Abstract
Replica exchange accelerated molecular dynamics (REXAMD) is a method that enhances conformational sampling while retaining at least one replica on the original potential, thus avoiding the statistical problems of exponential reweighting. In this paper we study three methods that can combine the data from the accelerated replicas to enhance the estimate of properties on the original potential: weighted histogram analysis method (WHAM), pairwise multistate Bennett acceptance ratio (PBAR), and multistate Bennett acceptance ratio (MBAR). We show that the method that makes the most efficient use of equilibrium data from REXAMD simulations is the MBAR method. This observation holds for both alchemical free energy and structural observable prediction. The combination of REXAMD and MBAR should allow for more efficient scaling of the REXAMD method to larger biopolymer systems.
Introduction
Free energy is the driving force behind biochemical problems of great importance, from drug binding to protein function. A computational approach to free energy calculation affords complete control over the system and method, as well as atomistic detail of the results. The use of computational free energy calculation can theoretically provide free energies for very specific processes that are difficult to isolate in experiment. Among the necessary conditions for accurate free energy prediction are accurate sampling and efficient use of the collected data.
The sampling of computational molecular dynamics has been practically limited due to the topography of the potential energy surface, which requires femtosecond timesteps when propagating the system, and the in the case of biopolymers, sampling many isolated regions of the potential energy. These issues mean that a large number of molecular dynamics steps need to be computed in order to simulate systems with micro-to millisecond relaxation times, but the most heroic brute force approaches have yielded only tens of microseconds.1–3 Accurate sampling can also be achieved through methodological developments that increase the sampling efficiency. For example, temperature replica exchange molecular dynamics (TREMD) has enjoyed a surge in popularity following the seminal publication of Sugita and Okamoto.4 TREMD overcomes local energy barriers by simultaneously simulating the dynamics of a set of non-interacting systems at different temperatures. Conformations sampled at high temperatures are more likely to overcome conformational barriers and exhibit higher sampling efficiency than low temperatures. The sampling efficiency passes between replicas through periodic Metropolis Monte Carlo attempts, which retain the detailed balance of the system. Despite the advantage of conformational exchange between temperatures, TREMD increases the energy of all degrees of freedom instead of activating those specifically important to conformational sampling.
Hamiltonian modification schemes selectively modify the system potential energy and can greatly enhance sampling over TREMD.5 For example, by applying a harmonic bias along a reaction coordinate of interest, umbrella sampling increases the number of conformations sampled local to the bias minimum.6,7 Accelerated molecular dynamics (AMD), on the other hand, typically applies a bias along the dihedral degrees of freedom, decreasing the potential barrier between states while retaining the general shape of potential basins.8 Unlike umbrella sampling AMD does not require prior knowledge of the reaction coordinate, although parameterization of the amount of bias should be done to optimize the acceleration. AMD has been used to reproduce NMR residual dipolar couplings, measures of molecular motion on up to the millisecond time scale, in the third IgG-binding domain of protein G.9 Despite the promise of AMD to increase the computational efficiency of conformational sampling, each observable must be exponentially reweighted in order to recover the unbiased ensemble average, which Shen and Hamelberg showed yields large statistical uncertainties when applying to highly accelerated simulations.10
In order to increase the statistical efficiency of AMD we recently developed a replica exchange accelerated molecular dynamics (REXAMD) scheme, which combines the selective activation of accelerated molecular dynamics and replica exchange.11 The resulting Hamiltonian replica exchange simultaneously simulates the dynamics on potentials of varying acceleration instead of varying temperature. We previously demonstrated the effectiveness of REXAMD for alchemical free energy calculations of small model systems.11 These free energy calculations only used the ∂V/∂λ values sampled from the ground, or unaccelerated, state. This analysis strategy makes no use of data generated in the accelerated states and limits the computational efficiency of the method as a result. Furthermore, the computational efficiency will decrease as the system size increases because the number of replicas required for an efficient replica exchange will increase.12 In order to mitigate this decrease in computational efficiency we will briefly introduce and compare the performance of three methods of recombining multi-state data from REXAMD simulations: WHAM, MBAR, and PBAR.
The most widely used method to combine different biased simulations and produce an estimate of the unbiased result is the weighted histogram method (WHAM).13 Central to this approach is the definition of an observable of interest, for example ∂V/∂λ. The bias from each of the multiple states is removed through an exponential reweighting of the biased probability density function of the observable. These unbiased probability density functions, one from each state, are then combined in a weighted sum at each value of the observable subject to the constraints that the weighted variance be minimized and the weights normalized. The WHAM formulation has been extended to REXAMD and applied to combine the (φ, ψ) distribution from accelerated potentials during a REXAMD simulation, yielding the unbiased (φ, ψ) distribution of various oligopeptides.14 While making use of the data generated in accelerated states improves the computational efficiency of the REXAMD method, WHAM suffers from an inherent systematic bias that plagues all histogram methods. The bias arises when the probability density of the observable varies greatly over the interval spanned by the bin, an observation rigorously derived by Kobrak.15 Decreasing the bin width attenuates the problem, but increases the statistical error of the histogram of the simulation data used to approximate the population density. The optimal bin width that balances these two effects should be found for each specific system, but the effects are always present.
The pairwise multistate Bennett acceptance ratio method (PBAR) method developed by Maragakis et al. extends the maximum likelihood derivation of the Bennett acceptance ratio method to handle multiple pairs of states simultaneously.16 The PBAR method is applicable to both equilibrium and non-equilibrium work data and requires that the work data between different pairs of states be independent. The independence criterion is a practical limitation when applying the PBAR method to the equilibrium samples generated from REMD simulations; the total amount of samples per state Ni must be split into independent samples for each pair of states Nij, thus reducing the statistical quality of the estimates. The PBAR method was validated on a gas-phase alchemical mutation of a capped amino acid simulated using TREMD and showed considerable precision and accuracy from a large data set (360 ns).
Shirts and Chodera developed a different multistate Bennett acceptance ratio method (MBAR).17 The MBAR approach requires equilibrium samples from each state and the energy of each sampled structure at every state. This lends itself naturally to a REMD approach when the temperature or Hamiltonian modification is straightforward and computing the energy of a structure at the different states is easy. In extreme cases post-processing of the equilibrium samples to compute the energy at different states can be expensive, such as when a soft-core alchemical potential is used.18,19 The MBAR analysis uses all of the equilibrium data for each pair of states, and therefore should scale better with the number of states than the PBAR method for equilibrium simulations. Validation was performed by constructing the potential of mean force of the extension of a DNA hairpin in an optical double trap from different constant force trajectories.
In this paper we will first describe the analytical model system and REXAMD simulation scheme. The WHAM, PBAR and MBAR techniques will be discussed in further depth, highlighting the main equations and practical implementation. The performance of WHAM and MBAR will then be compared against the previous ground state approach. The MBAR and PBAR methods will then be compared, and finally the extension of MBAR to computing equilibrium structural properties will be investigated.
Model System and Methods
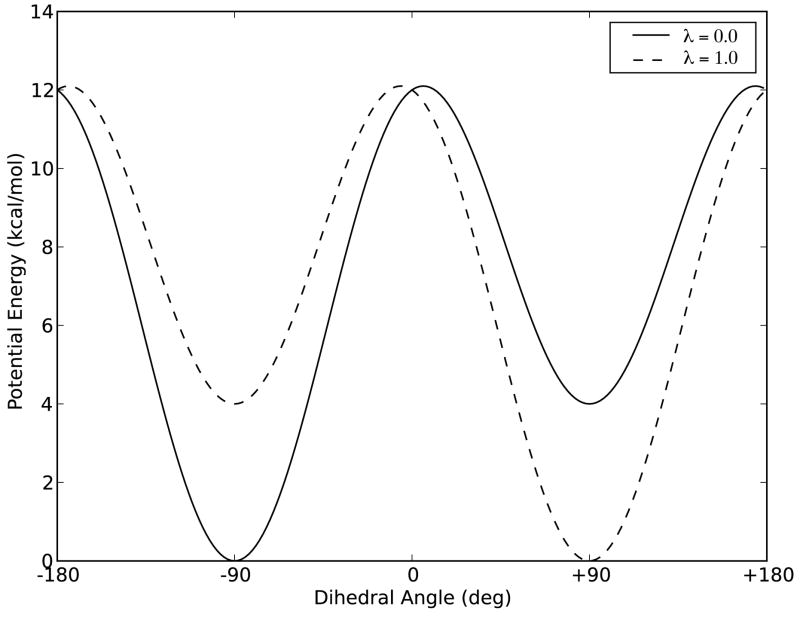
A simple, analytically solvable model system provides a wealth of information against which to compare the simulation performance. We selected a linear four-atom molecule (pseudo-butane) with no van der Waals or electrostatics forces, leaving the dihedral angle as the sole degree of freedom. There are two stable conformations at ± 90 degrees (p-form and m-form, respectively) with different relative depths shown in Figure I. The alchemical change progressed from the dominant p-form to the dominant m-form according to a linear scaling of the potentials (Equation 1), and due to the symmetry of the endpoints the alchemical free energy change is zero. The energetic barrier between the two conformations is at least 8 kcal/mol for all λi, and thus requires aggressive acceleration in order to generate an equilibrium distribution.
Figure 1.
| Equation 1 |
The REXAMD simulations have four states each, ranging from un-accelerated (s00) to an acceleration that results in a completely flat potential energy surface along the torsional degree of freedom (s03). The exchange rate between these states is higher than 50%, and thus efficient mixing of the replicas occurs on a very short timescale.20 The exchange period of the REXAMD simulations is 1 ps and the simulations are coupled to a 300K Langevin thermostat with a collision frequency of 50 ps−1. The instantaneous energy and dV/dλ values are taken from the exchanging structures. Systems corresponding to the endpoint lambda values (0.0 and 1.0) as well as the lambda values corresponding to a five-point Gaussian quadrature (0.04691, 0.23077, 0.50000, 0.76923, 0.95309) were simulated for five nanoseconds, and each λi REXAMD simulation was run four times.
The WHAM method applied to REXAMD relies on unbiasing the probability densities of a specific reaction coordinate for each state j through exponential reweighting (Equation 2). Equation 2 is exact in the limit of zero bin width, but must be approximated by a finite bin width histogram of the observable Q. Kobrak showed analytically that the discretization of a continuous observable Q required by WHAM results in a competition between systematic and statistical error.15 In the formalism of WHAM for REXAMD the systematic error arises from the approximation of the and from a finite bin width and increases with increasing bin width. The discretized and are estimated from histograms of the sampled data, which introduces a statistical error that decreases with increasing bin width. The from each state are then combined according to Equation 3 with a set of weights wj (Q) that minimize the variance at each Q. These conditions result in a set of self-consistent equations that can be iterated over until the desired level of precision is achieved (Equation A15 in Reference 14).
| Equation 2 |
| Equation 3 |
The MBAR method is rooted in the identity given in Equation 4 (Equation 5 in Reference 17) where qi(x) and qj(x) designate the un-normalized probability density functions of the configuration x in states i and j respectively, αij(x) is some arbitrary function, Zi and Zj are the configuration integrals from state i and j respectively, and Γ indicates that the integrals are evaluated over all configuration space. Approximating the expectation values as discrete averages of equilibrium data and summing over all states results in a set of K estimating equations (Equation 5), where K is the total number of states and Ni is the number of structures sampled at state i. The details of the selection of the function αij(x) are outside of the scope of this paper, but the selection exhibits the lowest variance of common reweighting estimators.17,21 The solution of Equation 5 yields estimates for the partition function Zi of each state.
| Equation 4 |
| Equation 5 |
The Python implementation of MBAR, PyMBAR (https://simtk.org/home/pymbar), was used for all MBAR analysis. The statistical uncertainty of the free energies and expectation values are based on an estimate of the asymptotic covariance matrix of the provided data, which requires an uncorrelated dataset. Correlation can be removed from the molecular dynamics data by subsampling the original data set at an interval greater than or equal to the equilibrium relaxation time of the molecular dynamics system. For this work we used the subsampling technique implemented in PyMBAR.17
The PBAR method extends the maximum-likelihood derivation of the Bennett acceptance ratio to multiple states. The log likelihood (Equation 6) involves the Fermi function f(x)=1/(1+ex)of the instantaneous work values between states Wij,, the free energy between states ΔFij, and the constant Mij = kBTln(Nij/Nji) that accounts for a different number of samples in the forward and reverse direction. The instantaneous work Wij is defined as the difference in potential energies of a specific configuration at states i and j, and these work values must be independent for Equation 6 to hold. This requires that a structure xni pulled from an equilibrium simulation of state I can only be used to calculate the work to go to a single other state. The entire set of xni must be separated into K non-overlapping sets of xnij, which greatly reduces the number of data points per state pair ij compared to MBAR.17 The log likelihood has well defined derivatives and thus any optimization method can be used to find the set of Zi that maximizes the likelihood function. We implemented the PBAR method in Python using a gradient descent optimization. In order to estimate the statistical uncertainty of the PBAR method the PBAR calculation was repeated multiple times using random subsets of the provided work data and we report the average and standard deviation of the results.16
| Equation 6 |
Combining Multistate Information for TI
The first application of the REXAMD method to alchemical free energy calculations used the instantaneous dV/dλ values taken from the un-accelerated, or ground state, to compute 〈dV/dλ〉λi for use with Gaussian quadrature thermodynamic integration (Equation 7).11 The ground state only represents 1/N of the total data, where N is the number of replicas per REXAMD, and therefore a significant fraction of the data is never used. Multiple REXAMD simulations at each λi are then used to estimate the statistical uncertainty of the TI Gaussian quadrature calculation. This was previously referred to as the reweighted runs strategy, but we will refer to this approach as TI-GS (Ground State) in this paper. The five-point Gaussian quadrature 〈dV/dλ〉λi from the four combined runs are summarized in Table I. The analytical results are not within the estimated statistical uncertainty of the TI-GS values, so there is room for the multistate methods to show improvement.
Table I.
Compiled Expectation Values of dV/dλ
| λi | Expectation Value of dV/dλ at λi (kcal/mol) | ||||
|---|---|---|---|---|---|
| TI-GS | TI-WHAM | Individual Lambda TI- MBAR | All Lambdas TI-MBAR | Analytical | |
| 0.04691 | +3.904 ± 0.004 | +3.138 ± 0.008 | +3.922 ± 0.001 | +3.922 ± 0.000 | +3.923 |
| 0.23077 | +3.69 ± 0.01 | +2.54 ± 0.02 | +3.714 ± 0.005 | +3.722 ± 0.002 | +3.722 |
| 0.50000 | −0.09 ± 0.08 | −0.08 ± 0.06 | −0.06 ± 0.04 | +0.01 ± 0.02 | +0.000 |
| 0.76923 | −3.703 ± 0.006 | −2.48 ± 0.02 | −3.710 ± 0.005 | −3.721 ± 0.002 | −3.722 |
| 0.95309 | −3.926 ± 0.003 | −3.125 ± 0.004 | −3.922 ± 0.001 | −3.922 ± 0.000 | −3.923 |
| Delta G | −0.03 ± 0.02 | −0.01 ± 0.02 | −0.02 ± 0.01 | +0.002 ± 0.006 | +0.000 |
| Equation 7 |
In TI-WHAM the four replicas of a REXAMD simulation at a specific λi are combined to estimate 〈dV/dλ〉λi. The instantaneous dV/dλλi values were separated into 8000 bins, which gave the optimal estimates of 〈dV/dλ〉λi. We then calculated the unbiased probability density function of dV/dλλi and subsequently the 〈dV/dλ〉λi shown in Table I. The WHAM estimates of 〈dV/dλ〉λi deviate strongly from the analytical results, and are actually worse than the TI-GS estimates. The steep curvature of the population density of dV/dλλi (not shown) forces a narrow bin width, which in turn increases the statistical error of the histogram approximation leading to a poor estimate of the biased probability density function and eventually 〈dV/dλ〉λi. The high accuracy of the computed free energy change from TI-WHAM is an artifact of using a symmetric alchemical change, and cannot be expected in realistic systems.
The Individual Lamda TI-MBAR also uses the four REXAMD replicas for each λi to compute 〈dV/dλ〉λi. This is exactly equivalent to the TI-WHAM method in the limit of zero bin width and should exhibit less bias.17 The 〈dV/dλ〉λi in Table I show improvement over both the TI-GS and TI-WHAM results, and are very close to the analytical results. The combined TI-GS result is calculated with four duplicate simulations of five nanoseconds each, resulting in a total of 20,000 structures per 〈dV/dλ〉λi. The Individual Lambda TI-MBAR runs utilize the four REXAMD states of five nanoseconds each for a similar 20,000 structures per 〈dV/dλ〉λi, and yield results that are quite comparable to the combined TI-GS result (Table II). In other words, these results imply that Individual Lambda TI-MBAR is able to calculate 〈dV/dλ〉λi to a comparable accuracy as TI-GS with only a quarter of the data required by TI-GS.
Table II.
Comparison of TI and TI-MBAR methods.
| Run | Alchemical Free Energy (kcal/mol) | ||
|---|---|---|---|
| TI-GS | Individual Lambda TI-MBAR 2 | All Lambdas TI-MBAR 3 | |
| 01 | −0.055 | −0.04 ± 0.02 | +0.02 ± 0.01 |
| 02 | −0.051 | −0.07 ± 0.03 | −0.02 ± 0.01 |
| 03 | +0.040 | +0.04 ± 0.03 | +0.02 ± 0.01 |
| 04 | −0.067 | −0.05 ± 0.03 | −0.02 ± 0.01 |
| Combined | −0.03 ± 0.021 | −0.02 ± 0.01 | +0.002 ± 0.006 |
The combined TI-GS results report the average and standard error from the four individual runs.
The individual lambda method only uses information from a specific lambda value to compute 〈dV/dλ〉λi at each λi.
The all lambdas method uses information from all the lambda values to compute 〈dV/dλ〉λi. at each λi.
A more efficient use of the data is to simultaneously use all four states from all five λi and thus utilize 100,000 structures per 〈dV/dλ〉λi. This All Lambdas TI-MBAR performs very well when considering the four individual five nanoseconds runs, but really shines when all four runs are combined (Table II). It is noteworthy that the MBAR estimate of the statistical uncertainty for a relatively low number of data points, namely the individual runs of All Lambdas TI-MBAR, is too low to account for the offset of the calculated free energy from the analytical result. An increase in the number of samples used does correct this, but this effect should be studied further to gain confidence in the MBAR uncertainty estimate as it applies to larger systems.
Direct Multistate Free Energy Estimates
The TI-MBAR method was helpful in comparing the advantage of MBAR over TI-WHAM, but the PBAR and MBAR methods were developed to directly estimate the free energy difference between states. The λi endpoints (0.0 and 1.0) need to be simulated in order for a direct estimate of the free energy difference from MBAR and PBAR, and the results in Table III include data from all seven λi. The addition of the endpoint data improves the combined runs TI-MBAR result from +0.002 ±0.006 to -0.002 ±0.005 kcal/mol. The direct MBAR method shows slightly better accuracy and precision than TI-MBAR for all of the runs as well as the combined data set. The performance gain of MBAR relative to TI-MBAR is expected to increase with the system size as the bias inherent the Gaussian quadrature process will be more evident in complex, non-symmetric alchemical changes.
Table III.
Comparison of TI-MBAR to MBAR and PBAR
| Run | Free Energy (kcal/mol) | |||
|---|---|---|---|---|
| TI-MBAR1 | MBAR1 | MBAR Subsets2 | PBAR3 | |
| 01 | +0.01 ± 0.01 | +0.005 ± 0.009 | −0.00 ± 0.03 | −0.01 ± 0.02 |
| 02 | −0.02 ± 0.01 | −0.016 ± 0.009 | −0.01 ± 0.03 | −0.02 ± 0.02 |
| 03 | +0.00 ± 0.01 | +0.000 ± 0.009 | +0.01 ± 0.03 | +0.00 ± 0.02 |
| 04 | +0.00 ± 0.01 | +0.001 ± 0.009 | −0.01 ± 0.03 | −0.01 ± 0.02 |
| Combined | −0.002 ± 0.005 | −0.001± 0.005 | −0.00 ± 0.01 | −0.007 ± 0.008 |
The uncertainty of the TI-MBAR and MBAR comes from the asymptotic covariance matrix estimator from Shirts and Chodera.17
The uncertainty in the MBAR Subsets results is estimated from the standard deviation from 1000 different subsets of 185 random reduced potential values per state for the individual runs, and 1000 different subsets of 740 random reduced potential values for the combined result.
The uncertainty in the PBAR results is estimated from the standard deviation from 1000 different subsets of 185 random work values per pair of states for the individual runs, and 1000 different subsets of 740 random work values for the combined result.
Interestingly, the direct MBAR method also outperforms the PBAR results. Shirts and Chodera predicted that because the PBAR method requires independent sets of work values between each pair of states MBAR would make better use of equilibrium data.17 To get independent sets of work values the Ni samples per state must be distributed into Nij sets that will be used to calculate the instantaneous work to go from state i to state j. With K total states and Ni structures per state each Nij can have at most Ni/K structures, and for large values of K the decrease in the number of available data points relative to the equilibrium number ni is large. For example, the four replicas and seven λi used in the model system PBAR calculation result in 27 pairs of states ij for a given state i. The 27 pairs of states reduce the 5000 Ni structures to only 185 Nij structures. In order to better use the equilibrium data set Maragakis et al. suggested repeating the PBAR analysis with different random subsets Nij and reporting the average and standard deviation.16 The average should be recovered with this method, but due to the small number of Nij in each PBAR calculation the standard deviation can be expected to be significantly higher in PBAR than MBAR, which is observed in Table III.
The MBAR Subsets results follow the procedure outlined above for sampling random subsets of the total Ni structures and reporting the average and standard deviation. The size of Ni for each MBAR Subset was limited to same size required by PBAR (185 for the individual runs, 740 for the combined runs), although for MBAR to work properly it must use the same 185 structures from Ni for each pair of states ij. The MBAR Subset results are comparable to the PBAR results, showing that the increased precision of the direct MBAR method is indeed due to the more efficient use of equilibrium data.
Equilibrium Conformations
The REXAMD method is not limited to free energy calculations, and has been used for determining equilibrium structural properties in conjunction with the WHAM method.14 The calculation of expectation values using MBAR instead of WHAM should avoid the bias introduced by discrete binning during the reconstruction process.15,17 The MBAR method naturally computes equilibrium expectation values, and this can be extended to computing population histograms and potential of mean force for observables. Figure II illustrates the distribution of the model system’s dihedral angle from a single simulation at λi= 0.5 where the potential energy surface is symmetric. The five thousand structures from the ground state method (Figure II) show an overpopulation of the p-form. All four REXAMD states at λi = 0.5 can be used to generate the unbiased probability density histogram using WHAM. The result does show a higher degree of symmetry than the ground state method, but the systematic error introduced by the large bin widths causes the result to deviate strongly from the analytical result. Decreasing the bin width does reduce this effect (data not shown), but finding the optimal bin width to balance the systematic and statistical errors can be difficult.15 If instead all four states of all seven λi are used by MBAR to compute the population histogram the probability density the analytical result is completely recovered (blue in Figure II). This finding reiterates the benefit of using MBAR over WHAM when combining equilibrium distributions from different biases.
Figure 2.
Many important equilibrium conformational analysis techniques can be expressed as expectation values, and thus would greatly benefit form the combination of REXAMD and MBAR. For example, the covariance matrix and the related principal components analysis and quasi-harmonic entropy (for the mass-weighted covariance matrix), which are very sensitive to sampling, are defined in terms of expectation values (Equation 8). The root mean squared deviation and the root mean squared fluctuation are also expectation values that are frequently used in conformational analysis.
| Equation 8 |
Conclusion
All REMD methods require an increasing number of replicas as the system size increases, and it becomes important to make efficient use of all of the replica data and not just the data at the desired temperature or acceleration. We set out to determine the best way to combine the biased data from REXAMD simulations to improve the accuracy of free energy calculations and structural analysis. The performance of WHAM and MBAR at computing 〈dV/dλ〉λi from multiple acceleration states at specific λi was compared to 〈dV/dλ〉λi from the ground state (no acceleration). TI-WHAM performed worse than the TI-GS, and this result was discussed in light of the competing errors when selecting the WHAM bin width. The Individual Lambda TI-MBAR, which is comparable to TI-WHAM in terms of number of structures used to compute each 〈dV/dλ〉λi, was very close to the analytical result. The asymptotic covariance matrix estimator was not able to cover the offset from the analytical results and indicates that MBAR will underestimate the statistical uncertainty when used with a relatively low number of samples. The All Lambdas TI-MBAR represented the most efficient method of calculating 〈dV/dλ〉λi with MBAR given the five intermediate λi and with this amount of data the estimates of 〈dV/dλ〉λi were both precise and accurate. The results were comparable to using four times as much ground state data, and show that the computational gain by combining the replica information from REXAMD is excellent.
The MBAR and PBAR methods were then compared against each other, which required simulating the alchemical endpoints. The MBAR method was approximately an order of magnitude more precise than the PBAR method. The inefficiency of PBAR relative to MBAR was shown to be due to the requirement of independent data sets for PBAR, which reduced the amount of data available for each pair of states during the analysis. The most efficient way of combining equilibrium samples of REXAMD data is conclusively MBAR. We then demonstrated the usefulness of MBAR in combining multiple states to generate unbiased structural quantities. The combination of REXAMD and MBAR should allow large system sizes to be efficiently sampled and analyzed.
Acknowledgments
We thank Michael Shirts and John Chodera for making the PyMBAR code publicly available (https://simtk.org/home/pymbar). This material is based upon work supported under a National Science Foundation Graduate Research Fellowship (to MF). Funding by NIH GM31749, NSF MCB-0506593 and MCA93S013 (to J. A. M.) also supports this work. We also gratefully acknowledge additional support from the Howard Hughes Medical Institute, the Center for Theoretical Biological Physics (NSF PHY-0822283), and the National Biomedical Computation Resource (NIH P 41 RR08605).
References
- 1.Freddolino PL, Liu F, Gruebele M, Schulten K. Biophys J. 2008;94(10):L75–77. doi: 10.1529/biophysj.108.131565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maragakis P, Lindorff-Larsen K, Eastwood MP, Dror RO, Klepeis JL, Arkin IT, Jensen MO, Xu H, Trbovic N, Friesner RA, Palmer AG, Shaw DE. The Journal of Physical Chemistry B. 2008;112(19):6155–6158. doi: 10.1021/jp077018h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mura C, McCammon JA. Nucleic Acids Res. 2008 Sep;36(15):4941–4955. doi: 10.1093/nar/gkn473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugita Y, Okamoto Y. Chemical Physics Letters. 1999;314(1–2):141–151. [Google Scholar]
- 5.Faraldo-Gomez JD, Roux B. J Comput Chem. 2007;28(10):1634–1647. doi: 10.1002/jcc.20652. [DOI] [PubMed] [Google Scholar]
- 6.Ciccotti G, Ferrario M, Hynes JT, Kapral R. Chemical Physics. 1989;129(2):241–251. [Google Scholar]
- 7.Torrie GM, Valleau JP. Journal of Computational Physics. 1977;23(2):187–199. [Google Scholar]
- 8.Hamelberg D, Mongan J, McCammon JA. J Chem Phys. 2004;120(24):11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- 9.Markwick PRL, Bouvignies G, Blackledge M. J Am Chem Soc. 2007;129(15):4724–4730. doi: 10.1021/ja0687668. [DOI] [PubMed] [Google Scholar]
- 10.Shen T, Hamelberg D. J Chem Phys 2008 Jul. 21;129(3):034103. doi: 10.1063/1.2944250. [DOI] [PubMed] [Google Scholar]
- 11.Fajer M, Hamelberg D, McCammon JA. Journal of Chemical Theory and Computation. 2008;4(10):1565–1569. doi: 10.1021/ct800250m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukunishi H, Watanabe O, Takada S. The Journal of Chemical Physics. 2002;116(20):9058–9067. [Google Scholar]
- 13.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. J Comput Chem. 1992;13(8):1011–1021. [Google Scholar]
- 14.Xu C, Wang J, Liu H. Journal of Chemical Theory and Computation. 2008;4(8):1348–1359. doi: 10.1021/ct7003534. [DOI] [PubMed] [Google Scholar]
- 15.Kobrak MN. J Comput Chem. 2003 Sep;24(12):1437–1446. doi: 10.1002/jcc.10313. [DOI] [PubMed] [Google Scholar]
- 16.Maragakis P, Spichty M, Karplus M. Phys Rev Lett 2006 Mar. 17;96(10):100602. doi: 10.1103/PhysRevLett.96.100602. [DOI] [PubMed] [Google Scholar]
- 17.Shirts MR, Chodera JD. The Journal of Chemical Physics. 2008;129(12):124105. doi: 10.1063/1.2978177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beutler TC, Mark AE, van Schaik RC, Gerber PR, van Gunsteren WF. Chemical Physics Letters. 1994;222(6):529–539. [Google Scholar]
- 19.Zacharias M, Straatsma TP, McCammon JA. The Journal of Chemical Physics. 1994;100(12):9025–9031. [Google Scholar]
- 20.Abraham MJ, Gready JE. Journal of Chemical Theory and Computation. 2008;4(7):1119–1128. doi: 10.1021/ct800016r. [DOI] [PubMed] [Google Scholar]
- 21.Tan Z. Journal of the American Statistical Association. 2004;99(468):1027–1036. doi: 10.1198/016214504000001033. [DOI] [PMC free article] [PubMed] [Google Scholar]