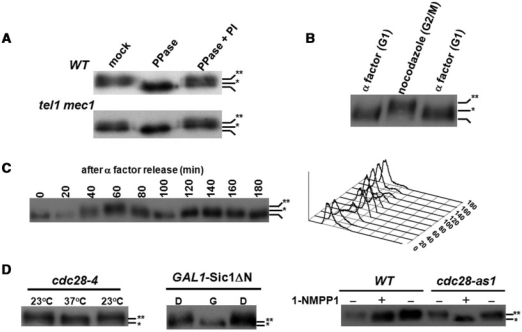
Under the examination of Tel1/Mec1-mediated phosphorylation, we observed a TEL1/MEC1-independent phosphorylation of Cdc13 (Figure 1A). In order to elucidate how these phosphorylations mediate other functions of Cdc13 and to identify the kinase that contributes to TEL1/MEC1-independent phosphorylation, we first investigated whether Cdc13 was phosphorylated in a cell cycle-dependent manner. Western blot analysis showed that Cdc13 is regulated in a cell cycle-dependent manner (Figure 1B) and hyperphosphorylated from late S (40 min in Figure 1C) to G2 (60 min in Figure 1C) stages. To determine whether Cdk1 is involved in this cell cycle-dependent phosphorylation, we examined Cdc13 phosphorylation in cdc28-4, SIC1 overexpressed and cdc28-as1 strains. All three strains can be manipulated to equip defective Cdk1 activity (37,38,40). The migration of Cdc13 in gels was faster in these Cdk1-crippling backgrounds (Figure 1D), suggesting that Cdc13 might be phosphorylated in a CDK1-dependent manner in vivo.
Figure 1.
Cdk1-dependent phosphorylation of Cdc13 in vivo. (A) MEC1/TEL1-independent phosphorylation of Cdc13. Cdc13 was chromosomally tagged with Myc9. Lysates from wild-type and mec1 tel1 strains were immunoprecipitated by a Myc antibody and incubated in PPase buffer, PPase buffer with PPase or PPase buffer containing PPase and PPase inhibitors (PI). Reactions were subjected to a 5.7% SDS–PAGE and transferred for western blot analysis using a Myc antibody. Non-, hypo- and hyper-phosphorylated Cdc13 are marked with none, single and double asterisks, respectively. (B) G2/M phase-specific phosphorylation of Cdc13. Lysates from α factor- or nocodazole-treated cells were extracted by TCA precipitation and Cdc13 phosphorylation was analyzed by western blot analysis. (C) Cell cycle-dependent phosphorylation of Cdc13. Overnight culture was grown to early log phase in YEPD, and arrested in G1 with α factor. When over 90% of the cells, as determined by microscopy, exhibited cell cycle arrest, cells were released into cell cycle. FACS analysis (showed at the right) of this sample indicated that cells had a 1N content of DNA. After release from α factor, cells were collected at 20-min intervals for 180 min and Cdc13 was analyzed by western blot analysis. (D) Cdk1-dependent phosphorylation of Cdc13. Cdc13 phosphorylation was analyzed in cdc28-4, SIC1 overexpressed and cdc28-as1 strains, which all can be manipulated to provide defective Cdk1 activities (37,38,40). cdc28-4 (a temperature sensitive mutant) was synchronized by nocodazole at 23°C and shifted to 37°C for 3 h; pGAL-Sic1 ΔN transformants were treated with dextrose (D) or galactose (G) for 3 h, and W303 (wild-type) and cdc28-as1 were treated with or without 1-NMPP1 for 3 h. Lysates were extracted and Cdc13 phosphorylation was analyzed by western blot analysis.
The telomerase recruitment domain of Cdc13 is phosphorylated by Cdk1 in vitro
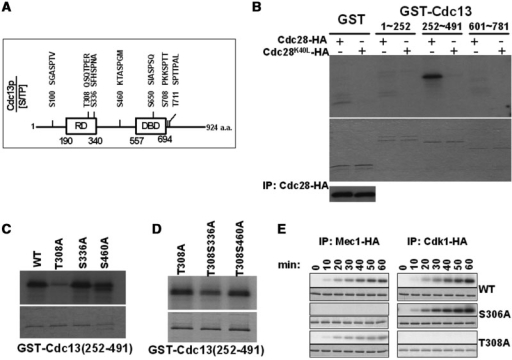
One apparent hypothesis for the CDK1-dependent Cdc13 phosphorylation is that Cdc13 may be directly phosphorylated by Cdk1. The CDK family of kinases has a preference for phosphorylation of serine in SP motifs or threonine in TP motifs (25). A total of seven such motifs were identified in Cdc13 (Figure 2A). Two of these sites in Cdc13 were found in the telomerase recruitment domain (T308 and S336) (Figure 2A). For the first step in determining whether Cdc13 is a direct substrate of Cdk1, we tested whether Cdk1 could phosphorylate Cdc13 in vitro. Cdk1 immunoprecipitates were incubated under in vitro kinase conditions in the presence of γ-[32P]-ATP and recombinant GST-Cdc13(1–252), GST-Cdc13(252–491) or GST-Cdc13(601–781) covering all seven Cdk1 consensus target sites. The reactions were subjected to SDS–PAGE and autoradiography (Figure 2B). The phosphorylated GST-Cdc13(252–491) subunit was observed, while neither GST alone nor other GST-Cdc13 proteins showed detectable phosphorylation by Cdk1 (Figure 2B). To confirm that Cdk1 kinase activity was indeed necessary for Cdc13 phosphorylation, assays were conducted using extracts isolated from a mutant strain expressing an HA-tagged Cdk1-kd protein with the amino acid lysine 40 mutated in its catalytic domain (41) (Figure 2B). Based on assays using equivalent amounts of protein, inactivation of Cdk1 kinase activity completely eliminated Cdc13 phosphorylation. These results demonstrated that Cdk1 kinase phosphorylates Cdc13 directly.
Figure 2.
Cdc13 is directly phosphorylated by Cdk1 in vitro. (A) Schematic diagram of Cdc13 illustrates its domain structure and potential S/T phosphorylation sites. The telomerase recruitment domain (RD) and DNA-binding domain (DBD) are indicated. (B) Cdk1-mediated phosphorylation of the telomerase recruitment domain of Cdc13 in vitro. Cdc28-HA and the kinase-dead Cdc28K40L-HA were purified by immunoprecipitation with an HA antibody. Immunoprecipitated kinases were analyzed by SDS–PAGE and western blotted with the HA antibody (shown at the bottom). GST-Cdc13(1–252), GST-Cdc13(252–491), GST-Cdc13(601–781) and GST alone were purified. IP-kinase assay was performed as described (30) with [γ-32P] ATP and phosphorylated proteins were detected by autoradiography (shown on top). The kinase reactions were also Coomassie stained (shown below) to confirm that lanes were equally loaded. (C) T308 is phosphorylated by Cdk1 in vitro. GST-Cdc13(252–491)-T308A, S336A and S460A were subjected to the IP-kinase assay as described in (B). (D) S336 is phosphorylated by Cdk1 in vitro. GST-Cdc13(252–491)-T308A, GST-Cdc13(252–491)-T308S336A and GST-Cdc13(252–491)-T308S460A were subjected to the IP-kinase assay as described in (B). (E) T308 is critical for the Mec1-mediated T306 phosphorylation in vitro. The IP-kinase assay was performed as described in (B) with equal amount of GST-Cdc13(276–332) wild-type, S306A and T308A using immunoprecipitated Cdc28-HA or Mec1-HA. Samples were collected at 10-min intervals for 60 min.
There are three S/TP sites (residue 308, 336 and 460) within the 225–491 fragment of Cdc13. To determine the exact phosphorylation sites, we purified mutant GST-Cdc13 containing single or double alanine (A) mutations at these sites. These substrates were then used in the IP-kinase assay. Compared to wild-type Cdc13, phosphorylation of the T308A Cdc13 subunit was drastically reduced (Figure 2C). These results imply that T308 is the major phosphorylation site of Cdc13. Double mutations at T308S336A, but not at T308S460A, caused a further reduction of phosphorylation, comparing to T308A alone (Figure 2D). Therefore, our data suggested that the phosphorylation sites of Cdc13 by Cdk1 are located at amino acids 308 and 336.
Since the Cdk1-mediated regulation on residue T308 is very close to a Mec1-mediated regulation on S306 of Cdc13 (30), we asked whether Cdk1 would modulate the action of Mec1 on Cdc13, or vice versa. As shown in Figure 2E, Cdk1 was able to phosphorylate Cdc13-S306A and Mec1 was also able to phosphorylate Cdc13-T308A. However, an attenuated Mec1-mediated phosphorylation was observed in Cdc13-T308A. On the other hand, the mutation on S306 made no major impact on the Cdk1-mediated modification on T308. These findings indicated that the Cdk1-mediated phosphorylation might be critical to promoting the action of Mec1 on Cdc13.
The telomerase recruitment domain of Cdc13 is phosphorylated by Cdk1 in vivo
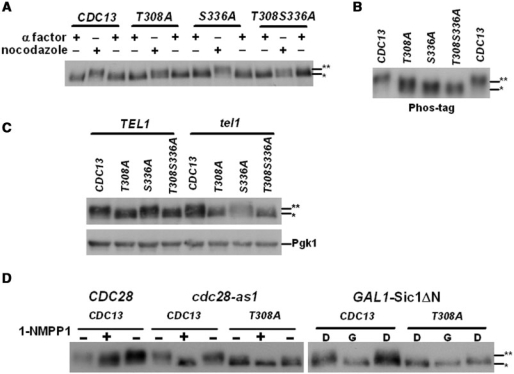
To verify whether residues 308 and 336 are indeed Cdk1-mediated phosphorylation sites in vivo, we created Cdc13 mutant strains at these sites. Compared to the wild-type strain, the cdc13-T308A, cdc13-S336A and cdc13-T308S336A mutants retained residual levels of phosphorylated Cdc13 isoforms (Figure 3A), which should be contributed from Tel1/Mec1-mediated phosphorylation. However, hyperphosphorylated forms of Cdc13 in cdc13-T308A and cdc13-T308S336A cells were reduced at the G2/M stage (Figure 3A), suggesting that T308 is phosphorylated in vivo. Due to the high molecular weight of Cdc13, the resolution of regular SDS–PAGE was not high enough to discern multiple phosphorylated forms of Cdc13. Therefore, a phosphor-specific gel using acrylamide-pendant Phos-tagTM (Phos-tag Consortium) (45) was applied to augment the difference of phosphorylation. Phosphorylation on T308 and S336 became more distinguishable in the Phos-tag gel (Figure 3B). Under the help of Phos-tag, increases of migration were detected in both T308A and S336 A (Figure 3B). These data indicated that both T308 and S336 are phosphorylated in vivo.
Figure 3.
Cdc13 is phosphorylated by Cdk1 in vivo. (A) Cdc13 is phosphorylated at the G2/M phase. CDC13 wild-type, T308A, S336A and T308S336A strains were synchronized with α factor or nocodazole. Lysates were extracted and the phosphorylated Cdc13 was analyzed by western blot analysis. Hypo- and hyper-phosphorylated Cdc13 are marked with single and double asterisks, respectively. (B) T308 and S336 were phosphorylated in vivo. Lysates of wild-type and Cdc13 mutants (T308A, S336A and T308S336A) were extracted from nocodazole synchronized cells. Phosphorylated Cdc13 was separated in a SDS–PAGE containing 10 μM of Phos-tag and analyzed by western blot analysis. (C) Cdc13 phosphorylation at T308 and S336 were Tel1-independent. Lysates of wild-type and Cdc13 mutants (T308A, S336A, T308S336A) were extracted from nocodazole synchronized TEL1 and tel1 strains. Phosphorylated Cdc13 was analyzed by western blot analysis. Pgk1 was served as a loading control. (D) Phosphorylation of Cdc13 at T308 is Cdk1-dependent. Wild-type and cdc28-as1 strains were subjected to 1-NMPP1 treatment and GAL1-Sic1 ΔN expressed cells were treated with dextrose (D) or galactose (G). Phosphorylation of Cdc13 was analyzed by western blot analysis.
Since previous studies demonstrated that Tel1 makes the major contribution to the activation of Cdc13 to recruit telomerase (31–33), to understand if the phosphorylation of T308 and S336 were Tel1-dependent, these mutants were introduced into the tel1 cells. As shown in Figure 3C, even in the absence of TEL1, reduction of the phosphorylation level was further observed in T308A mutation, comparing to the wild-type Cdc13. These findings argued that Cdk1-mediated phosphorylation does not rely on the action of Tel1-mediated phosphorylation on Cdc13.
We next examined whether T308 is authentically the Cdk1-phosphorylated site in vivo, T308A mutation was constructed in the cdc28-as1 strains. Compared to the normal cdc28-as1 strain, T308A mutation did not further convert the electrophoretic behavior of Cdc13 to a single sharper band with greater mobility (Figure 3D). Similar results were also observed in the Sic1-overexpressed strain (Figure 3D). Therefore, we concluded that Cdc13 is phosphorylated by Cdk1 in vivo at T308.
Cdk1 phosphorylates Cdc13 at the late S and G2 phases for proper cell cycle progression
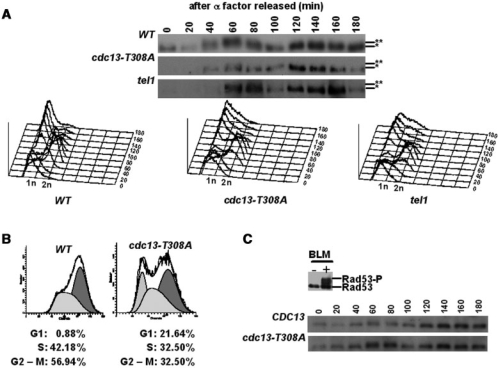
Tel1 is highly enriched at short telomeres from S through G2 phase (31–33). To determine whether Cdk1 targets Cdc13 at a similar timing to the action of Tel1, the status of Cdc13 phosphorylation was followed throughout the cell cycle. As observed in Figure 4, Cdc13 hyperphosphorylation was peaked at the late S and G2 phases, which is identical to the timing of the action of Tel1 as previously reported (31–33). This peak was diminished in the cdc13-T308A strain, further indicating that Cdk1 plays a critical role on the hyperphosphorylated peak from late S to G2 phase observed in the wild-type cells (Figure 4A). Following release from the arrest, a slight cell cycle delay was observed in the cdc13-T308A strain (Figure 4B, 40 min). In the absence of TEL1, a hyperphosphorylated Cdc13 peak at the late S and G2 phases was still observed (Figure 4A). This abnormal cell cycle progression was not due to activation of the checkpoint (Figure 4C). These results suggested that Cdk1-mediated phosphorylation of Cdc13 shares the same cell cycle stage with the Tel1/Mec1-mediated telomerase recruitment. However, this phosphorylation is Tel1-independent and required for accurate cell cycle progression.
Figure 4.
Cdk1 and Tel1 phosphorylate Cdc13 at late S and G2 phases. (A) After α factor arrest and release, lysates from wild-type, cdc13-T308A and tel1 cells were collected at 20-min intervals for 180 min and Cdc13 was analyzed by western blot analysis. Data of FACS analysis are shown at the bottom. Hypo- and hyper-phosphorylated Cdc13 are marked with single and double asterisks, respectively. (B) The FACS data of wild-type and cdc13-T308A at 40 min are shown. Percentages of cell cycle stages were quantified according to the manufactures instructions (Becton Dickinson). (C) Lack of checkpoint activation in cdc13-T308A cells. Lysates of wild-type and cdc13-T308A cells were collected and Rad53 phosphorylation was analyzed by western blot analysis using a Rad53 antibody. Bleomycin treatment (5 mU/ml, 3 h) was used as a positive control.
Telomere analysis of cdc13 mutants
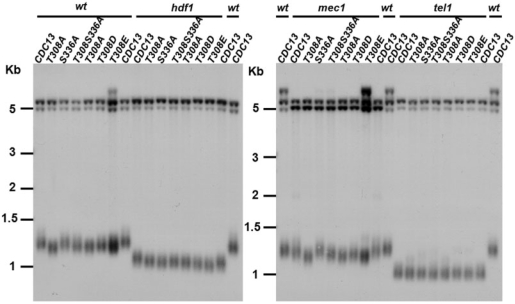
To identify potential cellular functions of Cdk1-mediated Cdc13 phosphorylation, we first analyzed whether mutations in these phosphorylation sites would affect the growth and replication of telomeres. In order to examine the phenotypes, point mutations were introduced into the untagged chromosomal copy of CDC13. As shown in Figure 5, single mutation at residue T308 to A caused telomere shortening. A slight growth defect was also observed in T308A cells, which was further reflected in its cell cycle profile (Figure 4). These findings indicated that phosphorylation on this residue is important for telomere maintenance. Moreover, single mutation S336A or double mutation T308S336A maintained similar telomere lengths to those of wild-type and T308A, respectively (Figure 5). These data indicated that the Cdk1-mediated regulation on S336 does not contribute to the telomere maintenance. While single mutation at residue 308 from T to A caused telomere shortening and a cell cycle delay, replacement of T308 with aspartic acid (D) or glutamic acid (E) partially recovered these phenotypes (Figures 5 and S1, and data not shown). These results further confirmed that the negative charge on this residue is important for telomere maintenance.
Figure 5.
Telomere analysis of cdc13 mutants. Cdk1 phosphorylation sites of Cdc13 were mutated from T or S to A, D or E, and mutations were created in wild-type, hdf1, mec1 and tel1 strains. DNA from each strain was digested with XhoI, separated in a 1% agarose gel, transferred to a nylon membrane and hybridized with a Y′ probe. Cells used in this experiment had pop-out the YIP cdc13 URA3 plasmid for at least 50 generations.
We next examined whether predisposed telomere damage would exacerbate the Cdk1-mediated defect on Cdc13. Cdc13 and Ku are both essential for telomere integrity. The Ku complex is telomere associated and helps to protect telomeres from degradation and from inappropriate recombination (50,51). Deletion of Ku causes telomere shortening. We measured the effect of T308A and S336A mutations on telomere length maintenance in the Ku mutant background. Mutations of T308A and S336A in the Ku mutant cells did not lead to further shortening or senescence phenotype (Figure 5 and data not shown). These results indicated that Cdk1-mediated Cdc13 phosphorylation no longer plays a role in modulating telomere homeostasis in the absence of Ku.
We then asked if Cdk1-mediated Cdc13 phosphorylation plays a synergistic effect with Tel1- or Mec1-mediated regulation on telomere lengths. Mutations of CDC13 were created in the tel1 and mec1 cells. Deletion of MEC1 leaded to mild telomere shortening, while deletion of TEL1 caused severe telomere shortening. A further reduction on telomere lengths was observed in mec1 cdc13-T308A and mec1 cdc13-T308S336A cells, but not in tel1 cdc13-T308A and tel1 cdc13-T308S336A cells (Figure 5). These results suggested that in the absence of Tel1-dependent phosphorylation on Cdc13 (and perhaps on other proteins), Cdk1-mediated phosphorylation of Cdc13 has no measurable effect on telomere length homeostasis.
The stability of Cdc13-T308A is increased
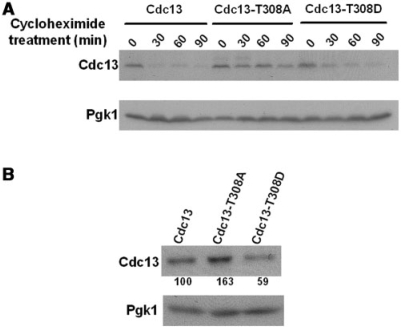
Many studies have shown that CDK promotes protein degradation, especially on cell cycle regulatory factors. Rapid turnover of Cln2 at the S phase is achieved by Cdk1-dependent phosphorylation (52). Proteolysis of the anaphase-promoting complex early mitotic inhibitor, Emi1, is induced by Cdk1 (53). In addition, TOP2-inhibition leads to Cdk1-dependent degradation of an ORC1/2 reservoir at mitosis (54). To examine whether Cdk1-mediated phosphorylation of Cdc13 also modulates the proteolysis of Cdc13, the relative Cdc13 stabilities in wild-type and mutant strains were measured. Wild-type, cdc13-T308A and cdc13-T308D strains were grown to log phase, and cycloheximide was added to turn off the production of Cdc13. The treatment was maintained for 90 min, and the Cdc13 protein levels were evaluated. As shown in Figure 6A, the half-life of Cdc13-T308A was extensively expanded, comparing to that of wild-type Cdc13. Furthermore, replacement of T308 with D could revert this delay of degradation. Since Cdc13 is phosphorylated by Cdk1 at late S/G2 phase, we also investigated the steady-state levels of Cdc13 after the action of Cdk1. Increased level of Cdc13-T308A was observed in G2/M-arrested cells (Figure 6B). These findings highlighted that, while Cdc13 protein is maintained in a cell cycle-dependent manner, Cdk1-mediated phosphorylation facilitates the degradation of Cdc13.
Figure 6.
Cdk1-mediated phosphorylation regulates the stability of Cdc13. Western blot analysis of the stability of Cdc13 was performed. (A) Wild-type, cdc13-T308A and cdc13-T308D strains were treated with cycloheximide. After cycloheximide treatment, cells were collected at 30-min intervals for 90 min. Lysates were TCA-precipitated, subjected to a 10% SDS–PAGE and transferred for western blot analysis using a Myc antibody. Pgk1 was served as a loading control. (B) Strains were arrested by nocodazole. Western blot analysis was performed as described in (A). The percentages of the expression levels of Cdc13 compared to that of the wild-type are shown below.
Cdk1-mediated phosphorylation of Cdc13 is not essential for the de novo telomere addition
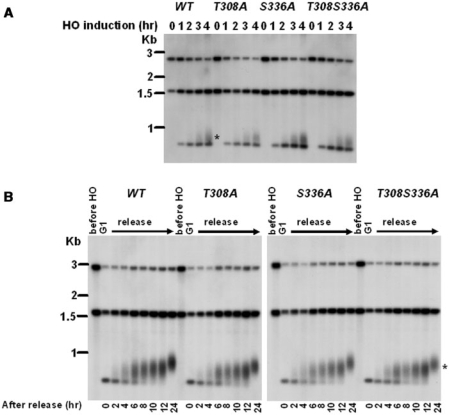
Lastly, we wish to address whether Cdk1-mediated Cdc13 phosphorylations are essential for de novo telomere addition. We tested the sudden telomere elongation by the de novo telomere addition assay developed by Diede and Gottschling (49). Strains were arrested at M phase by nocodazole treatment, and then subjected to induction of HO expression. Compared to wild-type cells, which showed telomere elongations at the HO cut site, the TG1–3/HO end was also elongated in T308A, S336A and T308AS336A cells (Figure 7A). We next investigated whether some partial defect in de novo telomere addition could be detected in these mutants. After cells were released into the cell cycle from α factor, both the control and the experimental strains proceeded with indistinguishable kinetics of telomere addition through the subsequent cell cycle (Figure 7B). These results indicated that phosphorylation of Cdc13 at amino acids 308 and 336 are not essential for de novo telomere addition.
Figure 7.
De novo telomere addition in cdc13 mutants. (A) cdc13 mutants were introduced into UCC5706. Strains were arrested by nocodazole. Galactose was added to arrested culture to induce the expression of HO endonuclease. DNA was isolated after 0, 1, 2, 3 and 4 h and digested with SpeI. Southern blot analysis was performed with a probe that recognizes sequence distal to the HO recognition site. De novo telomere addition is marked with an asterisk. (B) Strains were arrested in G1 by α factor, galactose was added to induce HO expression for 2 h, and cells were resuspended in prewarmed YEPD to release. Samples were taken at different time points for 24 h for analysis as described in (A).
DISCUSSION
Cdc13 recruits telomerase to its site of action through an electrostatic interaction with a telomerase component Est1 (22,23,55). We previously showed that Tel1 and Mec1 phosphorylate Cdc13 and govern the telomerase recruitment function of Cdc13 (30). Consistent with the recent finding that Cdk1 regulates Cdc13 (29), in this article we additionally demonstrated that Cdk1 phosphorylates Cdc13 at the telomerase recruitment domain. While it currently lacks structural information between Cdc13 and Est1 in the recruitment interface, we speculate that the hypernegatively charged domain of Cdc13 contributed by Tel1, Mec1 and Cdk1 may provide an optimal interface to recruit the potential positively charged domain near the amino acid 444 lysine residue of Est1 (56). This hypernegatively charged domain of Cdc13 forms at the late S and lasts to G2 phase, right at the timing for telomerase to function on telomeres.
An intriguing finding of this study is that Cdk1-mediated phosphorylation is required for, in addition to optimal telomerase recruitment, the proper turnover of Cdc13. While the protein level of Cdc13 is well regulated throughout the cell cycle, which peaks at late S/G2 phase and drops at the G1 phase, we found that phosphorylation at T308 is critical for the rapid degradation of Cdc13. What kind of disturbances would be caused by underdegraded Cdc13 existing in the nucleus after the G2 phase of the cell cycle is still unclear. Further identification of the proteolysis pathway of Cdc13 will be important to study this effect by uncoupling the function of tuning the protein level of Cdc13 from the function of telomerase recruitment of Cdc13 contributed by Cdk1.
Phosphoproteomics has been used to profile thousands of phosphorylation in cells. Using mass spectrometry to analyze phosphorylation sites in wild-type S. cerevisiae, both S306 and T308 of Cdc13 were found to be phosphorylated in vivo (57). In our previous study, mutations at the Tel1/Mec1 phosphorylation sites did not lead to an alteration of protein levels of Cdc13 (30). Interestingly, the T308 residue is very close to a Mec1-regulated site, S306, and T308 is critical for the full activity of Mec1 on S306, at least in our in vitro assay (Figure 2E). These results indicate that a conformational change at T308 might modulate Mec1-mediated Cdc13 phosphorylation. However, the Cdk1-mediated telomere shortening observed in the T308 strain was abolished in the absence of Tel1. Altogether these data suggest that Tel1 makes a greater impact, while Mec1 and Cdk1 kinases play supporting roles, to optimize the reaction of telomerase recruitment.
Previous studies showed that Cdk1 mutation could produce long single strand G-tails at telomeres (26,28), which was not observed in our T308S336A cells (data not shown). It remains possible, in addition to Cdc13, Cdk1 may target other proteins during telomere replication. In that case, the telomeric effect of Cdk1 on Cdc13 (26,28) should be masked in the cdc28-4 cells, that is, the cdc28-4 mutation causes a greater telomeric alteration than cdc13-T308A. Further studies to illustrate other targets of Cdk1 in telomere replication will be required in order to address this possibility.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
FUNDING
National Science Council (NRPGM-96-3112-B-002-023); National Health Research Institute (NHRI-EX98-9727BI to S.-C.T.). Funding for open access charge: National Health Research Institute of Taiwan.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Virginia Zakian and Huilin Zhou for the discussion and sharing of their unpublished data. We also thank Drs Steven Reed, David Morgan, Daniel Gottschling, Virginia Zakian, John Diffley, Orna Cohen-Fix and Katsunori Sugimoto for providing plasmids and strains.
REFERENCES
- 1.Zakian VA. Telomere functions: lessons from yeast. Trends Cell Biol. 1996;6:29–33. doi: 10.1016/0962-8924(96)81035-x. [DOI] [PubMed] [Google Scholar]
- 2.McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annu. Rev. Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 3.Shay JW, Zou Y, Hiyama E, Wright WE. Telomerase and cancer. Hum. Mol. Genet. 2001;10:677–685. doi: 10.1093/hmg/10.7.677. [DOI] [PubMed] [Google Scholar]
- 4.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 5.de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 6.Vega LR, Mateyak MK, Zakian VA. Getting to the end: telomerase access in yeast and humans. Nat. Rev. Mol. Cell Biol. 2003;4:948–959. doi: 10.1038/nrm1256. [DOI] [PubMed] [Google Scholar]
- 7.Artandi SE, DePinho RA. A critical role for telomeres in suppressing and facilitating carcinogenesis. Curr. Opin. Genet. Dev. 2000;10:39–46. doi: 10.1016/s0959-437x(99)00047-7. [DOI] [PubMed] [Google Scholar]
- 8.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 9.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat. Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 10.Reddel RR, Bryan TM, Colgin LM, Perrem KT, Yeager TR. Alternative lengthening of telomeres in human cells. Radiat. Res. 2001;155:194–200. doi: 10.1667/0033-7587(2001)155[0194:alotih]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 11.Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 13.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 14.Counter CM, Meyerson M, Eaton EN, Weinberg RA. The catalytic subunit of yeast telomerase. Proc. Natl Acad. Sci. USA. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin JJ, Zakian VA. An in vitro assay for Saccharomyces telomerase requires EST1. Cell. 1995;81:1127–1135. doi: 10.1016/s0092-8674(05)80017-0. [DOI] [PubMed] [Google Scholar]
- 16.Steiner BR, Hidaka K, Futcher B. Association of the Est1 protein with telomerase activity in yeast. Proc. Natl Acad. Sci. USA. 1996;93:2817–2821. doi: 10.1073/pnas.93.7.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Virta-Pearlman V, Morris DK, Lundblad V. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]
- 18.Zhou J, Hidaka K, Futcher B. The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol. Cell Biol. 2000;20:1947–1955. doi: 10.1128/mcb.20.6.1947-1955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 20.Lin JJ, Zakian VA. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl Acad. Sci. USA. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 22.Chandra A, Hughes TR, Nugent CI, Lundblad V. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 2001;15:404–414. doi: 10.1101/gad.861001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nugent CI, Lundblad V. The telomerase reverse transcriptase: components and regulation. Genes Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- 24.Henneke G, Koundrioukoff S, Hubscher U. Multiple roles for kinases in DNA replication. EMBO Rep. 2003;4:252–256. doi: 10.1038/sj.embor.embor774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 26.Frank CJ, Hyde M, Greider CW. Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol. Cell. 2006;24:423–432. doi: 10.1016/j.molcel.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 27.Teixeira MT, Gilson E. When CDK1 rides the telomere cycle. Mol. Cell. 2006;24:491–492. doi: 10.1016/j.molcel.2006.10.033. [DOI] [PubMed] [Google Scholar]
- 28.Vodenicharov MD, Wellinger RJ. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol. Cell. 2006;24:127–137. doi: 10.1016/j.molcel.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 29.Li S, Makovets S, Matsuguchi T, Blethrow J, Shah K, Blackburn EH. Cdk1-Dependent Phosphorylation of Cdc13 Coordinates Telomere Elongation during Cell-Cycle Progression. Cell. 2009;136:50–61. doi: 10.1016/j.cell.2008.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tseng SF, Lin JJ, Teng SC. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 2006;34:6327–6336. doi: 10.1093/nar/gkl786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arneric M, Lingner J. Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep. 2007;8:1080–1085. doi: 10.1038/sj.embor.7401082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bianchi A, Shore D. Increased association of telomerase with short telomeres in yeast. Genes Dev. 2007;21:1726–1730. doi: 10.1101/gad.438907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabourin M, Tuzon CT, Zakian VA. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol. Cell. 2007;27:550–561. doi: 10.1016/j.molcel.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1990. [Google Scholar]
- 35.Tsai YL, Tseng SF, Chang SH, Lin CC, Teng SC. Involvement of replicative polymerases, Tel1p, Mec1p, Cdc13p, and the Ku complex in telomere-telomere recombination. Mol. Cell Biol. 2002;22:5679–5687. doi: 10.1128/MCB.22.16.5679-5687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukamoto Y, Taggart AK, Zakian VA. The role of the Mre11-Rad50-Xrs2 complex in telomerase- mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 2001;11:1328–1335. doi: 10.1016/s0960-9822(01)00372-4. [DOI] [PubMed] [Google Scholar]
- 37.Lorincz AT, Reed SI. Sequence analysis of temperature-sensitive mutations in the Saccharomyces cerevisiae gene CDC28. Mol. Cell Biol. 1986;6:4099–4103. doi: 10.1128/mcb.6.11.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- 39.Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- 40.Noton E, Diffley JF. CDK inactivation is the only essential function of the APC/C and the mitotic exit network proteins for origin resetting during mitosis. Mol. Cell. 2000;5:85–95. doi: 10.1016/s1097-2765(00)80405-0. [DOI] [PubMed] [Google Scholar]
- 41.Agarwal R, Cohen-Fix O. Phosphorylation of the mitotic regulator Pds1/securin by Cdc28 is required for efficient nuclear localization of Esp1/separase. Genes Dev. 2002;16:1371–1382. doi: 10.1101/gad.971402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teng SC, Chang J, McCowan B, Zakian VA. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell. 2000;6:947–952. doi: 10.1016/s1097-2765(05)00094-8. [DOI] [PubMed] [Google Scholar]
- 43.Teng SC, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heinrich JN, Kwak SP, Howland DS, Chen J, Sturner S, Sullivan K, Lipinski K, Cheng KY, She Y, Lo F, et al. Disruption of ShcA signaling halts cell proliferation—characterization of ShcC residues that influence signaling pathways using yeast. Cell Signal. 2006;18:795–806. doi: 10.1016/j.cellsig.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 45.Kinoshita-Kikuta E, Aoki Y, Kinoshita E, Koike T. Label-free kinase profiling using phosphate affinity polyacrylamide gel electrophoresis. Mol. Cell Proteomics. 2007;6:356–366. doi: 10.1074/mcp.T600044-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Tseng SF, Chang CY, Wu KJ, Teng SC. Importin KPNA2 is required for proper nuclear localization and multiple functions of NBS1. J. Biol. Chem. 2005;280:39594–39600. doi: 10.1074/jbc.M508425200. [DOI] [PubMed] [Google Scholar]
- 47.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 48.Naiki T, Wakayama T, Nakada D, Matsumoto K, Sugimoto K. Association of Rad9 with double-strand breaks through a Mec1-dependent mechanism. Mol. Cell Biol. 2004;24:3277–3285. doi: 10.1128/MCB.24.8.3277-3285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diede SJ, Gottschling DE. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- 50.Gravel S, Larrivee M, Labrecque P, Wellinger RJ. Yeast Ku as a regulator of chromosomal DNA end structure. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- 51.Polotnianka RM, Li J, Lustig AJ. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 1998;8:831–834. doi: 10.1016/s0960-9822(98)70325-2. [DOI] [PubMed] [Google Scholar]
- 52.Lanker S, Valdivieso MH, Wittenberg C. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- 53.Margottin-Goguet F, Hsu JY, Loktev A, Hsieh HM, Reimann JD, Jackson PK. Prophase destruction of Emi1 by the SCF(betaTrCP/Slimb) ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev. Cell. 2003;4:813–826. doi: 10.1016/s1534-5807(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 54.Cuvier O, Stanojcic S, Lemaitre JM, Mechali M. A topoisomerase II-dependent mechanism for resetting replicons at the S-M-phase transition. Genes Dev. 2008;22:860–865. doi: 10.1101/gad.445108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bianchi A, Negrini S, Shore D. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol. Cell. 2004;16:139–146. doi: 10.1016/j.molcel.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 56.Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 57.Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl Acad. Sci. USA. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.