Abstract
TAL1 is a critical transcription factor required for hematopoiesis. However, perturbation of its activity often leads to T cell leukemia. Whether and how its transcriptional activities are regulated during hematopoiesis remains to be addressed. Here, we show that TAL1 is associated with histone demethylase complexes containing lysine-specific demethylase 1 (LSD1), RE1 silencing transcription factor corepressor (CoREST), histone deacetylase 1 (HDAC1), and histone deacetylase 2 in erythroleukemia and T cell leukemia cells. The enzymatic domain of LSD1 plays an important role in repressing the TAL1-directed transcription of GAL4 reporter linked to a thymidine kniase minimal promoter. Furthermore, we demonstrate that the TAL1-associated LSD1, HDAC1, and their enzymatic activities are coordinately down-regulated during the early phases of erythroid differentiation. Consistent with the rapid changes of TAL1–corepressor complex during differentiation, TAL1 recruits LSD1 to the silenced p4.2 promoter in undifferentiated, but not in differentiated, murine erythroleukemia (MEL) cells. Finally, shRNA-mediated knockdown of LSD1 in MEL cells resulted in derepression of the TAL1 target gene accompanied by increasing dimeH3K4 at the promoter region. Thus, our data revealed that histone lysine demethylase LSD1 may negatively regulate TAL1-mediated transcription and suggest that the dynamic regulation of TAL1-associated LSD1/HDAC1 complex may determine the onset of erythroid differentiation programs.
Keywords: H3K4 methylation, histone demethylation, transcriptional regulation, erythroid differentiation, stem cell leukemia (SCL)
The transcription factor TAL1 is a member of the basic helix–loop–helix (bHLH) family of transcription factors that is required for the development of all hematopoietic lineages (1, 2). Aberrant activation of this transcription factor is involved in up to 60% of T cell acute lymphoblastic leukemia (T-ALL) cases, indicating that misregulation of TAL1's activity may lead to the development of T cell leukemia (3–5). Its transcription activity is linked to either repression or activation of target genes during normal and malignant hematopoiesis (6). Importantly, TAL1 associates with many binding partners including E2A, GATA1, LMO2, LDB1, and ETO2, all of which are involved in modulating hematopoietic cell growth and differentiation (7–12). In particular, TAL1 differentially interacts with histone deacetylase 1/2 (HDAC1/2) and histone acetyltransferases p300 and p300/CBP-associated factor (PCAF), which can alter TAL1 function by activating or repressing the transcription of downstream target genes (13–15). Therefore, the data raise the possibility that TAL1 might regulate transcription by manipulating the histone code and local chromatin structure.
Methylation of Lys-4 of histone H3 tails (H3K4) correlates with transcriptionally-active euchromatin (16), and this methyl mark can be enzymatically removed by a newly-discovered histone demethylase (HDM), lysine-specific demethylase 1 (LSD1) (17). The core LSD1 complex consists of 4 subunits: LSD1, HDAC1/2, and RE1 silencing transcription factor corepressor (CoREST) (18, 19). CoREST and HDACs cooperatively stimulate LSD1 HDM activity by enhancing the binding of LSD1 to hypoacetylated nucleosomes (19–21). Interestingly, LSD1 can demethylate H3K4 methylation catalyzed by oncogenic mixed lineage leukemia; (MLL), whose gene is found to be frequently rearranged in leukemia. In this regard, LSD1 may be potentially involved in cancer development such as leukemogenesis. Furthermore, siRNA-mediated down-regulation of LSD1 in hematopoietic progenitors perturbs development of several hematopoietic lineages (22), suggesting that LSD1-mediated epigenetic modification may play an important role in hematopoietic development.
To further understand the mechanism by which TAL1-associated complexes influence hematopoietic cell growth and differentiation, and how they contribute to T cell leukemogenesis, we have purified TAL1-containing protein complexes from TAL1-expressing multipotent erythroleukemia K562 and T-ALL Jurkat cells. We found that TAL1 recruits LSD1 complexes and HDM activity to a TAL1 target promoter. It represses transcription of the target gene by altering the histone H3K4 methylation pattern on the promoter, which suggests that TAL1 might regulate transcription by manipulating the histone code. In support of this view, down-regulation of LSD1 specifically derepresses the TAL1 target gene by increasing H3K4 dimethylation and inhibits the erythroid differentiation program. The results suggest that the recruitment of epigenetic modifier LSD1 complexes controls TAL1 function in hematopoiesis.
Results
Purification of TAL1-Associated LSD1 Complex.
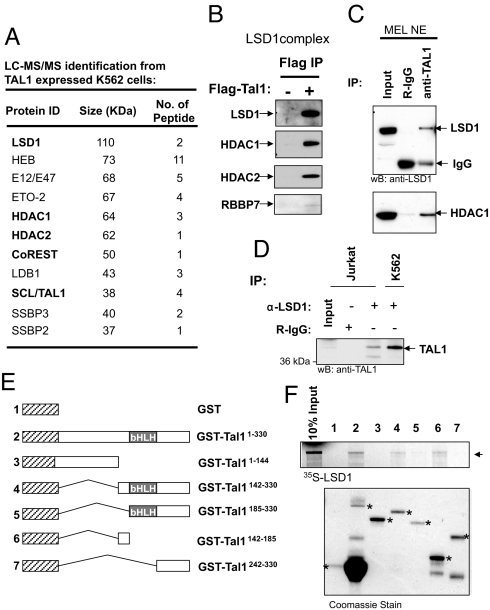
To understand the action of TAL1 in normal erythroid differentiation and leukemogenesis, we isolated TAL1-associated protein complexes through a Flag epitope-specific immunoaffinity column followed by anti-TAL1 immunopurification from Flag-TAL1-expressing K562 cells. The purified complexes were resolved by SDS/PAGE and identified by liquid chromatography tandem mass spectrometry (LC-MS/MS) (Fig. 1A). The majority of the polypeptides identified are involved in pathways that regulate chromatin structure and transcription. They fall into the following categories (Fig. 1A): (i) E12/E47 and HEB, known to heterodimerize with TAL1, (ii) LSD1 complex, possessing H3K4 HDM activity, (iii) transcription factors, some of them are known TAL1 binding partners, and (iv) ssDNA binding proteins, known as cofactors for LDB1 (23). We repeated the complex purification in Jurkat cells. Many of the TAL1-associated proteins identified match those purified from K562 cells (Fig. S1), suggesting that the complexes purified specifically interacted with TAL1. Among them, the most notable complex is the LSD1 complex that has not yet been reported as a TAL1-interacting partner (Fig. 1A). To validate TAL1 complexes containing both H3K4 demethylase LSD1 and HDAC1/2 (Fig. 1A and Fig. S1), we performed a WB using the Flag immunoaffinity purification material from the Jurkat nuclear extract. TAL1 specifically associates with the components of the core LSD1 complex, LSD1, HDAC1, HDAC2, and RBBP7 (Fig. 1B).
Fig. 1.
TAL1 associates with LSD1 complex in erythroid and T cell leukemia cells. (A) A partial list of TAL1-associated polypeptides identified by LC-MS/MS is indicated. (B) The Flag-purified complexes pulled down from Flag-Tal1 and mock-transduced Jurkat cells were analyzed by WB. (C) Nuclear extract from MEL cells was precipitated with TAL1 antibody and analyzed by WB using LSD1 and HDAC1 antibodies. (D) Nuclear extracts from Jurkat and K562 cells were precipitated with LSD1 antibody and analyzed by WB using TAL1 antibody. (E) Schematic representation of GST-TAL1 fusion proteins used in a GST pull-down assay. (F) 35S-labeled LSD1 was incubated with GST and GST-TAL1 fusion proteins preabsorbed to glutathione-Sepharose beads. (Upper) Bound LSD1 was visualized by fluorography after SDS/PAGE. (Lower) Coomassie-stained gel shows the relative protein loading.
To further address whether endogenous TAL1 interacts with the LSD1 complex, we performed immunoprecipitation assays in which murine erythroleukemia (MEL) nuclear extract was precipitated with TAL1 antibody and sequentially blotted with antibodies specific to LSD1 and HDAC1. Endogenous TAL1 interacts specifically with LSD1 and HDAC1 in MEL cells (Fig. 1C). In addition, the reverse immunoprecipitation reactions showed that the LSD1 antibody sufficiently pulled down TAL1 in both human erythroleukemia K562 cells and human T-ALL Jurkat cells (Fig. 1D). Thus, TAL1 associates with LSD1 complex in both T cell leukemia and erythroid progenitor cells.
TAL1 Physically Interacts with LSD1.
Next, we sought to determine whether TAL1 directly interacts with LSD1 and to characterize the domains involved in this interaction, TAL1 and its truncated mutants were expressed as GST fusion proteins (Fig. 1E), conjugated with glutathione-Sepharose beads, and incubated with [35S]methionine-labeled LSD1. Tal1 directly interacts with LSD1, and the interacting domain encompasses amino acids 142–185 proximal to the bHLH domain, which contains a Ser-172 site that becomes phosphorylated upon erythropoietin (EPO) induction (Fig. 1F, compare lanes 4 and 6 with 5) (24). Whether or not phosphorylation affects TAL1 and LSD1 interaction remains to be determined. Furthermore, the C-terminal amine oxidase domain of LSD1 required for HDM activity and transcription repression (17) is not essential for LSD1 to interact with TAL1 (Fig. S2A). Therefore, the in vitro pull-down assay confirmed that TAL1 directly interacts with LSD1.
TAL1 Recruits 2 Distinct LSD1 Complexes.
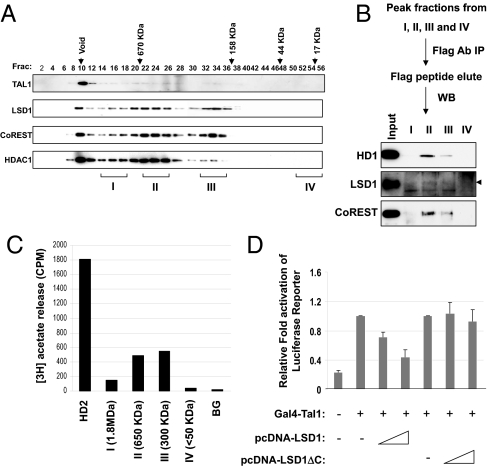
Given that the TAL1-associated LSD1 complex comprises multiple structural components for its enzymatic activities (19–21), we tested whether TAL1 recruits the entire LSD1 complex by using gel filtration analysis (Fig. 2A). TAL1, LSD1, CoREST, and HDAC1 comigrate in the ≈650- and ≈300-kDa fractions (Fig. 2A), suggesting that TAL1 and LSD1 form 2 complexes of 650 and 300 kDa. To confirm this finding, the peak fractions corresponding to the sizes of 1.8 MDa (I), 650 kDa (II), 300 kDa (III), and <50 kDa (IV) were collected separately, precipitated with anti-Flag conjugated agarose beads, and analyzed for the presence of LSD1 complex components, CoREST, LSD1, and HDAC1, by WB analysis. CoREST, LSD1, and HDAC1 are solely present in the 650 and 300-kDa fractions (Fig. 2B). These fractions also contain significant HDAC activity, whereas the 1.8-MDa (I) or 50-kDa (IV) size fractions have only minimum HDAC activity (Fig. 2C). Taken together, the data suggest that TAL1 forms at least 2 distinct LSD1 complexes.
Fig. 2.
TAL1 forms 2 distinct complexes with LSD1. (A) Nuclear extracts from Jurkat cells expressing Flag-Tal1 were fractionated through the Sephacryl S-300 HR column. The fractions were collected and analyzed by WB. (B) The peak fractions from 1.8 MDa (I), 650 kDa (II), 300 kDa (III), and <50 kDa (IV) were collected. The pooled fractions were precipitated with Flag antibody and analyzed by WB. (C) The peak fractions from 1.8 MDa (I), 650 kDa (II), 300 kDa (III), and <50 kDa (IV) were incubated with radiolabeled hyperacetylated histone. The amount of [3H]acetate released was quantitated by liquid scintillation counting. (D) Demethylase activity of LSD1 is required to repress TAL1-mediated transcription. HeLa cells were transfected with a Gal4-TK-luc reporter, an expression vector for GAL4-TAL1, and increased amounts of LSD1 or LSD1ΔC. A CMV-driven renilla luciferase plasmid was used as a transfection control. Transfected cells were cultured for 48 h and lysed for measurement of luciferase activity.
LSD1 Is Involved in TAL1-Mediated Transcriptional Repression.
LSD1 is a putative transcriptional repressor. We further tested whether LSD1 is required for TAL1-mediated transcriptional repression. A Tal1 cDNA fused to the DNA-binding domain of the yeast transcription factor GAL4 was cotransfected with the GAL4-TK-Luc reporter and increasing amounts of an LSD1-encoded plasmid into HeLa cells. The coexpression of LSD1 inhibited TAL1-mediated reporter activity in a dose-dependent fashion (Fig. 2D). In contrast, the deletion of the amine oxidase domain, which is essential for the HDM activity of LSD1, resulted in a complete loss of repression of TAL1-directed transcription (Fig. 2D), indicating that LSD1 represses TAL1-mediated transcriptional activity via its HDM activity. Given the role of TAL1 in hematopoiesis, the data also suggest that HDM activity of LSD1 may be required for TAL1-mediated transcriptional repression during hematopoiesis.
TAL1-Associated LSD1 and HDM Activity Are Dynamically Regulated During Hematopoiesis.
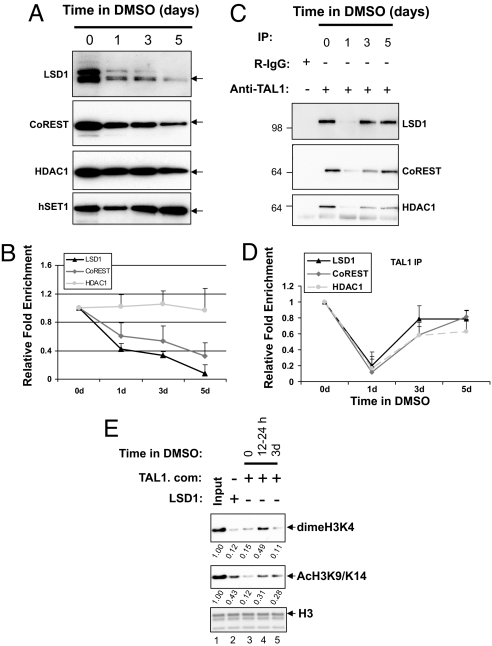
To access the role of LSD1 in hematopoiesis, we determined the expression levels of LSD1 core complex and hSET1 in DMSO-induced MEL cell differentiation. The levels of LSD1 and CoREST gradually decline during erythroid differentiation whereas HDAC1 remains relatively constant (Fig. 3 A and B). In contrast, hSET1 is significantly increased upon differentiation (Fig. 3A), suggesting that the LSD1 complex and hSET1 may differentially regulate hematopoietic cell development and that LSD1 plays a negative role in erythroid differentiation.
Fig. 3.
TAL1-associated LSD1 complex, HDM, and HDAC activities were subjected to regulation during erythroid differentiation. (A) Nuclear extracts from MEL cells treated with DMSO for the indicated periods of time were analyzed by WB. (B) The fold difference of the expressed proteins in MEL cells was determined from 4 independent experiments using image analysis of exposed X-ray films. (C) The same nuclear extracts were precipitated with TAL1 antibody and analyzed for the presence of LSD1, CoREST, and HDAC1 in the complex. (D) The fold difference of the immunoprecipitated proteins was determined from 3 independent experiments using image analysis of exposed X-ray films by ImageQuant 400. (E) The TAL1 complexes purified from MEL cells treated with DMSO for the indicated times were incubated with 1 μg of chicken core histone in the HDM buffer. The changes of diMeH3K4 and AcH3K9/K14 were analyzed by WB.
Given that the LSD1 complex interacts with and confers repressor activity to TAL1, and because its levels are greatly reduced during differentiation, we asked whether TAL1-associated LSD1 and its HDM activity are also changed throughout erythroid differentiation. To investigate this possibility, MEL nuclear extracts prepared 0, 1, 3, and 5 days after DMSO treatment were subjected to immunoprecipitation with TAL1 antibody. The associated proteins were identified by WB analysis. TAL1-associated LSD1, CoREST, and HDAC1 were decreased to a very low level at day 1 in the initial onset of differentiation and then came back at the later differentiation stage (Fig. 3 C and D). The significant decline of the TAL1–LSD1 complex coincides with the time period during which the MEL cells became committed to differentiation (25) and suggests that TAL1 was released from LSD1-containing repressor complexes in the early stages of erythroid differentiation.
Next, we investigated whether the HDM activity of the TAL1 complex correlated with the dynamic changes of association with LSD1. The TAL1-associated complexes purified from Flag-tagged TAL1-expressing MEL nuclear extracts treated with DMSO for 0, 1, and 3 days were incubated with chicken core histones as substrate, and HDM activity was analyzed by using antibodies specific to diMeH3K4 and AcH3K9/K14 (Fig. 3E). The TAL1 complex purified from MEL cells had a strong HDM activity toward diMeH3K4 compatible to the recombinant LSD1 protein (Fig. 3E Top, compare lanes 2 and 3). Upon DMSO-induced MEL differentiation, TAL1-containing complexes lost the H3K4 HDM activity in the 12- to 24-h period after the addition of DMSO (Fig. 3E Top, compare lane 3 and 4), which is correlated with a dramatic decrease in TAL1 and LSD1 complexes (Fig. 3D). However, in the late stage of differentiation, the TAL1-containing complexes regained H3K4 HDM activity (Fig. 3E Top, compare lanes 3–5). Parallel to the early decrease of HDM activity, TAL1-associated HDAC activity was also decreased upon DMSO-induced differentiation (Fig. 3E Middle, compare lanes 3–5). The HDAC activity did not recover by day 3 of differentiation (Fig. 3E Middle, compare lanes 4 and 5), even though the TAL1 and HDAC1 interaction had reappeared (Fig. 3D). Interestingly, although the LSD1 level gradually decreased during differentiation (Fig. 3B), the TAL1-containing LSD1 complex and HDM activity significantly increased by day 3 of differentiation compared with day 1 (Fig. 3 C and E), suggesting that the activity of TAL1 complexes may be differentially regulated during differentiation.
TAL1 Recruits LSD1 to Regulate Target Gene Expression During Erythroid Differentiation.
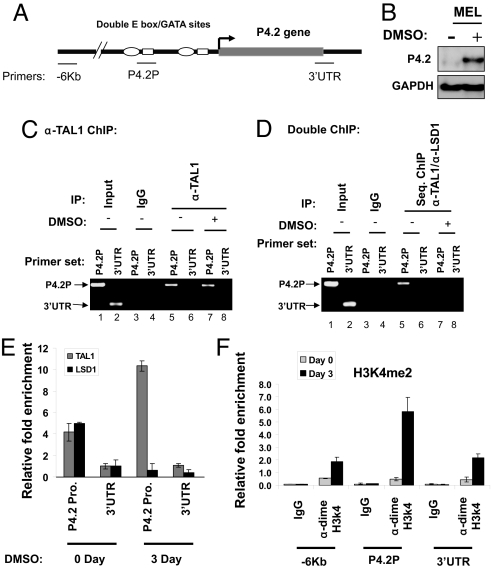
Erythroid protein EPB4.2 (P4.2) is an important component of the erythrocyte membrane and cytoskeleton structure that regulates stability and flexibility of erythrocytes (26). Within the P4.2 gene, 2 E box-GATA motifs are located at its proximal promoter (Fig. 4A). The binding of TAL1 to the promoter depends on the integrity of these motifs and is critical for P4.2 gene expression (12). Fig. 4B shows that the P4.2 gene is suppressed in undifferentiated MEL cells and induced upon DMSO treatment. We reasoned that TAL1 might target LSD1 to the P4.2 promoter to inhibit P4.2 expression in undifferentiated MEL cells and might dissociate from LSD1 after differentiation. To test this possibility, we carried out TAL1 ChIP and TAL1/LSD1 sequential ChIP to determine whether TAL1 and LSD1 colocalize at the P4.2 promoter before and after differentiation and whether colocalization results in the changes of promoter H3K4 methylation. As expected, TAL1 is specifically bound to the P4.2 promoter but not to the 3′ UTR (Fig. 4C). Although the binding of TAL1 alone at the P4.2 promoter did not change upon DMSO induction (Fig. 4C, compare lanes 5 and 7), TAL1 and LSD1 only cooccupied the P4.2 promoter when P4.2 was silenced and the P4.2 promoter-bound LSD1 was significantly decreased upon activation of P4.2 in differentiated cells (Fig. 4D, compare lanes 5 and 7). In addition, quantitative ChIP confirmed that the P4.2 promoter-bound LSD1 was markedly decreased upon differentiation, whereas TAL1 binding was increased (Fig. 4E). Furthermore, accompanying the loss of TAL1-associated LSD1 at the P4.2 promoter upon differentiation was a >11-fold increase in dimeH3K4 at the same region in differentiated MEL cells (Fig. 4F). Histone H3 and H4 acetylation were also up-regulated and depended on the TAL1 binding of the P4.2 promoter (Fig. S2B). The elevated dimeH3K4 and histone acetylation at the promoter correlated with the up-regulation of P4.2 transcription (Fig. 4B). The results suggest that the recruitment of LSD1 and its HDM activity by TAL1 may play a role in silencing the P4.2 gene in undifferentiated MEL cells, and decreased LSD1 occupancy at the promoter then allows TAL1 to activate P4.2 upon differentiation.
Fig. 4.
TAL1 recruits LSD1 to the p4.2 promoter in undifferentiated, but not differentiated, MEL cells. (A) Schematic representation of the mouse P4.2 gene locus. The primers are indicated. (B) mRNAs from WT or TAL1-transduced MEL cells treated with 1.5% DMSO were purified and analyzed by Northern blotting using the P4.2 probe. (C and D) Cross-linked chromatin from MEL cells in the presence and absence of 1.5% DMSO was precipitated with TAL1 antibody. The TAL1-chromatin complexes were reversed and the selected DNA was analyzed by PCR (C). (D) TAL1-DNA complexes from the first TAL1 ChIP were subjected to a second IP with anti-LSD1. The cross-links of these double IP protein/DNA complexes were reversed, and the DNA was analyzed by PCR. (E) Cross-linked chromatin from MEL cells in the presence and absence of 1.5% DMSO was precipitated with TAL1 and LSD1 antibodies. The selected chromatin complexes were reversed and analyzed by quantitative PCR. (F) The cross-linked chromatin from MEL cells treated with or without DMSO was precipitated with dimeH3K4 antibody. The precipitated DNA fragments were amplified by using primers specific for −6 Kb, the P4.2 promoter, and 3′ UTR regions by quantitative PCR.
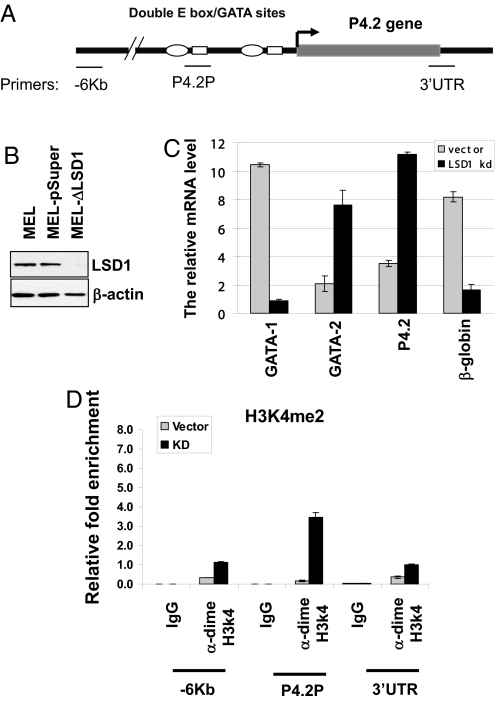
Knockdown (KD) of LSD1 in Erythroid Cells Leads to the Derepression of the TAL1 Target Genes.
If LSD1 is required for TAL1-mediated transcriptional repression, then the down-regulation of LSD1 by siRNA should derepress the TAL1 target gene. We directly addressed this notion by knocking down LSD1 expression using retroviral-mediated expression of LSD1 shRNA. The KD MEL cells displayed an ≈70% reduction in LSD1 protein levels compared with the parental and the vector control MEL cells (Fig. 5B).
Fig. 5.
The KD of LSD1 derepresses the expression of P4.2 gene. (A) Schematic representation of the mouse P4.2 gene locus. The primers are indicated. (B) WB analysis of whole-cell extracts from WT, pSuper vector control, and LSD1 KD MEL cells. (C) The RNA from the pSuper vector or LSD1 KD (shLSD1) MEL cells was isolated and quantitative RT-PCR was performed. (D) The cross-linked chromatin from the vector control and LSD1 KD MEL cells were precipitated with dimeH3K4 antibody. The precipitated DNA fragments were amplified by using primers specific for the −6 Kb, the p4.2 promoter, and 3′ UTR regions.
We then examined the effects of LSD1 KD on terminal erythroid differentiation by benzidine staining for the accumulation of hemoglobin. The benzidine-positive cells were significantly reduced in the LSD1 KD cells (Fig. S3A), reflecting the inhibition of erythroid differentiation. Importantly, the KD of LSD1 in MEL cells resulted in an increased expression of p4.2 and GATA-2, whereas the expression of β-globin and GATA-1 was significantly inhibited (Fig. 5C). To further confirm the role of LSD1 in hematopoiesis, we knocked down LSD1 in mouse ES cells that were differentiated into erythrocytes in the presence of EPO (Fig. S3B). The effects of LSD1 KD in ES cells resembled the MEL KD data (Fig. S3C). Interestingly, in both cells, GATA-2 is up-regulated, whereas GATA-1 is repressed, which suggests that LSD1 may regulate erythroid differentiation by controlling the interplay of GATA factors, although it is currently unclear whether LSD1 regulates GATA factors directly or indirectly. This should be the focus of future studies.
Next, we tested whether the derepression of the TAL1-targeted gene p4.2 upon the depletion of LSD1 was accompanied by an increase in dimeH3K4 at the p4.2 promoter (Fig. 5A). A ChIP assay was carried out by using antibody specific to dimeH3K4 in the vector control and LSD1 KD MEL cells. As we expected, the KD of LSD1 led to a 23-fold increase in dimeH3K4 level at the P4.2 proximal promoter (Fig. 5D). In contrast, the level of dimeH3K4 was moderately elevated at the −6 Kb (3.4-fold) and 3′ UTR (2.8-fold) regions upon LSD1 depletion (Fig. 5D). These data demonstrate that LSD1-mediated H3K4 demethylation plays a critical role in maintaining locally-repressive chromatin structure of the p4.2 gene in early hematopoietic progenitors, presumably targeted by TAL1. It is possible that the ectopic activation of this late terminal differentiation mark may perturb erythroid differentiation.
Discussion
Although several studies have revealed that the recruitment of coactivators and corepressors is the likely mechanism underlying TAL1-mediated transcriptional regulation (7, 9, 13–15), it remains unclear how TAL1 transcriptional activity is controlled during hematopoiesis. To exert transcriptional repression at certain stages of hematopoiesis, TAL1 can associate with several corepressors, including mSin3A, HDACs, and ETO-2 (7, 9, 14). LSD1 is a lysine-specific HDM that specifically removes monoH3K4 and dimeH3K4 (17). CoREST and HDAC1/2 can greatly potentiate LSD1 HDM activity (19–21). The recruitment of LSD1 by TAL1 suggests that the H3K4 HDM activity of LSD1 in part confers the repressive activity of TAL1 and may restrict TAL1 function in hematopoiesis (9, 14), therefore contributing to TAL1-induced leukemogenesis (27). This idea is supported by the observation that overexpression of LSD1 stimulates growth of erythroid progenitor cells (Fig. S4A).
LSD1 associates with CoREST, HDAC1/2, and other components that are required for its enzymatic activity and transcriptional repression (19–21). In T-ALL Jurkat cells, TAL1 forms 2 distinct complexes with LSD1 (Fig. 2 A and B). According to the predicted molecular mass, the 300-kDa complex apparently consists of the LSD1 core complex, whereas the 650-kDa complex might contain additional components. Although both complexes possess strong deacetylase and demethylase activities (Figs. 2C and 3E), the difference of these complexes in regulating TAL1 and LSD1 functions in normal or malignant hematopoiesis is currently unknown. Nevertheless, protein–protein interactions between LSD1 and its associated components control its enzymatic activity and substrate specificity. It is possible that the unknown components in the TAL1/LSD1 complex may be specifically required for controlling the action of TAL1 or LSD1 during hematopoiesis.
The deletion of LSD1 perturbs hematopoietic differentiation in both MEL and murine ES cells (Fig. 5C and Fig. S3), suggesting that the ubiquitously-expressed LSD1 controls tissue-specific differentiation processes, perhaps by interacting with tissue-specific transcription factors. Accordingly, LSD1 interacts with hematopoietic-specific repressor Gfi-1b to affect erythroid, megakaryocytic, and granulocytic differentiations (22). The data from Orkin's group (22) and our study suggest that LSD1 exerts its repressive effects with the guidance of hematopoietic-specific transcription factors, such as TAL1 and Gfi-1b, to control hematopoietic-restricted gene expression. Several lines of evidence suggest that TAL1 and Gfi-1b physically interact with each other and become part of the same transcriptional pathway. For example, the deletion of TAL1 and Gfi-1b in adult hematopoietic cells exhibits similar phenotypes (28–30). In addition, corepressor ETO-2 mediates the interaction between TAL1 and Gfi-1b in erythroid cells (9). Although ETO-2 is present in the TAL1/LSD1 complexes from our biochemical purifications from K562 and Jurkat cells (Fig. 1A and Fig. S1), it remains to be determined whether TAL1, Gfi-1b, ETO-2, and LSD1 are in the same complex.
It has been reported that TAL1 dynamically interacts with coactivators and corepressors during erythroid differentiation (9, 14, 15). Consistent with this view, we found that TAL1 associates with LSD1 and demethylase activity in undifferentiated MEL cells, but not in day 1 differentiated cells (Fig. 3 C and E), which coincides with the time period during which the MEL cells become committed to differentiation (25). However, association with the LSD1 complex was recovered during the late phases of differentiation (Fig. 3C). There are 2 possible scenarios for the dynamic changes of TAL1/LSD1 interaction during differentiation. First, in progenitor cells, LSD1 is recruited to restrict the ability of these cells responding to cellular differentiation program. After differentiation, LSD1 is then switched to repress TAL1 target genes that promote cellular proliferation. In supporting this view, LSD1 disappeared from the p4.2 promoter (Fig. 4 D and E) and the promoter became H3K4-hypermethylated and histone-hyperacetylated during differentiation (Fig. 4F and Fig. S2B). Another possibility is that the LSD1 protein complex changes its cofactor composition, which may alter its enzyme specificity. For example, when androgen receptor (AR) associates with LSD1, it changes the substrate specificity of LSD1 to demethylate repressive monoH3K9 or dimeH3K9 marks, which then promotes AR-dependent transcription (31). Therefore, LSD1 can repress or activate transcription by removing specific epigenetic makers and creating a condensed or relaxed chromatin environment, respectively. In supporting the possible dual functions of LSD1 during hematopoiesis, we found that TAL1 forms 2 LSD1 complexes in Jurkat cells (Fig. 2 A–C). Second, overexpression or down-regulation of LSD1 inhibits erythroid differentiation (Fig. S3 and Fig. S4), suggesting a dual role for LSD1 in erythroid differentiation.
What is the biological role of LSD1 in hematopoiesis? LSD1 regulates gene expression by modulating active or repressive chromatin domains (32, 33). It could potentially connect to human disease by perturbing normal gene expression patterns that are required for normal cell growth and differentiation. TAL1 is an oncoprotein whose activation is associated with a majority of T-ALL patients. The repressive function of TAL1 has been linked to leukemogenesis (9, 28). In T cell leukemia, TAL1 inhibits E2A/HEB transcriptional activity and perturbs cell cycle progression during the DN stage, where normal thymocytes undergo extensive cell division (6, 27). Thus, the possible function of corepressor LSD1 in TAL1-mediated transcriptional repression (Figs. 2D and 5 B–D) suggests that LSD1 may be involved in leukemogenesis by repressing E2A/HEB activity through its interaction with TAL1.
Materials and Methods
Cell Lines, Transfection, and shRNA KD.
K562, MEL, and Jurkat cells were maintained as described (14). Murine ES cells were cultured and induced to differentiate with EPO as described (34). Transfection, plasmid constructs, and shRNA-mediated KD are described in SI Text. The primer sequences are available in SI Text.
Purification of TAL1 Complexes.
TAL1 complexes were purified by a Flag immunoaffinity column from retroviral vector transduced K562 or Jurkat cells expressing a FLAG/HA double-tagged Tal1 by using a system developed by Nakatani and colleagues (35). The FLAG-purified materials were subsequently purified with TAL1-specific antibodies. The purified polypeptides were identified by LC-MS/MS at the Harvard Medical School Taplin Protein Sequence Facility, Boston.
Gel Filtration, GST Pull-Down, and HDM Assays.
Jurkat nuclear extract was fractionated on a Sephacryl S-300HR column (GE Healthcare). The eluted fractions were analyzed by WB and immunoprecipitation analysis. The GST pull-down (14) and histone demethylation (17) assays were carried out as described. Briefly, for the HDM assay, the HDM reaction mixture was analyzed by SDS/PAGE/WB by using dimeH3K4- or diacH3K9/K14-specific antibodies (Upstate).
RNA Isolation and Quantitative RT-PCR.
Total RNA isolated with a SV total RNA isolation kit (Promega) were reverse-transcribed by using oligo(dT) and SYBR Green quantitative PCR was performed as described (36).
ChIP and Double ChIP.
Real-time PCR quantitative ChIP assays were carried out as described (25) with slight modifications. Double ChIP was performed as described (37). LSD1 and CoREST antibodies were purchased from Upstate. TAL1 and HDAC1 were obtained from Santa Cruz Biotechnology.
Supplementary Material
Acknowledgments.
We thank Jörg Bungert (University of Florida, Gainesville), Yoshihiro Nakatani (Dana-Farber Cancer Institute, Boston), Yang Shi (Harvard Medical School, Boston), and Eric So (The Institute of Cancer Research, London) for reagents and Jörg Bungert and Lizi Wu for helpful suggestions and comments on the manuscript. This work was supported by National Institutes of Health Grants HL090589 and HL091929 (to S.H.) and grants from the Bankhead-Coley Cancer Research Program (to S.H. and Y.Q). C.N. is supported by the Intramural Research Program, National Institute of Diabetes, Digestive and Kidney Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900437106/DCSupplemental.
References
- 1.Porcher C, et al. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 2.Sánchez MJ, Bockamp EO, Miller J, Gambardella L, Green AR. Selective rescue of early haematopoietic progenitors in Scl−/− mice by expressing Scl under the control of a stem cell enhancer. Development. 2001;128:4815–4827. doi: 10.1242/dev.128.23.4815. [DOI] [PubMed] [Google Scholar]
- 3.Begley CG, et al. The gene SCL is expressed during early hematopoiesis and encodes a differentiation-related DNA-binding motif. Proc Natl Acad Sci USA. 1989;86:10128–10132. doi: 10.1073/pnas.86.24.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Q, et al. The tal1 gene undergoes chromosome translocation in T cell leukemia and potentially encodes a helix–loop–helix protein. EMBO J. 1990;9:415–424. doi: 10.1002/j.1460-2075.1990.tb08126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finger LR, et al. Involvement of the TCL5 gene on human chromosome 1 in T cell leukemia and melanoma. Proc Natl Acad Sci USA. 1989;86:5039–5043. doi: 10.1073/pnas.86.13.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palomero T, et al. Transcriptional regulatory networks downstream of TAL1/SCL in T cell acute lymphoblastic leukemia. Blood. 2006;108:986–992. doi: 10.1182/blood-2005-08-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goardon N, et al. ETO2 coordinates cellular proliferation and differentiation during erythropoiesis. EMBO J. 2006;25:357–366. doi: 10.1038/sj.emboj.7600934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ono Y, Fukuhara N, Yoshie O. Transcriptional activity of TAL1 in T cell acute lymphoblastic leukemia (T-ALL) requires RBTN1 or -2 and induces TALLA1, a highly specific tumor marker of T-ALL. J Biol Chem. 1997;272:4576–4581. doi: 10.1074/jbc.272.7.4576. [DOI] [PubMed] [Google Scholar]
- 9.Schuh AH, et al. ETO-2 associates with SCL in erythroid cells and megakaryocytes and provides repressor function in erythropoiesis. Mol Cell Biol. 2005;25:10235–10250. doi: 10.1128/MCB.25.23.10235-10250.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valge-Archer VE, et al. The LIM protein RBTN2 and the basic helix–loop–helix protein TAL1 are present in a complex in erythroid cell. Proc Natl Acad Sci USA. 1994;94:8617–8621. doi: 10.1073/pnas.91.18.8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wadman IA, et al. The LIM-only protein Lmo2 is abridging molecule assembling an erythroid DNA-binding complex which includes the TAL1, E47, GATA-1, and Ldb1/NL1 protein. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Z, Huang S, Chang L-S, Brandt SJ. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol Cell Biol. 2003;23:7585–7599. doi: 10.1128/MCB.23.21.7585-7599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S, Qiu Y, Stein R, Brandt SJ. p300 functions as a transcriptional coactivator for the Tal1/SCL oncoprotein. Oncogene. 1999;18:4958–4967. doi: 10.1038/sj.onc.1202889. [DOI] [PubMed] [Google Scholar]
- 14.Huang S, Brandt SJ. mSin3A regulates murine erythroleukemia cell differentiation through association with the TAL1 (or SCL) transcription factor. Mol Cell Biol. 2000a;20:2248–2259. doi: 10.1128/mcb.20.6.2248-2259.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang S, Qiu Y, Xu Z, Shi Y, Brandt SJ. P/CAF-mediated acetylation regulates the function of the basic helix–loop–helix transcription factor TAL1/SCL. EMBO J. 2000b;19:6792–6803. doi: 10.1093/emboj/19.24.6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stral B, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 17.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Humphrey GW, et al. Stable histone deacetylase complexes distinguished by the presence of SANT domain protein CoREST/kiaa0071 and Mta-L1. J Biol Chem. 2001;276:6817–6824. doi: 10.1074/jbc.M007372200. [DOI] [PubMed] [Google Scholar]
- 19.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for Corest in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 20.Shi Y, et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 21.Lee MG, et al. Functional interplay between histone demethylase and deacetylase enzymes. Mol Cell Biol. 2006;26:6395–6402. doi: 10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell. 2007;27:562–572. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- 23.Xu Z, et al. Single-stranded DNA-binding proteins regulate the abundance of LIM domain and LIM domain-binding proteins. Genes Dev. 2007;21:942–955. doi: 10.1101/gad.1528507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasad KSS, Jordan JE, Koury MJ, Bondurant MC, Brandt SJ. Erythropoietin stimulates transcription of the TAL1/SCL gene and phosphorylation of its protein products. J Biol Chem. 1995;270:11603–11611. doi: 10.1074/jbc.270.19.11603. [DOI] [PubMed] [Google Scholar]
- 25.Tsiftsoglou AS, Wong W. Molecular and cellular mechanisms of leukemic hemopoietic cell differentiation: An analysis of the Friend system. Anticancer Res. 1985;5:81–99. [PubMed] [Google Scholar]
- 26.Karacay B, Chang L-S. Induction of erythrocyte protein 4.2 gene expression during differentiation of murine erythroleukemia cells. Genomic. 1999;59:6–17. doi: 10.1006/geno.1999.5846. [DOI] [PubMed] [Google Scholar]
- 27.O'Neil J, Shank J, Cusson N, Murre C, Kelliher M. TAL1/SCL induces leukemia by inhibiting the transcriptional activity of E47/HEB. Cancer Cell. 2004;5:587–596. doi: 10.1016/j.ccr.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 28.Hall MA, et al. The critical regulator of embryonic hematopoiesis, SCL, is vital in the adult for megakaryopoiesis erythropoiesis, and lineage choice in CFU-S12. Proc Natl Acad Sci USA. 2003;100:992–997. doi: 10.1073/pnas.0237324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikkola HK, et al. Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukemia SCL/tal1 gene. Nature. 2003;421:547–551. doi: 10.1038/nature01345. [DOI] [PubMed] [Google Scholar]
- 30.Saleque S, Cameron S, Orkin SH. The zinc-finger protooncogene Gfi-1b is essential for development of the erythroid and megakaryotic lineages. Genes Dev. 2002;16:301–306. doi: 10.1101/gad.959102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metzger E, et al. LSD1 demethylates repressive histone marks to promote androgen receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 32.Lan F, et al. S. Pombe LSD1 homolog regulate heterochromatin progation and euchromatic gene transcription. Mol Cell. 2007;26:89–101. doi: 10.1016/j.molcel.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 33.Rudolph T, et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3–3. Mol Cell. 2007;26:103–115. doi: 10.1016/j.molcel.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 34.Kitajima K, Tanaka M, Zheng J, Sakai-Ogawa E, Nakano T. In vitro differentiation of mouse embryonic stem cells to hematopoietic cells on an OP9 stromal cell monolayer. Methods Enzymol. 2003;365:72–83. doi: 10.1016/s0076-6879(03)65005-6. [DOI] [PubMed] [Google Scholar]
- 35.Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 36.Huang S, Li X, Yusufzai TM, Qiu Y, Felsenfeld G. USF1 recruits histone modification complexes and is critical for maintenance of a chromatin barrier. Mol Cell Biol. 2007;27:7991–8002. doi: 10.1128/MCB.01326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crusselle-Davis VJ, Vieira KF, Zhou Z, Anantharaman A, Bungert J. Antagonistic regulation of β-globin gene expression by helix–loop–helix proteins USF and TFII-I. Mol Cell Biol. 2006;26:6832–6843. doi: 10.1128/MCB.01770-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.