Abstract
Transforming growth factor-beta 1 (TGF-β1) is an important growth inhibitor of epithelial cells and insensitivity to this cytokine results in uncontrolled cell proliferation and can contribute to tumorigenesis. TGF-β1 signals through the TGF-β type I and type II receptors, and activates the Smad pathway via phosphorylation of Smad2 and Smad3. Since little is known about the selective activation of Smad2 versus Smad3, we set out to identify novel Smad2 and Smad3 interacting proteins in epithelial cells. A nontransformed human cell line was transduced with Myc-His6-Smad2 or Myc-His6-Smad3-expressing retrovirus and was treated with TGF-β1. Myc-His6-Smad2 or Myc-His6-Smad3 was purified by tandem affinity purification, eluates were subject to SDS-PAGE and Colloidal Blue staining, and select protein bands were digested with trypsin. The resulting tryptic peptides were analyzed by liquid chromatography and tandem mass spectrometry and the SEQUEST algorithm was employed to identify proteins in the bands. A number of proteins that are known to interact with Smad2 or Smad3 were detected in the eluates. In addition, a number of putative novel Smad2 and Smad3 associated proteins were identified that have functions in cell proliferation, apoptosis, Actin cytoskeleton regulation, cell motility, transcription, and Ras or insulin signaling. Specifically, the interaction between Smad2/3 and the Cdc42 guanine nucleotide exchange factor, Zizimin1, was validated by co-immunoprecipitation. The discovery of these novel Smad2 and/or Smad3 associated proteins may reveal how Smad2 and Smad3 are regulated and/or uncover new functions of Smad2 and Smad3 in TGF-β1 signaling.
Keywords: TGF-β1, Smad, Mass spectrometry, Proteomics, Zizimin1
Transforming growth factor-beta 1 (TGF-β1) is an important growth regulator of many cell types to aid in maintaining tissue homeostasis. TGF-β1 can inhibit epithelial cell proliferation while promoting the growth of some fibroblasts. The activity of TGF-β1 occurs by signaling through the TGF-β type I (TβRI) and TGF-β type II (TβRII) transmembrane serine/threonine protein kinase receptors, and activates the Smad pathway through C-terminal phosphorylation of Smad2 and Smad3 on serine residues [Macias-Silva et al., 1996]. Activated Smad2 and Smad3 oligomerize with Smad4 and translocate to the nucleus where they interact with transcription factors, transcriptional co-repressors, or transcriptional co-activators to repress or activate a host of TGF-β1 responsive genes [Abdollah et al., 1997; de Caestecker et al., 2000; Heldin et al., 1997; Kloos et al., 2002; Massague, 2000; Massague and Wotton, 2000; Nakao et al., 1997; Souchelnytskyi et al., 1997; Ten Dijke et al., 2002; Wu et al., 1997]. Resistance to the antiproliferative effects of TGF-β1 is observed in many cancers including head and neck [Chen et al., 2001b; Garrigue-Antar et al., 1995; Munoz-Antonia et al., 1996], lung [Hougaard et al., 1999], gastric [Kang et al., 1999; Myeroff et al., 1995; Park et al., 1994], colon [Grady et al., 1999; Markowitz, 2000; Markowitz et al., 1995; Myeroff et al., 1995], pancreatic [Goggins et al., 1998], ovarian [Chen et al., 2001a; Wang et al., 2000], and some recurrent breast cancers [Chen et al., 1998; Lucke et al., 2001]. This loss of sensitivity to TGF-β1 is often due to mutations in the genes that encode Smad2 and Smad4, mothers against decapentaplegic homologue (MADH) 2 and MADH4 respectively [Eppert et al., 1996; Hahn et al., 1996; Salovaara et al., 2002; Yanagisawa et al., 2000].
Smad2 and Smad3 have an amino acid sequence identity of 91%; therefore it is often thought that Smad2 and Smad3 have redundant roles in TGF-β1 signaling. In spite of this similarity, data from Smad2 and Smad3 null mice and Smad2 and Smad3 silencing in epithelial cells, suggest that Smad2 and Smad3 have distinct roles, in addition to compensatory roles (reviewed in [Brown et al., 2007]). Microarray analysis of Smad2 and Smad3 null mouse embryonic fibroblasts (MEFs) and of epithelial cells in which Smad2 or Smad3 is silenced show an activation or repression of different and overlapping subsets of genes [Kretschmer et al., 2003; Yang et al., 2003]. Interestingly, silencing of Smad3, but not silencing of Smad2, blocks the growth inhibitory response of TGF-β1 in HaCaT cells indicating that Smad3 may have a more important role in TGF-β1-mediated cell cycle arrest than Smad2 [Kretschmer et al., 2003]. The cytostatic function of Smad3 over Smad2 in TGF-β1 signaling is also revealed in a study where Smad3 silencing results in inhibition of TGF-β1-mediated cell cycle arrest in a number of TGF-β1 sensitive cell lines [Kim et al., 2005]. Furthermore, raising the relative endogenous ratio of Smad3 to Smad2 by depleting Smad2 enhances the TGF-β1 cytostatic response and Smad3 activation and transcriptional activity of TGF-β1-treated cells [Kim et al., 2005]. In contrast, Yang et al. observe TGF-β1-mediated growth inhibition in Smad3 deficient mammary gland epithelial cells in culture [Yang et al., 2002]. Compensatory changes in protein levels or phosphorylation of Smad2 were not seen in the Smad3 deficient epithelium [Yang et al., 2002]. From these Smad2 or Smad3 depletion studies it is clear that in some cell types the roles of Smad2 and Smad3 are not simply redundant but are distinct.
Overexpression of Smad2 and Smad3 in human cells further elucidates the roles of Smad2 versus Smad3. Primary hepatic stellate cells that overexpress Smad3 have increased secretion of fibronectin and type I collagen, increased chemotaxis, decreased proliferation, more focal adhesions, and increased α-smooth muscle actin organization in actin stress fibers, compared to cells overexpressing Smad2 [Uemura et al., 2005]. In HepG2 human hepatoma cells, overexpression of Smad3/4 results in higher levels of transcriptional activation of the p21 promoter than overexpression of Smad2/4 [Moustakas and Kardassis, 1998]. In addition, overexpression of Smad3 induces apoptosis in lung epithelial cells, whereas overexpression of Smad2 does not induce apoptosis to the same extent [Yanagisawa et al., 1998]. These overexpression data also suggest that Smad2 and Smad3 have distinct functions.
The regulation of Smad2 and Smad3 is complex and can occur at the level of interaction with the TGF-β receptors, nuclear import and export, and/or at the transcriptional level. This regulation occurs through the interaction of Smad2 and Smad3 with a myriad of proteins in response to TGF-β1. However, little is known about the selective activation of Smad2 versus Smad3. Collectively, the studies described above indicate that Smad2 and Smad3 may play distinct roles in TGF-β1-mediated cytoskeleton rearrangement and cell cycle control. Therefore, we further investigated the roles and regulation of Smad2 and Smad3 in TGF-β1 signaling by identifying novel Smad2 and Smad3 associated proteins by affinity purification and liquid chromatography (LC) and tandem mass spectrometry (MS/MS). A number of putative novel Smad2 and Smad3 associated proteins were identified in addition to proteins that are known to interact with Smad2 and Smad3. Specifically, a Smad2 and Smad3 association with a novel interacting protein identified by LC-MS/MS, Zizimin1, was confirmed by co-immunoprecipitation experiments. These novel associated proteins have functions in cell proliferation, apoptosis, actin cytoskeleton regulation, cell motility, transcription, and Ras or insulin signaling and may reveal how Smad2 and Smad3 are regulated and/or uncover new functions of Smad2 and Smad3 in TGF-β1 signaling.
MATERIALS AND METHODS
Cell Lines and Culture Conditions
HaCaT cells [Boukamp et al., 1988] were provided by Dr. Petra Boukamp (Deutsches Krebsforschungszentrum, Heidelberg, Germany), the retrovirus Phoenix packaging cell line was a kind gift of Dr. Garry Nolan (Stanford University, Palo Alto, CA), and the human embryonic kidney cell line (293T) was purchased from American Type Culture Collection (Rockville, MD). All cell lines were maintained in Dulbecco’s Modified Eagle’s (DME)/high-glucose medium (GIBCO, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gemini Biosciences, Woodland Hills, CA) and 1% penicillin/streptomycin.
Antibodies and Other Reagents
TGF-β1 was from R&D Systems (Minneapolis, MN). Antibodies to Smad2/3 were from BD Transduction Laboratories (San Diego, CA), phosphorylated (Ser 465/467) Smad2 from Upstate Biotechnology, Smad3 from Zymed Laboratories, Inc. (San Francisco, CA), Smad4 and Myc (9E10) from Santa Cruz Biotechnology (Santa Cruz, CA), and actin and FLAG (M2) antibodies and streptavidin-Cy3 conjugate were obtained from Sigma (St. Louis, MO). Antibodies to phosphorylated Smad3 (Ser 423/425) were a generous gift of Dr. Ed Leof (Mayo Clinic, Rochester, MN).
DNA Expression Constructs
Myc-His6-Smad2 and Myc-His6-Smad3 were generated by polymerase chain reaction (PCR) using Smad2 and Smad3 cDNA as templates and the following primers: 5′-TTTTGAATTCATGGAGCAGAAGCTGATCAGCGAGGAGGACCTGCACCACCACCACCACCACTCGTCCATCTTGCCATTC-3′ and 5′-TTTTGAATTCTTATGACATGCTTGAGCA-3′ (Smad2) and 5′-TTTTGAATTCATGGAGCAGAAGCTGATCAGCGAGGAGGACCTGCACCACCACCACCACCACTCGTCCATCCTGCCTTTC-3′ and 5′-TTTG AATTCCTAAGACACACTGGAACA-3′ (Smad3). The resulting PCR fragments were digested with EcoRI and cloned into the EcoRI site of pCDNA3 (Invitrogen, Carlsbad, CA). The cloned Myc-His6-Smad2 and Myc-His6-Smad3 PCR products were then sequenced using T3 and T7 primers (Vanderbilt University DNA Sequencing Facility). To generate retroviral expression constructs, Myc-His6-Smad2 and Myc-His6-Smad3 were digested from pCDNA3 with EcoRI and cloned into the EcoRI site of pLZRS-MS-IRES-GFP resulting in LZRS-Myc-His6-Smad2 or LZRS-Myc-His6-Smad3. FLAG-Zizimin1 was generated as described previously [Meller et al., 2004]. The Smad2 cDNA was kindly provided by Drs. Jeff Wrana and Liliana Attisano (University of Toronto, Toronto, Canada) and the Smad3 cDNA was a generous gift of Dr. Ying Zhang (National Cancer Institute, Bethesda, MD). The LZRS retroviral construct was a generous gift of Dr. Garry Nolan (Stanford University, CA) and was modified as described [Ireton et al., 2002].
Luciferase Assays
HaCaT cells were transiently transfected with pCDNA3 expression vectors encoding Myc-His6-Smad2, Myc-His6-Smad3, or empty vector. For 12XCAGA-luciferase assays, cells were co-transfected with the pCAGA12-MLP-luciferase reporter vector (pGL3) previously described [Dennler et al., 1998]. For activin response element (ARE)-luciferase assays, cells were co-transfected with the pAR3-lux reporter vector (pGL2) and the FLAG-Forkhead domain protein-1 (FAST-1) expression vector (pCS) previously described [Hayashi et al., 1997; Yeo et al., 1999]. The 3XARE-luciferase reporter vector, the FLAG-FAST-1 expression vector, and the pCAGA12-MLP-luciferase reporter vector were kindly provided by Dr. Joan Massague (Memorial Sloan-Kettering Cancer Center, NY, NY), Dr. Malcom Whitman (Harvard University, Boston, MA), and Dr. Jean-Michel Gauthier (Hybrigenics, Paris, France), respectively. Forty-eight hours post-transfection the cells were treated with TGF-β1 (5 ng/ml) for 24 h and luciferase activity measurements were performed using the Dual-Luciferase® Reporter Assay System (Promega, Madison, WI).
Generation of Stable Cell Lines
Retrovirus was produced by transfecting Phoenix packaging cells with LZRS-Myc-His6-Smad2, LZRS-Myc-His6-Smad3, or LZRS empty vector (LZRS-Ø). HaCaT cells were transduced with the above retrovirus using 5 µg/ml hexadimethrine bromide (Sigma Chemical Company, St. Louis, MO) and 48 h post-transduction, HaCaT single cell clones expressing low levels of GFP were sorted using fluorescence activated cell sorting (FACS) by Dr. James Higginbotham at Vanderbilt University.
Covalent Attachment of Myc Antibodies to Protein G Sepharose
Protein G Sepharose (PGS) (Amersham Biosciences, Piscataway, NJ) was washed and resuspended in phosphate buffered saline (PBS). Myc antibodies were then conjugated to PGS for immunoprecipitation purposes as described. Myc antibodies were incubated with PGS [10 µg Myc antibodies/10 µl PGS slurry (50:50)] overnight at 4°C. After centrifugation at 12,000 rpm for 15 s, the supernatant was aspirated and the immunocomplexes were washed three times with 0.05 M sodium borate (pH 9.0) (EMD Chemicals, Gibbstown, NJ) and were incubated in 0.05 M sodium borate (pH 9.0) containing 100 mM dimethyl pimelimidate (DMP) (Pierce Biotechnology, Inc., Rockford, IL) at 25°C for 1 h. After 1 h, additional DMP was added for a final concentration of 200 mM DMP and incubated at 25°C for 1 h. Immunocomplexes were washed two times with 0.2 M ethanolamine (pH 8.0) (Mallinckrodt Baker, Inc., Phillipsburg, NJ) and were incubated in 0.2 M ethanolamine at 25°C for 2 h. Immunocomplexes were washed two times with PBS, one time with 8 M urea (Pharmacia, Uppsala, Sweden), two times with PBS, and were stored at 4°C in 0.1% sodium azide (Fisher Scientific, Fairlawn, NJ) in PBS.
Immunoprecipitation of Myc-His6-Smad2 and Myc-His6-Smad3
To detect phosphorylation of Myc-His6-Smad2 and Myc-His6-Smad3, HaCaT cell clones stably expressing Myc-His6-Smad2, Myc-His6-Smad3, or control cells were treated with or without TGF-β1 (5 ng/ml) for 1 h. Cell extracts were prepared by lysing in TALON lysis buffer (10 mM 3-[N-morpholino] propanesulfonic acid (MOPS) pH 7.0, 2% Triton X-100, 100 mM KCl, 10% glycerol, 3 mM imidazole, 5 mM MgCl2) supplemented with protease inhibitors (50 µg/ml PMSF, 10 µg/ml antipain, 10 µg/ml leupeptin, 10 µg/ml pepstatin A, 10 µg/ml chymostatin) and phosphatase inhibitors (4 mmol/l NaF, 0.1 mmol/l Na3VO4), and harvested by scraping. Myc-His6-Smad2 or Myc-His6-Smad3 was immunoprecipitated by incubating equal amounts (0.6 mg) of the resulting lysates with Myc:PGS at 4°C for 4 h. Immunocomplexes were washed three times with TALON lysis buffer and the proteins attached to the Myc:PGS were eluted by boiling in Laemmli sample buffer. Eluates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with phospho-Smad2 (Ser 465/467), phospho-Smad3 (Ser 423/425), or Myc antibodies (Santa Cruz) as described below.
Immunofluorescence Microscopy
Sub-confluent cells, grown on 22 mm2 glass cover slips, were treated for 2 h with 5 ng/ml TGF-β1. After treatment, cells were washed one time with PBS and fixed with 4% paraformaldehyde/PBS for 20 min at 25°C. Immunofluorescence staining using Myc antibodies diluted in 5% serum/PBS (1:200) was performed and nuclei were counterstained as previously described [Brown et al., 2004].
[3H]Thymidine Incorporation Assays
Sub-confluent HaCaT cell clones stably expressing Myc-His6-Smad2, Myc-His6-Smad3, or control cells were treated with TGF-β1 (5 ng/ml) for 48 h in 12-well plates and subject to [3H]thymidine incorporation assays as previously described [Brown et al., 2004].
Tandem Affinity Purification
Twenty 150 mm plates each of LZRS-Myc-His6-Smad2, LZRS-Myc-His6-Smad3, or LZRS-Ø-transduced HaCaT cell clones were grown to 90% confluence and treated with TGF-β1 (5 ng/ml) for 1 h. The plates were then placed on ice and the cells were washed twice with ice-cold PBS, lysed in 1 ml/plate with ice-cold TALON lysis buffer supplemented with protease and phosphatase inhibitors, and harvested by scraping. Myc-His6-Smad2, Myc-His6-Smad3, and their associated proteins were purified by incubating equal amounts (60 mg) of the resulting lysates with TALON metal affinity resin (BD Biosciences, Palo Alto, CA) at 4°C for 4 h. After 4 h, a sample of the proteins that did not attach to the TALON resin was saved as TALON flow through (FT). The TALON resin was washed 3 times with TALON lysis buffer and then washed 3 times with TALON wash buffer (TALON lysis buffer supplemented with 45 mM imidazole and protease and phosphatase inhibitors). Proteins bound to the TALON resin were eluted (TALON lysis buffer supplemented with 400 mM imidazole and protease and phosphatase inhibitors) and the resulting eluates were incubated overnight at 4°C with Myc agarose (9E10 AC) (Santa Cruz). After the overnight incubation, a sample of the proteins that did not attach to the Myc agarose was saved as Myc FT. The Myc agarose was washed 4 times with TALON lysis buffer and the proteins attached to the Myc agarose were eluted by boiling in Laemmli sample buffer.
Immunoblot Analyses
For expression analysis of the Myc-His6-Smad2 and Myc-His6-Smad3 constructs, HaCaT cells were transiently transfected with pCDNA3 expression vectors encoding Myc-His6-Smad2 or Myc-His6-Smad3, or empty vector and were treated with TGF-β1 (5 ng/ml) for 24 h. Cell lysates were prepared, protein concentrations were determined, and immunoblot analysis was performed as previously described [Brown et al., 2004].
Sample Preparation for Mass Spectrometry
A fraction of the proteins resulting from the tandem affinity purification were subject to 10% SDS-PAGE (0.75 mm) and Colloidal Blue (Invitrogen) staining. Protein bands visualized by Colloidal Blue staining of SDS-polyacrylamide gels that were unique to the eluates from cells transduced with Myc-His6-Smad2 or Myc-His6-Smad3 were excised and the proteins in those bands were subject to in-gel digestion with trypsin protease as described previously [Gharahdaghi et al., 1999; Shevchenko et al., 1996; Wilm et al., 1996]. Care was taken to avoid cutting out the antibody heavy chain from each lane as the signal intensity might inhibit the identification of proteins with less intense staining. Corresponding areas in the gel of the control cell eluate were also cut out as negative controls to identify any non-specific protein binding to the TALON resin and Myc agarose.
Mass Spectrometry
Liquid chromatography (LC) and tandem mass spectrometry (MS/MS) were used to analyze the tryptic peptides that resulted from the in-gel trypsin digestion. LC-MS analysis of the resulting tryptic peptides was performed using a ThermoFinnigan LTQ ion trap mass spectrometer equipped with a Thermo MicroAS autosampler and Thermo Surveyor high performance liquid chromatography (HPLC) pump, Nanospray source, and Xcalibur 1.4 instrument control. The peptides were separated on a packed capillary tip, 100 µm × 11 cm, with C18 resin (Jupiter C18, 5 micron, 300 angstrom, Phenomenex, Torrence, CA) using an inline solid phase extraction column that was 100 µm × 6 cm packed with the same C18 resin (using a frit generated with from liquid silicate Kasil 1 [Cortes, 1987] similar to that previously described [Licklider et al., 2002], except the flow from the HPLC pump was split prior to the injection valve. The flow rate during the solid phase extraction phase of the gradient was 1 µl/min and during the separation phase it was 700 µl/min. Mobile phase A was 0.1% formic acid and mobile phase B was acetonitrile with 0.1% formic acid. A 95 min gradient was performed with a 15 min washing period (100% A for the first 10 min followed by a gradient to 98% A at 15 min) to allow for solid phase extraction and removal on any residual salts. After the initial washing period, a 60 min gradient was performed where the first 35 min was a slow, linear gradient from 98% A to 75% A, followed by a faster gradient to 10% A at 65 min and an isocratic phase at 10% A to 75 min. MS/MS scans were acquired using an isolation width of 2 m/z, and activation time of 30 ms, and activation Q of 0.250 and 30% normalized collision energy using 1 microscan and ion time of 100 for each scan. The mass spectrometer was tuned prior to analysis using the synthetic peptide TpepK (AVAGKAGAR). Typical tune parameters were as follows: spray voltage of 1.8 KV, a capillary temperature of 150°C, a capillary voltage of 50 V and tube lens 100 V. The MS/MS spectra of the peptides was performed using data-dependent scanning in which one full MS spectra, using a full mass range of 400–2000 amu, was followed by 3 MS/MS spectra.
LC-MS/MS Data Analysis
Proteins were identified using the SEQUEST algorithm [Yates et al., 1995b] using a human International Protein Index (IPI) database (http://www.ebi.ac.uk/IPI/IPIhelp.html), v3151) (forward) with the same sequences in reverse (reverse) concatenated onto the database so the false-discovery rate could be predicted. Protein matches were preliminarily filtered using the following criteria: if the charge state of the peptide is 1, the xcorr is greater than or equal to 1, the RSp is less than or equal to 5, and the Sp is greater than or equal to 350. If the charge state is 2, the xcorr is greater than or equal to 1.8, the RSp is less than or equal to 5, and the Sp is greater than or equal to 350. If the charge state is 3, the xcorr is greater than or equal to 2.5, the RSp is less than or equal to 5, and the Sp is greater than or equal to 350. Once filtered based on these scores, all protein matches that had less than two peptide matches were eliminated from the results. Proteins with single peptides matches were only reported if they were previously identified as Smad2 and/or Smad3 associated proteins. The data were then compared to see if any proteins were unique to the control or experimental samples. Proteins that appeared in the control eluates were then subtracted from the eluates expressing Myc-His6-Smad2 and Myc-His6-Smad3. The filtering, databasing, and comparison of the SEQUEST results were achieved using Complete Hierarchical Integration of Protein Searches (CHIPS), a program developed in collaboration between the University of Arizona and Vanderbilt University. Only peptides identified from the forward database were then manually searched using the Protein Information Resource (http://pir.georgetown.edu/pirwww/search/peptide.shtml) to realize any multiple protein assignments. Peptides with multiple protein assignments were indicated in the supplemental data tables presented.
The false-discovery rates were determined by multiplying the number of peptides identified in the reverse database by two, dividing the quotient by the total number of peptides identified (forward and reverse), and multiplying the result by 100% [Elias et al., 2005]. The peptides identified from the reverse database were also subject to the filtering criteria outlined above. Proteins that were identified from the reverse database in the control eluates were subtracted from the proteins that were identified from the reverse database in the Myc-His6-Smad2 and Myc-His6-Smad3 eluates.
Tandem Affinity Purification and Co-Immunoprecipitation of Myc-His6-Smad2, Myc-His6-Smad3, and FLAG-Zizimin1
293T cells were transiently transfected with FLAG-Zizimin1 and/or Myc-His6-Smad2 or Myc-His6-Smad3 and treated with TGF-β1 (5 ng/ml) for 1 h. The interaction between FLAG-Zizimin1 and Myc-His6-Smad2 or Myc-His6-Smad3 was validated by two methods that included tandem affinity purification using TALON metal affinity resin and Myc antibodies in tandem and co-immunoprecipitation using FLAG or Myc antibodies. For the tandem affinity purification the cells were lysed in TALON lysis buffer and for the co-immunoprecipitation experiments, the cells were lysed in a high salt lysis buffer (10 mM MOPS pH 7.0, 2% Triton X-100, 425 mM KCl, 10% glycerol, 5 mM MgCl2) and both lysis buffers were supplemented with protease inhibitors and phosphatase inhibitors. Myc-His6-Smad2, Myc-His6-Smad3, and their associated proteins were purified by tandem affinity purification, as described above, using equal amounts (500 µg) of the resulting lysates. Co-immunoprecipitations were performed by incubating equal amounts (3 mg) of the resulting cell lysates with FLAG agarose (M2) (Sigma) or Myc agarose at 4°C overnight. After incubation, the FLAG and Myc agarose was washed 3 times with high salt lysis buffer and proteins bound to the agarose were eluted by boiling in Laemmli sample buffer.
RESULTS
Myc-His6-Smad2 and Myc-His6-Smad3 Expression in Mammalian Cells
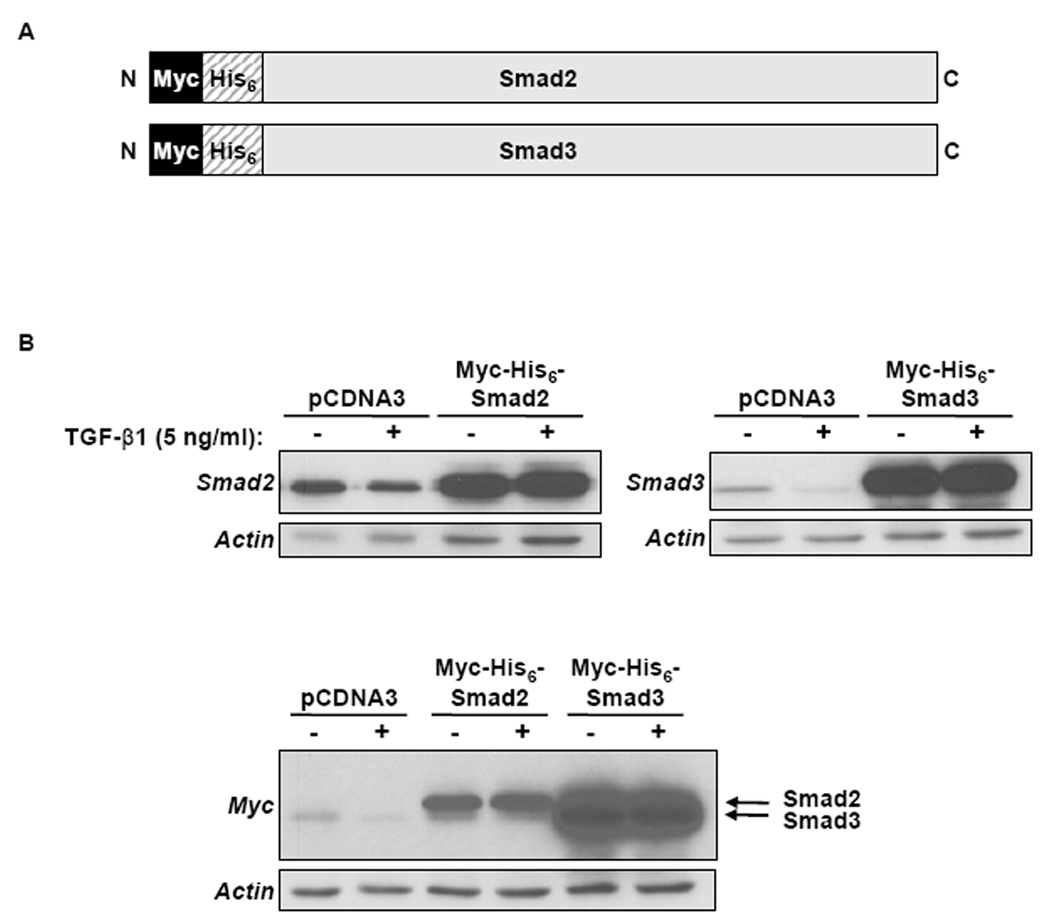
In the current study, epitope tagging and affinity purification were chosen to identify Smad2 and Smad3 interacting proteins because antibodies available for human Smad2 and Smad3 immunoprecipitation were cross-reactive with other proteins, therefore identification of novel interacting proteins with the endogenous Smad2 and Smad3 would not be possible. Myc and His6 epitope tags were affixed, in tandem, to the N-terminus of Smad2 and Smad3 by PCR (Fig. 1A). To test for expression of Myc-His6-Smad2 and Myc-His6-Smad3, the epitope-tagged Smads were transiently transfected into HaCaT cells and the cells were treated with TGF-β1. The cells were harvested 24 h after TGF-β1 treatment and immunoblot analysis was performed. Compared to vector control, high levels of Myc-His6-Smad2 and Myc-His6-Smad3 protein were detected by immunoblotting with Smad2, Smad3, and Myc antibodies (Fig. 1B). The cells were incubated with TGF-β1 for 24 h because endogenous Smad3 levels have been shown to decrease after extended TGF-β1 treatment times [Brown et al., 2004] and we wanted to ensure the overexpressed protein did not also decrease and affect the outcome of our assays. The levels of Myc-His6-Smad2 and Myc-His6-Smad3 protein did not change after TGF-β1 treatment.
Fig. 1.
Expression of Myc-His6-Smad2 or Myc-His6-Smad3 in HaCaT Cells. (A) N-terminally tagged Myc-His6-Smad2 and Myc-His6-Smad3 were generated by PCR, subcloned into the pCDNA3 vector, and the resulting products were sequenced. (B) Myc-His6-Smad2 or Myc-His6-Smad3 were transiently transfected into HaCaT cells and were treated with (+) or without (−) 5 ng/ml TGF-β1 for 24 h. Transfection with pCDNA3 was used as a control. Extracts from cells were prepared and subjected to immunoblot analyses using antibodies for Smad2, Smad3, or Myc. Actin was used as a loading control. Results are representative of at least three experiments.
Functional Activity of Ectopically Expressed Myc-His6-Smad2 and Myc-His6-Smad3
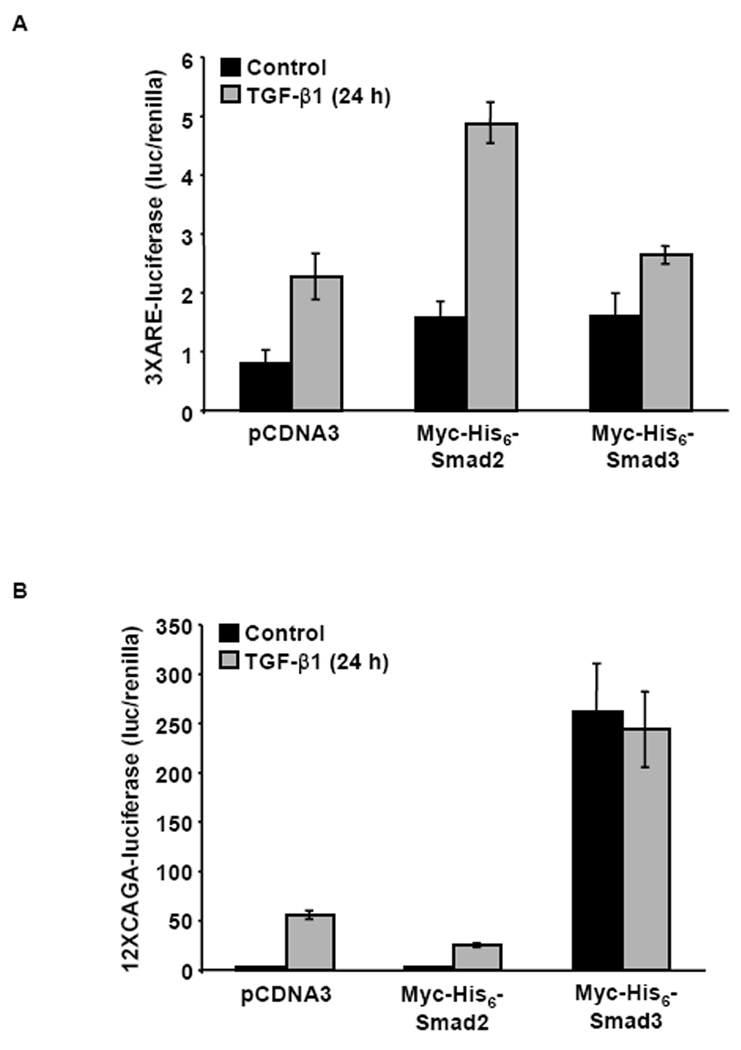
Smad2 and Smad3 can activate or repress a number of TGF-β1-responsive target genes. To ensure that ectopically expressed Myc-His6-Smad2 and Myc-His6-Smad3 were functional, we tested their transcriptional activity using luciferase reporter plasmids containing twelve “CAGA” repeats (12XCAGA), from the human PAI-1 gene promoter, or three repeats of the ARE (3XARE), from the Xenopus Mix.2 gene promoter. The 12XCAGA-luciferase reporter is activated by Smad3, but not Smad2 whereas activation of the 3XARE-luciferase reporter is dependent on expression of Smad2 and the transcription factor FAST-1, but not Smad3 and FAST-1 [Chen et al., 1997; Dennler et al., 1998; Piek et al., 2001; Zhou et al., 1998]. After transfection, cells were treated with TGF-β1 and harvested 24 h later for subsequent luciferase reporter assays. Treatment with TGF-β1 increased 3XARE-luciferase and 12XCAGA-luciferase activity in the control transfection and in HaCaT cells transfected with Myc-His6-Smad2 or Myc-His6-Smad3 (Fig. 2). Induction of luciferase activity in the control-transfected cells is attributed to endogenous Smad2 or Smad3 in the HaCaT cells. After TGF-β1 stimulation, a 3-fold increase in 3XARE-luciferase activity was observed in the control and Myc-His6-Smad2 transfected cells and a 2-fold increase in 3XARE-luciferase activity was seen in the Myc-His6-Smad3 transfected cells (Fig. 2A). After TGF-β1 stimulation, a 25-fold increase in 12XCAGA-luciferase activity was observed in the vector control cells and an 11-fold increase in 12XCAGA-luciferase activity was seen in the Myc-His6-Smad2 transfected cells (Fig. 2B). However, Myc-His6-Smad3 increased 12XCAGA-luciferase activity 120-fold in the presence or absence of TGF-β1 (Fig. 2B). These luciferase data indicate that the N-terminal epitope tags on Myc-His6-Smad2 and Myc-His6-Smad3 are functional in TGF-β1-mediated transcriptional responses.
Fig. 2.
TGF-β1 Stimulated ARE- or CAGA-Luciferase Activity in Myc-His6-Smad2 or Myc-His6-Smad3-Expressing HaCaT Cells. HaCaT cells were transiently transfected with (A) 3XARE-luciferase and FAST-1, or (B) 12XCAGA-luciferase, and Myc-His6-Smad2, Myc-His6-Smad3, or vector alone (pCDNA3). Cells were also co-transfected with renilla as a transfection control. Forty-eight hours post-transfection, cells were not treated (control) or treated with TGF-β1 for 24 h and 3XARE-luciferase, 12XCAGA-luciferase, and Renilla activities were measured. Results are presented as the average ratio of luciferase to renilla (luc/renilla) activity ± standard error of three replicates.
TGF-β1-Induced Myc-His6-Smad2 and Myc-His6-Smad3 Phosphorylation and Nuclear Translocation
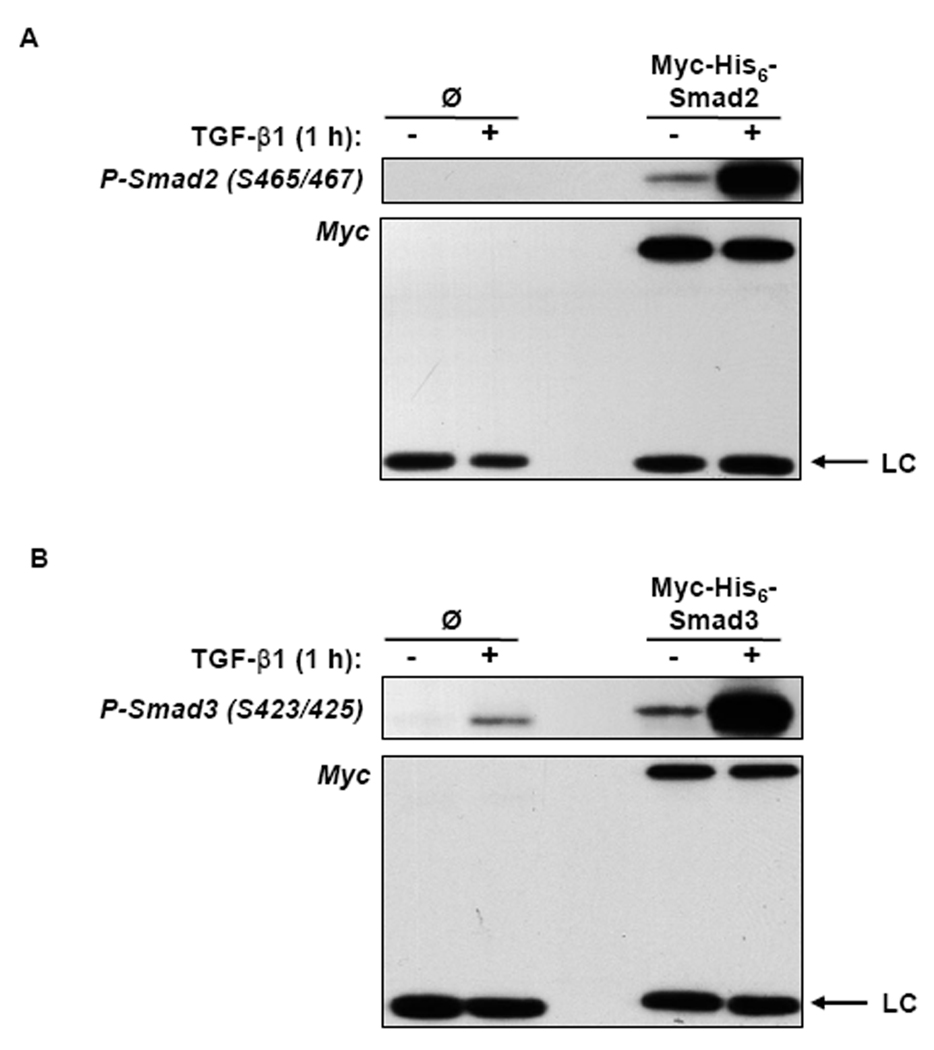
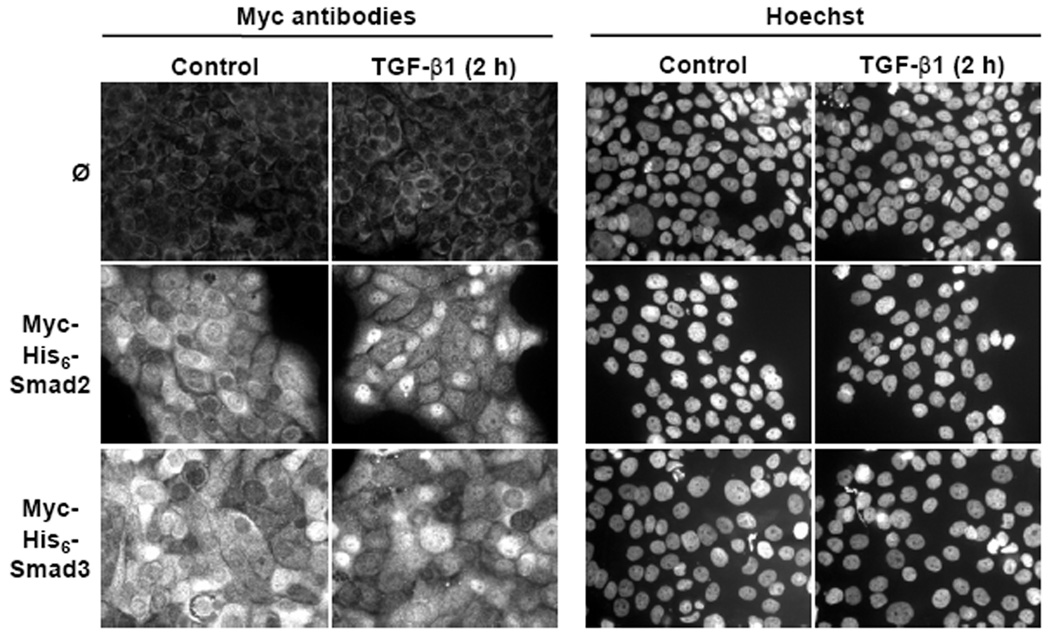
To determine if Myc-His6-Smad2 and Myc-His6-Smad3 were phosphorylated in response to TGF-β1 in the HaCaT cells, Myc-His6-Smad2 and Myc-His6-Smad3 were immunoprecipitated from lysates harvested from TGF-β1 treated cells. SDS-PAGE and immunoblot analyses revealed that after TGF-β1 treatment for 1 h, Myc-His6-Smad2 and Myc-His6-Smad3 were phosphorylated on serines 465/467 and serines 423/425, respectively (Fig. 3). A band appeared in the immunoprecipitate from the control cell lysate that was detected with phospho-Smad3 antibodies, however this band is non-specific as it did not migrate at the correct molecular weight. Myc-His6-Smad2 and Myc-His6-Smad3 did not immunoprecipitate from control cell lysates and total levels of Myc-His6-Smad2 and Myc-His6-Smad3 did not change in response to TGF-β1 (Fig. 3). Consistent with Smad phosphorylation, Myc-His6-Smad2 and Myc-His6-Smad3 accumulated in the nucleus after 2 h of TGF-β1 treatment, as demonstrated by immunofluorescence staining using Myc antibodies (Fig. 4). These immunofluorescence data, in addition to the phosphorylation data, show that the N-terminal epitope tags do not alter the ability of Myc-His6-Smad2 and Myc-His6-Smad3 to be C-terminally phosphorylated and translocate to the nucleus in response to TGF-β1.
Fig. 3.
TGF-β1 Induced Myc-His6-Smad2 and Myc-His6-Smad3 Phosphorylation. HaCaT cell clones expressing Myc-His6-Smad2, Myc-His6-Smad3, or the control clone (ø) were treated with (+) or without (−) 5 ng/ml TGF-β1 for 1 h and (A) Myc-His6-Smad2 or (B) Myc-His6-Smad3 was immunoprecipitated from total cell lysates using Myc antibodies. Phosphorylation of Myc-His6-Smad2 (Ser 465/467) and Myc-His6-Smad3 (Ser 423/425) was analyzed by SDS-PAGE and immunoblot analysis of cell lysates using phosphorylated Smad2 (P-Smad2) and Smad3 (P-Smad3) antibodies. Total Myc-His6-Smad2 and Myc-His6-Smad3 protein levels were detected by using Myc antibodies. Antibody light chain (LC) is indicated with arrows. Results are representative of two experiments.
Fig. 4.
TGF-β1 Induced Myc-His6-Smad2 and Myc-His6-Smad3 Nuclear Accumulation. HaCaT cell clones expressing Myc-His6-Smad2, Myc-His6-Smad3, or the control clone (ø) were treated with or without (control) 5 ng/ml TGF-β1 for 2 h and Myc-His6-Smad2 and Myc-His6-Smad3 localization was determined by immunofluorescence using Myc antibodies (left two columns). Hoechst was used to visualize cell nuclei (right two columns). Results are representative of two experiments.
Stable Myc-His6-Smad3 Expression in HaCaT Cells Resulted in Decreased Cell Proliferation
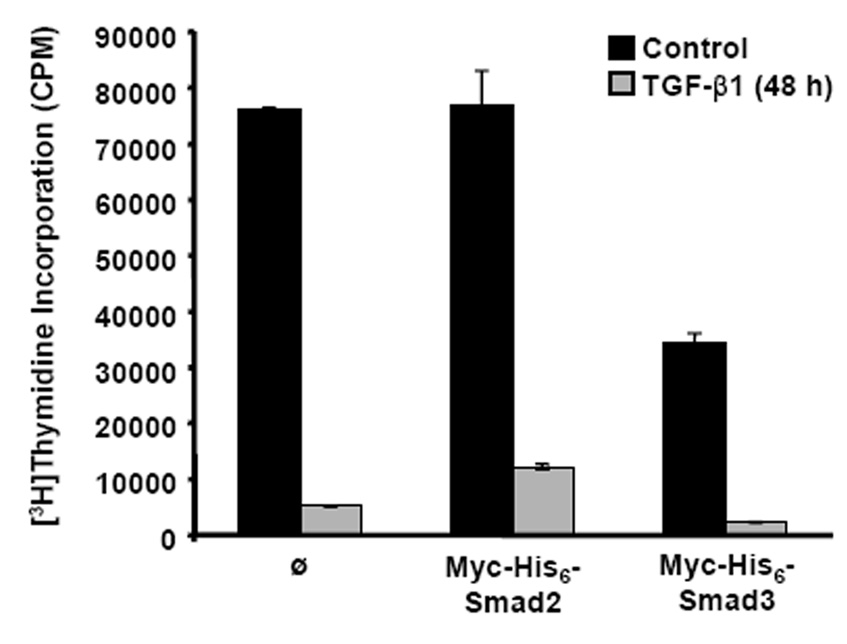
To determine if stable expression of Myc-His6-Smad2 and Myc-His6-Smad3 in HaCaT cells altered cell proliferation, [3H]thymidine incorporation assays were performed. Cells stably expressing Myc-His6-Smad3 exhibited a decrease in DNA synthesis (45% reduction) as compared to control cells and cells stably expressing Myc-His6-Smad2 (Fig. 5). After TGF-β1 treatment for 48 h, control cells and cells stably expressing Myc-His6-Smad2 or Myc-His6-Smad3 exhibited a decrease in DNA synthesis when compared to non-treated cells (Fig. 5). The control cells and Myc-His6-Smad3 expressing cells had a 94% reduction in [3H]thymidine incorporation whereas the cells expressing Myc-His6-Smad2 had an 84% reduction in [3H]thymidine incorporation.
Fig. 5.
Stable Expression of Myc-His6-Smad3 Reduced Cell Proliferation. HaCaT cell clones stably expressing Myc-His6-Smad2, Myc-His6-Smad3, or the control clone (Ø) were treated with 5 ng/ml TGF-β1 for 48 h, pulsed with [3H]thymidine for 2 h, and incorporated radioactivity was quantified. Results are presented as counts per minute (CPM) [3H]thymidine incorporation ± standard error of three replicates.
Tandem Affinity Purification of Myc-His6-Smad2 and Myc-His6-Smad3 and their Associated Proteins
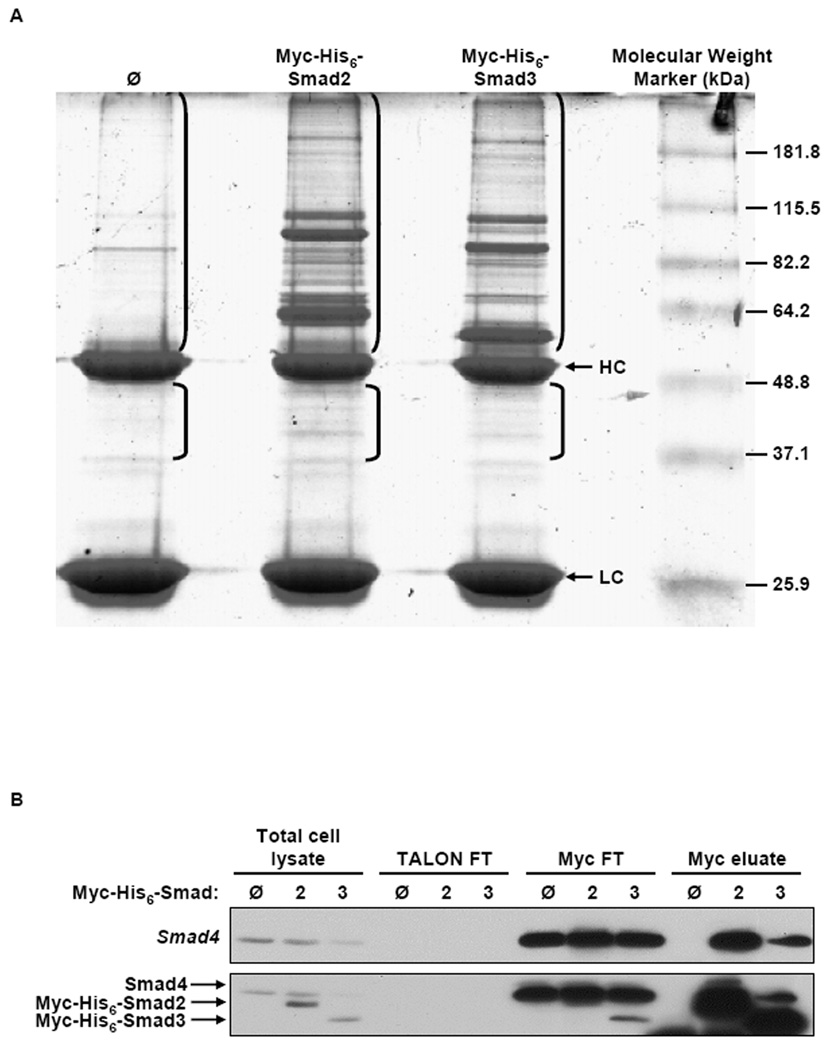
The strategy for tandem affinity purification of Myc-His6-Smad2 and Myc-His6-Smad3, and their associated proteins, is described in the Materials and Methods. Colloidal Blue staining of eluates from TGF-β1 treated, Myc-His6-Smad2 and Myc-His6-Smad3 expressing cells yielded a number of visible protein bands that were unique and not in the control eluate (Fig. 6A). These protein bands ranged in molecular weight from approximately 37 kDa to 240 kDa. SDS-PAGE and immunoblot analysis employing Myc antibodies confirmed expression of Myc-His6-Smad2 and Myc-His6-Smad3 in the appropriate total cell lysates and in the appropriate Myc eluates (Fig. 6B). Myc-His6-Smad2 and Myc-His6-Smad3 were not detected in the total cell lysate or in the Myc eluate from control cells (Fig. 6B). Myc-His6-Smad2 and Myc-His6-Smad3 were not detected in the TALON FT (Fig. 6B). In contrast, some Myc-His6-Smad2 and Myc-His6-Smad3 was detected in the Myc FT indicating that not all of the Myc-His6-Smad2 and Myc-His6-Smad3 bound to the Myc agarose (Fig. 6B). Co-immunoprecipitation of Smad4 in the Myc-His6-Smad2 and Myc-His6-Smad3 Myc eluates was also detected by immunoblotting with Smad4 antibodies (Fig. 6B). The identification of Smad4 in the Myc eluates from the Myc-His6-Smad2 and Myc-His6-Smad3 expressing cells indicated that the tandem affinity purification conditions allowed maintenance of Smad2 and Smad3 protein-protein interactions throughout the purification procedure.
Fig. 6.
Co-Purification of Smad2 and Smad3 Associated Proteins. HaCaT cells stably expressing Myc-His6-Smad2 (2), Myc-His6-Smad3 (3), or control cells (Ø) were treated with TGF-β1 (5 ng/ml) for 1 h, lysed and crude extracts (60 mg) were subjected to TALON IMAC and Myc immunoprecipitation in tandem. (A) Myc eluates were subject to SDS-PAGE and Colloidal Blue staining and sections of the gel outlined in the bracketed areas were cut from the gel and the proteins in those sections were identified by LC-MS/MS. Arrows indicate antibody heavy chain (HC) and light chain (LC). (B) Total cell lysate, Talon and Myc flow throughs (FT), and Myc eluates were subject to SDS-PAGE and immunoblot analysis using Smad4 (top) or Myc (bottom) antibodies, sequentially.
LC-MS/MS Identification of Smad2 and Smad3 Associated Proteins
In order to identify the Myc-His6-Smad2 or Myc-His6-Smad3 associated proteins from the tandem affinity purification eluates, LC-MS/MS was utilized. Sections of the Colloidal Blue stained gel containing visible proteins bands were excised from Myc-His6-Smad2 and Myc-His6-Smad3 expressing cell eluates and corresponding sections were excised from the control eluate (Fig. 6A). The proteins in these gel pieces were digested with trypsin, and the resulting tryptic peptides were analyzed by LC-MS/MS. The SEQUEST algorithm was employed to correlate fragmentation spectra of peptides with predicted amino acid sequences from protein databases [Yates et al., 1995a; Yates et al., 1995b]. In order to identify the proteins unique to the Myc-His6-Smad2 and Myc-His6-Smad3 expressing cell eluates, proteins that appeared in the control eluates were then subtracted from the eluates expressing Myc-His6-Smad2 and Myc-His6-Smad3.
Smad2 and Smad3 were identified in the eluates by LC-MS/MS from cells expressing Myc-His6-Smad2 or Myc-His6-Smad3, respectively (Table 1, Supplemental Table 1, and Supplemental Table 2). Smad2 and Smad3 were also identified in the same eluates by matrix assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (data not shown). Fourteen proteins were identified in the eluates from the cells expressing Myc-His6-Smad2 that are known to associate with Smad2 (Table 1 and Supplemental Table 1) [Abdollah et al., 1997; Akiyoshi et al., 1999; Barrios-Rodiles et al., 2005; Bonni et al., 2001; Hellemans et al., 2004; Kawabata et al., 1998; Kurisaki et al., 2003; Lagna et al., 1996; Leong et al., 2001; Lin et al., 2005; Lin et al., 2000; Luo et al., 1999; Luo et al., 2006; Nakao et al., 1997; Pan et al., 2005; Souchelnytskyi et al., 1997; Stroschein et al., 1999; Tsukazaki et al., 1998; Waddell et al., 2004; Warner et al., 2003; Zhang et al., 2001].These known interacting proteins had greater than or equal to one peptide associated with their identification. Thirteen proteins were identified in the eluates from the cells expressing Myc-His6-Smad3 that are known to associate with Smad3 (Table 1 and Supplemental Table 2). The identities of the known Smad3 associated proteins are the same as the Smad2 associated proteins except Praja1 [Saha et al., 2006] and ERK-2 [Kretzschmar et al., 1999] were identified only in the Myc-His6-Smad3 eluates and SnoN and Ski were not identified in the Myc-His6-Smad3 eluates [Wu et al., 1997]. Furthermore, DDX3Y was identified in both the Myc-His6-Smad2 and Myc-His6-Smad3 eluates but was only previously reported as a Smad2 interacting protein, as Smad3 was not studied in that report [Luo et al., 2006]. The IPI accession numbers of the proteins identified by LC-MS/MS and the total number of peptides assigned to each protein and the sequence coverage of those peptides are listed in Supplemental Table 1 and Supplemental Table 2. In addition, the tryptic peptide sequences that were used to identify the known interacting proteins and their cross correlation scores, ions/hits ratios, and the z scores, are listed in Supplemental Table 1 and Supplemental Table 2. Twelve tryptic peptides were identified in the Myc-His6-Smad2 and Myc-His6-Smad3 eluates that are found in both Smad2 and Smad3 and, therefore, cannot be used to distinguish between Smad2 and Smad3 (Supplemental Table 1 and Supplemental Table 2). However, more than four unique tryptic peptides were detected by LC-MS/MS that positively identified Smad2 and Smad3 in the eluates. The identification of these known Smad2 and Smad3 interacting proteins by LC-MS/MS validate the current method of tandem affinity purification used to identify novel Smad2 and Smad3 associated proteins.
Table 1.
Identification of Previously Reported Smad2 and/or Smad3 Associated Proteins by LC-MS/MS and SEQUEST
| # of peptidesa | ||||
|---|---|---|---|---|
| Protein | Description | Smad2 | Smad3 | References |
| Smad2 | Receptor-activated Smad | 23 | 16 | [Abdollah et al.,1997; Kawabata et al.,1998; Nakao et al.,1997; Souchelnytskyi et al.,1997] |
| Smad3 | Receptor-activated Smad | 10 | 23 | [Kawabata et al.,1998; Nakao et al.,1997] |
| Smad4 | Common mediator Smad | 15 | 4 | [Lagna et al.,1996; Nakao et al.,1997; Wu et al., 1997] |
| CKIδ | Casein kinase I δ | 2 | 2 | [Waddell et al., 2004] |
| DDX3Yb | ATP-dependent RNA helicase | 2 | 2 | [Luo et al., 2006] |
| Erbin | ErbB2 interacting protein | 11 | 12 | [Warner et al., 2003] |
| ERK-2c | Mitogen-activated protein kinase 1 | 0 | 2 | [Kretzchmar et al.,1999] |
| Hsp70 | Heat shock 70 kDa protein | 6 | 6 | [Knuesel et al., 2003; Luo et al., 2006] |
| Man1 | Inner nuclear membrane protein | 44 | 43 | [Hellemans et al., 2004; Lin et al., 2005; Pan et al., 2005] |
| Praja1 | E3 ubiquitin ligase | 0 | 1 | [Saha et al., 2006] |
| SARA | Smad anchor for receptor activation | 5 | 4 | [Tsukazaki et al., 1998] |
| Skid | Transcriptional co-repressor | 1 | 0 | [Akiyoshi et al., 1999; Luo et al., 1999] |
| Skip | Transcriptional co-activator | 1 | 1 | [Leong et al., 2001] |
| SMURF2 | E3 ubiquitin ligase | 20 | 20 | [Bonni et al., 2001; Lin et al.,2000; Zhang et al., 2001] |
| SnoNd | Transcriptional co-repressor | 1 | 0 | [Stroschein et al., 1999] |
| WWP2 | E3 ubiquitin ligase | 3 | 1 | [Barrios-Rodiles, et al., 2005] |
| YY1 | Transcriptional co-repressor | 2 | 1 | [Kurisaki et al., 2003] |
The number of unique tryptic peptides that correspond to each previously reported protein identified by LC-MS/MS in the tandem affinity purification from the Myc-His6-Smad2 (Smad2) or Myc-His6-Smad3 (Smad3) expressing HaCaT cells from the SDS-polyacrylamide gel shown in Fig. 6. The complete results are included in Supplementary Table 1 and Supplementary Table 2.
DDX3Y has been reported to interact with Smad2, but the interaction with Smad3 was not investigated in the reference indicated.
ERK-2 has been previously reported to interact with Smad2.
Ski and SnoN have been previously reported to interact with Smad3.
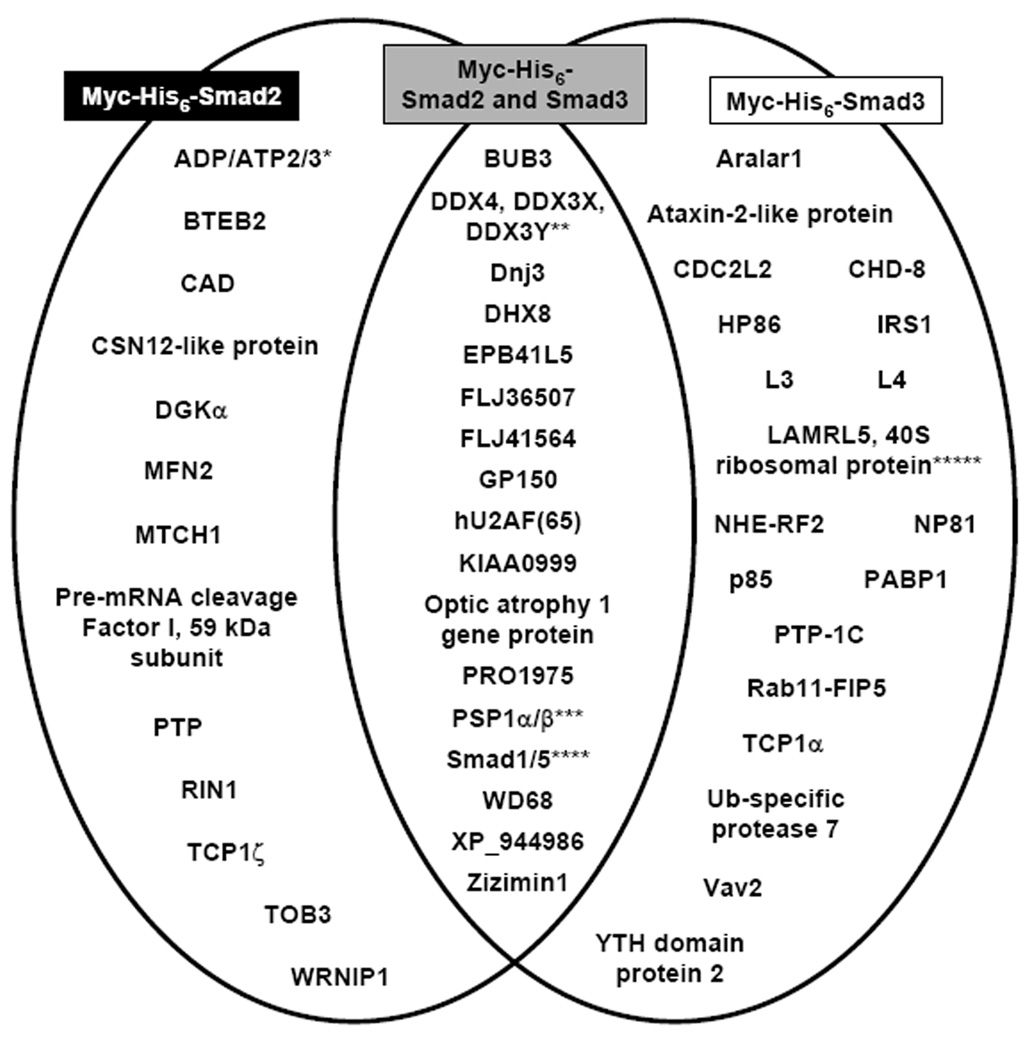
In addition to the known interacting proteins, several novel Smad2 and Smad3 associated proteins were identified by tandem affinity purification and LC-MS/MS (Supplemental Table 3 and Supplemental Table 4). The proteins listed had at least two or more peptides associated with their identification, although a number of proteins were identified with only one peptide that was utilized in their identification (data not shown). The false-discovery rates of the proteins identified from the Smad2 and Smad3 analyses were calculated to be 1.8% and 0%, respectively (Supplemental Table 5). Fourteen proteins were identified to uniquely associate with Smad2, twenty proteins were identified to uniquely associate with Smad3, and twenty-one proteins were identified to associate with both Smad2 and Smad3 (Supplemental Table 3, Supplemental Table 4, and Fig. 7). The IPI accession numbers of the proteins identified by LC-MS/MS and the total number of peptides assigned to each protein and the sequence coverage of those peptides are listed in Supplemental Table 3 and Supplemental Table 4. In addition, the tryptic peptide sequences that were used to identify the known interacting proteins and their cross correlation scores, ions/hits ratios, and the z scores, are listed in Supplemental Table 3 and Supplemental Table 4.
Fig. 7.
Identification of Novel Smad2 and Smad3 Associated Proteins. Venn diagram exhibiting the novel Smad2 and Smad3 associated proteins identified by LC-MS/MS unique to the Myc-His6-Smad2 (black box) and Myc-His6-Smad3 (white box) expressing HaCaT cells, or common to both Myc-His6-Smad2 and Myc-His6-Smad3 (gray box). These proteins were not identified in the control cells and had greater than two or more peptides matched to the protein. Thirteen proteins were identified that uniquely associated with Smad2, twenty proteins were identified that uniquely associated with Smad3, and nineteen proteins were identified that associated with both Smad2 and Smad3. *, **, ***, ****, ***** Analysis of the peptides associated with these protein identifications does not allow discrimination between ADP/ATP2 and 3, DDX4, DDX3X, and DDX3Y, PSP1α and β, Smad1 and Smad5, and LAMRL5 and 40S ribosomal protein due to high protein homology (see Supplemental Table 3 and Supplemental Table 4).
FLAG-Zizimin1 Associated with Myc-His6-Smad2 and Myc-His6-Smad3
In the tandem affinity purification experiments, dedicator of cytokinesis protein 9 (IPI Accession # IPI00151888), also known as Zizimin1, was identified to associate with Myc-His6-Smad2 and Myc-His6-Smad3. The Rho family of small GTPases, including Rac, Rho and Cdc42 are important regulators of multiple cellular activities and, most notably, reorganization of the actin cytoskeleton. Zizimin1 is a 236 kDa protein that defines a novel family of GEFs for Rho proteins. The new family, CZH proteins, contains 11 mammalian members including DOCK180 proteins. Zizimin1 was shown to activate Cdc42 yet signaling pathways involving Zizimin1 are currently unknown [Meller et al., 2005]. The identification of Zizimin1 was facilitated by the detection of six tryptic peptides in both the Myc-His6-Smad2 and Myc-His6-Smad3 eluates (Supplementary Table 3 and Supplementary Table 4). Five of the six tryptic peptides were identified in both Myc-His6-Smad2 and Myc-His6-Smad3 eluates and one unique peptide was identified in each Myc-His6-Smad2 and Myc-His6-Smad3 eluate (Fig. 8). Zizimin1 tryptic peptides were not observed in the control eluate. Since there were a large number of Zizimin1 peptides that were identified in the Smad2 and Smad3 eluates and there were reagents available to further characterize the interaction, we chose Zizimin1 to perform follow-up studies with Smad2 and Smad3.
Fig. 8.
Zizimin1 Amino Acid Sequence and the Tryptic Peptides Identified in Smad2 and Smad3 Eluates. Associated Smad2 and Smad3 proteins were purified from TGF-β1 treated HaCaT cells stably expressing Myc-His6-Smad2 or Myc-His6-Smad3 and were identified by TALON IMAC and Myc immunoprecipitation in tandem and LC-MS/MS. SEQUEST and CHIPS analysis of tryptic peptides from the tandem affinity purification and LC-MS-MS identified Zizimin1 as a novel Smad2 and Smad3 associated protein. Amino acid sequence of Zizimin1 and the six tryptic peptides identified in the Smad2 and Smad3 eluates are shown. Bold - peptides identified in the Smad2 and Smad3 eluates; Underline - peptide identified in the Smad2 eluate; Italics - peptide identified in the Smad2 eluate.
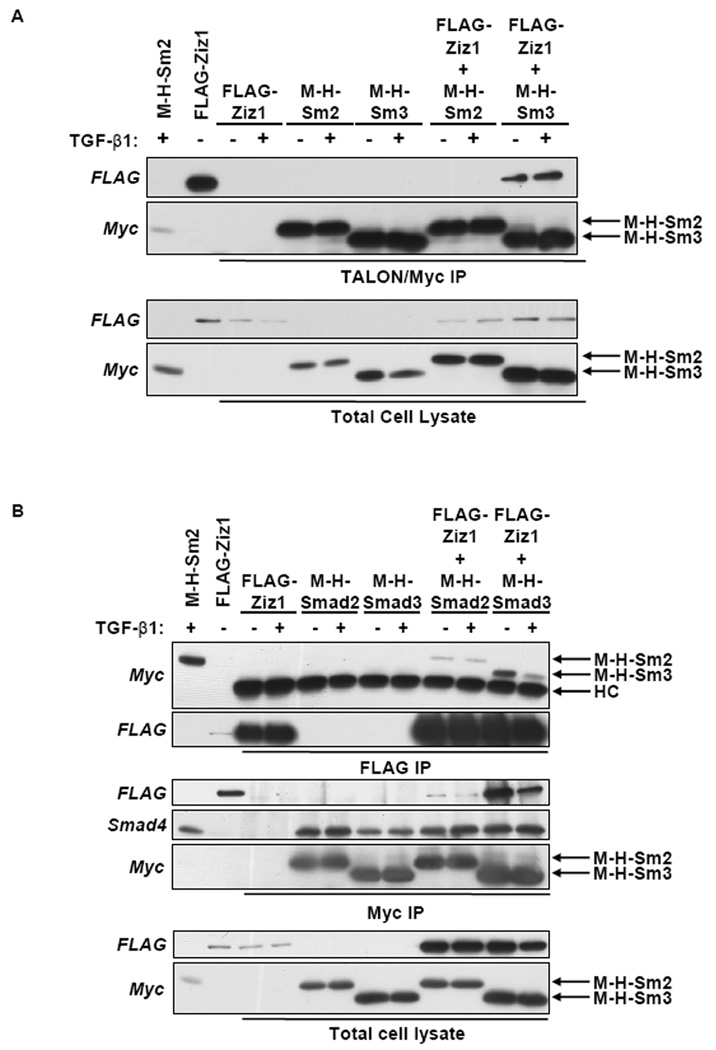
The interaction between Zizimin1 and Smad2 or Smad3 was first confirmed by tandem affinity purification of Myc-His6-Smad2 or Myc-His6-Smad3 from 293T cells that expressed FLAG-Zizimin1 and/or Myc-His6-Smad2 or Myc-His6-Smad3 and incubated with or without TGF-β1. In this experiment Myc-His6-Smad2 or Myc-His6-Smad3 was subject to TALON immobilized metal affinity chromatography (IMAC) and Myc immunoprecipitation in tandem and co-immunoprecipitation of FLAG-Zizimin1 was visualized by immunoblotting with FLAG antibodies. FLAG-Zizimin1 co-purified with Myc-His6-Smad3 regardless of TGF-β1 stimulation (Fig. 9A). After longer film exposure or immunoprecipitation of more protein, we observed co-purification of Myc-His6-Smad2 with FLAG-Zizimin1 (data not shown). However the amount of FLAG-Zizimin1 that co-purified with Myc-His6-Smad2 was far less than the amount of FLAG-Zizimin1 that co-purified with Myc-His6-Smad3. As controls, immunoprecipitations in 293T cells that only expressed FLAG-Zizimin1, Myc-His6-Smad2 or Myc-His6-Smad3 were performed. Smad4 was used as a positive control for Smad2 and Smad3 interacting proteins, and was shown to co-immunoprecipitate with Myc-His6-Smad2 and Myc-His6-Smad3 in the Myc immunoprecipitations (data not shown).
Fig. 9.
FLAG-Zizimin1 Associated with Myc-His6-Smad2 and Myc-His6-Smad3. 293T cells expressing FLAG-Zizimin1, Myc-His6-Smad2 (M-H-Sm2), or Myc-His6-Smad3 (M-H-Sm3) were treated with (+) or without (−) TGF-β1 (5 ng/ml) for 1 h, lysed, and crude extracts (3 mg) were subject to (A) TALON IMAC and Myc immunoprecipitation (IP) in tandem or (B) Myc or FLAG IP. Eluates were analyzed by SDS-PAGE and immunoblotting with FLAG, Myc, or Smad4 antibodies. A portion of each total cell lysate was analyzed as a control for protein expression levels. Extracts from HaCaT cells expressing Myc-His6-Smad2 and 293T cells expressing FLAG-Zizimin1 were used as positive controls for immunoblotting. HC, antibody heavy chain.
The association between Zizimin1 and Smad2 and Smad3 was further confirmed by co-immunoprecipitation experiments of Myc-His6-Smad2, Myc-His6-Smad3, and/or FLAG-Zizimin1 in 293T cells incubated with or without TGF-β1. Consistent with the tandem affinity co-purification results, Myc-His6-Smad2 and Myc-His6-Smad3 co-immunoprecipitated with FLAG-Zizimin1 when FLAG antibodies were used (Fig. 9B). More Myc-His6-Smad3 protein co-immunoprecipitated with FLAG-Zizimin1 than Myc-His6-Smad2. Similarly, FLAG-Zizimin1 co-immunoprecipitated with Myc-His6-Smad2 and Myc-His6-Smad3 when Myc antibodies were used in the immunoprecipitation (Fig. 9B). Although the FLAG-Zizimin1 and Myc-His6-Smad3 association decreased after incubation with TGF-β1 in this experiment this decrease was not reproducible. Smad4 was used as a positive control for Smad2 and Smad3 interacting proteins, and was shown to co-immunoprecipitate with Myc-His6-Smad2 and Myc-His6-Smad3 in the Myc immunoprecipitations. As negative controls, immunoprecipitations in 293T cells that only expressed FLAG-Zizimin1, Myc-His6-Smad2, or Myc-His6-Smad3 were performed.
DISCUSSION
In order to further understand Smad2 and Smad3 function and regulation, we identified novel Smad2 and Smad3 interacting proteins using tandem affinity purification and LC-MS/MS. Previous reports have identified novel Smad2 and/or Smad3 protein-protein interactions with techniques including yeast two-hybrid assays [Akiyoshi et al., 1999; Chen et al., 2002b; Colland et al., 2004; Lin et al., 2000; Verschueren et al., 1999; Warner et al., 2003; Wicks et al., 2005], luminescence-based mammalian interactome mapping [Barrios-Rodiles et al., 2005], and immunoprecipitation of endogenous [Luo et al., 2006] and epitope-tagged Smad proteins [Grimsby et al., 2004; Knuesel et al., 2003; Stroschein et al., 1999] combined with mass spectrometry, however our study has many advantages over some of these previous reports. Our approach using epitope tags in tandem allowed for robust purification of Myc-His6-Smad2, Myc-His6-Smad3, and their associated proteins with few proteins purified from the control cells (Fig. 6 and unpublished data). Supporting this idea was the identification of a number of bona fide Smad2 and Smad3 interacting proteins that have physiological significance in TGF-β1 signaling (Table 1). Other advantages of our study included using side-by-side experiments that allowed direct comparison of Smad2 and Smad3 associated proteins in human epithelial cells that were treated with TGF-β1 prior to purifying Smad-containing complexes. Most other research papers examined either Smad2 or Smad3 in non-human and non-TGF-β1-treated cells. Treating the cells with TGF-β1 prior to harvest allowed for improved validation of our purification technique as a greater number of previously reported Smad2 and Smad3 interacting proteins were identified than if the cells were not incubated with TGF-β1 (data not shown). Employing human epithelial cells generated data more relevant to human disease and the use of TGF-β1-treated cells allowed for inclusion of another subset of Smad2 and/or Smad3 interacting proteins that were not identified in previous studies. As a result of the experimental differences presented, our data are substantially different from earlier reports and a number of previously unreported Smad protein-protein interactions were identified in the current investigation.
The cell lysis and tandem affinity purification method used in this study liberated a number of previously reported Smad2 and Smad3 interacting proteins (Table 1). The previously identified Smad3 associated proteins were the same as the Smad2 associated proteins except SnoN and Ski were not detected in the Myc-His6-Smad3 eluate and ERK-2 was not observed in the Myc-His6-Smad2 eluate, although they have been shown to be Smad3 or Smad interacting proteins, respectively [Kretzschmar et al., 1999; Stroschein et al., 1999; Sun et al., 1999]. The lack of identification of SnoN and Ski-associated tryptic peptides in the Myc-His6-Smad3 eluate and ERK-2 in the Myc-His6-Smad2 eluate could be due to their masking by other proteins with high abundance. As a result of this phenomenon, it cannot be said from this LC-MS/MS analysis, alone, that a protein associates with only Smad2 or Smad3 if tryptic peptides are identified in one eluate and not in the other. The identification of these known Smad2 and Smad3 interacting proteins validate the current method of tandem affinity purification used to identify novel Smad2 and Smad3 associated proteins.
In addition to the previously reported Smad2 and Smad3 interacting proteins, a number of novel Smad2 and Smad3 associated proteins were identified in our study (Supplemental Table 3 and Supplemental Table 4). Additional experiments are required to determine if the novel Smad2 and Smad3 associated proteins identified in this study were directly interacting proteins, or if their association with Smad2 and Smad3 were mediated by one or more proteins. In addition, it is not clear if all of the protein interactions identified were dependent on TGF-β1 stimulation. The functions of the novel Smad2 and Smad3 associated proteins identified in the current report are broad and are either new to or have been previously associated with TGF-β1 signaling. Functions that were characterized by the identification of multiple proteins include transcriptional regulation, actin cytoskeleton regulation, cell motility, cell proliferation, apoptosis, mitochondrial functions, and bone morphogenic protein, Ras, and insulin signaling. It is well known that TGF-β1 can function to regulate the actin cytoskeleton, cell motility, and cell proliferation. TGF-β1-mediated growth inhibition is directly regulated through the Smad proteins [Chen et al., 2002a; Feng et al., 2000; Liu et al., 1997; Seoane et al., 2001; Yagi et al., 2002] and evidence also exists that Smad2 and Smad3 play a role in TGF-β1-mediated cell motility [Ashcroft and Roberts, 2000; Ashcroft et al., 1999; Hosokawa et al., 2005]. Therefore, it was not unexpected that a number of the novel Smad2 and Smad3 associated proteins identified in this study have functions involved in cell proliferation, actin cytoskeleton regulation, and cell motility.
Further authenticating the novel Smad2 and Smad3 interacting proteins identified by mass spectrometry was the validation of Zizimin1 as a novel Smad2 and Smad3 associated protein, by tandem affinity purification and co-immunoprecipitation experiments (Fig. 9). Zizimin1 is a Cdc42 GEF that induces Cdc42 activation and filopodia formation upon overexpression in NIH3T3 fibroblasts [Meller et al., 2002]. The formation of filopodia is dependent on the activation of Cdc42 by Zizimin1 [Meller et al., 2002]. Cdc42 is a member of the Rho family of GTPases, which also includes Rac and Rho, and mediates filopodia formation, gene expression, cell-cycle progression, cell polarity, and cell-cell contacts. As Cdc42 can activate a number of transcription factors [Aznar and Lacal, 2001], it is possible that Zizimin1 plays a role in the activation of Cdc42 and affect TGF-β1-mediated Smad-dependent transcriptional responses. Alternatively, Zizimin1 and Smad2/3 interaction may maintain or disrupt the actin cytoskeleton and cell polarity, or be involved in TGF-β1-mediated epithelial to mesenchymal transition. It is well known that TGF- β1 can function to regulate the actin cytoskeleton and cell motility. Evidence also exists that Smad2 and Smad3 play a role in TGF-β1-mediated cell motility [Ashcroft and Roberts, 2000; Ashcroft et al., 1999; Hosokawa et al., 2005]. Furthermore, in the prostate carcinoma cell line (PC-3U) and the breast carcinoma cell line (MDA-MB-468), TGF-β1 stimulation activated Cdc42 and this activation was shown to be necessary for actin cytoskeleton reorganization in response to TGF-β1 and was dependent on Smad4 [Edlund et al., 2002; Edlund et al., 2004]. A role for Smad2 and Smad3 in mediating Cdc42 activation has not been reported, however it is possible that Zizimin1 mediates TGF-β1 activation of Cdc42 in these cell lines.
Our primary effort in the present study was to isolate Smad 2 and Smad3 interacting proteins. Interestingly, we identified proteins that interacted with Smad2 or Smad3 and not with both of them. This discovery may lead to the functional characterization of proteins that regulate distinct Smad2 or Smad3 activities. Of note, we observed that stable expression of Myc-His6-Smad3, and not Myc-His6-Smad2, in HaCaT cells resulted in a decrease in non-stimulated cell proliferation (Fig. 5). These data are consistent with previous reports that suggest Smad3, not Smad2, is the primary modulator of TGF-β1 growth inhibitory responses. Silencing of Smad3, but not Smad2 silencing, blocks the growth inhibitory response of TGF-β1 in human epithelial cells indicating that Smad3 may have a more important role in TGF-β1-mediated cell cycle arrest than Smad2 [Kim et al., 2005; Kretschmer et al., 2003]. In addition, raising the relative endogenous ratio of Smad3 to Smad2 by depleting Smad2 enhances the TGF-β1 cytostatic response [Kim et al., 2005]. The mass spectrometry and Smad2/3 overexpression data, together, support the idea that Smad2 and Smad3 may have separate, in addition to overlapping, roles in TGF-β signaling. This area of investigation warrants further attention.
TGF-β1 induction of Smad2 and Smad3 phosphorylation results in a number of Smad2 and Smad3 protein interactions that regulate Smad2 and Smad3 signaling. These protein-protein interactions occur in the nucleus, cytoplasm, or at the mitochondria and may aid in selectively activating Smad2 versus Smad3, modulating their functions to activate or repress TGF-β1 target genes, or in regulating yet unknown functions of Smad2 and Smad3. The current study unveiled previously unreported Smad2 and Smad3 interacting proteins that will reveal potentially unique roles for Smad2 and Smad3-mediated signaling and/or novel regulators of Smad2 and/or Smad3. These novel interacting proteins and pathways involved in Smad2 and Smad3 signaling act as a platform from which to perform further experiments, as the physiological relevance of select novel protein-protein interactions is currently under investigation in our laboratory. Furthermore, our data may show that Smad2 and Smad3 have distinct, in addition to overlapping, functions in TGF-β1 signaling.
Supplementary Material
ACKNOWLEDGMENTS
We thank Salisha Hill and Jade Johnston for their technical assistance in the Proteomics Laboratory in the Mass Spectrometry Research Center and Dr. Martin Schwartz for use of Zizimin1 reagents and guidance. We would also like to thank Roger Dahlman for designing and implementing an application to determine coverage of proteins from peptide sequences.
Contract grant sponsor: NIH; Contract grant number: R01CA085492 and R01CA102162 (H.L.M.)
Contract grant sponsor: NIH; Contract grant number: T32CA09592 (K.A.B.)
Contract grant sponsor: NIH; Contract grant number: P30ES000267 and P30CA68485 (Core Services)
REFERENCES
- Abdollah S, Macias-Silva M, Tsukazaki T, Hayashi H, Attisano L, Wrana JL. TbetaRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J Biol Chem. 1997;272:27678–27685. doi: 10.1074/jbc.272.44.27678. [DOI] [PubMed] [Google Scholar]
- Akiyoshi S, Inoue H, Hanai J, Kusanagi K, Nemoto N, Miyazono K, Kawabata M. c-Ski acts as a transcriptional co-repressor in transforming growth factor-beta signaling through interaction with smads. J Biol Chem. 1999;274:35269–35277. doi: 10.1074/jbc.274.49.35269. [DOI] [PubMed] [Google Scholar]
- Ashcroft GS, Roberts AB. Loss of Smad3 modulates wound healing. Cytokine Growth Factor Rev. 2000;11:125–131. doi: 10.1016/s1359-6101(99)00036-2. [DOI] [PubMed] [Google Scholar]
- Ashcroft GS, Yang X, Glick AB, Weinstein M, Letterio JL, Mizel DE, Anzano M, Greenwell-Wild T, Wahl SM, Deng C, Roberts AB. Mice lacking Smad3 show accelerated wound healing and an impaired local inflammatory response. Nat Cell Biol. 1999;1:260–266. doi: 10.1038/12971. [DOI] [PubMed] [Google Scholar]
- Aznar S, Lacal JC. Rho signals to cell growth and apoptosis. Cancer Lett. 2001;165:1–10. doi: 10.1016/s0304-3835(01)00412-8. [DOI] [PubMed] [Google Scholar]
- Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, Luga V, Przulj N, Robinson M, Suzuki H, Hayashizaki Y, Jurisica I, Wrana JL. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- Bonni S, Wang HR, Causing CG, Kavsak P, Stroschein SL, Luo K, Wrana JL. TGF-beta induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat Cell Biol. 2001;3:587–595. doi: 10.1038/35078562. [DOI] [PubMed] [Google Scholar]
- Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KA, Aakre ME, Gorska AE, Price JO, Eltom SE, Pietenpol JA, Moses HL. Induction by transforming growth factor-beta1 of epithelial to mesenchymal transition is a rare event in vitro. Breast Cancer Res. 2004;6:R215–R231. doi: 10.1186/bcr778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KA, Pietenpol JA, Moses HL. A tale of two proteins: differential roles and regulation of Smad2 and Smad3 in TGF-beta signaling. J Cell Biochem. 2007;101:9–33. doi: 10.1002/jcb.21255. [DOI] [PubMed] [Google Scholar]
- Chen CR, Kang Y, Siegel PM, Massague J. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell. 2002a;110:19–32. doi: 10.1016/s0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- Chen F, Ogawa K, Liu X, Stringfield TM, Chen Y. Repression of Smad2 and Smad3 transactivating activity by association with a novel splice variant of CCAAT-binding factor C subunit. Biochem J. 2002b;364:571–577. doi: 10.1042/BJ20011703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Carter D, Garrigue-Antar L, Reiss M. Transforming growth factor beta type I receptor kinase mutant associated with metastatic breast cancer. Cancer Res. 1998;58:4805–4810. [PubMed] [Google Scholar]
- Chen T, Triplett J, Dehner B, Hurst B, Colligan B, Pemberton J, Graff JR, Carter JH. Transforming growth factor-beta receptor type I gene is frequently mutated in ovarian carcinomas. Cancer Res. 2001a;61:4679–4682. [PubMed] [Google Scholar]
- Chen T, Yan W, Wells RG, Rimm DL, McNiff J, Leffell D, Reiss M. Novel inactivating mutations of transforming growth factor-beta type I receptor gene in head-and-neck cancer metastases. Int J Cancer. 2001b;93:653–661. doi: 10.1002/ijc.1381. [DOI] [PubMed] [Google Scholar]
- Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- Colland F, Jacq X, Trouplin V, Mougin C, Groizeleau C, Hamburger A, Meil A, Wojcik J, Legrain P, Gauthier JM. Functional proteomics mapping of a human signaling pathway. Genome Res. 2004;14:1324–1332. doi: 10.1101/gr.2334104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes HJ, Pfeiffer C, Richter B, Stevens T. Porous ceramic bed supports for fused silica packed capillary columns used in liquid chromatography. High Resolution Chromatography & Chromatography Communications. 1987;10:446–448. [Google Scholar]
- de Caestecker MP, Piek E, Roberts AB. Role of transforming growth factor-beta signaling in cancer. J Natl Cancer Inst. 2000;92:1388–1402. doi: 10.1093/jnci/92.17.1388. [DOI] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. Embo J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund S, Landstrom M, Heldin CH, Aspenstrom P. Transforming growth factor-beta-induced mobilization of actin cytoskeleton requires signaling by small GTPases Cdc42 and RhoA. Mol Biol Cell. 2002;13:902–914. doi: 10.1091/mbc.01-08-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund S, Landstrom M, Heldin CH, Aspenstrom P. Smad7 is required for TGF-beta-induced activation of the small GTPase Cdc42. J Cell Sci. 2004;117:1835–1847. doi: 10.1242/jcs.01036. [DOI] [PubMed] [Google Scholar]
- Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui LC, Bapat B, Gallinger S, Andrulis IL, Thomsen GH, Wrana JL, Attisano L. MADR2 maps to 18q21 and encodes a TGFbeta-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- Feng XH, Lin X, Derynck R. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-beta. Embo J. 2000;19:5178–5193. doi: 10.1093/emboj/19.19.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigue-Antar L, Munoz-Antonia T, Antonia SJ, Gesmonde J, Vellucci VF, Reiss M. Missense mutations of the transforming growth factor beta type II receptor in human head and neck squamous carcinoma cells. Cancer Res. 1995;55:3982–3987. [PubMed] [Google Scholar]
- Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999;20:601–605. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Goggins M, Shekher M, Turnacioglu K, Yeo CJ, Hruban RH, Kern SE. Genetic alterations of the transforming growth factor beta receptor genes in pancreatic and biliary adenocarcinomas. Cancer Res. 1998;58:5329–5332. [PubMed] [Google Scholar]
- Grady WM, Myeroff LL, Swinler SE, Rajput A, Thiagalingam S, Lutterbaugh JD, Neumann A, Brattain MG, Chang J, Kim SJ, Kinzler KW, Vogelstein B, Willson JK, Markowitz S. Mutational inactivation of transforming growth factor beta receptor type II in microsatellite stable colon cancers. Cancer Res. 1999;59:320–324. [PubMed] [Google Scholar]
- Grimsby S, Jaensson H, Dubrovska A, Lomnytska M, Hellman U, Souchelnytskyi S. Proteomics-based identification of proteins interacting with Smad3: SREBP-2 forms a complex with Smad3 and inhibits its transcriptional activity. FEBS Lett. 2004;577:93–100. doi: 10.1016/j.febslet.2004.09.069. [DOI] [PubMed] [Google Scholar]
- Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA, Topper JN, Gimbrone MA, Jr, Wrana JL, Falb D. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Hellemans J, Preobrazhenska O, Willaert A, Debeer P, Verdonk PC, Costa T, Janssens K, Menten B, Roy NV, Vermeulen SJ, Savarirayan R, Hul WV, Vanhoenacker F, Huylebroeck D, Paepe AD, Naeyaert JM, Vandesompele J, Speleman F, Verschueren K, Coucke PJ, Mortier GR. Loss-of-function mutations in LEMD3 result in osteopoikilosis, Buschke-Ollendorff syndrome and melorheostosis. Nat Genet. 2004;36:1213–1218. doi: 10.1038/ng1453. [DOI] [PubMed] [Google Scholar]
- Hosokawa R, Urata MM, Ito Y, Bringas P, Jr, Chai Y. Functional significance of Smad2 in regulating basal keratinocyte migration during wound healing. J Invest Dermatol. 2005;125:1302–1309. doi: 10.1111/j.0022-202X.2005.23963.x. [DOI] [PubMed] [Google Scholar]
- Hougaard S, Norgaard P, Abrahamsen N, Moses HL, Spang-Thomsen M, Skovgaard Poulsen H. Inactivation of the transforming growth factor beta type II receptor in human small cell lung cancer cell lines. Br J Cancer. 1999;79:1005–1011. doi: 10.1038/sj.bjc.6690161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton RC, Davis MA, van Hengel J, Mariner DJ, Barnes K, Thoreson MA, Anastasiadis PZ, Matrisian L, Bundy LM, Sealy L, Gilbert B, van Roy F, Reynolds AB. A novel role for p120 catenin in E-cadherin function. J Cell Biol. 2002;159:465–476. doi: 10.1083/jcb.200205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SH, Bang YJ, Im YH, Yang HK, Lee DA, Lee HY, Lee HS, Kim NK, Kim SJ. Transcriptional repression of the transforming growth factor-beta type I receptor gene by DNA methylation results in the development of TGF-beta resistance in human gastric cancer. Oncogene. 1999;18:7280–7286. doi: 10.1038/sj.onc.1203146. [DOI] [PubMed] [Google Scholar]
- Kawabata M, Inoue H, Hanyu A, Imamura T, Miyazono K. Smad proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. Embo J. 1998;17:4056–4065. doi: 10.1093/emboj/17.14.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SG, Kim HA, Jong HS, Park JH, Kim NK, Hong SH, Kim TY, Bang YJ. The endogenous ratio of Smad2 and Smad3 influences the cytostatic function of Smad3. Mol Biol Cell. 2005;16:4672–4683. doi: 10.1091/mbc.E05-01-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloos DU, Choi C, Wingender E. The TGF-beta--Smad network: introducing bioinformatic tools. Trends Genet. 2002;18:96–103. doi: 10.1016/s0168-9525(02)02556-8. [DOI] [PubMed] [Google Scholar]
- Knuesel M, Wan Y, Xiao Z, Holinger E, Lowe N, Wang W, Liu X. Identification of novel protein-protein interactions using a versatile mammalian tandem affinity purification expression system. Mol Cell Proteomics. 2003;2:1225–1233. doi: 10.1074/mcp.T300007-MCP200. [DOI] [PubMed] [Google Scholar]
- Kretschmer A, Moepert K, Dames S, Sternberger M, Kaufmann J, Klippel A. Differential regulation of TGF-beta signaling through Smad2, Smad3 and Smad4. Oncogene. 2003;22:6748–6763. doi: 10.1038/sj.onc.1206791. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Doody J, Timokhina I, Massague J. A mechanism of repression of TGFbeta/ Smad signaling by oncogenic Ras. Genes Dev. 1999;13:804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisaki K, Kurisaki A, Valcourt U, Terentiev AA, Pardali K, Ten Dijke P, Heldin CH, Ericsson J, Moustakas A. Nuclear factor YY1 inhibits transforming growth factor beta- and bone morphogenetic protein-induced cell differentiation. Mol Cell Biol. 2003;23:4494–4510. doi: 10.1128/MCB.23.13.4494-4510.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Partnership between DPC4 and SMAD proteins in TGF-beta signalling pathways. Nature. 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- Leong GM, Subramaniam N, Figueroa J, Flanagan JL, Hayman MJ, Eisman JA, Kouzmenko AP. Ski-interacting protein interacts with Smad proteins to augment transforming growth factor-beta-dependent transcription. J Biol Chem. 2001;276:18243–18248. doi: 10.1074/jbc.M010815200. [DOI] [PubMed] [Google Scholar]
- Licklider LJ, Thoreen CC, Peng J, Gygi SP. Automation of nanoscale microcapillary liquid chromatography-tandem mass spectrometry with a vented column. Anal Chem. 2002;74:3076–3083. doi: 10.1021/ac025529o. [DOI] [PubMed] [Google Scholar]
- Lin F, Morrison JM, Wu W, Worman HJ. MAN1, an integral protein of the inner nuclear membrane, binds Smad2 and Smad3 and antagonizes transforming growth factor-beta signaling. Hum Mol Genet. 2005;14:437–445. doi: 10.1093/hmg/ddi040. [DOI] [PubMed] [Google Scholar]
- Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- Liu X, Sun Y, Constantinescu SN, Karam E, Weinberg RA, Lodish HF. Transforming growth factor beta-induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc Natl Acad Sci U S A. 1997;94:10669–10674. doi: 10.1073/pnas.94.20.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucke CD, Philpott A, Metcalfe JC, Thompson AM, Hughes-Davies L, Kemp PR, Hesketh R. Inhibiting mutations in the transforming growth factor beta type 2 receptor in recurrent human breast cancer. Cancer Res. 2001;61:482–485. [PubMed] [Google Scholar]
- Luo K, Stroschein SL, Wang W, Chen D, Martens E, Zhou S, Zhou Q. The Ski oncoprotein interacts with the Smad proteins to repress TGFbeta signaling. Genes Dev. 1999;13:2196–2206. doi: 10.1101/gad.13.17.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q, Nieves E, Kzhyshkowska J, Angeletti RH. Endogenous Transforming Growth Factor-{beta} Receptor-mediated Smad Signaling Complexes Analyzed by Mass Spectrometry. Mol Cell Proteomics. 2006;5:1245–1260. doi: 10.1074/mcp.M600065-MCP200. [DOI] [PubMed] [Google Scholar]
- Macias-Silva M, Abdollah S, Hoodless PA, Pirone R, Attisano L, Wrana JL. MADR2 is a substrate of the TGFbeta receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- Markowitz S. TGF-beta receptors and DNA repair genes, coupled targets in a pathway of human colon carcinogenesis. Biochim Biophys Acta. 2000;1470:M13–M20. doi: 10.1016/s0304-419x(99)00031-1. [DOI] [PubMed] [Google Scholar]
- Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- Massague J, Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. Embo J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller N, Irani-Tehrani M, Kiosses WB, Del Pozo MA, Schwartz MA. Zizimin1, a novel Cdc42 activator, reveals a new GEF domain for Rho proteins. Nat Cell Biol. 2002;4:639–647. doi: 10.1038/ncb835. [DOI] [PubMed] [Google Scholar]
- Meller N, Irani-Tehrani M, Ratnikov BI, Paschal BM, Schwartz MA. The novel Cdc42 guanine nucleotide exchange factor, zizimin1, dimerizes via the Cdc42-binding CZH2 domain. J Biol Chem. 2004;279:37470–37476. doi: 10.1074/jbc.M404535200. [DOI] [PubMed] [Google Scholar]
- Meller N, Merlot S, Guda C. CZH proteins: a new family of Rho-GEFs. J Cell Sci. 2005;118:4937–4946. doi: 10.1242/jcs.02671. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Kardassis D. Regulation of the human p21/WAF1/Cip1 promoter in hepatic cells by functional interactions between Sp1 and Smad family members. Proc Natl Acad Sci U S A. 1998;95:6733–6738. doi: 10.1073/pnas.95.12.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Antonia T, Li X, Reiss M, Jackson R, Antonia S. A mutation in the transforming growth factor beta type II receptor gene promoter associated with loss of gene expression. Cancer Res. 1996;56:4831–4835. [PubMed] [Google Scholar]
- Myeroff LL, Parsons R, Kim SJ, Hedrick L, Cho KR, Orth K, Mathis M, Kinzler KW, Lutterbaugh J, Park K, et al. A transforming growth factor beta receptor type II gene mutation common in colon and gastric but rare in endometrial cancers with microsatellite instability. Cancer Res. 1995;55:5545–5547. [PubMed] [Google Scholar]
- Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin CH, Miyazono K, ten Dijke P. TGF-beta receptor-mediated signalling through Smad2, Smad3 and Smad4. Embo J. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Estevez-Salmeron LD, Stroschein SL, Zhu X, He J, Zhou S, Luo K. The integral inner nuclear membrane protein MAN1 physically interacts with the R-Smad proteins to repress signaling by the transforming growth factor-{beta} superfamily of cytokines. J Biol Chem. 2005;280:15992–16001. doi: 10.1074/jbc.M411234200. [DOI] [PubMed] [Google Scholar]
- Park K, Kim SJ, Bang YJ, Park JG, Kim NK, Roberts AB, Sporn MB. Genetic changes in the transforming growth factor beta (TGF-beta) type II receptor gene in human gastric cancer cells: correlation with sensitivity to growth inhibition by TGF-beta. Proc Natl Acad Sci U S A. 1994;91:8772–8776. doi: 10.1073/pnas.91.19.8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piek E, Ju WJ, Heyer J, Escalante-Alcalde D, Stewart CL, Weinstein M, Deng C, Kucherlapati R, Bottinger EP, Roberts AB. Functional characterization of transforming growth factor beta signaling in Smad2- and Smad3-deficient fibroblasts. J Biol Chem. 2001;276:19945–19953. doi: 10.1074/jbc.M102382200. [DOI] [PubMed] [Google Scholar]
- Saha T, Vardhini D, Tang Y, Katuri V, Jogunoori W, Volpe EA, Haines D, Sidawy A, Zhou X, Gallicano I, Schlegel R, Mishra B, Mishra L. RING finger-dependent ubiquitination by PRAJA is dependent on TGF-beta and potentially defines the functional status of the tumor suppressor ELF. Oncogene. 2006;25:693–705. doi: 10.1038/sj.onc.1209123. [DOI] [PubMed] [Google Scholar]
- Salovaara R, Roth S, Loukola A, Launonen V, Sistonen P, Avizienyte E, Kristo P, Jarvinen H, Souchelnytskyi S, Sarlomo-Rikala M, Aaltonen LA. Frequent loss of SMAD4/DPC4 protein in colorectal cancers. Mol Pathol. 2002;55:385–388. doi: 10.1136/gut.51.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, Mortensen P, Shevchenko A, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci U S A. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souchelnytskyi S, Tamaki K, Engstrom U, Wernstedt C, ten Dijke P, Heldin CH. Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-beta signaling. J Biol Chem. 1997;272:28107–28115. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- Stroschein SL, Wang W, Zhou S, Zhou Q, Luo K. Negative feedback regulation of TGF-beta signaling by the SnoN oncoprotein. Science. 1999;286:771–774. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- Sun Y, Liu X, Eaton EN, Lane WS, Lodish HF, Weinberg RA. Interaction of the Ski oncoprotein with Smad3 regulates TGF-beta signaling. Mol Cell. 1999;4:499–509. doi: 10.1016/s1097-2765(00)80201-4. [DOI] [PubMed] [Google Scholar]
- Ten Dijke P, Goumans MJ, Itoh F, Itoh S. Regulation of cell proliferation by Smad proteins. J Cell Physiol. 2002;191:1–16. doi: 10.1002/jcp.10066. [DOI] [PubMed] [Google Scholar]
- Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- Uemura M, Swenson ES, Gaca MD, Giordano FJ, Reiss M, Wells RG. Smad2 and Smad3 play different roles in rat hepatic stellate cell function and alpha-smooth muscle actin organization. Mol Biol Cell. 2005;16:4214–4224. doi: 10.1091/mbc.E05-02-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschueren K, Remacle JE, Collart C, Kraft H, Baker BS, Tylzanowski P, Nelles L, Wuytens G, Su MT, Bodmer R, Smith JC, Huylebroeck D. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5'-CACCT sequences in candidate target genes. J Biol Chem. 1999;274:20489–20498. doi: 10.1074/jbc.274.29.20489. [DOI] [PubMed] [Google Scholar]
- Waddell DS, Liberati NT, Guo X, Frederick JP, Wang XF. Casein kinase Iepsilon plays a functional role in the transforming growth factor-beta signaling pathway. J Biol Chem. 2004;279:29236–29246. doi: 10.1074/jbc.M400880200. [DOI] [PubMed] [Google Scholar]
- Wang D, Kanuma T, Mizunuma H, Takama F, Ibuki Y, Wake N, Mogi A, Shitara Y, Takenoshita S. Analysis of specific gene mutations in the transforming growth factor-beta signal transduction pathway in human ovarian cancer. Cancer Res. 2000;60:4507–4512. [PubMed] [Google Scholar]
- Warner DR, Pisano MM, Roberts EA, Greene RM. Identification of three novel Smad binding proteins involved in cell polarity. FEBS Lett. 2003;539:167–173. doi: 10.1016/s0014-5793(03)00155-8. [DOI] [PubMed] [Google Scholar]
- Wicks SJ, Haros K, Maillard M, Song L, Cohen RE, Dijke PT, Chantry A. The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF-beta signalling. Oncogene. 2005;24:8080–8084. doi: 10.1038/sj.onc.1208944. [DOI] [PubMed] [Google Scholar]
- Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- Wu RY, Zhang Y, Feng XH, Derynck R. Heteromeric and homomeric interactions correlate with signaling activity and functional cooperativity of Smad3 and Smad4/DPC4. Mol Cell Biol. 1997;17:2521–2528. doi: 10.1128/mcb.17.5.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi K, Furuhashi M, Aoki H, Goto D, Kuwano H, Sugamura K, Miyazono K, Kato M. c-myc is a downstream target of the Smad pathway. J Biol Chem. 2002;277:854–861. doi: 10.1074/jbc.M104170200. [DOI] [PubMed] [Google Scholar]
- Yanagisawa K, Osada H, Masuda A, Kondo M, Saito T, Yatabe Y, Takagi K, Takahashi T. Induction of apoptosis by Smad3 and down-regulation of Smad3 expression in response to TGF-beta in human normal lung epithelial cells. Oncogene. 1998;17:1743–1747. doi: 10.1038/sj.onc.1202052. [DOI] [PubMed] [Google Scholar]
- Yanagisawa K, Uchida K, Nagatake M, Masuda A, Sugiyama M, Saito T, Yamaki K, Takahashi T, Osada H. Heterogeneities in the biological and biochemical functions of Smad2 and Smad4 mutants naturally occurring in human lung cancers. Oncogene. 2000;19:2305–2311. doi: 10.1038/sj.onc.1203591. [DOI] [PubMed] [Google Scholar]
- Yang YA, Tang B, Robinson G, Hennighausen L, Brodie SG, Deng CX, Wakefield LM. Smad3 in the mammary epithelium has a nonredundant role in the induction of apoptosis, but not in the regulation of proliferation or differentiation by transforming growth factor-beta. Cell Growth Differ. 2002;13:123–130. [PubMed] [Google Scholar]
- Yang YC, Piek E, Zavadil J, Liang D, Xie D, Heyer J, Pavlidis P, Kucherlapati R, Roberts AB, Bottinger EP. Hierarchical model of gene regulation by transforming growth factor beta. Proc Natl Acad Sci U S A. 2003;100:10269–10274. doi: 10.1073/pnas.1834070100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JR, 3rd, Eng JK, McCormack AL. Mining genomes: correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal Chem. 1995a;67:3202–3210. doi: 10.1021/ac00114a016. [DOI] [PubMed] [Google Scholar]
- Yates JR, 3rd, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995b;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- Yeo CY, Chen X, Whitman M. The role of FAST-1 and Smads in transcriptional regulation by activin during early Xenopus embryogenesis. J Biol Chem. 1999;274:26584–26590. doi: 10.1074/jbc.274.37.26584. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2001;98:974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Zawel L, Lengauer C, Kinzler KW, Vogelstein B. Characterization of human FAST-1, a TGF beta and activin signal transducer. Mol Cell. 1998;2:121–127. doi: 10.1016/s1097-2765(00)80120-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.