Abstract
Many proteins possess intrinsic disorder (ID) and lack a rigid three-dimensional structure in at least part of their sequence. ID has been hypothesized to influence protein-protein and protein-ligand interactions. We calculated ID for nearly 400 vertebrate and invertebrate members of the biomedically important nuclear hormone receptor (NHR) superfamily, including all 48 known human NHRs. The predictions correctly identified regions in 20 of the 23 NHRs suggested as disordered based on published X-ray and NMR structures. Of the four major NHR domains (N-terminal domain, DNA-binding domain, D-domain, and ligand-binding domain), we found ID to be highest in the D-domain, a region of NHRs critical in DNA recognition and heterodimerization, coactivator/corepressor interactions and protein-protein interactions. ID in the D-domain and LBD was significantly higher in “hub” human NHRs that have 10 or more downstream proteins in their interaction networks compared to “non-hub” NHRs that interact with fewer than 10 downstream proteins. ID in the D-domain and LBD was also higher in classic, ligand-activated NHRs than in orphan, ligand-independent NHRs in human. The correlation between ID in human and mouse NHRs was high. Less correlation was found for ID between mammalian and non-mammalian vertebrate NHRs. For some invertebrate species, particularly sea squirts (Ciona), marked differences were observed in ID between invertebrate NHRs and their vertebrate orthologs. Our results indicate that variability of ID within NHRs, particularly in the D-domain and LBD, is likely an important evolutionary force in shaping protein-protein interactions and NHR function. This information enables further understanding of these therapeutic targets.
Keywords: D-domain, Ingenuity Pathways Analysis, intrinsic disorder, nuclear hormone receptor, orphan receptor, ligand binding domain, ligand-protein interactions, PONDR, protein-protein interactions, X-ray structure
INTRODUCTION
Many proteins possess intrinsic disorder (ID) in some part of their sequence 1. Such disordered areas of proteins do not have a rigid three-dimensional structure and frequently are not resolved in X-ray or NMR structures. At the same time these regions can perform various biological functions 2 such as DNA binding, protein-protein interactions 3, cell signaling 4, and protein-ligand interactions 5. It has also been suggested that the level of disorder in well connected proteins in a network was expected to correlate with the number of interacting partners, as lack of folded structure might enable binding to multiple targets 4. A survey of 26 diverse protein families possessing at least one member with experimentally confirmed disordered sequence of 30 or more amino acids (AA), indicated that these disordered proteins evolve more rapidly than ordered proteins 6.
Several methods for prediction of protein disorder are widely available that assume the absence of rigid structure is encoded in the primary amino acid sequence, and these include reliable, experimentally validated software incorporating neural network and machine learning approaches 1, 2, 5, 7, 8. Most published studies assess full length protein ID even in multi-domain proteins 9–11, and although there is no consensus, several studies have focused on disordered regions of ≥ 30 AA as predictions for smaller regions may be less relevant. For example two studies 4, 12 looked at disorder in units of 10 AA from 30–100 AA, another study 1 focused on regions greater or equal to 50 AA, while a fourth study 13 looked at windows from 10 – 100 AA. Analysis of nearly one million proteins for conserved predicted disorder focused on windows of at least 20 AA and found that less than 10% were of greater than 30 AA in length 14. A genome wide survey of human proteins focused on intrinsically disordered loops longer than 50 residues (with disorder in greater than 30 residues) and found these disordered regions were generally on the surface of the protein 15.
As biology is complex, attempts to understand it frequently focus on scale-free network approaches 16–18. The role of ID in protein interaction networks 5, cell signaling and cancer associated proteins has been determined using an ID predictor 4. Proteins that are highly connected (also known as “hubs”) are frequently seen in such networks and in 4 eukaryotic genomes, such hub proteins with 10 or more interacting partners were significantly more disordered than other non-hub or “end” proteins found at the termini of networks 12. Although not widely studied in humans and other species, hub proteins tend to be involved in regulation, transcription and development in yeast 12. In another study analysis of transcriptional factors indicated over 80% of them have extended regions of ID (≥ 10 AA) relative to less than 54.5% of other types of proteins, and the degree of disorder was significantly higher in eukaryotes than prokaryotes 13. It has also been suggested that ID in transcription factors may contribute to their localization in the nucleus. Disease-related mutations have been shown to map to disordered regions 9. Protein disorder has also been associated with alternative splicing in human genes that may function to enable regulatory and functional diversity, facilitating more rapid evolution 19.
Nuclear hormone receptors (NHRs) are modular domain proteins that possess: 1. a structurally flexible N-terminal domain (NTD; also called A/B domain) involved in transactivation and protein-protein interactions; 2. a conserved zinc finger DNA-binding domain (DBD; also called C-domain) involved in DNA recognition and binding; 3. a poorly conserved D-domain (‘hinge’ domain) involved in DNA recognition and heterodimerization 20, coactivator/corepressor interactions 21 and protein-protein interactions 22 and 4. a ligand-binding domain (LBD; also called E-domain) which can bind agonists or antagonists and serves as an ‘interior interaction surface’23. There is also an F-domain that is not clearly defined and is absent in many NHRs; therefore we have considered it as part of the LBD. In combination these domains allow this diverse superfamily of key transcriptional regulators to be involved in important physiological functions 24 affecting endogenous metabolism, cell growth, xenobiotic protection, proliferation and oxidative stress.
Some NHRs, termed “orphan receptors”, do not have identified ligand activators and likely function as ligand-independent transcription factors. Most NHRs, however, bind ligands, which include a large array of endogenous and exogenous molecules and these ligand-activated NHRs are often referred to as “classic” NHRs. Classic NHRs include a group of receptors with narrow ligand specificity directed towards endogenous hormones or vitamins: thyroid hormone receptors (TRα and TRβ representing families NR1A1 and NR1A2, respectively), vitamin D receptor (VDR; family NR1I1), estrogen receptors (ERα and ERβ representing families NR3A1 and NR3A2, respectively), glucocorticoid receptor (GR; family NR3C1), mineralocorticoid receptor (MR; family NR3C2), progesterone receptor (PR; family NR3C3), and androgen receptor (AR; family NR3C4). Some NHRs are activated by multiple ligands, in some cases with uncertainty as to the identity of the physiologically important ligands as compounds like phospholipids were found in the LBD by X-ray crystallography but are of unknown significance. These receptors, sometimes termed ‘adopted orphans’ 25 include the peroxisome proliferators-activated receptors (PPARα, δ, and γ representing families NR1C1, NR1C2, and NR1C3, respectively), liver X receptors (LXRα, LXRβ representing families NR1H3 and NRH2, respectively which are also know as ‘oxysterol receptors’), farnesoid X receptor (FXR; family NR1H4 which is also known as the ‘bile acid receptor’), pregnane X receptor (PXR; family NR1I2), and constitutive androstane receptor (CAR; family NR1I3). NHRs themselves represent important targets for modulation of a variety of diseases 25–30
The functional importance of ID in NHRs has not been well-studied. Structural studies of several receptors in the NR1H and NR1I subfamilies have demonstrated significant stretches of disordered AA in the LBD. For example, PXR (NR1I2) is a transcriptional regulator of many enzymes and transporters involved in the transport, metabolism and biosynthesis of bile acids and a wide array of xenobiotics31, 32. PXR activators include prescription drugs, herbal compounds, and possibly fat-soluble vitamins which may cause adverse effects by altering the metabolism and transport of medications and hormones 33. X-ray crystallography of the LBD of human PXR 34 includes regions that generally cannot be resolved at residues 178–197 35. Binding of the agonist rifampicin induces structural disorder in human PXR at residues 178–209, 229–235, 310–317 36. Disorder is also observed to some degree in other NHRs. The LXRβ interloop region between helix-1 and helix-3 is difficult to resolve crystallographically due to unclear electron density 37–39. Like PXR, VDR has an extensive disordered interloop region between helices (H) 1 and 3 that is poorly conserved across species. This region is also unresolved crystallographically in zebrafish VDR 40. Published crystal structures of human VDR excised this disordered interloop region because the presence of it interferes with stable crystal formation 41. The human FXR crystal structure also shows disorder in the region between H1 and H3 consisting of a 15 residue insertion domain 42.
The aims of the current study were therefore to determine the extent of predicted ID across the classical and orphan 24 NHR families in different species (vertebrates and invertebrates) and relate this to observed experimental disorder, extent of protein-protein interactions, the co-evolution with ligands, and overall function. We also sought to determine how ID varies between NHR domains and the conservation patterns of ID across different animal species for a given NHR.
METHODS
Calculation of protein structural disorder
NHR sequences were identified and downloaded from either Genbank (http://www.ncbi.nlm.nih.gov/Genbank/) or Ensembl (http://www.ensembl.org/). Protein sequences were used for all analyses. Fragmented sequences were not used. Where possible, RefSeq sequences were used as these have undergone review and annotation by the National Center for Biotechnology Information (NCBI) staff. The orthology of sequences to the human sequence was verified by reciprocal BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) analysis using protein sequences. For some invertebrate receptors, the exact orthology to vertebrate NHRs is not clear. In these cases, the receptors were simply assigned to a subfamily. There is no consensus definition on the exact boundaries for the LBD and DBD for NHRs. Therefore, we defined the DBD and LBD based on published crystallographic structures. Most of the DBD crystallographic studies defined a ‘core DBD structure’; this was what was used to define the DBD boundaries. The NTD was defined as all sequence N-terminal of the DBD. The D-domain was defined as all sequence between the DBD and LBD. Although some NHRs are said to have a short C-terminal F-domain 43, this domain was not considered in the analysis as it is variably defined and not present in many NHRs. The LBD for all NHRs continued to the end of the C-terminus. To align domains across NHRs within a family, protein sequences were aligned by ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and refined as needed by T-Coffee (http://www.ebi.ac.uk/t-coffee/).
For each NHR family, one representative crystal structure served as the template to define the DBD and LBD boundaries for the other NHR sequences in that family. Given the absence of crystallographic structures of NR6 and NR0 receptors, NR5 sequences were used as the guide to define domains in NR6 and NR0 receptors. The following crystallographic structures were used as templates: NR1 DBD – human NR1A2 44; NR1 LBD – human NR1C3 45; NR2 DBD - human NR2B2 44; NR2 LBD - mouse NR2B1 LBD 46; NR3 DBD - human NR3A1 47; NR3 LBD - human NR3A1 48; NR4 LBD – human NR4A2 49; NR 4/5/6 DBD - mouse NR5A1 50; NR5/6/0 LBD – mouse NR5A1 51.
Disorder prediction of protein sequences were performed using the previously experimentally validated and widely used PONDR VL3H algorithm 8 available at http://www.ist.temple.edu/disprot/predictor/php. Sequences were input as a text file and were summarized by the fraction of residues with probability of disorder greater than 50% across the full length protein 9–11. Disorder probabilities were also analyzed by domain (NTD, DBD, D-domain, and LBD) or total protein sequence. The longest disorder stretch (longest continuous stretch of 30 or more AA residues with disorder probability ≥0.5), and fraction of NTD, DBD, D-domain, and LBD or total protein in disorder blocks ≥30 AA were also calculated 4, 12. The disorder calculations for each amino acid residue are available as Supplemental Table 1–5.
Calculation of Nuclear Hormone Receptor Networks
Ingenuity Pathways Analysis 5.0 (IPA, Ingenuity Systems, Redwood City, CA) software 52 was used to calculate the up- and downstream direct interactions with each human NHR individually. It has been previously reported that the definition of a hub protein, in terms of a number of interacting partners, has not been definitively defined 12. Previously, others have used ten interactions as a cutoff value for hubs and we have also used the same value in this study.
Statistics
Correlations were assessed between the predicted ID data for human NHR and the up-stream, down-stream and total number of direct interactions derived from the Ingenuity software. Mean ± SD values were calculated for the predicted ID data across species. Differences that were greater than 2 ± SD were considered significantly different from mean values determined for vertebrates. The human disorder data was statistically analyzed with SPSS 14.0 (SPSS Inc. Chicago IL) or JMP 7.0.1 (SAS, Cary, NC) and mean ±SD values for classic and orphan receptors, 10 or less down-stream protein interactions or >10 interactions, and all species versus human NHR were compared with the t-test.
RESULTS
Quantitative analysis of NHR domains by species
ID was calculated for nearly 400 NHR (Supplemental Table 1–5) and analyzed by the NTD, DBD, D-domain, LBD and across the full sequence (Supplemental Table 6). Analysis of ID across the full length protein of all NHRs for all species revealed (Table 1) nearly 2.7 fold higher mean ID probability in the NTD, DBD, and D-domain compared with the LBD, while full length (average) disorder is intermediate and relatively consistent. Nearly identical results were obtained when analysis was focused only on either the human or mouse receptors. The rank order between human NHRs and all species is similar; however, ID in the DBD, LBD and D-domain is statistically significantly lower for human NHRs compared with all species (Table 1). Even though the average ID probability in the LBD is significantly lower than that for the other domains, there is substantial inter-receptor variability in ID in the LBD. Average ID probability between human and mouse (r2 for DBD = 0.92, LBD = 0.79 and full length = 0.88) and human and rat (r2 for DBD = 0.92, LBD = 0.70 and full length = 0.89) receptors correlated tightly. The correlations between human and chicken (r2 for DBD = 0.90, LBD = 0.61 and full length = 0.65), human and African clawed frog (r2 for DBD = 0.58, LBD = 0.60 and full length = 0.57) and human and zebrafish (r2 for DBD = 0.85, LBD = 0.38 and full length = 0.57) receptors generally decreased as the evolutionary distance increased.
Table 1.
Analysis of domains by species using full length disorder. Numbers in parentheses represent number of NHRs.
| Mean Intrinsic Disorder ± SD (number of NHR) | |||||
|---|---|---|---|---|---|
| NHR Domain | NTD | DBD | D-Domain | LBD | Total |
| All species | 0.69 ± 0.13 | 0.67 ± 0.14 | 0.73 ± 0.13 | 0.25 ± 0.12 | 0.49 ± 0.10 |
| (382) | (383) | (383) | (397) | (393) | |
| Human | 0.69 ± 0.14 | 0.65* ± 0.14 | 0.69** ± 0.14 | 0.23* ± 0.13 | 0.49 ± 0.11 |
| (46) | (46) | (46) | (48) | (48) | |
T-test p < 0.002
T-test p < 0.0001
As several published studies 4, 12 assessed disordered regions of greater or equal to 30 AA, we analyzed the NTD, DBD, D-domain, LBD and full sequence for stretches of 30 or more consecutive AA residues that each had a disorder probability of 0.5 or higher. We will refer to these as “predicted disorder blocks.” We calculated fraction disorder in this way, across all species and then for human alone. The fraction of sequence predicted to be in disorder blocks in the NTD, DBD and D-Domain was 5–7 fold higher than in the LBD, with total fraction disordered intermediate (Table 2). Similar results were obtained when analysis was confined only to human receptors; in addition, the percent sequence in disordered blocks in the D-domain is statistically significantly lower for human NHR compared to other species (Table 2).
Table 2.
Analysis of domains by species using fraction disorder (>30 amino acids). Numbers in parentheses represent number of NHRs.
| Mean Intrinsic Disorder ± SD (number of NHR) | |||||
|---|---|---|---|---|---|
| NHR Domain | NTD | DBD | D-Domain | LBD | Total |
| All species | 0.57 ± 0.38 | 0.55 ± 0.13 | 0.74 ± 0.34 | 0.10 ± 0.13 | 0.37 ± 0.13 |
| (382) | (383) | (393) | (393) | (393) | |
| Human | 0.55 ± 0.39 | 0.55 ± 0.12 | 0.66* ± 0.35 | 0.10 ± 0.13 | 0.36 ± 0.14 |
| (46) | (46) | (48) | (48) | (48) | |
T-test p < 0.0001
Quantitative analysis of NHR protein-protein interaction networks and relationship to disorder
It has been suggested that comparison of protein-protein interaction networks across species could have potential for studying evolution 5, namely understanding those proteins that are the major hubs in the networks with many connections. Network building tools and algorithms have been used to connect NHRs to each other to visualize the current state of knowledge and understand some of the complex interactions they have with other proteins and ligands 53, 54. Human NHRs were evaluated in this study using the IPA software which contains over a million manually curated findings 52, to determine the number of published direct interactions with each protein (Figure 1) and relate this to the predicted ID (Supplemental Table 7). Preliminary analyses of 6 NHRs (LXRα, LXRβ, VDR, PXR, FXR and CAR) indicated that IPA contains more direct interaction data than the HPRD or BOND (previously BIND) databases (data not shown) and was therefore used for all human NHRs. Analysis of the data for 46 human NHRs indicated that those with more upstream interactions tended to also have more downstream interactions (Table 3) (r2 = 0.81, number of upstream interactions = 1.56 + 0.57* number of downstream interactions, p < 0.0001). This correlation may reflect that some proteins are favored over others for research 55, resulting in more published interactions discovered. Alternatively, it is possibly an artifact of the content acquisition process in the software. If papers mention on average n-times as many downstream interactions of a receptor than upstream interactions, this average is roughly independent of the receptor, and there is a sufficiently large number of independent papers, then this would lead to the observed correlation because of the central limit theorem. The notion of upstream/downstream is not always clearly defined in databases and in IPA it reflects the direction of the interaction regardless of its nature (Andreas Krämer, personal communication). However for our purposes in this study we generated direct networks around each NHR identically to determine any relationship between ID in the different domains and upstream and downstream connections.
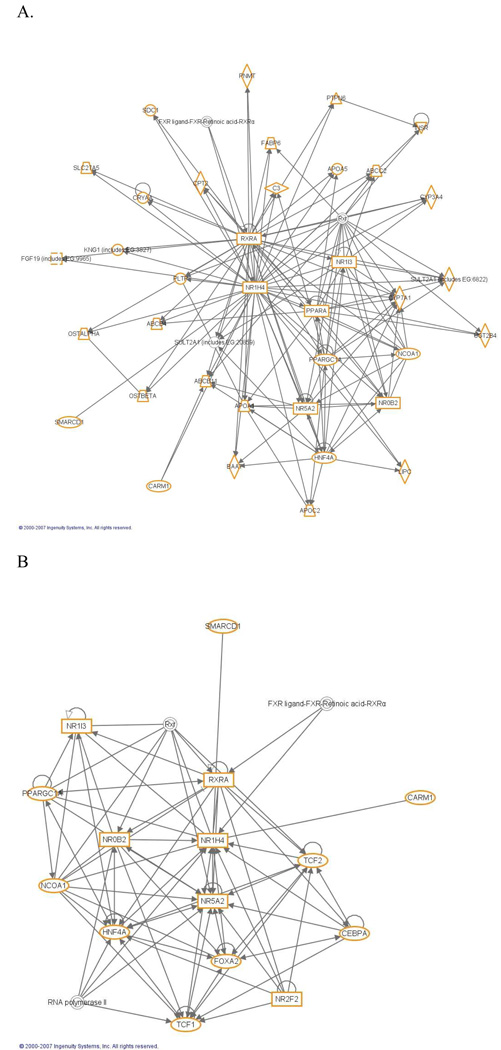
Figure 1.
A. Downstream and B. Upstream direct interaction networks for human FXR (NR1H4) showing connected genes using Ingenuity Pathways Analysis 5.0 (Ingenuity Systems). Human receptors (rectangles), transporters (trapezoid) and enzymes (diamonds) are shown connected to FXR.
Table 3.
Direct interactions for human NHR obtained from Ingenuity Pathways Analysis.
| NHR family | NHR | Upstream direct interactions | Downstream direct interactions | Total direct interactions |
|---|---|---|---|---|
| 1A1 | TR α | 32 | 69 | 101 |
| 1A2 | TR β | 59 | 114 | 173 |
| 1B1 | RAR α | 72 | 106 | 178 |
| 1B2 | RAR β | 46 | 42 | 88 |
| 1B3 | RAR γ | 12 | 29 | 41 |
| 1C1 | PPAR α | 65 | 127 | 192 |
| 1C2 | PPAR β/δ | 28 | 46 | 74 |
| 1C3 | PPAR γ | 84 | 179 | 263 |
| 1D1 | Rev-Erb α | 8 | 6 | 14 |
| 1D2 | Rev-Erb β | 2 | 4 | 6 |
| 1F1 | ROR α | 9 | 14 | 23 |
| 1F2 | ROR β | N/A | N/A | N/A |
| 1F3 | ROR γ | N/A | N/A | N/A |
| 1H2 | LXR β | 9 | 19 | 28 |
| 1H3 | LXR α | 18 | 37 | 55 |
| 1H4 | FXR | 14 | 36 | 50 |
| 1I1 | VDR | 47 | 69 | 116 |
| 1I2 | PXR | 17 | 37 | 54 |
| 1I3 | CAR | 18 | 35 | 53 |
| 2E1 | TLX | 1 | 3 | 4 |
| 2E3 | PNR | 0# | 0# | 0# |
| 2A1 | HNF4 α | 49 | 163 | 212 |
| 2A2 | HNF4 γ | 1 | 4 | 5 |
| 2B1 | RXR α | 94 | 226 | 320 |
| 2B2 | RXR β | 24 | 49 | 73 |
| 2B3 | RXR γ | 15 | 32 | 47 |
| 2C1 | TR2 | 9 | 10 | 19 |
| 2C2 | TR4 | 8 | 20 | 28 |
| 2F1 | COUP TFI | 20 | 43 | 63 |
| 2F2 | COUP TFII | 13 | 37 | 50 |
| 2F6 | EAR 2 | 5 | 6 | 11 |
| 3A1 | ER α | 162 | 198 | 360 |
| 3A2 | ER β | 54 | 73 | 127 |
| 3B1 | ERR α | 12 | 55 | 67 |
| 3B2 | ERR β | 6 | 7 | 13 |
| 3B3 | ERR γ | 8 | 8 | 16 |
| 3C1 | GR | 109 | 178 | 287 |
| 3C2 | MR | 16 | 21 | 37 |
| 3C3 | PR | 49 | 51 | 100 |
| 3C4 | AR | 148 | 163 | 311 |
| 4A1 | Nur77 | 29 | 34 | 63 |
| 4A2 | Nurr1 | 9 | 9 | 18 |
| 4A3 | NOR-1 | 13 | 10 | 23 |
| 5A1 | SF1 | 38 | 55 | 93 |
| 5A2 | FTZ-F1b | 28 | 45 | 73 |
| 6A1 | GCNF | 2 | 6 | 8 |
| NR0B1 | DAX1 | 15 | 17 | 32 |
| NR0B2 | SHP | 35 | 44 | 79 |
| Mean (± SD) | 33 ± 37 | 55 ± 59 |
N/A data not available for these individual receptors.
This nuclear receptor is in the database but there are no reported interactions.
When the ID data was split into those NHR (non-hubs or end proteins) with 10 or less interactions (n = 12) or (hubs 12) >10 interactions (n = 32) (Table 4 and Table 5), for the D-domain and LBD, the mean ID values were statistically significantly lower for those NHR with 10 or less downstream interactions compared with those with >10 downstream interactions, respectively (Table 4). This would indicate that predicted ID in the D-domain and LBD specifically may relate to human NHR promiscuity in that increased ID correlates with the number of experimentally verified and published direct down-stream interactions available in the IPA database. These NHR with >10 downstream interactions may also bind a wider array of ligands and in turn, potentially be activated by them.
Table 4.
Analysis of human NHR downstream protein-protein interaction networks and relationship to full length disorder. Numbers in parentheses represent number of NHRs.
| Mean Intrinsic Disorder ± SD (number of NHR) | |||||
|---|---|---|---|---|---|
| NHR Domain | NTD | DBD | D-Domain | LBD | Total |
| ≤ 10 direct | 0.69 ± 0.12 | 0.65 ± 0.13 | 0.64 ± 0.08 | 0.20 ± 0.15 | 0.48 ± 0.13 |
| interactions | (12) | (12) | (12) | (12) | (12) |
| > 10 direct | 0.69 ± 0.14 | 0.66 ± 0.14 | 0.72** ± 0.15 | 0.24* ± 0.12 | 0.50 ± 0.10 |
| interactions | (32) | (32) | (34) | (34) | (34) |
T-test p < 0.01
T-test p < 0.002
Table 5.
Analysis of human NHR downstream protein-protein interaction networks and relationship to fraction of sequence in disorder blocks (>30 amino acids). Numbers in parentheses represent number of NHRs.
| Mean Intrinsic Disorder ± SD (number of NHR) | |||||
|---|---|---|---|---|---|
| NHR Domain | NTD | DBD | D-Domain | LBD | Total |
| ≤ 10 direct | 0.60 ± 0.39 | 0.56 ± 0.03 | 0.65 ± 0.26 | 0.08 ± 0.11 | 0.37 ± 0.15 |
| interactions | (12) | (12) | (12) | (12) | (12) |
| > 10 direct | 0.56 ± 0.38 | 0.54 ± 0.14 | 0.67 ± 0.39 | 0.11 ± 0.14 | 0.36 ± 0.14 |
| interactions | (32) | (32) | (34) | (34) | (34) |
Quantitative analysis of classic versus orphan receptors and relationship to disorder
Classifying the receptors as either classic or orphan 24 and comparing the mean ID (Supplemental Table 8) indicated the difference between the means for full length disorder for classic and orphan were statistically significant. The ID in the LBD and D-domain was higher in the classic NHR (Table 6). When the data were analyzed in blocks of greater or equal to 30 amino acids, again the LBD and D-domain were also statistically significant between classic and orphan receptors (Table 7). This indicates that the LBD and D-domain may be responsible for the aforementioned difference in the total full length disorder.
Table 6.
Analysis of human classic versus orphan NHR and relationship to full length disorder. Numbers in parentheses represent number of NHRs.
| Mean Intrinsic Disorder ± SD (number of NHR) | |||||
|---|---|---|---|---|---|
| NHR Domain | NTD | DBD | D-Domain | LBD | Total |
| Classic NHR | 0.69 ± 0.13 | 0.64 ± 0.14 | 0.76** ± 0.14 | 0.26* ± 0.13 | 0.52* ± 0.10 |
| (23) | (23) | (23) | (23) | (23) | |
| Orphan NHR | 0.68 ± 0.15 | 0.66 ± 0.13 | 0.63 ± 0.10 | 0.20 ± 0.13 | 0.47 ± 0.12 |
| (23) | (23) | (23) | (25) | (25) | |
T-test p < 0.02
T-test p < 0.0001
Table 7.
Analysis of human classic versus orphan NHR and relationship to fraction disorder (>30 amino acids). Numbers in parentheses represent number of NHRs.
| Mean Intrinsic Disorder ± SD (number of NHR) | |||||
|---|---|---|---|---|---|
| NHR Domain | NTD | DBD | D-Domain | LBD | Total |
| Classic | 0.56 ± 0.36 | 0.54 ± 0.16 | 0.73* ± 0.37 | 0.13* ± 0.16 | 0.38 ± 0.14 |
| (22) | (23) | (22) | (23) | (22) | |
| Orphan | 0.53 ± 0.42 | 0.55 ± 0.04 | 0.59 ± 0.33 | 0.07 ± 0.10 | 0.35 ± 0.14 |
| (23) | (23) | (25) | (25) | (25) | |
T-test p < 0.04
We also evaluated the published structures for NHR looking for evidence of disorder, unstructured regions, poor or no electron density reported in the original publications. We have shown that nearly half (23) of the NHR with regions of predicted ID have been shown to have some regions of disorder in the X-ray or NMR structures (Supplemental Table 9). For 20 of these 23 NHR (87 %) there was an excellent agreement for predicted versus experimentally determined disordered residues or regions. While these published studies did not generally analyze for ID across the full sequence, which would enable a direct comparison with our predictions, we did however find that there are many more studies of the LBD, some on DBDs, and little structural information on NTD or D-domain to date.
Qualitative analysis by NHR across species
To assess general patterns of ID, we analyzed all NHR sequences for stretches of 30 or more consecutive AA residues that each had a disorder probability of 0.5 or higher, as described above. Data for all human NHRs are plotted as bar graphs in Figure 2, showing the NHR domains NTD, DBD, D-domain and LBD, from left to right. The black bars indicate likely “ordered” sequence and grey bars indicate predicted disordered blocks. Due to the variability in length of the NTDs for the various NHRs, sequences were aligned to the beginning of the DBD to allow for better visual comparison between domains.
Figure 2.
Patterns of predicted ID in human NHRs. In the bar graphs, stretches of 30 or more consecutive amino acids that each had a disorder probability of 0.5 or higher (“predicted disorder blocks”) are colored grey. Black bars indicate “ordered” sequence. The NHRs are aligned to the beginning of the DBD to allow for easier comparison between domains. The DBD and LBD domains are annotated. The NTD and D-domain regions are to the left of the DBD and LBD, respectively.
Several general patterns are readily noticeable in comparing across the human NHRs. First, predicted ID in the NTD is highly variable (Figure 2), adding a further layer of complexity to the substantial differences in NTD sequence lengths. Twelve human NHRs (NR1C3, 1F3, 1H4, 1I1, 1I2, 1I3, 2A1, 2A2, 2C1, 2E1, 5A1 and 5A2) have no predicted disorder blocks in the NTD while the NTDs of eleven human NHRs (1A1, 1C2, 1D1, 1D2, 1F1, 1H2, 2B1, 2F1, 2F2, 2F6 and 3B1) are predicted to be entirely or nearly entirely disordered. Predicted ID within the NTD is not even necessarily conserved within a subfamily. For example, the NTD of human NR1H2 (LXRβ) is predicted to be entirely disordered whereas that of human NR1H4 (FXR) is predicted to be ordered. In contrast, the entire NTDs of all three human NR2F (2F1, 2F2, and 2F6) receptors are predicted to be disordered.
Second, in every human NHR that has a DBD (i.e., except the ‘single domain’ NHRs NR0B1 and 0B2), at least a portion of the C-terminal end of the DBD is predicted to be disordered. A common pattern is for the N-terminal half of the DBD to be ordered and the C-terminal half of the DBD to be disordered (Figure 2). Typically, the C-terminal half of the DBD and an adjacent portion of the D-domain are predicted to be part of a continuous disorder block.
Third, for 25 of the 48 human NHRs (including both classic and orphan NHRs), no disorder blocks were predicted in the LBD (Figure 2). For those NHRs that had predicted disorder blocks in the LBD, the blocks were often in the far C-terminal end or in discrete inter-helical domains known to be disordered (e.g., H1-H3 helix of NR1H and 1I receptors) from biochemical experiments or structural studies 35, 38, 42, 56.
We also compared patterns of disorder across species for each of the NHRs. With few exceptions, we generally found that predicted disorder patterns were conserved between vertebrate species but could be dramatically different when comparing vertebrate with invertebrate orthologs. Focusing in detail on some of these NHRs will illustrate these observations.
FXR (NR1H4) has been termed the ‘bile acid receptor’ and regulates enzymes and transporters involved in bile acid biosynthesis and transport 42, 57. In humans and mice, FXRs are activated best by primary bile acids. We have recently shown that FXRs have different ligand specificities across species, in a pattern that likely reflects adaptation to cross-species differences in bile salt structures 58. The predicted ID patterns of FXRs are generally similar across vertebrate species with the exception of variability in the NTD (Figure 3). The most extreme contrast in the FXRs was the ortholog from the sea squirt Ciona intestinalis, an invertebrate urochordate. The sea squirt FXR is predicted to have an entirely disordered NTD compared to the vertebrate FXRs, where less than half of the NTD is predicted to be ordered. This may reflect an evolutionary shift in function for this receptor. Ciona and other invertebrate animals do not utilize bile salts, implying that the sea squirt FXR has different ligands and functions from vertebrate FXRs which we have recently confirmed 58.
Figure 3.
Cross-species comparisons of predicted ID for the FXRs. See Figure 2 for details on the color coding and annotation of the plots. The NTD of the sea squirt FXR, which has a predicted ID pattern different from vertebrate FXRs, is indicated by *.
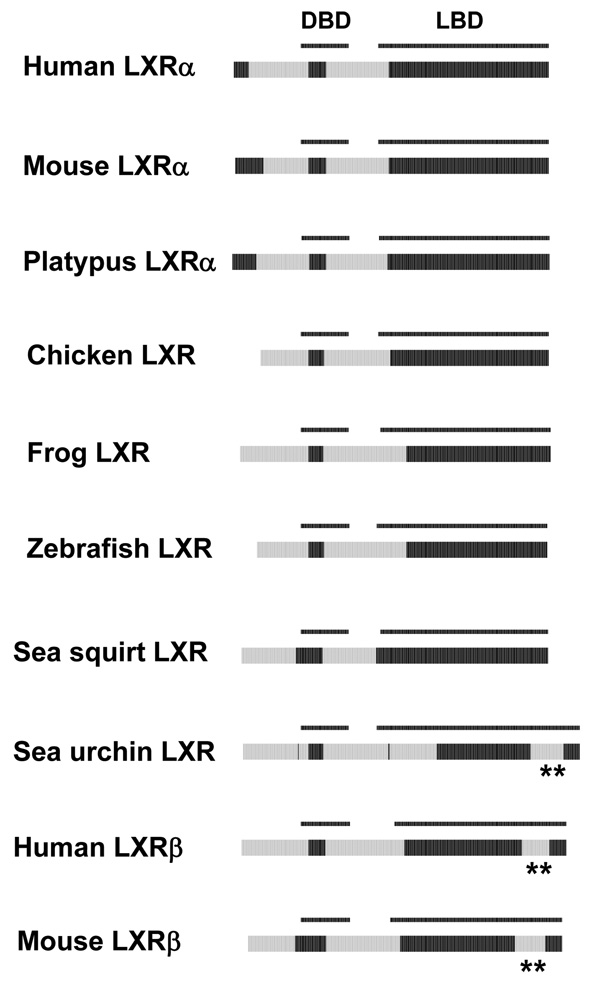
LXRs are closely related to FXRs but are activated instead by oxysterols 57. Mammalian LXRα and LXRβ have very similar ligand specificities but do differ in tissue expression patterns. Most non-mammalian vertebrates have a single LXR. For non-mammalian species, the pufferfish LXR has been the subject of the most detailed study 59. Pufferfish LXR is more closely related to mammalian LXRα by sequence similarity, although it more closely resembles mammalian LXRβ in the ubiquity of tissue expression. The current sequence data suggest that a single LXR gene duplicated before or early in mammalian evolution 59, 60. If this hypothesis is correct, then one of the duplicated genes maintained the ubiquitous tissue expression (LXRβ) while the other (LXRα) carried out specific roles in cholesterol and lipid metabolism with more restricted expression in adipose tissue, liver, and macrophages. The predicted ID patterns of three non-mammalian vertebrate LXRs (chicken, African clawed frog, and zebrafish) are intermediate between mammalian LXRα and LXRβ (Figure 4). Human and mouse LXRβ have a predicted disorder block involving helices 9, 10, and 11 of the LBD not found in LXRαs or the non-mammalian vertebrate LXRs (indicated by ** in Figure 4). Mammalian LXRαs have a longer NTD than the other vertebrate LXRs with the N-terminal end of the NTD predicted as being ordered. Interestingly, two putative invertebrate LXR orthologs have contrasting predicted disorder patterns. The sea squirt LXR has a disorder pattern nearly identical to non-mammalian vertebrate LXRs while the purple sea urchin LXR has a pattern of disorder very similar to mammalian LXRβ, including the predicted disordered sequence in helices 9–11 of the LBD.
Figure 4.
Cross-species comparisons of predicted ID for the LXRs. See Figure 2 for details on the color coding and annotation of the plots. A segment of LBD predicted as disordered in some LXRs is indicated by **.
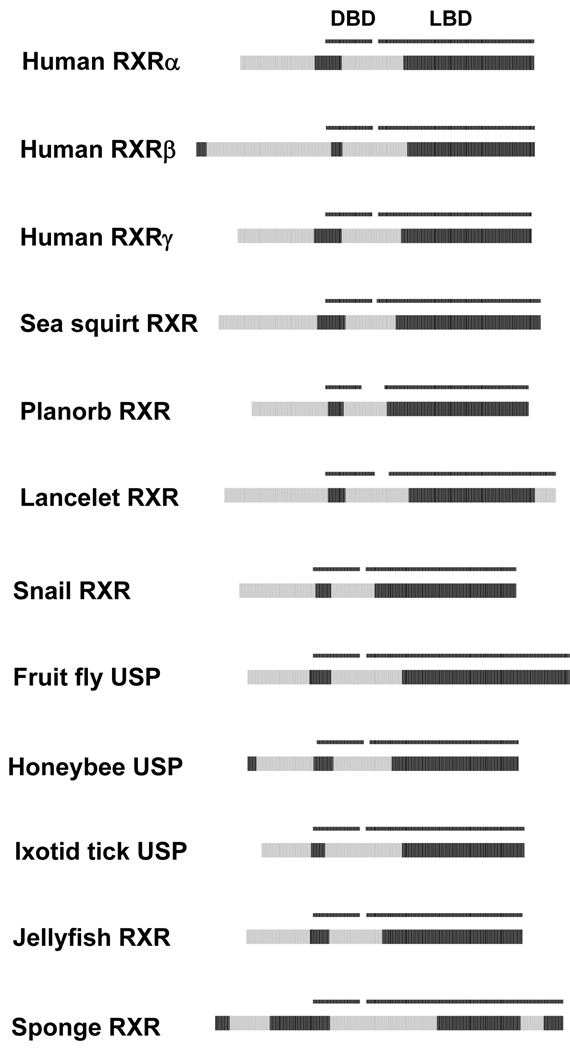
The NR2B subfamilies contain what are thought to be the evolutionarily oldest NHRs 61, 62. In some invertebrates, these receptors are termed Ultraspiracle receptors (USP). In other animals, including all vertebrates, the NR2B receptors are termed retinoid X receptors (RXRs). We compared the predicted ID patterns of RXRs and USPs across a wide range of vertebrate and invertebrate species. Consistent with the high degree of sequence conservation of RXRs and USPs across species, the disorder patterns of RXRs and USPs are remarkably similar among nearly all vertebrate and invertebrate species analyzed (Figure 5). The only exception was the RXR from the marine sponge 63 which has two predicted disorder blocks in the LBD comprising over a third of the LBD sequence length (one in the N-terminal portion and the other involving helices 9 and 10). There was also less predicted ID in the NTD of the sponge RXR relative to the RXRs and USPs from other species. The high degree of conservation of the RXRs/USPs may reflect strong negative evolutionary selection of these essential hub proteins (e.g. human RXRα was calculated to have 226 downstream interactions, Table 3). For example, RXRs/USPs are obligate heterodimers for NR1 receptors. The function of NR2B receptors as heterodimeric partners for NR1 receptors is therefore likely to have a very long evolutionary history, as RXRs and USPs have been shown to subserve this function in a phylogenetically diverse array of vertebrate and invertebrate animals 61, 62. To our knowledge the only published study of the sponge RXR indicated that this receptor mediated retinoic acid-induced morphogenetic functions but heterodimeric complexes were not analyzed 63.
Figure 5.
Cross-species comparisons of predicted ID for the USPs and RXRs. See Figure 2 for details on the color coding and annotation of the plots.
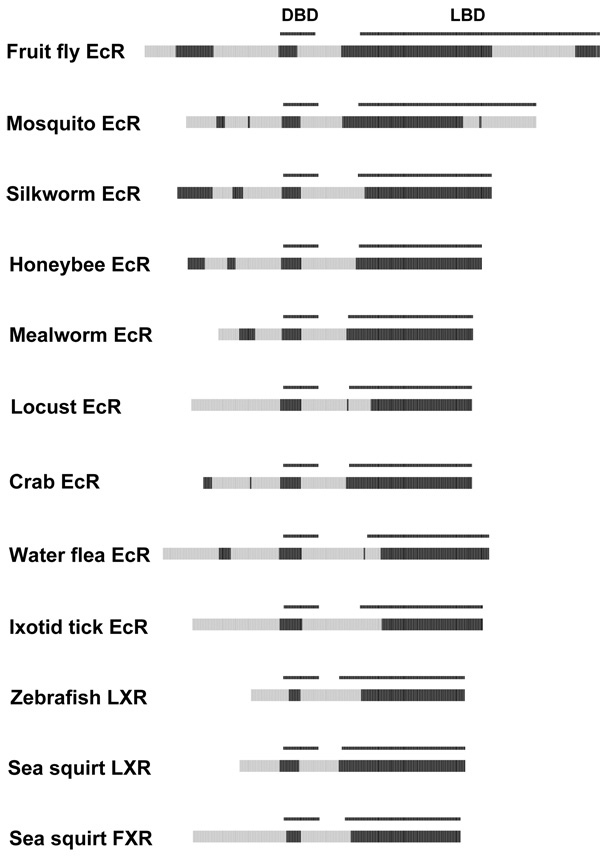
The invertebrate ecdysone receptors (EcRs, NR1H1) mediate a variety of developmental functions in invertebrates 64 and showed much more variability in disorder patterns than their NR2B heterodimeric patterns (Figure 6). The predicted disorder of NTD of the EcRs varied across species. The Dipteran EcRs also differed from the rest in having a predicted disorder block in the last C-terminal third of the LBD. The variability in predicted ID for EcRs is consistent with both phylogenetic and functional studies demonstrating evolutionary divergence of EcRs 65, 66.
Figure 6.
Cross-species comparisons of predicted ID for the ecdysone receptors (EcRs). See Figure 2 for details on the color coding and annotation of the plots.
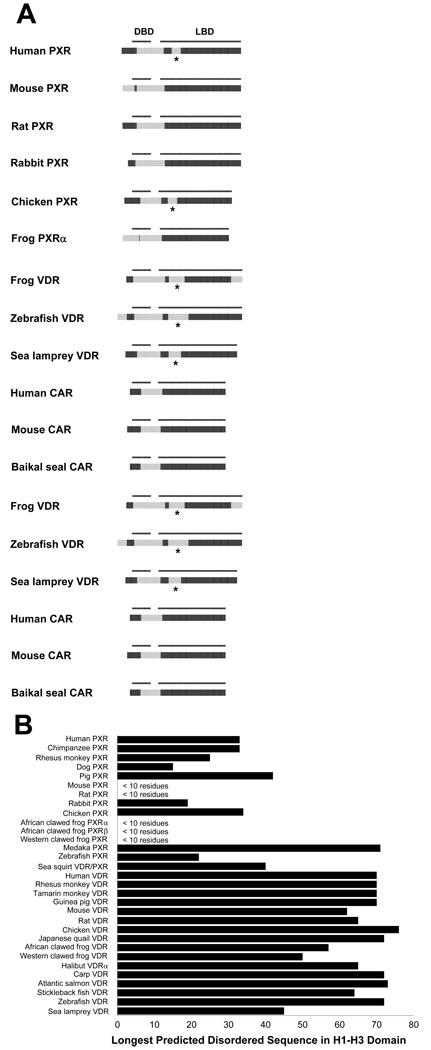
The NR1I subfamily (VDR, PXR, and CAR) showed the highest variability in predicted ID across species for any NHR subfamily. As discussed in the Introduction, the H1–H3 interhelical domain of human VDR, zebrafish VDR, and human PXR has been shown to be disordered based on biochemical and/or structural studies. We found that the H1–H3 interhelical domain had a predicted disorder block in every VDR analyzed, including the putative sea squirt VDR/PXR (Figure 7a), a receptor that we have preliminarily classified as a VDR 67. Interestingly, this same domain was not predicted to be disordered in some of the PXRs. In the case of the African clawed frog and Western clawed frog PXRs, this is a result of a much shorter H1–H3 interhelical domain relative to other PXRs (these are similar in length to CARs which also lack any predicted disorder block between H1 and H3). But for mouse and rat PXRs, no stretch of 10 or more residues in the H1–H3 interhelical domain was predicted to be disordered, despite the H1–H3 interhelical domain being the same length in these receptors as that for human PXR (Figure 7b). In pig, rabbit, and zebrafish PXRs, the possibly disordered sequence length was only approximately 20 residues, shorter than the predicted disorder blocks in the H1–H3 interhelical domain of human, pig, and medaka PXRs (Figure 7b). It is also worth noting that multiple residues within the H1–H3 interhelical domain were shown to have nucleotide variation consistent with positive evolutionary selection in a previous published phylogenetic analysis of the vertebrate NHR superfamily 68. Consequently, variability in ID may be a contributing factor to the marked differences in pharmacology between PXRs from different species 69–71. In contrast to VDRs and PXRs, all of the CARs analyzed were predicted to have no disorder blocks in the LBD (Figure 7).
Figure 7.
Cross-species comparisons of predicted ID for the NR1I (VDR, PXR, and CAR) receptors. (A) Cross-species comparisons of predicted ID for the NR1I receptors. See Figure 2 for details on the color coding and annotation of the plots. Disordered sequence in the H1–H3 interhelical domain is indicated by *. (B) Comparison of the longest predicted disorder block in the H1–H3 interhelical domain of VDRs and PXRs. For the mouse, rat, African clawed frog, and Western clawed frog PXRs, no stretch of 10 or more amino acid residues were identified in the H1–H3 interhelical domain that had disorder probabilities of 0.5 or more.
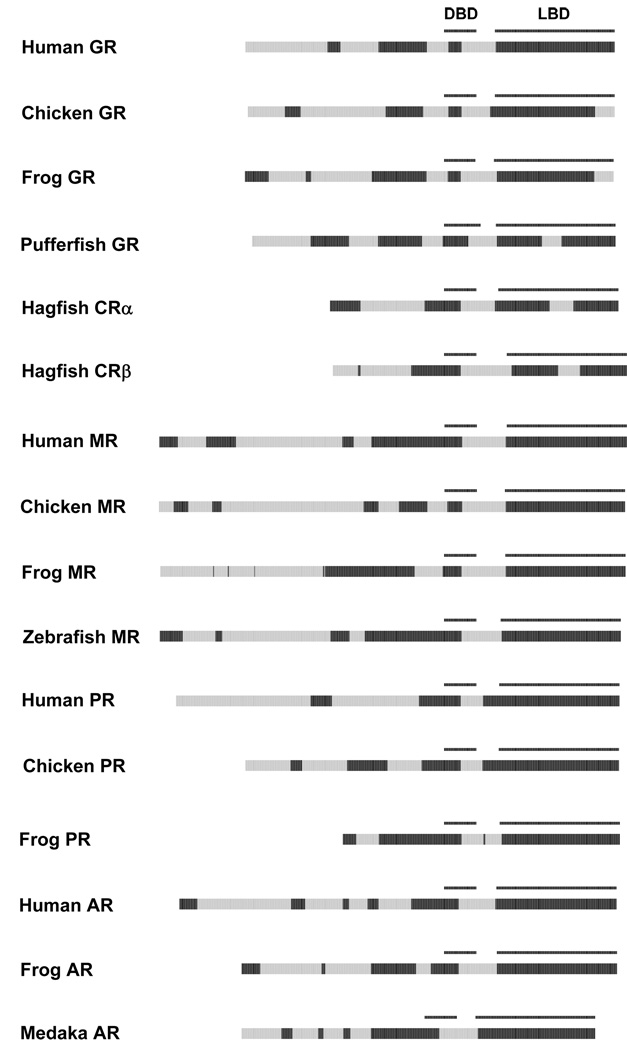
A previous study has examined ID in the NTD of NR3C receptors (GR, MR, PR, and AR) 10. As shown in Figure 8, the NTDs of the NR3C receptors differ in length and predicted disorder blocks. The LBDs of the NR3C receptors in general were predicted to be ordered. The exceptions occurred in the non-mammalian GRs. The extreme C-terminal portion of the chicken and African clawed frog GRs were predicted to be disordered. In addition, the pufferfish GR and two splice variants of a single Atlantic hagfish (Myxine glutinosa) corticoid receptor (CRα and CRβ; 72) had a predicted disorder block in the region where helices 6, 7, and 8 are located in the mammalian NR3C receptors. Phylogenetic analysis indicates that GRs and MRs are likely the result of duplication and then divergence of a single CR gene (as found in Agnathan species like hagfish and the sea lamprey) 72. It is interesting to find that pufferfish GR is predicted to have retained the predicted disordered sequence involving helices 6 to 8 found in hagfish CRα and CRβ while other GRs and the MRs do not have any predicted ID in this region of the LBD. These results may reflect evolutionary changes in ID during the co-evolution of the NR3C receptors and their cognate ligands 72.
Figure 8.
Cross-species comparisons of predicted ID for the NR3C (GR, MR, PR, and AR) receptors. See Figure 2 for details on the color coding and annotation of the plots.
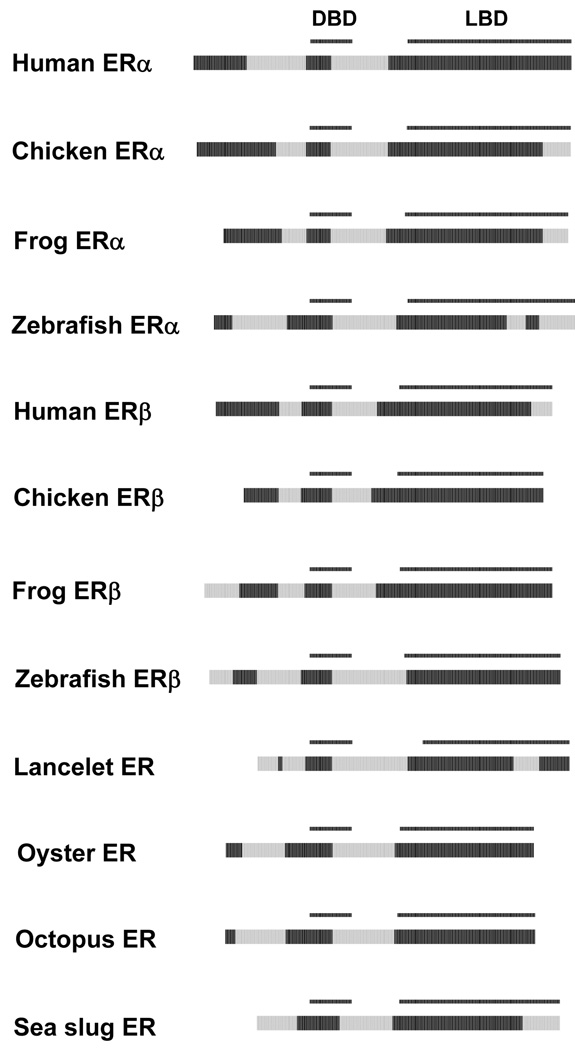
Similar to the NR3C receptors, the ERs (NR3A1 and 3A2) showed variability in predicted ID in the NTD, with human ERα and ERβ being predicted as more ordered than most of the other ERs (Figure 9). As with the chicken and African clawed frog GRs, the extreme C-terminal portion of the chicken ERα, Western clawed frog ERα, zebrafish ERα, human ERβ, and California sea slug (Aplysia californica) ERα was predicted to be disordered. In addition, the zebrafish ERα and Florida lancelet (Branchiostoma floridae) ERα both had a predicted disorder block in the region of helices 9, 10, and 11.
Figure 9.
Cross-species comparisons of predicted ID for the ERs. See Figure 2 for details on the color coding and annotation of the plots.
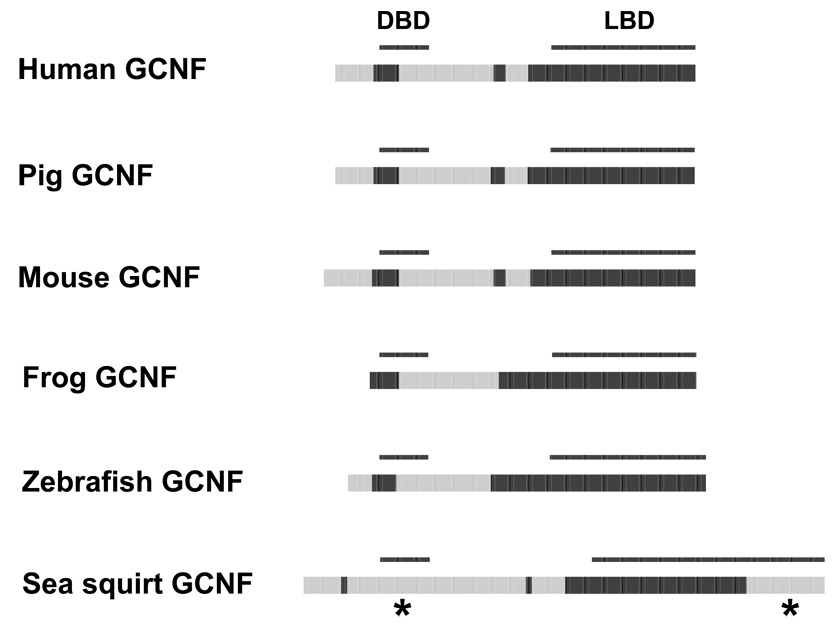
The predicted disorder patterns of NHRs in families 4, 5, 6, and 0 did not vary noticeably across species. One of the more extreme variations was however seen in comparing the sea squirt NR6A1 (germ cell nuclear factor, GCNF) to the vertebrates GCNFs. The sea squirt GCNF had a predicted disorder block of more than 100 amino acid residues in the C-terminal end of the LBD whereas no ID was predicted in any of the vertebrate GCNFs analyzed (Figure 10).
Figure 10.
Cross-species comparisons of predicted ID for the GCNFs. See Figure 2 for details on the color coding and annotation of the plots. Differences in the predicted ID for the sea squirt NTD and LBD compared to vertebrate GCNFs are indicated by *.
DISCUSSION
The increasing generation of biological data using high throughput methods in drug discovery increasingly necessitates the application of computational technologies. Computational models are also becoming widely available for individual proteins known to have some relationship to toxicity 73 such as drug metabolizing enzymes, transporters, ion channels and receptors 74. Many of these proteins are known to be regulated by NHRs which represent a large diverse superfamily of key transcriptional regulators involved in important physiological functions affecting endogenous metabolism, cell growth, proliferation, xenobiotic protection, and oxidative stress 75, 76. Our knowledge and understanding of NHRs is however in the early stages. This ultimately has implications for our ability to predict and modulate the transcription of key genes in humans for therapeutic advantage.
With the exception of one other study focused on NR3C receptors only (discussed below), to date there has been little evaluation of predicted ID for other NHRs. The analyses performed in this study therefore represents a novel approach to understanding the changes in ID in different regions of the protein that may impact protein-protein interactions, DNA-binding, ligand binding, co-activator/co-repressor recruitment and binding, while also reflecting NHR co-evolution driven by the selectivity or promiscuity of ligand interactions.
In this study we have quantitatively determined the extent of predicted ID across the NHR families in different vertebrate and invertebrate species. This was achieved by focusing on measures of ID for the NHR, namely using the NTD, DBD, D-domain, LBD and full length protein. We have observed some significant differences between these measures of disorder across NHR and across species for nearly all NHRs.
Analysis of the complete NHR for all species and the human NHR datasets both showed approximately 2.7 fold higher disorder in the NTD, DBD or D-domain compared with the LBD while full length disorder is intermediate. ID in the DBD, LBD and D-domain is statistically significantly lower for human NHR compared with all species. It was also noted that there was more variability in the disorder in the LBD across species and across NHRs for human. These differences were magnified when we focused on the fraction of sequence in disorder blocks ≥30 AA residues, with ID in the NTD, DBD or D-domain being 5–7 fold higher than for the LBD. Only ID in the D-domain was statistically significantly lower in human compared with other species. The significance of lower ID in this domain of human NHR is unclear at present. The analysis of disorder blocks also drew our attention to the generally completely ordered (by this measure) LBD of the NR3 receptors (GR, AR, PR, MR and ERα). We speculate that the physiological important roles of these receptors require selectivity in ligand binding and therefore limiting ID may prevent conformational flexibility in the LBD that could enable promiscuity and more diverse ligand binding, thereby impacting its evolution 23. These receptors do, however, have significant disorder in the NTD 77.
We are aware of one other study that has looked at ID in these steroid hormone receptors, 78 specifically at the NTD in AR and GR and showed predicted ID in the NTD (AF-1 region) that was also indicated by the experimental mutational studies of regions controlling gene expression performed by others 10 78. These authors suggested that plasticity in the AF-1 region due to ID was important for its function. It has also been suggested that ID facilitates more rapid evolution of gene products (and may result in alternative splicing) as these sequences themselves evolve more rapidly than ordered ones due to fewer structural constraints requiring limited AA substitutions 9, 19. Within a NHR family it would appear that different members alternate the level of disorder in each domain and this may drive the evolution in different directions depending on the function of the NHR. For example, increased ID in the LBD may increase promiscuity of ligand binding while increased disorder in the D-domain may alter DNA recognition and heterodimerization 20, coactivator interactions 21, corepressor and protein-protein interactions.
One of our hypotheses was that ID could impact function and protein interactions. Network building tools and algorithms have been used to connect human NHR to each other to understand some of the complex interactions with other proteins and ligands 53, 54, and compare protein-protein interaction networks across species 5. This has enabled further understanding of those proteins that are the major hubs in the networks with many connections. In our study human NHR were further evaluated to determine the number of published direct interactions with each protein upstream and downstream (for example Figure 1) and relate this to the average predicted ID (Supplemental Table 7).
We found that for 46 human NHRs with data in the IPA database, after separating NHRs with 10 or less down-stream interactions (likely end proteins) and those with >10 down-stream interactions (likely hub proteins 12), then the D-domain and LBD in those NHRs with 10 or less down-stream interactions had significantly lower ID. Our observations in human would tend to agree with other studies of transcription factors showing those with more partners or target genes tended to be more disordered in yeast 11, 12 and the degree of disorder was significantly higher in transcription factors in eukaryotes than prokaryotes 13. To our knowledge this is the first such analysis with human NHR only. It should be noted that the definition of a distinct cut-off for protein interactions that specifies a “hub” is ill defined 12. In this study we used 10 as a cutoff which had been previously used 12. Others have used the top 20% of proteins when looking at a heterogeneous set of yeast transcriptional factors 11 based on the assumption that protein-protein interactions maintain a scale free architecture. It has been suggested that there is no conclusive proof that protein-protein interactions are indeed scale free throughout their full interactome as all studies have been reliant on simulations 5. We therefore propose that 10 represents a good cutoff. For example the estrogen receptor α has previously been suggested as a hub with between 12–116 interactions (depending on the database protein interaction database used) 5 and in our current study this protein has 198 downstream interactions (Table 3).
Orphan receptors do not have identified ligand activators and likely function as ligand-independent transcription factors while classic receptors are ligand-activated NHRs. When we divided the human NHRs using this classification we found the ID in the LBD and D-domain was higher in the classic NHRs when using disorder calculated as either full length or in blocks of greater or equal to 30 AA (Table 6 and Table 7). The importance of the ID in the LBD and D-domain may relate to a degree of plasticity or flexibility with functional importance for ligand activation as described previously for the classic NHR (NTD plasticity in AR and GR 10, 78). Whereas disorder in the orphan receptors may be less useful if they function as ligand-independent transcription factors and may not require structural flexibility for ligand binding or DNA binding. A previous study has looked at human proteins and found that the intrinsically disordered loop region of the vitamin D receptor was on the surface of the protein 15. We have not investigated this further for the human NHR in this study but we would add that PXR is known to have an experimentally disordered region on the surface 36.
The differences we have observed in ID in this study are most frequent in the D-domain. It is interesting also to note that several of the published studies (Supplemental Table 9) also found experimentally determined disorder in the D-domain e.g. TRα 22, RARβ 79 and RXR α 80 and these NHR have 69, 42 and 226 downstream protein interactions in the human IPA database, therefore clearly representing hub proteins. The NTD of NHR are less conserved than the other regions 81. While others have shown experimentally and theoretically the role of disorder in the AF-1 domain (residing in the NTD), for interactions with transcription factors 10, 82 this would appear to be reasonably comparable across NHR which serve as hubs or non-hubs (ends) based on our study, and therefore does not enable discrimination. Previous ID analysis of transcription factors suggested the DBD may either be well structured (ordered) or highly unstructured (disordered) in regions such as AT-hooks and basic motifs 13. Our results comparing classic and orphan NHRs indicate that ID in the LBD may have a relationship with the structural diversity of molecules that bind to the receptor though further study is needed.
We have also shown by a thorough literature analysis that at least 23 different NHR have been determined experimentally by either X-ray crystallography, NMR and/or circular dichroism spectroscopy, to possess some disordered regions frequently between 10–15 AA in length and in some cases longer (Supplemental Table 9). Other studies have shown regions to be disordered in solution. The disorder predictor used in the present study is also known to be capable of identifying experimentally determined regions of disorder 2, 4, 7, 8. Our results therefore provide further validation of this PONDR algorithm and highlight its utility for defining disorder in NHRs. Other studies have not focused on intra-domain differences in transcription factors to a large extent and instead looked mainly at comparisons between species 9, 11. The closest studies focused on a narrow subset of human NHRs like the steroid receptors, and in particular the function of disorder in the NTD 77, 78, 82 while our studies greatly expand upon these.
Conclusions
The results in this study indicate ID may have had a key role in the evolutionary shifts of NHR. Future studies may relate this ID to other proteins and their co-evolution with diverse ligands, extent of protein-protein interactions and overall function across species. This could also have potential implications for studying evolution of these proteins and their associated networks of interactions with therapeutic targets in human. This would suggest a broader application of the techniques for calculation of ID across protein classes both in human and other species.
Supplementary Material
The following supplemental files are available free of charge via the Internet at http://pubs.acs.org.
Supplemental Table 1 – NR1A-1F intrinsic disorder data
Supplemental Table 2 – NR1H-1I intrinsic disorder data
Supplemental Table 3 – NR2 intrinsic disorder data
Supplemental Table 4 – NR3 intrinsic disorder data
Supplemental Table 5 – NR4, 5, 6 intrinsic disorder data
Supplemental Table 6 - Intrinsic disorder calculated for the NTD, DBD, D-domain, LBD and across the full sequence.
Supplemental Table 7 – Analysis of human NHR disorder compared with IPA upstream and down stream connections.
Supplemental Table 8 – Analysis of human NHR disorder by orphan and classic designation
Supplemental Table 9 - Analysis of published NHR structures with observed structural disorder
Acknowledgement
SE acknowledges Dr. David Lawson for providing information on the disorder analysis approach and Dr’s Andreas Krämer and Dana L. Abramovitz at Ingenuity Inc are thanked for kindly providing their software and advice. Dr. Maggie A.Z. Hupcey kindly provided encouragement and comments on this study. M.D.K. was supported by National Institutes of Health Grant K08-GM074238 and the Competitive Medical Research Fund from the University of Pittsburgh Medical Center.
Abbreviations
- AR
Androgen receptor
- CAR
constitutive androstane receptor
- DBD
DNA binding domain
- EcR
ecdysone receptors
- ER
estrogen receptor
- FXR
farnesoid X receptor
- GCNF
germ cell nuclear factor
- GR
glucocorticoid receptor
- ID
intrinsic disorder
- LBD
ligand binding domain
- LXR
liver X receptors
- MR
mineralocorticoid receptor
- NHR
nuclear hormone receptor
- NTD
N-terminal domain
- PPAR
peroxisome proliferator-activated receptors
- PR
progesterone receptor
- PXR
pregnane X receptor
- RXR
retinoid X receptors
- TR
thyroid hormone receptor
- USP
ultraspiracle receptors
- VDR
vitamin D receptor
References
- 1.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19(1):26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 2.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41(21):6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 3.Chen JW, Romero P, Uversky VN, Dunker AK. Conservation of intrinsic disorder in protein domains and families: II. functions of conserved disorder. J Proteome Res. 2006;5(4):888–898. doi: 10.1021/pr060049p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323(3):573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 5.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. Febs J. 2005;272(20):5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 6.Brown CJ, Takayama S, Campen AM, Vise P, Marshall TW, Oldfield CJ, Williams CJ, Dunker AK. Evolutionary rate heterogeneity in proteins with long disordered regions. J Mol Evol. 2002;55(1):104–110. doi: 10.1007/s00239-001-2309-6. [DOI] [PubMed] [Google Scholar]
- 7.Dunker AK, Brown CJ, Obradovic Z. Identification and functions of usefully disordered proteins. Adv Protein Chem. 2002;62:25–49. doi: 10.1016/s0065-3233(02)62004-2. [DOI] [PubMed] [Google Scholar]
- 8.Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK, Obradovic Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J Bioinform Comput Biol. 2005;3(1):35–60. doi: 10.1142/s0219720005000886. [DOI] [PubMed] [Google Scholar]
- 9.Minezaki Y, Homma K, Kinjo AR, Nishikawa K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J Mol Biol. 2006;359(4):1137–1149. doi: 10.1016/j.jmb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 10.McEwan IJ, Lavery D, Fischer K, Watt K. Natural disordered sequences in the amino terminal domain of nuclear receptors: lessons from the androgen and glucocorticoid receptors. Nucl Recept Signal. 2007;5:e001. doi: 10.1621/nrs.05001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh GP, Dash D. Intrinsic disorder in yeast transcriptional regulatory network. Proteins. 2007;68(3):602–605. doi: 10.1002/prot.21497. [DOI] [PubMed] [Google Scholar]
- 12.Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput Biol. 2006;2(8):e100. doi: 10.1371/journal.pcbi.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45(22):6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen JW, Romero P, Uversky VN, Dunker AK. Conservation of intrinsic disorder in protein domains and families: I. A database of conserved predicted disordered regions. J Proteome Res. 2006;5(4):879–887. doi: 10.1021/pr060048x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuchi S, Homma K, Minezaki Y, Nishikawa K. Intrinsically disordered loops inserted into the structural domains of human proteins. J Mol Biol. 2006;355(4):845–857. doi: 10.1016/j.jmb.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 16.Ekins S, Bugrim A, Nikolsky Y, Nikolskaya T. Systems biology: applications in drug discovery. In: Gad S, editor. Drug discovery handbook. New York: Wiley; 2005. pp. 123–183. [Google Scholar]
- 17.Ekins S, Nikolsky Y, Bugrim A, Kirillov E, Nikolskaya T. Pathway mapping tools for analysis of high content data. Methods Mol Biol. 2007;356:319–350. doi: 10.1385/1-59745-217-3:319. [DOI] [PubMed] [Google Scholar]
- 18.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. The large-scale organization of metabolic networks. Nature. 2000;407(6804):651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 19.Romero PR, Zaidi S, Fang YY, Uversky VN, Radivojac P, Oldfield CJ, Cortese MS, Sickmeier M, LeGall T, Obradovic Z, Dunker AK. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc Natl Acad Sci U S A. 2006;103(22):8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyamoto T, Kakizawa T, Ichikawa K, Nishio S, Takeda T, Suzuki S, Kaneko A, Kumagai M, Mori J, Yamashita K, Sakuma T, Hashizume K. The role of hinge domain in heterodimerization and specific DNA recognition by nuclear receptors. Mol Cell Endocrinol. 2001;181(1–2):229–238. doi: 10.1016/s0303-7207(01)00483-x. [DOI] [PubMed] [Google Scholar]
- 21.Iordanidou P, Aggelidou E, Demetriades C, Hadzopoulou-Cladaras M. Distinct amino acid residues may be involved in coactivator and ligand interactions in hepatocyte nuclear factor-4alpha. J Biol Chem. 2005;280(23):21810–21819. doi: 10.1074/jbc.M501221200. [DOI] [PubMed] [Google Scholar]
- 22.Nascimento AS, Dias SM, Nunes FM, Aparicio R, Ambrosio AL, Bleicher L, Figueira AC, Santos MA, de Oliveira Neto M, Fischer H, Togashi M, Craievich AF, Garratt RC, Baxter JD, Webb P, Polikarpov I. Structural rearrangements in the thyroid hormone receptor hinge domain and their putative role in the receptor function. J Mol Biol. 2006;360(3):586–598. doi: 10.1016/j.jmb.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 23.Nagl SB, Freeman J, Smith TF. Evolutionary constraint networks in ligand-binding domains: an information-theoretic approach. Pac Symp Biocomput. 1999:90–101. doi: 10.1142/9789814447300_0009. [DOI] [PubMed] [Google Scholar]
- 24.Benoit G, Cooney A, Giguere V, Ingraham H, Lazar M, Muscat G, Perlmann T, Renaud JP, Schwabe J, Sladek F, Tsai MJ, Laudet V. International Union of Pharmacology. LXVI. Orphan nuclear receptors. Pharmacol Rev. 2006;58(4):798–836. doi: 10.1124/pr.58.4.10. [DOI] [PubMed] [Google Scholar]
- 25.Kliewer SA, Lehmann JM, Willson TM. Orphan nuclear receptors: shifting endocrinology into reverse. Science. 1999;284(5415):757–760. doi: 10.1126/science.284.5415.757. [DOI] [PubMed] [Google Scholar]
- 26.Pellicciari R, Costantino G, Fiorucci S. Farnesoid X receptor: from structure to potential clinical applications. J Med Chem. 2005;48(17):5383–5403. doi: 10.1021/jm0582221. [DOI] [PubMed] [Google Scholar]
- 27.Bourguet W, Germain P, Gronemeyer H. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci. 2000;21(10):381–388. doi: 10.1016/s0165-6147(00)01548-0. [DOI] [PubMed] [Google Scholar]
- 28.Kliewer SA, Lehmann JM, Milburn MV, Willson TM. The PPARs and PXRs: nuclear xenobiotic receptors that define novel hormone signaling pathways. Recent Prog Horm Res. 1999;54:345–367. discussion 367–8. [PubMed] [Google Scholar]
- 29.Berkenstam A, Gustafsson JA. Nuclear receptors and their relevance to diseases related to lipid metabolism. Curr Opin Pharmacol. 2005;5(2):171–176. doi: 10.1016/j.coph.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Mohan R, Heyman RA. Orphan nuclear receptor modulators. Curr Top Med Chem. 2003;3(14):1637–1647. doi: 10.2174/1568026033451709. [DOI] [PubMed] [Google Scholar]
- 31.Staudinger J, Liu Y, Madan A, Habeebu S, Klaassen CD. Coordinate regulation of xenobiotic and bile acid homeostasis by pregnane X receptor. Drug Metab Dispos. 2001;29(11):1467–1472. [PubMed] [Google Scholar]
- 32.Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, Willson TM, Koller BH, Kliewer SA. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A. 2001;98(6):3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tabb MM, Kholodovych V, Grun F, Zhou C, Welsh WJ, Blumberg B. Highly chlorinated PCBs inhibit the human xenobiotic response mediated by the steroid and xenobiotic receptor (SXR) Environ Health Perspect. 2004;112(2):163–169. doi: 10.1289/ehp.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carnahan VE, Redinbo MR. Structure and function of the human nuclear xenobiotic receptor PXR. Curr Drug Metab. 2005;6(4):357–367. doi: 10.2174/1389200054633844. [DOI] [PubMed] [Google Scholar]
- 35.Watkins RE, Davis-Searles PR, Lambert MH, Redinbo MR. Coactivator binding promotes the specific interaction between ligand and the pregnane X receptor. J Mol Biol. 2003;331:815–828. doi: 10.1016/s0022-2836(03)00795-2. [DOI] [PubMed] [Google Scholar]
- 36.Chrencik JE, Orans J, Moore LB, Xue Y, Peng L, Collins JL, Wisely GB, Lambert MH, Kliewer SA, Redinbo MR. Structural disorder in the complex of human pregnane X receptor and the macrolide antibiotic rifampicin. Mol Endocrinol. 2005;19(5):1125–1134. doi: 10.1210/me.2004-0346. [DOI] [PubMed] [Google Scholar]
- 37.Farnegardh M, Bonn T, Sun S, Ljunggren J, Ahola H, Wilhelmsson A, Gustafsson JA, Carlquist M. The three-dimensional structure of the liver X receptor beta reveals a flexible ligand-binding pocket that can accommodate fundamentally different ligands. J Biol Chem. 2003;278(40):38821–38828. doi: 10.1074/jbc.M304842200. [DOI] [PubMed] [Google Scholar]
- 38.Svensson S, Ostberg T, Jacobsson M, Norstrom C, Stefansson K, Hallen D, Johansson IC, Zachrisson K, Ogg D, Jendeberg L. Crystal structure of the heteroimeric complex of LXRalpha and RXRbeta ligand-binding domains in a fully agonistic conformation. Embo J. 2003;22(18):4625–4633. doi: 10.1093/emboj/cdg456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoerer S, Schmid A, Heckel A, Budzinski RM, Nar H. Crystal structure of the human liver X receptor beta ligand-binding domain in complex with a synthetic agonist. J Mol Biol. 2003;334(5):853–861. doi: 10.1016/j.jmb.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 40.Ciesielski F, Rochel N, Moras D. Adaptability of the Vitamin D nuclear receptor to the synthetic ligand Gemini: remodelling the LBP with one side chain rotation. J Steroid Biochem Mol Biol. 2007;103(3–5):235–242. doi: 10.1016/j.jsbmb.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Rochel N, Wurtz JM, Mitschler A, Klaholz B, Moras D. The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol Cell. 2000;5(1):173–179. doi: 10.1016/s1097-2765(00)80413-x. [DOI] [PubMed] [Google Scholar]
- 42.Downes M, Verdecia MA, Roecker AJ, Hughes R, Hogenesch JB, Kast-Woelbern HR, Bowman ME, Ferrer JL, Anisfeld AM, Edwards PA, Rosenfeld JM, Alvarez JG, Noel JP, Nicolaou KC, Evans RM. A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol Cell. 2003;11(4):1079–1092. doi: 10.1016/s1097-2765(03)00104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gronemeyer H, Laudet V. Transcription factors 3: nuclear receptors. Protein Profile. 1995;2(11):1173–1308. [PubMed] [Google Scholar]
- 44.Rastinejad F, Perlmann T, Evans RM, Sigler PB. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995 May 18;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 45.Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature. 1998 September 10;395(6698) doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 46.Bourgeut W, Vivat V, Wurtz J-M, Chambon P, Gronemeyer H, Moras D. Crystal structure of a heterodimer complex of RAR and RXR ligand-binding domains. Molecular Cell. 2000;5:289–298. doi: 10.1016/s1097-2765(00)80424-4. [DOI] [PubMed] [Google Scholar]
- 47.Schwabe JWR, Chapman L, Finch JT, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- 48.Brzozowski AM, Pike ACW, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson J-Å, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997 October 16;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 49.Flaig R, Greschik H, Peluso-Iltis C, Moras D. Structural basis for the cell-specific activities of the NGFI-B and the Nurr1 ligand-binding domain. Journal of Biological Chemistry. 2005;280(19):19250–19258. doi: 10.1074/jbc.M413175200. [DOI] [PubMed] [Google Scholar]
- 50.Little TH, Zhang Y, Matulis CK, Weck J, Zhang Z, Ramachandran A, Mayo KE, Radhakrishnan I. Sequence-specific deoxyribonucleic acid (DNA) recognition by steroidogenic factor 1: a helix at the carboxy terminus of the DNA binding domain is necessary for complex stability. Molecular Endocrinology. 2006;20(4):831–843. doi: 10.1210/me.2005-0384. [DOI] [PubMed] [Google Scholar]
- 51.Li Y, Choi M, Cavey G, Daugherty J, Suino K, Kovach A, Bingham NC, Kliewer SA, Xu HE. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Molecular Cell. 2005;17:491–502. doi: 10.1016/j.molcel.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 52.Krämer A, Richards DR, Bowlby JO, Felciano RM. Functional modularity in a large-scale mammalian molecular interaction network. Proceedings of the 2005 IEEE Computational Systems Bioinformatics Conference, Stanford, 2005; Stanford: 2005. p. 161. [Google Scholar]
- 53.Ekins S, Kirillov E, Rakhmatulin EA, Nikolskaya T. A Novel Method for Visualizing Nuclear Hormone Receptor Networks Relevant to Drug Metabolism. Drug Metab Dispos. 2005;33:474–481. doi: 10.1124/dmd.104.002717. [DOI] [PubMed] [Google Scholar]
- 54.Apic G, Ignjatovic T, Boyer S, Russell RB. Illuminating drug discovery with biological pathways. FEBS Lett. 2005;579(8):1872–1877. doi: 10.1016/j.febslet.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 55.Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296(5569):910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- 56.Adachi R, Shulman AI, Yamamoto K, Shimomura I, Yamada S, Mangelsdorf DJ, Makishima M. Structural determinants for vitamin D receptor response to endocrine and xenobiotic signals. Mol Endocrinol. 2004;18(1):43–52. doi: 10.1210/me.2003-0244. [DOI] [PubMed] [Google Scholar]
- 57.Kalaany NY, Mangelsdorf DJ. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu Rev Physiol. 2006;68:159–191. doi: 10.1146/annurev.physiol.68.033104.152158. [DOI] [PubMed] [Google Scholar]
- 58.Reschly EJ, Ai N, Ekins S, Welsh WJ, Hagey LR, Hoffman AF, Krasowski MD. Evolution of the bile salt nuclear receptor FXR in vertebrates. J Lipid Res. 2008 doi: 10.1194/jlr.M800138-JLR200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maglich JM, Caravella JA, Lambert MH, Willson TM, Moore JT, Ramamurthy L. The first completed genome sequence from a teleost fish (Fugu rubripes) adds significant diversity to the nuclear receptor superfamily. Nucleic Acids Res. 2003;31(14):4051–4058. doi: 10.1093/nar/gkg444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reschly EJ, Ai N, Welsh WJ, Ekins S, Hagey LR, Krasowski MD. Ligand specificity and evolution of liver X receptors. J Steroid Biochem Mol Biol. 2008 doi: 10.1016/j.jsbmb.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Escriva H, Safi R, Hanni C, Langlois MC, Saumitou-Laprade P, Stehelin D, Capron A, Pierce R, Laudet V. Ligand binding was acquired during evolution of nuclear receptors. Proc Natl Acad Sci U S A. 1997;94(13):6803–6808. doi: 10.1073/pnas.94.13.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laudet V. Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor. J Mol Endocrinol. 1997;19(3):207–226. doi: 10.1677/jme.0.0190207. [DOI] [PubMed] [Google Scholar]
- 63.Wiens M, Batel R, Korzhev M, Muller WE. Retinoid X receptor and retinoic acid response in the marine sponge Suberites domuncula. J Exp Biol. 2003;206(Pt 18):3261–3271. doi: 10.1242/jeb.00541. [DOI] [PubMed] [Google Scholar]
- 64.Riddiford LM, Cherbas P, Truman JW. Ecdysone receptors and their biological actions. Vitam Horm. 2000;60:1–73. doi: 10.1016/s0083-6729(00)60016-x. [DOI] [PubMed] [Google Scholar]
- 65.Bonneton F, Zelus D, Iwema T, Robinson-Rechavi M, Laudet V. Rapid divergence of the ecdysone receptor in Diptera and Lepidoptera suggests coevolution between ECR and USP-RXR. Mol Biol Evol. 2003;20(4):541–553. doi: 10.1093/molbev/msg054. [DOI] [PubMed] [Google Scholar]
- 66.Iwema T, Billas IM, Beck Y, Bonneton F, Nierengarten H, Chaumot A, Richards G, Laudet V, Moras D. Structural and functional characterization of a novel type of ligand-independent RXR-USP receptor. Embo J. 2007;26(16):3770–3782. doi: 10.1038/sj.emboj.7601810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reschly EJ, Bainy AC, Mattos JJ, Hagey LR, Bahary N, Mada SR, Ou J, Venkataramanan R, Krasowski MD. Functional evolution of the vitamin D and pregnane X receptors. BMC Evol Biol. 2007;7(1):222. doi: 10.1186/1471-2148-7-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krasowski MD, Yasuda K, Hagey LR, Schuetz EG. Evolutionary selection across the nuclear hormone receptor superfamily with a focus on the NR1I subfamily (vitamin D, pregnane X, and constitutive androstane receptors) Nucl Recept. 2005;3:2. doi: 10.1186/1478-1336-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moore LB, Maglich JM, McKee DD, Wisely B, Willson TM, Kliewer SA, Lambert MH, Moore JT. Pregnane X receptor (PXR), constitutive androstane receptor (CAR), and benzoate X receptor (BXR) define three pharmacologically distinct classes of nuclear receptors. Mol Endocrinol. 2002;16(5):977–986. doi: 10.1210/mend.16.5.0828. [DOI] [PubMed] [Google Scholar]
- 70.Krasowski MD, Yasuda K, Hagey LR, Schuetz EG. Evolution of the pregnane X receptor: adaptation to cross-species differences in biliary bile salts. Mol Endocrinol. 2005;19(7):1720–1739. doi: 10.1210/me.2004-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ekins S, EJ R, Hagey LR, Krasowski MD. Evolution of pharmacologic specificity in the Pregane X Receptor. BMC Evol Biol. 2008;8:103. doi: 10.1186/1471-2148-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bridgham JT, Carroll SM, Thornton JW. Evolution of hormone-receptor complexity by molecular exploitation. Science. 2006;312(5770):97–101. doi: 10.1126/science.1123348. [DOI] [PubMed] [Google Scholar]
- 73.Ekins S. Computational Toxicology: risk assessment for pharmaceutical and environmental chemicals. Hoboken, NJ: John Wiley and Sons; 2007. [Google Scholar]
- 74.Ekins S, Swaan PW. Computational models for enzymes, transporters, channels and receptors relevant to ADME/TOX. Rev Comp Chem. 2004;20:333–415. [Google Scholar]
- 75.Ulrich RG. The toxicogenomics of nuclear receptor agonists. Curr Opin Chem Biol. 2003;7(4):505–510. doi: 10.1016/s1367-5931(03)00080-2. [DOI] [PubMed] [Google Scholar]
- 76.Ulrich RG, Rockett JC, Gibson GG, Pettit SD. Overview of an interlaboratory collaboration on evaluating the effects of model hepatotoxicants on hepatic gene expression. Environ Health Perspect. 2004;112(4):423–427. doi: 10.1289/ehp.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lavery DN, McEwan IJ. Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. Biochem J. 2005;391(Pt 3):449–464. doi: 10.1042/BJ20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lavery DN, McEwan IJ. Structural Characterization of the Native NH2-Terminal Transactivation Domain of the Human Androgen Receptor: A Collapsed Disordered Conformation Underlies Structural Plasticity and Protein-Induced Folding. Biochemistry. 2008;47(11):3360–3369. doi: 10.1021/bi702221e. [DOI] [PubMed] [Google Scholar]
- 79.Pogenberg V, Guichou JF, Vivat-Hannah V, Kammerer S, Perez E, Germain P, de Lera AR, Gronemeyer H, Royer CA, Bourguet W. Characterization of the interaction between retinoic acid receptor/retinoid X receptor (RAR/RXR) heterodimers and transcriptional coactivators through structural and fluorescence anisotropy studies. J Biol Chem. 2005;280(2):1625–1633. doi: 10.1074/jbc.M409302200. [DOI] [PubMed] [Google Scholar]
- 80.Holmbeck SM, Foster MP, Casimiro DR, Sem DS, Dyson HJ, Wright PE. High-resolution solution structure of the retinoid X receptor DNA-binding domain. J Mol Biol. 1998;281(2):271–284. doi: 10.1006/jmbi.1998.1908. [DOI] [PubMed] [Google Scholar]
- 81.Warnmark A, Wikstrom A, Wright AP, Gustafsson JA, Hard T. The N-terminal regions of estrogen receptor alpha and beta are unstructured in vitro and show different TBP binding properties. J Biol Chem. 2001;276(49):45939–45944. doi: 10.1074/jbc.M107875200. [DOI] [PubMed] [Google Scholar]
- 82.Lavery DN, McEwan IJ. The human androgen receptor AF1 transactivation domain: interactions with transcription factor IIF and molten-globule-like structural characteristics. Biochem Soc Trans. 2006;34(Pt 6):1054–1057. doi: 10.1042/BST0341054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following supplemental files are available free of charge via the Internet at http://pubs.acs.org.
Supplemental Table 1 – NR1A-1F intrinsic disorder data
Supplemental Table 2 – NR1H-1I intrinsic disorder data
Supplemental Table 3 – NR2 intrinsic disorder data
Supplemental Table 4 – NR3 intrinsic disorder data
Supplemental Table 5 – NR4, 5, 6 intrinsic disorder data
Supplemental Table 6 - Intrinsic disorder calculated for the NTD, DBD, D-domain, LBD and across the full sequence.
Supplemental Table 7 – Analysis of human NHR disorder compared with IPA upstream and down stream connections.
Supplemental Table 8 – Analysis of human NHR disorder by orphan and classic designation
Supplemental Table 9 - Analysis of published NHR structures with observed structural disorder