Abstract
Soluble epoxide hydrolase (sEH) is a multifunctional protein encoded by the EPHX2 gene. The biological functions and enzyme kinetics of sEH have been extensively investigated, however, little is known about its transcriptional regulation and mechanisms of tissue specific expression. Here, a luciferase gene based reporter assay was used to identify the minimal promoter and cis regulatory elements of EPHX2. The minimal promoter was identified as a GC-rich region between nts −374 to +28 with respect to the putative transcriptional start site. A reporter plasmid carrying this minimal promoter showed higher or similar activities in comparison to a reporter plasmid carrying nts −5,974 to +28 of EPHX2 in 9 human cell lines that were tested. Sp1 binding sites that are involved in augmenting the minimal promoter activity of EPHX2 were identified by nested deletion analysis, site-specific mutation, electrophoretic mobility shift assay, and chromatin immunoprecipitation assay.
Keywords: epoxide hydrolase, EPHX2, promoter, transcription
1. Introduction
Soluble epoxide hydrolase (sEH; E.C.3.3.2.3) is encoded by a gene, EPHX2, that is found on human chromosome 8, region 8p21-12 [1]. EPHX2 has 19 exons that encode a mature protein of 555 amino acid residues. In humans, sEH seems largely involved in the metabolism of epoxyfatty acids. An example is the hydration of epoxy-eicosatrienoic acids (EETs) to their dihydroxy eicosatrienoic acid derivatives (DHETs) [2, 3]. EETs are important signaling molecules derived from arachidonic acid through metabolism by cytochrome P450s (including CYP2C8, CYP2C9, and/or CYP2J2) [4–7]. EETs have biological roles in cardiovascular biology, renal function, inflammation response and pain [6, 8, 9]. The stabilization of EETs through the inhibition of sEH has beneficial effects on blood pressure, reduction of inflammation, and reduction of organ damage, and analgesia [10–16]. These results underline the importance of sEH in human health.
The biologically active form of sEH is a homodimer of two 62.5 kDa subunits [17]. sEH is a bifunctional enzyme with epoxide hydrolase activity (C-terminal region) that acts on epoxyfatty acids and a Mg2+-dependent phosphatase activity (N-terminal region) that acts on lipid phosphates [18, 19]. However, there may be other endogenous substrates, and the biological role of the phosphatase activity is unknown [19]. sEH activity is highest in mammalian liver and kidney [20–23], but it has also been found in many other tissues. In humans and rodents sEH expression occurs in liver, kidney, lung, heart, gut, brain, placenta, bladder, prostate, testis, spleen, skin, ovary, vascular endothelium, and some smooth muscle [13, 17, 24–32]. Although the distribution and tissue-enriched expression of sEH have been studied, the molecular mechanisms of sEH regulation are poorly understood. Several studies have shown that sEH can be regulated by endogenous chemical mediators and by some xenobiotics. 1) The sEH is induced by the administration of agonists of peroxisome proliferator activated receptor alpha (PPAR-alpha) in rodents [23, 31, 33]. Most organisms appear to respond to these agonists with a 2- to 3-fold increase in hepatic sEH activity. 2) Male mice show 55% and 283% higher sEH activities in liver and kidney, respectively, in comparison to female mice [31] suggesting the involvement of sex hormones in sEH regulation. Additionally, ovariectomy increases sEH activities in both the liver and kidney of mice suggesting a suppressive role of female sex hormones such as estrogen [31]. 3) sEH activity appears closely tied to the renin-angiotensin system. Angiotensin II (Ang II) appears to increase sEH protein levels and catalytic activity in rat renal cortical tissue [10]. Furthermore, Ang II has recently been shown to directly upregulate EPHX2 expression [34]. 4) In humans, cigarette smoke transiently reduces sEH activity in the lung [24]. 5) Exposure to gamma radiation (12 h) increases the levels of EPHX2 mRNAs in human cell lines [35]. This increase appears to result from NFκB induction. Interestingly, a splice variant of sEH that is found in mouse ovary shows differences in epoxide hydrolase activity [32]. In this splice variant, the first 44 residues of the N-terminal region are encoded by sequences that form part of the 2nd intron of the mouse Ephx2 gene. This splice variant seems to have a role in ovarian function in mice. To our knowledge, such a splice variant has not been found for the human EPHX2 gene.
In this study, we characterize the transcriptional regulation of EPHX2 by transient reporter assay, electrophoretic mobility shift assay (EMSA), and chromatin immunoprecipitation (ChIP) assay. A luciferase-based reporter assay was developed and used to determine the minimal promoter of EPHX2 as well as enhancing and inhibitory regions that are found upstream of EPHX2 in 9 different human cell lines. The identification of responsive elements within the promoter of EPHX2 or cellular transcription factors that specifically interact with these responsive elements may lead to novel drug targets for high blood pressure, inflammation, and other diseases that are associated with sEH.
2. Materials and methods
2.1. In silico analysis and 5′-prime rapid amplification of cDNA ends (5′-RACE)
Sequence information of the 5′-flanking region of the EPHX2 gene was obtained from the human genome database at the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/). EPHX2 is located on chromosome 8 of the assembled sequence NT_023666. The computer programs Database of Transcriptional Start Sites (DBTSS, http://dbtss.hgc.jp/) and Dragon GC+ Promoter Finder (http://research.i2r.a-star.edu.sg/DRAGON/) were used to identify the putative transcriptional start site of EPHX2. The GC content of the 5′-flanking region (nts −6,000 to +500 with respect to the putative transcriptional start site) of EPHX2 was calculated on the basis of a 200 nts-long, running window. The computer program CpG Island Searcher (http://www.cpgislands.com/) was used to identify CpG islands within the 5′-flanking region (nts −10,000 to +10,000) of EPHX2. The computer program Transfac (http://www.gene-regulation.com/) was used to identify potential binding sites of transcription factors.
One μg of human liver total RNA (Stratagene) was used as a template to produce 5′-terminal enriched cDNAs using a Smart cDNA Synthesis kit (Clontech). Universal primer A Mix, supplied in the kit, and a gene specific reverse primer (5′-TCCGGTCACTTTCTCCAGTTC-3′) were used to amplify the 5′-terminous of EPHX2. The amplification was performed as follows: 94°C for 1 min, 63°C for 1 min, 68°C for 1 min for 25 cycles with KOD Hot Start DNA Polymerase (Novagen). The PCR product was ligated into the pCR Blunt II-TOPO vector (Invitrogen), and the sequences of the cDNA inserts of 20 randomly isolated plasmids were determined (UC Davis Division of Biological Sciences Sequencing Facility).
2.2. Cell cultures
The human cell lines used in this study are described in Table 1. Most of these cell lines were generously provided as gifts. DU-145 [36] was obtained from Alan Epstein of the Keck School of Medicine, Univ. of Southern California, Los Angeles, CA, USA and Gerald DeNardo of the Univ. of California Health Center, Sacramento, CA, USA. SW480 [37]; Caco-2 [38]; 293T [39] and HeLa [40]; and HuH-7 [41], ACHN [42], and SN12C [43] were obtained from Ken Kaplan; Bo Lonnerdal; Martin L. Privalsky; and Robert Weiss respectively, all from the Univ. of California, Davis, CA, USA. HepG2 [44] was purchased from ATCC (Manassas, VA, USA). All of the cell lines were cultured in RPMI1640 medium (Mediatech) supplemented with 10% fetal bovine serum (v/v) (BioWhittaker) under a 5% CO2 atmosphere at 37°C. The cells were passaged at 90–100% confluency by washing with PBS and trypsinization (0.05% trypsin, 0.53 mM EDTA) for 3–5 min at 37°C prior to subculture. This trypsinization procedure was also used to harvest cells for the luciferase-based reporter assay and sEH activity assay. Following trypsinization, the cell suspension was centrifuged at 400 × g for 5 min at 5°C, and resuspended in fresh RPM1640 medium.
Table 1.
sEH transcription and expression and transfection efficiency in human cell lines
| Cell Line | Ref. | Origin | Disease | sEH specific activity (pmol/min/mg)a | Relative EPHX2 mRNA (EPHX2/GAPDH)a | Renilla luciferase activitya (RLU/1×104 cells)b |
|---|---|---|---|---|---|---|
| HepG2 | [44] | liver | carcinoma | 310±50 | 0.18±0.03 | 600±30 |
| HuH-7 | [41] | 940±40 | 0.99±0.20 | 130±30 | ||
|
| ||||||
| 293T | [39] | kidney | nonec | 210±10 | 0.54±0.08 | 98000±11000 |
| ACHN | [42] | adenocarcinoma | 31±15 | 0.38±0.04 | 30±20 | |
| SN12C | [43] | carcinoma | n.d.d | 0.08±0.02 | 240±60 | |
|
| ||||||
| Caco-2 | [38] | colon | adenocarcinoma | 120±10 | 0.33±0.02 | 780±70 |
| SW480 | [37] | 130±30 | 0.68±0.08 | 7000±700 | ||
|
| ||||||
| DU 145 | [36] | prostate | carcinoma | 440±10 | 0.39±0.03 | 9500±400 |
|
| ||||||
| HeLa | [40] | cervix | adenocarcinoma | 52±15 | 0.14±0.05 | 3400±200 |
The control pRL-CMV plasmid contains a luciferase gene from Renilla reniformis under the control of a CMV promoter.
Values are mean ± SD from three different experiments.
Human cell line, derived from embryo kidney, expressing SV40 large T antigen abundantly.
n.d. indicates not detectable under the condition of our assay.
2.3. Generation of reporter plasmid constructs containing the 5′-flanking region of EPHX2
In order to generate the reporter plasmids, nts −5,974 to +28 (−5974/+28 construct), nts −5,974 to −325 (−5974/−325 construct), and nts −374 to +28 (−374/+28 construct) of the 5′-flanking region of EPHX2 were amplified by PCR and cloned immediately upstream of the firefly luciferase gene in the pGL3-Basic reporter plasmid (Promega). PCR was performed with KOD Hot Start DNA Polymerase in the presence of 5% DMSO according to the manufacturer’s protocol. Genomic template DNAs were isolated from 1 ×107 HepG2 cells following standard procedures [45]. Forward and reverse primers (F1: 5′-CGCGGTACCCAAGGAGGGGAGAGAACACTGAGCATT-3′, F2: 5′-CGCGGTACCGCATTCCAAGTCCAGCAAGT-3′, R1: 5′-CGCCTCGAGACGCAGCTAACCTGGGAGAT-3′, and R2: 5′-CGCCTCGAGCTTCCTGGACGGACTGACA-3′) were synthesized (Sigma Genosys) on the basis of the sequence of the EPHX2 gene (GeneBank accession no. NM_001979). Restriction endonuclease recognition sites (underlined) for KpnI and XhoI were incorporated into the forward and reverse primers, respectively. Following PCR, the products were digested with KpnI and XhoI and inserted immediately upstream (at the KpnI and XhoI sites) of the firefly luciferase gene of pGL3-Basic. The −5974/+28, −5974/−325, and −374/+28 constructs were generated using the primer sets F1+R1, F1+R2, and F2+R1, respectively. Additional reporter plasmids, containing nested deletions or a point mutation of the −374 to +28 nts region of EPHX2 were generated by PCR using the template DNAs and primers described in Table 2. The authenticity of each of the constructs was confirmed by nucleotide sequencing.
Table 2.
Primers and templates used to generate the reporter plasmids
| Primer (5′ - 3′) | ||
|---|---|---|
| F3: GGGCATAAGAGACTTTGGACTTT | ||
| F4: CGTTACATCCAGAGGCGAGA | ||
| F5: TCTGACTCCCTTTCCTGTGC | ||
| F6: TTTCCTGTGCCCCTCCCCC | ||
| F7: CCCCTCCCCCTGCCTCTTTC | ||
| F8: TGCCTCTTTCCCGGCCAGAG | ||
| F9: GCCAGAGTCCAGCCTTAACC | ||
| F10: CCGGGCAGAGGGCGGAGTC | ||
| F11: CCCGTTAAGGGGGTGTGGGG | ||
| F12: AGGCGGGGCCAGGGCAGG | ||
| F13: GGGCAGAGCCGGGCCAAG | ||
| F14: GCTGGGCGGGTCATGCGC | ||
| F15: TGGCCTTCGCGCATCTCCC | ||
| F16: GGGGGTGTGGGGAGGGGGCAGAGCCGGGCCAAG | ||
| F17: TCGGGGCAGAGCCGGGCC | ||
| R3: GGTACCTATCGATAGAGAAATGTTCTGGC | ||
| R4: GTTAAGGCTGGACTCTGGCC | ||
| R5: CCTCCCCACACCCCCTTAAC | ||
| R6: TTAACGGGGTTAAGGCTGGACTCTGGCC | ||
| R7: ACCTGCCCTGGCCCCGCC | ||
| Construct | Primer set | Template used |
|
| ||
| −272/+28 | F3+R3 | −374/+28 |
| −230/+28 | F4+R3 | −374/+28 |
| −165/+28 | F5+R3 | −374/+28 |
| −155/+28 | F6+R3 | −374/+28 |
| −146/+28 | F7+R3 | −374/+28 |
| −136/+28 | F8+R3 | −374/+28 |
| −123/+28 | F9+R3 | −374/+28 |
| −104/+28 | F10+R3 | −374/+28 |
| −86/+28 | F11+R3 | −374/+28 |
| −63/+28 | F12+R3 | −374/+28 |
| −41/+28 | F13+R3 | −374/+28 |
| −24/+28 | F14+R3 | −374/+28 |
| −4/+28 | F15+R3 | −374/+28 |
| −374/+28Δ(−104/−87) | F11+R4 | −374/+28 |
| −374/+28Δ(−63/−42) | F13+R5 | −374/+28 |
| −374/+28Δ(−104/−87,−63/−42) | F16+R6 | −374/+28 |
| −374/+28Δ(−104/−42) | F13+R4 | −374/+28 |
| −63/+28TT | F17+R7 | −63/+28 |
| −374/+28TT | F17+R7 | −374/+28 |
2.4. Luciferase activity assay
The human cell lines used in these assays are described above and in Table 1. Cells from each of the human cell lines were seeded into 24-well tissue culture plates (Falcon) at a density of 5×104 cells/well, allowed to attach and recover for 24 h, and then transfected with 200 ng of each plasmid using FuGENE6 transfection reagent (Roche). All of the transfections were carried out according to the protocol provided by the manufacturer using 0.6 μl of the transfection reagent per well. For each transfection, 4 ng of a control plasmid, pRL-CMV (Promega), expressing luciferase of Renilla reniformis (renilla luciferase) under control of a cytomegalovirus (CMV) promoter, was added as an internal control reporter. After a 48 h-long incubation at 37°C, the cells were lysed with 100 μl/well (500 μl/well for 293T) of Triton Lysis Buffer (TLB: 100 mM potassium phosphate, pH 7.8; containing 0.2% Triton X-100). The luciferase assay was carried out by means of the Dual-Glo Luciferase Reporter Assay System (Promega) using 20 μl of the cell lysate. Luciferase activities were determined using a Spectra Fluor Plus microplate reader (TECAN). Firefly luciferase activity was normalized with respect to renilla luciferase activity, and shown as relative luciferase activity (relative luciferase activity = firefly luciferase activity/renilla luciferase activity). The transfection efficiency (using pRL-CMV) of each cell line is shown as relative light units (RLU) of renilla luciferase per 1 × 104 cells (Table 1). The Pearson correlation coefficient was used to determine statistically significant correlations. This coefficient was determined using the computer program R (http://www.r-project.org/).
2.5. sEH activity assay
sEH was released from 1 × 106 cells by sonication (Sonic Dismembrator Model 100, Fisher Scientific) for 3 sec in 500 μl of cell lysis buffer (20 mM sodium phosphate, pH 7.4; 5 mM EDTA; 1 mM DTT; 1 mM PMSF; and 0.01% Tween-20). Cell debris was removed by centrifugation (10,000 × g) for 15 min at 4°C and the supernatant was stored at −80°C until used. sEH activity was measured in a 100 μl reaction volume containing 0.05 mM of racemic [3H]-trans-1,3-diphenylpropene oxide (t-DPPO) as substrate as described previously [46]. The reaction mixture was incubated at 30°C for 30 min. The reaction was then quenched by the addition of 60 μl of methanol, and 200 μl of isooctane was added in order to extract the remaining epoxide from the aqueous phase. Parallel reactions were performed in which 1-hexanol (200 μl) was used for extraction (instead of the isooctane) in order to measure potential glutathione S-transferase activity which can also transform the substrate [46]. Diol (or glutathione conjugate) in the aqueous phase was quantified using a scintillation counter (Wallac Model 1409, Wellesley, MA).
2.6. Semi-quantitative RT-PCR
The cells were released from the washed culture dish by use of trypsin (0.05% trypsin, 0.53 mM EDTA) and subsequently washed three times with PBS (centrifugation at 1,000 × g for 5 min at 5°C). Total RNA was extracted from the cells using an RNeasy Mini kit (Qiagen). Total RNA (1 μg) was converted to cDNA using Superscript II reverse transcriptase (Invitrogen). The primers and the PCR conditions employed are as follows: GAPDH: 5′-CAGCCTCAAGATCATCAGCA-3′ (sense) and 5′-TTCTAGACGGCAGGTCAGGT-3′ (antisense), EPHX2: 5′-GTGTTCATTGGCCATGACTG-3′ (sense) and 5′-CTCAGTGACCATCCTGCTGA-3′ (antisense); 28 cycles composed of steps at 95°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec. The products of the PCR were separated on a 2% agarose gel containing ethidium bromide and visualized with UV light. Relative mRNA abundance was quantified by densitometry using ImageJ software (http://rsb.info.nih.gov/ij/) with normalization to GAPDH (EPHX2/GAPDH).
2.7. Electrophoretic mobility shift assay (EMSA)
EMSA was used to measure the ability of nuclear extracts to bind to nts −54 to −33 (Oligo 1: 5′-CAGGGCAGGGGCGGGGCAGAGC-3′), −74 to −47 (Oligo 2: 5′-GTGTGGGGAGGAGGCGGGGCCAGGGCAG-3′), −86 to −64 (Oligo 3: 5′-CCCGTTAAGGGGGTGTGGGGAGG-3′), or −102 to −80 (Oligo 4: 5′-GGGCAGAGGGCGGAGTCCCGTTA-3′) of the 5′-flanking region of EPHX2. The single-stranded oligonucleotides (Oligos 1 to 4 above) were labeled at the 3′-end with biotin (3′ End Labeling kit, Pierce), then annealed to their respective antisense strands to form labeled double-stranded DNAs. The binding reaction (20 min at room temperature) was carried out in a final volume of 20 μl of binding buffer (10 mM Hepes, pH 7.5, 2.5 mM MgCl2, 50 mM NaCl, 0.5 mM DTT, 4% glycerol, 1 μg of poly dI-dC, 1 μg of BSA) containing the biotin-labeled probe (20 fmol), with or without nuclear extract from the 293T cell line. The nuclear extract was prepared with NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce) according to the manufacture’s protocol. In the supershift experiments, the nuclear extract was preincubated with anti-Sp1 antibody (Santa Cruz Biotechnology) for 30 min on ice. DNA-nuclear extracts and/or anti-Sp1 antibody complexes were separated on 6% nondenaturing polyacrylamide gels and visualized by the LightShift Chemiluminescent EMSA kit (Pierce) following the manufacturer’s protocol.
2.8 Chromatin immunoprecipitation (ChIP) assay
ChIP assays were carried out using a ChIP assay kit (Upstate Biotechnology) according to the manufacturer’s protocol. 293T cells (1 × 106 cells) were seeded into 10 cm-diameter tissue culture plates (Falcon) then incubated for 3 days after which the cells were crosslinked with 1% formaldehyde at 37°C for 10 min. Following fixation the cells were rinsed with ice-cold PBS containing 1x complete, EDTA-free, protease inhibitor cocktail (Roche), then detached from culture surface using a Teflon policeman, and then centrifuged (400 × g for 4 min at 4°C) to pellet the cells. SDS lysis buffer (400 μl) was added to the cell pellet and the cells were lysed by incubation for 10 min on ice. The lysate was sonicated 4 times for 10 sec (Sonic Dismembrator Model 100, Fisher Scientific) in order break the chromosomal DNA into fragments of less than 1 kb. This solution was then centrifuged using a microcentrifuge (10,000 × g for 10 min at 4°C) and the supernatant was diluted 10-fold in ChIP dilution buffer. The diluted supernatant (20 μl) was used as the “Input” for immunoprecipitation and subsequent PCR.
Immunoprecipitation was performed overnight at 4°C with 1 μl of anti-Sp1 antibody (2 μg/μl) or 2 μl of control rabbit IgG (1 μg/μl). Immuno-complexes were isolated from the overnight immunoprecipitation reaction by the addition of 30 μl of a salmon sperm DNA/protein A agarose slurry (supplied in the kit), and incubation for 1 h at 4°C. The precipitates were washed sequentially with 1 ml each of low-salt wash buffer, high-salt wash buffer, and LiCl wash buffer, then twice with TE buffer. The immuno-complexes were eluted by adding 500 μl of elution buffer (1% SDS, 0.1 M NaHCO3) and incubation for 15 min at room temperature. In order to remove the DNA crosslinks, 20 μl of 5 M NaCl was added to the eluates and the solution was heated at 65°C for 6 h. Following heating 20 μl of 1 M Tris-HCl (pH 6.5) and 2 μl of 10 mg/ml proteinase K, were added and the solution was incubated at 45°C for 1 h. The released DNA fragments were purified with a Gel Extraction kit (Qiagen) in a final volume of 50 μl. The DNA fragments were used as a template for PCR using forward (5′-GAGATTCTAGCCTGGGGTCC-3′) and reverse (5′-ACGCAGCTAACCTGGGAGAT-3′) primers that amplified the −214 to +28 nts region of the 5′-flanking region of EPHX2. PCR was performed following the semi-quantitative RT-PCR method described above for 28, 33 or 38 cycles of amplification. The resulting DNA was resolved on a 1% agarose gel and stained with ethidium bromide.
3. Results
3.1. Sequencing of the −5974/+28 construct
The nucleotide sequence of the 5′-flanking region of EPHX2 from the HepG2 cells (in the −5974/+28 construct) was nearly identical to that found on human chromosome 8 nts 5,716,979 to 5,722,980 (GenBank accession no. NT_023666). There were three differences between our sequence and the NT_023666 sequence. Our sequence had an extra CTCTT repeat at nt 5,719,222, and lacked T and CACACACA sequences at nts 5,721,164 and 5,722,557, respectively.
3.2. Putative transcriptional start site of EPHX2
The transcriptional start site of EPHX2 was predicted by two methods: in silico analysis of the 5′-flanking region (to 5,974 nts upstream) and 5′-RACE. The Dragon GC+ Promoter Finder program predicted transcription initiation at the first T in the sequence CTTCGC, i.e., nt 5,722,954 of the NT_023666 sequence. In contrast, DBTSS analysis of 210 human sEH-encoding ESTs showed that transcription initiates at the first C (nt 5,722,953) in the CTTCGC sequence in 40 of these ESTs (~20%); whereas, transcription initiates at the first T in CTTCGC in only 2 of the 210 ESTs (<1%). In comparison, sequencing of 5′-RACE products identified numerous 5′-prime ends, which corresponded to nts 5,722,945, 5,722,957, 5,722,968, 5,722,973, 5,722,977, 5,722,981, 5,722,997, and 5,722,999 of the NT_023666 sequence. Consistent with these findings, multiple transcriptional start sites have been shown to be associated with a TATA-less, GC-rich sequences [47, 48]. In this study, transcription initiation was predicted to occur at the first C of the CTTCGC sequence (nt 5,722,953) of the 5′-flanking region of EPHX2.
3.3. Relative transcriptional activity of the 5′-flanking region of EPHX2
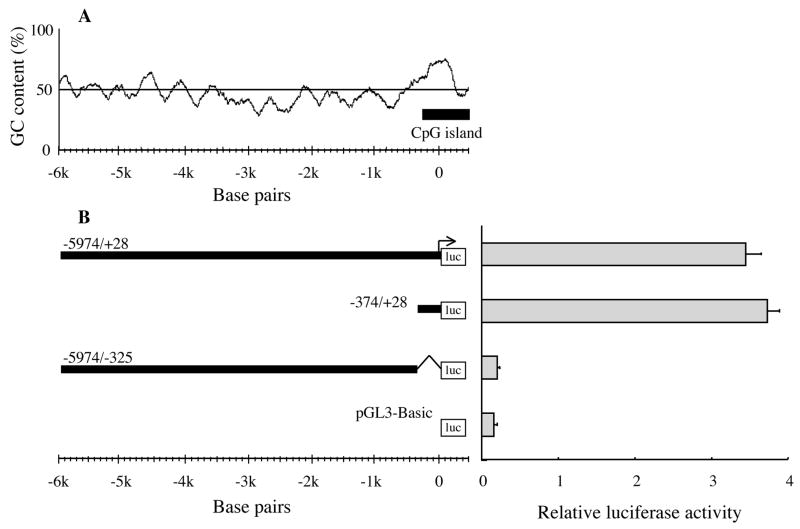
The in silico analysis of the 5′-flanking region (to 5,974 nts) of EPHX2 did not identify a consensus TATA box or CAAT sequences. However, the in silico analysis did identify a 751 nts-long region (nts −344 to +407) with high GC content (~64%) near the predicted transcriptional start site (Fig. 1). A CpG island was also identified around nts −291 to +507 (Fig. 1A). In order to test the in silico predictions, a transient expression assay based on a reporter firefly luciferase gene was established. In the human liver cancer-derived HepG2 cell line, the reporter plasmid carrying nts −374 to +28 of EPHX2 (−374/+28 construct) gave the same relative luciferase activity as a plasmid carrying nts −5,974 to +28 (−5974/+28 construct) indicating that the −374 to +28 sequence was the minimal essential promoter (Fig. 1B). However, the relatively low transfection efficiency of HepG2 cells (i.e., only 1/160th the efficiency of 293T cells, see below) made it difficult to measure relative luciferase activity when reporter plasmids carrying DNA fragments shorter than the −374 to +28 region were tested.
Figure 1.
GC-content and putative minimal essential promoter analysis of the 5′-flanking region of EPHX2. (A) GC-content (%) of the 5′-flanking region of EPHX2 from 6,000 nts upstream to 500 nts downstream of the putative transcriptional start site. A moving window of 200 nts was used to calculate the GC content. The bar indicates a putative CpG island (−291 to +507). (B) The minimal promoter sequence for expression of a reporter firefly luciferase (luc) gene in HepG2 cells was found from 374 nts upstream to 28 nts downstream (−374/+28 construct) of the putative transcriptional start site of EPHX2. The −5974/−325 construct lacks 352 nts (from 324 nts upstream to 28 nts downstream) of the −5974/+28 construct. The pGL3-Basic construct contains no EPHX2-derived sequence. The transcriptional activity of these constructs is shown in terms of relative luciferase activity (i.e., firefly luciferase activity/control renilla luciferase activity). Error bars indicate the standard deviation of three independent experiments. The arrow indicates the putative transcriptional start site of EPHX2.
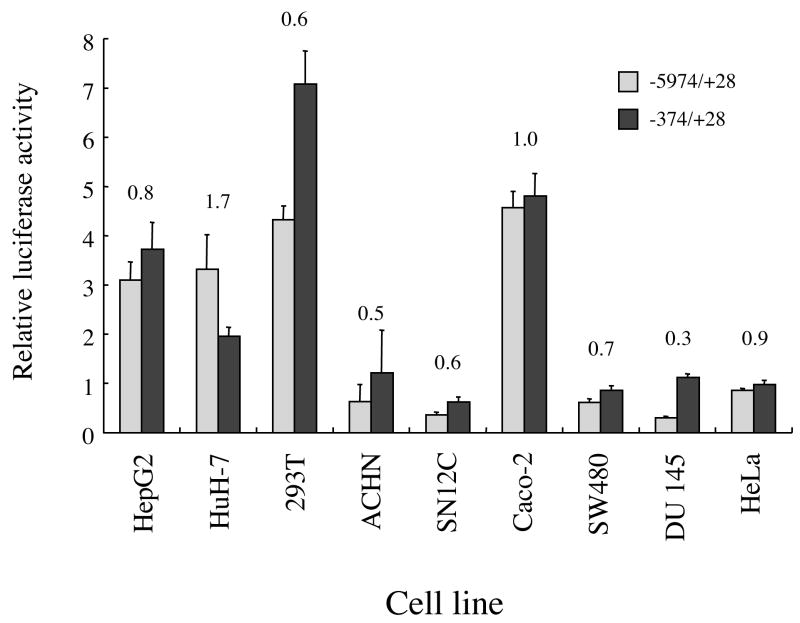
In order to identify a cell line which supported higher transfection efficiency and transcriptional activity, 9 human cell lines (Table 1) were screened. Initially, endogenous sEH activity and relative mRNA abundance of EPHX2 (Table 1) were measured in these cell lines under the assumption that high sEH activity would directly correlate to high levels of EPHX2 mRNAs. A good correlation (r = 0.73, p<0.05) was found between sEH activity and relative mRNA abundance. All of the cell lines tested showed lower sEH activities in comparison to that reported from a S9 fraction prepared from human kidney using the same t-DPPO substrate (1,200 to 5,500 pmol/min/mg) [21]. Of the 9 cell lines tested, HuH-7 showed the highest sEH activity (940 pmol/min/mg) and relative mRNA abundance (EPHX2/GAPDH = 0.99). sEH activity was not detected in the SN12C cell line under the conditions used in this study, even though the cell line showed low levels of relative mRNA abundance of EPHX2 (Table 1).
The ability of the −5974/+28 and −374/+28 constructs to induce relative luciferase activity was tested in the 9 human cell lines in which sEH activity was screened (Fig. 2). No correlation was found between sEH activity and relative luciferase activity of the −5974/+28 and −374/+28 constructs {r = 0.32 (p = 0.4) and 0.04 (p = 0.9), respectively}. There was, however, a weak correlation between sEH activity and the ratio of the relative luciferase activity of the −5974/+28 construct with respect to the −374/+28 construct {(−5974/+28)/(−374/+28)} (r = 0.66, p = 0.05). The shorter −374/+28 construct showed the same or higher relative luciferase activity in comparison to the longer −5974/+28 construct in 8 of the 9 cell lines tested (Fig. 2). This suggested that the −5974 to −375 region may contain “inhibitory sequences”. In contrast, the −5974/+28 construct showed 1.7-fold higher relative luciferase activity in comparison to the −374/+28 construct in the HuH-7 cells.
Figure 2.
Relative luciferase activity of two reporter constructs carrying 6.0 kb (−5974/+28 construct) or 0.4 kb (−374/+28 construct) of the 5′-flanking region of EPHX2 in 9 human cell lines (described in Table 1). The results are expressed as relative luciferase activity (i.e., firefly luciferase activity/control renilla luciferase activity). The numbers above the bars indicate the ratio of the relative luciferase activity of the −5974/+28 construct divided by that of the −374/+28 construct. Error bars indicate the standard deviation of three independent experiments.
The transfection efficiency (shown as RLU of renilla luciferase per 1 × 104 cells in Table 1) of the pRL-CMV plasmid was more than 10-fold higher in 293T cells than in the 8 other cell lines tested. Transfection efficiency in the 293T cell line was about 160-fold higher than that of HepG2 cells.
3.4. Transcriptional analysis of the GC-rich region of the 5′-flanking region of EPHX2
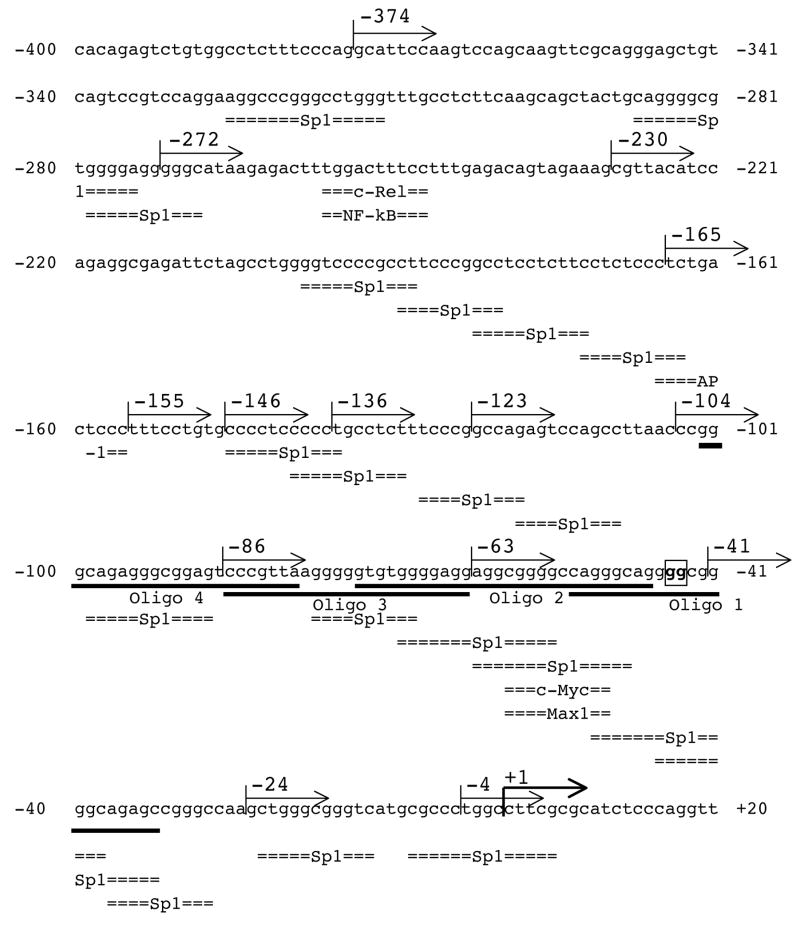
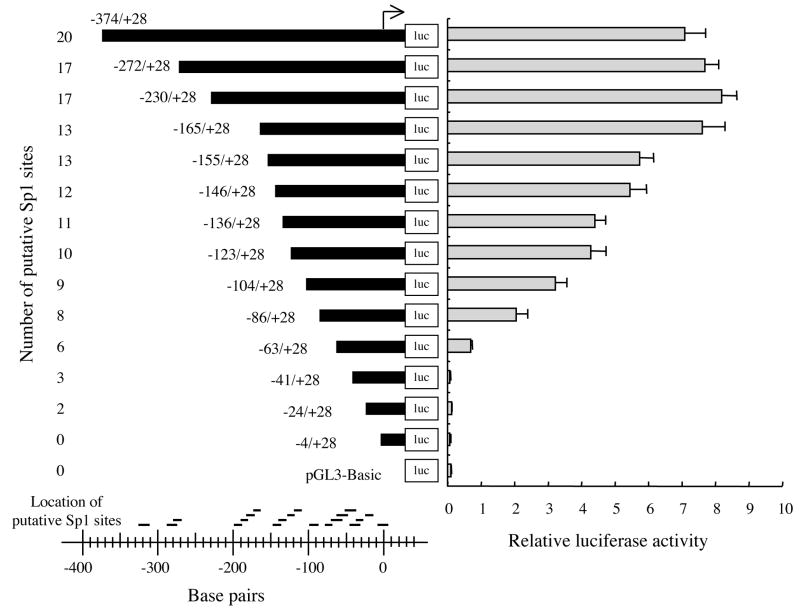
The in silico analysis identified numerous potential cis-elements within the GC-rich region (nts −400 to +20) of the 5′-flanking region of EPHX2 (Fig. 3). These potential regulatory elements include c-Rel/NFκB, AP-1, and cMyc/Max1 elements, as well as 20 potential Sp1 binding sites. Sp1 sites are also referred to as GC boxes [49–51]. In order to identify functional regulatory elements within this region, nested deletions were generated and the ability of the resulting fragments to activate the firefly luciferase gene in pGL3-Basic was determined in 293T cells (Fig. 4). Reporter plasmids carrying nts −374 to −165 of the 5′-flanking region of EPHX2 showed identical relative luciferase activities. Relative luciferase activity gradually declined in reporter plasmids carrying shorter lengths of the 5′-flanking region (from nts −165 to −63). Relative luciferase activity was not detected in reporter plasmids containing less than 63 bp of the 5′-flanking region. Statistically significant (p<0.01) differences were found in reporter plasmids carrying deletions of nts −165 to −155 (AP-1), −104 to −86 (Sp1), −86 to −63 (Sp1), and −63 to −41 (Sp1s and cMyc/Max1). Potential regulatory elements found in these regions are indicated within the parentheses.
Figure 3.
Nucleotide sequence of the 5′-flanking region of EPHX2 from −400 nts upstream to 20 nts downstream of the putative transcriptional start site. The sequence corresponds to nts 5,722,553 to 5,722,972 of human chromosome 8 (GenBank accession no. NT_023666). The putative transcriptional start site C is marked by +1 and indicated by a bold arrow. Numbers to the left and right indicate nucleotides upstream (−) and downstream (+) of the putative transcriptional start site. Putative transcription factor binding sites (Sp1, c-Rel/NFκB, AP-1, and c-Myc/Max1) are indicated below the sequence. The 5′ ends of the nested deletions (−272 to −3) used in the promoter analysis (see Fig. 4) are indicated by arrows above the sequence. The underlined text indicates the oligonucleotides (Oligos 1 to 4) used for EMSA in Fig. 7A. The GG to TT mutation site of the −374/+28TT and −63/+28TT constructs is boxed and indicated by bold.
Figure 4.
Relative luciferase activity following transfection of human 293T cells with reporter plasmids carrying nested deletions of the 5−-flanking region (from 374 nts upstream to 28 nts downstream) of the putative transcriptional start site of EPHX2. The nucleotide sequence of the 5′ ends of the nested deletions are shown in Fig. 3. The results are expressed as the relative luciferase activities (i.e., firefly luciferase activity/control renilla luciferase activity). Error bars indicate the standard deviation of three independent experiments. The number and relative locations of the putative Sp1 binding sites within are shown to the left and bottom, respectively. The arrow indicates the putative transcriptional start site.
3.5. Transcriptional regulation of the GC-rich region of the 5′-flanking region of EPHX2
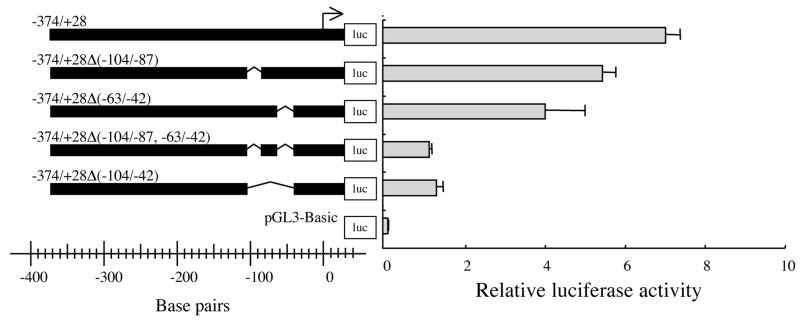
Reporter plasmids carrying nested deletions of the 5′-flanking region of EPHX2 indicated that the −165 to +28 nts region is important for transcriptional activity (Fig. 4). In order to test whether specific regions within the −165 to +28 nts region were important for full promoter activity, deletion of nts −104 to −87 or −63 to −42 from the −374/+28 construct were generated (Fig. 5). The −374/+28Δ(−104/−87) and −374/+28Δ(−63/−42) constructs resulted in 20% and 40% reductions, respectively, of relative luciferase activity in comparison to the −374/+28 construct. Deletions of both regions, the −374/+28Δ(−104/−87, −63/−42) construct, resulted in an 80% drop in relative luciferase activity. Deletion of the entire −104 to −42 region −374/+28Δ(−104/−42) construct, also showed an approximately 80% drop in relative luciferase activity (Fig. 5).
Figure 5.
Relative luciferase activity was measured following transfection of human 293T with reporter plasmids carrying various deletions of the −374/+28 construct {−374/+28Δ(−104/−87), −374/+28Δ(−63/−42), −374/+28Δ(−104/−87, −63/−42), and −374/+28Δ(−104/−42)}. The results are expressed as relative luciferase activity (i.e., firefly luciferase activity/control renilla luciferase activity). Error bars indicate the standard deviation of three independent experiments. The arrow indicates the putative transcriptional start site.
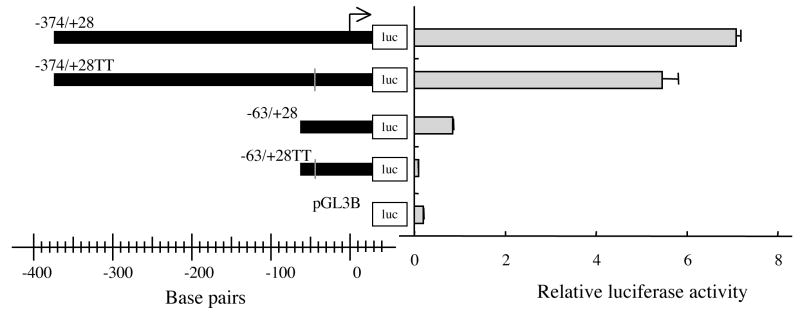
The results of luciferase reporter assay (Figs. 4 and 5) indicated that the −63 to −42 nts region is essential for EPHX2 transcription. This 22 nts-long region contained or overlapped six potential regulatory elements including four Sp1, one c-Myc, and one Max1 sites (Fig. 3). Only the putative Sp1 site that starts at nt −47 showed complete identity to the Sp1 consensus sequence GGGGCGGGGC [52]. Thus, reporter constructs, −63/+28TT and −374/+28TT, in which the conserved Sp1 site was mutated from GGGGCGGGGC to GGTTCGGGGC were tested in the luciferase reporter assay. No relative luciferase activity (p<0.01) was observed with the −63/+28TT construct, whereas, the −374/+28TT construct showed a 23% reduction in relative luciferase activity (p<0.01) (Fig. 6). The transcriptional activity of the −374/+28TT construct (5.4 ± 0.4) was slightly higher than that of the −374/+28Δ(−63/−42) construct (4.3 ± 0.7), in which the Sp1 consensus sequence was completely removed (Fig. 5).
Figure 6.
Mutational analysis of the −374 to +28 and −63 to +28 nts regions of the 5′-flanking region of EPHX2. Relative luciferase activity following transfection of human 293T with reporter plasmids carrying GG to TT mutations of the −374/+28 (−374/+28TT) and −63/+28 (−63/+28TT) constructs. The mutations are described in the text and shown in Fig. 3. The results are expressed as relative luciferase activity (i.e., firefly luciferase activity/control renilla luciferase activity). Error bars indicate the standard deviation of three independent experiments. The arrow indicates the putative transcriptional start site.
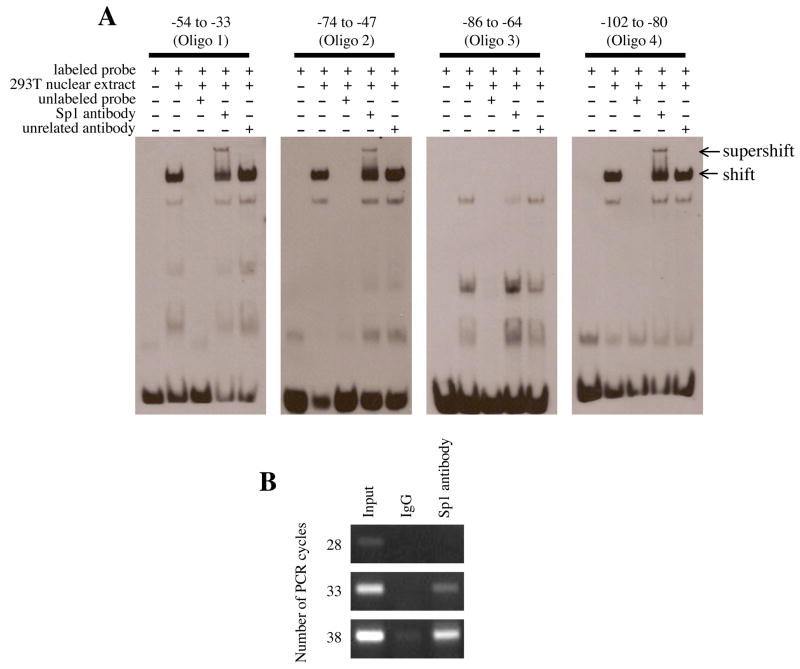
3.6. EMSA and ChIP assay
Electrophoretic mobility assays were performed in order to test whether the putative Sp1 sites that are located within nts −102 to −33 of the 5′-flanking region of EPHX2 can bind to proteins in the nuclear extract of 293T cells (Fig. 7A). On the basis of the supershift with anti-Sp1 antibody, Sp1 in the nuclear extract of 293T cells bound to nts −54 to −33 (Oligo 1), −74 to −47 (Oligo 2), and −102 to −80 (Oligo 4). However, Sp1 appeared not to bind to a DNA fragment containing nts −86 to −64 (Oligo 3). Additionally, ChIP assays were performed to further test whether Sp1 binds to the EPHX2 flanking region (Fig. 7B). The chromosomal DNA-protein complexes from 293T cells were specifically recognized by the anti-Sp1 antibody, and the subsequent PCR using a pair of oligonucleotide primers that targeted the −214 to +28 region of the EPHX2 confirmed that Sp1 was binding to the Sp1 binding site within this region.
Figure 7.
EMSA and ChIP assay of the 5′-flanking region of EPHX2. (A) The electrophoretic mobility shift assay was performed using biotin-labeled probes (Oligo 1: −54 to −33, Oligo 2: −74 to −47, Oligo 3: −86 to −64, and Oligo 4: −102 to −80). Following incubation with nuclear extract and/or anti-Sp1 antibody, the reaction was separated by 6% native PAGE. A 100-fold molar excess of unlabeled probe was used in the competition assays. Arrows at the right indicate DNA-nuclear extract complex (shift) or DNA-nuclear extract- anti-Sp1 antibody complex (supershift). (B) The chromatin immunoprecipitation assay was performed using chromosomal DNA fragments (Input) from 293T cells. Immunoprecipitation was performed with control IgG or anti-Sp1 antibody. Following heat and proteinase K treatment, the released chromosomal DNA fragments were used as template for PCR using primers that specifically amplified the −214 to +28 region of the 5′-flanking region of EPHX2. The number of PCR cycles is shown to the left.
4. Discussion
Soluble epoxide hydrolase is a key enzyme in the arachidonic acid pathway in mammals [3]. Inhibition of sEH has been associated with improvements in various disease models such as high blood pressure, atherosclerosis, and kidney failure presumably by increasing EETs and possibly other fatty acid epoxides [10, 13, 15, 53, 54]. On the other hand, our observations also suggest that increased expression of EPHX2 could lead to cardiovascular diseases. In contrast to the large body of work that has been done on the enzymatic properties and biological roles of sEH, the transcriptional regulation and tissue-enriched expression of EPHX2 is poorly defined. Here we characterize the essential and enhancing elements of the promoter of EPHX2. Transcriptional repressors of EPHX2, analogous to compounds that inhibit enzymatic activity, could be potential targets for therapeutic application. Alternatively, enhancement of EPHX2 transcription may offer beneficial effects in certain cases [55]. For example, increased EPHX2 transcription could reduce or inhibit angiogenesis that is caused by EETs in cancerous tissues [56]. In agreement with this, sEH expression is generally reduced in human malignant tissues [57]. Furthermore, a variety of lipid epoxides including both fatty acids and terpenes [58] are excellent substrates for sEH. This suggests that sEH may catalyze other important chemical mediators that are involved in disease induction.
The in silico analysis of the 5′-flanking region of the EPHX2 did not identify typical TATA or CAAT box motifs. However, a GC-rich region was found between nts −374 and +28 of EPHX2 (Fig. 1A). This approximately 0.4 kb-long region was found to contain the minimal essential sequence for full relative luciferase activity (Fig. 1B). Androgen {e.g., GG(A/T)ACANNNTGTTCT [59]} and PPAR-alpha {e.g., AGGTCA(A/T)AGGTCA [60] responsive elements were not found in the 5′-flanking region (nts −5,974 to +28) of EPHX2. This was somewhat surprising since PPAR-alpha agonist such as clofibrate[23, 31, 33] and testosterone [31] have been shown to increase hepatic sEH activity in rodents. These response elements may, however, be found in other regions or they may be different from the response elements in rodents. Furthermore, there may be inherent differences between human and rodent models such that the extrapolation of potential enzyme induction effects observed with PPAR ligands in the rodent model to humans should be done with caution. These observations also illustrate the need for similar reporter analysis systems for EPHX2 in rodents and other non-human model species.
Our analysis of the 5′-flanking region of EPHX2 indicated that the GC-rich region (−374 to +28) of EPHX2 is involved in the basic transcriptional regulation of EPHX2. To further evaluate the role of this region as a core promoter, we evaluated the ability of the −374/+28 construct to induce luciferase activity in 9 human cell lines. We found that the transfection efficiency of these cell lines varied by more than 1,000-fold. Furthermore, correlations between sEH activity and relative luciferase activity for each construct (−5974/+28 and −374/+28) were not found. There was, however, a weak correlation (r = 0.66, p = 0.05) between sEH activity and a ratio of the relative luciferase activity of the −5974/+28 construct with respect to the −374/+28 construct {(−5974/+28)/(−374/+28)} (Fig. 2). These findings suggested that in addition to promoter effects, tissue-enriched factors are also important. Additionally, these findings suggested that the mechanisms of transcriptional regulation among these cells could be different. Using the cell line (293T) with the highest transfection efficiency, we found that the −165 to +28 nts region is the minimal sequence necessary for full luciferase activity (Fig. 4). Furthermore, luciferase activity gradually declined in reporter plasmids carrying shorter lengths of the 5′-flanking region (from nts −165 to −63). This suggested that one or more of the four regulatory elements located in this region {−165 to −155 (AP-1), −104 to −86 (Sp1), −86 to −63 (Sp1), and −63 to −41 (Sp1s and cMyc/Max1)} are functionally active.
Transcriptional regulation of genes that are expressed under TATA-less promoters often involves sequences that are found within GC-rich regions [48]. Sp1 binding sites are also commonly located within CpG islands in order to maintain the appropriate methylation pattern of downstream genes [61–63]. Through analysis of the minimal essential sequence (nts −165 to +28) required for full expression, we found numerous potential regulatory elements, especially Sp1 sites in this −165 to +28 region (Fig. 3). We showed that nts −63 to −42 are more important for EPHX2 transcription than nts −104 to −87 (Fig. 5). Electrophoretic mobility assays also showed that Sp1 binds to nts −54 to −33, nts −74 to −47, and nts −102 to −80 but not nts −86 to −64 (Fig. 7A). Furthermore, the ChIP assay showed that Sp1 binds to the −214 to +28 region of the EPHX2 (Fig. 7B). Only one completely conserved Sp1 consensus sequence motif, GGGGCGGGGC (nts −47 to −38), was found within the −165 to +28 nts region. Mutation of this sequence to GGTTCGGGGC resulted in a loss of transcriptional activity (Fig. 6). This suggested that in EPXH2, the Sp1 site immediately upstream of the putative transcriptional start site is essential for expression. Because Sp1 expression is generally ubiquitous [64] such a promoter system could explain the ubiquitous expression of sEH in numerous tissues in mammals [17, 20, 21, 23, 25–30, 65–67]. However, it does not explain the regulation of EPHX2 by PPAR-alpha agonists and testosterone as predicted from in vivo induction studies in rodents. As discussed earlier, this discrepancy could be due to the effects of clofibrate and testosterone being indirect rather than direct, but it also could illustrate a difference between man and rodents.
Ang II is known to increase sEH activity and sEH protein levels in kidney [10] and vascular endothelium [34]. Ang II induces the promoter activity of EPHX2 in transient transfection assays, through the binding of c-Jun to the AP-1 binding site of the promoter. These data tie the regulation of sEH and EETs to the rennin-angiotensin dependent regulation of blood pressure and inflammation [34]. The molecular basis of the high basal levels of sEH expression in renal, hepatic and a few other tissues as well as its regulation by endogenous compounds and xenobiotics awaits further work.
Acknowledgments
Support for this work was provided by NIEHS R37 ES02710, NIEHS P42 ES04699, NIH/NHLBI R01 HL59699-06A1, and NIEHS P30 ES05707. We thank Drs. Michael L. Goodson and Michael S. Denison from the University of California at Davis for technical advice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Larsson C, White I, Johansson C, Stark A, Meijer J. Localization of the human soluble epoxide hydrolase gene (EPHX2) to chromosomal region 8p21-p12. Hum Genet. 1995;95:356–358. doi: 10.1007/BF00225209. [DOI] [PubMed] [Google Scholar]
- 2.Morisseau C, Hammock BD. Epoxide hydrolases: mechanisms, inhibitor designs, and biological roles. Annu Rev Pharmacol Toxicol. 2005;45:311–333. doi: 10.1146/annurev.pharmtox.45.120403.095920. [DOI] [PubMed] [Google Scholar]
- 3.Newman JW, Morisseau C, Hammock BD. Epoxide hydrolases: their roles and interactions with lipid metabolism. Prog Lipid Res. 2005;44:1–51. doi: 10.1016/j.plipres.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Carroll MA, Schwartzman M, Capdevila J, Falck JR, McGiff JC. Vasoactivity of arachidonic acid epoxides. Eur J Pharmacol. 1987;138:281–283. doi: 10.1016/0014-2999(87)90445-6. [DOI] [PubMed] [Google Scholar]
- 5.Medhora M, Harder D. Functional role of epoxyeicosatrienoic acids and their production in astrocytes: approaches for gene transfer and therapy (review) Int J Mol Med. 1998;2:661–669. doi: 10.3892/ijmm.2.6.661. [DOI] [PubMed] [Google Scholar]
- 6.Roman RJ. P-450 metabolites of arachidonic acid in the control of cardiovascular function. Physiol Rev. 2002;82:131–185. doi: 10.1152/physrev.00021.2001. [DOI] [PubMed] [Google Scholar]
- 7.Spiecker M, Liao JK. Vascular protective effects of cytochrome p450 epoxygenase-derived eicosanoids. Arch Biochem Biophys. 2005;433:413–420. doi: 10.1016/j.abb.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Capdevila JH, Nakagawa K, Holla V. The CYP P450 arachidonate monooxygenases: enzymatic relays for the control of kidney function and blood pressure. Adv Exp Med Biol. 2003;525:39–46. doi: 10.1007/978-1-4419-9194-2_9. [DOI] [PubMed] [Google Scholar]
- 9.Sun J, Sui X, Bradbury JA, Zeldin DC, Conte MS, Liao JK. Inhibition of vascular smooth muscle cell migration by cytochrome p450 epoxygenase-derived eicosanoids. Circ Res. 2002;90:1020–1027. doi: 10.1161/01.res.0000017727.35930.33. [DOI] [PubMed] [Google Scholar]
- 10.Imig JD, Zhao X, Capdevila JH, Morisseau C, Hammock BD. Soluble epoxide hydrolase inhibition lowers arterial blood pressure in angiotensin II hypertension. Hypertension. 2002;39:690–694. doi: 10.1161/hy0202.103788. [DOI] [PubMed] [Google Scholar]
- 11.Imig JD, Zhao X, Zaharis CZ, Olearczyk JJ, Pollock DM, Newman JW, Kim IH, Watanabe T, Hammock BD. An orally active epoxide hydrolase inhibitor lowers blood pressure and provides renal protection in salt-sensitive hypertension. Hypertension. 2005;46:975–981. doi: 10.1161/01.HYP.0000176237.74820.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Node K, Huo Y, Ruan X, Yang B, Spiecker M, Ley K, Zeldin DC, Liao JK. Anti-inflammatory properties of cytochrome P450 epoxygenase-derived eicosanoids. Science. 1999;285:1276–1279. doi: 10.1126/science.285.5431.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Z, Xu F, Huse LM, Morisseau C, Draper AJ, Newman JW, Parker C, Graham L, Engler MM, Hammock BD, Zeldin DC, Kroetz DL. Soluble epoxide hydrolase regulates hydrolysis of vasoactive epoxyeicosatrienoic acids. Circ Res. 2000;87:992–998. doi: 10.1161/01.res.87.11.992. [DOI] [PubMed] [Google Scholar]
- 14.Schmelzer KR, Kubala L, Newman JW, Kim IH, Eiserich JP, Hammock BD. Soluble epoxide hydrolase is a therapeutic target for acute inflammation. Proc Natl Acad Sci USA. 2005;102:9772–9777. doi: 10.1073/pnas.0503279102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao X, Yamamoto T, Newman JW, Kim IH, Watanabe T, Hammock BD, Stewart J, Pollock JS, Pollock DM, Imig JD. Soluble epoxide hydrolase inhibition protects the kidney from hypertension-induced damage. J Am Soc Nephrol. 2004;15:1244–1253. [PubMed] [Google Scholar]
- 16.Inceoglu B, Jinks SL, Schmelzer KR, Waite T, Kim IH, Hammock BD. Inhibition of soluble epoxide hydrolase reduces LPS-induced thermal hyperalgesia and mechanical allodynia in a rat model of inflammatory pain. Life Sci. 2006;79:2311–2319. doi: 10.1016/j.lfs.2006.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammock BD, Storms DH, Grant DF. Epoxide Hydrolases. In: Guengerich FP, editor. Comprehensive Toxicology. Pergamon; Oxford: 1997. pp. 283–305. [Google Scholar]
- 18.Newman JW, Morisseau C, Harris TR, Hammock BD. The soluble epoxide hydrolase encoded by EPXH2 is a bifunctional enzyme with novel lipid phosphate phosphatase activity. Proc Natl Acad Sci USA. 2003;100:1558–1563. doi: 10.1073/pnas.0437724100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tran KL, Aronov PA, Tanaka H, Newman JW, Hammock BD, Morisseau C. Lipid sulfates and sulfonates are allosteric competitive inhibitors of the N-terminal phosphatase activity of the mammalian soluble epoxide hydrolase. Biochemistry. 2005;44:12179–12187. doi: 10.1021/bi050842g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang P, Meijer J, Guengerich FP. Purification of human liver cytosolic epoxide hydrolase and comparison to the microsomal enzyme. Biochemistry. 1982;21:5769–5776. doi: 10.1021/bi00266a007. [DOI] [PubMed] [Google Scholar]
- 21.Yu Z, Davis BB, Morisseau C, Hammock BD, Olson JL, Kroetz DL, Weiss RH. Vascular localization of soluble epoxide hydrolase in the human kidney. Am J Physiol Renal Physiol. 2004;286:F720–726. doi: 10.1152/ajprenal.00165.2003. [DOI] [PubMed] [Google Scholar]
- 22.Waechter F, Bentley P, Bieri F, Muakkassah-Kelly S, Staubli W, Villermain M. Organ distribution of epoxide hydrolases in cytosolic and microsomal fractions of normal and nafenopin-treated male DBA/2 mice. Biochem Pharmacol. 1988;37:3897–3903. doi: 10.1016/0006-2952(88)90071-8. [DOI] [PubMed] [Google Scholar]
- 23.Oesch F, Schladt L, Hartmann R, Timms C, Worner W. Rat cytosolic epoxide hydrolase. Adv Exp Med Biol. 1986;197:195–201. doi: 10.1007/978-1-4684-5134-4_16. [DOI] [PubMed] [Google Scholar]
- 24.Petruzzelli S, Franchi M, Gronchi L, Janni A, Oesch F, Pacifici GM, Giuntini C. Cigarette smoke inhibits cytosolic but not microsomal epoxide hydrolase of human lung. Hum Exp Toxicol. 1992;11:99–103. doi: 10.1177/096032719201100207. [DOI] [PubMed] [Google Scholar]
- 25.Pacifici GM, Temellini A, Giuliani L, Rane A, Thomas H, Oesch F. Cytosolic epoxide hydrolase in humans: development and tissue distribution. Arch Toxicol. 1988;62:254–257. doi: 10.1007/BF00332483. [DOI] [PubMed] [Google Scholar]
- 26.Wixtrom RN, Silva MH, Hammock BD. Cytosolic epoxide hydrolase in human placenta. Placenta. 1988;9:559–563. doi: 10.1016/0143-4004(88)90028-8. [DOI] [PubMed] [Google Scholar]
- 27.Enayetallah AE, French RA, Thibodeau MS, Grant DF. Distribution of soluble epoxide hydrolase and of cytochrome P450 2C8, 2C9, and 2J2 in human tissues. J Histochem Cytochem. 2004;52:447–454. doi: 10.1177/002215540405200403. [DOI] [PubMed] [Google Scholar]
- 28.Sevanian A, Stein RA, Mead JF. Lipid epoxide hydrolase in rat lung preparations. Biochim Biophys Acta. 1980;614:489–500. doi: 10.1016/0005-2744(80)90238-7. [DOI] [PubMed] [Google Scholar]
- 29.Zheng J, Plopper CG, Lakritz J, Storms DH, Hammock BD. Leukotoxin-diol: a putative toxic mediator involved in acute respiratory distress syndrome. Am J Respir Cell Mol Biol. 2001;25:434–438. doi: 10.1165/ajrcmb.25.4.4104. [DOI] [PubMed] [Google Scholar]
- 30.Pham MA, Magdalou J, Totis M, Fournel-Gigleux S, Siest G, Hammock BD. Characterization of distinct forms of cytochromes P-450, epoxide metabolizing enzymes and UDP-glucuronosyltransferases in rat skin. Biochem Pharmacol. 1989;38:2187–2194. doi: 10.1016/0006-2952(89)90075-0. [DOI] [PubMed] [Google Scholar]
- 31.Pinot F, Grant DF, Spearow JL, Parker AG, Hammock BD. Differential regulation of soluble epoxide hydrolase by clofibrate and sexual hormones in the liver and kidneys of mice. Biochem Pharmacol. 1995;50:501–508. doi: 10.1016/0006-2952(95)00167-x. [DOI] [PubMed] [Google Scholar]
- 32.Hennebold JD, Mah K, Perez W, Vance JE, Stouffer RL, Morisseau C, Hammock BD, Adashi EY. Identification and characterization of an ovary-selective isoform of epoxide hydrolase. Biol Reprod. 2005;72:968–975. doi: 10.1095/biolreprod.104.035899. [DOI] [PubMed] [Google Scholar]
- 33.Hammock BD, Ota K. Differential induction of cytosolic epoxide hydrolase, microsomal epoxide hydrolase, and glutathione S-transferase activities. Toxicol Appl Pharmacol. 1983;71:254–265. doi: 10.1016/0041-008x(83)90342-3. [DOI] [PubMed] [Google Scholar]
- 34.Ai D, Fu Y, Guo D, Tanaka H, Wang N, Tang C, Hammock BD, Shyy JY, Zhu Y. Angiotensin II up-regulates soluble epoxide hydrolase in vascular endothelium in vitro and in vivo. Proc Natl Acad Sci USA. 2007;104:9018–9023. doi: 10.1073/pnas.0703229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park WY, Hwang CI, Im CN, Kang MJ, Woo JH, Kim JH, Kim YS, Kim H, Kim KA, Yu HJ, Lee SJ, Lee YS, Seo JS. Identification of radiation-specific responses from gene expression profile. Oncogene. 2002;21:8521–8528. doi: 10.1038/sj.onc.1205977. [DOI] [PubMed] [Google Scholar]
- 36.Stone KR, Mickey DD, Wunderli H, Mickey GH, Paulson DF. Isolation of a human prostate carcinoma cell line (DU 145) Int J Cancer. 1978;21:274–281. doi: 10.1002/ijc.2910210305. [DOI] [PubMed] [Google Scholar]
- 37.Leibovitz A, Stinson JC, McCombs WB, 3rd, McCoy CE, Mazur KC, Mabry ND. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976;36:4562–4569. [PubMed] [Google Scholar]
- 38.Fogh J, Wright WC, Loveless JD. Absence of HeLa cell contamination in 169 cell lines derived from human tumors. J Natl Cancer Inst. 1977;58:209–214. doi: 10.1093/jnci/58.2.209. [DOI] [PubMed] [Google Scholar]
- 39.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldstein MN, Slotnick IJ, Journey LJ. In vitro studies with HeLa cell line sensitive and resistant to actinomycin D. Ann N Y Acad Sci. 1960;89:474–483. doi: 10.1111/j.1749-6632.1960.tb20171.x. [DOI] [PubMed] [Google Scholar]
- 41.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 42.Borden EC, Hogan TF, Voelkel JG. Comparative antiproliferative activity in vitro of natural interferons α and α for diploid and transformed human cells. Cancer Res. 1982;42:4948–4953. [PubMed] [Google Scholar]
- 43.Naito S, von Eschenbach AC, Giavazzi R, Fidler IJ. Growth and metastasis of tumor cells isolated from a human renal cell carcinoma implanted into different organs of nude mice. Cancer Res. 1986;46:4109–4115. [PubMed] [Google Scholar]
- 44.Aden DP, Fogel A, Plotkin S, Damjanov I, Knowles BB. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature. 1979;282:615–616. doi: 10.1038/282615a0. [DOI] [PubMed] [Google Scholar]
- 45.Gross-Bellard M, Oudet P, Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973;36:32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- 46.Borhan B, Mebrahtu T, Nazarian S, Kurth MJ, Hammock BD. Improved radiolabeled substrates for soluble epoxide hydrolase. Anal Biochem. 1995;231:188–200. doi: 10.1006/abio.1995.1520. [DOI] [PubMed] [Google Scholar]
- 47.Smale ST, Schmidt MC, Berk AJ, Baltimore D. Transcriptional activation by Sp1 as directed through TATA or initiator: specific requirement for mammalian transcription factor IID. 1990;87:4509–4513. doi: 10.1073/pnas.87.12.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smale ST. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- 49.Dynan WS, Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- 50.Kadonaga JT, Carner KR, Masiarz FR, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 51.Kadonaga JT, Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci USA. 1986;83:5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Briggs MR, Kadonaga JT, Bell SP, Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986;234:47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- 53.Davis BB, Thompson DA, Howard LL, Morisseau C, Hammock BD, Weiss RH. Inhibitors of soluble epoxide hydrolase attenuate vascular smooth muscle cell proliferation. Proc Natl Acad Sci USA. 2002;99:2222–2227. doi: 10.1073/pnas.261710799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu D, Li N, He Y, Timofeyev V, Lu L, Tsai HJ, Kim IH, Tuteja D, Mateo RK, Singapuri A, Davis BB, Low R, Hammock BD, Chiamvimonvat N. Prevention and reversal of cardiac hypertrophy by soluble epoxide hydrolase inhibitors. Proc Natl Acad Sci USA. 2006;103:18733–18738. doi: 10.1073/pnas.0609158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hutchens MP, Nakano T, Dunlap J, Traystman RJ, Hurn PD, Alkayed NJ. Soluble epoxide hydrolase gene deletion reduces survival after cardiac arrest and cardiopulmonary resuscitation. Resuscitation. 2007 doi: 10.1016/j.resuscitation.2007.06.031. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Michaelis UR, Fisslthaler B, Medhora M, Harder D, Fleming I, Busse R. Cytochrome P450 2C9-derived epoxyeicosatrienoic acids induce angiogenesis via crosstalk with the epidermal growth factor receptor (EGFR) FASEB J. 2003;17:770–772. doi: 10.1096/fj.02-0640fje. [DOI] [PubMed] [Google Scholar]
- 57.Enayetallah AE, French RA, Grant DF. Distribution of soluble epoxide hydrolase, cytochrome P450 2C8, 2C9 and 2J2 in human malignant neoplasms. J Mol Histol. 2006;37:133–141. doi: 10.1007/s10735-006-9050-9. [DOI] [PubMed] [Google Scholar]
- 58.Hammock BD, Gill SJ, Mumby SM, Ota K. Comparison of epoxide hydrases in the soluble and microsomal fractions of mammalian liver. In: Bhatnagar RS, editor. Molecular basis of environmental toxicity. Ann Arbor Science; Ann Arbor: 1980. pp. 229–272. [Google Scholar]
- 59.Roche PJ, Hoare SA, Parker MG. A consensus DNA-binding site for the androgen receptor. Mol Endocrinol. 1992;6:2229–2235. doi: 10.1210/mend.6.12.1491700. [DOI] [PubMed] [Google Scholar]
- 60.Juge-Aubry C, Pernin A, Favez T, Burger AG, Wahli W, Meier CA, Desvergne B. DNA binding properties of peroxisome proliferator-activated receptor subtypes on various natural peroxisome proliferator response elements. Importance of the 5′-flanking region. J Biol Chem. 1997;272:25252–25259. doi: 10.1074/jbc.272.40.25252. [DOI] [PubMed] [Google Scholar]
- 61.Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 62.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 63.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 64.Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pichare MM, Gill SS. The regulation of cytosolic epoxide hydrolase in mice. Biochem Biophys Res Commun. 1985;133:233–238. doi: 10.1016/0006-291x(85)91866-2. [DOI] [PubMed] [Google Scholar]
- 66.VanRollins M, Kaduce TL, Knapp HR, Spector AA. 14, 15-Epoxyeicosatrienoic acid metabolism in endothelial cells. J Lipid Res. 1993;34:1931–1942. [PubMed] [Google Scholar]
- 67.Enayetallah AE, French RA, Barber M, Grant DF. Cell-specific subcellular localization of soluble epoxide hydrolase in human tissues. J Histochem Cytochem. 2006;54:329–335. doi: 10.1369/jhc.5A6808.2005. [DOI] [PubMed] [Google Scholar]