Abstract
Telomeres are essential for maintaining cellular proliferative capacity and their loss has been implicated in aging. A key regulator in telomere maintenance is the telomeric protein TRF1, which was also identified as Pin2 in a same screen for Pin1. Pin1 is a unique prolyl isomerase that regulates protein conformation and function after phosphorylation. However, little is known about the role of Pin1 in telomere regulation and the modulation of TRF1 by upstream signals. Here we identify TRF1 as a major conserved substrate for Pin1 during telomere maintenance and aging. Pin1 inhibition renders TRF1 resistant to protein degradation, enhances TRF1 binding on telomeres, and leads to gradual telomere loss in human cells and in mice. Pin1-deficient mice also display widespread premature aging phenotypes within just one generation similar to those in telomerase-deficient mice after 4–5 consecutive generations. Thus, Pin1 is an essential novel regulator of TRF1 stability, telomere maintenance and aging.
Telomeres cap chromosome ends and their loss has been implicated in aging1–3. Indeed, telomerase-deficient mice eventually display accelerated telomere loss and premature aging after many generations4,5. A key regulator in maintaining the optimal telomere length is the telomeric protein TRF16–8. Indeed, TRF1 and its interacting proteins affect telomere length8–12. TRF1 is also regulated by multiple mechanisms including the PinX3/Fbx4-mediated proteasome pathway12,13, but little is known about its regulation by upstream signals.
TRF1 was also identified as Pin2 in the same mitotic screen for Pin114,15. Pin1 is a unique prolyl isomerase that binds to and isomerizes specific phosphorylated Ser/Thr-Pro motifs in certain proteins14,16–23. Notably, Pro-directed phosphorylation is a central mechanism in diverse cellular processes including cell growth and stress response. Importantly, Pin1-catalyzed postphosphorylation conformational changes can have profound effects on many key proteins in diverse cellular processes23. Pin1 is tightly regulated and often functions as a molecular timer to act on multiple targets to synergistically drive certain cellular processes towards one direction under given conditions23. For example, in response to growth stimulation, Pin1 is transcriptionally activated, which then positively acts on multiple signaling steps to promote cell division24. Significantly, Pin1 inhibition results in several age-dependent phenotypes20,22,25 and both Pin1 and TRF1 are involved in mitotic regulation18,23,26–29. However, nothing is known about the role of Pin1 in regulating TRF1 and telomere maintenance.
To examine whether TRF1 is a Pin1 substrate, we first examined TRF1 phosphorylation during the cell cycle because Pin1 binds to many mitotic phosphoproteins16,17,23, and both Pin1 and TRF1 have important mitotic function18,23,26–29. Endogenous TRF1 in mitotic cells had a slower electrophoretic mobility than that in G1 cells, as detected by two different antibodies (Fig. 1a, S1a). This slower mobility form was recognized by the pThr-Pro-specific monoclonal antibody (mAb) (Fig. 1a). An even more complete mitosis-specific TRF1 mobility shift was observed when in vitro synthesized 35S-TRF1 was incubated with Xenopus cell cycle extracts (Fig. S1b). Thus, TRF1 is phosphorylated in mitotic cells likely on Thr-Pro motifs.
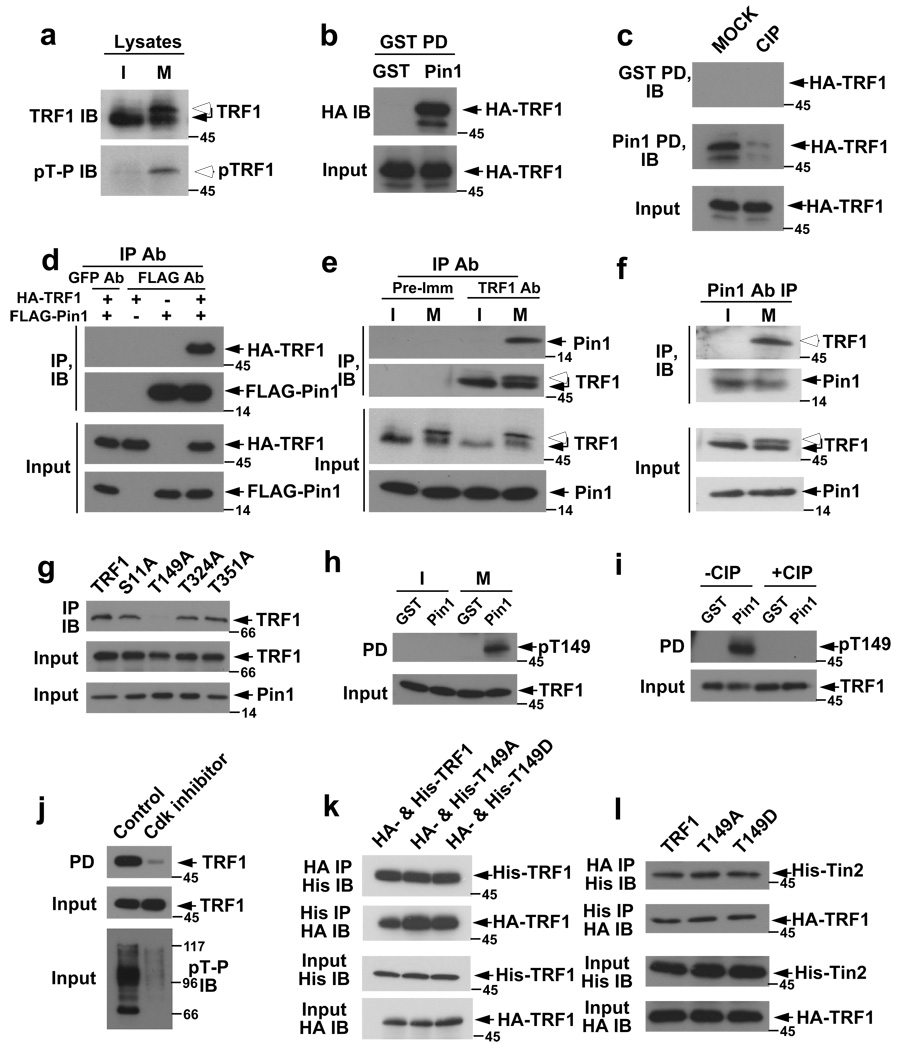
Figure 1. Pin1 binds to the pThr149-Pro motif in TRF1 in a phosphorylation-dependent and mitosis-specific manner.
(a) Phosphorylation of TRF1 on Thr-Pro motifs in mitosis. Interphase (I) and mitotic (M) HeLa cell extracts (upper) or their anti-TRF1 immunoprecipitates (lower) were immunoblotted with anti-full length TRF1 or pThr-Pro mAb. (b) Pin1 and TRF1 interaction in vitro. HeLa cells expressing HA-TRF1 were subjected to GST pulldown, and immunoblotted with anti-HA 12CA5 mAb. (c) Phosphorylation-dependent Pin1 and TRF1 interaction. Cells expressing TRF1 were incubated with or without CIP phosphatase before GST pulldown assay. (d) Co-immunoprecipitation of expressed Pin1 and TRF1. HeLa cells expressing FLAG-Pin1 and HA-TRF1 were immunoprecipitated with anti-FLAG or control GFP mAb, and immunoblotted with 12CA5 mAb. (e, f) Reciprocal co-immunoprecipitation of endogenous Pin1 and TRF1. Interphase and mitotic HeLa cell extracts were subjected to immunoprecipitation and then immunoblot with indicated antibodies. (g) Co-immunoprecipitation of Pin1 with TRF1, but not TRF1T149A in cells. Cells expressing HA-Pin1 and GFP-TRF1 or its mutant were immunoprecipitated with 12CA5 mAb, followed by immunoblot with anti-TRF1 antibodies. (h, i) Phosphorylation of TRF1 on Thr149. Interphase or mitotic TRF1-expressing cells were subjected to GST-Pin1 pulldown, followed by immunoblotting with anti-pThr149 TRF1 antibodies (h) or after CIP treatment (i). (j) Abolishment of TRF1 binding to Pin1 by Cdk inhibition. TRF1-transfected cells were treated with Roscovitine, followed by immunoblot with pThr-Pro mAb (lower) or GST-Pin1 pulldown assay (upper). (k, l) No effects of Thr149 mutations on the ability of TRF1 to interact with itself or Tin2. Cells were co-transfected with TRF1 or its mutants in different tags (k), or with TRF1 and Tin2 in different tags (l), followed by reciprocal immunoprecipitation and immunoblot. Full scans for f and i are presented in Fig. S12.
To detect Pin1 binding to TRF1, we expressed TRF1 in cells, followed by GST-Pin1 pulldown assay, as described16,18,19. GST-Pin1, but not control GST, pulled down to HA-TRF1 from lysates (Fig. 1b), but this binding was abolished by a phosphatase treatment (Fig. 1c), suggesting a phosphorylation-dependent interaction. To examine the Pin1 and TRF1 interaction in vivo, we performed reciprocal co-immunoprecipitation experiments on transfected or endogenous proteins. HA-TRF1 was immunoprecipitated by anti-FLAG-Pin1, but not anti-GFP antibodies (Fig. 1d). Moreover, endogenous Pin1 and TRF1 were reciprocally co-immunoprecipitated only in mitotic, but not interphase cells (Fig. 1e, f). Thus, Pin1 interacts with TRF1 in vivo in a phosphorylation-dependent and mitosis-specific manner.
To map the domains required for the interaction, a series of Pin1 and TRF1 mutants were used in binding assays. TRF1 bound only to the WW domain, but not the PPIase domain of Pin1 (Fig. S2a). The TRF1 fragment from 48 to 268, but not from 1 to 47, 269 to 316 or 317 to 419, bound to Pin1 (Fig. S2b), indicating that the Pin1-binding site is located at the TRF homology domain. This was further confirmed by introducing Ala substitution into each of the total four possible Pin1-binding Ser/Thr-Pro motifs in TRF1. Pin1 bound to TRF1, TRF1S11A, TRF1T324A, and TRF1S351A, but not TRF1T149A, as shown by GST-Pin1 pulldown assay (Fig. S2c) or co-immunoprecipitation experiments (Fig. 1g). These results indicate that the Thr149-Pro motif is necessary for Pin1 binding and might be phosphorylated in cells.
To detect Thr149 phosphorylation, we raised pThr149-specific TRF1 antibodies, which recognized TRF1 that was pulled down by Pin1 from mitotic cells (Fig. 1h) and this recognition was abolished by dephosphorylation of Pin1-bound TRF1 with CIP (Fig. 1i). To identify possible Thr149 kinases, we used compounds that selectively inhibited 7 classes of protein kinases. Only the Cdk inhibitor Roscovitine abolished the ability of TRF1 to interact with Pin1 (Fig. 1j, S3). Since the TRF homology domain mediates TRF1 homodimerization15 and also interacts with other proteins such as Tin210, we examined whether these functions were affected by Thr149 mutations. Neither Thr149Ala nor Thr149Asp mutation affected TRF1 binding to itself (Fig. 1k) or Tin2 (Fig. 1l), as assayed by co-immunoprecipitation experiments. These results together indicate that Pin1 interacts with the pThr149-Pro motif in TRF1 in a phosphorylation-dependent and mitosis-specific manner.
Given the specific interaction between Pin1 and TRF1, the central question is whether this interaction has any biological significance or pathological consequence. The facts that Pin1 regulates turnover of many proteins23 and that TRF1 turnover is regulated12,13 suggest that Pin1 might regulate TRF1 protein stability. To examine this possibility, we stably knocked down endogenous Pin1 in human cells using a retrovirus-mediated RNA interference targeting Pin1 (Pin1-shRNA)30, followed by monitoring endogenous TRF1 protein stability using a cycloheximide chase12. In contrast to vector control cells where TRF1 was unstable12,13, TRF1 was almost completely stable in Pin1-silenced cells (Fig. 2a, b). To confirm these results, we used a Pin1 dominant-negative mutant, GFP-S16A Pin1 WW domain mutant to inhibit Pin1 function in cells. TRF1 was also stable in GFP-Pin1 S16A cells, but not in vector cells (Fig. 2c, d). Therefore, Pin1 inhibition renders TRF1 resistant to degradation in human cells.
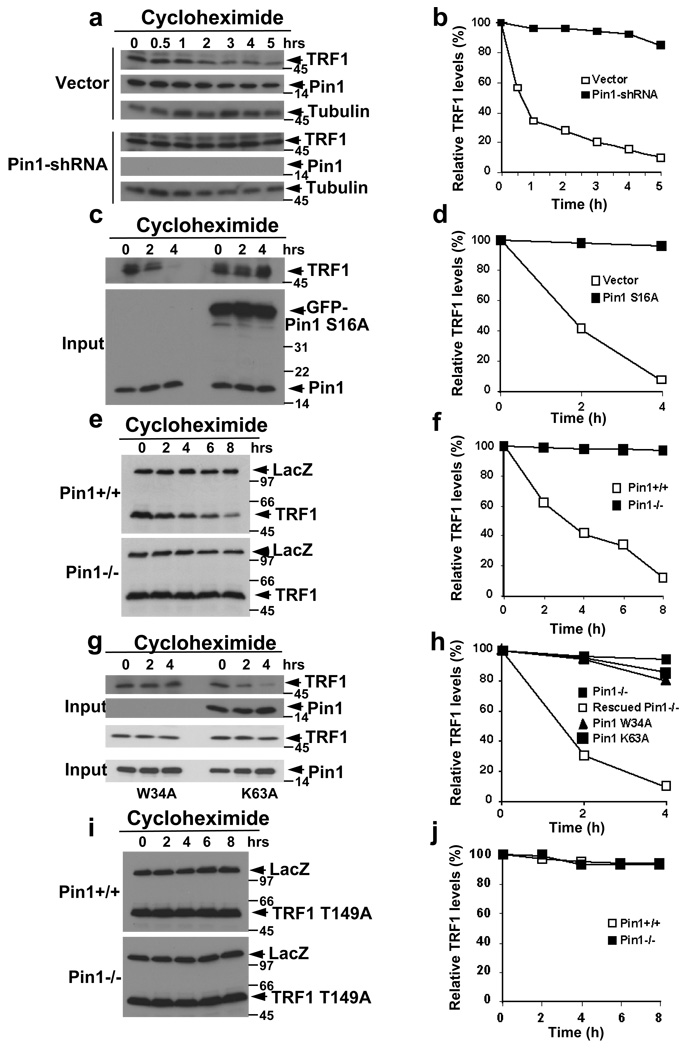
Figure 2. Pin1 inhibition via three approaches renders TRF1 resistant to degradation, and the Pin1 action depends on its ability to act on the pThr149-Pro motif in TRF1.
(a, b) Pin1 knockdown increases endogenous TRF1 protein stability. MCF7 cells stably expressing-shRNA directed at Pin1 were treated with cycloheximide for indicated time points, followed by immunoblotting analysis with anti-TRF1, anti-tubulin or anti-Pin1 antibodies (a) and semi-quantification using tubulin as a loading control and relative TRF1 levels at time 0 as 100% (b). (c, d) Dominant-negative mutant Pin1 S16A renders TRF1 resistant to degradation in human cells. HeLa cells were transfected with plasmid encoding GFP or GFP-Pin1 S16A WW domain, followed by determining TRF1 stability using the cycloheximide chase. (e, f) Pin1 knockout increases expressed TRF1 protein stability. Pin1+/+ and Pin1−/− mouse breast cancer cells were co-transfected with Xpress-TRF1 and control Xpress-LacZ, followed by determining TRF1 stability using the cycloheximide chase. (g, h) Rescuing the TRF1 degradation in Pin1−/− cells by re-expression of Pin1, but not its WW domain or PPIase domain mutant. Pin1−/− cells were transfected with Pin1 or its mutants, followed by determining TRF1 stability using the cycloheximide chase. (i, j) Inability of Pin1 to affect the stability of TRF1 T149A. Pin1+/+ and Pin1−/− cells were co-transfected with Xpress-TRF1 T149A and control Xpress-LacZ, followed by determining TRF1 stability using the cycloheximide chase. Full scans for e and i are presented in Fig. S12.
Since both Pin1 and its binding site Thr149-Pro motif in TRF1 are conserved in mice6,14,15, we examined the effects of Pin1 on TRF1 turnover in mouse cells using wild-type (Pin1+/+, WT) or Pin1 knockout (Pin1−/−, KO) mouse breast cancer cells26. Xpress-TRF1 was unstable in Pin1+/+ cells, but was nearly completely stable in Pin1−/− cells (Fig. 2e, f), although stability of co-transfected Xpress-LacZ was not affected (Fig. 2e). To ensure that this defect is due to loss of Pin1 and its direct action on TRF1, we re-expressed Pin1, or its binding mutant (W34A) or isomerase-inactive mutant (K63A) into Pin1−/− cells, followed by monitoring TRF1 stability. Re-expression of only Pin1, but not its mutants, fully rescued the defective TRF1 degradation in Pin1−/− cells (Fig. 2g, h), indicating the requirements for both binding and isomerizing activities of Pin1. We also examined stability of the TRF1 mutant (TRF1 T149A) that cannot bind to Pin1 (Fig. 1g, S2c). This TRF1 mutant was stable in Pin1+/+ and Pin1−/− cells (Fig. 2i, j), indicating the essential role of the Pin1-binding site in TRF1. Thus, Pin1 is a central regulator of TRF1 turnover by binding to the conserved pThr149-Pro motif in TRF1 and presumably inducing conformational changes. Given that the pThr149-Pro motif is located in the TRF1 homology domain that also interacts with many proteins such as Fbx412, Pin1-catalyzed conformational changes might regulate Fbx4-mediated degradation of TRF1.
Given the tight control of Pin1 over TRF1 turnover, the next important question is whether Pin1 affects the cellular function of TRF1. Two of the most well accepted functions of TRF1 is its ability to bind on telomeres and to negatively regulate telomere length in human HT1080 cells7,8. To examine the effects of Pin1 on TRF1 binding on telomeres, we stably silenced Pin1 in HT1080 cells with the Pin1-shRNA construct, followed by telomere Chip analysis using two different TRF1 antibodies. Endogenous TRF1 was very stable in multiple Pin1-shRNA stable cell lines (Fig. S4). Importantly, the amounts of TRF1 bound on telomeres were increased by 2~2.5 folds in Pin1-inhibited cells (Fig. 3a, b). Thus, inhibition of Pin1 renders TRF1 resistant to degradation, and also increases its binding on telomeres.
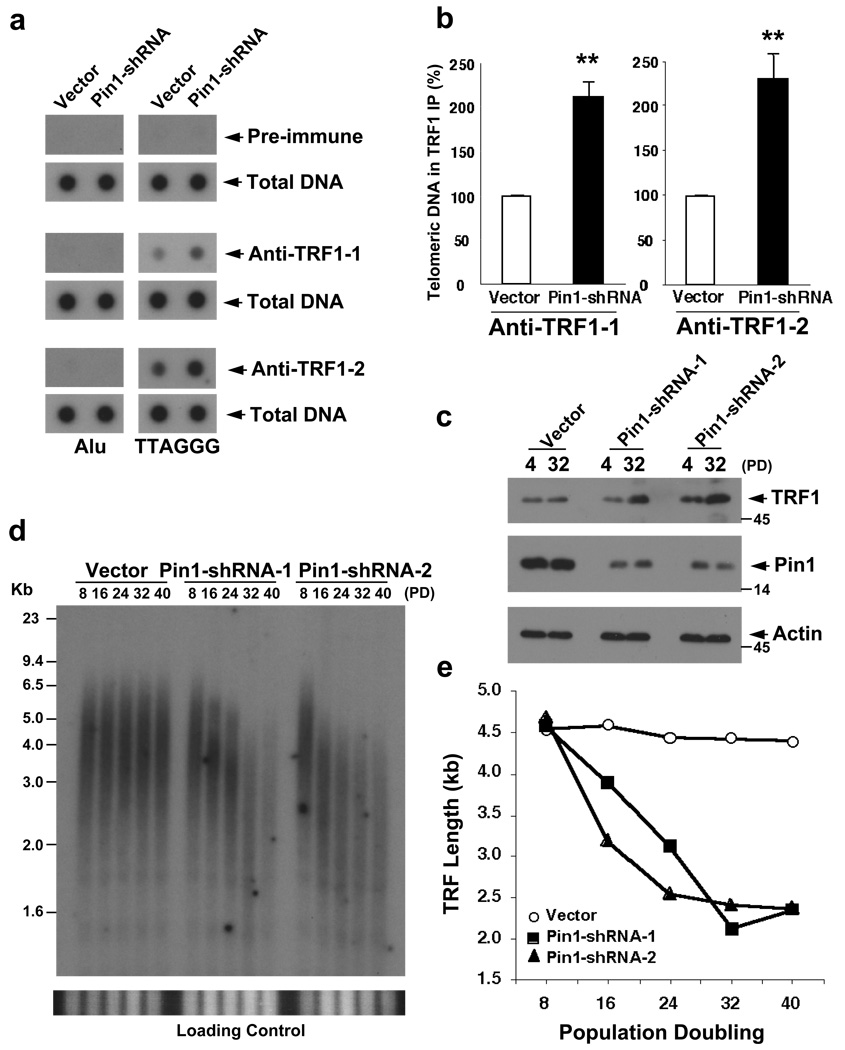
Figure 3. Pin1 inhibition increases binding of TRF1 on telomeres and also leads to gradual and progressive telomere shortening in human cells.
(a, b) Pin1 knockdown increases binding of TRF1 on telomeres. HT1080 cells were infected with Pin1-shRNA or control virus and stable cell lines were selected and confirmed using immunoblot with anti-Pin1 or anti-actin antibodies (Fig. S4). These stable cells were subjected to telomere Chip with anti-TRF1 antibodies and the presence of TTAGGG repeats and Alu repeats in the precipitated DNA was analyzed by Southern blotting (a), followed by quantifications using ImageQuant analysis (b). The graph shows mean±SD of three experiments. **, P<0.01. (c–e) Pin1 knockdown increases TRF1 levels and leads to a gradual and progressive telomere shortening in cells. Multiple independent Pin1-shRNA and vector control HT1080 cells were maintained continuously in culture, and harvested at different PDs, followed by measuring TRF1 levels using immunoblotting (c) and telomere lengths using genomic Southern blotting analysis, with the gels being stained with ethidium bromide to insure equal loading of genomic DNA prior to hybridization (lower panels) (d). Average TRF length versus PD number was quantified using ImageQuant (e). Similar telomere shortening were also observed in other independent Pin1-shRNA stable cell lines (Fig. S6).
To examine whether Pin1 also affects the ability of endogenous TRF1 to regulate telomere length, we continuously cultured multiple Pin1-shRNA and vector stable HT1080 cell lines, followed by measuring TRF1 levels and telomere restriction fragment (TRF) lengths at different population doublings (PDs). TRF1 levels remained higher (Fig. 3c) and TRF lengths were gradually and progressively shortened, as evidenced from reduced TTAGGG hybridization signal and shortened average TRF length (Fig. 3d, e, S6). Thus, inhibition of Pin1 not only increases TRF1 stability and its binding on telomeres, but also leads to gradual and progressive telomere shortening, as does overexpresson of TRF17 or inhibition of the Fbx4-mediated degradation of TRF112.
The next important question is how important TRF1 is in mediating the Pin1-dependent telomere regulation. TRF1 inhibits telomere elongation without affecting telomerase activity itself7,8. If TRF1 would mediate the effects of Pin1 on telomere maintenance, we would expect 1) that Pin1 would not affect telomerase activity in telomerase-positive cells, 2) that Pin1 would not affect telomere length in telomerase-negative cells, 3) that Pin1 would not affect telomere elongation induced by overexpression of TERT and TER because telomere elongation under this condition is not affected by telomere-bound TRF131, and 4) that the concomitant ablation of TRF1 in Pin1-silenced cells would correct the observed telomere shortening.
To examine whether Pin1 inhibition affects telomerase activity in HT1080 cells, we measured telomerase activity in Pin1-inhibited cells where telomere shortening was observed using the TRAP assay. Pin1 knockdown did not inhibit telomerase activity (Fig. 4a, b), indicating that Pin1 inhibition induces gradual telomere shortening without affecting telomerase activity. To examine the effects of Pin1 on telomere length in telomerase-negative cells, we used Pin1-shRNA lentiviruses or retroviruses to stably infect telomerase-negative human diploid WI-38 dermal fibroblasts at passage 1432 or immortalized GM847 cells that maintain long but heterogeneous telomeres using the ALT mechanism33. Although Pin1-shRNA effectively knocked down Pin1 in both cell lines, there was not detectable difference in telomere lengths between Pin1-shRNA and vector cells (Fig. 4d, e, S5a, 7), indicating that Pin1 knockdown does not induce telomere shortening in telomerase-negative cells.
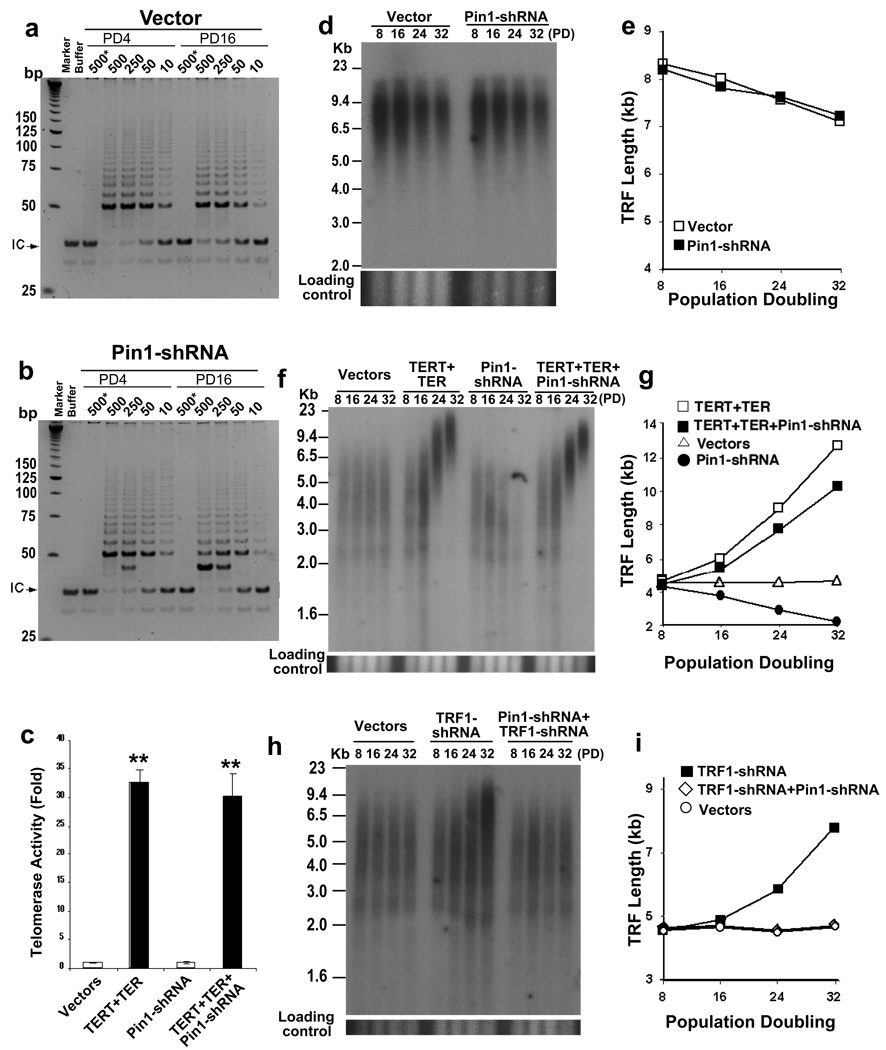
Figure 4. Pin1 regulates telomere maintenance by acting through the TRF1-dependent mechanism.
(a, b) No effects of Pin1 knockdown on telomerase activity. Telomerase-enriched nuclear extracts isolated from control (a) and Pin1-shRNA (b) stable cells at PD4 and PD16 were subjected to TRAP assay. Buffer or boiled lysates (marked by *) was included in one assay as a negative control. Arrows point to the internal control (IC). (d, e) No effects of Pin1 knockdown on telomere length in telomerase-negative human cells. WI38 cell pools stably infected with Pin1-shRNA lentiviruses and vector viruses were harvested at different PDs, followed by measuring telomere lengths using genomic Southern blotting analysis (d). Average TRF length versus PD number was quantified using ImageQuant (e). Similar results were also obtained in GM847 cells that maintain telomere using the ALT mechanism (Fig. S7). (c, f, g) No effects of Pin1 knockdown on telomere elongation induced by overexpression of TERT and TER. HT1080 cells were infected with TERT retroviruses and TER lentiviruses, and then with Pin1-shRNA lentiviruses, or control vectors and triple stable cell pools were selected, followed by, measuring telomerase activity by TRAP assay (c) and measuring telomere lengths at different PDs using genomic Southern blotting analysis (f, g). The graph shows mean±SD of three experiments. **, P<0.01. (h, i) Inability of Pin1 knockdown to induce telomere shortening in TRF1 silenced cells. HT1080 cells were infected with TRF1-shRNA retroviruses and then with Pin1-shRNA lentiviruses, or control vectors and double infected stable cell pools were selected, followed by measuring telomere lengths at different PDs using genomic Southern blotting analysis (h, i). Similar results were also observed in other independent Pin1-shRNA and TRF1-shRNA double stable cell pools (Fig. S8).
To examine the effects of Pin1 inhibition on telomere lengthening rate induced by telomerase overexpression, we sequentially infected HT1080 cells with TERT retroviruses and TER lentiviruses, and then with Pin1-shRNA lentiviruses or control vectors, followed by selection of triple stable cell pools. Stable overexpression of both TERT and TER increased telomerase activity by ~30 folds and led to continuous telomere elongation over time (Fig. 4c, f, g, S5b,), as shown31. However, neither increased telomerase activity nor telomere lengthening rate was inhibited by Pin1 knockdown (Fig. 4c, f, g, S5b). These results indicate that Pin1 inhibition does not affect telomerase activity or suppress its ability to elongate telomeres.
To examine the effects of TRF1 knockdown on telomere shortening induced by Pin1 inhibition, we stably infected HT1080 cells with TRF1-shRNA retroviruses and then with Pin1-shRNA lentiviruses. Both TRF1 and Pin1 were significantly reduced in double stable cells (Fig. S5c). Importantly, whereas telomeres were gradually elongated with time in single TRF1 knockdown cells, telomere lengths remained unchanged in TRF1 and Pin1 double knockdown cells (Fig. 4h, i, S8). The inability of TRF1 knockdown to elongate telomeres in Pin1-inhibited cells might be due to TRF1 stabilization when Pin1 is inhibited (Fig. 2). Indeed, TRF1 levels were significantly higher in TRF1 and Pin1 double knockdown cells than those in single TRF1 knockdown cells (Fig. S5c). Thus, Pin1 inhibition cannot induce telomere shortening when TRF1 is also concomitantly knocked down. These results together indicate that Pin1 regulates telomere maintenance by acting through the TRF1-dependent regulatory mechanism.
TRF1 negatively regulates telomere elongation in human cells7,8, but its effects on telomere maintenance in mouse cells is unknown. Given the conserved effects of Pin1 on TRF1 stability and obvious effects of Pin1 on telomere length in human cells, we examined whether TRF1 knockdown affects telomere length in mouse cells, and whether Pin1 knockout affects TRF1 levels and telomere maintenance in mice.
To determine the effects of TRF1 knockdown on telomere length in mouse cells, we used TRF1-shRNA lentiviruses to stably knock down TRF1 in MEFs (data not shown), followed by measuring telomere length at 40 PDs using quantitative telomere FISH (qFISH). TRF1-shRNA cells contained longer telomeres than vector cells, making the telomere distribution curve shifting to the right (Fig. 5a, b, S9), indicate a conserved function for TRF1 in regulating telomere length in mouse cells.
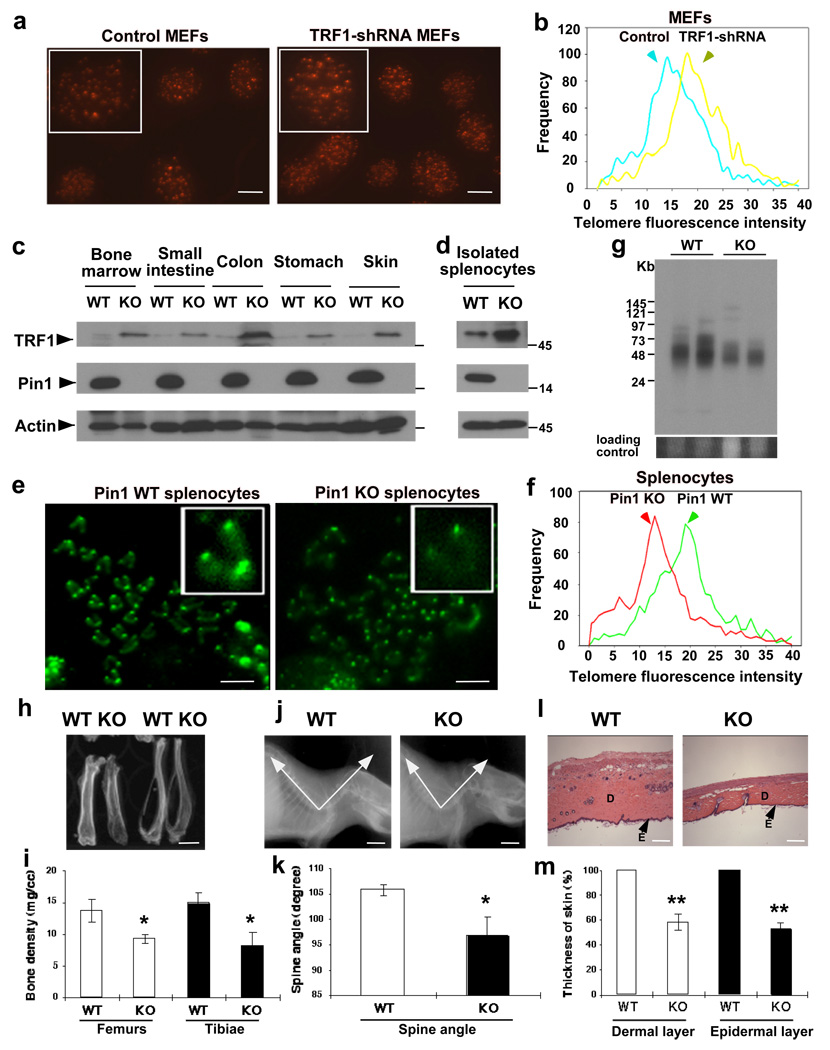
Figure 5. Pin1 knockout results in elevated TRF1 levels, induces accelerated telomere loss and leads to a wide range of premature aging phenotypes in mice.
(a–b) Elongated telomeres in TRF1-shRNA MEFs. MEFs stably infected with TRF1-shRNA lentiviruses or control viruses were immunoblotted with anti-TRF1 or -actin antibodies (data not shown) and after 40 PDs, and subjected to telomere FISH (a), followed by quantifying their telomere fluorescence intensity (b, Fig. S9). (c, d) Elevated TRF1 in Pin1−/− tissues or isolated splenocytes. Pin1+/+ or Pin1−/− tissues or isolated splenocytes were homogenized and immunoblotted with indicated antibodies. (e, f) Shortened telomeres in Pin1−/− mice. Pin1+/+ and Pin1−/− splenocytes were stimulated with LPS and metaphase spreads subjected to telomere FISH (e), followed by quantifying their telomere fluorescence intensity (f, Fig. S11). Similar shorted telomeres were also observed in total splenocytes or isolated splenic B cells, as assayed by flow-FISH (Fig. S10). (g) Reduced TRF length in Pin1−/− mice. Pin1+/+ and Pin1−/− splenocytes were cast into agarose plugs, followed by in-gel digestion and hybridization with a 32P-(TTAGGG)4 probe. The average TRF lengths in Pin1+/+ and Pin1−/− splenocytes were 48.5± 12.3 and 31.2±5.9 kb (n=4, p<0.01), respectively. (h, i) Reduced bone density in Pin1−/− mice. (h) Representative bone radiographs of femurs and tibiae from Pin1+/+ and Pin1−/− littermates at ~18 months. (i) A comparison of the average and standard deviations of four mice. (j, k) Lordokyphosis in Pin1−/− mice. (j) Representative X-ray radiographs of Pin1+/+ and Pin1−/−mice. (k) A comparison of the average and standard deviations of spine angles of four mice. (l, m) Skin atrophy in Pin1−/− mice. (l) Representative skin sections of Pin1+/+ and Pin1−/− littermate stained with hematoxylins and eosin. E, epidermal layer; D, dermal layer. (m) A comparison of the relative skin thickness of four mice. The graph shows mean±SD of four mice. *, P<0.05; **, P<0.01. Scale bars represent 10 µm in a, 5 µm in e, 0.5 cm in h and j, and 100 µm in l. Full scans for c are presented in Fig. S12.
To determine the effects of Pin1 on TRF1 levels and telomere length in mice, we compared TRF1 levels and telomere length in Pin1+/+ or Pin1−/− littermates. While TRF1 was barely detectable in most tissues or readily detectable in isolated splenocytes, its levels were much higher in corresponding Pin1−/− tissues or cells (Fig. 5c, d), supporting that Pin1 inhibition stabilizes TRF1 protein in vivo. We then assayed the effects of Pin1 knockout on telomere length in mice using three different methods. Flow cytometry FISH (Flow-FISH) revealed that the fluorescence signals in total splenocytes (Fig. S10a) or sorted splenic B cells (Fig. S10b) were lower in Pin1−/− mice than in Pin1+/+ mice, making telomere fluorescence distribution curves shifting to the left. qFISH on metaphase chromosome spreads prepared from LPS-stimulated splenocytes confirmed that Pin1−/− cells contained much shorter telomeres than Pin1+/+ cells, with the average telomere fluorescence intensity being reduced by 34.7% (Fig. 5e, f, S11). Genomic Southern analysis showed that the average TRF length in Pin1−/− splenocytes was shorter by 35.7%, as compared with Pin1+/+ controls (Fig. 5g). Telomere loss was also evidenced from decreased TTAGGG hybridization signals (Fig. 5g). Thus Pin1 deletion leads to accelerated telomere loss in mice within just one generation to the similar extents in telomerase knockout mice after 4–5 consecutive generations4,5.
Accelerated telomere loss has been implicated in premature aging1–3. Given accelerated telomere loss in Pin1−/− mice within one generation, we examined aging phenotypes in these mice. Indeed, as compared with Pin1+/+ controls, Pin1−/− mice showed decreased bone radiodensity (Fig. 5h, i), reduced spine angle (Fig. 5j, k) and thinner dermal and epidermal layers (Fig. 5l, m). In addition, previous reports show several other accelerated aging phenotypes in Pin1−/− mice, including overall appearance, motor coordination defects, neuron degeneration, retina degeneration, testis atrophy and breast atrophy20,22,25. All these premature aging phenotypes bear a resemblance to those in humans. Thus, these accelerated telomere loss and premature aging indicate an essential regulatory role for Pin1 in telomere maintenance and aging.
Although TRF1 was independently isolated as Pin2 in the same screen for Pin114, their relationship is unknown. Our results have shown that Pin1 binds to the conserved pThr149-Pro motif in TRF1, promotes TRF1 turnover and inhibits its function to regulate telomere length in humans and mice. Since Pin1 is a growth-regulated gene that plays a critical role in multiple growth signal pathways23,24, Pin1-dependent regulation of TRF1 might provide a link between cell growth signaling and telomere regulation.
The findings that accelerated telomere loss and a wide range of premature aging phenotypes in Pin1−/− mice within a single generation are remarkably similar to those in telomerase-deficient mice after 4–5 consecutive generations4,5 indicate a pivotal role of Pin1 for preventing aging. Since Pin1 often acts on multiple targets to help drive certain cellular processes, as shown in cancer and Alzheimer’s disease (AD)18,22,23,26, Pin1 likely acts on other substrates to regulate aging. Indeed, Pin1 regulates other aging-related proteins, including p5321 and p66Shc34. However, given the critical role of telomere length in aging1–3 and inhibition of Pin1 by multiple mechanisms including oxidative modification23,35, loss of Pin1-dependent regulation of TRF1 and telomere maintenance plays a key role in aging and age-related disease, most notably AD. Pin1 regulates several mitotic phosphoproteins including tau and APP that have well-known roles in AD, but is inhibited in diseased neurons18,22,23,26,35. Interestingly, telomere shortening correlates with the AD status36, suggesting that Pin1-dependent regulation of TRF1 and telomeres might also contribute to the AD pathogenesis.
In summary, our results demonstrate for the first time an essential and conserved role for Pin1 in regulating TRF1 stability, telomere maintenance and aging. Given the central role of Prodirected phosphorylation and Pin1 in diverse cellular processes and inhibition of Pin1 by oxidative damage, Pin1-catalyzed prolyl isomerization is a novel mechanism to help coordinate various cellular processes needed for preventing aging, and its deregulation contributes to age-related disease. Further studies how Pin1-dependent regulation is coordinated with other aging mechanisms and how Pin1 deregulation contributes to aging and age-related disease should help elucidate the molecular mechanisms of aging and might lead to new interventions for preventing or treating age-related disease.
METHODS
Reagents
TRF1 and truncated mutants expression vectors were produced as described previously12. To generate TRF1S11A, T149A, T324A, or T351A, missense mutations in the TRF1 cDNA were introduced by the QuikChangeTM site-directed mutagenesis kit (Stratagene) according to manufacturer’s instructions and confirmed by sequencing. Recombinant proteins were expressed and purified from bacteria or synthesized using in vitro TNT system as described previously18,19. Protein kinase inhibitors are purchased from Calbiochem and used at the highest concentrations for inhibiting selective kinases, as recommended by the manufacturer. To detect endogenous TRF1, we used two different polyclonal antibodies: one was raised against N-terminal 319 residues, which worked well for immunoprecipitation15 and the other against full length TRF protein, which worked well for immunoblotting analysis, as shown12. Phosphorylated Thr149-specific TRF1 antibodies were raised by immunizing rabbits with a KLH-coupled pThr149-containing TRF1 peptide. (FENDERIpTLESALMI) (Proteintech Group). Phosphorylated Thr-Pro-specific monoclonal antibody was purchased from Cell signaling.
GST pulldown, immunoprecipitation and immunoblotting
GST pulldown, immunoprecipitation, and immunoblotting analyses were performed as described11,18. Briefly, relevant proteins were expressed in HeLa or HT1080 cells by transient transfection, or translated in vitro using the TNT coupled transcription/translation kit in the presence of 35S-Met, followed by lysis or dilution in a buffer containing 50 mM HEPES, pH 7.5, 150 mM NaCl, 100 mM NaF, 1 mM sodium orthovanadate, 10% glycerol, 1% Triton X100, 10 µg/ml aprotinin, 10 µg/ml leupeptin, 50 µg/ml phenylmethylsulfonyl fluoride and 1 mM DTT. The cellular supernatants were incubated with 1 µM GST or GST fusion proteins or specific antibodies for 1 hr at 4°C and 15 µl of glutathione agarose beads or protein A agarose were then added, followed by further incubation for 2 h at 4°C. The precipitated proteins were washed 4–6 times in the same buffer and subjected to immunoblotting analysis.
Protein stability assay
For protein stability assay, cells were transfected stably or transiently with expression plasmids as indicated. Cycloheximide (100 µg/ml) was added to the media to block new protein synthesis. Cells were harvested at each time points, and total lysates were analyzed by immunoblot with anti-TRF1, anti-Pin1, anti-tubulin, or anti-b-actin antibodies. The blots were scanned and semi-quantitated by using the software NIH image 1.6.2, as described previously12. The results are plotted such that the protein levels at 0 h time point are set at 100%.
Establishment of stable cell lines
MCF7, HT1080, WI38 and GM847 cells were infected with Pin1-shRNA or control constructs and multiple independent stable clones were selected using 2 µg/ml puromycin, as described previously30. To double knock down TRF1 and Pin1 in HT1080 cells, cells were first infected with TRF1-shRNA retroviruses and a day later with Pin1-shRNA lentiviruses, followed by selection of double stable cell pools. To overexpress TERT and TER and to also knock down Pin1 expression in HT1080 cells, cells were sequentially infected with TERT retroviruses and TER lentiviruses and a day later with Pin1-shRNA lentiviruses or control vectors, followed by selection of triple stable cell pools. Stable cell clones or pools were checked for protein expression by immunoblotting analysis with various antibodies to confirm the expected protein expression. We maintained stable cell lines continuously in culture, splitting on every fourth day and seeding at the concentration of 6×105 cells per 10 cm culture dish.
Telomere Chip analysis
Telomere Chip (chromatin immunoprecipitation) analysis was carried out using two different anti-TRF1 polyclonal antibodies, as described37.
Pin1 knockout mouse strains
The genetic background of Pin1−/− mice is mixed 129/Sv and C57L/B620. Since Pin1+/− heterozygous mice are indistinguishable from Pin1+/+ mice20, we focused on the phenotypes on Pin1−/− mice.
Flow-FISH, qFISH and TRF length assay
Telomeric flow-FISH was carried out, described38. Briefly, spleens from Pin1+/+ and Pin1−/− littermates, were used to prepare single-cell suspensions by mincing samples through a 70-µm nylon cell strainer (BD Bioscience). Erythrocytes were removed from the cell suspensions by lysis with ACK lysing buffer. Viable cells were counted and resuspended in phosphate-buffered saline (PBS)−0.1% bovine serum albumin (BSA) to the concentration of 1×106 cells/ml. Cells were pelleted and resuspended in 500 µl of hybridization solution (70% formamide, 20 mM Tris, pH 7.0, 1% BSA, 0.3 µg/ml fluorescent isothiocyanate-conjugated (CCCTAA)3 peptide nucleic acid (PNA) probe (Applied Biosystems). DNA was denatured at 87°C for 15 min, and the samples were hybridized for 2h at room temperature. Cells were washed five times in wash solution 1 (70% formamide, 10 mM Tris, pH 7.0, 1% BSA, 0.1% Tween 20) and one time in wash solution 2 (PBS, 0.1% BSA, 0.1% Tween20). For DNA counterstaining, cells were resuspended in 500 µl staining solution (PBS, 0.1% BSA, 10 µg/ml RNase A, 0.01 µg/ml LDS751) for 2h. For B cell staining, cells were resuspended in 500 µl staining solution (PBS, 2% FCS, CD45R/B220) for 30 min before acquisition on the FACSan flow cytometry (Becton Dickinson). Mean telomere fluorescence of cells gated at G0/G1 was analyzed with CellQuest software.
qFISH was performed as described with the following modifications39. Briefly, primary splenocytes were isolated from Pin1+/+ and Pin1−/− littermates at 12 months old and stimulated with LPS to trigger cell cycle re-entry, followed by arresting cells at metaphase using Colcemid treatment, while interphase MEFs grown on overslips were fixed with 4% PFA. Cells were fixed with 3:1 methanol-acetic acid and dropped on slides, followed by hybridization with a FITC- or Cy3-labeled (CCCTAA)3 PNA probe, and DNA were stained with DAPI. After PNA hybridization, fluorescence signals at telomeres were visualized under an epifluorescence microscope (Axiovert 200M; Zeiss) equipped with a computer piloted filter wheel. After localization of metaphases, blue (DAPI) and green (FITC) fluorescence signals were captured. A flat field template was used to correct for unevenness in field illumination. Fixed exposure times of 4 or 5 s were sufficient to reveal all telomere signals in a metaphase spread. Original images were saved and used for quantitative analysis. Telomere fluorescence units were calculated with AxioVision 4.5 software (Zeiss). Over 1000 telomere spots in interphase nuclei or metaphase chromosomes from each sample were used to collect telomere fluorescent-signal data.
TRF length was assayed using genomic Southern analysis, as previously described40. Briefly, splenocytes isolated from Pin1+/+ and Pin1−/− littermates at 12 months old were combined with an equal volume of 2% low melting point (LMP) agarose and cast into 100 µl plug molds. Cells were lysed in a lithium dodecyl sulfate (LDS) solution, followed by digestion with HaeIII and HinfI restriction enzymes. Plugs were run on a 1.2% pulsed field grade agarose (Bio-Rad) gel and stained with EtBr, followed by drying down on filter paper before being subjected to hybridization with. The gels were dried and hybridized with a 32P-(TTAGGG)4 oligonucleotide probe and average TRF length was calculated by quantifying the hybridization signals using ImageQuant, as described11.
X-ray and histological examinations
For bone density, femurs and tibiae were harvested from Pin1+/+ and Pin1−/− mice and were analyzed with GE Locus eXplore µCT. Mouse tissues were fixed in 10% para-formaldehyde and embedded in paraffin. Sections (5 µm) of three different regions of skin from Pin1+/+ and Pin1−/− mice were stained with Harris haematoxylin and eosin according to standard procedures.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to J. Frangioni and R. Bronson for the assistance in determining bone-related and aging phenotypes in Pin1 knockout mice and W. Hahn for TERT and TER expression vectors. J. L. is a Human Frontiers Science Program Fellow, C.Y.S. is a Predoctoral Fellow of the DOD Breast Cancer Research Program and L. P. is a recipient of Mentored Research Scientist Development Award from NIH (K01). The work was supported by NIH grants R01CA122434 to X. Z. Z., and R01GM058556 and AG017870 to K.P.L.
REFERENCES
- 1.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.Rodier F, Kim SH, Nijjar T, Yaswen P, Campisi J. Cancer and aging: the importance of telomeres in genome maintenance. Int J Biochem Cell Biol. 2005;37:977–990. doi: 10.1016/j.biocel.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 3.Blasco MA. Telomere length, stem cells and aging. Nat Chem Biol. 2007;3:640–649. doi: 10.1038/nchembio.2007.38. [DOI] [PubMed] [Google Scholar]
- 4.Blasco MA, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 5.Lee HW, et al. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- 6.Chong L, et al. A human telomeric protein. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 7.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein Trf1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 8.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 9.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 10.Kim SH, Kaminker P, Campisi J. TIN2, a new regulator of telomere length in human cells. Nat Genet. 1999;23:405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou XZ, Lu KP. The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell. 2001;107:347–359. doi: 10.1016/s0092-8674(01)00538-4. [DOI] [PubMed] [Google Scholar]
- 12.Lee TH, Perrem K, Harper JW, Lu KP, Zhou XZ. F-box protein Fbx4 targets Pin2/TRF1 for ubiquitin-mediated degradation and regulates telomere maintenance. J Biol Chem. 2006;281:759–768. doi: 10.1074/jbc.M509855200. [DOI] [PubMed] [Google Scholar]
- 13.Chang W, Dynek JN, Smith S. TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev. 2003;17:1328–1333. doi: 10.1101/gad.1077103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 15.Shen M, Haggblom C, Vogt M, Hunter T, Lu KP. Characterization and cell cycle regulation of related human telomeric proteins Pin2 and TRF1 suggest a role in mitosis. Proc. Natl. Acad. Sci. USA. 1997;94:13618–13623. doi: 10.1073/pnas.94.25.13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yaffe MB, et al. Sequence-specific and phosphorylation-dependent proline isomerization: A potential mitotic regulatory mechanism. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 17.Shen M, Stukenberg PT, Kirschner MW, Lu KP. The essential mitotic peptidylprolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins. Genes Dev. 1998;12:706–720. doi: 10.1101/gad.12.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu PJ, Wulf G, Zhou XZ, Davies P, Lu KP. The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature. 1999;399:784–788. doi: 10.1038/21650. [DOI] [PubMed] [Google Scholar]
- 19.Lu PJ, Zhou XZ, Shen M, Lu KP. A function of WW domains as phosphoserine-or phosphothreonine-binding modules. Science. 1999;283:1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 20.Liou YC, et al. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc. Natl. Acad. Sci. USA. 2002;99:1335–1340. doi: 10.1073/pnas.032404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng H, et al. The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature. 2002;419:849–853. doi: 10.1038/nature01116. [DOI] [PubMed] [Google Scholar]
- 22.Liou Y-C, et al. Role of the prolyl isomerase Pin1 in protecting against age-dependent neurodegeneration. Nature. 2003;424:556–561. doi: 10.1038/nature01832. [DOI] [PubMed] [Google Scholar]
- 23.Lu KP, Zhou XZ. The prolyl isomerase Pin1: a pivotal new twist in phosphorylation signalling and human disease. Nat Rev Mol Cell Biol. 2007;8:904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- 24.Ryo A, et al. Pin1 is an E2F target gene essential for the Neu/Ras-induced transformation of mammary epithelial cells. Mol Cell Biol. 2002;22:5281–5295. doi: 10.1128/MCB.22.15.5281-5295.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atchison FW, Capel B, Means AR. Pin1 regulates the timing of mammalian primordial germ cell proliferation. Development. 2003;130:3579–3586. doi: 10.1242/dev.00584. [DOI] [PubMed] [Google Scholar]
- 26.Pastorino L, et al. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-beta production. Nature. 2006;440:528–534. doi: 10.1038/nature04543. [DOI] [PubMed] [Google Scholar]
- 27.Kishi S, Wulf G, Nakamura M, Lu KP. Telomeric protein Pin2/TRF1 induces mitotic entry and apoptosis in cells containing short telomeres and is down-regulated in breast tumors. Oncogene. 2001;20:1497–1508. doi: 10.1038/sj.onc.1204229. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura M, et al. A specific interaction between the telomeric protein Pin2/TRF1 and the mitotic spindle. Curr Biol. 2001;11:1512–1516. doi: 10.1016/s0960-9822(01)00456-0. [DOI] [PubMed] [Google Scholar]
- 29.Zhou XZ, Perrem K, Lu KP. Role of Pin2/TRF1 in telomere maintenance and cell cycle control. J. Cell. Biochem. 2003;89:19–37. doi: 10.1002/jcb.10496. [DOI] [PubMed] [Google Scholar]
- 30.Lim J, et al. Pin1 has opposite effects on wild-type and P301L tau stability and tauopathy. J. Clin. Invest. 2008;118:1877–1889. doi: 10.1172/JCI34308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. Embo J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milyavsky M, et al. Prolonged culture of telomerase-immortalized human fibroblasts leads to a premalignant phenotype. Cancer Res. 2003;63:7147–7157. [PubMed] [Google Scholar]
- 33.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. Embo J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinton P, et al. Protein kinase C beta and prolyl isomerase 1 regulate mitochondrial effects of the life-span determinant p66Shc. Science. 2007;315:659–663. doi: 10.1126/science.1135380. [DOI] [PubMed] [Google Scholar]
- 35.Sultana R, et al. Oxidative modification and down-regulation of Pin1 in Alzheimer's disease hippocampus: A redox proteomics analysis. Neurobiol Aging. 2006;27:918–925. doi: 10.1016/j.neurobiolaging.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Panossian LA, et al. Telomere shortening in T cells correlates with Alzheimer's disease status. Neurobiol Aging. 2003;24:77–84. doi: 10.1016/s0197-4580(02)00043-x. [DOI] [PubMed] [Google Scholar]
- 37.Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 38.Rufer N, Dragowska W, Thornbury G, Roosnek E, Lansdorp PM. Telomere length dynamics in human lymphocyte subpopulations measured by flow cytometry. Nat Biotechnol. 1998;16:743–747. doi: 10.1038/nbt0898-743. [DOI] [PubMed] [Google Scholar]
- 39.Chiang YJ, et al. Expression of telomerase RNA template, but not telomerase reverse transcriptase, is limiting for telomere length maintenance in vivo. Mol Cell Biol. 2004;24:7024–7031. doi: 10.1128/MCB.24.16.7024-7031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hemann MT, Greider CW. G-strand overhangs on telomeres in telomerase-deficient mouse cells. Nucleic Acids Res. 1999;27:3964–3969. doi: 10.1093/nar/27.20.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.