Abstract
The human genome is a highly dynamic structure that shows a wide range of genetic polymorphic variation. Unlike other types of structural variation, little is known about inversion variants within normal individuals because such events are typically balanced and are difficult to detect and analyze by standard molecular approaches. Using sequence-based, cytogenetic and genotyping approaches, we characterized six large inversion polymorphisms that map to regions associated with genomic disorders with complex segmental duplications mapping at the breakpoints. We developed a metaphase FISH-based assay to genotype inversions and analyzed the chromosomes of 27 individuals from three HapMap populations. In this subset, we find that these inversions are less frequent or absent in Asians when compared with European and Yoruban populations. Analyzing multiple individuals from outgroup species of great apes, we show that most of these large inversion polymorphisms are specific to the human lineage with two exceptions, 17q21.31 and 8p23 inversions, which are found to be similarly polymorphic in other great ape species and where the inverted allele represents the ancestral state. Investigating linkage disequilibrium relationships with genotyped SNPs, we provide evidence that most of these inversions appear to have arisen on at least two different haplotype backgrounds. In these cases, discovery and genotyping methods based on SNPs may be confounded and molecular cytogenetics remains the only method to genotype these inversions.
INTRODUCTION
Knowledge about structural variation in the human genome has grown rapidly over the last few years. The advent of genome-scanning technologies based on comparative genomic hybridization has enabled the discovery of thousands of copy-number polymorphisms in ‘normal’ human individuals (1–6). In contrast to copy-number variation, in the absence of a similar high-throughput discovery method for inversions, fewer inversion polymorphisms have been detected and characterized. Most known examples have come indirectly from studies of human diseases where inversion polymorphisms have been identified because of their association with susceptibilities to recurrent genomic rearrangements (7–19).
A sequence-based methodology focused on the mapping of paired-end sequences has been developed to systematically detect such balanced events (20–22). This clone-based approach allows the detection and characterization of all types of variants (>5 kbp in length), including balanced chromosomal rearrangements such as inversions. Using this fosmid, clone-based analysis of one and eight individuals respectively, Tuzun et al. (20) and Kidd et al. (21) identified globally 1695 regions of structural variation, including 217 inversions validated by fingerprint, sequence analysis or fluorescence in situ hybridization (FISH). Of these inversions, 67% show evidence of large blocks of sequence homology at the breakpoints, with the remainder mediated by shorter common repeat sequences, making them difficult to characterize by standard molecular approaches (21).
Although useful for the discovery and refinement of variants, this method does not easily scale to the number of samples needed to characterize common inversions in larger groups of individuals. Additionally, the presence of duplicated and repeated sequences near the inversion breakpoints complicates other molecular genotyping approaches. Therefore, we developed a metaphase FISH-based assay to genotype inversions with complex repeats at the inversion breakpoints. Using this approach, we characterized six of the larger inversion polymorphisms in three HapMap populations [Yoruba Nigerian, n = 11; CEPH European, n = 8; Japanese/Han Chinese (Asian), n = 8]. This includes three previously described events—a 970 kbp inversion on 17q21.31, a 4.7 Mbp inversion on 8p23 and a 2 Mbp inversion on 15q13.3—and three novel fosmid detected inversions: a 1.2 Mbp inversion on 15q24, a 1.5 Mbp inversion on 17q12 and a 1.9 Mbp inversion on 3q29 (Table 1). For clarity, we use throughout this manuscript the convention developed by Stefansson et al. (13) and refer to the two structural configurations relative to the order represented in the human genome reference assembly. Thus, H2 indicates a configuration opposite from that in the assembly, and H1 indicates the same configuration as the assembly (build35). We present insight into the worldwide distribution, evolution and recurrence of these seemingly intractable human polymorphisms.
Table 1.
Human disease-associated inversion polymorphisms characterized in the present study
| Cytogenetic location | Inversion size | Mapping (UCSC May 2004) | Disease-causing rearrangement at location |
|---|---|---|---|
| 3q29 | 1.9 Mb | chr3:196886879–198874600 | 3q29 microdeletion syndrome |
| 8p23 | 4.7 Mb | chr8:7225962–12487029 | inv dup(8p), +der(8)(pterp23.1::p23.2-pter) and del(8)(p23.1;p23.2) |
| 15q13.3 | 2 Mb | chr15:28524207–30602466 | 15q13.3 microdeletion (mental retardation, epilepsy, schizophrenia and autism) |
| 15q24 | 1.2 Mb | chr15:72151413–73356183 | 15q24 microdeletion syndrome |
| 17q12 | 1.5 Mb | chr17:31888441–33393152 | RCAD syndrome |
| 17q21.31 | 900 kb | chr17:40899921–41989253 | 17q21.31 microdeletion syndrome |
RESULTS
Sequence analysis of inversion breakpoints
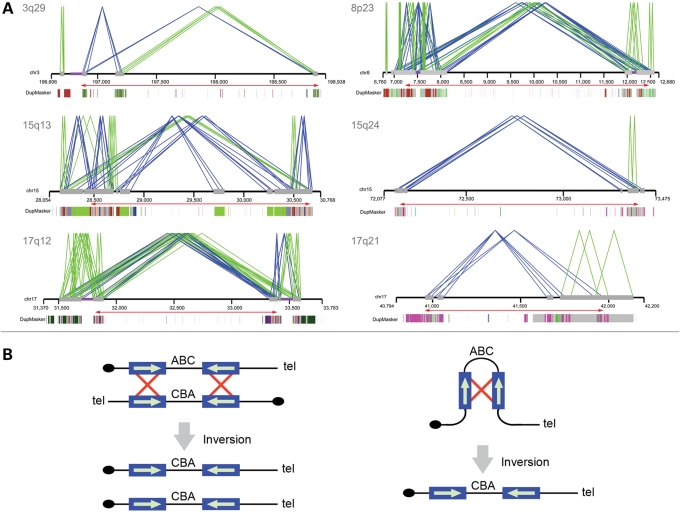
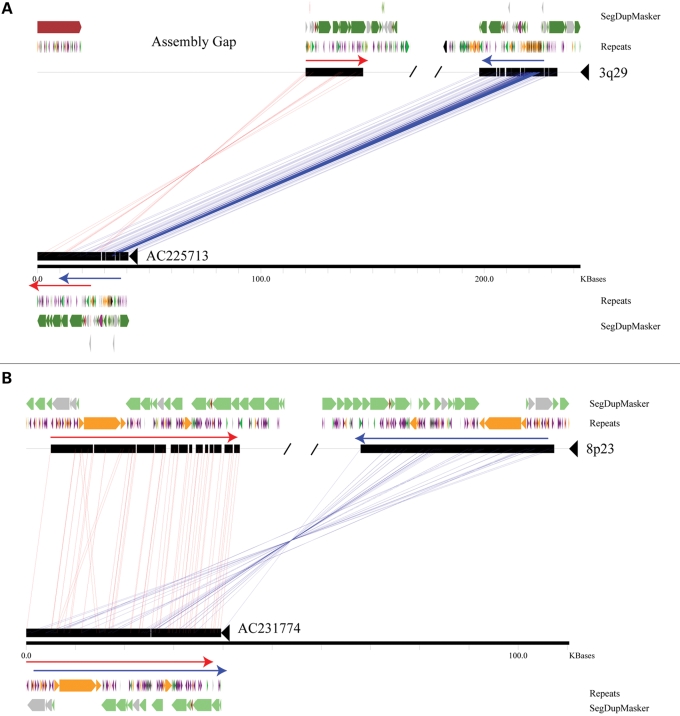
Segmental duplications have been shown to be highly overrepresented near sites of structural variation in the human genome (1,3,20,21). We analyzed the duplication architecture near the breakpoints of each inversion by analyzing the human genome assembly and clones that captured the breakpoints of the inverted allele in HapMap samples. We found that all of the inversions have pairs of highly homologous segmental duplications near both the breakpoints (Fig. 1A). In each of the six cases, the relevant pair of segmental duplications flanking the region of inversion is oriented in an inverted configuration consistent with non-allelic homologous recombination as the predominant mechanism for their origin (Supplementary Material, Table S1; Fig. 1B). Previous analyses utilizing sequenced BAC clones (23) of the 17q21.31 inversion localized the inversion breakpoints to homologous copies of the LRRC37 core duplicon (24). Analysis of an individual fosmid clone (AC225713) from Yoruba sample GM19129 indicates that the 3q29 inversion breakpoints are localized within 15.5 kbp stretches of homologous sequence (chr3:196868578–196884133 and chr3:198832975–198848521, hg17, 98.5% identity) (Fig. 2A). A more precise definition is not possible since the proximal breakpoint co-localizes with a gap in the human genome reference assembly, an artifact that may be a consequence of the structural polymorphism at this locus (25). Analysis of a clone (AC231774) from CHB sample GM18555 indicates that the 8p23 inversion breakpoints occur within 133 kbp segments having a 97.8% identity (chr8:8001218–8134378 and chr8:12321711–12461735 on hg17) (Fig. 2B). The sequenced fosmid is completely duplicated, but subtle, single nucleotide differences between the proximal and distal duplications are consistent with the clone actually spanning the inversion breakpoint. Sequenced clones are not available for the 15q13, 15q24 and 17q12 inversions, but each event is supported by multiple fosmid clones that span at least one of the inversion breakpoints and have end sequences mapping within duplicated sequences. More complete investigation of the breakpoints of these structural rearrangements will require the use of larger insert clones (such as BACs) and reliable assemblies derived from a single haplotype.
Figure 1.
Duplication architecture of inversion breakpoint regions. (A) Paralogy between large (≥10 kb), highly identical (≥95%) segmental duplications (gray bars) is shown. Direct (green lines) or inverted (blue lines) orientations of pairwise alignments of segmental duplications are indicated. A relaxed threshold (size ≥5 kbp and sequence identity ≥90%) was applied for the 15q24 inversion. The underlying ancestral duplication (duplicon) (24) composition of each duplication as determined by DupMasker (45) is represented as color-coded blocks. Red arrows show the extent of the inverted region. (B) Interchromosomal, intrachromosomal or intrachromatid non-allelic homologous recombination (NAHR) between inverted repeats causes inversion of the intervening sequence. Repeat sequences are indicated as blue boxes, with their orientation indicated by green arrows and recombination is shown by red crosses. Adapted in part from Sharp et al. (48).
Figure 2.
Sequence resolution of inversion breakpoints. A miropeats comparison (49) between sequenced fosmids corresponding to the 3q29 (A) and 8p23 (B) inversions and the build35 reference are shown. For simplicity, only the regions around the breakpoints are depicted. Both inversion breakpoints map within highly identical stretches of duplicated sequence found in an inverted configuration (red and blue arrows), with the 3q29 inversion coincident with a gap in the reference assembly. Segmental duplications were predicted using DupMasker (45) and colored based upon the position of each ancestral duplicon. The locations of common repeats are also indicated: green: LINEs; purple: SINEs; orange: LTR elements; pink: DNA; gray/black: other/low complexity regions.
Inversion analysis by fluorescence in situ hybridization
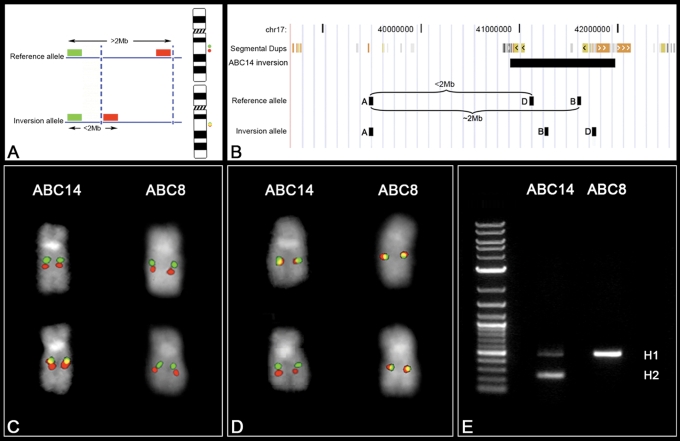
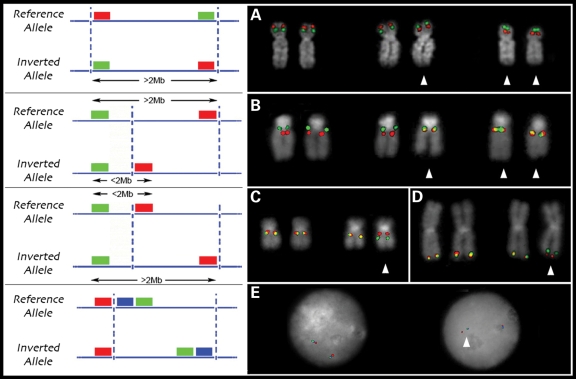
Inversions larger than 2 Mbp are typically assayed singly by metaphase FISH, using two probes located more than 2 Mbp apart inside the inverted region. When the physical distance between DNA probes is <2 Mbp, their position relative to one another on metaphase chromosomes becomes more difficult to assess. Smaller inversions (<2 Mbp) can be resolved by three-color interphase FISH. Unfortunately, the extended chromatin of interphase nuclei frequently loops back making it difficult to correctly order probes separated by more than 100 kbp, especially when breakpoints map to duplicated sequences hundreds of kbp in length. To address this deficit, we developed a FISH-based assay exploiting the limits of metaphase resolution. In order to directly visualize most of the inversion polymorphisms in this study, we selected one probe located inside and one outside the inverted region. The separation of the probes in the reference assembly is >2 Mbp, enough to visualize these as two separate signals on metaphase chromosomes. Inversion of the region repositions the two probes within close proximity to each other, visualized as overlapping yellow signals on metaphase chromosomes (Fig. 3A).
Figure 3.
FISH inversion assay. (A) A schematic showing human genomic probes labeled in green and red mapping >2 Mb apart in the non-inverted state that appears as two distinct signals on chromosomal metaphase spreads. In the inverted state, the two probes map less the 2 Mb apart and appear as a merged yellow signal. Dashed blue lines indicate the inversion breakpoints. (B) We applied the FISH assay to distinguish the orientation of the 17q21.31 region on metaphase chromosomes. Human genomic fosmid probes A and B map >2 Mb apart in the non-inverted state and appear as two distinct signals (red and green) on chromosomal metaphase spreads. In contrast, probes A and B map less the 2 Mb apart in the inverted state and appear as a merged (red+green = yellow) signal (C). A reciprocal assay on the same samples using probes A and D (non-inverted=yellow; inverted=red+green) confirm the specificity of the assay (D). An analysis of 27 HapMap cell lines using this assay showed 100% correspondence between the H1/H2 haplotype and the non-inverted/inverted status as determined by PCR (E) and SNP genotyping.
As a test of our method, we examined the 17q21.31 inversion polymorphism. The 17q21.31 inversion represents one of the most structurally complex and evolutionarily dynamic regions of the genome (13,23,26). This locus occurs in humans as two haplotypes, H1 (direct orientation based on hg17) and H2 (inverted orientation), which show no recombination between them over a region of 1.5 Mbp (13). The whole region shows an extended pattern of linkage disequilibrium due to this suppression of recombination. Notably, the H2 inverted haplotype is enriched in European populations and is predisposed to recurrent microdeletions associated with the 17q21.31 microdeletion syndrome. In every patient examined to date, the rearrangement occurs on chromosomes bearing the inversion of that same region (27). The 970 kbp inversion is too small to be validated by two-color metaphase FISH using two probes inside the inversion and is not easy to characterize by three-color interphase FISH due to the large blocks of highly homologous segmental duplications flanking the event. FISH analyses of the 17q21.31 inversions were performed using the metaphase FISH assay shown in Figure 3B in 27 individuals from three HapMap populations. The inversion was detected in three European individuals (Fig. 3C) and inversion genotype status was confirmed using a reciprocal assay (Fig. 3D). All samples were genotyped molecularly by polymerase chain reaction (PCR) for the presence of the intronic 238 bp deletion that identifies the H2 haplotype (28) (Fig. 3E) and haplotypes were also assigned to the H1 or H2 class based on two diagnostic SNPs as described previously (13). All 27 human samples (3 H2 and 51 H1 chromosomes) were 100% concordant between FISH, PCR and SNP genotyping (Table 2, Supplementary Material, Table S2).
Table 2.
Summary of FISH results
| Population | 3q29 inversion | 8p23 inversion | 15q13.3 inversion | 15q24 inversion | 17q12 inversion | 17q21 inversion | All inversions |
|---|---|---|---|---|---|---|---|
| CEPH European | 12.5% (2/16) | 50% (8/16) | 25% (4/16) | 0% (0/16) | 6.2% (1/16) | 18.7% (3/16) | 18.7% (18/96) |
| Yoruba Nigerian | 9.1% (2/22) | 59.1% (13/22) | 22.7% (5/22) | 0% (0/22) | 9.1% (2/22) | 0% (0/22) | 16.7% (22/132) |
| Japanese/Han Chinese (Asian) | 0% (0/16) | 12.5% (2/16) | 12.5% (2/16) | 6.2% (1/16) | 0% (0/16) | 0% (0/16) | 5.2% (5/96) |
| All populations | 7.4% (4/54) | 42.6% (23/54) | 20.4% (11/54) | 1.9% (1/54) | 5.5% (3/54) | 5.5% (3/54) |
Inversions frequencies in three HapMap populations are shown. The number of inverted chromosomes over the total chromosomes is indicated in brackets.
Using this approach, we characterized five additional large inversion polymorphisms in three HapMap populations (Fig. 4, Table 2 and Supplementary Material, Table S2). This includes a ∼4.7 Mbp inversion at 8p23, a polymorphic inversion that occurs at frequencies of ∼26% in Europeans (7,15) and ∼27% in Japanese individuals (29). The polymorphism is found to be present in the transmitting parents of individuals with inverted duplications, marker chromosomes and deletions of 8p23 (7,15). A heterozygous inversion of the 1.5 Mbp microdeletion syndrome region on chromosome 15q13.3 was predicted by fosmid paired-end analysis and observed in the parents of individuals with a 15q13.3 microdeletion syndrome (19). The chromosome 17q12 inversion was identified through the analysis of discordant fosmid paired ends. Interestingly, this inversion maps precisely to a deletion region associated with renal cysts and diabetes (RCAD) syndrome (30). Once again this 1.5 Mbp inversion is flanked by two large blocks of highly homologous segmental duplications. Finally, we examined two other regions, corresponding to sites of recurrent microdeletion associated with human disease—a 1.9 Mbp inversion on chromosome 3q29, predicted by fosmid paired-end analysis in both European and Yoruban populations, and a 1.2 Mbp inversion on chromosome 15q24 detected by fosmids in a single Chinese individual (31–34). The frequencies observed for each inversion in the three HapMap populations are summarized in Table 2.
Figure 4.
FISH genotyping of inversion polymorphisms. (A) Metaphase FISH validation of the 8p23 inversion using two probes located inside of the inversion. Metaphase FISH-based assay to resolve inversions <2 Mb using one probe located inside and one outside the inverted region is shown for the 15q13.3 (B), 17q12 (C) and 3q29 (D) inversions. Interphase triple color FISH validation was used for the 15q24 inversion (E). Arrows indicate inverted chromosomes.
In general, with the exception of the 17q21.31 inversion that is enriched in European and Middle Eastern populations, we identified no significant differences for the frequency of inversion alleles between Caucasians (19%) and Yorubans (17%). However, we did observe a lower frequency of these six inversions within the Asian population (5%) (P < 0.01, Fisher’s exact test) (Table 2). We found that the common 8p23 and 15q13.3 inversions are the only inversions present in all three populations, although they show a lower frequency in Asians with respect to the other groups.
Analysis of the inversion breakpoints by arrayCGH
Using oligonucleotide comparative genomic hybridization, we assessed copy number of segmental duplications at the breakpoints of the inversions in eight DNA samples (35) to determine whether differences in duplication architecture might correlate with the inversion status. Although inversion status and copy number of flanking segmental duplications did not correlate in most cases, there were two interesting exceptions: 17q21.31 and 3q29. In the case of 17q21.31, array comparative genomic hybridization (arrayCGH) experiments showed that two European individuals, GM12156 (heterozygous H1/H2) and GM12878 (homozygous H1/H1), have a 75 kbp extended duplication at the distal breakpoint of the 17q21.31 inversion not found in the other individuals (Supplementary Material, Fig. S1). This duplication was previously described as a copy-number polymorphism in the human population (1,3,6,13). Notably, FISH experiments with the probe G248P85784G12 in the 27 HapMap individuals used in this study showed that the duplication is present at a frequency of 50% in European population, but is absent in the Yoruba and Asian individuals. This apparently European-specific duplication segregates with all H2 haplotypes and 38% of the H1 haplotypes (Supplementary Material, Table S3)—thus, while all H2 haplotypes carry this duplication, it is not specific to H2. The 17q21 inversion and duplication show complete linkage disequilibrium (D′ = 1) but, because of their relative frequencies, inversion status cannot be accurately predicted using duplication status. Similarly, analysis of the segmental duplications at the breakpoint of the 3q29 inversion shows that carriers for the inverted allele (3q29H2) show a 25 kbp copy-number loss within the duplicated sequence (Supplementary Material, Fig. S2). These data suggest that these regions are highly variable and that different segmental duplication organization at the breakpoints may be related to the inverted chromosome architecture.
Tagging inversion alleles using HapMap SNPs
Given their potential impact on suppressing recombination, large, non-recurrent inversions in the human population may be associated with extended blocks of linkage disequilibrium. Characteristic SNP-genotype patterns have been used to discover inversion events (36) and develop SNP-tagging approaches to more effectively genotype inversion status. We compared the inversion genotypes determined by FISH in the 27 samples analyzed with SNP genotypes from the HapMap project (rel 23) (37) (Supplementary Material, Fig. S3). With the exception of the known 17q21.31 haplotype, none of the inversions showed a striking pattern of extended linkage disequilibrium. However, several loci, including 15q24, 17q12 and 8p23, showed suggestive signals of SNP-allele correlation. It is possible that these apparent associations could be artifacts caused by chance correlations between SNP and inversion alleles, which are present at low frequencies in the sampled individuals. In order to further investigate this possibility, we attempted to identify other HapMap individuals likely to carry at least one inversion allele. We first identified a set of SNPs for each population that showed high correlation for the inverted allele (r2 values >0.7; Supplementary Material, Table S4). Based on the genotypes at the most highly correlated SNPs for each inversion, we then selected additional HapMap individuals predicted to carry an inverted chromosome. We directly tested inversion status for 21 of these samples using the FISH assays described above (Table 3). Correspondence between predicted and observed inversion genotypes was found for the 8p23 and 17q12 inversions (Table 3). The 8p23 inversion is found worldwide at a high frequency (where it is actually the major allele in some populations) and has a complex evolutionary history (see below). We recalculated r2 values after incorporating these additional inversion genotypes (Supplementary Material, Fig. S4). For 17q12, the correspondence is surprising since the predictions were based on a single CEU inverted chromosome (for YRI, where a distinct set of SNP loci have high r2 values, one sample, GM19200 is predicted to also be inverted, but this sample has not been directly tested). Three SNP positions remain potentially associated with the inversion in the CEU population (Supplementary Material, Fig. S4: the T allele of rs8074144, the G allele of rs4074770 and the C allele of rs12449449). However, examination of the genotypes for these three SNPs in the relevant trios indicates that this apparent association may be an artifact caused by errors in our SNP phase assignments. Further clarification of these potential tag SNPs will require the direct determination of the SNP and inversion haplotypes.
Table 3.
Inversion status assessed by FISH in 21 individuals predicted to be inverted by SNP genotyping
| Inversion | Cell line | Population | FISH genotyping |
|---|---|---|---|
| 3q29 | GM11881 | CEU | Inverted, heterozygous |
| 3q29 | GM11992 | CEU | Not inverted |
| 3q29 | GM18501 | YRI | Not inverted |
| 3q29 | GM18505 | YRI | Not inverted |
| 8p23 | GM07022 | CEU | Inverted, homozygous |
| 8p23 | GM12264 | CEU | Inverted, homozygous |
| 8p23 | GM18852 | YRI | Inverted, homozygous |
| 8p23 | GM18853 | YRI | Inverted, homozygous |
| 8p23 | GM18529 | CHB | Inverted, heterozygous |
| 8p23 | GM18571 | CHB | Inverted, heterozygous |
| 8p23 | GM18966 | JPT | Inverted, heterozygous |
| 15q13 | GM18501 | YRI | Not inverted |
| 15q13 | GM18853 | YRI | Not inverted |
| 15q13 | GM19130 | YRI | Not inverted |
| 15q13 | GM19238 | YRI | Inverted, heterozygous |
| 15q24 | GM18542 | CHB | Not inverted |
| 15q24 | GM18563 | CHB | Not inverted |
| 15q24 | GM18948 | JPT | Not inverted |
| 15q24 | GM18953 | JPT | Not inverted |
| 17q12 | GM07022 | CEU | Inverted, heterozygous |
| 17q12 | GM12264 | CEU | Inverted, heterozygous |
Haplotype analyses
Since recombination between inverted alleles is reduced, inversion polymorphisms may lead to unusually long genomic segments having a deeper than average coalescence time. This scenario would result in long stretches of increased levels of nucleotide diversity. We searched for such signals utilizing fosmid-end sequences derived from the eight HapMap individuals used to discover the inversion variants. For each locus, we aligned all end-sequence pairs against the human reference assembly (build35) and calculated the percent identity across the entire inversion interval (Supplementary Material, Fig. S5; Supplementary Material, Table S5). This approach combines sequence information from both haplotypes present in each sample. As expected, for the 17q21.31 locus a clear signal of decreased sequence identity is observed for GM12156, an individual known to be heterozygous for this inversion. A clear pattern of reduced sequence identity associated with inversion status is not observed for any of the other loci. The larger 4.7 Mbp 8p23 inversion does show evidence of increased divergence with respect to the genome reference (38), but this excess divergence is not restricted to the inversion allele.
The lack of clearly increased sequence difference is somewhat surprising since several of the inversions are present at moderate frequencies and are therefore presumed to be fairly ancient alleles. Several possibilities may account for both this observation and the paucity of successful SNP tags. First, rather than representing a single event that has been maintained in human populations, the inversions may have occurred at multiple times as a result of recurrent mutation events involving the duplicated sequences. Second, the recombination barrier limiting the exchange of sequence between inverted and non-inverted alleles may not be as strong as generally supposed. For example, the inversions we studied are large (up to 4.7 Mbp in size), and double recombination events or long stretches of gene conversion may have frequently occurred within the inversion interval. We constructed median-joining haplotype networks using phased HapMap SNPs (rel 21) to further explore these possibilities. Reasoning that any exchange between alleles is likely to be suppressed nearer to the inversion breakpoints (39), we considered each breakpoint separately and focused on SNPs located inside of the inversion interval but near each breakpoint (Supplementary Material, Fig. S6). For the 3q29 and 17q12 inversions, the networks were drawn using SNPs located within 100 kbp of the inversion breakpoints. Because of the extent of duplications and reduced SNP density, SNPs up to 400 kbp away from the breakpoints were used for 15q13.3 (Supplementary Material, Table S6).
At each breakpoint for the 3q29 inversion, the three heterozygous CEU samples each contained at least a single haplotype that clustered comparatively close to each other within the haplotype network, while at least one of the two YRI samples heterozygous for the inversion contained haplotypes located in a distinctly separate clade. This observation is consistent with separate occurrences for the inversion in the CEU and YRI populations (Supplementary Material, Fig. S6). Haplotype networks for the 17q12 locus show a pattern similar to that observed for 3q29. However, at the 3′ breakpoint, it is possible that both the YRI and CEU inversion haplotypes are related, as GM19240 contains a haplotype closely related to the inversion-carrying haplogroup 8 in Supplementary Material, Figure S6. Likewise, the pattern observed for 15q13.3 is also consistent with independent occurrences for the inversion. At this locus, note that sample GM12878 is homozygous for the inversion but has SNP genotypes near the 3′ breakpoint that belong to distinct haplotype groups. Likewise, at the 5′ breakpoint sample GM12004 is heterozygous for the inversion but homozygous for haplogroup 15, which is distinct from the haplotypes associated with the inversion carriers (Supplementary Material, Fig. S6). These findings should not be considered definitive since this analysis relies on an ascertained set of SNPs as well as inferred haplotypes.
Evolutionary analyses
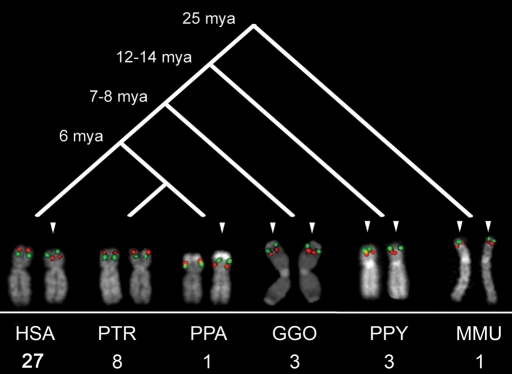
We determined whether the inversion represents the derived or ancestral state based on comparisons with outgroup non-human primate species. We tested for the presence of the inversions by examining lymphoblastoid cell lines from a diverse panel of great ape cell lines using the FISH assays established using the HapMap individuals. Zody et al. (23) previously examined the evolutionary history of the 17q21.31 inversion. Remarkably, they found this inversion to be widely polymorphic within the chimpanzee population (56% allele frequency), and they also found a polymorphic inversion of the same region in orangutan, suggesting that the region is subject to recurrent inversions. Their analysis favors the inverted orientation as the likely great ape and human ancestral state (23). Analysis of the 8p23 inversion in a large sample of eight chimpanzees from the same species (Pan troglodytes) revealed that all of them were in the H1 orientation. Surprisingly, analysis of a single Pan paniscus (Bonobo) individual found that it was heterozygous for the inversion (H1/H2). All gorillas (n = 3, Gorilla), orangutans (n = 3, Pongo pygmaeus) and one macaque (n = 1, Macaca mulatta) were homozygous for the H2 configuration, indicating that although this locus has a complicated evolutionary history, the 8p23 H2 haplotype is likely the ancestral state (Fig. 5). We examined all the other inversions (15q13.3, 17p12, 3q29 and 15q24) in a single chimpanzee, a single bonobo, a single gorilla and a single orangutan and found none of these individuals to be inverted, suggesting that the non-inverted haplotype represents the ancestral configuration (Supplementary Material, Fig. S7). Our metaphase FISH-based assay could not be used to resolve the 17q12 inversion in orangutan as the fosmid probe G248P87121H7 maps inside a larger evolutionary paracentric inversion specific of orangutan (26).
Figure 5.
Comparative analysis of the 8p23 inversion. FISH validation of 8p23 inversion in 27 HapMap individuals, eight chimpanzees, one bonobo, three gorillas, three orangutans and one macaque showed the inverted orientation (H2) as the most likely ancestral state. The inversion polymorphism might have occurred in the shared human and chimpanzee lineage and still be polymorphic in human and bonobo, but fixed in the chimpanzee population. Arrows indicate inverted chromosomes.
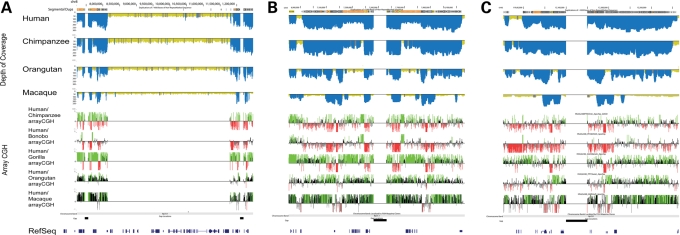
Since the duplications play a pivotal role in origin of the inversions, we compared the extent of segmental duplication at the breakpoints of the inversions among four different primate species. In order to reconstruct the evolutionary origins of those segmental duplications, we used whole-genome shotgun (WGS) sequences from human, chimpanzee, orangutan and macaque to detect regions of excess read depth against the human genome reference assembly (build35) (35). Ancient duplications, which date to the root of the primate phylogeny, are found at the breakpoints for all six inversions we studied (Fig. 6; Supplementary Material, Figs S8 and S9). We also found that in most cases, the breakpoints contain more basepairs that are duplicated in humans relative to the other primate species (Fig. 6; Supplementary Material, Figs S8 and S10; Supplementary Material, Table S7). This suggests that more complex architectures might have been acquired around the breakpoints relatively recently in human evolution. We note, however, exceptions to this trend, especially 15q24 in which the breakpoint regions are more duplicated in macaque than in human. We complemented this computational analysis with comparative genomic hybridization using a targeted oligonucleotide array (arrayCGH) (Fig. 6; Supplementary Material, Fig. S8; Supplementary Material, Table S8). Comparison of a single human versus a single individual from other primate species indicated that humans generally have more copies of the duplications at the breakpoints. Interestingly, bonobo is the only African ape with a heterozygous status of the 8p23 inversion, and it has more copies of the duplications than the reference human sample (Fig. 6). FISH experiments confirmed that bonobo contains additional copies at the 8p23 inversion locus (Supplementary Material, Fig. S11).
Figure 6.
(A) Comparative segmental duplication analysis of the 8p23 inversion region. The top panel shows the computationally predicted regions of segmental duplications [excess depth of coverage (blue) of aligned human, chimpanzee, orangutan and macaque WGS sequence to the human reference genome (build35)]. The lower panel shows the results of arrayCGH experiments for chimpanzee, bonobo, gorilla, orangutan and macaque using human as test. Enlargement of the distal and proximal breakpoints are shown in (B) and (C), respectively.
DISCUSSION
Using molecular cytogenetic, genomic and comparative approaches, we characterized the distribution and evolutionary history of six human inversion polymorphisms in a subset of samples from the HapMap collection. In each case, sequence analysis showed highly identical intrachromosomal duplications located at the breakpoints of the inversions. Without exception, these flanking duplications map in an inverted orientation implicating non-allelic homologous recombination as the mechanism of origin for each inversion. We developed a FISH-based assay to directly genotype inversions smaller than 2 Mbp, taking advantage of the limits of metaphase chromosome resolution. Using the 17q21.31 inversion as a test case (13,23), we showed excellent correspondence between molecular and cytogenetic genotypes and used this assay to directly genotype additional inversion polymorphisms. Our analysis provides the first directly observed genotype and frequency data regarding these six inversion polymorphisms of the human genome. Comparative primate analyses showed that four of the inversions were restricted to the human species with the H1 (or direct) orientation representing the ancestral state. In contrast, comparative primate analyses suggest that the H2 (or inverted) orientation represents the ancestral state for the 8p23 and 17q21.31 loci. In both of these latter cases, we find the inversion polymorphisms in at least one other great-ape population.
A central question with respect to these inversion polymorphisms is whether they have occurred once during human history or recurrently on different genetic backgrounds. Recent studies utilizing new array platforms and the denser HapMap phase II SNP genotypes have found that upwards of 80% of common CNVs are effectively tagged by existing SNPs (40,41). Similarly, analysis of HapMap samples using fosmid end-sequencing indicated that 24% of the variants predicted in at least two individuals may be present on different haplotypes (21). These results likely reflect both the recurrent nature of a subset of CNVs and the comparative deficit of ascertained SNPs near CNV loci. Unlike the 17q21.31 inversion, we did not find strong evidence of extended linkage disequilibrium or increased sequence diversity for the other inversion loci. Haplotype network analysis allowed us to infer that inverted and non-inverted alleles can map to the same haplotype implying that both the chromosomal configurations may occur on the same pre-existing haplotype background. In several cases, we deduced that inversions must have occurred on different human haplotypes providing further evidence of recurrence. Nevertheless, integrating the cytogenetic inversion status and SNP haplotype information for these samples allowed us to accurately and prospectively predict additional inversion alleles for the 3q29, 17q21.31 and 8p23 inversions, but not the other inversions (Table 3). This suggests that the SNP analysis may define a common haplotype representing many inversion chromosomes, but such an analysis may misclassify some alleles and thus underestimate the true inversion frequency. Copy-number polymorphisms, such as the 75 kbp European-specific duplication at the 17q21 locus, although informative could not be used to unambiguously predict inversion status. In total, these findings suggest that inversions where breakpoints occur within large, highly homologous segmental duplications may be recurrent variants. In these cases, molecular cytogenetics remains the only method, at present, to genotype these inversions.
It remains possible that other, multiple SNP-tagging or more sophisticated principal-components based approaches may be successful in predicting inversion status. Results for the 8p23 locus, however, indicate that such methods should also be interpreted cautiously. Using a principal-components and neighbor-joining analysis of SNP genotypes, Deng et al. (42) estimated that the 8p23 inversion was almost absent from the Asian (ASN) HapMap population (2/178 chromosomes, ∼1% allele frequency). In contrast, using direct FISH genotyping of a subset of the same samples, we have identified two individuals (Supplementary Material, Table S2) heterozygous for the inversion, leading to an estimated frequency of 12.5% (2/16 chromosomes) (Table 2). Moreover, using SNP genotyping followed by FISH validation, we were able to confirm three additional heterozygous individuals carriers for the inversion for a total of five Asian individuals with the inverted haplotype (5/22 chromosomes) (Table 3). Our findings are consistent with a previous estimate based on direct inversion genotyping of the general Japanese population (29). Taken together, these observations suggest that inversions associated with large blocks of highly identical segmental duplications are likely to be recurrent in humans and that discovery and genotyping methods based on SNPs may underestimate the true frequency of these events.
MATERIALS AND METHODS
FISH analysis
Metaphase spreads were obtained from lymphoblast cell lines from 27 human HapMap individuals (Coriell Cell Repository, Camden, NJ, USA), eight chimpanzees (Clint, Marcus, Douglas, Virginia, Cochise, PTR7, PTR12 and PTR13), one bonobo (PPA2), three gorillas (GGO5, GGO8 and GGO13), three orangutans (PPY1, PPY6 and PPY9) and one subspecies of macaque (MMU, Macaca mulatta). FISH experiments were performed using fosmid clones (Supplementary Material, Table S9) directly labeled by nick-translation with Cy3-dUTP (Perkin-Elmer), Cy5-dUTP (Perkin-Elmer) and fluorescein-dUTP (Enzo) as described by Lichter et al. (43), with minor modifications. Briefly: 300 ng of labeled probe were used for the FISH experiments; hybridization was performed at 37°C in 2xSSC, 50% (v/v) formamide, 10% (w/v) dextran sulphate and 3 µg sonicated salmon sperm DNA, in a volume of 10 µL. Posthybridization washing was at 60°C in 0.1xSSC (three times, high stringency). Nuclei were simultaneously DAPI stained. Digital images were obtained using a Leica DMRXA2 epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). DAPI, Cy3, Cy5 and fluorescein fluorescence signals, detected with specific filters, were recorded separately as grayscale images. Pseudocoloring and merging of images were performed using Adobe Photoshop software. A minimum of 25 metaphases and 50 interphase cells were scored for each inversion to statistically determine the orientation of the examined region.
Segmental duplication analysis
Human segmental duplication content of inversion breakpoints was determined using the whole genome assembly comparison approach (44). We displayed the duplication content for large and highly identical duplication pairs [size ≥10 kb, sequence identity ≥95% with the exception of the 15q24 interval where we used a relaxed threshold (size ≥5 kb and sequence identity ≥90%)]. Non-redundant ancestral duplication loci (duplicons) were determined as described previously (24,45). The duplication content of human, bonobo, chimpanzee, orangutan and macaque was determined using the whole-genome shotgun sequence detection (WSSD) method (35,46). We also assessed copy-number differences in shared duplications by interspecific array comparative genomic hybridization as previously reported (35) (GEO Accession: GSE13885). We performed cross-species arrayCGH with human, Coriell GM15510 as a reference (GEO accession number: GSE13884) using chimpanzee (Clint, Coriell S006006), bonobo (LB502), gorilla (Bahati), orangutan (Susie, ISIS #71) and macaque (ID17573) samples. A total of eight intra-specific experiments were performed and the log2 relative hybridization intensity was calculated for each probe. These experiments included the following genomic DNA comparisons: one human (Coriell GM15510) versus eight HapMap individuals (GM18517, GM18507, GM18956, GM19240, GM18555, GM12878, GM19129 and GM12156). The inter-specific experiments were performed with a standard replicate dye-swap experimental design (reverse labeling of test and reference samples). We further restricted our analysis to those regions that were greater than 20 kbp in length and contained at least 20 probes. We used a heuristic approach to calculate log2 thresholds of significance for each comparison, where we dynamically adjusted the thresholds for each hybridization to result in a false discovery rate of <1% in the control regions (35).
Haplotype analysis
Pairwise r2 values were calculated between HapMap single nucleotide polymorphisms (SNPs) (release 23) and inversion genotypes for the 27 individuals examined (Supplementary Material, Fig. S3). Since the phase of the inversion allele cannot be unambiguously deduced with respect to SNPs, we calculated r2 between unphased SNP and inversion genotypes assuming that the minor alleles occur on the same haplotype. This assumption serves to inflate apparent values of r2, increasing the likelihood of detecting true linkage-disequilibrium (at the cost of an increased false positive rate). Samples predicted to have an inversion allele based on the genotypes of SNPs with the highest r2 values were chosen for follow-up genotyping (Table 3) and additional r2 plots incorporating these added genotypes were created (Supplementary Material, Fig. S4). We assessed the sequence diversity of the unique portion of each inversion haplotype by comparing sequence data from individuals carrying the inverted allele against individuals without the inversion allele. For this purpose, we first determined the inversion genotype status both molecularly and by FISH for eight HapMap genomes. Individual clone end sequences were mapped against the genome reference as previously described (20,21). For each library, sequence identity was defined as the total number of Q20 nucleotide differences divided by the total number of aligned Q20 nucleotides. The number of high-quality aligned nucleotides for each locus is given in Supplementary Material, Table S5. Median-joining haplotype networks were constructed from phased HapMap SNP genotypes (release 21) using Nexus (47). Analysis was limited to those individuals who have been directly genotyped using the described FISH assays. The SNP positions used to construct each network are given in Supplementary Material, Table S6. Inverted haplotypes should cluster together into distinct clades (perhaps intermingled with non-inverted haplotypes) if the inversion events occurred once and if there was no exchange of genetic information between inverted and non-inverted chromosomes. Within the network diagrams, we identified haplotypes belonging to heterozygous individuals (dashed lines) or to homozygotes (solid lines) and searched for exceptions to this expectation.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by a National Science Foundation Graduate Research Fellowship to [J.M.K.], a Marie Curie fellowship to [T.M-B.] and by the National Institutes of Health [HG004120 to E.E.E.]. Funding to pay the Open Access charge was provided by the Howard Hughes Medical Institute.
ACKNOWLEDGEMENTS
We thank T. Brown for comments during the preparation of this manuscript. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 2.Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 3.Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong K.K., deLeeuw R.J., Dosanjh N.S., Kimm L.R., Cheng Z., Horsman D.E., MacAulay C., Ng R.T., Brown C.J., Eichler E.E., et al. A comprehensive analysis of common copy-number variations in the human genome. Am. J. Hum. Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perry G.H., Ben-Dor A., Tsalenko A., Sampas N., Rodriguez-Revenga L., Tran C.W., Scheffer A., Steinfeld I., Tsang P., Yamada N.A., et al. The fine-scale and complex architecture of human copy-number variation. Am. J. Hum. Genet. 2008;82:685–695. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giglio S., Calvari V., Gregato G., Gimelli G., Camanini S., Giorda R., Ragusa A., Guerneri S., Selicorni A., Stumm M., et al. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am. J. Hum. Genet. 2002;71:276–285. doi: 10.1086/341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osborne L.R., Li M., Pober B., Chitayat D., Bodurtha J., Mandel A., Costa T., Grebe T., Cox S., Tsui L.C., et al. A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat. Genet. 2001;29:321–325. doi: 10.1038/ng753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gimelli G., Pujana M.A., Patricelli M.G., Russo S., Giardino D., Larizza L., Cheung J., Armengol L., Schinzel A., Estivill X., et al. Genomic inversions of human chromosome 15q11-q13 in mothers of Angelman syndrome patients with class II (BP2/3) deletions. Hum. Mol. Genet. 2003;12:849–858. doi: 10.1093/hmg/ddg101. [DOI] [PubMed] [Google Scholar]
- 10.Sharp A.J., Hansen S., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 11.Koolen D.A., Vissers L.E., Pfundt R., de Leeuw N., Knight S.J., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat. Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 12.Shaw-Smith C., Pittman A.M., Willatt L., Martin H., Rickman L., Gribble S., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat. Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 13.Stefansson H., Helgason A., Thorleifsson G., Steinthorsdottir V., Masson G., Barnard J., Baker A., Jonasdottir A., Ingason A., Gudnadottir V.G., et al. A common inversion under selection in Europeans. Nat. Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 14.Visser R., Shimokawa O., Harada N., Kinoshita A., Ohta T., Niikawa N., Matsumoto N. Identification of a 3.0-kb major recombination hotspot in patients with Sotos syndrome who carry a common 1.9-Mb microdeletion. Am. J. Hum. Genet. 2005;76:52–67. doi: 10.1086/426950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giglio S., Broman K.W., Matsumoto N., Calvari V., Gimelli G., Neumann T., Ohashi H., Voullaire L., Larizza D., Giorda R., et al. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am. J. Hum. Genet. 2001;68:874–883. doi: 10.1086/319506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirchhoff M., Bisgaard A.M., Duno M., Hansen F.J., Schwartz M. A 17q21.31 microduplication, reciprocal to the newly described 17q21.31 microdeletion, in a girl with severe psychomotor developmental delay and dysmorphic craniofacial features. Eur. J. Med. Genet. 2007;50:256–263. doi: 10.1016/j.ejmg.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Kurotaki N., Stankiewicz P., Wakui K., Niikawa N., Lupski J.R. Sotos syndrome common deletion is mediated by directly oriented subunits within inverted Sos-REP low-copy repeats. Hum. Mol. Genet. 2005;14:535–542. doi: 10.1093/hmg/ddi050. [DOI] [PubMed] [Google Scholar]
- 18.Giorda R., Ciccone R., Gimelli G., Pramparo T., Beri S., Bonaglia M.C., Giglio S., Genuardi M., Argente J., Rocchi M., et al. Two classes of low-copy repeats comediate a new recurrent rearrangement consisting of duplication at 8p23.1 and triplication at 8p23.2. Hum. Mutat. 2007;28:459–468. doi: 10.1002/humu.20465. [DOI] [PubMed] [Google Scholar]
- 19.Sharp A.J., Mefford H.C., Li K., Baker C., Skinner C., Stevenson R.E., Schroer R.J., Novara F., De Gregori M., Ciccone R., et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuzun E., Sharp A.J., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 21.Kidd J.M., Cooper G.M., Donahue W.F., Hayden H.S., Sampas N., Graves T., Hansen N., Teague B., Alkan C., Antonacci F., et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korbel J.O., Urban A.E., Affourtit J.P., Godwin B., Grubert F., Simons J.F., Kim P.M., Palejev D., Carriero N.J., Du L., et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zody M.C., Jiang Z., Fung H.C., Antonacci F., Hillier L.W., Cardone M.F., Graves T.A., Kidd J.M., Cheng Z., Abouelleil A., et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat. Genet. 2008;40:1076–1083. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Z., Tang H., Ventura M., Cardone M.F., Marques-Bonet T., She X., Pevzner P.A., Eichler E.E. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat. Genet. 2007;39:1361–1368. doi: 10.1038/ng.2007.9. [DOI] [PubMed] [Google Scholar]
- 25.Bovee D., Zhou Y., Haugen E., Wu Z., Hayden H.S., Gillett W., Tuzun E., Cooper G.M., Sampas N., Phelps K., et al. Closing gaps in the human genome with fosmid resources generated from multiple individuals. Nat. Genet. 2008;40:96–101. doi: 10.1038/ng.2007.34. [DOI] [PubMed] [Google Scholar]
- 26.Cardone M.F., Jiang Z., D’Addabbo P., Archidiacono N., Rocchi M., Eichler E.E., Ventura M. Hominoid chromosomal rearrangements on 17q map to complex regions of segmental duplication. Genome Biol. 2008;9:R28. doi: 10.1186/gb-2008-9-2-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koolen D.A., Sharp A.J., Hurst J.A., Firth H.V., Knight S.J., Goldenberg A., Saugier-Veber P., Pfundt R., Vissers L.E., Destree A., et al. Clinical and molecular delineation of the 17q21.31 microdeletion syndrome. J. Med. Genet. 2008;45:710–720. doi: 10.1136/jmg.2008.058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baker M., Litvan I., Houlden H., Adamson J., Dickson D., Perez-Tur J., Hardy J., Lynch T., Bigio E., Hutton M. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum. Mol. Genet. 1999;8:711–715. doi: 10.1093/hmg/8.4.711. [DOI] [PubMed] [Google Scholar]
- 29.Sugawara H., Harada N., Ida T., Ishida T., Ledbetter D.H., Yoshiura K., Ohta T., Kishino T., Niikawa N., Matsumoto N. Complex low-copy repeats associated with a common polymorphic inversion at human chromosome 8p23. Genomics. 2003;82:238–244. doi: 10.1016/s0888-7543(03)00108-3. [DOI] [PubMed] [Google Scholar]
- 30.Mefford H.C., Clauin S., Sharp A.J., Moller R.S., Ullmann R., Kapur R., Pinkel D., Cooper G.M., Ventura M., Ropers H.H., et al. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am. J. Hum. Genet. 2007;81:1057–1069. doi: 10.1086/522591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willatt L., Cox J., Barber J., Cabanas E.D., Collins A., Donnai D., FitzPatrick D.R., Maher E., Martin H., Parnau J., et al. 3q29 microdeletion syndrome: clinical and molecular characterization of a new syndrome. Am. J. Hum. Genet. 2005;77:154–160. doi: 10.1086/431653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baynam G., Goldblatt J., Townshend S. A case of 3q29 microdeletion with novel features and a review of cytogenetically visible terminal 3q deletions. Clin. Dysmorphol. 2006;15:145–148. doi: 10.1097/01.mcd.0000198934.55071.ee. [DOI] [PubMed] [Google Scholar]
- 33.Ballif B.C., Theisen A., Coppinger J., Gowans G.C., Hersh J.H., Madan-Khetarpal S., Schmidt K.R., Tervo R., Escobar L.F., Friedrich C.A., et al. Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Mol. Cytogenet. 2008;1:8. doi: 10.1186/1755-8166-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharp A.J., Selzer R.R., Veltman J.A., Gimelli S., Gimelli G., Striano P., Coppola A., Regan R., Price S.M., Knoers N.V., et al. Characterization of a recurrent 15q24 microdeletion syndrome. Hum. Mol. Genet. 2007;16:567–572. doi: 10.1093/hmg/ddm016. [DOI] [PubMed] [Google Scholar]
- 35.Marques-Bonet T., Kidd J.M., Ventura M., Graves T.A., Cheng Z., Hillier L.W., Jiang Z., Baker C., Malfavon-Borja R., Fulton L.A., et al. A burst of segmental duplications in the genome of the African great ape ancestor. Nature. 2009;457:877–881. doi: 10.1038/nature07744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bansal V., Bashir A., Bafna V. Evidence for large inversion polymorphisms in the human genome from HapMap data. Genome Res. 2007;17:219–230. doi: 10.1101/gr.5774507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nusbaum C., Mikkelsen T.S., Zody M.C., Asakawa S., Taudien S., Garber M., Kodira C.D., Schueler M.G., Shimizu A., Whittaker C.A., et al. DNA sequence and analysis of human chromosome 8. Nature. 2006;439:331–335. doi: 10.1038/nature04406. [DOI] [PubMed] [Google Scholar]
- 39.Roberts P.A. The genetics of chromosome aberration. In: Ashburner M., Novistski E., editors. The Genetics and Biology of Drosophila. New York: Academic Press; 1976. pp. 68–184. [Google Scholar]
- 40.McCarroll S.A., Kuruvilla F.G., Korn J.M., Cawley S., Nemesh J., Wysoker A., Shapero M.H., de Bakker P.I., Maller J.B., Kirby A., et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 41.Cooper G.M., Zerr T., Kidd J.M., Eichler E.E., Nickerson D.A. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat. Genet. 2008;40:1199–1203. doi: 10.1038/ng.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng L., Zhang Y., Kang J., Liu T., Zhao H., Gao Y., Li C., Pan H., Tang X., Wang D., et al. An unusual haplotype structure on human chromosome 8p23 derived from the inversion polymorphism. Hum. Mutat. 2008;29:1209–1216. doi: 10.1002/humu.20775. [DOI] [PubMed] [Google Scholar]
- 43.Lichter P., Tang C.J., Call K., Hermanson G., Evans G.A., Housman D., Ward D.C. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- 44.Bailey J.A., Yavor A.M., Massa H.F., Trask B.J., Eichler E.E. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang Z., Hubley R., Smit A., Eichler E.E. DupMasker: a tool for annotating primate segmental duplications. Genome Res. 2008;18:1362–1368. doi: 10.1101/gr.078477.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 47.Bandelt H.J., Forster P., Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 48.Sharp A.J., Cheng Z., Eichler E.E. Structural variation of the human genome. Annu. Rev. Genomics Hum. Genet. 2006;7:407–442. doi: 10.1146/annurev.genom.7.080505.115618. [DOI] [PubMed] [Google Scholar]
- 49.Parsons J.D. Miropeats: graphical DNA sequence comparisons. Comput. Appl. Biosci. 1995;11:615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.