Abstract
Mammalian signaling networks contain an abundance of negative feedback regulators that may have overlapping (“fail-safe”) or specific functions. Within the NF-κB signaling module, IκBα is known as a negative feedback regulator, but the newly characterized inhibitor IκBδ is also inducibly expressed in response to inflammatory stimuli. To examine IκBδ's roles in inflammatory signaling, we mathematically modeled the 4-IκB-containing NF-κB signaling module and developed a computational phenotyping methodology of general applicability. We found that IκBδ, like IκBα, can provide negative feedback, but each functions stimulus-specifically. Whereas IκBδ attenuates persistent, pathogen-triggered signals mediated by TLRs, the more prominent IκBα does not. Instead, IκBα, which functions more rapidly, is primarily involved in determining the temporal profile of NF-κB signaling in response to cytokines that serve intercellular communication. Indeed, when removing the inducing cytokine stimulus by compound deficiency of the tnf gene, we found that the lethality of iκbα−/− mouse was rescued. Finally, we found that IκBδ provides signaling memory owing to its long half-life; it integrates the inflammatory history of the cell to dampen NF-κB responsiveness during sequential stimulation events.
Keywords: inflammation, mathematical modeling, NF-kappaB, pathogen
Cellular responses to the environment are the result of complex signaling events involving multiple signaling protein isoforms encoded in gene families. The isoforms' distinct functions are generally thought to be due to differences in their intermolecular interaction characteristics. However, emerging evidence indicates that in signaling the precise dynamics are physiologically important and are mediated by an increasing number of feedback regulators. Their specific functions may in principle be determined by alternate kinetic control of synthesis and degradation.
NF-κB is an inducible transcription factor involved in diverse physiological scenarios. The primary NF-κB transcription factor of inflammatory responses is the ubiquitous RelA:p50 dimer, which forms via a dimerization domain within the NF-κB-distinctive Rel-homology domain (RHD). In unstimulated cells, it is held in an inactive state by proteins that have inhibitory, or IκB, activity. Three canonical IκB proteins, IκBα, -β and -ε have been described. Upon inflammatory stimulation via TNF receptor (TNFR) or TLR-mediated pathogenic signals, they are phosphorylated on specific N-terminal serine residues by IKK2 containing kinase complexes and subsequently degraded via the 26S proteasome pathway, which releases RelA:p50 DNA binding activity. Developmental signals such as B-cell activator factor (BAFF) and lymphotoxin-β (LTβ), members of TNF superfamliy, trigger the IKK1-dependent inactivation of a 4th IκB activity, termed IκBδ, allowing for activation of the RelA:p50 dimer (1). IκBδ consists of a homodimer of nfkb2/p100 proteins, in which at least 1 inhibitor domain is available for binding RelA:p50 (2). This homodimer is contained within a multimeric 500-kDa complex that may also contain nfkb1/p105 proteins (3).
Precise regulation of RelA:p50 is required to maintain healthy inflammatory signaling responses (4) and 2 of the 3 canonical IκB proteins have been found to provide negative feedback on to RelA:p50 (5–7). In response to transient TNF stimulation, for example, IκBα limits NF-κB responses but IκBε functions effectively when IκBα does not. This overlapping function represents a “fail-safe” mechanism.
With the identification of a 4th IκB, we wondered whether this IκBδ activity may also play a role in inflammatory signaling mediated by canonical IKK. If IκBδ has a function in regulating inflammatory responses, is it distinct or overlapping with the other IκB isoforms? As inflammatory signaling is highly dynamic, we constructed a mathematical model of the NF-κB signaling module that incorporates IκBδ. Using an integrated approach of computational and experimental analyses, we found that IκBδ is a negative feedback regulator of RelA:p50 in inflammatory signaling and, remarkably, that it attenuates RelA:p50 activity stimulus-specifically. Furthermore, we identified the kinetic rate constants that confer this stimulus specificity.
Results
IκBδ Feedback and Mathematical Modeling.
To investigate the potential roles of the IκBδ activity in inflammatory signaling, we first examined p100/IκBδ protein expression. Consistent with previous studies (8–10), p100/IκBδ protein was found to be up-regulated in response to all inflammatory stimuli tested (Fig. 1A, and Fig. S1A). We now determined that this gene activation is NF-κB-dependent, suggesting a potential negative feedback loop.
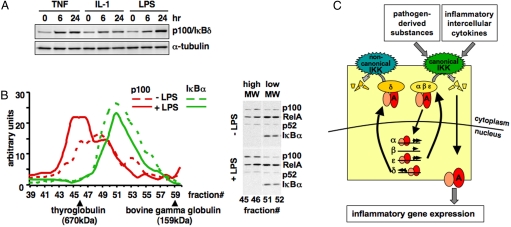
Fig. 1.
Characterization of IκBδ as a negative feedback regulator. (A) Western blots of whole cell extracts prepared from MEFs stimulated chronically with indicated inflammatory agents, TNF (1 ng/mL), IL-1 (1 ng/mL), LPS (100 ng/mL). Protein levels of p100/IκBδ increase whereas α-tubulin does not. (B) Chromatogram of gel-filtration studies used to separate LPS-stimulated or -unstimulated MEF cytosolic extracts. The graph shows the results from quantitated Western blots against p100 and IκBα. Thyroglobulin (670 kDa) and bovine gamma globulin (159 kDa) served as molecular size standards. (Right) A sample Western blot with indicated gel filtration fractions with antibodies against the indicated NF-κB/IκB proteins. (C) A diagram of the 4 IκB-containing NF-κB signaling module. Both pathogen-derived substances and inflammatory cytokines lead to canonical IKK-mediated NF-κB activation via the inducible degradation of 3 IκB proteins IκBα, IκBβ, and IκBε. Postinduction attenuation is mediated by an overlapping set of IκB proteins: IκBα, IκBε, and IκBδ.
There are 2 p100-containing complexes, the self-inhibited dimeric complex RelA:p100 and the high molecular weight ternary complex that binds RelA:p50 dimers and contains the IκBδ activity (2). To distinguish between the 2 p100-containing complexes, we performed gel filtration chromatography on cytosolic extracts. In resting cells, p100 was found to be present in both high (fractions 45 and 46) and low (fractions 51 and 52) molecular weight-containing fractions. In contrast, IκBα was found only in low molecular weight-containing fractions. Interestingly, LPS stimulation for 24 h led to increases of p100 and RelA in the fractions containing high molecular weight complexes, suggesting LPS-induced p100 protein primarily assembles into the ternary complex, termed IκBδ (Fig. 1B). Similar conclusions could be drawn from detergent sensitivity studies (Fig. S1 B–D), whose specificity was established in refs. 2 and 11.
To investigate the role of IκBδ in the highly dynamic signaling events triggered by inflammatory stimuli, we mathematically modeled the NF-κB signaling module (Fig. 1C) not only with the 3 canonical IκB proteins (12), but—with an expanded reaction network (Fig. S1E)—also the inducible IκBδ activity. To that end, we determined some of the relevant parameters that describe its metabolism by quantitatively measuring mRNA and protein expression. We noted that, whereas IκBα mRNA was induced highly and rapidly within 30 min, nfκb2 mRNA increased more slowly (Fig. S1F). By measuring mRNA levels during a time course with the mRNA synthesis inhibitor, actinomycin D (actD), we estimated that the half-life of the IκBα mRNA was ≈20 min, whereas the half-life of the nfκb2 mRNA was ≈6 h (Fig. S1G). The resulting mathematical model represents a tool to investigate the dynamic regulation of NF-κB by the 4 IκB proteins in response to inflammatory stimuli functioning through IKK2 (this study) or developmental stimuli functioning through IKK1 (2).
Computational Phenotyping of Negative Feedback Regulators.
Different stimuli activate canonical IKK activity with different temporal profiles (12). To explore NF-κB regulation in response to diverse canonical IKK dynamics, we generated a library of canonical IKK curves (Fig. 2A Top), using an algorithm that controls the amplitudes and durations of several activity phases (Fig. S2A). The resulting 1,044 canonical IKK curves were used as inputs for repeated simulations of NF-κB activation in wild-type, IκBδ- or IκBα-deficient models (Fig. 2A Lower). By calculating the difference in NF-κB activities generated by wild-type and mutant modules for a given IKK input profile, we assessed the functional importance of each IκB.
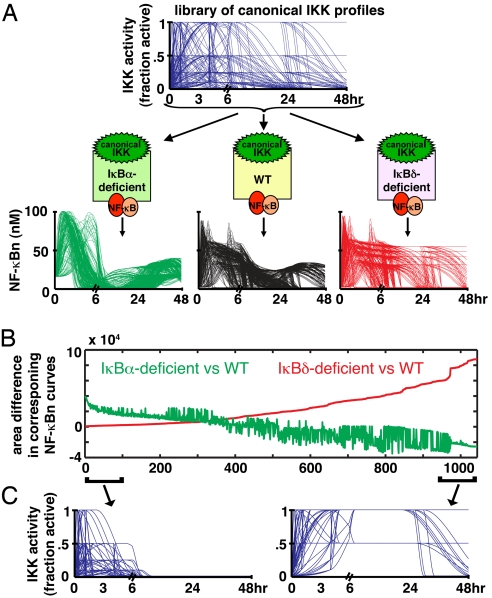
Fig. 2.
Phenotyping negative feedback regulators within signaling dynamics. (A) A library of 1,044 hypothetical canonical IKK input curves (Top) over a 48-h time course (see Fig. S2A for details) was generated and fed into wild-type, IκBα-, or IκBδ-deficient computational models (Middle) to calculate the corresponding 1044 NF-κB “outputs” for each model (Bottom). (B) A plot of the quantitated NF-κB phenotype for each canonical IKK curve in IκBα or IκBδ-deficient cells, as predicted by model simulations. Differences of NF-κB activity between wild-type and knockout cells (“the phenotype”) were calculated for each canonical IKK profile and plotted after sorting the canonical IKK profiles for increasing NF-κB phenotypes in IκBδ-deficient models. (C) Time course plots of the canonical IKK activity profiles that gave rise to the least or maximum IκBδ-phenotype, as defined by the bottom and top 10% of the curves found in B.
At first glance, the simulation data indicated that IKK curves for which IκBδ-mediated feedback is functionally important do not generally require regulation by IκBα (Fig. S2B). Sorting these simulation results in order of increasing sensitivity to IκBδ revealed that IκBδ- and IκBα-mediated feedback are important for attenuating NF-κB activity in response to different types of canonical IKK profiles (Fig. 2B). Plotting the 2 groups of IKK curves showed that IκBδ-deficient systems causes misregulation of NF-κB in response to prolonged canonical IKK stimuli, whereas IκBα-deficiency causes misregulation in response to transient stimuli (Fig. 2C). Thus, the computational analysis suggested that the relative importance of different negative feedback regulators is determined by the temporal profiles of IKK activity and, specifically, that IκBδ may be important in providing negative feedback on NF-κB in response to stimuli that elicit sustained canonical IKK activity.
IκBδ Attenuates NF-κB Activation Triggered by Pathogens.
TNF and lipopolysaccharide (LPS) have been shown to activate canonical IKK activity with different dynamics within a 2-h time course (12). In extended time course experiments, we found that TNF stimulation triggered a transient canonical IKK activity that decreased after 1 h and reached the basal level before 24 h (Fig. 3A and Fig. S3A). In contrast, LPS stimulation led to delayed canonical IKK activation that peaked at 1 h and was sustained at ≈40% of the peak activity even at the 48-h time point. Based on the computational phenotyping methodology, we predicted that IκBδ-deficient cells would be more defective in controlling the LPS- than TNF-induced NF-κB activity profiles.
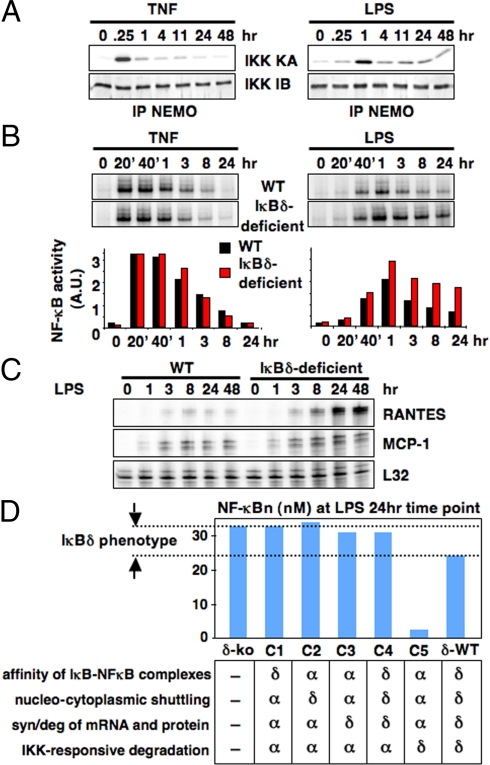
Fig. 3.
IκBδ mediates NF-κB attenuation stimulus-specifically. (A) IKK kinase activity assays (IKK KA) from MEFs chronically stimulated with TNF (1 ng/mL) or LPS (100 ng/mL). NEMO-associated kinase complex immunoprecipitated from extracts prepared at the indicated times, and used for a kinase assay with GST-IκBα (1–54) as a substrate. Equal loading was confirmed by immunoblotting against IKK1 (IKK IB). (B) NF-κB DNA binding activities induced by chronic TNF (1 ng/mL) or LPS (100 ng/mL) stimulation in wild-type and IκBδ-deficient MEFs were monitored by EMSA. In the case of TNF, the first phase (20′, 40′ and 1-h time points) and second phase (3-, 8-, and 24-h time points) are separated by a transient trough in activity, as shown in ref. 7. In the case of LPS, an elongated NF-κB activity spans the entire time course. Signals were quantitated and graphed relative to resting cells. (C) RPA to monitor the expression of NF-κB-responsive inflammatory genes chronically induced by LPS (100 ng/mL) stimulation in wild-type and IκBδ-deficient MEFs. (D) Computational analyses of in silico chimeras reveal the relative importance of kinetic rate constants for IκBδ-mediated attenuation of LPS signaling. Chimeric mutants were made by swapping rate constants of IκBα to those of IκBδ as indicated. NF-κB activities induced by 24-h LPS stimulation were plotted for each chimera.
To test the model predictions, wild-type and IκBδ-deficient cells were stimulated with TNF or LPS for 24 h. As predicted, TNF-induced NF-κB activation was similar in wild-type and IκBδ-deficient cells whereas in response to LPS, prolonged NF-κB activation was observed in IκBδ-deficient cells (Fig. 3B). Furthermore, the misregulation of the RelA:p50 dimer in LPS-stimulated IκBδ-deficient cells resulted in enhanced LPS-induced gene expression: induction of inflammatory gene RANTES was increased in IκBδ-deficient cells after LPS stimulation whereas MCP-1 gene expression was less affected (Fig. 3C). The misregulation of inflammatory gene expression and NF-κB activity in IκBδ-deficient cells were also seen when a lower dose of LPS (Fig. S3B) or the TLR3 ligand poly(I:C) (Fig. S4) was used. These results indicate that IκBδ dampens NF-κB activity stimulus-specifically such that a sustained canonical IKK activity is more susceptible to its feedback mechanism.
What is the molecular basis for IκBδ feedback's specificity for TLR-mediated signals? Using chimeric IκBα-IκBδ mutants in silico, we found that neither IκBδ's binding affinities for NF-κB dimers (chimera C1) nor its shuttling rate constants (chimera C2) were sufficient to dampen pathogen-induced NF-κB activity. However, mRNA and protein synthesis and degradation rate constants of IκBδ are critical in conjunction with its unresponsiveness to canonical IKK signals: When controlling the expression of an IκBα protein, which is responsive to canonical IKK degradative signals, chimeras with IκBδ synthesis and degradation rates provide some dampening (chimeras C3 and C4); in the context of a canonical IKK-unresponsive IκBδ-protein, IκBα synthesis and degradation rates are predicted to dramatically over-attenuate, not allowing for any late NF-κB activity (chimera C5). These analyses suggest that the combination of IκBδ's unresponsiveness to the degradative signaling of canonical IKK and relatively low synthesis and degradation rates, allows for a slow build-up of IκBδ inhibitory activity even in the context of long-lasting canonical IKK activity; these are the critical determinants of the stimulus-specific function of IκBδ-mediated feedback to TLR-mediated signaling.
IκBα Attenuates NF-κB Activation Triggered by Cytokines, but Not Pathogens.
Our combined computational and experimental studies suggest different physiological roles for IκBα and IκBδ (Fig. 4A). We propose that IκBα-mediated negative feedback regulates NF-κB primarily in response to inflammatory cytokines such as TNF and IL-1, which produce transient canonical IKK activities. In contrast, long-lasting canonical IKK activities that render IκBδ functionally important are a hallmark of pathogen signaling via TLRs (Fig. 3 and Figs. S3 and S4) (12, 13).
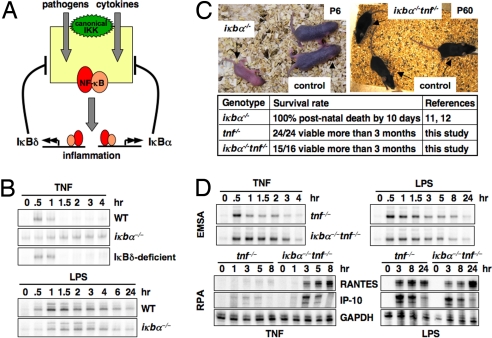
Fig. 4.
Distinct functions of IκBα and IκBδ in pathogenic and cytokine signaling. (A) IκBα and IκBδ have distinct functions in regulating NF-κB activation: We propose that IκBα is primarily involved in providing negative feedback in response to cytokine-induced signaling; IκBδ is important in mediating negative feedback upon stimulation by pathogens. (B) NF-κB DNA binding activities induced by 15-min transient TNF (1 ng/mL) or chronic LPS (100 ng/mL) stimulation in wild-type and mutant MEFs were monitored by EMSA. (C) Six-day-old iκbα−/− mice were moribund and died at 7 days. The 60-day-old iκbα−/−tnf−/− mice were indistinguishable from littermate control mice with no observable inflammation and organ abnormalities. (D) NF-κB DNA binding activities (Upper) and expression of NF-κB-responsive inflammatory genes (Lower) induced by 15-min transient TNF (1 ng/mL) or chronic LPS (100 ng/mL) stimulation in iκbα−/−tnf−/− and tnf−/− BMDMs were monitored by EMSA or RPA, respectively.
To test the IκBα side of this hypothesis, wild-type and mutant MEFs were stimulated with TNF or IL-1. We found that NF-κB activation was not affected by IκBδ deficiency, but was hyperinduced in IκBα-deficient cells, suggesting the essential role of IκBα, but not IκBδ, in terminating cytokine-induced responses. In contrast, IκBα seemed to play little role in attenuating LPS-induced NF-κB activity (Fig. 4B and Fig. S5A).
The essential function of IκBα in limiting inflammatory responses was demonstrated by the peri-natal lethality of IκBα-deficient mice due to multiorgan inflammation (14, 15). Indeed, we also found that IκBα deficiency resulted in premature death 7 days after birth. To test the in vivo specificity of the IκBα negative feedback loop for cytokine signaling, we generated IκBα knockout mice that were also deficient in the ubiquitous inducing cytokine stimulus TNF. Remarkably, the lethality of IκBα-deficient mice was rescued by the compound deficiency (Fig. 4C) with no evidence of inflammation or secondary lymphoid organ abnormalities in adult animals (Fig. 4C and data not shown). Furthermore, we examined macrophages derived from these mice. We found that BMDMs deficient in IκBα fail to appropriately terminate NF-κB activity in response to TNF stimulation but do not show hyperactive NF-κB in response to LPS stimulation (Fig. 4D Upper). Interestingly, misregulation of TNF-induced NF-κB activity also resulted in enhanced inflammatory gene expression: RANTES and IP-10 gene expression were hyperinduced in iκbα−/−tnf−/− BMDMs. In contrast, in response to LPS, IP-10 and RANTES gene expression were similar in the 2 genotypes, except that early time points were repressed in iκbα−/−tnf−/− BMDMs, consistent with a reduced LPS-induced NF-κB activation observed at early time points (Fig. 4D Lower and Fig. S5B). Overall, these genetic and molecular studies support the hypothesis that IκBα is primarily involved in regulating TNF-induced signaling.
IκBδ Controls the Fraction of Stimulus-Responsive NF-κB.
Unlike canonical IκB proteins (IκBα, -β, -ε), IκBδ is not subject to canonical IKK-triggered degradation, allowing it to accumulate during a prolonged time course. In conjunction with a long half-life, IκBδ may determine the fraction or balance of cellular NF-κB dimers that are responsive to inflammatory signals (Fig. 5A). Whereas in naïve fibroblasts, RelA-containing dimers are mostly bound to canonical IκBs and IκBδ only constitutes 5–10% of the total dimer pool (2), we hypothesized that IκBδ feedback control would shift the balance sufficiently toward a new steady state in which a smaller fraction of NF-κB is activatable by subsequent inflammatory signals.
Fig. 5.
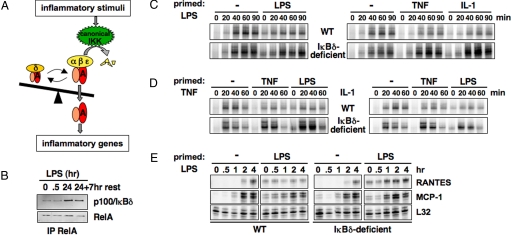
IκBδ integrates inflammatory history to limit NF-κB activation. (A) A schematic model to describe that the balance between IKK-responsive and -unresponsive RelA:p50 complexes determines the outcome of inflammatory exposure. (B) Western blots of immunoprecipitates to monitor p100/IκBδ associated with RelA during an LPS stimulation time course. RelA was immunoprecipitated from MEF whole cell extracts prepared at indicated time points. The last time point follows a 7-h rest period in endotoxin-free medium subsequent to PBS washes. (C) NF-κB activity induced by chronic LPS (100 ng/mL) in inflammation-primed wild-type and IκBδ-deficient MEFs. Cells were primed as in B. Nuclear extracts were prepared and NF-κB DNA binding activities were compared between naïve and primed cells by EMSA. (D) NF-κB activity induced by chronic TNF (1 ng/mL) and IL-1 (1 ng/mL) in inflammation-primed wild-type and IκBδ-deficient cells. Experiments were designed as in C. (E) NF-κB target gene expression monitored by RPA to assess the functional consequence of priming. Wild type and IκBδ-deficient MEFs were primed with LPS as in C, and LPS-induced (100 ng/mL) inflammatory gene expression was analyzed in naïve and primed cells at indicated time points.
To test this hypothesis, we first examined complexes of IκBδ and RelA proteins by coimmunoprecipitation. LPS increased the amount of IκBδ bound to RelA and this interaction was stable after the removal of stimulus (Fig. 5B). Furthermore, repeat stimulation experiments in wild-type and IκBδ-deficient cells with different combinations of stimuli revealed an IκBδ memory function. A priming phase of 24 h with LPS did affect the subsequent early phase of LPS-induced NF-κB activation (Fig. 5C). This attenuation effect was not stimulus-specific: Priming with cytokines also reduced subsequent LPS activation of NF-κB (Fig. 5D), and furthermore, priming with either stimulus affected subsequent TNF or IL-1 activation of NF-κB. In contrast, no decrease, but a slight increase in NF-κB activation was observed in primed IκBδ-deficient cells. That may be due to elevated expression of 1 or several NF-κB inducible intracellular signaling proteins (www.nf-kb.org), emphasizing the potential importance of long-lasting IκBδ to limit the amount of activatable NF-κB. In addition, we found that the attenuation effect on NF-κB increased with the duration of the priming phase, and decreased with longer rest period (Fig. S6), correlating with the slow IκBδ synthesis and degradation rate constants. Attenuation of inflammatory signaling by prior inflammatory exposure also had effects on NF-κB target gene expression: LPS priming in wild-type cells led to decreased induction of the chemokine RANTES gene expression, but not in IκBδ-deficient controls. In contrast, attenuation of MCP-1 gene expression in primed cells is IκBδ-independent (Fig. 5E).
Discussion
Inflammatory activation of NF-κB is highly dynamic and several negative feedback regulators have been identified. With IκBα being thought of as the primary negative feedback regulator, IκBε was found to provide for a “fail-safe” mechanism in resting or transiently stimulated cells (5, 6). The highly inducible de-ubiquitinase A20, which functions upstream of IKK, was found to function primarily as a rheostat that mediates signaling cross-talk rather than an obligate negative feedback regulator (16). In contrast, in the present study, IκBδ was revealed as a bona fide negative feedback regulator, albeit one that is stimulus-specific for pathogen-derived TLR-mediated signals. In fact, contrary to prior assumptions, IκBα's function was also shown to be restricted to a subset of inflammatory stimuli, specifically cytokines mediating intercellular communication. It has little role in the attenuation of TLR-mediated inflammatory signals. Indeed, the finding that the perinatal lethality of iκbα−/− mice was rescued by compound tnf deficiency supports this model. The previous mention that compound deficiency by tnfr1 does not rescue the lethality (15) was not documented, but, if corroborated, may indicate a role for tnfr2 in this process.
The molecular basis for functional specificity of regulatory proteins is often sought in their interaction specificity or affinities, described by equilibrium interactions parameters (Kd). Within the context of dynamic signaling events, functional specificity may, however, be conferred by kinetic rate constants (kass, kdiss, or ksyn, kdeg). Discovering the functional roles of kinetic control requires different tools than when equilibrium interaction characteristics are the focus. Mathematical modeling with differential equations is one prerequisite. In addition, we have developed a computational phenotyping tool to characterize the specific roles of IκBα and IκBδ within the context of dynamic signaling events, by exploiting the apparent modularity of the inflammatory signaling network (17). Focusing on the IκB-NF-κB signaling module that has a defined input (IKK activity) and output (nuclear NF-κB), we could examine signal processing in response to diverse physiological stimuli. By abstracting these into a library of hypothetical IKK profiles (Fig. 2), we explored the signal processing space comprehensively, unbiased by the limitations of currently known stimuli. Mapping our insights back to actual TLR and cytokine receptor-mediated signals, we were able to experimentally examine the computational predictions (Fig. 3). We suggest that the computational phenotyping methodology we present here is applicable to other signaling systems.
The contrasting functions of IκBα and IκBδ are rooted in differences in the kinetic rate constants governing their NF-κB-inducible synthesis, half-life, and canonical IKK-inducible degradation. Our combined computational modeling and experimental studies revealed that these characteristics are important in the context of stimulus-specific temporal control of canonical IKK activities: pathogen-derived substances are only slowly metabolized and activate canonical IKK for extended durations in part by triggering autocrine positive feedback loops (12), whereas cytokines have short half-lives and induce receptor down-regulation. Our studies suggest that even the relatively low induction of the nfkb2 gene leads to slowly accumulating IκBδ during inflammatory signaling because IκBδ is not degraded in response to canonical IKK signals. However, IκBα is much more highly inducible allowing it to turn off transient NF-κB activities when canonical IKK signaling has ceased. But, because IκBα's degradation is induced by canonical IKK, it is less efficient in attenuating NF-κB during long-lasting IKK signaling.
Although the nfkb2 gene is often associated with developmental NF-κB signaling, a role in controlling acute phase responses has also been noted (18) but has remained mechanistically unclear. With the realization that developmental signaling via the noncanonical pathway not only involves the generation of p52 containing dimers, but also the inactivation of a ternary complex containing a homodimeric p100, termed IκBδ (2), mechanistic studies of the nfkb2 gene products must distinguish between multiple molecular species. Here, we have used gel-filtration and detergent sensitivity analyses to study the high molecular weight IκBδ-containing complex.
Because of its relatively long half-life (≈8 h), IκBδ also functions constitutively to determine the cellular steady state, integrating the recent history of inflammatory exposure that may limit subsequent NF-κB activation (Fig. 5). As a mediator of inflammatory tolerance, it may regulate RelA activities during T cell activation (19, 20), osteoclastogenesis (21) and lymph node formation (22). Similarly, IκBδ is likely to play a role in providing a brake for cancer-associated chronically-elevated IKK2 signaling (23). Indeed, mutations that cause C-terminal truncations of p100 are found in some B and T cell malignancies (24), and signals impinging on IKK1 that are able to relieve such attenuation of RelA:p50 play a role in some Hodgkin lymphoma cells (25) and are found to be elevated via mutations found in multiple myeloma (26, 27). Moreover, LPS-induced IκBδ processing in macrophages or B-cells may similarly relieve its inhibitory function (10). However, when not subject to degradation signals, the ternary IκBδ complex may interconvert to self-inhibited dimeric RelA:p100 dimers limiting RelA availability for signaling. Modeling this process will require accounting for NF-κB monomer expression, dimerization and higher order complex formation to investigate signaling during longer term developmental and disease processes.
Materials and Methods
Animals, Cell Culture, and Reagents.
Wild type and gene-deficient C57BL/6 mice were maintained in accordance with the Animal Care Program at UCSD. Primary and 3T3-immortalized MEFs were generated from E12.5–14.5 embryos (12). Bone marrow-derived macrophages (BMDMs) were made from BM suspensions prepared from mouse femurs. A total of 1–2 × 106 BM cells were cultured for 1 week with L929-conditioned DMEM on 10-cm plates and MEFs were at 90% confluence at stimulation. We used recombinant murine 1 ng/mL TNFα (Roche), 1 ng/mL IL-1 (EMD Biosciences), 100 ng/mL LPS (Sigma, B5:055), 50 μg/mL poly(I:C) (Amersham Biosciences). Cycloheximide and actinomycinD were from Sigma. Antibodies against RelA/p65 (sc-372), RelB (sc-226), IκBα (sc-371) were from Santa Cruz Biotechnology. p100 (1495) antibody was from National Cancer Institute, Biological Resources Branch, Frederick, MD.
Experimental Analyses.
Whole cell extracts were prepared in RIPA buffer and normalized for total protein before immunoblot analysis. Nuclear extracts from BMDM were prepared by high salt extraction buffer. Immunoprecipitation-Western analysis, kinase assay, electrophoretic mobility shift assay (EMSA), deoxycholate (DOC) assays, and RNase Protection Assays (RPA) were performed as described in refs. 2, 6, and 12. A key advance in the current study is that we were able to focus on RelA:p50 activity by using biochemical and genetic strategies to remove deregulated RelB activity documented in ref. 28. In EMSAs comparing wild-type and nfkb2−/− MEF (Fig. 3B and Fig. S4C), nuclear extracts were ablated of RelB-containing DNA binding activity by preincubating them with RelB antibody whose specificity was confirmed (Fig. S7A). In gene expression studies (Figs. 3C and 5E and Fig. S3B), IκBδ-deficient cells were nfκb2−/−relb−/− cells ensuring that the hyperactivated gene expression was due to RelA rather than deregulated RelB (Fig. S7B). For gel-filtration studies, cytoplasmic extracts prepared from LPS-treated (24 h) or untreated MEF were injected into Superose6 10/300GL column (Amersham Pharmacia, column volume 24 mL) developed with 140 mM NaCl; 25 mM Tris·HCl, pH7.5; 1mMDTT at flow rate 0.5 mL/min, using AKTA purifier (GE Healthcare). Fractions were collected and analyzed by Western blotting with a mixture of the indicated antibodies and developed with ECL reagent (Perkin-Elmer). Films were scanned at 300 dpi resolution and analyzed with National Institutes of Health ImageJ software. Background-subtracted, normalized band Intensities were plotted against fraction numbers with Microsoft Excel. All results shown are representative of at least 3 independent experiments.
Computational Modeling.
The model was based on the 3 IκB-containing NF-κB signaling module (12) to which reactions pertaining to IκBδ were added (Fig. S1E). Based on the work described here, a version of this model was used to explore the role of IκBδ in developmental signaling (2). The model contains 32 species (Table S1) and 100 reactions described as ordinary differential equations with parameter values based on experimental measurements (Table S2). The model uses numerical input curves for IKK2 activation to compute the resulting NF-κB activity (defined as free nuclear NF-κB) over time, using MATLAB version R2008a (The MathWorks, Inc.) with subroutine ode15s, a variable order, multistep solver. The newly developed Computational Phenotyping Tool (Fig. 2) consists of 2 parts: (i) a set of inputs to probe signal processing characteristics of the signaling module and (ii) a metric to quantitate the effects of negative feedback knockouts. A library of theoretical canonical IKK input curves was generated using a simple algorithm (Fig. S2A) and PCHIP. The phenotyping metric applied here was the cumulative difference at every time point between NF-κB activity produced in knockout and wild-type cells for a specific IKK curve (Fig. S2B). The in silico chimera analysis was performed by starting with an IκBδ-deficient model into which the duplicated IκBα negative feedback was introduced (Fig. 3D). Then, the indicated kinetic rate constants of IκBα were swapped to those of IκBδ, as indicated, and 24 h of LPS stimulation was simulated.
Supplementary Material
Acknowledgments.
We thank C. Lynch, H. Kim, S. Werner, and M. Asagiri for technical advice, and Santa Cruz Biotechnology for antibodies. This study was supported by National Institutes of Health Grants GM071573 and GM071862 (to A.H.) and AI064326 (to G.G.) and an American Heart Association Predoctoral Fellowship (to J.D.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.T. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812367106/DCSupplemental.
References
- 1.Hoffmann A, Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 2.Basak S, et al. A fourth IkappaB protein within the NF-kappaB signaling module. Cell. 2007;128:369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savinova OV, Hoffmann A, Ghosh G. Mol Cell. 2009. The Nfkb1 and Nfkb2 proteins p105 and p100 function as the core of heterogeneous NF-kBsomes. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 5.O'Dea EL, et al. A homeostatic model of IkappaB metabolism to control constitutive NF-kappaB activity. Mol Syst Biol. 2007;3:111. doi: 10.1038/msb4100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kearns JD, Basak S, Werner SL, Huang CS, Hoffmann A. IkappaBepsilon provides negative feedback to control NF-kappaB oscillations, signaling dynamics, and inflammatory gene expression. J Cell Biol. 2006;173:659–664. doi: 10.1083/jcb.200510155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: Temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 8.Liptay S, Schmid RM, Nabel EG, Nabel GJ. Transcriptional regulation of NF-kappa B2: Evidence for kappa B-mediated positive and negative autoregulation. Mol Cell Biol. 1994;14:7695–7703. doi: 10.1128/mcb.14.12.7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lombardi L, et al. Structural and functional characterization of the promoter regions of the NFKB2 gene. Nucleic Acids Res. 1995;23:2328–2336. doi: 10.1093/nar/23.12.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mordmuller B, Krappmann D, Esen M, Wegener E, Scheidereit C. Lymphotoxin and lipopolysaccharide induce NF-kappaB-p52 generation by a co-translational mechanism. EMBO Rep. 2003;4:82–87. doi: 10.1038/sj.embor.embor710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baeuerle PA, Baltimore D. I kappa B: A specific inhibitor of the NF-kappa B transcription factor. Science. 1988;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- 12.Werner SL, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005;309:1857–1861. doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
- 13.Covert MW, Leung TH, Gaston JE, Baltimore D. Achieving stability of lipopolysaccharide-induced NF-kappaB activation. Science. 2005;309:1854–1857. doi: 10.1126/science.1112304. [DOI] [PubMed] [Google Scholar]
- 14.Beg AA, Sha WC, Bronson RT, Baltimore D. Constitutive NF-kappa B activation, enhanced granulopoiesis, and neonatal lethality in I kappa B alpha-deficient mice. Genes Dev. 1995;9:2736–2746. doi: 10.1101/gad.9.22.2736. [DOI] [PubMed] [Google Scholar]
- 15.Klement JF, et al. IkappaBalpha deficiency results in a sustained NF-kappaB response and severe widespread dermatitis in mice. Mol Cell Biol. 1996;16:2341–2349. doi: 10.1128/mcb.16.5.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Werner SL, et al. Encoding NF-kappaB temporal control in response to TNF: Distinct roles for the negative regulators IkappaBalpha and A20. Genes Dev. 2008;22:2093–2101. doi: 10.1101/gad.1680708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 18.Mercurio F, DiDonato JA, Rosette C, Karin M. p105 and p98 precursor proteins play an active role in NF-kappa B-mediated signal transduction. Genes Dev. 1993;7:705–718. doi: 10.1101/gad.7.4.705. [DOI] [PubMed] [Google Scholar]
- 19.Ishimaru N, Kishimoto H, Hayashi Y, Sprent J. Regulation of naive T cell function by the NF-kappaB2 pathway. Nat Immunol. 2006;7:763–772. doi: 10.1038/ni1351. [DOI] [PubMed] [Google Scholar]
- 20.Legarda-Addison D, Ting AT. Negative regulation of TCR signaling by NF-kappaB2/p100. J Immunol. 2007;178:7767–7778. doi: 10.4049/jimmunol.178.12.7767. [DOI] [PubMed] [Google Scholar]
- 21.Novack DV, et al. The IkappaB function of NF-kappaB2 p100 controls stimulated osteoclastogenesis. J Exp Med. 2003;198:771–781. doi: 10.1084/jem.20030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tucker E, et al. A novel mutation in the Nfkb2 gene generates an NF-κB2 “super repressor.”. J Immunol. 2007;179:7514–7522. doi: 10.4049/jimmunol.179.11.7514. [DOI] [PubMed] [Google Scholar]
- 23.Basak S, Hoffmann A. Crosstalk via the NF-kappaB signaling system. Cytokine Growth Factor Rev. 2008;19(3–4):187–197. doi: 10.1016/j.cytogfr.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Courtois G, Gilmore TD. Mutations in the NF-kappaB signaling pathway: Implications for human disease. Oncogene. 2006;25:6831–6843. doi: 10.1038/sj.onc.1209939. [DOI] [PubMed] [Google Scholar]
- 25.Chiu A, et al. Hodgkin lymphoma cells express TACI and BCMA receptors and generate survival and proliferation signals in response to BAFF and APRIL. Blood. 2007;109:729–739. doi: 10.1182/blood-2006-04-015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keats JJ, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Annunziata CM, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basak S, Shih VF, Hoffmann A. Generation and activation of multiple dimeric transcription factors within the NF-kappaB signaling system. Mol Cell Biol. 2008;28:3139–3150. doi: 10.1128/MCB.01469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.