Abstract
The SRY-related, HMG box SoxB1 transcription factors are highly homologous, evolutionarily conserved proteins that are expressed in neuroepithelial cells throughout neural development. SoxB1 genes are down-regulated as cells exit the cell-cycle to differentiate and are considered functionally redundant in maintaining neural precursor populations. However, little is known about Sox3 function and its mode of action during primary neurogenesis. Using gain and loss-of-function studies, we analyzed Sox3 function in detail in Xenopus early neural development and compared it to that of Sox2. Through these studies we identified the first targets of a SoxB1 protein during primary neurogenesis. Sox3 functions as an activator to induce expression of the early neural genes, sox2 and geminin in the absence of protein synthesis and to indirectly inhibit the Bmp target Xvent2. As a result, Sox3 increases cell proliferation, delays neurogenesis and inhibits epidermal and neural crest formation to expand the neural plate. Our studies indicate that Sox3 and 2 have many similar functions in this process including the ability to activate expression of geminin in naïve ectodermal explants. However, there are some differences; Sox3 activates the expression of sox2, while Sox2 does not activate expression of sox3 and sox3 is uniquely expressed throughout the ectoderm prior to neural induction suggesting a role in neural competence. With morpholino-mediated knockdown of Sox3, we demonstrate that it is required for induction of neural tissue by BMP inhibition. Together these data indicate that Sox3 has multiple roles in early neural development including as a factor required for noggin-mediated neural induction.
Keywords: Sox3, Sox2, Neural progenitors, Neural induction, Geminin
1. Introduction
Neural development progresses through multiple steps, commencing when naïve ectoderm is specified by signals from the underlying mesoderm to form neuroectodermal precursors (Hemmati-Brivanlou and Melton, 1994, 1997). These stem-cell like precursors proliferate until cued to withdraw from the cell-cycle and differentiate into various neural cell types dependent upon their position in the neural tube and time of cell-cycle exit (Cremisi et al., 2003; Edlund and Jessell, 1999). A proliferating progenitor population must be maintained for a specified time period to allow for a sufficient number of neural cells and for the development of all types of neural tissue. Accumulating evidence indicates that this process is controlled in part by members of the Sox transcription factor family (Pevny and Placzek, 2005; Wegner, 1999; Wegner and Stolt, 2005).
As members of the High Mobility Group (HMG) box super family of DNA-binding proteins, Sox proteins are grouped by their HMG homology (Bowles et al., 2000). The 10 subfamilies of Sox proteins are involved in a variety of different developmental events (Bowles et al., 2000; Wegner, 1999), but it is the Groups B, C, and E proteins that play various roles in the development of the nervous system (Pevny and Placzek, 2005; Sasai, 2001; Wegner and Stolt, 2005). Important to this study, the soxB1 group genes, sox1, 2, and 3, are expressed broadly in the dividing neuroepithelial cells throughout development and their expression is reduced when these cells differentiate (Collignon et al., 1996; Pevny et al., 1998; Uchikawa et al., 1999; Wood and Episkopou, 1999). In this way soxB1 genes mark neural progenitors (Aubert et al., 2003; Ying et al., 2003). Indeed, numerous studies indicate that the SoxB1 proteins, Sox2 and Sox3 are necessary for the maintenance of a neural progenitor population (Pevny and Placzek, 2005; Uchikawa et al., 1999; Wegner and Stolt, 2005; Wilson and Koopman, 2002). Furthermore, Sox1, 2 and 3 repress expression of pan-neuronal markers in the chick spinal cord indicating that they maintain undifferentiated, proliferating progenitors by counteracting neurogenesis (Bylund et al., 2003; Graham et al., 2003; Schlosser et al., 2008). Consistent with its role in maintaining a proliferating neural progenitor population, Sox2 is also expressed in embryonic stem-cells (Boiani and Scholer, 2005; Brandenberger et al., 2004) and is one of four key proteins required to convert differentiated adult fibroblasts to pluripotent stem-cells (Takahashi and Yamanaka, 2006). Together these data indicate an important role for SoxB1 proteins in the maintenance of neural progenitor cells.
SoxB1 genes have overlapping expression patterns and there is significant evidence that these highly homologous proteins have redundant roles in nervous system development. These studies indicate that phenotypes for each SoxB1 member are restricted to tissues where only this specific sox gene is expressed. For example, mutations in Sox2 in humans result in retinal defects presenting as microphthalmia to anophthalmia (Fantes et al., 2003; Williamson et al., 2006) in which the severity is Sox2 dose-dependent (Taranova et al., 2006). Sox1 null mice exhibit lens fiber defects (Nishiguchi et al., 1998) and spontaneous seizures due to the loss of neurons and disorganization in the ventral striatum (Ekonomou et al., 2005; Malas et al., 2003; Nishiguchi et al., 1998). Sox3 null mice have pituitary and craniofacial defects (Rizzoti et al., 2004; Rizzoti and Lovell-Badge, 2007). While these loss-of-function studies demonstrate unique functions for each protein in organ formation, they leave unanswered the exact role of the SoxB1 proteins independently or in combination during neural induction and primary neurogenesis.
At the onset of neural induction, the three Xenopus SoxB1 genes have distinct expression patterns (Koyano et al., 1997; Nitta et al., 2006; Penzel et al., 1997; Zhang et al., 2003) indicating that they also have distinct roles at this time. Specifically, only sox3 is maternally supplied with expression throughout the ectoderm prior to gastrulation and restricted to the presumptive neural plate by mid-gastrula. In contrast, sox2 expression begins at the onset of gastrulation in the neuroectoderm and sox1 is not expressed strongly until after gastrulation (stage 13) (Pevny and Placek, 2005; Rogers et al., 2008). Maternal Sox3 has been shown to repress endoderm formation to facilitate normal germ layer formation (Zhang et al., 2003; Zhang and Klymkowsky, 2007) and it has been demonstrated that Sox2 and Sox3 are required for neural development (Dee et al., 2008; Mizuseki et al., 1998a) and that Sox2 with FGF and another unknown factor can induce neural tissue (Mizuseki et al., 1998a). Yet little is known about the role of Sox3 at the time of neural induction and questions remain: Does its unique expression pattern throughout the ectoderm in early gastrulae indicate a distinct function in neural induction? If like Sox2, it is required for neural determination, what is the mechanism?
Here, we investigate the role of Sox3 in neural induction and formation in Xenopus laevis. Gain-of-function studies demonstrate that Sox3 inhibits Xvent2 expression indirectly but activates the expression of the early neural genes sox2 and geminin in the absence of proteins synthesis and in naïve ectodermal explants thereby increasing cell proliferation and inhibiting epidermal and neural crest formation. While Sox3 functions as a repressor to inhibit mesendoderm formation, we show that in the nervous system, Sox3 functions as an activator to expand the neural plate. Even though Sox2 overexpression causes similar changes, suggesting that both Sox3 and Sox2 are functionally redundant during early neural development, Sox3 is required uniquely for Noggin-mediated induction.
2. Materials and methods
2.1. Embryo culturing and manipulations
Xenopus laevis eggs were isolated and fertilized using standard methods (Sive et al., 2000) and staged according to Nieuwkoop and Faber (1994). Animal ectodermal explants were isolated from stage 9 embryos, cultured in 75% NAM (Peng, 1991; Slack, 1984) and collected according to sibling embryos.
2.2. Plasmid construction
Sox3ΔC-Engrailed was constructed in the same manner as described (Zhang et al., 2003). Amino acids 1-290 of the Sox3 protein was amplified via PCR adding an XhoI and ClaI site to the 5′ and 3′ ends of the gene respectively, and deleting the last 20 amino acids. This fragment was then fused in frame to a fragment encoding amino acids 2-298 of the Drosophila engrailed protein (Conlon et al., 1996; Badiani et al., 1994; Evan and Hancock, 1985) and the resulting Sox3ΔC-Engrailed was subcloned into the pCS2+ expression vector. The plasmid sox3mut was made by site-directed mutagenesis. Nucleotide changes were made to correspond to the sequence flanking cSox1 (Fig. S3A) (Kamachi et al., 1998): The sox3mut forward primer is 5′-GCTCCAGATGTACAGCATGATGATGGAGACCGACCAGAGCCCCG-3′ and the sox3mut reverse primer is 5′-CGGGGCTCTGGTCGGTCTCCATCATCATGCTGTACATCTGGAGC-3′ (mutated nucleotides are underlined).
2.3. Preparation of RNAs and microinjection
Synthetic capped mRNAs were made by in vitro transcription using mMessage mMachine (Ambion). sox3, sox3-VP16 (Zhang et al., 2003), sox3-GR (Kishi et al., 2000), sox2 (Mizuseki et al., 1998a), or sox2-GR (Kishi et al., 2000), mRNA was injected as described in the figure legends. For ectodermal explant assays, 2.5 pg noggin mRNA (Geng et al., 2003; Knecht et al., 1995), 5 pmol Sox3MO (TGTCGGTGTCCAACATGCTATACAT), and 400 pg sox3mut mRNA were injected into the animal pole of 1-cell stage embryos.
2.4. Whole mount in situ hybridization and β-galactosidase assay
Whole mount in situ hybridization (WISH) was performed as described (Harland, 1991; Hemmati-Brivanlou et al., 1990). For lineage tracing, β-galactosidase activity was visualized with Red-gal (Research Organics) or X-gal. We used the following genes as in situ probes: sox2 (Mizuseki et al., 1998a), geminin (Kroll et al., 1998), zic1 (Mizuseki et al., 1998a), soxD (Mizuseki et al., 1998b), epi-keratin (Jonas et al., 1985), Xp63 (Lu et al., 2001), bmp4 (Fainsod et al., 1994; Nishimatsu et al., 1992), Xvent2 (Ladher et al., 1996; Onichtchouk et al., 1996; Papalopulu and Kintner, 1996; Schmidt et al., 1996), slug (Mayor et al., 1995), N-cam (Kintner and Melton, 1987), sox9 (Lee and Saint-Jeannet, 2003), Xtwist (Hopwood et al., 1989) and sox10 (Aoki et al., 2003).
2.5. Luciferase assay
Luciferase assays were performed as described (Casey et al., 1999) using the Dual Luciferase Reporter Assay System (Promega). Xenopus embryos were injected with 5 pg of pRL-CMV and pGL2-basic as a control or 50 pg BRE (1243/-191)x4-luc (BRE-luciferase; (Hata et al., 2000). Inhibition by Sox3 or Sox2 was tested by injecting 400 pg sox mRNA with pRL-CMV and BRE-luciferase. As a control, either 400 pg sox11 (Hiraoka et al., 1997) or 50 pg of constitutively active BMP-2/4 type I receptor (CABR) (Candia et al., 1997) was co-injected. Pools of ten stage 11 embryos were collected in triplicate for each injection mixture. Embryos were mixed with 1 ml of 1× Passive Lysis Buffer and 10 μl or 20 μl of this supernatant was assayed. Experiments were repeated at least twice and showed similar trends.
2.6. Clearing and cartilage staining
Cartilage staining was performed as described (Bellmeyer et al., 2003) with the following modifications: tadpoles were fixed with MEMFA, stained for 3-days with 0.4% alcian blue staining solution and washed and cleared in 2% KOH.
2.7. Cell proliferation assay
Embryos were injected with 200 pg sox3 and 100 pg lacZ mRNA into one dorsal cell of an eight-cell embryo. At stage 13, embryos were fixed with MEMFA and assayed for β-galactosidase activity. Proliferating cells were detected with anti-phospho-Histone H3 (Ser10) antibody (Upstate/Millipore) and HRP-conjugated goat, anti-rabbit IgG (Upstate/Millipore) as described (Brivanlou and Harland, 1989; Dent et al., 1989).
2.8. Cycloheximide experiments
Embryos were injected with 400 pg of sox3-GR or sox2-GR mRNA. Those analyzed for sox2 expression were incubated in 10 μg/ml cycloheximide at stage 9 and induced with 1 μM Dexamethasone (Dex) 30 min later, while embryos analyzed for geminin and sox3 were incubated in cycloheximide at stage 11 and induced with Dex 30 min later. All embryos were collected at stages 12 and 12.5 and analyzed for β-galactosidase activity and then gene expression by WISH.
2.9. Terminal transferase assay
Whole mount TUNEL assay was performed after (Hensey and Gautier, 1998) with some changes: NEB reagents (TdT enzyme MO252S, NEB Buffer 4) used, Roche blocking agent (BMB). Labeled cells were visualized as described for WISH as described by (Harland, 1991).
2.10. RT-PCR
Semi-quantitative RT-PCR was performed as described (Wilson and Hemmati-Brivanlou, 1995). Total RNA was extracted from 10 or 20 ectodermal explants and cDNA was generated using random hexamer primers. Primers are: ef1α (XMMR, http://xenbase.org/xenbase/original/WWW/Marker_pages/primers.html), epi-keratin (XMMR), N-cam (XMMR), zic1 (Sasai et al., 2001), and geminin (XMMR), sox2 forward: CTTACATGAACGGCTCGCC, reverse: CCCCAGGTAGGTACATGC, annealing 58 degrees and 28 cycles. The RT-PCR results were also confirmed by quantifying band intensity with AlphaEase FC (AlphaInnotech; Fig. S3C).
3. Results
3.1. Sox2 and Sox3 induce neural progenitors and delay differentiation
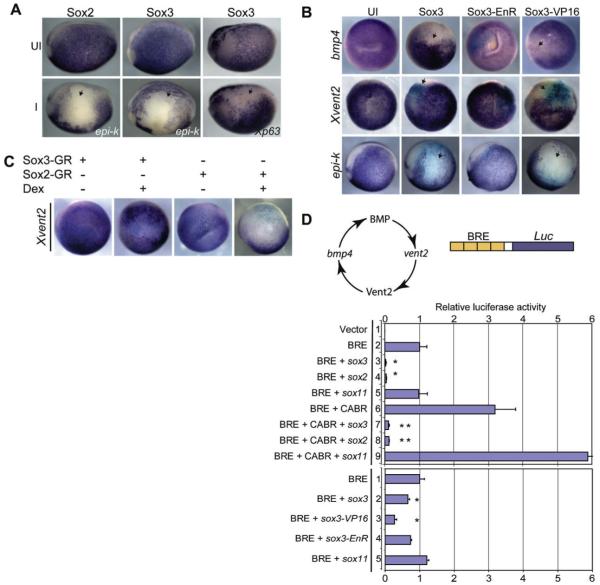
A role for maternal Sox3 in axis formation was defined in part by injection of sox3 mRNA into the equatorial region of dorsal cells (Zhang et al., 2003). This misexpressed Sox3 inhibited expression of Xenopus nodal (Xnr5) thereby ventralizing the embryo. To define the function of Sox3 during early neural development, we injected sox3 mRNA into the animal pole of 1 of 2-cells and analyzed the expression of the early neural genes sox2, geminin, soxD and zic1 which, like sox3, are known to be induced in response to BMP antagonists (Mizuseki et al., 1998a; Rogers et al., 2008; Taylor et al., 2006; Tropepe et al., 2006). Overexpression of sox3 or sox2 expanded the neural plate, as indicated by expanded expression of sox2 (n = 42/71), geminin (n = 37/44), soxD (n = 28/28) and zic1 (n = 126/155) in neurula embryos (Fig. 1A). To determine the effect on neuronal differentiation, embryos were collected at early neurula when neuronal differentiation is initiated (Bellefroid et al., 1996) and at tailbud when primary neurogenesis is complete. Overexpression of sox3 inhibited the expression of the pan-neuronal and proneural markers n-tubulin (n = 88/92) and ngnr-1 (n = 30/32), respectively in stage 14 embryos (Fig. 1B). However, in tailbud embryos n-tubulin and neurogenin r-1 expression was expanded (Fig. 1B, n-tub, n = 109/125, ngnr-1, n = 95/126) indicating that neuronal differentiation was delayed. While overexpression of sox2 also induced cells expressing n-tubulin (n = 24/32) outside of the normal neuronal boundary at tailbud stage, ngnr-1 was unaltered (n = 32/36). To rule out the unlikely possibility that these responses to sox3-overexpression were due to alterations in axis formation in the blastula, we injected RNA encoding a hormone inducible form of Sox3 (Sox3-GR) and activated this protein after the mid-blastula transition (MBT) at stage 9.5 with dexamethasone (Dex). Sox3-GR expanded expression of sox2, geminin, and soxD in neurula stage embryos similarly to Sox3 and delayed and expanded expression of n-tubulin and ngnr-1 (compare Fig. 1 and Fig. S1). Both Sox2 and Sox3 inhibited the formation of the trigeminal placode in both stages studied (Fig. 1B, Sox3, n-tub n = 72/179, ngnr-1, n = 28/42; Sox2, n-tub, n = 21/53, ngnr-1, n = 31/53).
Fig. 1.
Sox3 expands the expression of early neural markers and delays the expression of proneural and neuronal markers. (A) WISH of stage 17 neurulae for early neural markers as indicated in lower right corner. (B) WISH of stage 14 neurulae and stage 23 tailbud embryos for the pan-neuronal marker: n-tub and pro-neural marker: neurogenin r-1 ngn. Embryos are injected with sox2 or sox3 (400 or 500 pg) and lacZ (100 pg) mRNA in 1 of 2-cells. Images are a dorsal view with anterior to the bottom and the injected side on the right marked by a black asterisk in the first panel.
3.2. Sox3 activates geminin and sox2 expression in the absence of protein synthesis
While it is generally reported that SoxB1 proteins function as activators, Sox3 functions as a repressor to inhibit Xnr5 and alter axis formation (Zhang et al., 2003; Zhang and Klymkowsky, 2007). To determine the transcriptional activity of Sox3 in early neural development, we injected mRNA coding for either Sox3, Sox3-VP16, a dominant active form, or Sox3-EnR, a dominant repressor form, into the animal pole and assayed for the expression of gem and sox2 in gastrula embryos. Sox3 expanded expression of sox2 (n = 62/67) and gem (n = 93/108), while Sox3-VP16 induced both genes ectopically (sox2, n = 33/36 and gem, n = 40/44) on the ventral side of the embryos (arrows, Fig. 2A). In contrast, Sox3-EnR repressed sox2 expression in gastrula embryos (n = 33/37, Fig. S2A), induced gem (n = 38/40, Fig. 2A, Fig. S2B) and repressed n-tubulin expression (Fig. S2C). Therefore, these results suggest that Sox3 functions as an activator in early neural ectoderm determination but may also function as a repressor to indirectly drive the activation of gem.
Fig. 2.
Sox3 activates geminin and sox2 expression in the absence of protein synthesis. (A) WISH of stage 12 embryos injected with sox3, sox3-VP16, or sox3-EnR mRNA (200-800 pg) at the 1-cell stage. Sox3 and Sox3-VP16 expand expression of sox2 and gem (arrows) and Sox3-EnR induces gem in stage 12 embryos. Images are ventral view with the anterior to the top. (B) High levels of Sox2 and Sox3 induce expression of gem in ectodermal explants. RT-PCR analysis of ectodermal explants for sox2, gem and ef1α expression from embryos injected with noggin (25 pg), sox2, sox3 (500 or 800 pg) and sox3-VP16 (200 pg). Sibling embryos were stage 12. (C) Sox3-GR induces expression of sox2 (Dex, n = 22/30; Dex + CHX, n = 12/20) and gem (Dex, n = 31/35; Dex + CHX, n = 46/54). (D) Sox2-GR induces gem in the absence of protein synthesis (Dex, n = 25/43; Dex + CHX, n = 35/47). WISH of embryos injected with either Sox2-GR or Sox3-GR (400 pg) and incubated without or with Dex (1 μM), CHX (10 μM), or Dex + CHX at either stage 9.5 or 11 and collected at stage 12. Hatched rectangle indicates region of flat mount high magnification shown below whole embryo images.
It has been reported that Sox2 alone was insufficient to induce the expression of neural, proneural or neuronal markers in ectodermal explants (Mizuseki et al., 1998a). To establish whether Sox3 activates either sox2 or gem in the absence of neural inducers, ectodermal explants from embryos injected with sox3, sox2,or sox3-VP16 were assayed by semi-quantitative RT-PCR for their expression (Fig. 2B). As a positive control, explants from embryos injected with noggin mRNA were also assayed. At high concentrations, Sox3 induced sox2 expression weakly and both Sox2 (S2H), and Sox3 (S3H) were able to induce expression of gem. Additionally, Sox3-VP16 increased expression of gem and sox2 relative to uninjected explants, and similarly to noggin-overexpression. Thus, Sox3 alone and Sox3-VP16 can activate expression of sox2 and gem in naïve ectoderm.
To date, no direct targets of Sox3 or Sox2 involved in primary neurogenesis have been identified in Xenopus. To determine if Sox2 or Sox3 directly regulates the expression of gem or each other during neural induction, both Sox2-GR and Sox3-GR were induced with Dex in the presence of the protein synthesis inhibitor cycloheximide (CHX; Fig. 2C and D). As a control, injected embryos were also incubated with CHX in the absence of Dex. Due to the ubiquitous ectodermal expression of sox3 and gem until mid-gastrula (data not shown), these embryos were treated with CHX at stage 11 to allow for the restriction of their expression to the dorsal side and then induced with Dex at st. 11.5. In embryos incubated in CHX alone, Sox3-GR did not alter expression of sox2 or gem. However, when incubated with Dex alone (sox2, n = 22/30, gem, n = 31/35), or Dex and CHX (sox2, n = 12/20, gem, n = 46/54), Sox3-GR induced expression of sox2 and gem demonstrating that Sox3 activates expression of these genes in the absence of protein synthesis (Fig. 2C). Interestingly, although Sox3-GR is able to directly induce expression of sox2, Sox2-GR did not upregulate expression of sox3 when induced at stage 11 with Dex (n = 31/41), or with Dex and CHX (Fig. 3B, n = 39/39). However, Sox2-GR did activate expression of gem in the presence of Dex alone (n = 25/43) and Dex and CHX (n = 35/47, Fig. 2D).
Fig. 3.
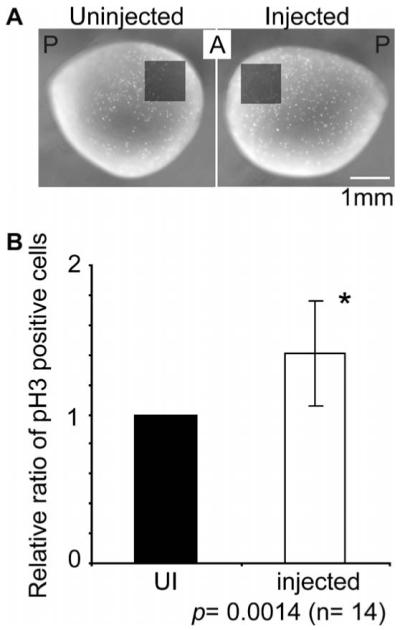
Sox3 expands the neural plate in part by increasing cell proliferation. (A) Diagram of 1 mm2 region in which pH3 positive cells were counted. (B) Relative ratio of pH3 positive cells on the injected side to the uninjected (UI) side. Vertical bars represent standard deviations. *p = 0.014, Student’s t-test.
3.3. Cell proliferation is increased by Sox3-overexpression
We next asked whether Sox3 expands the neural plate by increasing cell proliferation of existing progenitors. To address this, we injected sox3 mRNA with lacZ as a tracer into one animal, dorsal cell of 8-cell embryos. The majority of the descendants of these dorsal cells (D1.1 and D1.2 at 16-cell stage) develop into neural cells (Moody, 1987). At stage 13 (early neural plate stage), the embryos were fixed and assayed for β-galactosidase activity and subsequently immunohistostained with anti-phosphorylated histone H3 (pH3) antibody. pH3 positive cells in the anterior neural plate were counted (1 mm2) (Fig. 3A). As a control, the same position on the uninjected side of the embryo was counted. The sox3 injected area showed a 1.4 fold increase in cell proliferation over the control side (Fig. 3B, n = 14, p = 0.0014). There was no difference between the number of pH3 positive cells on the left and right side of uninjected embryos (n = 7, p = 0.4257, data not shown). This result suggests that expansion of the neural plate is in part due to increased cell proliferation.
3.4. Sox3 inhibits epidermal development via repression of the BMP signaling cascade
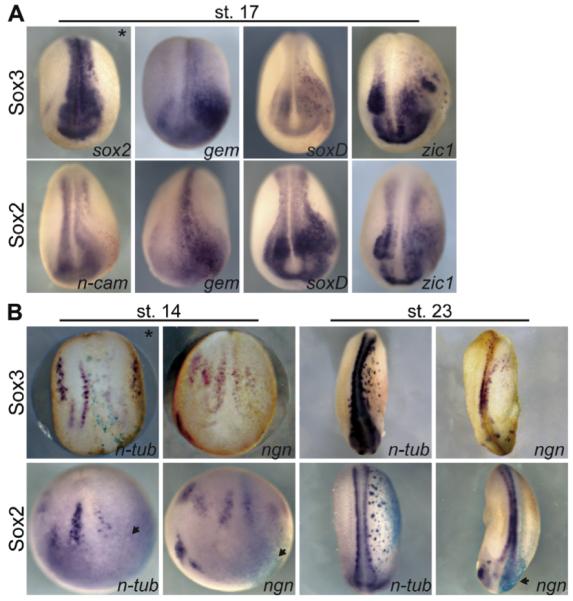
To determine if expansion of the neural plate is also in part due to the conversion of non-neural ectoderm cells to a neural fate, we analyzed the expression pattern of the epidermal markers, epidermal-keratin (epi-k) and Xp63 (Fig. 4A) in embryos injected with sox3 or sox2 and lacZ mRNA into the animal pole of one blastomere of two-cell embryos. Expression of epi-k (n = 132/152) and Xp63 (n = 23/23) was inhibited on the injected side in both the dorso-lateral and ventral regions (Fig. 4A). Since epidermal formation requires BMP signaling (Hawley et al., 1995; Wilson and Hemmati-Brivanlou, 1995), we tested whether sox3-overexpression blocked the formation of epidermis by inhibiting expression of bmp4, or its direct target, Xvent2 (Fig. 4B-D) and, whether it functioned as an activator or repressor to do so. The expression of bmp4, Xvent2 and epi-k was reduced by overexpression of sox3 (bmp4, n = 64/69, Xvent2, n = 52/52, epi-k, n = 26/28) or sox3-VP16 (bmp4, n = 35/45; Xvent2, n = 50/50, epi-k, n = 13/13) in gastrula (st. 12, bmp, Xvent2) and neurula (st. 17, epi-k) embryos, while Sox3-EnR had no effect on expression of these genes. These experiments demonstrate that Sox3 functions as an activator to inhibit expression of bmp4 and Xvent2 and epidermis formation (Fig. 4B).
Fig. 4.
Sox3, Sox2 and Sox3-VP16 inhibit Xvent2 expression and epidermal formation. (A) WISH for epi-k and Xp63 in embryos injected with 400-800 pg of sox3 or sox2 mRNA with lacZ (100 pg) mRNA into one of 2-cells. Uninjected (UI) side in top row, injected (I) side in bottom with arrow indicating injected area. Anterior is to the left and dorsal to the top. (B) Sox3, sox3-EnR, or sox3-VP16 mRNA (200-400 pg) was injected with lacZ mRNA into one of 2 blastomeres and gastrula or neurula embryos were assayed for bmp4, Xvent2 or epi-k expression by WISH. Bmp4 and Xvent2 (st. 12) embryos are animal pole view with the dorsal side to the top and epi-k embryos (st. 17) are a lateral view with anterior to the left. (C) WISH of stage 12 embryos injected with Sox3-GR into one blastomere of 2-cell embryos and untreated or treated with Dex at stage 9.5. Embryos are an animal pole views with dorsal to the top. (D) Diagram of BMP positive feedback loop, BRE-luciferase construct and relative luciferase activities in bar graph form. One-cell embryos were injected in the animal pole with vector or BRE-luciferase with sox3, sox2, sox11, sox3-EnR (400 pg), or sox3-VP16 mRNA (200 pg) and 50 pg CABR mRNA (top: lanes 3-9, and bottom: 2-5). The graph represents one experiment done in triplicate. Horizontal lines represent standard deviations. *p < 0.05 indicates values that differ significantly from lane 2 and **p < 0.01 indicates values that differ significantly from lane 6, Student’s t-test.
Previous studies showed that maternal Sox3 acts as a repressor, therefore to verify that Sox3 represses Xvent2 independent of maternal repressor activity. We overexpressed the hormone inducible Sox3-GR or Sox2-GR and incubated embryos with or without Dex (Fig. 4C). In embryos expressing Sox2-GR (n = 36/40) or Sox3-GR (n = 31/31) and treated with Dex, Xvent2 expression was inhibited. This supports a role for zygotic Sox3 in acting to repress epidermis as well as inducing neural gene expression.
There is a positive feedback loop in BMP signaling (Fig. 4D) in which BMP activates expression of Xvent2 and, once translated, XVent2 protein activates bmp4 transcription (Ladher et al., 1996; Onichtchouk et al., 1996). To determine where Sox3 is affecting this signaling loop, we tested the effect of Sox3 overexpression on the activity of a BMP response element (BRE)-luciferase construct (Hata et al., 2000) which contains four copies of the 152 bp BRE from the Xvent2 regulatory region. The BRE contains a Smad consensus binding site and enhances transcription in response to activated BMP signaling. There is no consensus Sox binding site. BRE-luciferase was injected with or without sox3 mRNA into one to two-cell embryos which were analyzed for luciferase activity as gastrulae (stage 11 and 12.5; Fig. 4D, lanes 1-3, data not shown). Like Xvent2, BRE-luciferase was expressed in gastrula embryos and decreased in response to Sox3. Since this decrease in expression could be due to inhibition of bmp expression, we co-injected a constitutively active form of the BMP receptor (CABR) and BRE-luciferase with or without Sox3. The presence of CABR led to an increase in luciferase activity, however this activation was also blocked by Sox3 (Fig. 4D, lanes 6 and 7). Moreover, this inhibition is specific to Sox3 since injection of the SoxE gene, sox11, did not show a reproducible or significant effect on BRE-luciferase activity (Fig. 4D, lanes 5, 9). To confirm that Sox3 repressed Xvent2 while functioning as an activator, BRE-luciferase assays were performed in the presence of Sox3, Sox3-VP16 and Sox3-EnR (Fig. 4D, lanes 1A-6A). Both Sox3 (p = .03) and Sox3-VP16 (p = .02) repressed BRE activity and although Sox3-EnR showed a trend of repressing the BRE, it was not statistically significant using a student’s t-test (p = 0.16).
3.5. Sox3 is required for noggin-mediated neural induction
There are differences in the expression patterns of the soxB1 subgroup genes, with only sox3 expressed prior to neural induction in frog. This raised the possibility that Sox3 is required as a competence factor for ectodermal cells to respond to neural induction signals. To test this hypothesis, we used Morpholino oligos (MO) (Fig. S3) to knock-down Sox3 translation in ectodermal explants from embryos injected with noggin mRNA. BMP is present in ectodermal explants, thus they differentiate into epidermis. However, in the presence of BMP antagonists, such as Noggin, explants differentiate into neural tissue (Wilson and Hemmati-Brivanlou, 1995). If Sox3 is required for ectoderm to respond to neural induction signals, Noggin would be unable to induce formation of neural tissue in the absence of Sox3. We first tested the Sox3MO specificity in vitro. As expected, Sox3 translation was blocked completely by Sox3MO while chick Sox3 translation was not altered significantly (Fig. S3B). Next, ectodermal explants were dissected from embryos injected with noggin mRNA and Sox3MO, cultured until sibling embryos completed gastrulation (stage 13-14), and assayed by RT-PCR for the expression of various neural and epidermal markers (Fig. 5). The neural markers, zic1, gem, and N-cam were induced by Noggin and this was reduced by Sox3MO co-injection. Moreover, inhibition of epi-k by Noggin was released by Sox3MO injection. We were able to rescue this phenotype by injecting sox3 mRNA in which the Sox3MO annealing sequence was mutated (sox3mut, Section 2 and Fig. S3A). These results suggest that Sox3, which is expressed prior to BMP antagonists such as Noggin, is necessary for the inhibition of BMP signaling by Noggin, and therefore, neural induction.
Fig. 5.
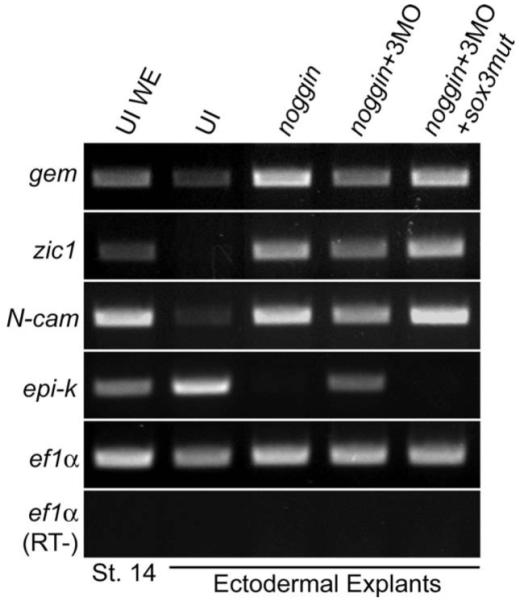
Sox3 is required for neural induction by Noggin in ectodermal explants. RT-PCR analysis of stage 14 ectodermal explants from embryos injected at the one cell stage with noggin mRNA with or without Sox3MO and sox3mut mRNA. The ectodermal explants were dissected from blastula embryos (stages 8, 9) and cultured until siblings reached neurula stage (stages 13-14). Lane 1, uninjected whole embryo (UI WE); 2, uninjected ectodermal explants (UI); 3, 2.5 pg noggin mRNA; 4, 2.5 pg noggin mRNA and 5 pmol Sox3MO; 5, 2.5 pg noggin mRNA with 5 pmol Sox3MO and 400 pg sox3mut mRNA.
3.6. Sox3 inhibits neural crest cell development
Sox3 and Sox3-GR injected embryos exhibited two phenotypes: ectopic pigment in all stages tested (Fig. 6A) and a kinked head in tadpole embryos (Fig. 6C). Excess pigment was induced as early as gastrula stage. To determine if the pigmented cells were either ectopic cement gland cells or premature neural crest-derived pigment cells, we performed WISH for the cement gland marker, Xag-1, and neural crest specifiers, slug, sox9, and sox10. Although the cement gland as marked by the expression of Xag-1 was expanded laterally, the pigment cells in the injected embryos were not Xag-1 positive (Fig. 6A, n = 49/77). Additionally, markers for neural crest were not induced ectopically in the pigmented cells, and instead, expression of slug (n = 36/57), sox9 (n = 47/68) and sox10 (n= 46/56) was reduced in early neurula and early tailbud embryos (Fig. 6B) and this was phenocopied by overexpression of Sox3-EnR (Fig. S4). These data suggest that the excess pigment induced by Sox3 is not due to premature differentiation of neural crest-derived melanocytes or ectopic cement gland and in fact, neural crest formation is inhibited in neurulae by Sox3 and Sox3-EnR.
Fig. 6.
Sox3 interferes with neural crest formation and migration. (A) Overexpression of sox3-GR induces ectopic pigment (129/181) and expands the cement gland but does not induce ectopic cement gland as indicated by WISH of Xag-1 (n = 49/77) in two right panels. (B) WISH of sox9 (n = 47/68), slug (n = 36/57), and sox10 (n = 46/56) in embryos injected with Sox3-GR and lacZ mRNA. Dorsal view of stages 13-21 embryos with anterior to the top (top row). Asterisk marks injected side. After stage 21, expression is expanded (bottom row) (sox9, n = 33/38, slug, n = 50/52, sox10, n = 50/50), but migration is inhibited (Xtwist, n = 96/130). Uninjected and injected sides of same embryo shown. (C) Development of branchial cartilage is disrupted by Sox3 and Sox3-VP16 as demonstrated by cartilage staining using Alcian blue. Embryos are dorsal view with anterior to the top and cartilage is ventral view with anterior to the top. (D) Two representative embryos demonstrating that Sox3 induces apoptosis in the head region as marked by TUNEL staining (n = 13/22). Left embryo injected in the left side and right embryo injected in the right side to account for potential asymmetric cell death. All embryos injected in 1 of 2 cells with sox3 (400 pg), sox3-VP16 (100 pg) or sox3-GR (400 pg) and lacZ (100-200 pg) or GFP (300 pg) mRNA.
In situ hybridization analysis of Sox-GR injected embryos revealed that while slug, sox9 and sox10 were initially repressed, in stage 23 embryos, slug (n = 50/52), sox9 (n = 33/38), sox10 (n = 50/50), and hairyA2 expression was expanded (Fig. 6B and data not shown). In contrast, neural crest marker expression does not “rebound” in sox3-EnR injected embryos (Fig. S4). Therefore, Sox3-overexpression causes a delay and then abnormal expansion of neural crest specifier genes.
To investigate the effect of delayed neural crest formation, we analyzed the phenotype of late tailbud and tadpole embryos. By stage 32 the embryos had a kinked head phenotype which was more severe in Sox3-VP16 injected embryos and absent in Sox-EnR embryos (Fig. 6C and Fig. S5). Alcian blue staining of cartilage revealed that the ceratobranchials, neural crest derivatives (Sadaghiani and Thiebaud, 1987), were severely reduced in Sox3 (n = 10/15) and Sox3-VP16 (n = 3/5) injected embryos (Fig. 6C). The expansion of neural crest cells in mid-tailbud embryos and subsequent loss of branchial cartilage indicates that the late induced neural crest cells are unable to differentiate or complete normal migration. To determine whether the loss of the craniofacial structures was due to a lack of migration or cell death, embryos were injected with Sox3-GR, induced at stage 9.5 and then assayed for a marker of neural crest migration (Xtwist). Expression of Xtwist was disorganized and did not extend as far ventrally as controls (Fig. 6B, n = 96/130) specifically in the second, third and fourth neural crest streams that give rise to the branchial cartilage (Schuff et al., 2007). Anterior migrating neural crest streams were unaffected in the majority of embryos (Fig. S6, n = 62/96). This indicates that overexpression of Sox3 prevents normal migration of neural crest cells by altering the timing of induction. This is supported by the fact that Sox3-GR affected neural crest migration only if induced prior to neurula stage (Fig. S6, n = 27/35). To determine the fate of the late induced, non-migratory neural crest cells, Tunel assays were performed. In a majority of embryos overexpressing Sox3, there was a two-fold increase in apoptosis in the head region (Fig. 6D, n = 13/22). Therefore, overexpression of Sox3 delays neural crest development resulting in reduced migration and increased cell death which leads to a loss of craniofacial cartilage.
4. Discussion
Sox3, one of the earliest neural genes expressed, has been implicated in the maintenance of a neural progenitor population (Bylund et al., 2003; Graham et al., 2003) and is required for the formation of the nervous system (Dee et al., 2008). Yet, the mechanism for its activity and its role in neural induction has not been determined; to date, no direct targets for SoxB1 proteins in primary neurogenesis have been identified. In this report, we investigate the mechanism by which Sox3 functions in primary neurogenesis in Xenopus with both gain- and loss-of-function analysis and compare the effect of sox2-overexpression to that of sox3. With these studies we discovered that overexpression of sox3 increases the neural progenitor population to expand the neural plate via ‘direct’ activation of geminin and sox2 expression, increased cell proliferation, inhibition of BMP signaling and disruption of neural crest formation. Furthermore, even though Sox2 and Sox3 are insufficient to induce ectopic neural tissue, we demonstrate that Sox2 and Sox3 are both sufficient to induce gem expression in naïve ectodermal explants. Overexpression of dominant active and dominant negative forms of Sox3 in combination with CHX studies, indicated that these effects were the result of Sox3 functioning as an activator. During neural development, Sox2 shared all of these functions withSox3 indicating that they share similar roles at this stage in neural development. However, sox3 is expressed prior to sox1, sox2 and neural inducers such as noggin in the early gastrula embryo, and accordingly Sox3 appears to be required for Noggin-mediated neural induction and the subsequent induction of sox2 expression.
4.1. Sox3 activates the neural progenitor markers sox2 and geminin in the absence of protein synthesis
With gain-of-function studies, we show that Sox3 induced the expression of the neural progenitor markers, sox2 and geminin, expanded the expression of the neuralizing factors, zic1 and soxD, and inhibited expression of neuronal and proneural genes n-tubulin and ngnr-1, respectively. These changes in gene expression led to increased cell proliferation and facilitated the expansion of the neural plate. This data is consistent with that of other overexpression studies in the chick spinal cord (Bylund et al., 2003; Graham et al., 2003) and in zebrafish (Dee et al., 2008). However, we further demonstrate that: (1) Sox3 and Sox2 activate geminin in the absence of protein synthesis; (2) Sox3 ‘directly’ activates sox2 but not vice versa and (3) neurogenesis is not inhibited by Sox3, but rather delayed. The delayed expression of proneural and neuronal markers may be due to the decay of injected sox3 mRNA by early tailbud. However, this is unlikely because neuronal differentiation eventually occurs at the same in embryos in which various concentrations of sox2, sox3 or sox3-GR mRNA was injected suggesting that the presence of neither Sox2 nor Sox3 is sufficient to permanently block differentiation. Together, these data indicate that during primary neurogenesis, Sox3 induces the formation of neural progenitor cells and maintains neural cells in an undifferentiated state for a longer time period leading to a delay in neurogenesis.
While overexpression of sox3 expands the expression of a variety of neural markers in whole embryos, it can only induce ectopic expression of geminin and does not induce ectopic neural tissue in the ventral ectoderm of neurulae or in naïve ectodermal explants at gastrula stage (Fig. 1A, 2B, D). Thus, additional factor(s) are required for induction of neural tissue. This is consistent with studies by Mizuseki et al. demonstrating that Sox2 induces neural tissue in ectodermal caps from gastrulating embryos only in combination with FGF (Mizuseki et al., 1998a). It is also supported by our studies demonstrating that when injected into the a4 cell of a 32-cell embryo (fated to become epidermis (Moody, 1987)) neither Sox3 alone nor Sox2 and 3 in combination induced slug or any early neural markers in neurulae, while Sox3-VP16 induced both (data not shown). Furthermore, high levels of Sox2 or Sox3 and low levels of Sox3-VP16 are sufficient to induce expression of geminin but not of any other neural marker tested (Fig. 2B). In these experiments, Sox3-VP16 is more effective at inducing geminin but it does not activate expression of other neural or neuronal makers. Thus, it is likely that a partner protein or a second factor possibly activated by FGF signaling is required for expansion of the neural plate.
Even though Sox3 does not effectively induce neural tissue when overexpressed in naïve or non-neural ectoderm, it does inhibit expression of epidermal markers (Fig. 4) indicating that Sox3 alone prevents ventral and lateral ectodermal cells from differentiating into epidermis. Investigation of the mechanism of inhibition of epidermis revealed that Sox3 and Sox3-VP16 but not Sox3-EnR repress BRE-luc and Xvent expression. This, in turn, leads to the down-regulation of bmp4 and the subsequent loss of epidermis. Together these data suggest that Sox3 functions as an activator to induce sox2 and geminin and that repression of Xvent2 expression is not due to Sox3 functioning as a repressor, but rather indirect via the action of a Sox3 target.
We have identified the first targets of SoxB1 proteins involved in primary neurogenesis and showed that Sox3 acts as an activator to induce expansion of the early neural markers. However, SoxB1 proteins can function as either activators or repressors depending on the context. For example, Sox2 activates expression of δ-crystallin, Nestin, and Pax6 (Ambrosetti et al., 1997; Donner et al., 2007; Kamachi et al., 2000; Kamachi et al., 2001; Tanaka et al., 2004; Wilson and Koopman, 2002) and both activates and represses expression of itself, Oct-4, FGF-4, Nanog, and UTF1 to control the stem-cell state (Boer et al., 2007; Botquin et al., 1998). In Xenopus, Sox3 directly represses expression of Xnr5 and Xnr6 in the blastula embryo (Zhang et al., 2003; Zhang and Klymkowsky, 2007), and in this report, we present evidence that Sox3 functions as a repressor to inhibit neurogenesis (Fig. S2B). Although it is unknown how Sox3 activity is modulated, there are several possibilities, including interaction with different partner proteins (Kamachi et al., 2000; Wilson and Koopman, 2002) and post-translational modification, such as SUMOylation. SUMOylation of the fly SoxB1 homologue SoxN reduces its ability to function as an activator (Savare et al., 2005) and a similar effect was seen for the Xenopus SoxE protein (Taylor and Labonne, 2005). Furthermore, Sox3 has a conserved consensus SUMO modification site, thereby strengthening the possibility that SUMOylation modulates activity. Further experiments need to be conducted to understand the dynamics of Sox3 function.
4.2. Sox3 is required for neural induction
In Xenopus, sox3 is expressed maternally with zygotic expression occurring prior to the onset of gastrulation (Penzel et al., 1997; Rogers et al., 2008). In contrast, sox1 and sox2 expression commence with gastrulation (Mizuseki et al., 1998a; Nitta et al., 2006) coinciding with the expression of BMP-inhibitors and neural induction. Functional analysis of Sox3 demonstrated that maternal Sox3 represses mesendoderm development (Zhang et al., 2003; Zhang and Klymkowsky, 2007), and here we show that after neural induction, Sox3 plays a role in maintaining an undifferentiated progenitor population. To determine if Sox3 has a role after axis formation and before neurogenesis (in particular neural induction), we tested whether inhibition of BMP by Noggin could induce neural tissue in ectodermal explants when Sox3 levels are reduced by a morpholino. With reduced Sox3, inhibition of BMP by Noggin does not effectively block epidermogenesis or induce neural tissue. One possibility is that neural induction by Noggin was only partially blocked in these experiments because maternal Sox3 protein or other factors such as XOct-25 that facilitate neural induction were present (Takebayashi-Suzuki et al., 2007). The relationship between such factors and Sox3 is unknown and they may work in conjunction or be redundant with Sox3.
4.3. Sox3 and neural crest development
Sox3 overexpression induces ectopic pigment cells of unknown heritage similar to the phenotype caused by overexpression of multiple Zic family members (Fujimi et al., 2006). Although Sox3 expands expression of zic1, the data presented in this study indicates that the pigment cells are not derived from the neural crest as neural crest gene expression is reduced in these cells. Additionally, although Sox3 expands the cement gland laterally, Xag-1 is not induced ectopically at the site of injection, indicating that these cells are not ectopic cement glands. Future studies will attempt to identify the fate and the molecular mechanism of induction of the pigmented cells.
Since overexpression of sox3 increased pigment formation and repressed bmp4 expression levels, we further investigated the effect of Sox3 on neural crest. Sox3 repressed slug, sox9, and sox10 expression in gastrula, neurula and early tailbud embryos, and it dispersed and expanded these markers in mid-late tailbud embryos (Fig. 6B). It is unclear why the expression of the genes rebounded. Again it is possible that expression increases as injected mRNA decays. However, it is also possible that a neural crest induction signal increases in early tailbud or that the expansion of the neural crest is in response to increased zic1 expression (Mizuseki et al., 1998a; Nakata et al., 1998). We have shown that these slug, sox9, and sox10 expressing cells did not develop into neural crest derivatives as indicated by the lack of migration, increased apoptosis and eventual loss of branchial cartilage in tadpoles. These data are consistent with studies in chick in which misexpression of Sox2 decreased slug expression in early embryos that later failed to undergo epithelial to mesenchymal transition (Wakamatsu et al., 2004) and delay of induction or decreased migration of the avian trunk neural crest cells led to a high density of cells and then increased cell death and decreased neurogenesis (Maynard et al., 2000; Vogel et al., 1993; Vogel and Weston, 1988).
4.4. Shared functions of Sox3 and Sox2
Sox2 shares high homology with Sox3, is expressed in overlapping domains during neural development (Nitta et al., 2006; Rogers et al., 2008; Uchikawa et al., 1999), and shares the ability to counteract neurogenesis in the chick neural tube (Bylund et al., 2003). It has been proposed that Sox2 and Sox3 are functionally redundant during early neural development and that both are required to maintain a neural progenitor population (Avilion et al., 2003). Furthermore, mouse Sox1 and Sox3 single mutants do not show any broad CNS defects, and in Sox2 mutants levels of Sox3 are elevated (Miyagi et al., 2008), indicating that Sox1, 2, and 3 can compensate for each other during central nervous system (CNS) development (Episkopou, 2005; Nishiguchi et al., 1998; Zhao et al., 2004). To begin to determine if they share the same function in early neural development in gastrula and neurula embryos, we did a side by side comparison of the effect of sox2-overexpression to that of sox3. In most experiments, Sox2 behaved in an identical manner to Sox3. As seen for sox3, overexpression of sox2 expanded the expression of neural markers (Fig. 1A) including ‘directly’ activating gem expression, and delayed (Fig. 2C) and then expanded the expression of the pan-neuronal marker n-tubulin (Fig. 1B). Additionally, Sox2 inhibited expression of epikeratin (Fig. 4A), bmp4 (data not shown), Xvent2, BRE-luciferase (Fig. 4C, lanes 4, 8) and slug (data not shown) in neurula embryos. One difference between the two proteins was that Sox2-GR was more effective than Sox3-GR at inducing gem and inhibiting Xvent2 expression, but more surprising was the inability of Sox2 to induce sox3 while Sox3-GR was able to induce sox2 (Fig. 2B, C). Together these results suggest that Sox2 and 3 both expand the neural plate by inducing the expression of geminin and repress the epidermal fate by inhibition of Xvent2, and that sox2 may in fact be downstream of Sox3.
In toto, this research defined the roles and mode of action of Sox3 in primary neurogenesis. Sox3: (1) inhibits Xvent expression indirectly and therefore, BMP signaling and epidermogenesis, (2) activates sox2 and geminin expression in the absence of protein synthesis thereby maintaining cells in an undifferentiated, proliferating state and delaying neurogenesis and (3) is required for Noggin-mediated induction of neural tissue suggesting a role in providing competence to ectoderm to respond to neural induction signals. In the future, isolation of co-factor(s) and analysis of post-translational modification events will help us to understand the molecular steps required for neural development and neural stem-cell formation.
Supplementary Material
Acknowledgments
We thank Y. Sasai, T.D. Sargent, K.L. Kroll, P.D. Vize, A.H. Brivanlou, D. Kimelman, M.W. Klymkowsky, and P.J. Scotting for plasmids. We also thank I.B. Dawid for the pH3 anti-bodies and T. Grammer for providing Sox3MO sequences. This work was funded by grants from the NIH (NSO48918) to ESC.
REFERENCES
- Ambrosetti DC, Basilico C, Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol. 1997;17:6321–6329. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Saint-Germain N, Gyda M, Magner-Fink E, Lee YH, Credidio C, Saint-Jeannet JP. Sox10 regulates the development of neural crest-derived melanocytes in Xenopus. Dev. Biol. 2003;259:19–33. doi: 10.1016/s0012-1606(03)00161-1. [DOI] [PubMed] [Google Scholar]
- Aubert J, Stavridis MP, Tweedie S, O’Reilly M, Vierlinger K, Li M, Ghazal P, Pratt T, Mason JO, Roy D, Smith A. Screening for mammalian neural genes via fluorescence-activated cell sorter purification of neural precursors from Sox1-gfp knock-in mice. Proc. Natl. Acad. Sci. USA. 2003;100(Suppl 1):11836–11841. doi: 10.1073/pnas.1734197100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badiani P, Corbella P, Kioussis D, Marvel J, Weston K. Dominant interfering alleles define a role for c-Myb in T-cell development. Genes Dev. 1994;8:770–782. doi: 10.1101/gad.8.7.770. [DOI] [PubMed] [Google Scholar]
- Bellefroid EJ, Bourguignon C, Hollemann T, Ma Q, Anderson DJ, Kintner C, Pieler T. X-MyT1, a Xenopus C2HC-type zinc finger protein with a regulatory function in neuronal differentiation. Cell. 1996;87:1191–1202. doi: 10.1016/s0092-8674(00)81815-2. [DOI] [PubMed] [Google Scholar]
- Bellmeyer A, Krase J, Lindgren J, LaBonne C. The protooncogene c-myc is an essential regulator of neural crest formation in Xenopus. Dev. Cell. 2003;4:827–839. doi: 10.1016/s1534-5807(03)00160-6. [DOI] [PubMed] [Google Scholar]
- Boer B, Kopp J, Mallanna S, Desler M, Chakravarthy H, Wilder PJ, Bernadt C, Rizzino A. Elevating the levels of Sox2 in embryonal carcinoma cells and embryonic stem cells inhibits the expression of Sox2:Oct-3/4 target genes. Nucleic Acids Res. 2007;35:1773–1786. doi: 10.1093/nar/gkm059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiani M, Scholer HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat. Rev. Mol. Cell. Biol. 2005;6:872–884. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- Botquin V, Hess H, Fuhrmann G, Anastassiadis C, Gross MK, Vriend G, Scholer HR. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev. 1998;12:2073–2090. doi: 10.1101/gad.12.13.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- Brandenberger R, Khrebtukova I, Thies RS, Miura T, Jingli C, Puri R, Vasicek T, Lebkowski J, Rao M. MPSS profiling of human embryonic stem cells. BMC Dev. Biol. 2004;4:10. doi: 10.1186/1471-213X-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brivanlou AH, Harland RM. Expression of an engrailed-related protein is induced in the anterior neural ectoderm of early Xenopus embryos. Development. 1989;106:611–617. doi: 10.1242/dev.106.3.611. [DOI] [PubMed] [Google Scholar]
- Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat. Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Candia AF, Watabe T, Hawley SH, Onichtchouk D, Zhang Y, Derynck R, Niehrs C, Cho KW. Cellular interpretation of multiple TGF-beta signals: intracellular antagonism between activin/BVg1 and BMP-2/4 signaling mediated by Smads. Development. 1997;124:4467–4480. doi: 10.1242/dev.124.22.4467. [DOI] [PubMed] [Google Scholar]
- Casey ES, Tada M, Fairclough L, Wylie CC, Heasman J, Smith JC. Bix4 is activated directly by VegT and mediates endoderm formation in Xenopus development. Development. 1999;126:4193–4200. doi: 10.1242/dev.126.19.4193. [DOI] [PubMed] [Google Scholar]
- Collignon J, Sockanathan S, Hacker A, Cohen-Tannoudji M, Norris D, Rastan S, Stevanovic M, Goodfellow PN, Lovell-Badge R. A comparison of the properties of Sox-3 with Sry and two related genes, Sox-1 and Sox-2. Development. 1996;122:509–520. doi: 10.1242/dev.122.2.509. [DOI] [PubMed] [Google Scholar]
- Conlon FL, Sedgwick SG, Weston KM, Smith JC. Inhibition of Xbra transcription activation causes defects in mesodermal patterning and reveals autoregulation of Xbra in dorsal mesoderm. Development. 1996;122:2427–2435. doi: 10.1242/dev.122.8.2427. [DOI] [PubMed] [Google Scholar]
- Cremisi F, Philpott A, Ohnuma S. Cell cycle and cell fate interactions in neural development. Curr. Opin. Neurobiol. 2003;13:26–33. doi: 10.1016/s0959-4388(03)00005-9. [DOI] [PubMed] [Google Scholar]
- Dee CT, Hirst CS, Shih YH, Tripathi VB, Patient RK, Scotting PJ. Sox3 regulates both neural fate and differentiation in the zebrafish ectoderm. Dev. Biol. 2008;320:289–301. doi: 10.1016/j.ydbio.2008.05.542. [DOI] [PubMed] [Google Scholar]
- Dent JA, Polson AG, Klymkowsky MW. A whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. Development. 1989;105:61–74. doi: 10.1242/dev.105.1.61. [DOI] [PubMed] [Google Scholar]
- Donner AL, Episkopou V, Maas RL. Sox2 and Pou2f1 interact to control lens and olfactory placode development. Dev. Biol. 2007;303:784–799. doi: 10.1016/j.ydbio.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund T, Jessell TM. Progression from extrinsic to intrinsic signaling in cell fate specification: a view from the nervous system. Cell. 1999;96:211–224. doi: 10.1016/s0092-8674(00)80561-9. [DOI] [PubMed] [Google Scholar]
- Ekonomou A, Kazanis I, Malas S, Wood H, Alifragis P, Denaxa M, Karagogeos D, Constanti A, Lovell-Badge R, Episkopou V. Neuronal migration and ventral subtype identity in the telencephalon depend on SOX1. PLoS Biol. 2005;3:e186. doi: 10.1371/journal.pbio.0030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Episkopou V. SOX2 functions in adult neural stem cells. Trends Neurosci. 2005;28:219–221. doi: 10.1016/j.tins.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Evan GI, Hancock DC. Studies on the interaction of the human c-myc protein with cell nuclei: p62c-myc as a member of a discrete subset of nuclear proteins. Cell. 1985;43:253–261. doi: 10.1016/0092-8674(85)90030-3. [DOI] [PubMed] [Google Scholar]
- Fainsod A, Steinbeisser H, De Robertis EM. On the function of BMP-4 in patterning the marginal zone of the Xenopus embryo. EMBO J. 1994;13:5015–5025. doi: 10.1002/j.1460-2075.1994.tb06830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes J, Ragge NK, Lynch SA, McGill NI, Collin JR, Howard-Peebles PN, Hayward C, Vivian AJ, Williamson K, van Heyningen V, FitzPatrick DR. Mutations in SOX2 cause anophthalmia. Nat. Genet. 2003;33:461–463. doi: 10.1038/ng1120. [DOI] [PubMed] [Google Scholar]
- Fujimi TJ, Mikoshiba K, Aruga J. Xenopus Zic4: conservation and diversification of expression profiles and protein function among the Xenopus Zic family. Dev. Dyn. 2006;235:3379–3386. doi: 10.1002/dvdy.20906. [DOI] [PubMed] [Google Scholar]
- Geng X, Xiao L, Tao Q, Hu R, Rupp RA, Ding X. The Xenopus noggin promoter drives roof-plate specific transcription. Neuroreport. 2003;14:2163–2166. doi: 10.1097/00001756-200312020-00006. [DOI] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hata A, Seoane J, Lagna G, Montalvo E, Hemmati-Brivanlou A, Massague J. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell. 2000;100:229–240. doi: 10.1016/s0092-8674(00)81561-5. [DOI] [PubMed] [Google Scholar]
- Hawley SH, Wunnenberg-Stapleton K, Hashimoto C, Laurent MN, Watabe T, Blumberg BW, Cho KW. Disruption of BMP signals in embryonic Xenopus ectoderm leads to direct neural induction. Genes Dev. 1995;9:2923–2935. doi: 10.1101/gad.9.23.2923. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Frank D, Bolce ME, Brown BD, Sive HL, Harland RM. Localization of specific mRNAs in Xenopus embryos by whole-mount in situ hybridization. Development. 1990;110:325–330. doi: 10.1242/dev.110.2.325. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Melton D. Vertebrate embryonic cells will become nerve cells unless told otherwise. Cell. 1997;88:13–17. doi: 10.1016/s0092-8674(00)81853-x. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Melton DA. Inhibition of activin receptor signaling promotes neuralization in Xenopus. Cell. 1994;77:273–281. doi: 10.1016/0092-8674(94)90319-0. [DOI] [PubMed] [Google Scholar]
- Hensey C, Gautier J. Programmed cell death during Xenopus development: a spatio-temporal analysis. Dev. Biol. 1998;203:36–48. doi: 10.1006/dbio.1998.9028. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Komatsu N, Sakai Y, Ogawa M, Shiozawa M, Aiso S. XLS13A and XLS13B: SRY-related genes of Xenopus laevis. Gene. 1997;197:65–71. doi: 10.1016/s0378-1119(97)00242-4. [DOI] [PubMed] [Google Scholar]
- Hopwood ND, Pluck A, Gurdon JB. A Xenopus mRNA related to Drosophila twist is expressed in response to induction in the mesoderm and the neural crest. Cell. 1989;59:893–903. doi: 10.1016/0092-8674(89)90612-0. [DOI] [PubMed] [Google Scholar]
- Jonas E, Sargent TD, Dawid IB. Epidermal keratin gene expressed in embryos of Xenopus laevis. Proc. Natl. Acad. Sci. USA. 1985;82:5413–5417. doi: 10.1073/pnas.82.16.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y, Uchikawa M, Collignon J, Lovell-Badge R, Kondoh H. Involvement of Sox1, 2 and 3 in the early and subsequent molecular events of lens induction. Development. 1998;125:2521–2532. doi: 10.1242/dev.125.13.2521. [DOI] [PubMed] [Google Scholar]
- Kamachi Y, Uchikawa M, Kondoh H. Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet. 2000;16:182–187. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev. 2001;15:1272–1286. doi: 10.1101/gad.887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintner CR, Melton DA. Expression of Xenopus N-CAM RNA in ectoderm is an early response to neural induction. Development. 1987;99:311–325. doi: 10.1242/dev.99.3.311. [DOI] [PubMed] [Google Scholar]
- Kishi M, Mizuseki K, Sasai N, Yamazaki H, Shiota K, Nakanishi S, Sasai Y. Requirement of Sox2-mediated signaling for differentiation of early Xenopus neuroectoderm. Development. 2000;127:791–800. doi: 10.1242/dev.127.4.791. [DOI] [PubMed] [Google Scholar]
- Knecht AK, Good PJ, Dawid IB, Harland RM. Dorsal-ventral patterning and differentiation of noggin-induced neural tissue in the absence of mesoderm. Development. 1995;121:1927–1935. doi: 10.1242/dev.121.6.1927. [DOI] [PubMed] [Google Scholar]
- Koyano S, Ito M, Takamatsu N, Takiguchi S, Shiba T. The Xenopus Sox3 gene expressed in oocytes of early stages. Gene. 1997;188:101–107. doi: 10.1016/s0378-1119(96)00790-1. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Salic AN, Evans LM, Kirschner MW. Geminin, a neutralizing molecule that demarcates the future neural plate at the onset of gastrulation. Development. 1998;125:3247–3258. doi: 10.1242/dev.125.16.3247. [DOI] [PubMed] [Google Scholar]
- Ladher R, Mohun TJ, Smith JC, Snape AM. Xom: a Xenopus homeobox gene that mediates the early effects of BMP-4. Development. 1996;122:2385–2394. doi: 10.1242/dev.122.8.2385. [DOI] [PubMed] [Google Scholar]
- Lee YH, Saint-Jeannet JP. Sox9, a novel pancreatic marker in Xenopus. Int. J. Dev. Biol. 2003;47:459–462. [PubMed] [Google Scholar]
- Lu P, Barad M, Vize PD. Xenopus p63 expression in early ectoderm and neurectoderm. Mech. Dev. 2001;102:275–278. doi: 10.1016/s0925-4773(01)00315-x. [DOI] [PubMed] [Google Scholar]
- Malas S, Postlethwaite M, Ekonomou A, Whalley B, Nishiguchi S, Wood H, Meldrum B, Constanti A, Episkopou V. Sox1-deficient mice suffer from epilepsy associated with abnormal ventral forebrain development and olfactory cortex hyperexcitability. Neuroscience. 2003;119:421–432. doi: 10.1016/s0306-4522(03)00158-1. [DOI] [PubMed] [Google Scholar]
- Maynard TM, Wakamatsu Y, Weston JA. Cell interactions within nascent neural crest cell populations transiently promote death of neurogenic precursors. Development. 2000;127:4561–4572. doi: 10.1242/dev.127.21.4561. [DOI] [PubMed] [Google Scholar]
- Mayor R, Morgan R, Sargent MG. Induction of the prospective neural crest of Xenopus. Development. 1995;121:767–777. doi: 10.1242/dev.121.3.767. [DOI] [PubMed] [Google Scholar]
- Miyagi S, Masui S, Niwa H, Saito T, Shimazaki T, Okano H, Nishimoto M, Muramatsu M, Iwama A, Okuda A. Consequence of the loss of Sox2 in the developing brain of the mouse. FEBS Lett. 2008;582:2811–2815. doi: 10.1016/j.febslet.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development. 1998a;125:579–587. doi: 10.1242/dev.125.4.579. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Shiota K, Nakanishi S, Sasai Y. SoxD: an essential mediator of induction of anterior neural tissues in Xenopus embryos. Neuron. 1998b;21:77–85. doi: 10.1016/s0896-6273(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Moody SA. Fates of the blastomeres of the 16-cell stage Xenopus embryo. Dev. Biol. 1987;119:560–578. doi: 10.1016/0012-1606(87)90059-5. [DOI] [PubMed] [Google Scholar]
- Nakata K, Nagai T, Aruga J, Mikoshiba K. Xenopus Zic family and its role in neural and neural crest development. Mech. Dev. 1998;75:43–51. doi: 10.1016/s0925-4773(98)00073-2. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) Garland Publishing Inc; New York: 1994. [Google Scholar]
- Nishiguchi S, Wood H, Kondoh H, Lovell-Badge R, Episkopou V. Sox1 directly regulates the gamma-crystallin genes and is essential for lens development in mice. Genes Dev. 1998;12:776–781. doi: 10.1101/gad.12.6.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimatsu S, Suzuki A, Shoda A, Murakami K, Ueno N. Genes for bone morphogenetic proteins are differentially transcribed in early amphibian embryos. Biochem. Biophys. Res. Commun. 1992;186:1487–1495. doi: 10.1016/s0006-291x(05)81574-8. [DOI] [PubMed] [Google Scholar]
- Nitta KR, Takahashi S, Haramoto Y, Fukuda M, Onuma Y, Asashima M. Expression of Sox1 during Xenopus early embryogenesis. Biochem. Biophys. Res. Commun. 2006;351:287–293. doi: 10.1016/j.bbrc.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Onichtchouk D, Gawantka V, Dosch R, Delius H, Hirschfeld K, Blumenstock C, Niehrs C. The Xvent-2 homeobox gene is part of the BMP-4 signalling pathway controlling [correction of controlling] dorsoventral patterning of Xenopus mesoderm. Development. 1996;122:3045–3053. doi: 10.1242/dev.122.10.3045. [DOI] [PubMed] [Google Scholar]
- Papalopulu N, Kintner C. A Xenopus gene, Xbr-1, defines a novel class of homeobox genes and is expressed in the dorsal ciliary margin of the eye. Dev. Biol. 1996;174:104–114. doi: 10.1006/dbio.1996.0055. [DOI] [PubMed] [Google Scholar]
- Peng HB. Xenopus laevis: practical uses in cell and molecular biology. Solutions and protocols. Methods Cell Biol. 1991;36:657–662. [PubMed] [Google Scholar]
- Penzel R, Oschwald R, Chen Y, Tacke L, Grunz H. Characterization and early embryonic expression of a neural specific transcription factor xSOX3 in Xenopus laevis. Int. J. Dev. Biol. 1997;41:667–677. [PubMed] [Google Scholar]
- Pevny L, Placzek M. SOX genes and neural progenitor identity. Curr. Opin. Neurobiol. 2005;15:7–13. doi: 10.1016/j.conb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Pevny LH, Sockanathan S, Placzek M, Lovell-Badge R. A role for SOX1 in neural determination. Development. 1998;125:1967–1978. doi: 10.1242/dev.125.10.1967. [DOI] [PubMed] [Google Scholar]
- Rizzoti K, Brunelli S, Carmignac D, Thomas PQ, Robinson IC, Lovell-Badge R. SOX3 is required during the formation of the hypothalamo-pituitary axis. Nat. Genet. 2004;36:247–255. doi: 10.1038/ng1309. [DOI] [PubMed] [Google Scholar]
- Rizzoti K, Lovell-Badge R. SOX3 activity during pharyngeal segmentation is required for craniofacial morphogenesis. Development. 2007;134:3437–3448. doi: 10.1242/dev.007906. [DOI] [PubMed] [Google Scholar]
- Rogers CD, Archer TC, Cunningham DD, Grammer TC, Silva Casey EM. Sox3 expression is maintained by FGF signaling and restricted to the neural plate by Vent proteins in the Xenopus embryo. Dev. Biol. 2008;313:307–319. doi: 10.1016/j.ydbio.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaghiani B, Thiebaud CH. Neural crest development in the Xenopus laevis embryo, studied by interspecific transplantation and scanning electron microscopy. Dev. Biol. 1987;124:91–110. doi: 10.1016/0012-1606(87)90463-5. [DOI] [PubMed] [Google Scholar]
- Sasai N, Mizuseki K, Sasai Y. Requirement of FoxD3-class signaling for neural crest determination in Xenopus. Development. 2001;128:2525–2536. doi: 10.1242/dev.128.13.2525. [DOI] [PubMed] [Google Scholar]
- Sasai Y. Roles of Sox factors in neural determination: conserved signaling in evolution? Int. J. Dev. Biol. 2001;45:321–326. [PubMed] [Google Scholar]
- Savare J, Bonneaud N, Girard F. SUMO represses transcriptional activity of the Drosophila SoxNeuro and human Sox3 central nervous system-specific transcription factors. Mol. Biol. Cell. 2005;16:2660–2669. doi: 10.1091/mbc.E04-12-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser G, Awtry T, Brugmann SA, Jensen ED, Neilson K, Ruan G, Stammler A, Voelker D, Yan B, Zhang C, Klymkowsky MW, Moody SA. Eya1 and Six1 promote neurogenesis in the cranial placodes in a SoxB1-dependent fashion. Dev. Biol. 2008;320:199–214. doi: 10.1016/j.ydbio.2008.05.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JE, von Dassow G, Kimelman D. Regulation of dorsal-ventral patterning: the ventralizing effects of the novel Xenopus homeobox gene Vox. Development. 1996;122:1711–1721. doi: 10.1242/dev.122.6.1711. [DOI] [PubMed] [Google Scholar]
- Schuff M, Rossner A, Wacker SA, Donow C, Gessert S, Knochel W. FoxN3 is required for craniofacial and eye development of Xenopus laevis. Dev. Dyn. 2007;236:226–239. doi: 10.1002/dvdy.21007. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Early Development of Xenopus laevis: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2000. [Google Scholar]
- Slack JM. Regional biosynthetic markers in the early amphibian embryo. J. Embryol. Exp. Morphol. 1984;80:289–319. [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takebayashi-Suzuki K, Arita N, Murasaki E, Suzuki A. The Xenopus POU class V transcription factor XOct-25 inhibits ectodermal competence to respond to bone morphogenetic protein-mediated embryonic induction. Mech. Dev. 2007;124:840–855. doi: 10.1016/j.mod.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kamachi Y, Tanouchi A, Hamada H, Jing N, Kondoh H. Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells. Mol. Cell. Biol. 2004;24:8834–8846. doi: 10.1128/MCB.24.20.8834-8846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taranova OV, Magness ST, Fagan BM, Wu Y, Surzenko N, Hutton SR, Pevny LH. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JJ, Wang T, Kroll KL. Tcf- and Vent-binding sites regulate neural-specific geminin expression in the gastrula embryo. Dev. Biol. 2006;289:494–506. doi: 10.1016/j.ydbio.2005.10.047. [DOI] [PubMed] [Google Scholar]
- Taylor KM, Labonne C. SoxE factors function equivalently during neural crest and inner ear development and their activity is regulated by SUMOylation. Dev. Cell. 2005;9:593–603. doi: 10.1016/j.devcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Tropepe V, Li S, Dickinson A, Gamse JT, Sive HL. Identification of a BMP inhibitor-responsive promoter module required for expression of the early neural gene zic1. Dev. Biol. 2006;289:517–529. doi: 10.1016/j.ydbio.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Uchikawa M, Kamachi Y, Kondoh H. Two distinct subgroups of Group B Sox genes for transcriptional activators and repressors: their expression during embryonic organogenesis of the chicken. Mech. Dev. 1999;84:103–120. doi: 10.1016/s0925-4773(99)00083-0. [DOI] [PubMed] [Google Scholar]
- Vogel KS, Marusich MF, Weston JA. Restriction of neurogenic ability during neural crest cell differentiation. J. Neurobiol. 1993;24:162–171. doi: 10.1002/neu.480240204. [DOI] [PubMed] [Google Scholar]
- Vogel KS, Weston JA. A subpopulation of cultured avian neural crest cells has transient neurogenic potential. Neuron. 1988;1:569–577. doi: 10.1016/0896-6273(88)90106-7. [DOI] [PubMed] [Google Scholar]
- Wakamatsu Y, Endo Y, Osumi N, Weston JA. Multiple roles of Sox2, an HMG-box transcription factor in avian neural crest development. Dev. Dyn. 2004;229:74–86. doi: 10.1002/dvdy.10498. [DOI] [PubMed] [Google Scholar]
- Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner M, Stolt CC. From stem cells to neurons and glia: a Soxist’s view of neural development. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Williamson KA, Hever AM, Rainger J, Rogers RC, Magee A, Fiedler Z, Keng WT, Sharkey FH, McGill N, Hill CJ, Schneider A, Messina M, Turnpenny PD, Fantes JA, van Heyningen V, FitzPatrick DR. Mutations in SOX2 cause anophthalmia-esophageal-genital (AEG) syndrome. Hum. Mol. Genet. 2006;15:1413–1422. doi: 10.1093/hmg/ddl064. [DOI] [PubMed] [Google Scholar]
- Wilson M, Koopman P. Matching SOX: partner proteins and co-factors of the SOX family of transcriptional regulators. Curr. Opin. Genet. Dev. 2002;12:441–446. doi: 10.1016/s0959-437x(02)00323-4. [DOI] [PubMed] [Google Scholar]
- Wilson PA, Hemmati-Brivanlou A. Induction of epidermis and inhibition of neural fate by Bmp-4. Nature. 1995;376:331–333. doi: 10.1038/376331a0. [DOI] [PubMed] [Google Scholar]
- Wood HB, Episkopou V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech. Dev. 1999;86:197–201. doi: 10.1016/s0925-4773(99)00116-1. [DOI] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Zhang C, Basta T, Jensen ED, Klymkowsky MW. The beta-catenin/VegT-regulated early zygotic gene Xnr5 is a direct target of SOX3 regulation. Development. 2003;130:5609–5624. doi: 10.1242/dev.00798. [DOI] [PubMed] [Google Scholar]
- Zhang C, Klymkowsky MW. The Sox axis, Nodal signaling, and germ layer specification. Differentiation. 2007;75:536–545. doi: 10.1111/j.1432-0436.2007.00190.x. [DOI] [PubMed] [Google Scholar]
- Zhao S, Nichols J, Smith AG, Li M. SoxB transcription factors specify neuroectodermal lineage choice in ES cells. Mol. Cell. Neurosci. 2004;27:332–342. doi: 10.1016/j.mcn.2004.08.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.