Abstract
The emergence of the human brain is one of evolution’s most compelling mysteries. With its singular importance and astounding complexity, understanding the forces that gave rise to the human brain is a major undertaking. Recently, the identification and publication of the complete genomic sequence of humans, mice, chimpanzees, and macaques has allowed for large-scale studies looking for the genic substrates of this natural selection. These investigations into positive selection, however, have generally produced incongruous results. Here we consider some of these studies and their differences in methodologies with an eye towards how they affect the results. We also clarify the strengths and weaknesses of each of these approaches and discuss how these can be synthesized to develop a more complete understanding of the genetic correlates behind the human brain and the selective events that have acted upon them.
Keywords: Hominid, Primate, Human evolution, Brain evolution, Neurogenetics, Molecular evolution
Introduction
The human brain is wondrously large and complex. It, perhaps more than any other feature, defines what it means to be human. Understanding how the human brain evolved and the genetic differences underlying this evolution offers not only insight into the unique biology and neurological diseases affecting humans, but also scratches the philosophical itch of what makes us human. In recent years advances in sequencing technology have offered scientists the opportunity to begin thorough investigations of the question. With complete genomes now available for human, mouse, rat, chimpanzee, dog, and rhesus macaque and lower coverage genomes for additional species, it has become possible to consider these questions on a genomic scale.
There have been many attempts to do just that: to use genomic data to identify the genetic signatures of positive selection on the human genome, some broadly and some specifically on the brain. Although some advances have been made in looking at regulatory changes or changes in transcriptomics, and without a doubt this will prove to be an important component to the brain evolution story, the vast majority of studies have focused on protein coding changes.
This has been the case for several reasons. First, protein coding changes are simple to identify. Regulatory regions are evolutionarily labile, at least more so than protein coding regions, and still cannot be easily notated in genomic sequence. This also ties into the second major point; functionality is more easily assessed bioinformatically for proteins than for regulatory sequence. Our understanding of the physiochemical properties of the amino acids and the structure and function of proteins allows for protein changes to be placed into context. By way of contrast, changes even in known regulatory regions almost always require in vitro or in vivo studies for any functionality to be assessed. Finally, there exist well established metrics, such as KA/KS (or dN/dS or ω) for the detection of positive selection in protein sequences.
KA/KS has long been used to identify selection from divergence data. KS is the ratio of synonymous mutations per synonymous site and is generally thought to be representative of the neutral rate of mutation at least in mammals where synonymous sites are generally understood to be under no selective constraint. KA is representative of the number of amino acid replacement substitutions per possible replacement site. Taken together, KA/KS values equal to one indicate neutrality, whereas KA/KS values significantly different from one reflect the effects of selection. Most genes show KA/KS values significantly less than one (often around 0.2) reflecting the general selective constraint on established proteins. KA/KS values greater than one are much less common and reflect the effects of positive selection in driving amino acid changing mutations to fixation at rates more rapidly than expected by neutrality.
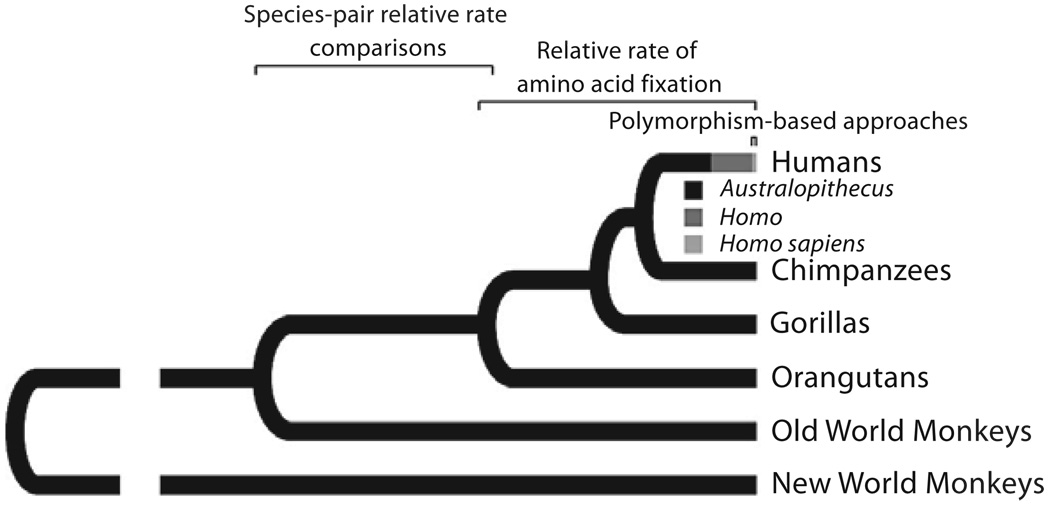
More recent positive selection can also be identified elsewhere in the genome through polymorphism-based methods (fig. 1). Some of these methods rely on deviations from the neutral expectation of the allele frequency spectrum which reflects the relative proportions of single nucleotide polymorphisms occurring at various frequencies, such as Tajima’s D [Tajima, 1989] and Fay and Wu’s H [Fay and Wu, 2000], and others compare rates of polymorphism to rates of divergence, such as the McDonald-Kreitman [McDonald and Kreitman, 1991] and Hudson-Kreitman-Aguade test [Hudson et al., 1987]. Increasingly sophisticated tests continue to be developed with bases elsewhere, such as the sizes of linkage disequilibrium blocks.
Fig. 1.
Simplified primate phylogeny.
These methods, however, focus on relatively recent selection; signatures of ancient selective events are eliminated over evolutionary time. Recent positive selection can and has been identified in regulatory regions as well as protein-coding regions through a dearth of high frequency segregating sites (or balancing selection through an excess of these sites), but there exists little in the way of statistical methodology to identify selective events once the equilibrium they disturb has been reestablished. Other methodologies using different metrics obtained from polymorphism data suffer from the same fate.
Yet these same issues that have so far stymied investigations into regulatory sequence evolution still play a role in our understanding of protein evolution underlying the emergence of the human brain. Many large gene or genomic studies have been undertaken to look for positive selection in human brain evolution. Although some general patterns have emerged, such as the widespread loss or change of olfactory genes, often these studies have returned different results. Most find evidence of selection somewhere, but the lists of genes often do not overlap. Some of the reasons for these differences are methodological or result from different starting data sets or levels of quality control. What is important to note, however, is that some of the differences that are observed do not simply represent incongruities among studies, but actually represent different answers to biologically different questions.
Here we will consider the breadth of human brain evolution covered by these studies, focusing on the different questions they ask and assumptions they make. This will hopefully offer insight into the diverse results found in the studies and lead to a more complete understanding not only of human brain evolution, but also the investigations themselves.
Recent Human Selection
There have been a significant number of studies over the past several years focusing on recent human selection, meaning selection operating over the last two hundred thousand years and roughly correlating to the emergence of anatomically modern Homo sapiens from Africa. This has been fueled by two major advancements in the field. First, the publication of the results of the HapMap survey made available large amounts of polymorphism data to the community [The International HapMap Consortium, 2005; Frazer et al., 2007]. Second, the emergence of new methodologies and statistics for the study of recent positive selection has allowed researchers new and novel ways to interpret the data [Sabeti et al., 2006; Thornton et al., 2007].
There have been several instances of candidate genes involved in brain development and function showing evidence for positive selection in humans, specifically the microcephaly-associated genes MCPH1 [Evans et al., 2005] and ASPM [Mekel-Bobrov et al., 2005]. These genes, whose evolution includes evidence of more ancient positive selection discussed further below, were initially of interest because mutations therein caused primary microcephaly, a brain developmental disorder resulting in smaller than normal brain sizes without concomitant physical abnormalities. Regions of high frequency extended haplotypes were identified for these genes which were unable to be modeled through neutral evolution following standard demographic assumptions. This remains an outstanding question, however, as other studies have suggested that these haplotype patterns might not be out of the ordinary in human genomes [Currat et al., 2006; Yu et al., 2007]. The patterns of haplotypic diversity seen in ASPM and MCPH1 do not seem to be unique to these genes. Although formally possible that numerous genes have undergone significant positive selection akin to these two, it seems more likely that there exists some less understood and underappreciated demographic scenario that accounts for the findings. What is certain is that there is some underlying mechanism that we do not fully understand.
Relevant to these discussions, however, is the role of assumptions in coloring our perceptions of the studies. As stated above, these two genes were originally targeted for study because of their association with the microcephaly brain development disorder, and although it is tempting to associate selective events with these phenotypes, it is not a given. Indeed, numerous recent studies have since shown that the haplotypes identified in these genes are not associated with IQ or brain size [Woods et al., 2006; Dobson-Stone et al., 2007; Mekel-Bobrov et al., 2007; Rushton et al., 2007; Timpson et al., 2007]. This, in and of itself, does not invalidate findings of positive selection, and in fact it is possible that positive selection is acting on another phenotypic character, but it casts a long shadow over any and all interpretations. The finding itself, though, is independent of phenotypic understanding. This fact must be kept at the forefront when evaluating the conclusions drawn from the data.
Apart from these candidate gene studies, numerous whole genome scans for positive selection have been undertaken [The International HapMap Consortium, 2005; Carlson et al., 2005; Kelley et al., 2006; Voight et al., 2006; Sabeti et al., 2007; Williamson et al., 2007]. Although these studies do find similar results among genes with very strong selective signatures (the lactase gene and genes involved in skin pigmentation for instance), there remains substantial variability. Only one of these studies identifies nervous system development genes as particularly well represented [Williamson et al., 2007] and several specifically exclude the candidate genes ASPM and MCPH1. So what information can be taken from this?
First, there are the general issues that exist within the field. Demography is problematic, especially in humans where population bottlenecks exist not only in the species as a whole, but also within sub-populations. The improvement of methodologies and increasing access to genomic levels of variation will allow for greater empirical comparisons between genes, but the pervasiveness of positive selection remains elusive and will affect our understanding of what constitutes an outlier relative to the neutral expectation. This has been illustrated particularly in the case of ASPM and MCPH1. Ascertainment bias is also a potential problem. Many of the current SNPs available for the whole genome studies were previously identified in specific subpopulations which might affect the interpretation of results. Further, many rare variants could be missing from these studies, affecting tests that focus on deviations from the allele frequency spectrum. Additionally, variation in recombination rates across the genome might render certain gene-gene comparisons invalid. The effects of recombination not only on variation, but on the tests themselves, have yet to be fully explored.
But apart from the methodological difficulties that will eventually be resolved, there remains variability because the nature of the tests themselves is different. Although some test statistics focus on completed sweeps, others focus on those sweeps which are ongoing, necessarily identifying different subsets of genes. Within those focusing on completed sweeps, some identify sweeps from the more ancient past (two hundred thousand years) whereas others identify more recent events (fifty thousand years). Although these differences might sound semantic, the evolutionary milieu was greatly variable among the time periods, and the genes and phenotypes under selection were likely very different.
This is particularly relevant as it relates to brain evolution. As stated above, anatomically modern humans are believed to have emerged between 100,000 and 200,000 years ago; the ‘human’ brain was already established at this point. It is possible that additional genetic changes might have occurred imparting greater cognitive or linguistic abilities, but the large-scale anatomical changes that are the hallmark of the human brain has been static since then. Indeed the brain size of Homo heidelbergensis, who lived five hundred thousand years ago and is thought to be the direct ancestor of Homo sapiens, was only slightly smaller than that of an average human living today [Neill, 2007]. This, coupled with drastic changes between Australopithecus and early Homo approximately two million years ago, suggests that perhaps the most salient genetic sweeps of positive selection affecting the emergence of the modern human brain are outside the scope of detection by polymorphism based approaches.
Divergence from Primates
Because of the inability of polymorphism scans to detect the selective sweeps of five hundred thousand years ago or more that led to the human brain, much work has focused on the differences between humans and chimpanzees. The role of protein coding changes in the human-chimpanzee divergence began to take a back seat to regulatory changes long ago [King and Wilson, 1975], but recently this has been revisited as a number of studies have focused on amino acid altering mutations in this terminal human lineage.
When the chimpanzee genome was published, only the human, mouse, and rat genomes were publicly available with the dog following shortly thereafter. Because of this, early studies focusing on the differences between humans and chimpanzees used one or more of these distantly related species as an outgroup [Clark et al., 2003; The Chimpanzee Sequencing and Analysis Consortium, 2005; Bustamante et al., 2005; Khaitovich et al., 2005; Nielsen et al., 2005; Arbiza et al., 2006]. The evolutionary time separating human and chimpanzee (roughly 5 million years) is at least one fifteenth that separating the other, non-primate, mammalian species. Because of this, questions arose as to the identification of orthologs between the species as well as improper alignments and multiple mutations at the same position. It was because of these potential confounds that additional studies were undertaken when the rhesus genome was published [Gibbs et al., 2007].
These early studies produced interesting lists of genes possibly evolving more rapidly in humans, but they largely failed to overlap. Certain categories and genes, particularly those expected to be under very strong positive selection, such as spermatogenesis genes and immune response genes, do show up regularly, but the overlap is nevertheless much less than perhaps is anticipated. The reasons for this can be legion but most seem focused on methodological rather than biological differences. In particular, studies have varied in whether they consider the human-chimpanzee branch as a whole or attempt to partition changes between the two hominoid terminal branches. In the case of the latter, variation in methods for ancestral sequence reconstruction might play a role. Another source of variation among the studies is in which chimpanzee sequence is used and how strict quality control standards are applied. Because of the short evolutionary time, a few genotyping errors, or even slightly deleterious polymorphisms appearing as fixed differences, can have major effects.
As we have seen, studies hoping to identify positive selection on the human genome by KA/KS values greater than one (indicative of positive selection) on the human lineage since the divergence of chimpanzees or on the combined terminal lineages of humans and chimpanzees have largely been ineffectual in identifying genes related to brain differences, although not entirely. One study that focused on brain-expressed genes found a correlation between brain expression and higher evolutionary rates in the human terminal branch compared to the chimpanzee terminal branch [Yu et al., 2006]. This study suggests that some of the disparity observed among the previous studies is the result of improper reconstruction of ancestral sequences as a consequence of long divergent outgroups such as rodents. This remains unproven, however, and it remains to be determined which is the best or correct method.
Although there are many differences among the studies, there are also some similarities. Genes involved in sensory perception, and more specifically chemosensory perception, consistently appear to be overrepresented in these studies of positive selection. In particular, it is well established that large changes in olfactory genes are widespread in humans, chimpanzees, and more broadly across primates [Gimelbrant et al., 2004; Gilad et al., 2005]. Nevertheless, the failure to identify large numbers of brain genes under positive selection between humans and chimpanzees has raised questions. The phenotypic difference in the brains of the two species is unmistakable, so what possibilities exist to explain the genetic findings?
Three primary explanations immediately present themselves. The first is that the phenotypic differences in the brains of human and chimpanzees are the result not of many changes, but rather of a very few, yet quite significant, differences. In this scenario only a handful of genes need be positively selected whereas the remainder of the brain-associated genes remain largely unchanged. This is unfulfilling for several reasons not the least of which is the seemingly impossible task of a single or few genes accomplishing changes of this magnitude. Nevertheless, it remains a possibility, although unlikely.
The second possibility is that in fact protein-coding changes are of greatly diminished importance relative to regulatory changes. As mentioned previously, this has long been considered the case, largely because the magnitude of the changes was thought to be incongruous with the high levels of similarity between the two genomes. More recently this understanding has been altered to emphasize the complexity of the system and epistatic protein networks. The general argument is that even small protein changes have such far reaching consequences that it is unlikely that they could be selected for without significantly disturbing the brain as a whole.
The final possibility is simply that current methods lack the power to detect positive selection between humans and chimpanzees. Indeed it might not even be methodological deficits, but rather a simple sample-size-related statistical power failure. The crux of this argument is that so little evolutionary time has elapsed since the divergence of humans and chimpanzees that there are simply not enough mutations to separate the signal from the noise in these studies. Mutation rates are such that it is not unlikely for human-chimpanzee orthologs to have no differences even at synonymous sites. The random and stochastic nature of genetic drift means that the difference between zero, one, or two changes is not particularly great, and yet with so few changes expected between humans and chimpanzees they take on greater importance.
Because of this, a gene evolving under neutral, or even significantly negative, selection might result in a KA/KS value greater than one simply because stochastic chance has resulted in a KS value that is zero or close to it. At the same time, a gene under positive selection might appear to have a KA/KS value at or less than one simply because stochastic chance has over-inflated the KS value. In short, the problem with the human-chimpanzee comparison, and other short lineage comparisons, is that the effect of stochastic noise is disproportionately large relative to the desired signal.
One way in which this failure of the human-chimpanzee comparison can be overcome is to use more divergent lineages. In practice, this has been done by comparing humans to old world monkeys, in particular the rhesus macaque [Gibbs et al., 2007]. The power issue is overcome as the evolutionary time is increased to a degree such that signal reasserts itself over noise; but rather than identify genes important since the human-chimpanzee divergence, this comparison identifies genes under positive selection as far back as the ape-old world monkey divergence.
There have been relatively few studies that incorporate the rhesus genome alongside the human and chimpanzee genomes and a non-primate outgroup. Those that have been done, however, tend to show a greater number of genes under positive selection in the lineage leading from the catarrhine ancestor to the hominoid ancestor and include a number of brain genes of interest. This is perhaps unsurprising as there are substantial phenotypic differences between the brain of an old world monkey and that of an ape. In fact, the importance of the monkey to ape transition and its relevance to human evolution is largely understated. Nevertheless, the genes thus identified are not necessarily those that lead to the uniquely human phenotype; they are rather the genes (some of the genes) responsible for the ape phenotype. These are important when considering non-human primate models of disease, which generally tend to be old- or new-world monkeys, and might provide interesting candidate genes, but the evidence does not provide proof that these genes lead to uniquely human characters.
Differences across Species in Levels of Selection
There is one additional general failure of the KA/KS metric in assessing positive selection which we have neglected. That is its ability to be reduced to less than one, despite bouts of positive selection, due to the diluting effects of negative selection either by time or across the gene itself. Periods of evolutionary time in which a gene is under negative selection or neutrality can overwhelm bouts of positive selection. Indeed this is likely to be the case in the human-rhesus comparison for those genes that have undergone positive selection since the human-chimpanzee divergence. Although KA is elevated as a result of selective forces for only this short time, the KS denominator remains constant. The net effect is a perhaps slightly higher KA/KS value than would have been observed had there been no positive selection event, but still a value less than one is indicative of overall negative selection across the complete time frame studied.
The same effect can be seen if only one part of a protein is under positive selection while the rest remains under strong selective constraint. In this case the selected region is overwhelmed by the rest of the protein and the net result again is an overall KA/KS value less than one and the appearance of negative selection. It is not hard to imagine scenarios for which this situation might be plausible; for instance, positive selection acting on ligand binding moieties while the rest of the protein remains fixed. Indeed this observation has been made for the major histocompatibility complex (MHC), which is generally regarded as one of the prime examples of positive selection. In the MHC the antigen binding regions are under extreme diversifying selection whereas other structural components remain more stable. In the case of the MHC, the areas under positive selection overwhelm those under constraining selection, but the MHC is unusual in this regard. Similar patterns of selective differences within a gene are also seen in membrane-bound receptors wherein transmembrane domains tend to be evolving at slower rates than either intracellular or extracellular regions.
Regardless of the mechanism, the result is the same: a KA/KS value which is slightly greater than it would have been had the positive selection event not occurred, but still not great enough to be detected as relevant in the absence of other data. What can be done to ameliorate these problems? To address the dilution effects of time, shorter intervals can be used. This nevertheless suffers from either the loss of statistical power as time decreases, as the human-chimpanzee examples illustrate, or a complete inability to determine if an intermediate sequence is unavailable. For instance in the divergence of apes from old world monkeys there is no intermediary sequence information between the old world monkeys and the divergence of the gibbons and siamangs. This time period is thus irreducible.
More can be done to address heterogeneity across physical space. Rather than focus on entire genes as the unit of study, predefined domains can be used. Alternatively, primary sequence sliding windows or tertiary sequence defined regions can be used assuming that multiple testing is appropriately accounted for. This suffers from the same loss of statistical power as the size under study decreases. Whether there are fewer mutational events because there is not enough time or because there are not enough physical locations, the paucity of mutations results in a lack of power.
One approach that can be taken is to look for differences in KA/KS values between species pairs. The null expectation here is that levels of selection reflected by KA/KS are the same between the species pairs under study. Deviations from the null can be tested for and used to identify differences in selective regimes. One outstanding difficulty with this approach, however, is that the nature of this difference is ambiguous. A difference could be the result of one species pair experiencing increased negative selection, one species pair experiencing positive selection, or one species pair undergoing a relaxation of selective constraint. The last category is particularly important as relaxation of selective constraint is not affected solely by selective pressures, but also is influenced by demographic factors including effective population size. Previous studies have attempted to control for these demographic effects by attempting to identify a baseline level of relaxation of constraint from either a control set of genes or, more broadly, from the whole genome.
The earliest study, predating the publication of the rhesus genome, focused on the differences in KA/KS values between humans-macaques and mouse-rat [Dorus et al., 2004]. This study found an increase in the KA/KS values of brain genes that was not seen in a control set of housekeeping genes. Moreover, the brain genes, when subdivided by functional class, showed a significant acceleration among those genes involved in developmental processes. (In these cases, ‘acceleration’ refers to an increase in Ka/Ks values or the fixation rate of amino acid mutations scaled to that of synonymous mutations). The difference in KA/KS values between the brain set and the control set ruled out the effects of population size and demographics in generating the pattern, but it remained formally possible simply that rodent brains had increased purifying selection or that primate brains had undergone selective relaxation. Although not excluded by the genetic data, these interpretations seem unlikely given our current understanding of primate and rodent neurology.
This study garnered two particularly relevant criticisms. The first involved the identification of ‘brain genes.’ Although this list contained genes generally believed to be relevant to brain function and development, it was by no means exhaustive. Similarly, there was no bias in the selection of brain genes, but nevertheless it remains possible that these genes are not representative of brain genes as a whole and that the results should not be generalized. The second criticism goes hand in hand with the first, suggesting that the control genes used are not generally appropriate and that different or additional genes would have produced more certain results. Both of these criticisms are appropriate and reflect limitations of the study at the time it was undertaken. They both can be ameliorated somewhat by the introduction of more recently published genomic data.
Similar studies were undertaken during the preparation of the rhesus and chimpanzee genomes for publication. In the rhesus, genes with higher KA/KS values in primates compared to rodents were overrepresented in categories of taste and smell sensory perception as well as the broad category of transcription factors [Gibbs et al., 2007]. This forms perhaps the most direct comparison as the species pairs used are the same. The same caveat that was noted above, however, applies to these studies as well. Although gaining statistical power through the use of a more distantly related species (old world monkey), genes that are identified might have undergone positive selection at some point during a longer evolutionary time frame. As mentioned before, it seems notable that many individual examples seem to indicate that these genes are particularly enhanced for positive selection during the lineage leading from monkeys to apes rather than the uniquely human terminal lineage since the chimpanzee divergence.
Although not directly comparable to the earlier study, this confound was removed by studies using the chimpanzee genome [The Chimpanzee Sequencing and Analysis Consortium, 2005; Khaitovich et al., 2005]. Using the human-chimpanzee comparison against the mouse-rat, these studies found that brain genes as a whole showed an acceleration in hominoids relative to rodents. This was not seen for genes representative of other organs including heart, kidney, liver, and even testis. Although not quite significantly faster than genes as a whole (p = 0.08), this was a significant acceleration compared to any other organ (p < 0.05). More work is obviously needed and the caveats regarding power to detect selection in short lineages still apply, but this is perhaps the most promising indication that protein changes on a large scale might have played a role in the emergence of the human brain.
Other studies, however, have reached opposite conclusions from the same basic premise. One study found no evidence of a human acceleration in brain-specific genes when compared to other genes in the genome [Shi et al., 2006]. This study compared the rate of evolution of genes on the human terminal branch to the chimpanzee terminal branch using the rhesus macaque sequence as an outgroup to parse changes. This comparison raised two important points for these comparisons. The first is the conceptually simple, but practically difficult, issue of defining ‘brain genes.’ This difficulty is pervasive and beyond the scope of this discussion. The second, however, is apropos and relates to the relative importance of the chimpanzee error rate in any study using the data. The chimpanzee genome appears to have sequencing error rates approaching 0.07% [Taudien et al., 2006], 50–100 times higher than the human genome and only an order of magnitude less than anticipated divergence levels between humans and chimpanzees. The differences in results thus are perhaps suggestive of the strong effect of these error rates and how they are treated by, or not treated by, the various studies.
An additional study which used a separately derived macaque sequence, in this case the sister species of rhesus, Macaca fascicularis, also failed to find an acceleration in brain-expressed genes compared to other genes in the genome [Wang et al., 2007]. The authors offer several reasons for the discordant findings including unintended biases towards slowly evolving, highly expressed genes or particularly slow evolving categories of genes, but discount these possibilities after additional examinations of the data. They also raise the specter of definitions, arguing possible differences between ‘brain-expressed’ and ‘brain-specific’ genes. Although they fail to find a difference in their data between the two, the points raised are valid and are likely to impact other studies.
So what can be made of these discrepancies? Perhaps the clearest interpretation is that from the outset defining both the experimental data set, the ‘brain genes’, and the control data set must be defined clearly. Differing definitions of brain genes are likely to cause differences in results. This problem is difficult to address as consensus is unlikely to be reached and the concept is simple enough to appear intuitive. The importance of this problem is highlighted in studies that found highly discordant lists of ‘brain genes’ depending on how the term is defined [Shi et al., 2006]. Control data sets contain not only these problems, but also problems in defining which set of data is itself is best. Other tissue-specific genes might better reflect the constraint experienced simply by being a tissue-specific gene, but selective forces are hardly uniform and if we have learned nothing else, it is that selection might be acting on genes and traits in ways of which we are largely unaware. Perhaps the most appropriate way to address control gene sets is simply to use multiple sets and compare the differing results.
One final point needs be made about these studies. As stated at the outset, the basic premise of the studies is that positive selection will cause an elevation in KA/KS from the usual selective pressure for that gene. Because we have no way of knowing what the natural selective pressure for that gene is, we depend upon other species to offer direction, and control gene sets to inform us regarding the relevance of those species. This is all well and good, and might indeed represent the best that can be achieved at this time, but the underlying assumption is still a fairly large one and one that needs be examined before drawing conclusions that might be too definitive.
Exceptions That Prove the Rule?
Previously we have discussed some of the numerous genome-wide or otherwise large scale studies that have purported to search for positive selection in the human genome. The picture painted is a muddied one that offers no clear conclusions especially as it relates to genes involved with the brain. Are there many genes whose protein changes are responsible for the human phenotype? Is the evolution of the human brain represented in widespread signatures of selection in the genome? We still do not know. What is becoming clear, however, is that for a subset of genes at least, evidence for selection exists. And although the exact phenotypic traits upon which selection is acting is still unproven, for many of these genes a role in the evolution of the human brain remains a likely possibility.
Above we touched upon studies of the microcephaly-associated genes MCPH1 and ASPM in regards to ongoing selection in humans, but there is strong evidence for evolution during the primate history of these genes as well [Zhang, 2003; Evans et al., 2004b; Kouprina et al., 2004]. ASPM shows KA/KS values greater than one, indicative of positive selection in both the lineage separating the lesser apes, gibbons and siamangs, from the great apes as well as in the human terminal branch since divergence from the chimpanzee. These values are significantly different from other species pairs and tend to cluster discretely within the gene.
MCPH1 shows a pattern that has become more familiar to studies of primate brain gene evolution [Evans et al., 2004a; Wang and Su, 2004]. As with ASPM, primate lineages leading to human (from the catarrhine ancestor) are evolving at rates greater than other species. Also similar to ASPM, the protein changing mutations tend to be clustered heterogeneously within the protein. Unlike ASPM, however, a KA/KS value greater than one is not seen in the human terminal branch. Indeed, the human terminal KA/KS value is not significantly different from the chimpanzee, gorilla, orangutan, or gibbon terminal branch. Rather, the bout of positive selection in MCPH1 appears to have occurred in the lineage leading from the catarrhine ancestor to the great ape ancestor, perhaps reflecting the emergence of the ape brain rather than specifically that of the human.
GLUD2, which, in primates, encodes the brain-specific isoform of glutamate dehydrogenase (GDH), a protein responsible for the catabolism of the neurotransmitter glutamate, is another example of a brain-specific gene that shows a signature of positive selection [Burki and Kaessmann, 2004]. GLUD2 arose from a duplication event of GLUD1 after the divergence of the lineage that would become the apes from the catarrhine ancestor. Shortly after its emergence the new gene underwent a bout of positive selection (as newly emerged duplicates are wont to do). As with MCPH1, this period of positive selection was confined to the lineage between its birth at roughly the time of the catarrhine ancestor and the divergence of the great apes. It thus again seems likely that it represents a gene responsible for the ape brain phenotype.
The gene that started it all was FOXP2. Originally identified as the source of a linguistic disorder, the molecular evolution of FOXP2 was found to show evidence for strong positive selection in the human terminal branch [Enard et al., 2002; Zhang et al., 2002]. This gene is a good example of both the shortcomings of the human-chimpanzee comparison as well as how they can be overcome. Despite the apparent effects of selection, the human terminal branch does not offer a KA/KS value significantly greater than one. This is due solely to the lack of statistical power as this branch contains two amino acid changes and zero synonymous changes. Nevertheless, a diagnosis of positive selection can be made because this is such an extreme departure from the amino acid mutation rate seen in other species (only one amino acid difference separates chimpanzees from mice).
From these and other examples, it is clear that there exist genes for which positive selection can unequivocally be deduced and who likely have played some role in the emergence of the human brain. What remains undetermined is the pervasiveness of these effects and their relative importance compared to other mechanisms shaping the emergence of the human brain, including the evolution of non-coding and regulatory regions. It is further unclear exactly what the different phenotypes imparted by the changes are and whether they exist in isolation or only in the context of other changes occurring at the same time. Further studies will certainly be forthcoming and with them a greater understanding of how the human brain has emerged.
A Footnote on Anthropocentrism
In the course of these studies, it becomes inevitable that there is talk of anthropocentrism. Several factors should be made clear: First, there is no scientific reason to think that the lineage leading to humans is privileged or otherwise different from the lineages leading to other species. It might be the case that the mechanisms driving the emergence of the human phenotype vary somewhat from other species, but this is probably not true and no evidence has been presented indicating that this is the case. Second, that positive selection was at work on the human brain should not come as a surprise or otherwise set it apart from other phenotypes. Indeed, we are interested in the brain because, as humans, it is such a major part of who we are. Behaviors, psychiatric disorders, emotions, language, all of these intrigue us and warrant the study of the brain. Other traits unique to humans, such as the changes in body hair and sweat glands related to a novel thermoregulatory strategy, are equally important and warrant study. Finally, the same studies can, and likely will, be done for any species. We can legitimately ask what makes a mouse so ‘mousy’ or a cat so ‘catty’ The methodologies will be largely similar and we will expect to see the same sorts of results. That these studies generally take a back seat in visibility to those in humans does not reflect on the science itself, but rather on the priorities of our human society.
References
- Arbiza L, Dopazo J, Dopazo H. Positive selection, relaxation, and acceleration in the evolution of the human and chimp genome. PLoS Comput Biol. 2006;2:e38. doi: 10.1371/journal.pcbi.0020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Kaessmann H. Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter flux. Nat Genet. 2004;36:1061–1063. doi: 10.1038/ng1431. [DOI] [PubMed] [Google Scholar]
- Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, Civello D, Adams MD, Cargill M, Clark AG. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Thomas DJ, Eberle MA, Swanson JE, Livingston RJ, Rieder MJ, Nickerson DA. Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res. 2005;15:1553–1565. doi: 10.1101/gr.4326505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Glanowski S, Nielsen R, Thomas PD, Kejariwal A, Todd MA, Tanenbaum DM, Civello D, Lu F, Murphy B, Ferriera S, Wang G, Zheng X, White TJ, Sninsky JJ, Adams MD, Cargill M. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Currat M, Excoffier L, Maddison W, Otto SP, Ray N, Whitlock MC, Yeaman S. Comment on ‘Ongoing adaptive evolution of ASPM, a brain size determinant in Homo sapiens’ and ‘Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans’. Science. 2006;313:172. doi: 10.1126/science.1122822. author reply 172. [DOI] [PubMed] [Google Scholar]
- Dobson-Stone C, Gatt JM, Kuan SA, Grieve SM, Gordon E, Williams LM, Schofield PR. Investigation of MCPH1 G37995C and ASPM A44871G polymorphisms and brain size in a healthy cohort. Neuroimage. 2007;37:394–400. doi: 10.1016/j.neuroimage.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Dorus S, Vallender EJ, Evans PD, Anderson JR, Gilbert SL, Mahowald M, Wyckoff GJ, Malcom CM, Lahn BT. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- Enard W, Przeworski M, Fisher SE, Lai CS, Wiebe V, Kitano T, Monaco AP, Paabo S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- Evans PD, Anderson JR, Vallender EJ, Choi SS, Lahn BT. Reconstructing the evolutionary history of microcephalin, a gene controlling human brain size. Hum Mol Genet. 2004a;13:1139–1145. doi: 10.1093/hmg/ddh126. [DOI] [PubMed] [Google Scholar]
- Evans PD, Anderson JR, Vallender EJ, Gilbert SL, Malcom CM, Dorus S, Lahn BT. Adaptive evolution of ASPM, a major determinant of cerebral cortical size in humans. Hum Mol Genet. 2004b;13:489–494. doi: 10.1093/hmg/ddh055. [DOI] [PubMed] [Google Scholar]
- Evans PD, Gilbert SL, Mekel-Bobrov N, Vallender EJ, Anderson JR, Vaez-Azizi LM, Tishkoff SA, Hudson RR, Lahn BT. Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans. Science. 2005;309:1717–1720. doi: 10.1126/science.1113722. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Man O, Glusman G. A comparison of the human and chimpanzee olfactory receptor gene repertoires. Genome Res. 2005;15:224–230. doi: 10.1101/gr.2846405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelbrant AA, Skaletsky H, Chess A. Selective pressures on the olfactory receptor repertoire since the human-chimpanzee divergence. Proc Natl Acad Sci USA. 2004;101:9019–9022. doi: 10.1073/pnas.0401566101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JL, Madeoy J, Calhoun JC, Swanson W, Akey JM. Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res. 2006;16:980–989. doi: 10.1101/gr.5157306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Paabo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Kouprina N, Pavlicek A, Mochida GH, Solomon G, Gersch W, Yoon YH, Collura R, Ruvolo M, Barrett JC, Woods CG, Walsh CA, Jurka J, Larionov V. Accelerated evolution of the ASPM gene controlling brain size begins prior to human brain expansion. PLoS Biol. 2004;2:E126. doi: 10.1371/journal.pbio.0020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Mekel-Bobrov N, Gilbert SL, Evans PD, Vallender EJ, Anderson JR, Hudson RR, Tishkoff SA, Lahn BT. Ongoing adaptive evolution of ASPM, a brain size determinant in Homo sapiens. Science. 2005;309:1720–1722. doi: 10.1126/science.1116815. [DOI] [PubMed] [Google Scholar]
- Mekel-Bobrov N, Posthuma D, Gilbert SL, Lind P, Gosso MF, Luciano M, Harris SE, Bates TC, Polderman TJ, Whalley LJ, Fox H, Starr JM, Evans PD, Montgomery GW, Fernandes C, Heutink P, Martin NG, Boomsma DI, Deary IJ, Wright MJ, de Geus EJ, Lahn BT. The ongoing adaptive evolution of ASPM and Microcephalin is not explained by increased intelligence. Hum Mol Genet. 2007;16:600–608. doi: 10.1093/hmg/ddl487. [DOI] [PubMed] [Google Scholar]
- Neill D. Cortical evolution and human behaviour. Brain Res Bull. 2007;74:191–205. doi: 10.1016/j.brainresbull.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Bustamante C, Clark AG, Glanowski S, Sackton TB, Hubisz MJ, Fledel-Alon A, Tanenbaum DM, Civello D, White TJ, J JS, Adams MD, Cargill M. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton JP, Vernon PA, Bons TA. No evidence that polymorphisms of brain regulator genes Microcephalin and ASPM are associated with general mental ability, head circumference or altruism. Biol Lett. 2007;3:157–160. doi: 10.1098/rsbl.2006.0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ES. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P, Bakewell MA, Zhang J. Did brain-specific genes evolve faster in humans than in chimpanzees? Trends Genet. 2006;22:608–613. doi: 10.1016/j.tig.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taudien S, Ebersberger I, Glockner G, Platzer M. Should the draft chimpanzee sequence be finished? Trends Genet. 2006;22:122–125. doi: 10.1016/j.tig.2005.12.007. [DOI] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton KR, Jensen JD, Becquet C, Andolfatto P. Progress and prospects in mapping recent selection in the genome. Heredity. 2007;98:340–348. doi: 10.1038/sj.hdy.6800967. [DOI] [PubMed] [Google Scholar]
- Timpson N, Heron J, Smith GD, Enard W. Comment on papers by Evans et al., and Mekel-Bobrov et al, on Evidence for Positive Selection of MCPH1 and ASPM. Science. 2007;317:1036. doi: 10.1126/science.1141705. author reply 1036. [DOI] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Chien HC, Osada N, Hashimoto K, Sugano S, Gojobori T, Chou CK, Tsai SF, Wu CI, Shen CK. Rate of evolution in brain-expressed genes in humans and other primates. PLoS Biol. 2007;5:e13. doi: 10.1371/journal.pbio.0050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YQ, Su B. Molecular evolution of microcephalin, a gene determining human brain size. Hum Mol Genet. 2004;13:1131–1137. doi: 10.1093/hmg/ddh127. [DOI] [PubMed] [Google Scholar]
- Williamson SH, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, Nielsen R. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods RP, Freimer NB, De Young JA, Fears SC, Sicotte NL, Service SK, Valentino DJ, Toga AW, Mazziotta JC. Normal variants of Microcephalin and ASPM do not account for brain size variability. Hum Mol Genet. 2006;15:2025–2029. doi: 10.1093/hmg/ddl126. [DOI] [PubMed] [Google Scholar]
- Yu F, Hill RS, Schaffner SF, Sabeti PC, Wang ET, Mignault AA, Ferland RJ, Moyzis RK, Walsh CA, Reich D. Comment on ‘Ongoing adaptive evolution of ASPM, a brain size determinant in Homo sapiens’. Science. 2007;316:370. doi: 10.1126/science.316.5823.370a. [DOI] [PubMed] [Google Scholar]
- Yu XJ, Zheng HK, Wang J, Wang W, Su B. Detecting lineage-specific adaptive evolution of brain-expressed genes in human using rhesus macaque as outgroup. Genomics. 2006;88:745–751. doi: 10.1016/j.ygeno.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Zhang J. Evolution of the human ASPM gene, a major determinant of brain size. Genetics. 2003;165:2063–2070. doi: 10.1093/genetics/165.4.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Webb DM, Podlaha O. Accelerated protein evolution and origins of human-specific features: Foxp2 as an example. Genetics. 2002;162:1825–1835. doi: 10.1093/genetics/162.4.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]