Abstract
Human lysosomal enzymes acid-β-glucosidase (GCase) and acid-α-galactosidase (α-Gal A) hydrolyze, respectively, the sphingolipids glucosyl- and globotriaosyl- ceramide, and mutations in these enzymes lead to the lipid metabolism disorders Gaucher and Fabry disease. We have investigated the structure and stability of GCase and α-Gal A at the neutral-pH environment reflective of the endoplasmic reticulum and the acidic-pH environment reflective of the lysosome. These details are important for the development of pharmacological chaperone therapy for Gaucher and Fabry disease, in which small molecules bind mutant enzymes in the ER to enable the mutant enzyme to meet quality control requirements for lysosomal trafficking. We report crystal structures of apo GCase at pH 4.5, pH 5.5, and in complex with the pharmacological chaperone isofagomine (IFG) at pH 7.5. We also present thermostability analysis of GCase at pH 7.4 and pH 5.2 using differential scanning calorimetry. We compare our results with analogous experiments using α-Gal A and the chaperone 1-deoxygalactonijirimycin (DGJ), including the first structure of α-Gal A with DGJ. Both GCase and α-Gal A are more stable at lysosomal pH with and without their respective iminosugars bound, and notably, the GCase/IFG complex stability is pH sensitive. We show that the conformations of the active site loops in GCase are sensitive to ligand binding but not pH, whereas analogous galactose- or DGJ- dependent conformational changes in α-Gal A are not seen. Thermodynamic parameters obtained from α-Gal A unfolding indicate two-state, van't-Hoff unfolding in the absence of the iminosugar at neutral and lysosomal pH, and non two-state unfolding in the presence of DGJ. Taken together, these results provide insight into how GCase and α-Gal A are thermodynamically stabilized by iminosugars, and suggest strategies for the development of new pharmacological chaperones for lysosomal storage disorders.
Glycoside hydrolases are involved in turnover of intracellular substrates in the acidic environment (pH ∼5.2 (1)) of the lysosome. These enzymes are synthesized and folded in the neutral pH environment of the endoplasmic reticulum (ER), exported to the Golgi apparatus for sorting, and subsequently trafficked to lysosomes. Inherited genetic mutations in specific glycoside hydrolases result in enzyme deficiency in the lysosome and the family of diseases called lysosomal storage disorders, which include Anderson-Fabry, Gaucher, Tay-Sachs, and Sandhoff diseases (2, 3). Many of the mutations associated with these diseases are missense mutations that result in single amino acid substitutions remote from the active site of these enzymes, however. Such mutant enzymes retain at least partial catalytic activity in vitro (4, 5) but exhibit impaired cellular trafficking (6-8). Only correctly folded proteins in the ER are efficiently transported to the Golgi apparatus and then to their intended cellular destinations. Aberrantly-folded molecules are recognized by the ER quality control system and are either retained in this compartment, sent out of the ER for proteolytic degradation via the proteasome, or a combination of the above (9, 10). Thus, pathophysiology in lysosomal storage disorders is not linked directly to abolished intrinsic enzyme activity, but first to a defect in protein stability, then to a defect in cellular trafficking, and finally to reduced lysosomal hydrolase activity (6-8). The mutant protein accumulates either in the ER, the Golgi, or endocytic vesicles, is then targeted for degradation, and a reduced level of active enzyme reaches the lysosome (7).
The current standard of care for individuals with lysosomal storage diseases is enzyme replacement therapy (ERT) (11, 12), in which an intravenous infusion of recombinant wild type human enzyme is internalized by affected cells via cell surface receptors and delivered to lysosomes to break down accumulated substrate. Another FDA-approved treatment approach for certain types of Gaucher Disease, called substrate reduction therapy (SRT), utilizes a small molecule inhibitor of glycolipid synthesis to reduce substrate production (13, 14). These treatments address substrate accumulation but do not address the protein folding defects and their potential contributions such as release of pro-inflammatory cytokines (15) that may play a role in the pathophysiology of these diseases.
Pharmacological chaperones are a promising new treatment strategy for diseases of protein misfolding and mistrafficking, and compounds are currently being evaluated for lyososomal storage disorders (LSDs) (3, 16-19), among others (20). In this approach, a tailored small molecule inhibitor of a target lysosomal enzyme binds to the protein during initial biosynthesis and folding in the ER. Different competitive inhibitors are in development as candidate chaperones for various LSDs (7, 21-33). Although somewhat counterintuitive, enzyme inhibitors can increase steady-state lysosomal levels of active enzymes by enabling the mutant protein to meet quality control standards in the ER and restore trafficking (34).
The mechanism by which pharmacological chaperones function is not known but two scenarios can be envisioned. The structure of a folded mutant enzyme that retains partial catalytic activity is likely to be very similar to the native, wild-type structure. Thus, pharmacological chaperones might bind to a fully folded mutant protein target to stabilize a native-like conformation. Alternatively, mutations may affect the folding pathway by stabilizing a non-native, less stable intermediate state. In this case, the pharmacological chaperone may bind to a non-native conformation and accelerate the transition to the native-like state. Once in the final subcellular destination and in the presence of substrate, the inhibitor dissociates from the mutant enzyme, and the enzyme can then perform its function. Clinically, the dissociation of the inhibitor can be optimized through the dosing regimen, which can be tailored to the cellular lifetime of the enzyme of interest. For example, lysosomal hydrolases have half-lives on the order of a few days (35).
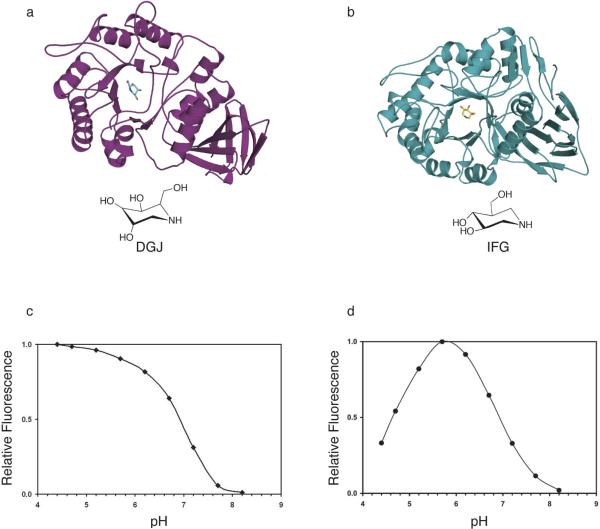
In this paper, we have characterized the effects of pH and pharmacological chaperone binding on the structure and thermostability of acid-α-galactosidase (α-Gal A) and acid-•-glucosidase (GCase), enzymes whose mutations lead to the lysosomal storage disorders Anderson-Fabry (12) and Gaucher disease (36, 37), respectively. Despite exhibiting very low sequence similarity, the three-dimensional structures of GCase (E.C. 3.2.1.45, CaZY (38) glycoside hydrolase family 30, ∼60kDa monomer) (23, 39) and α-Gal A (E.C. 3.2.1.22, CaZY glycoside hydrolase family 27, ∼50kDa monomer) (40) (Fig. 1) share a TIM-barrel ((•/•)8) motif in the catalytic domain, and an immunoglobulin fold in the same relative position (overall rmsd for Cα atoms ∼ 4.5 Å). In α-Gal A, the N-terminal ∼300 residues comprise the TIM-barrel, and the C-terminal ∼100 residues make up the predominantly •-sheet-containing immunoglobulin-like domain (40) (Fig. 1a). In the larger enzyme, GCase, the connectivity between participating elements of secondary structure is different from that of α-Gal A. Each tertiary structural element in GCase is composed of contributions from disparate regions of the sequence, and the additional three-stranded •-sheet brings together in space, and stabilizes, the N- and C-terminal ends (39) (Fig. 1b). Whereas α-Gal A is a dimer (40-44), the oligomeric state of GCase has been shown to be a monomer when isolated from normal spleens (45-47) but a dimer when isolated from Gaucher patients (45, 47). Both enzymes retain the stereochemistry of their respective substrate and product, and the catalytic nucleophile and proton donor are either two aspartate residues (α-Gal A) (40, 48) or two glutamate residues (GCase) (4, 49). Finally, both enzymes are activated by saposins. Globosyltriosylceramide, the substrate for α-Gal A involved in Fabry disease, binds saposin B prior to presentation to α-Gal A (50). Compared to globotriosylceramide, the substrate for GCase, glucosylceramide, is considerably more hydrophobic, yet glucosylceramide is hydrolyzed by GCase directly without the need of the activator. GCase activity is enhanced by saposin C, however (51, 52).
Fig. 1.
Cartoon representation of (a) α-Gal A with DGJ (this work) (b) GCase with IFG (PDB code 2NSX). DGJ and IFG are pictured as ball-and-stick, and their chemical structures are depicted below their respective cartoons. (c) Activity profile of α-Gal A as a function of pH and (d) activity profile of GCase as a function of pH.
Recently, we demonstrated that the potent iminosugar inhibitor isofagomine (IFG) acts as a pharmacological chaperone by improving protein trafficking for the N370S GCase mutant enzyme, resulting in increased cellular GCase levels in patient-derived fibroblasts (23). The structure of IFG-bound GCase at pH 4.5 (Fig. 1b, PDB code 2NSX) revealed a substantial rearrangement of one region (loop 1, residues 311−319) from an extended loop to an •-helical conformation, at the mouth of the active site. This induced fit has several important ramifications. First, a new surface topology is created, including two hydrophobic grooves extending from the site of catalysis in the triosphosphate isomerate (TIM) barrel domain. Second, these hydrophobic surfaces are suitable for binding of the alkyl chains of glucosylceramide. Third, Asn 370, located on the interior of GCase on a stable helix and the most common mutation site that causes Type 1 Gaucher disease, plays a direct role in stabilizing the configuration of loop 1 in the new conformation. Lastly, Tyr 313, located on loop 1, both stabilizes loop 1 in the •-helical conformation and alters its H-bonding pattern between the proton donor and catalytic nucleophile in the active site. We now extend our investigations to include a comparison of GCase structures at different pHs to elucidate the details of the active site loop conformations and stability conferred by inhibitor binding. Interestingly, analogous studies of α-Gal A with the pharmacological chaperone deoxygalactonijirimycin (DGJ) (16, 22, 53-55) do not reveal major conformational changes upon pH, inhibitor, or product binding, suggesting that, unlike GCase, α-Gal A may bind its subtrate like a “lock-and-key”. Taken together, our data support the model of pharmacological chaperoning whereby the inhibitor binds to and stabilizes the native-like state of the target enzyme.
EXPERIMENTAL PROCEDURES
Crystallization, data collection, structure determination, and refinement
We purchased lyophilized, recombinant human acid-•-glucosidase (Cerezyme®) and acid-•-galactosidase and Fabrazyme® (Genzyme Corp) from Brigham & Women's Hospital pharmacy. Cerezyme® and Fabrazyme® were reconstituted with phosphate-buffered saline (Sigma) to ∼1 mg/ml and partially deglycosylated the enzymes for 3 days using N-glycosidase F (Glyko, Novato, CA) as described previously (39). The deglycosylation step improves crystal quality but does not affect protein structure (56). We solved crystal structures of IFG-bound GCase at pH 7.5 (C1), apo GCase at pH 5.5 (C2), two structures of apo GCase at pH 4.5 (C3 and C4), as well as apo α-Gal A (L1), galactose-bound α-Gal A (L2), and DGJ-bound α-Gal A at pH 7.5 (L3). A summary of conditions appears in Table 1.
Table 1.
Summary of structures and their roles in this investigation
| Crystal | Crystallization condition | Role in investigation | PDB code |
|---|---|---|---|
| C1 | Na/K H2PO4, Hepes pH 7.5 Soak: IFG Cryo: glycerol |
pH-dependence of IFG binding on GCase structure | 3GXF |
| C2 | Na/K H2PO4, citrate buffer pH 5.5 Cryo: Li2SO4 |
Apo GCase structure at high limit of lysosomal pH | 3GXI |
| C3 | (NH4)2(SO4), acetate buffer pH 4.5, guanidinium hydrochloride, KCl Cryo: Li2SO4 |
Apo GCase structure at low limit of lysosomal pH Effect of crystallization cocktail (compare to C4) |
3GXM |
| C4 | Same as C2 Soak: acetate buffer pH 4.5 Cryo: Li2SO4 |
Apo structure at low limit of lysosomal pH Effect of crystallization cocktail (compare to C3) |
3GXD |
| L1 | Acetate buffer pH 4.5, PEG 4000, ammonium sulfate Cryo: paratone-n |
Apo α-GAL A, lysosomal pH | 3GXN |
| L2 | Acetate buffer pH 4.5, PEG 4000, ammonium sulfate Soak: galactose Cryo: ethylene glycol |
Galactose-bound α-GAL A, lysosomal pH | 3GXP |
| L3 | Acetate buffer pH 4.5, PEG 4000, ammonium sulfate Soak: DGJ Cryo: ethylene glycol |
DGJ-bound α-GAL A, lysosomal pH | 3GXT |
GCase was crystallized by hanging drop vapor diffusion, similar to methods reported previously, with some modification described below. GCase crystals at pH 4.5 (C3) were grown by hanging-drop vapor diffusion using a cocktail containing 1M ammonium sulfate, 0.1M acetate buffer pH 4.5, 0.17 M guanidinium hydrochloride, 0.02 M KCl (39). The crystals were then harvested in a solution of 0.1 M Acetate buffer pH 4.5, 1.8 M Li2SO4 just prior to flash-cooling in liquid nitrogen. GCase crystals at pH 4.5 (C4) were also obtained by soaking C2 crystals grown by vapor diffusion over a solution containing 0.8 M NaH2PO4, 0.8 M KH2PO4, 0.1 M citrate buffer pH 5.5 for 10 min in a solution containing 0.1 M acetate buffer pH 4.5, 1.8 M Li2SO4. The crystals were then flash-cooled in liquid nitrogen. The C1 crystals were obtained by soaking GCase crystals grown at pH 7.5 (23) in mother liquor containing 500 •M IFG for 10 min. C1 crystals were cryoprotected with mother liquor plus 20% glycerol.
α-Gal A was crystallized as described previously (40) with some modifications. After deglycosylation, we exchanged α-Gal A into 20 mM Tris pH 7.5 using concentration devices with a molecular weight cutoff of 10,000 kDa (Millipore). Crystallization trials were conducted with 10, 20, and 40 mg/ml deglycosylated α-Gal A. Independent of the protein concentration, crystals grew using hanging drop vapor diffusion from a cocktail containing 0.1 M acetate buffer pH 4.5, 20−25 % PEG 4000, and 0.15−0.22 M ammonium sulfate. Crystals appeared after 1 week (40 mg/ml) or after 2 months (10 mg/ml) and ranged in size from 0.05 × 0.05 × 0.1 mm (40 mg/ml) to 0.4 × 0.4 × 1 mm (10, 20 mg/ml). Crystals were soaked for 1 h with 200 •M DGJ (L3) in mother liquor, or 0.1 M galactose (L2) in mother liquor. Prior to cooling, crystals were transferred to a solution containing mother liquor (no DGJ, no galactose) supplemented with 30% ethylene glycol. Apo α-Gal A (L1) crystals were flash-cooled after cryoprotection with paratone-n.
Crystallographic data were collected at the GM/CA-CAT (C1, C2, C4, L1, L2, L3) and Bio-CARS (C3) beamlines at the Advanced Photon Source (Darien, IL) and processed with XDS (57) (L3) or HKL2000 (58) (C1-C4, L1, L2). The structures of GCase were solved by rigid body refinement in Refmac5 (59) using a protein model derived from PDB code 2NT0 after deleting loop 1 (residues 312−319) and loop 2 (residues 342−350) from all four copies in the asymmetric unit, as well as all N-linked carbohydrates, phosphate anions and waters. The structures of α-Gal A were solved by molecular replacement with the program Molrep (59) using a monomeric search model derived from PDB code 1R46 (40). We fit the atomic models into their respective electron density maps using Coot (60) and refined them using Refmac5 (59). We used medium non-crystallographic symmetry restraints for α-Gal A structures, generated topology and geometry restraints for IFG and DGJ using PRODRG2 (61), and identified bound water molecules using Coot (60). The percentage of residues in the most favored and additional allowed regions of the Ramachandran plot are 99.4−100% for all data sets, and crystallographic statistics are shown in Table 2. The Secondary Structure Matching (SSM) algorithm (62) from Coot was used for superposition, and figures were generated using Pymol (DeLano Scientific, San Carlo, CA).
Table 2.
Data collection and refinement statistics
| GCase+IFG, pH 7.5 (C1) | GCase, pH 5.5 (C2) | GCase pH 4.5 (C3) | GCase pH 4.5 soaked (C4) | α-Gal A (L1) | α-Gal A + GAL (L2) | α-Gal A + DGJ (L3) | |
|---|---|---|---|---|---|---|---|
| Data collection | |||||||
| Space group | P21 | P21 | P21 | P21 | P3221 | P3221 | P3221 |
| Cell dimensions | |||||||
| a, b, c (Å) | 110.3, 92.0, 152.4 | 110.05, 91.7, 152.3 | 110.5, 91.8, 152.8 | 109.3, 91.7, 152.5 | 90.4, 90.4, 217.5 | 89.5, 89.5, 215.9 | 89.6, 89.6, 216.6 |
| α, β, γ (°) | 90. 111.2, 90 | 90, 111.1, 90 | 90, 111.2, 90 | 90, 111.0, 90 | 90, 90,120 | 90, 90,120 | 90, 90, 120 |
| Resolution (Å)* | 32.0−2.40 (2.49−2.40) | 141.4−1.85 (1.92−1.85) | 34−2.2 (2.28−2.20) | 142−2.3 (2.38−2.30) | 78.3−3.0 (3.11−3.0) | 77.6−2.2 (2.28−2.20) | 77.6−2.7 (2.85−2.70) |
| Rsym | 11.3(37.5) | 9.1(44.9) | 10.3(47.3) | 9.1(44.2) | 10.3(62.9) | 9.4(40.2) | 9.7(42.8) |
| I / σI | 8.7(2.1) | 23(2.5) | 9.8(2.2) | 21.8(2.1) | 15.3(1.9) | 17.7(2.6) | 6.4(1.8) |
| Completeness (%)+ | 93.2(79.8) | 93.5(76.0) | 96.4(91.2) | 80.8(58.9) | 88.7(66.0) | 81.9(42.7) | 97.7(98.7) |
| Redundancy | 3.1 | 5.4 | 2.5 | 6.0 | 6.3 | 6.3 | 2.4 |
| Refinement | |||||||
| Resolution (Å) | 32−2.4 | 141.4−1.8 | 20−2.2 | 42−2.5 | 78.3−3.0 | 77.6−2.2 | 77.6.2−2.7 |
| No. reflections | 98363 | 216309 | 131814 | 81268 | 17705 | 40251 | 26223 |
| Rwork / Rfree* | 17.8/24.5 | 19.0/23.1 | 22.0/27.6 | 20.4/27.5 | 23.6/30.0 | 21.3/26.9 | 23.7/30.3 |
| No. molecules | |||||||
| Protein residues | 1988 | 1988 | 1988 | 1988 | 780 | 781 | 781 |
| N-acetyl-glucosamine | 4 | 4 | 4 | 6 | 4 | 12 | 11 |
| (NAG) | |||||||
| Mannose (MNN) | 2 | 5 | |||||
| Sulfate anion (SO42--) | 28 | 1 | 4 | 5 | |||
| Phosphate anion (PO43−) | 64 | 63 | 50 | ||||
| Glycerol (GOL) | 5 | ||||||
| Chaperone | 2 (IFG) | 2 (DGJ) | |||||
| Tris | 1 | ||||||
| Galactose | 2 | ||||||
| Water | 923 | 1699 | 1181 | 222 | 162 | 73 | |
| B-factors | |||||||
| Protein | 26.2 | 26.1 | 24.6 | 49.2 | 121.0 | 71.1 | 63.4 |
| NAG | 34.8 | 33.7 | 30.9 | 62.3 | 139.6 | 93.0 | 80.8 |
| MNN | 199.9 | 88.5 | |||||
| SO42-- | 46.7 | 126.9 | 91.0 | ||||
| PO43− | 74.6 | 63.4 | 79.4 | ||||
| GOL | 43.9 | ||||||
| Chaperone | 28.2 | 58.5 | |||||
| Tris | 58.7 | ||||||
| Galactose | 55.6 | ||||||
| Water | 29.0 | 37.7 | 28.5 | 43.3 | 54.3 | 45.0 | |
| R.m.s deviations | |||||||
| Bond lengths (Å) | 0.02 | 0.017 | 0.016 | 0.016 | 0.018 | 0.020 | 0.016 |
| Bond angles (°) | 1.957 | 1.683 | 1.682 | 1.706 | 1.844 | 2.049 | 1.912 |
Highest resolution shell is shown in parenthesis. 5% of reflections were selected for Rfree.
GCase crystals are shaped as thin plates that diffract anisotropically along their thin edge, and resulting in incomplete data in the highest resolution bin. α-Gal A crystals are very radiation-sensitive rods, so some data were not collected in the highest resolution bin.
Differential scanning calorimetry (DSC)
Scanning calorimetric experiments were performed with a VP-DSC (Microcal Inc., Northampton, MA) microcalorimeter. Samples of lyophilized α-Gal A (Fabrazyme®) and GCase (Cerezyme®) were reconstituted in the appropriate buffer and dialyzed for 6−16h using a Slide-a-lyzer (Pierce, 10,000 kDa molecular weight cutoff) and degassed prior to the experiment. For α-Gal A (∼50 •M), thermal denaturation curves were measured at pH 7.5 using phosphate-buffered saline (Sigma, 10 mM phosphate, 0.0027 M KCl, 0.138 M NaCl) and at pH 5.2 using 10 mM acetate, 0.0027 M KCl, 0.138 M NaCl. Buffers used for GCase (∼25 •M) were 30 mM phosphate, 0.15 M NaCl, pH 7.5 and 30 mM Acetate, 0.15 M NaCl, pH 5.15. For inhibitor studies, GCase or α-Gal A was incubated 0.05−2.5 mM IFG or with 0.05− 3.8 mM DGJ at room temperature, respectively. All samples were prepared and measured in duplicate or triplicate.
To study the concentration dependence of the melting temperature, we recorded DSC thermograms at 47.5 •M and 8.6 •M for α-Gal A and 25 •M, 9.6 or 14 •M for GCase. We observed no concentration-dependent change in melting temperature (Tm) for either apo α-Gal A or apo GCase (data not shown), suggesting that neither protein undergoes a classic dissociation mechanism upon unfolding under the experimental conditions. Thermal denaturation curves for for GCase were irreversible due to precipitation at high temperature, and could not be analyzed using equilibrium thermodynamics. To determine conditions for microscopic reversibility of unfolding for α-Gal A, we analyzed the scan-rate dependence of the thermograms. We saw no difference between melting temperatures obtained by scans measured with a rate of 1 and 1.5 °C/min, and all subsequent scans were measured at a rate of 1 °C/min. Thus, we were able to apply equilibrium thermodynamics analysis to the α-Gal A system.
DSC data were analyzed with Microcal Origin 7. Scans were first corrected by subtracting the sample trace from that of the buffer alone, and the concentrations were normalized (63). In the case of GCase, severe aggregation upon melting precluded further fitting to a thermodynamic model. As a measure of protein stability, our analysis is limited to differences in Tm of the normalized data. For α-Gal A, data were fit to a non two-state model, which provides the calorimetric (•HCal) and the van't Hoff (•HvH) enthalpies. •HCal is the measured enthalpy during protein unfolding, whereas •HvH is the theoretical enthalphy of the transition assuming a two-state model. The cooperativity unit is defined by the ratio •HCal/•HvH. Sturtevant analysis was carried out as described (64-66).
Enzyme Activity Assays
The results of enzyme activity assays were kindly provided by Amicus Therapeutics. In brief, enzymatic activities of α-Gal A and GCase (50 ng prepared in 0.1M citrate, 0.2M phosphate buffer pH 5.2 containing 0.1% Triton X-100 and 0.25% sodium taurocholate) were determined by incubating the enzymes in the presence of 3 mM of 4-methylumbelliferyl-α-D-galactopyranoside or 4-methylumbelliferyl-β-D-glucopyranoside, respectively, in reaction buffer (50 mM sodium phosphate, 150 mM sodium chloride) adjusted to pHs 4.4−8.2. After incubation at 37°C for one hour, the reactions were stopped by the addition of an equal volume of 0.5 M sodium carbonate (pH 10.8). Fluorescence of liberated substrate was read on plate reader (Ex. 355 nm; Em. 460 nm), and α-Gal A or GCase activity was plotted as a function of pH.
RESULTS
Enzyme activity profile
The enzyme activity of α-Gal A and GCase were measured in the range of pH 4.5−8.2. Consistent with their lysosomal function, α-Gal A and GCase exhibit pH-sensitive enzyme activity (Fig 1c, d). Activity profiles of GCase and α-Gal A indicate that maximal activity is observed at lysosomal pH of below 6 and most enzyme activity is abolished at the neutral pH of the endoplasmic reticulum (pH 7.4). These results from recombinant enzymes are consistent with previous activity studies of α-Gal A (67) and GCase (68) isolated and purified from human placenta and spleen, respectively, and correlate with thermostability (see below).
Structural comparisons
GCase
Previously, we observed variability in the active site region of independent monomers in the asymmetric unit of apo GCase at pH 7.5 (PDB code 2NT1)(23). Specifically, loop 1 was found in an •-helical conformation in two of the four crystallographic monomers in the asymmetric unit, and extended in the remaining two monomers. Because these conformations were seen in the absence of ligand, we suggested that the two conformations of loop 1 trapped crystallographically are sampled upon folding in the ER (23). In our previous study, we presumed that the pH 4.5 glycerol-bound (23) or sulfate-bound (39) GCase structures grown from ammonium sulfate were identical to apo GCase, because only one conformation, namely that where loop 1 is in an extended conformation, had been observed in those structures.
To systematically examine the effects of pH and crystallization cocktail on these observed loop conformations, and address the conformational variability of the active site loops with pH and inhibitor binding, we have now solved the crystal structures of IFG-bound GCase at pH 7.5 (C1) cryoprotected with glycerol, and apo GCase at pH 5.5 (C2) and pH 4.5 (C3, C4, Table 1, 2). All apo structures were flash cooled in 1.8 M Li2SO4 and buffer, omitting glycerol and ethylene glycol that are typically used as cryoprotectants and could bind in the active site. C3 and C4 differ in the method used for crystallization: C3 was grown from ammonium sulfate and frozen with Li2SO4 directly whereas C4 was obtained by taking crystals grown from phosphate salts at pH 5.5 and soaking them in a suitable solution at pH 4.5 prior to cryoprotection. A comparison of C3 and C4 therefore enables the identification of any effects of crystallization cocktail at pH 4.5. C2 and C4 bracket the reported lysosomal pH range of 4.7−5.5 (1).
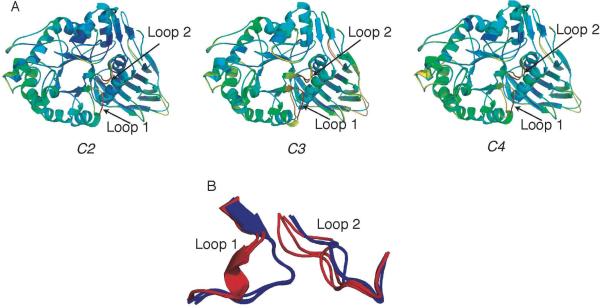
The three apo GCase structures are nearly identical (rmsd ∼0.45 Å over all pairs of crystallographically observed monomers), each with two α-helical and two extended loop 1 conformations observed among the four independent monomers in the crystallographic asymmetric unit. These apo structures are also nearly indistinguishable from 2NT1. In comparison to the rest of the enzyme, the loop 1 and loop 2 regions have the highest thermal B-factors (Fig. 2a). We observed a correlation between interpretable, yet comparatively poor, 2Fo-Fc electron density for the extended conformation for loop 1 paired with excellent electron density for loop 2, and vice versa. Thus, there appears to be no crystallization cocktail or pH bias for these loop conformations (Fig 2b). Given that the structures are solved to similar resolution limits, the high thermal factors of loops 1 and 2 suggest that in the low pH environment observed in the lysosome, these loops are likely to be sampling several conformations. Thus, for the GCase polypeptide alone, the full active site, which comprises the catalytic center and hydrophobic subsites created by loops 1 and 2, does not appear to be preorganized for substrate binding. The native catalytic environment of GCase, which involves proximity to the lysosomal membrane and binding to saposin C in a poorly understood fashion (69), may also influence the conformation of the GCase active site.
Fig. 2.
Structures of apo GCase grown under different conditions. Overlay of two monomers in asymmetric unit exhibiting distinct loop 1 structures (α-helical or extended). (A) Left C2, pH 5.5; middle C3, pH 4.5; right C4, pH 4.5 (soaked). Ribbon diagrams are colored using a rainbow corresponding to increasing thermal factor from blue (low B-factor) to red (high B-factor). (B) Superposition of structures in (A)-(C) in loop 1, loop 2 region. Red: α-helical loop 1, blue: extended loop 1conformation.
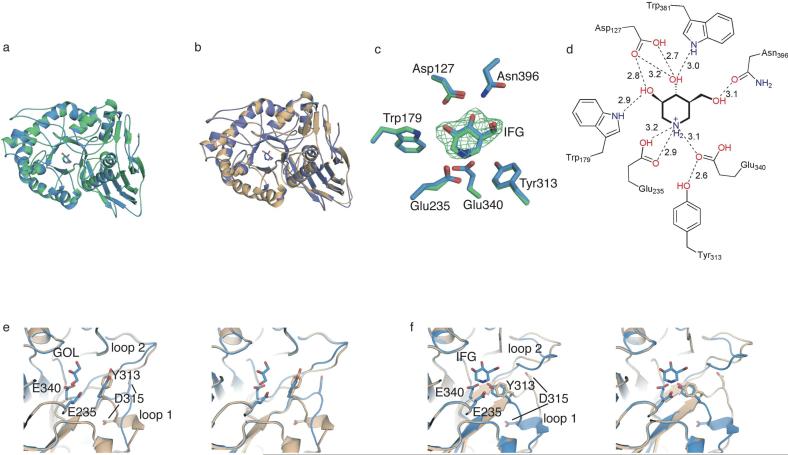
The overall structure of IFG-bound GCase at pH 7.5 (C1) is similar to our previously determined structures of IFG-bound GCase at pH 4.5 (PDB code 2NSX, rmsd 0.19 Å, Fig 3a) and of apo GCase in the α-helical conformation (2NT1, rmsd 0.38 Å, Fig 3b)(23). Like for 2NSX, IFG is bound in C1 in two of the four monomers in the asymmetric unit, and glycerol is found in the remaining two monomers. The catalytic centers of 2NSX and C1 (Fig 3c) are nearly indistinguishable: the active site residues are locked into the same conformation and IFG is held in place by the same interacting residues at comparable distances (Fig 3d). This result suggests that the protonation states of acidic active site residues are unchanged and it is consistent with: (a) the pKa of 8.4 for IFG (70), and (b) the low pKa of the catalytic/acid-base residues. Alternatively, the protonation state of IFG may not alter the structure of GCase.
Fig. 3.
GCase structures at various pHs and in complex with IFG. (A) Overlay of GCase structure with IFG at pH 4.5 (2NSX, green) and pH 7.5 (C1, this work, blue). (B) Overlay of IFG-bound C1 (blue) with with apo GCase at pH 5.5 (C2, beige) exhibiting α-helical loop 1. (C) Superposition of active site residues from overlay shown in (A). Fo-Fc difference density from initial rigid body refinement is contoured at 3 •. (D) Schematic diagram of hydrogen bonding interactions involved in stabilizing IFG in the active site of C1. Distances are in Å. (E) Active-site stereo view of glycerol-bound monomer of C1 superimposed with corresponding monomer from apo pH 7.5 (2NT1). (F) Active-site stereo view of IFG-bound monomer of C1 superimposed with corresponding monomer from apo GCase pH 7.5 (2NT1). For (E)-(F), the bound ligand, nucleophilic (Glu 340) and basic (Glu 235) residues, and two key loop 1 residues (Tyr 313, Asp 315) are shown in ball-and-stick representation.
Surprisingly, IFG binds to the monomers in the asymmetric unit where loop 1 is observed in the extended conformation for apo GCase (comparison here is to the apo structures C2-C4 and 2NT1), resulting in a rearrangement to the •-helical conformation, and glycerol is found in the active site of the remaining monomers where loop 1 is unwound (Fig e,f). Glycerol and IFG bind in the active site and appear to induce GCase into a particular conformation, but only IFG can thermally stabilize GCase (see below) and is a pharmacological chaperone. It would seem that the particular induced fit of IFG-bound GCase is the substrate-ready conformation of GCase, and this configuration is important for pharmacological chaperoning of GCase. However, given that the oligomeric state of GCase in vivo not fully understood (45, 46, 68), further experiments will be required to rule out the possibility that GCase exists as a cooperative dimer where the conformations of loop 1 in one monomer is coordinated with that of the second monomer. For example, IFG binding in the active site of one GCase monomer may induce the unwinding of loop 1 in the second GCase monomer, which in turn enables glycerol to bind in the active site, or vice versa. The lifetime of the conformational change induced by IFG after exchange for substrate in vivo remains an open question as well.
α-Gal A
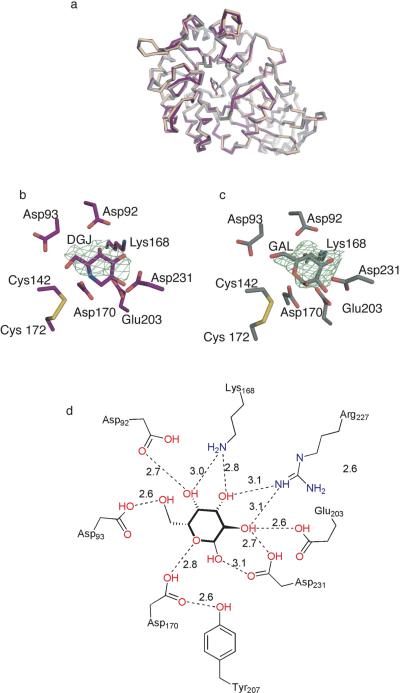
To elucidate pH-dependent features of α-Gal A, we solved structures of apo (3.0 Å resolution), galactose-soaked (2.2 Å resolution), and DGJ-soaked (2.7 Å resolution) α-Gal A derived from a Chinese Hamster Ovary cell line (71) at pH 4.5. The asymmetric unit contains one α-Gal A dimer, and although we subjected our protein samples to deglycosylation with N-glycosidase F, numerous carbohydrate linkages are visible in the crystal structures. This is the first report of DGJ-bound α-Gal A, and we compared our structures with previously determined structures of fully glycosylated α-Gal A derived from a human cell line complexed with ethylene glycol and galactose at pH 7.5 (PDB codes 1R46, 1R47, respectively) (40). The three-dimensional structures of α-Gal A complexed with the hydrolysis product, galactose, or the product analog inhibitor DGJ show no polypeptide backbone or active site changes compared to apo α-Gal A (Fig 4a, rmsd for C• atoms of ∼0.4 Å) but we observe considerably higher B-factors for the lowest resolution apo structure compared to DGJ- or galactose-bound α-Gal A. Similarly, the three-dimensional structure of the protein does not change as a function of the pH, presumably because the disulfide bonds at the mouth of the active site and elsewhere in the protein preclude such a dramatic rearrangement. Galactose and DGJ bind in the α-Gal A active site with similar distorted chair conformations (Fig 4b,c), with the methoxy arm of DGJ (Fig. 1a) stabilized through interactions with Asp 93. Extensive hydrogen bonding and electrostatic interactions are observed to stabilize these ligands in the α-Gal A active site, but the 2.7 Å resolution of the structure of DGJ-bound α-Gal A precludes precise measurement of H-bonding distances for the chaperone. The galactose-bound structure is the only α-Gal A structure where significant numbers of water molecules are visible, and because of its higher resolution (2.2 Å) we can report hydrogen bonding distances (Fig 4d). The higher resolution of the galactose-bound complex was enabled by the large size of the crystal. In addition, a strong peak in the difference (Fo-Fc) electron density map is found at the dimer interface. This peak was modeled as a Tris molecule, presumably from the protein buffer. The functional significance of a bound ligand in this interface is currently unknown, but could be important for design of pharmacological chaperones that bind to regions remote from the active site of α-Gal A and stabilize the functional dimer (see below). Notably, like IFG-bound GCase but not apo or glycerol-bound GCase, a tyrosine residue, Tyr 207, is H-bonded with a 2.6 Å distance to the α-Gal A nucleophile Asp 170. Thus, in contrast to GCase, apo α-Gal A appears pre-organized for catalysis and its apo structure likely represents the active conformation. This prearrangement is consistent with the broad range of α-galactose-containing substrates that can be hydrolyzed by α-Gal A (72).
Fig. 4.
α-Gal A structures. (A) Superposition of apo α-Gal A (beige), galactose-bound α-Gal A (silver), and DGJ-bound α-Gal A (purple). (B) Active site region of DGJ-bound α-Gal A. Fo-Fc difference density immediately following original molecular replacement solution is contoured at 2.5 •. (C) Active site region of galactose-bound α-Gal A. Fo-Fc difference density immediately following original molecular replacement solution at 3 •. (D) Schematic diagram of hydrogen bonding interactions involved in stabilizing galactose in the active site of DGJ.
Stability comparisons
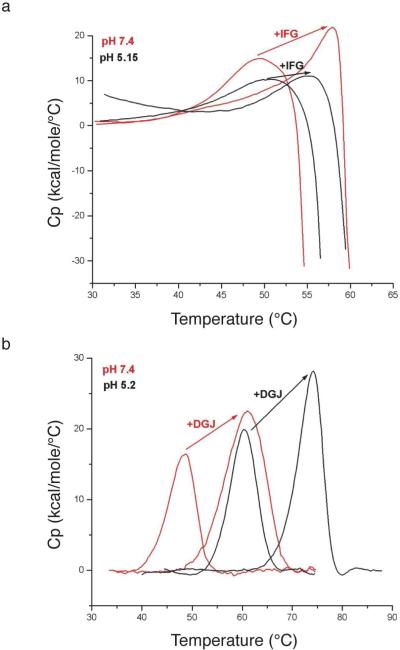
To study the stability conferred by pharmacological chaperones on GCase and α-Gal A, we used differential scanning calorimetry (DSC). We recorded denaturation thermograms for GCase and α-Gal A at pH 5.2 and pH 7.4 in the presence and absence of their respective pharmacological chaperones (Fig. 5).
Fig. 5.
Differential scanning calorimetry. (A) Thermal denaturation of GCase upon addition of IFG at pH 7.4 (red) and pH 5.2 (black). (B) Thermal denaturation for α-Gal A upon addition of DGJ at pH 7.4 (red), and pH 5.2 (black).
GCase
GCase precipitates immediately after the folding-unfolding transition has occurred, which is observed by the heat capacity falloff in DSC (Fig. 5a). Thus, we cannot extract thermodynamic parameters from these data, and are limited to comparing relative melting temperatures and shapes of the melting curves. A modest increase in the melting temperature (Tm) of GCase is observed for the acidic pH environment when compared to neutral pH (49.3 °C at pH 7.4 to 51.7 °C at pH 5.2, Fig 5a, Table 2A). A broad transition is observed for apo GCase at both pHs (Fig 5a), indicating low cooperativity and possibly a population of intermediate states during unfolding. One hypothesis to explain this behavior is that the domains of GCase are not composed of distinct regions of the amino acid chain (see above) and thus the unfolding pathway may involve many intermediate conformational variations.
In the presence of slight molar excess of IFG, the melting temperature of GCase measured by DSC increases reproducibly to 58 °C at pH 7.4 and to 55.4 °C at pH 5.2 (Fig. 5a, Table 2A). Thus, the binding of IFG to GCase confers more stability to GCase at a neutral pH reflective of the ER than at a pH of the lysosome. This effect is presumably dictated by the nanomolar binding affinity of IFG (23). In addition, binding IFG to GCase sharpens the folding-unfolding transition of the denaturation curves, suggesting a more cooperative unfolding transition when IFG is bound. It is likely that IFG binding to the TIM barrel of GCase reduces its conformational flexibility, leading to increased thermostability and cooperativity of unfolding.
α-Gal A
α-Gal A undergoes thermal unfolding without aggregation (Fig. 5b), but we conducted the α-Gal A unfolding experiments under conditions of microscopic reversibility (see Experimental Section). The stability of apo α-Gal A exhibits considerable pH sensitivity: the Tm is 48 °C at pH 7.4 and 60.2 °C at pH 5.2 (Table 2b). Thermal unfolding of apo α-Gal A at both pHs is accompanied by an enthalpy of ∼100 kcal/mol dimer that is fit well to a 2-state model (Table 3). These results suggest that dissociation of the dimer and unfolding of each monomer occurs simultaneously (73) and hence, the folded monomer is not stable. The lack of concentration dependence on Tm further supports the model that no dissociation of the dimer occurs prior to unfolding.
Table 3.
Summary of melting temperatures from differential scanning calorimetry. (A) GCase with and without IFG at pH 7.4 and pH 5.15. (B) α-Gal A with and DGJ at pH 7.4 and pH 5.2
| A |
|
Tm (°C) |
B |
|
Tm (°C) |
|---|---|---|---|---|---|
| GCase | 49.3 | α-Gal A | 48.0 | ||
| pH 7.4 | GCase+IFG | 58.0 | pH 7.4 | α-Gal A +DGJ | 60.6 |
| GCase | 51.7 | α-Gal A | 60.2 | ||
| pH 5.2 | GCase+IFG | 55.4 | pH 5.2 | α-Gal A +DGJ | 73.5 |
The addition of DGJ to α-Gal A at both pHs increases the Tm ∼13°C, to 60.6°C at pH 7.4 to 73.5°C at pH 5.2 (Table 2b). In contrast to GCase with IFG, the increase in stability of α-Gal A with DGJ bound is approximately the same at low and high pH. The calorimetric enthalpy of unfolding of α-Gal A in the presence of DGJ is ∼ 200 kcal/mol per dimer, which corresponds to twice the calculated van't-Hoff (2-state) enthalpy (Table 3). van't-Hoff analysis suggests that ligand binding in this case results in a non-2-state unfolding transition with lower cooperativity. One possibility is that ligand binding leads to the preferential stabilization of the TIM barrel domain where binding takes places. Specifically, the TIM barrel, stabilized by the binding of DGJ, and β-barrel domains within each α-Gal A monomer, unfold independently but at the same temperature. Two lines of evidence support this inter-domain cooperativity for unfolding of the α-Gal A with DGJ. First, the half inhibitory concentration (IC50) is in the nM range for the inhibition of α-Gal A by DGJ (54), and we have shown that DGJ binds in the active site, located in the TIM barrel. With a slight excess of DGJ, all available active sites are expected to be saturated with inhibitor, and no other binding sites have been identified for DGJ on α-Gal A. Second, Sturtevant plot analysis, which uses the slope of the plot of 1/Tm and ln [ligand] to determine the number of bound ligands per molecule, indicates 1.7 DGJ molecules, or close to 2, are bound per dimer for α-Gal A. This analysis uses the slope of the plot of 1/Tm and ln [ligand] to determine the number of bound ligands per molecule. No plateau is expected in a plot of Tm versus ligand concentration if the ligand binds only to the folded state of the enzyme. This behavior is well documented, and a result of the interplay between temperature dependent (a) equilibrium between the folded and unfolded protein conformations; (b) the binding constant of the ligand to the folded enzyme; and (c) unfolding coupled to ligand dissociation, which together result in a linear relationship between 1/Tm and ln[Ligand] (64-66).
DISCUSSION
In the ER, numerous protein chaperones aid the folding of a nascent polypeptide, influence the timing of folding, or recognize features or flaws of the newly folded protein, such as extent of glycosylation or disulfide bond formation (9, 10, 74). These quality control systems are highly redundant and have evolved to prevent proteins compromised in any way from continuing on the path to maturation. Primary quality control in the ER is thought to be governed by certain biophysical properties of the folded protein, namely reduced thermal stability as detected by incorrectly exposed hydrophobic patches or mobile loops. Proteins recognized as misfolded can be refolded and recovered by the ER-assisted folding (ERAF) pathway or degraded by the ER-associated degradation (ERAD) pathway (75). Conditions that destabilize proteins, such as missense mutations or changes in pH, can shift the fate toward ERAD and reduced ER export. Because the newly folded protein is not tested for partial function, many mutant proteins, which exhibit properties different from the wild type protein, do not have the opportunity to perform their cellular function before they are degraded. The lack of activity due to ER degradation often leads to disease states, such as those described here for Gaucher and Fabry disease.
Conversely, conditions that stabilize proteins, such as the presence of pharmacological chaperones or osmolytes, or reduced temperature of cell growth, can increase the fraction of newly folded enzyme capable of ER export. The goal of pharmacological chaperones is to bind specifically to a partially-active mutant enzyme target to pass detection by ER quality control machinery and promote proper cellular trafficking. There are several ways that a pharmacological chaperone may help trafficked proteins, both wild-type and mutant, pass the ER quality control system and reach their correct destinations: the chaperone may (1) interact while the protein is folding and decrease the amount of time it takes to fold, thereby reducing the chance that it will be recognized as unfolded; (2) bind once the protein is fully folded and stabilize it so that it is less likely to unfold or to be recognized as misfolded; (3) stabilize the protein and promote normal post-translational modification (e.g., glycosylation) and processing within the ER lumen; (4) stabilize the protein in environments that would otherwise promote unfolding or degradation such as the low pH of the endosome and lysosome or the protease-rich environment of the lysosome; (5) induce or stabilize a conformation that promotes interaction with a binding partner required for proper trafficking; or (6) all or some combination of the above.
For Gaucher and Fabry disease specifically, understanding the structural properties of GCase and α-Gal A in the neutral pH of the ER to the lower pH environment of the lysosome provides valuable insight. We hypothesized that DGJ and IFG promote export from the ER at least in part by stabilizing these enzymes in the neutral pH environment of the ER. Since α-Gal A and GCase have little catalytic activity at neutral pH (Fig. 1c, d), it was not clear if the active site of these enzymes was preserved. In this study, we used X-ray crystallography to explain how the structures of GCase and α-Gal A respond to pH changes and ligand binding. The organic solvents and other components of the crystallization mother liquor for GCase have no effect on the behavior of loop 1, providing further evidence that the induced conformation with IFG is functionally relevant. The α-helical conformation in loop 1 is present at low and high pH but unwinds in the presence of glycerol, which is known not to be an effective stabilizer even though it binds to the catalytic center. In addition, the conformation of loop 2 in GCase is sensitive to ligand binding. By contrast, induction of a specific conformation is not a feature of the action of DGJ as a chaperone for α-Gal A; therefore, we have shown that conformational changes are not a necessary feature of an effective chaperone. We also used differential scanning calorimetry to show that pharmacological chaperones enhance the stability of GCase and α-Gal A. Both GCase and α-Gal A are more stable at low pH where they are most active, and the addition of the respective iminosugar increases the stability of both enzymes at neutral pH. However, the stability conferred to GCase by IFG is notably greater at neutral pH than acidic pH, an effect not observed for DGJ/α-Gal A. Notably, although several studies have addressed GCase stability with pharmacological chaperones by circular dichroism (28, 30) or fluorescence (6), these techniques do not provide insight into the unfolding pathway. To the best of our knowledge, this thermodynamic stability analysis is the first such study for α-Gal A. Our experiments were performed with recombinant wild-type human GCase and α-Gal A, but similar binding events and protein stabilization likely occur for missense mutant versions of these lysosomal enzymes that retain catalytic activity in vitro. We (23) and other researchers (6, 29, 30, 76) have shown that small molecule inhibitors, including IFG, improve GCase export from the ER and increase lysosomal levels of both wild-type and mutant GCase in patient derived cell lines. From a clinical standpoint, our results are directly applicable to dual therapy involving (wild-type) enzyme replacement therapy with pharmacological chaperones. This approach may improve the efficacy of enzyme replacement therapy by increasing the stability and lifetime of recombinant GCase in Gaucher patients.
In summary, we have shown that the biophysical properties of GCase and α-Gal A are consistent with their known function in the low pH environment of the lysosome, and these properties may be useful for the development of new pharmacological chaperones. With the knowledge that an increase in stability, but not necessarily induction of a particular conformational change, may be key for an effective chaperone, it should now be possible to design a pharmacological chaperone remote from the active site or at a dimer interface. A remote binder would confer stability to the mutant enzyme and enable proper trafficking, and would eliminate competition with the substrate for the active site once in the lysosome. High throughput screening has identified compounds that enhance GCase activity, yet are unlikely to bind in the active site (77). The binding site(s) for these compounds are not known, but some non-active site hot spots for ligand binding on GCase have been identified recently experimentally and by in silico methods (78). Lastly, it is not known whether α-Gal A dimerizes or interacts with other proteins to facilitate export from the ER. Although crystallographic studies did not reveal a change in α-Gal A conformation upon binding of DGJ, complex folding pathways are known for dimeric proteins (79). For GCase, new evidence implicates the lysosomal integral membrane II (LIMP-2) as its lysosomal trafficking partner (72), although this trafficking mechanism may be tissue-specific (80). Successful export of GCase from the ER is likely conditional on the binding of LIMP-2 to GCase, which may depend on the specific conformation of GCase and its stability. We are currently investigating these possibilities.
Table 4.
Thermodynamic parameters for thermal denaturation of α-Gal A with and without DGJ at pH 7.4 and pH 5.2.
| ΔH × 105 cal | ΔHV × 105 cal | χ2 × 106 | Fit analysis | ||
|---|---|---|---|---|---|
| pH 7.4 | GLA |
1.18 |
1.16 |
1.9 |
2-state |
| |
GLA+DGJ |
2.11 |
0.91 |
2.1 |
Non 2-state |
| pH 5.2 | GLA |
1.41 |
1.31 |
0.33 |
2-state |
| GLA+DGJ |
2.12 |
1.32 |
1.47 |
Non 2-state |
ACKNOWLEDGMENT
This work was supported by NIH fellowship F32AG027647 to R.L.L., as well as the McKnight Endowment for Neuroscience (D.R., G. A. P). We are grateful to Amicus Therapeutics for IFG and DGJ, conducting the activity assays (John J. Flanagan), as well as valuable discussions and critical review of the manuscript (Brandon Wustman, David Lockhart, and Hung Do). We are grateful to the Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems at MIT (National Science Foundation Grant NSF-0070319 and NIH Grant GM68762) for calorimetric measurements. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. W-31-109-ENG-38. GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the BioCARS Sector 14 was supported by the National Institutes of Health, National Center for Research Resources, under grant number RR007707.
Abbreviations
- GCase
acid-β-glucosidase
- α-Gal A
acid-α-galactosidase
- IFG
isofagomine
- DGJ
1-deoxygalactonijirimycin
- ER
endoplasmic reticulum
- ERT
enzyme replacement therapy
- SRT
substrate reduction therapy
- LSDs
lysosomal storage disorders
- TIM
triosphosphate isomerase
- SSM
secondary structure matching
- DSC
differential scanning calorimetry
- Tm
melting temperature
- ΔHcal
calorimetric enthalpy
- ΔHvH
van't Hoff enthalpy
- IC50
half inhibitory concentration
- ERAF
ER-assisted folding
- ERAD
ER-associated degradation
- LIMP-2
lysosomal integral membrane II
- PDB
protein data bank
REFERENCES
- 1.Anderson RG, Orci L. A view of acidic intracellular compartments. J. Cell. Biol. 1988;106:539–543. doi: 10.1083/jcb.106.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Futerman AH, van Meer G. The cell biology of lysosomal storage disorders. Nat. Rev. Mol. Cell. Biol. 2004;5:554–565. doi: 10.1038/nrm1423. [DOI] [PubMed] [Google Scholar]
- 3.Sawkar AR, D'Haeze W, Kelly JW. Therapeutic strategies to ameliorate lysosomal storage disorders--a focus on Gaucher disease. Cell. Mol. Life Sci. 2006;63:1179–1192. doi: 10.1007/s00018-005-5437-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grace ME, Newman KM, Scheinker V, Berg-Fussman A, Grabowski GA. Analysis of human acid beta-glucosidase by site-directed mutagenesis and heterologous expression. J. Biol. Chem. 1994;269:2283–2291. [PubMed] [Google Scholar]
- 5.Liou B, Kazimierczuk A, Zhang M, Scott CR, Hegde RS, Grabowski GA. Analyses of variant acid beta -glucosidases: effects of gaucher disease mutations. J. Biol. Chem. 2006;281:4242–4253. doi: 10.1074/jbc.M511110200. [DOI] [PubMed] [Google Scholar]
- 6.Kornhaber GJ, Tropak MB, Maegawa GH, Tuske SJ, Coales SJ, Mahuran DJ, Hamuro Y. Isofagomine induced stabilization of glucocerebrosidase. Chembiochem. 2008;9:2643–2649. doi: 10.1002/cbic.200800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steet RA, Chung S, Wustman B, Powe A, Do H, Kornfeld SA. The iminosugar isofagomine increases the activity of N370S mutant acid beta-glucosidase in Gaucher fibroblasts by several mechanisms. Proc. Natl. Acad. Sci. U.S.A. 2006;103:13813–13818. doi: 10.1073/pnas.0605928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ron I, Horowitz M. ER retention and degradation as the molecular basis underlying Gaucher disease heterogeneity. Hum. Mol. Genet. 2005;14:2387–2398. doi: 10.1093/hmg/ddi240. [DOI] [PubMed] [Google Scholar]
- 9.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell. Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 10.Ellgaard L, Molinari M, Helenius A. Setting the standards: quality control in the secretory pathway. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- 11.Barton NW, Brady RO, Dambrosia JM, Di Bisceglie AM, Doppelt SH, Hill SC, Mankin HJ, Murray GJ, Parker RI, Argoff CE, et al. Replacement therapy for inherited enzyme deficiency--macrophage-targeted glucocerebrosidase for Gaucher's disease. N. Engl. J. Med. 1991;324:1464–1470. doi: 10.1056/NEJM199105233242104. [DOI] [PubMed] [Google Scholar]
- 12.Zarate YA, Hopkin RJ. Fabry's disease. Lancet. 2008;372:1427–1435. doi: 10.1016/S0140-6736(08)61589-5. [DOI] [PubMed] [Google Scholar]
- 13.Elstein D, Hollak C, Aerts JM, van Weely S, Maas M, Cox TM, Lachmann RH, Hrebicek M, Platt FM, Butters TD, Dwek RA, Zimran A. Sustained therapeutic effects of oral miglustat (Zavesca, N-butyldeoxynojirimycin, OGT 918) in type I Gaucher disease. J. Inherit. Metab. Dis. 2004;27:757–766. doi: 10.1023/B:BOLI.0000045756.54006.17. [DOI] [PubMed] [Google Scholar]
- 14.Pastores GM, Barnett NL, Kolodny EH. An open-label, noncomparative study of miglustat in type I Gaucher disease: Efficacy and tolerability over 24 months of treatment. Clin. Ther. 2005;27:1215–1227. doi: 10.1016/j.clinthera.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Allen MJ, Myer BJ, Khokher AM, Rushton N, Cox TM. Pro-inflammatory cytokines and the pathogenesis of Gaucher's disease: increased release of interleukin-6 and interleukin-10. Quart. J. Med. 1997;90:19–25. doi: 10.1093/qjmed/90.1.19. [DOI] [PubMed] [Google Scholar]
- 16.Fan JQ, Ishii S. Active-site-specific chaperone therapy for Fabry disease. FEBS J. 2007;274:4962–4971. doi: 10.1111/j.1742-4658.2007.06041.x. [DOI] [PubMed] [Google Scholar]
- 17.Kolter T, Wendeler M. Chemical chaperones--a new concept in drug research. Chembiochem. 2003;4:260–264. doi: 10.1002/cbic.200390045. [DOI] [PubMed] [Google Scholar]
- 18.Yu Z, Sawkar AR, Kelly JW. Pharmacologic chaperoning as a strategy to treat Gaucher disease. FEBS J. 2007;274:4944–4950. doi: 10.1111/j.1742-4658.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 19.Yam GH, Bosshard N, Zuber C, Steinmann B, Roth J. Pharmacological chaperone corrects lysosomal storage in Fabry disease caused by trafficking-incompetent variants. Am. J. Physiol. Cell. Physiol. 2006;290:C1076–1082. doi: 10.1152/ajpcell.00426.2005. [DOI] [PubMed] [Google Scholar]
- 20.Ulloa-Aguirre A, Janovick JA, Brothers SP, Conn PM. Pharmacologic rescue of conformationally-defective proteins: implications for the treatment of human disease. Traffic. 2004;5:821–837. doi: 10.1111/j.1600-0854.2004.00232.x. [DOI] [PubMed] [Google Scholar]
- 21.Compain P, Martin OR, Boucheron C, Godin G, Yu L, Ikeda K, Asano N. Design and synthesis of highly potent and selective pharmacological chaperones for the treatment of Gaucher's disease. Chembiochem. 2006;7:1356–1359. doi: 10.1002/cbic.200600217. [DOI] [PubMed] [Google Scholar]
- 22.Ishii S, Chang HH, Kawasaki K, Yasuda K, Wu HL, Garman SC, Fan JQ. Mutant alpha-galactosidase A enzymes identified in Fabry disease patients with residual enzyme activity: biochemical characterization and restoration of normal intracellular processing by 1-deoxygalactonojirimycin. Biochem J. 2007;406:285–295. doi: 10.1042/BJ20070479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lieberman RL, Wustman BA, Huertas P, Powe AC, Jr., Pine CW, Khanna R, Schlossmacher MG, Ringe D, Petsko GA. Structure of acid beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease. Nat. Chem. Biol. 2007;3:101–107. doi: 10.1038/nchembio850. [DOI] [PubMed] [Google Scholar]
- 24.Lin H, Sugimoto Y, Ohsaki Y, Ninomiya H, Oka A, Taniguchi M, Ida H, Eto Y, Ogawa S, Matsuzaki Y, Sawa M, Inoue T, Higaki K, Nanba E, Ohno K, Suzuki Y. N-octyl-beta-valienamine up-regulates activity of F213I mutant beta-glucosidase in cultured cells: a potential chemical chaperone therapy for Gaucher disease. Biochim. Biophys. Acta. 2004;1689:219–228. doi: 10.1016/j.bbadis.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Maegawa GH, Tropak M, Buttner J, Stockley T, Kok F, Clarke JT, Mahuran DJ. Pyrimethamine as a potential pharmacological chaperone for late-onset forms of GM2 gangliosidosis. J. Biol. Chem. 2007;282:9150–9161. doi: 10.1074/jbc.M609304200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okumiya T, Kroos MA, Vliet LV, Takeuchi H, Van der Ploeg AT, Reuser AJ. Chemical chaperones improve transport and enhance stability of mutant alpha-glucosidases in glycogen storage disease type II. Mol. Genet. Metab. 2007;90:49–57. doi: 10.1016/j.ymgme.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 27.Parenti G, Zuppaldi A, Gabriela Pittis M, Rosaria Tuzzi M, Annunziata I, Meroni G, Porto C, Donaudy F, Rossi B, Rossi M, Filocamo M, Donati A, Bembi B, Ballabio A, Andria G. Pharmacological enhancement of mutated alpha-glucosidase activity in fibroblasts from patients with Pompe disease. Mol. Ther. 2007;15:508–514. doi: 10.1038/sj.mt.6300074. [DOI] [PubMed] [Google Scholar]
- 28.Sawkar AR, Adamski-Werner SL, Cheng WC, Wong CH, Beutler E, Zimmer KP, Kelly JW. Gaucher disease-associated glucocerebrosidases show mutation-dependent chemical chaperoning profiles. Chem. Biol. 2005;12:1235–1244. doi: 10.1016/j.chembiol.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Sawkar AR, Cheng WC, Beutler E, Wong CH, Balch WE, Kelly JW. Chemical chaperones increase the cellular activity of N370S beta -glucosidase: a therapeutic strategy for Gaucher disease. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15428–15433. doi: 10.1073/pnas.192582899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawkar AR, Schmitz M, Zimmer KP, Reczek D, Edmunds T, Balch WE, Kelly JW. Chemical chaperones and permissive temperatures alter localization of Gaucher disease associated glucocerebrosidase variants. ACS Chem. Biol. 2006;1:235–251. doi: 10.1021/cb600187q. [DOI] [PubMed] [Google Scholar]
- 31.Tropak MB, Blanchard JE, Withers SG, Brown ED, Mahuran D. High-throughput screening for human lysosomal beta-N-Acetyl hexosaminidase inhibitors acting as pharmacological chaperones. Chem. Biol. 2007;14:153–164. doi: 10.1016/j.chembiol.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tropak MB, Reid SP, Guiral M, Withers SG, Mahuran D. Pharmacological enhancement of beta-hexosaminidase activity in fibroblasts from adult Tay-Sachs and Sandhoff patients. J. Biol. Chem. 2004;279:13478–13487. doi: 10.1074/jbc.M308523200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu L, Ikeda K, Kato A, Adachi I, Godin G, Compain P, Martin O, Asano N. Alpha-1-C-octyl-1-deoxynojirimycin as a pharmacological chaperone for Gaucher disease. Bioorg. Med. Chem. 2006;14:7736–7744. doi: 10.1016/j.bmc.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Fan JQ. A contradictory treatment for lysosomal storage disorders: inhibitors enhance mutant enzyme activity. Trends Pharmacol. Sci. 2003;24:355–360. doi: 10.1016/S0165-6147(03)00158-5. [DOI] [PubMed] [Google Scholar]
- 35.Jonsson LM, Murray GJ, Sorrell SH, Strijland A, Aerts JF, Ginns EI, Barranger JA, Tager JM, Schram AW. Biosynthesis and maturation of glucocerebrosidase in Gaucher fibroblasts. Eur. J. Biochem. 1987;164:171–179. doi: 10.1111/j.1432-1033.1987.tb11008.x. [DOI] [PubMed] [Google Scholar]
- 36.Butters TD. Gaucher disease. Curr. Opin. Chem. Biol. 2007;11:412–418. doi: 10.1016/j.cbpa.2007.05.035. [DOI] [PubMed] [Google Scholar]
- 37.Beutler E, Gelbart T. Glucocerebrosidase (Gaucher disease) Hum. Mutat. 1996;8:207–213. doi: 10.1002/(SICI)1098-1004(1996)8:3<207::AID-HUMU2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 38.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dvir H, Harel M, McCarthy AA, Toker L, Silman I, Futerman AH, Sussman JL. X-ray structure of human acid-beta-glucosidase, the defective enzyme in Gaucher disease. EMBO Rep. 2003;4:704–709. doi: 10.1038/sj.embor.embor873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garman SC, Garboczi DN. The molecular defect leading to Fabry disease: structure of human alpha-galactosidase. J. Mol. Biol. 2004;337:319–335. doi: 10.1016/j.jmb.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 41.Dean KJ, Sweeley CC. Studies on human liver alpha-galactosidases. II. Purification and enzymatic properties of alpha-galactosidase B (alpha-N-acetylgalactosaminidase) J. Biol. Chem. 1979;254:10001–10005. [PubMed] [Google Scholar]
- 42.Dean KJ, Sweeley CC. Studies on human liver alpha-galactosidases. I. Purification of alpha-galactosidase A and its enzymatic properties with glycolipid and oligosaccharide substrates. J. Biol. Chem. 1979;254:9994–10000. [PubMed] [Google Scholar]
- 43.Bishop DF, Desnick RJ. Affinity purification of alpha-galactosidase A from human spleen, placenta, and plasma with elimination of pyrogen contamination. Properties of the purified splenic enzyme compared to other forms. J. Biol. Chem. 1981;256:1307–1316. [PubMed] [Google Scholar]
- 44.Mayes JS, Beutler E. alpha-galactosidase A from human placenta. Stability and subunit size. Biochim. Biophys. Acta. 1977;484:408–416. doi: 10.1016/0005-2744(77)90096-1. [DOI] [PubMed] [Google Scholar]
- 45.Choy FY, Woo M, Potier M. In situ radiation-inactivation size of fibroblast membrane-bound acid beta-glucosidase in Gaucher type 1, type 2 and type 3 disease. Biochim. Biophys. Acta. 1986;870:76–81. doi: 10.1016/0167-4838(86)90010-5. [DOI] [PubMed] [Google Scholar]
- 46.Dawson G, Ellory JC. Functional lysosomal hydrolase size as determined by radiation inactivation analysis. Biochem. J. 1985;226:283–288. doi: 10.1042/bj2260283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maret A, Potier M, Salvayre R, Douste-Blazy L. Modification of subunit interaction in membrane-bound acid beta-glucosidase from Gaucher disease. FEBS Lett. 1983;160:93–97. doi: 10.1016/0014-5793(83)80943-0. [DOI] [PubMed] [Google Scholar]
- 48.Hart DO, He S, Chany CJ, 2nd, Withers SG, Sims PF, Sinnott ML, Brumer H., 3rd. Identification of Asp-130 as the catalytic nucleophile in the main alpha-galactosidase from Phanerochaete chrysosporium, a family 27 glycosyl hydrolase. Biochemistry. 2000;39:9826–9836. doi: 10.1021/bi0008074. [DOI] [PubMed] [Google Scholar]
- 49.Premkumar L, Sawkar AR, Boldin-Adamsky S, Toker L, Silman I, Kelly JW, Futerman AH, Sussman JL. X-ray structure of human acid-beta-glucosidase covalently bound to conduritol-B-epoxide. Implications for Gaucher disease. J. Biol. Chem. 2005;280:23815–23819. doi: 10.1074/jbc.M502799200. [DOI] [PubMed] [Google Scholar]
- 50.Sandhoff K, Kolter T. Topology of glycosphingolipid degradation. Trends Cell. Biol. 1996;6:98–103. doi: 10.1016/0962-8924(96)80999-8. [DOI] [PubMed] [Google Scholar]
- 51.de Alba E, Weiler S, Tjandra N. Solution structure of human saposin C: pH-dependent interaction with phospholipid vesicles. Biochemistry. 2003;42:14729–14740. doi: 10.1021/bi0301338. [DOI] [PubMed] [Google Scholar]
- 52.Ho MW, O'Brien JS. Gaucher's disease: deficiency of ‘acid’ -glucosidase and reconstitution of enzyme activity in vitro. Proc. Natl. Acad. Sci. U S A. 1971;68:2810–2813. doi: 10.1073/pnas.68.11.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yam GH, Zuber C, Roth J. A synthetic chaperone corrects the trafficking defect and disease phenotype in a protein misfolding disorder. FASEB J. 2005;19:12–18. doi: 10.1096/fj.04-2375com. [DOI] [PubMed] [Google Scholar]
- 54.Fan JQ, Ishii S, Asano N, Suzuki Y. Accelerated transport and maturation of lysosomal alpha-galactosidase A in Fabry lymphoblasts by an enzyme inhibitor. Nat. Med. 1999;5:112–115. doi: 10.1038/4801. [DOI] [PubMed] [Google Scholar]
- 55.Asano N, Ishii S, Kizu H, Ikeda K, Yasuda K, Kato A, Martin OR, Fan JQ. In vitro inhibition and intracellular enhancement of lysosomal alpha-galactosidase A activity in Fabry lymphoblasts by 1-deoxygalactonojirimycin and its derivatives. Eur. J. Biochem. 2000;267:4179–4186. doi: 10.1046/j.1432-1327.2000.01457.x. [DOI] [PubMed] [Google Scholar]
- 56.Brumshtein B, Wormald MR, Silman I, Futerman AH, Sussman JL. Structural comparison of differently glycosylated forms of acid-beta-glucosidase, the defective enzyme in Gaucher disease. Acta Crystallogr. 2006;D62:1458–1465. doi: 10.1107/S0907444906038303. [DOI] [PubMed] [Google Scholar]
- 57.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- 58.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 59.Collaborative Computational Project Number 4 Acta Crystallogr. 1994;D50:760. [Google Scholar]
- 60.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 61.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. 2004;D60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 62.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. 2004;D60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 63.Freire E. Thermal denaturation methods in the study of protein folding. Methods Enzymol. 1995;259:144–168. doi: 10.1016/0076-6879(95)59042-0. [DOI] [PubMed] [Google Scholar]
- 64.Sanchez-Ruiz JM. Ligand effects on protein thermodynamic stability. Biophys. Chem. 2007;126:43–49. doi: 10.1016/j.bpc.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 65.Fukada H, Sturtevant JM, Quiocho FA. Thermodynamics of the binding of L-arabinose and of D-galactose to the L-arabinose-binding protein of Escherichia coli. J. Biol. Chem. 1983;258:13193–13198. [PubMed] [Google Scholar]
- 66.Manly SP, Matthews KS, Sturtevant JM. Thermal denaturation of the core protein of lac repressor. Biochemistry. 1985;24:3842–3846. doi: 10.1021/bi00336a004. [DOI] [PubMed] [Google Scholar]
- 67.Beutler E, Kuhl W. Purification and properties of human alpha-galactosidases. J. Biol. Chem. 1972;247:7195–7200. [PubMed] [Google Scholar]
- 68.Maret A, Salvayre R, Negre A, Douste-Blazy L. Properties of the molecular forms of beta-glucosidase and beta-glucocerebrosidase from normal human and Gaucher disease spleen. Eur. J. Biochem. 1981;115:455–461. [PubMed] [Google Scholar]
- 69.Alattia JR, Shaw JE, Yip CM, Prive GG. Molecular imaging of membrane interfaces reveals mode of beta-glucosidase activation by saposin C. Proc. Natl. Acad. Sci. U S A. 2007;104:17394–17399. doi: 10.1073/pnas.0704998104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jensen HH, Lyngbye L, Bols M. A Free-Energy Relationship between the Rate of Acidic Hydrolysis of Glycosides and the pK(a) of Isofagomines. Angew. Chem. Int. Ed. Engl. 2001;40:3447–3449. doi: 10.1002/1521-3773(20010917)40:18<3447::aid-anie3447>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 71.Eng CM, Guffon N, Wilcox WR, Germain DP, Lee P, Waldek S, Caplan L, Linthorst GE, Desnick RJ. Safety and efficacy of recombinant human alpha-galactosidase A--replacement therapy in Fabry's disease. N. Engl. J. Med. 2001;345:9–16. doi: 10.1056/NEJM200107053450102. [DOI] [PubMed] [Google Scholar]
- 72.Garman SC. Structural studies on a-GAL and a-GALAL: The atomic basis of Fabry and Schindler diseases. Biocatal. and Biotranform. 2006;24:129–136. [Google Scholar]
- 73.Ragone R. How the protein concentration affects unfolding curves of oligomers. Biopolymers. 2000;53:221–225. doi: 10.1002/(SICI)1097-0282(200003)53:3<221::AID-BIP1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 74.van Anken E, Braakman I. Versatility of the endoplasmic reticulum protein folding factory. Crit. Rev. Biochem. Mol. Biol. 2005;40:191–228. doi: 10.1080/10409230591008161. [DOI] [PubMed] [Google Scholar]
- 75.Wiseman RL, Powers ET, Buxbaum JN, Kelly JW, Balch WE. An adaptable standard for protein export from the endoplasmic reticulum. Cell. 2007;131:809–821. doi: 10.1016/j.cell.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 76.Street TO, Bolen DW, Rose GD. A molecular mechanism for osmolyte-induced protein stability. Proc. Natl. Acad. Sci. U S A. 2006;103:13997–14002. doi: 10.1073/pnas.0606236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zheng W, Padia J, Urban DJ, Jadhav A, Goker-Alpan O, Simeonov A, Goldin E, Auld D, LaMarca ME, Inglese J, Austin CP, Sidransky E. Three classes of glucocerebrosidase inhibitors identified by quantitative high-throughput screening are chaperone leads for Gaucher disease. Proc. Natl. Acad. Sci. U S A. 2007;104:13192–13197. doi: 10.1073/pnas.0705637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Landon MR, Lieberman RL, Hoang QQ, Kozakov D, Ju S, Orwig SD, Brenke R, Chuang G-Y, Beglov D, Vajda S, Petsko GA, D. R. Detection of non-catalytic binding hot spots on protein surfaces via fragment-based methods: application of targets of neurological disorders. J. Comp. Aid. Mol. Design accepted. 2009 doi: 10.1007/s10822-009-9283-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rumfeldt JA, Galvagnion C, Vassall KA, Meiering EM. Conformational stability and folding mechanisms of dimeric proteins. Prog. Biophys. Mol. Biol. 2008;98:61–84. doi: 10.1016/j.pbiomolbio.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 80.Balreira A, Gaspar P, Caiola D, Chaves J, Beirao I, Lima JL, Azevedo JE, Miranda MC. A nonsense mutation in the LIMP-2 gene associated with progressive myoclonic epilepsy and nephrotic syndrome. Hum. Mol. Genet. 2008;17:2238–2243. doi: 10.1093/hmg/ddn124. [DOI] [PubMed] [Google Scholar]