Abstract
The prevalence of CD36 deficiency in East Asian and African populations suggests that the causal variants are under selection by severe malaria. Previous analysis of data from the International HapMap Project indicated that a CD36 haplotype bearing a nonsense mutation (T1264G; rs3211938) had undergone recent positive selection in the Yoruba of Nigeria. To investigate the global distribution of this putative selection event, we genotyped T1264G in 3420 individuals from 66 populations. We confirmed the high frequency of 1264G in the Yoruba (26%). However, the 1264G allele is less common in other African populations and absent from all non-African populations without recent African admixture. Using long-range linkage disequilibrium, we studied two West African groups in depth. Evidence for recent positive selection at the locus was demonstrable in the Yoruba, although not in Gambians. We screened 70 variants from across CD36 for an association with severe malaria phenotypes, employing a case–control study of 1350 subjects and a family study of 1288 parent–offspring trios. No marker was significantly associated with severe malaria. We focused on T1264G, genotyping 10 922 samples from four African populations. The nonsense allele was not associated with severe malaria (pooled allelic odds ratio 1.0; 95% confidence interval 0.89–1.12; P = 0.98). These results suggest a range of possible explanations including the existence of alternative selection pressures on CD36, co-evolution between host and parasite or confounding caused by allelic heterogeneity of CD36 deficiency.
INTRODUCTION
Erythrocytes infected with mature forms of the Plasmodium falciparum parasite adhere to endothelium, platelets, leucocytes and uninfected erythrocytes, a behaviour considered key to the pathogenesis of severe falciparum malaria (1). The majority of P. falciparum clinical isolates bind CD36 (1–4). A family of variant molecules called P. falciparum erythrocyte membrane protein 1 (PfEMP1) is responsible for binding CD36 and other host antigens (5). PfEMP1 is expressed by the parasite onto the surface of infected red blood cells (iRBCs) and is subject to switching during the course of an infection. CD36 is found on a range of cell types including platelets, dendritic cells and endothelium (6). Adhesion of iRBCs to endothelial CD36 helps the parasite avoid splenic passage, contributes to microcirculatory occlusion and promotes local inflammatory responses (1). CD36-mediated binding of iRBCs to dendritic cells inhibits their maturation and function (7), and CD36 on platelets is required for the formation of platelet-mediated clumps, which are associated with severe disease (8).
CD36 deficiency (or Naka-negative blood group) has been reported in Japanese (9), Thais (10) and African-Americans (11). The prevalence of CD36 deficiency in East Asian and African populations raises the possibility that the responsible variants have been selected for by malaria. The molecular basis of CD36 deficiency in African-Americans is distinct from that found in East Asia (12). Common alleles reported in East Asia include a proline to serine missense mutation at codon 90 of the CD36 gene (C478T) and a frameshift mutation at codon 317 (1159insA) (13). The commonest reported African CD36 deficiency allele is a nonsense mutation in exon 10 of CD36 (T1264G; rs3211938), which terminates the polypeptide before the second transmembrane domain (12).
Further evidence that evolutionary selection has shaped genetic variation at the CD36 locus emerged from phase 1 of the International Haplotype Map (HapMap) project (14). The HapMap project performed whole-genome high-resolution genotyping in samples from four populations including 30 parent–offspring trios from the Yoruba ethnic group from Ibadan in southwestern Nigeria, 30 parent–offspring trios from Utah (of northern and western European ancestry), 45 unrelated Han Chinese from Beijing and 45 unrelated Japanese from Tokyo. The CD36 nonsense allele, 1264G, was common in the Yoruba (∼25%) but not detected in other populations. The 1264G allele was present on haplotypes found to be unusually similar over hundreds of kilobases (see Supplementary Material, Fig. S1). This signal, termed extended haplotype homozygosity (EHH), suggested that 1264G had been under recent positive evolutionary selection (15,16).
Genetic epidemiological studies of severe falciparum malaria have examined whether CD36 deficiency alleles affect susceptibility, but have not produced consistent answers (Table 1). An initial case–control study of 1359 Gambian and Kenyan children found CD36 deficiency appeared to be associated with susceptibility to severe disease, leading the authors to propose that another infectious pathogen was responsible for the frequency of CD36 deficiency in East Asians and Africans (12). In contrast, a matched case–control study of 693 pairs of Kenyan children found that 1264G heterozygotes were protected from malaria, particularly from having multiple syndromes of malaria (17). A third study of 223 children from Ibadan in Nigeria was unable to detect a significant difference in T1264G frequency between children with severe disease and those with asymptomatic parasitaemia (18). However, a recent study of 913 Kenyan children has, once again, suggested that 1264G is associated with susceptibility to malaria (19).
Table 1.
Published risk estimates of severe malaria phenotypes associated with CD36 deficiency alleles
| Publication | Population | Cases | Controls | Phenotype | Allele/genotype | OR | 95% CI | P-value |
|---|---|---|---|---|---|---|---|---|
| Aitman et al. (2000) (12) | Gambia and Kenya | 598 | 761 | All cases | def versus wt | 1.53 | 1.09–2.15 | 0.01a |
| (388)b | 761 | CM | def versus wt | 1.49 | 1.01–2.20 | 0.04 | ||
| (97) | (331) | CM | GG versus rest | — | — | 0.04 | ||
| Pain et al. (2001) (17) | Kenya | 693 | 693 | All cases | GG and GT versus TT | 0.74 | 0.55–0.99 | 0.036a |
| (104) | 693 | Three or four syndromes | GG and GT versus TT | 0.47 | 0.22–0.95 | 0.024 | ||
| (589) | 693 | One or two syndromes | GG and GT versus TT | 0.79 | 0.58–1.07 | 0.11 | ||
| (413) | 693 | CM | GG and GT versus TT | 0.84 | 0.60–1.18 | 0.27 | ||
| (250) | 693 | Respiratory distress | GG and GT versus TT | 0.67 | 0.43–1.04 | 0.051 | ||
| (304) | 693 | SA | GG and GT versus TT | 0.62 | 0.41–0.94 | 0.017 | ||
| (146) | 693 | Hypoglycaemia | GG and GT versus TT | 0.54 | 0.30–0.96 | 0.026 | ||
| Amodu et al. (2005) (18) | Nigeria | 101 | 53 | Uncomplicated versus asymptomatic | G versus T | — | — | 0.062c |
| 69 | 53 | Severe versus asymptomatic | G versus T | — | — | 0.326 | ||
| Ayodo et al. (2007) (19) | Luo (Kenya) | 456 | 457 | All cases | GT versus rest | 1.5 | 1.03–2.18 | 0.03 |
| 456 | 457 | All cases | GG versus GT versus TT | — | — | 0.061 | ||
| 456 | 457 | All cases | G versus T | — | — | 0.0023d | ||
| 456 | 457 | All cases | G versus T | — | — | 0.00043e |
OR, odds ratio; CI, confidence interval; def, CD36 deficiency alleles 1264G and 1439C pooled; wt, CD36 wild-type allele; the remaining alleles and genotypes refer to CD36 T1264G. CM, cerebral malaria; SA, severe anaemia; Severe, all severe cases; Uncomplicated, non-severe malaria.
aMantel–Haenszel stratified analysis.
bSample numbers in brackets refer to subsets of the studies’ cases or controls.
cMultivariate analysis.
dWeighted for population differentiation (Yoruba versus Masai).
eWeighted for population differentiation (Yoruba versus Kikuyu).
We set out to analyse in detail the relationship between CD36 variation and severe malaria. Our aims were as follows.
Determine the distribution of the putative T1264G selection event in a range of African and non-African populations.
Replicate the CD36 EHH signal in the Yoruba and, if possible, in additional populations.
Test single-nucleotide polymorphisms (SNPs) across CD36 for associations with severe malaria. Previous genetic association studies have focused on T1264G, and only a fraction of variation across the gene has been analysed. Other SNPs in or near CD36 may demonstrate novel disease associations and could assist the fine-mapping of functional changes.
Employ a series of population- and family-based studies of severe malaria phenotypes to refine the risk estimate associated with T1264G.
RESULTS
A global survey of CD36 haplotypes and T1264G
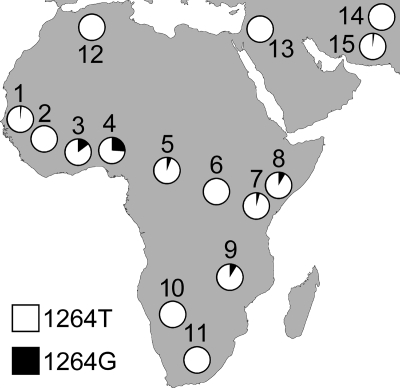
We genotyped the T1264G polymorphism in 3420 individuals from 66 ethnic groups. These included 974 individuals from the Human Genome Diversity Project (HGDP)-Centre d'Etude du Polymorphisme Humain (CEPH) Diversity Panel (51 ethnic groups) and 2446 additional Gambian, Malawian, Kenyan and Ghanaian cord blood samples and population controls (15 ethnic groups). The 1264G allele was at a frequency of 26% in the Yoruba of Nigeria, a finding consistent with the HapMap Yoruba (24.2%) and another recent study of Nigerian children (18). The 1264G allele was present in populations across sub-Saharan Africa, but was generally less frequent than in the Yoruba (Fig. 1). The 1264G allele was not seen in two African populations living in the regions of lower malaria risk, the South African Bantu and the Namibian San. However, their limited sample sizes may mean the allele is present at low frequencies but undetected. The pygmies of central Africa, such as the Biaka (Central African Republic) and the Mbuti (Democratic Republic of Congo), are relatively genetically isolated from other sub-Saharan populations. There is, however, evidence that the Biaka have experienced admixture with neighbouring groups (20). This is consistent with the presence of 1264G alleles in the Biaka (5.8%) but not in the Mbuti (0%). The 1264G allele was absent from almost all non-African ethnic groups. The Makrani of Pakistan were the only group outside of Africa (and the only Pakistani ethnic group) to have 1264G (2.1%). The presence of 1264G is consistent with past African admixture in the Makrani (21). For a full breakdown of samples genotyped and 1264G allele frequencies, see Supplementary Material, Table S1.
Figure 1.
Frequencies of the CD36 1264G nonsense allele in a range of African and Middle Eastern populations. For each population, the country of origin, number of individuals samples and 1264G allele frequency are given (in parentheses) as follows: 1, Fula, Jola, Mandinka, Manjago, Serehuli, Serere and Wolloff (The Gambia, 582, 2%); 2, Mandenka (Senegal, 21, 0%); 3, Kasem, Nankan and Buli (Ghana, 737, 14%); 4, Yoruba (Nigeria, 25, 26%); 5, Biaka Pygmy (Central African Republic, 26, 5.8%); 6, Mbuti Pygmy (Democratic Republic of Congo, 7, 0%); 7, northeastern Bantu (Kenya, 11, 4.5%); 8, Chonyi, Duruma, Giriama and Kauma (Kenya, 722, 8.6%); 9, Malawians (Malawi, 405, 8.9%); 10, San (Namibia, 6, 0%); 11, southeastern and southwestern Bantu (South Africa, 7, 0%); 12, Mozabite (Mzab region of Algeria, 23, 0%); 13, Palestinian, Druze and Bedouin (Israel, 138, 0%); 14, Sindhi, Pathan, Kalash, Hazara, Burusho, Brahui and Balochi (Pakistan, 158, 0%); 15, Makrani (Pakistan, 24, 2.1%). The 1264G allele is absent from all other populations assayed but not shown in this figure.
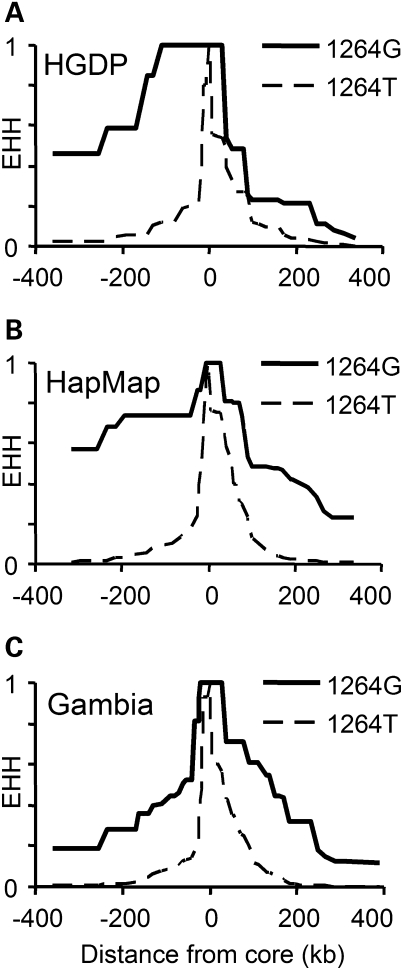
We genotyped 53 additional SNPs, in and around CD36, in the HGDP-CEPH Diversity Panel. These markers comprised 12 intragenic SNPs (average density 1 SNP/6.3 kb) and 41 SNPs extending 300 kb up- and downstream of CD36 (average density 1 SNP/15.3 kb). We compared haplotypes carrying 1264G with those bearing the ancestral 1264T, calculating the EHH surrounding the core SNP (T1264G) (see Materials and Methods). EHH reflects the probability that two haplotypes, chosen at random from a population, will share identical markers, from the core SNP to a specified test position. Haplotypes carrying the 1264G allele were more similar over longer distances than those with the ancestral T-allele (Fig. 2), a common trend when comparing ancestral and derived alleles (22). Measured ∼100 kb upstream from the core SNP (at rs1851937), the 1264G haplotype EHH was still 1.0 compared with 0.117 for 1264T, while at ∼300 kb upstream (rs304775), 1264G EHH was 0.462 versus 0.026 for 1264T. Downstream, however, EHH values were lower, and there was less difference between the two haplotype subtypes; at ∼100 kb (rs12669309), EHH was 0.231 (G) versus 0.12 (T), and 0.064 (G) versus 0.006 (T) at ∼300 kb (rs2367090). To assess the significance of these differences, we measured relative EHH (REHH, the ratio between EHH on haplotypes marked by 1264G and haplotypes bearing the T-allele) at a distance of ∼0.25 cM from the core SNP and compared these measurements with the empirical genome-wide distribution of the long-range haplotype (LRH) test (14)). The REHH was 17.6 (5′) and 3.2 (3′). In other words, the EHH was 17-fold greater on haplotypes bearing 1264G at 0.25 cM upstream. This 5′ value is >99th centile compared with the empirical distribution of REHH and similar to (or greater than) previous reports of alleles under selection in Africans [e.g. G6PD deficiency, haplotype frequency 18%, REHH 7 (15)]. However, the 3′ REHH of 3.2 was not significant.
Figure 2.
EHH decay surrounding T1264G in HGDP Yoruba, HapMap Yoruba and Gambian trios. Breakdown of extended haplotype homozygosity (EHH) with distance, on haplotypes partitioned by the alleles of T1264G (rs3211938; the ‘core’ SNP). We compared (A) 25 HGDP-CEPH Yoruba individuals, (B) HapMap genotypes from 60 Yoruba parents and (C) 202 Gambian parents.
For comparison, we downloaded phased haplotypes from the HapMap Yoruba, paring down the data set to the same 54 SNPs (including T1264G) (Fig. 2). As expected, EHH measurements were greater on 1264G haplotypes. Upstream EHH was, at ∼100 kb, 0.741 (G) versus 0.089 (T); at ∼300 kb, 0.571 (G) versus 0.012 (T); downstream, at ∼100 kb, 0.483 (G) versus 0.129 (T); and at ∼300 kb, 0.227 (G) versus 0.005 (T). The HapMap Yoruba had REHH values of 29.7 (5′) and 14.6 (3′) [both >99th centile and consistent with the original observation (14)]. REHH values for our HGDP Yoruba are lower than those for the HapMap Yoruba, particularly downstream of T1264G. There are a range of explanations for these differences: (i) the HapMap data had a slightly lower missing genotype rate (1 versus 3% for HGDP); (ii) the HapMap haplotypes were inferred from higher density marker data; (iii) our HGDP sample size was smaller (50 haplotypes versus 120 HapMap haplotypes); and (iv) the HapMap haplotypes were inferred from parent–offspring trios. Offspring genotypes can be used to unambiguously resolve phase at some parental markers. To test the impact of phasing accuracy on EHH, we obtained Yoruba genotypes from the HapMap project and phased them without utilizing offspring data (treating the parents as 60 unrelated individuals). The resulting haplotypes had an REHH of 29.6 upstream, but only 5.1 downstream (Supplementary Material, Fig. S2). This suggests that the asymmetry in HGDP Yoruba REHH, between 5′ and 3′ regions, may reflect technical artefacts affecting haplotype estimation downstream (e.g. assay performance, phasing accuracy) rather than a biological signal (e.g. selection acting on sequences upstream of T1264G).
Too few 1264G haplotypes were sampled from the Makrani, the Biaka of the Central African Republic or the northeastern Bantu for a valid assessment of EHH to be made. To study 1264G haplotypes in detail, we selected 101 parent–offspring trios from our Gambian family study (see next section). Twenty-six trios were selected because one or both parents had the 1264G allele. The remaining 75 were randomly chosen from the family study. Samples were genotyped and analysed in a manner similar to the HGDP-CEPH Diversity Panel. Offspring genotypes were used to assist phasing. In total, 30 1264G haplotypes were compared with 374 1264T haplotypes (Fig. 2). EHH values on Gambian CD36 1264G haplotypes were raised. Upstream EHH was, at ∼100 kb, 0.405 (G) versus 0.082 (T); at ∼300 kb, 0.184 (G) versus 0.006 (T); downstream, at ∼100 kb, 0.607 (G) versus 0.114 (T); and at ∼300 kb, 0.122 (G) versus 0.003 (T). REHH was 19.1 upstream and 12.8 downstream. However, EHH is often raised around low frequency, derived alleles. The significance of these results was determined using phased genome-wide SNP data from 658 Gambian parent–offspring trios genotyped on an Illumina 650Y BeadArray platform (manuscript in preparation). The offspring in these trios were malaria cases; so to minimize bias, we analysed only untransmitted parental chromosomes. We calculated the REHH for 15 807 derived alleles of frequency 1–3%, where REHH could be assessed at 0.25 ± 0.01 cM from the core SNP. Comparison with this empirical, genome-wide distribution of the LRH test suggests our Gambian REHH values are not significantly unusual, ∼59th centile (12.8) and ∼77th centile (19.1). This finding highlights that low-frequency alleles are often surrounded by long haplotypes, limiting the power of EHH testing.
Genetic association studies with severe malaria phenotypes
Seventy CD36 SNPs, including T1264G and G1439C [a missense mutation in strong linkage disequilibrium in Gambians with the 1444delA frameshift mutation, another CD36 deficiency allele (12)], were tested for disease associations in Gambian case–control and family studies (see Material and Methods for further details of SNP selection). The family study comprised 1288 children affected by severe malaria and both parents [these trios included 508 cases of cerebral malaria (CM) and 333 cases of severe malarial anaemia (SA)]. The case–control study was separate from the family study and comprised 727 cases of severe malaria (339 CM, 226 SA) and 623 population controls. Both studies were powered to detect realistic effect sizes at a significance threshold of P <0.05. Only one marker, rs4728191, an intronic SNP, exceeded this threshold in both studies. The minor allele was associated with protection on both occasions [case–control, severe malaria, allelic odds ratio (OR) 0.76; 95% confidence interval (CI) 0.61–0.96, P = 0.019; family study, SA, allelic OR 0.70; 95% CI 0.5–0.99, P = 0.041]. This finding needs cautious interpretation, as the associations were with different phenotypes and the Hardy–Weinberg P-value for rs4728191 in cord blood controls was marginal (P = 0.006). All results from the case–control and family studies with P <0.05 are documented in Supplementary Material, Table S2.
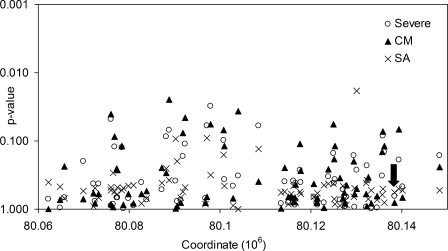
Using the UNPHASED application, we combined the case–control and family data into a single analysis (Fig. 3). Given that 70 SNPs have been tested, the formal significance threshold, with conservative Bonferroni correction, would be roughly 0.0007 (0.05/70). No marker in CD36 had P <0.002 for any genetic model or phenotype. Rs4728191 was not significantly associated with severe malaria phenotypes in the combined analysis (severe malaria, allelic OR 0.89; 95% CI 0.79–1.01, P = 0.06; SA allelic OR 0.84; 95% CI 0.67–1.06, P = 0.13). Neither T1264G (severe malaria, allelic OR 0.93; 95% CI 0.67–1.29, P = 0.65), nor G1439C (severe malaria, allelic OR 0.90; 95% CI 0.68–1.19, P = 0.46), nor both deficiency alleles considered together (severe malaria, allelic OR 0.91; 95% CI 0.73–1.13, P = 0.39) were significantly associated with severe malaria phenotypes. Complete results from the pooled analysis are documented in Supplementary Material, Table S3.
Figure 3.
P-values for the pooled Gambian case–control and family studies. Allelic model P-values derived from UNPHASED analysis of 70 SNPs across CD36, for severe falciparum malaria (Severe), and the subphenotypes of cerebral malaria (CM) and severe malarial anaemia (SA). The arrow marks the position of T1264G (rs3211938). Chromosome 7 coordinates from NCBI build 36/dbSNP 126.
Given the previous report of protection associated with 1264G heterozygotes (17), we specifically checked for such an effect. Deficiency allele heterozygotes did tend to be underrepresented among cases (versus wild-type homozygotes, OR 0.83; 95% CI 0.66–1.05). In addition, no CD36-deficient individual (e.g. 1264G homozygote, 1439C homozygote or compound heterozygote) was found among the 727 cord blood controls, although several were seen among the cases (three in the case–control study and eight in the family study) (deficiency allele homozygotes versus wild-type homozygotes OR 1.59; 95% CI 0.74–3.39). Together these results suggested a non-significant trend towards heterozygote advantage (overdominant model P = 0.09). However, the low frequency of 1264G (1.9%) and 1439C (1.8%) in Gambians limited the statistical power. To increase the power, we genotyped T1264G and G1439C in five further sample sets derived from populations where 1264G is more common (8.2% in Kenyan controls, 9.0% in Malawian controls and 14.2% in Ghanaian controls). These additional studies comprised:
718 Malawian cases and 405 controls (640 CM, 101 SA);
708 Kenyan cases and 902 controls (216 CM, 270 SA);
225 Malawi trios (216 CM, 39 SA);
234 Kenyan trios (114 CM, 85 SA);
792 Ghanaian cases and 806 controls (44 CM, 296 SA).
The 1439C allele was at a frequency of 0.1% in Ghanaian controls, but absent in Kenya and Malawi.
T1264G was not associated with severe malarial phenotypes in the additional five studies individually. Pooling data across all seven studies (5681 cases and controls, and 1747 nuclear family trios) using the UNPHASED application, we found no association between T1264G (severe malaria, allelic OR 1.0; 95% CI 0.89–1.12, P = 0.98) or deficiency alleles (1264G and 1439C pooled, severe malaria, allelic OR 0.98; 95% CI 0.89–1.09, P = 0.76) and severe malaria (Table 2). The putative trend towards a heterozygous advantage, suggested by the Gambian data, was not supported by this larger data set (deficiency allele heterozygotes versus wild-type homozygotes, OR 0.97; 95% CI 0.86–1.09; deficiency allele homozygotes versus wild-type homozygotes, OR 1.09; 95% CI 0.74–1.6; overdominant model P = 0.53) (Table 3).
Table 2.
Estimated risk for CD36 T1264G in severe malaria
| Population | Cases | Controls | OR | 95% CI | P-value |
|---|---|---|---|---|---|
| Population-based studies | |||||
| Gambia | 727 | 623 | 0.74 | 0.4–1.37 | 0.34 |
| Malawi | 718 | 405 | 0.91 | 0.67–1.25 | 0.57 |
| Kenya | 708 | 902 | 1.20 | 0.93–1.55 | 0.15 |
| Ghana | 792 | 806 | 1.05 | 0.86–1.29 | 0.63 |
| Family-based studies | |||||
| Gambia | 1288 | 2576 | 0.92 | 0.59–1.45 | 0.73 |
| Malawi | 225 | 450 | 1.00 | 0.62–1.62 | 1.00 |
| Kenya | 234 | 468 | 0.86 | 0.55–1.34 | 0.50 |
| Pooled | 4692 | 6230 | 1.00 | 0.89–1.12 | 0.98 |
Allelic odds ratios (OR) and 95% confidence intervals (CI) for CD36 1264G compared with the T-allele, in severe malaria. P-values were derived from logistic regression analysis with covariates of ethnic group, gender and HbS genotype (population-based studies), case–pseudo-control analysis (family studies) or UNPHASED analysis (pooled data across all studies).
Table 3.
Pooled analysis of CD36 deficiency alleles and severe malaria phenotypes
| Phenotype | Marker | Genotype | OR | 95% CI | rec | over | dom | allelic |
|---|---|---|---|---|---|---|---|---|
| Severe | T1264G | GT versus TT | 1.00 | 0.88–1.14 | — | — | — | — |
| — | GG versus TT | 0.98 | 0.64–1.52 | — | — | — | — | |
| 0.94 | 0.99 | 0.99 | 0.98 | |||||
| def | het versus wt | 0.97 | 0.86–1.09 | — | — | — | — | |
| — | hom versus wt | 1.09 | 0.74–1.61 | — | — | — | — | |
| 0.62 | 0.53 | 0.64 | 0.76 | |||||
| SA | T1264G | GT versus TT | 1.05 | 0.86–1.28 | — | — | — | — |
| — | GG versus TT | 0.79 | 0.35–1.74 | |||||
| 0.53 | 0.62 | 0.75 | 0.89 | |||||
| def | het versus wt | 1.05 | 0.87–1.27 | — | — | — | — | |
| — | hom versus wt | 0.89 | 0.43–1.84 | — | — | — | — | |
| 0.76 | 0.60 | 0.67 | 0.75 | |||||
| CM | T1264G | GT versus TT | 1.09 | 0.91–1.31 | — | — | — | — |
| — | GG versus TT | 0.78 | 0.37–1.64 | — | — | — | — | |
| 0.47 | 0.34 | 0.45 | 0.60 | |||||
| def | het versus wt | 1.03 | 0.87–1.22 | — | — | — | — | |
| — | hom versus wt | 1.03 | 0.55–1.93 | — | — | — | — | |
| 0.95 | 0.74 | 0.73 | 0.74 |
Pooled analysis performed from our seven studies using the UNPHASED application. Phenotypes: Severe, severe falciparum malaria; SA, severe malarial anaemia; CM, cerebral malaria. Markers: T1264G or both CD36 deficiency (def) alleles pooled (G1439C + 1444delA and T1264G). T1264G genotypes: GG, 1264G homozygotes; GT, heterozygotes; TT, 1264T homozygotes; het, deficiency allele heterozygotes; hom, deficiency allele homozygotes; wt, wild-type homozygotes; OR, odds ratio; CI, confidence interval; P-values are reported for four genetic models rec(essive), over(dominant), dom(inant) and allelic.
DISCUSSION
We set out to analyse the distribution of 1264G haplotypes in a range of global populations, to screen variation across the gene for association with severe malaria and to target 1264G with well-powered tests of disease association. It is important to stress that the initial observation of raised EHH on 1264G haplotypes was only briefly noted in previous work, with little discussion of its biological significance (14,16). This study represents the first detailed description of the CD36 EHH signal found in the HapMap Yoruba. In addition, we presented novel data from the HGDP collection exploring the frequency of the nonsense allele across Africa and replicating the EHH signal in the Yoruba. Our association study of severe malaria and T1264G employed a sample size more than twice that of all previous studies combined (Table 1). Given our relative statistical power, the marginal significance of past results and their inconsistent outcomes, our data suggest that CD36 T1264G is not associated with severe malaria. Furthermore, no CD36 marker tested appeared to be significantly associated with disease susceptibility.
The reason for the expansion of the 1264G allele in West-Central Africa is unclear. Nigeria has a high prevalence of malaria infection, however, so does the Gambia (1264G frequency 2%) and costal regions of Kenya (9%). The high frequency in Nigeria might indicate a recent origin for the allele in that region with subsequent migration. The lower allele frequency outside Nigeria could simply reflect selective sweeps in progress, but at earlier stages. There are a range of plausible explanations for the presence of selection in the absence of a malaria association. It is possible that the signal of selection detected around 1264G could be spurious; or that host–parasite co-evolution has eliminated the advantage of this allele, subsequent to an initial selective sweep. Alternatively, the failure to detect an association might represent type II error. The 1264G allele could provide a modest degree of protection from severe malaria (e.g. an OR between 0.95 and 1); protect from an infrequent but life-threatening malaria phenotype; or only offer protection to the rare deficiency allele homozygotes. This association study would have only limited power to detect an effect in these situations. A small effect size might be sufficient to maintain the allele at low frequency in populations. Hypothetically, the expansion of 1264G alleles in the Yoruba could represent a local selection event on top of a low-level background of selection by malaria. The additional Yoruba event could also be malaria-related, for example the effects of a P. falciparum strain specific to southwestern Nigeria. However, CD36 operates in a range of key biological processes including thrombostasis (23), glucose metabolism (24), lipid handling (25), immune function (26), angiogenesis (27) and possibly taste (28). Therefore, other environmental factors need to be considered as selective pressures.
Combining evidence for natural selection with disease association statistics has been proposed as a technique to increase power to detect disease associations. Our data highlights a potential pitfall for this approach. In recently published work, an association between T1264G and severe malaria (P = 0.03) was combined with the substantial allele frequency difference between the Yoruba (a population exposed to endemic malaria) and the Kikuyu and Masai (populations at low malaria risk). The combined P-values reported were highly significant (up to P = 0.00043) (19). However, as we have seen, there is a substantial T1264G allele frequency difference between the Yoruba and most African populations, including others exposed to endemic malaria. This highlights the possibility that marginal evidence of a genetic association with disease A (e.g. severe malaria) could be conflated with strong evidence of natural selection caused by an unrelated process B (e.g. a novel dietary source). Combined analysis remains an exciting approach, but ensuring a signal of selection relates to the same disease as association data (or gauging the degree of confidence) will be a challenge.
It is possible that CD36 deficiency alleles cause deleterious consequences, particularly in the homozygous state, that balance any hypothetical advantages. These include neonatal immune thrombocytopenia (29), altered aerobic exercise capacity (30) and dysregulation of lipid metabolism (25). There is also, ironically, evidence that absence of CD36 is disadvantageous to the host during malaria infection. IRBCs are recognized and phagocytosed by monocytes and macrophages following CD36 binding (31,32). One disease association study based in Thailand (475 adult patients with severe or mild malaria) reported an intronic dinucleotide repeat allele in CD36 [in3(TG)12] associated with protection from severe malaria (33). The authors showed that other alleles of the same microsatellite were associated with the production of a short CD36 isoform altering the P. falciparum-binding epitope. In contrast, the protective in3(TG)12 allele was associated with a full-length transcript, leading the authors to suggest that iRBC binding to intact CD36 could facilitate efficient clearance. The majority of P. falciparum isolates bind CD36 (1–4) yet only a fraction of infections lead to life-threatening consequences. In vitro binding studies suggest that parasite strains from asymptomatic controls with parasitaemia have high levels of CD36 binding, whereas strains from individuals with non-severe, uncomplicated disease have moderate levels of CD36 binding. The lowest levels of CD36 binding are found in isolates from individuals with severe phenotypes such as CM, and particularly SA (2,3,34,35). CD36 (also known as fatty acid translocase) is highly expressed in adipose tissue and skeletal muscle (6). Absence of CD36 as a target for sequestration on these large capillary beds may promote splenic passage and clearance of CD36-binding parasites. This could select for parasite clones which have switched to alternative host ligands. Post-mortem immunohistochemistry studies show relatively low expression of CD36 on brain endothelium without increased expression during malaria (1,36). In contrast, other host ligands of PfEMP1, such as intercellular adhesion molecule-1 and E-selectin, are commonly expressed on cerebral endothelium and induced by inflammation (36,37).
CD36 deficiency alleles have been reported to modulate expression in a tissue-specific way. Some variants prevent expression on platelets (CD36 deficiency type II), whereas other alleles abolish CD36 expression on a wider range of cells including platelets, macrophages, monocytes and probably other cell types (type I) (38,39). The 1264G allele is considered to be a type I allele, but little is known about expression patterns (if any) on non-haematopoietic cells. In addition to T1264G and 1444delA, a range of CD36 deficiency alleles have been reported in populations of African origin. These include 990delG and 1530ins14bp (12). Sequencing in Gambians has suggested that 1264G and 1444delA are the commonest deficiency alleles in this population, with others being <1% frequency (12). However, caution is required when extrapolating these findings across Africa. Allelic heterogeneity, particularly when unrecognized, has the potential to impair power in association analysis (40,41) and could also be complicating our population genetic study. The frequency distribution of CD36 deficiency across Africa could be quite different than suggested by typing just 1264G and 1439C+1444delA. Resequencing of CD36 in a range of African and Asian populations is needed to describe the full repertoire and geographic distribution of CD36 deficiency alleles.
In conclusion, the relationship between CD36 and malaria is complex. The putative selection event associated with the CD36 1264G nonsense allele appears to be focused in West-Central Africa, although this and other CD36 deficiency alleles are present across sub-Saharan and Asian populations. A number of factors may explain the lack of an association between CD36 variation and severe malaria phenotypes. However, the existence of alternative (or additional) selection pressures on CD36 deficiency alleles is an interesting possibility that needs further investigation. Additional field- and in vitro work is required to define the phenotypic consequences of CD36 deficiency alleles in health and disease.
MATERIALS AND METHODS
Human Genome Diversity Panel
Sample preparation and genotyping.
DNA was derived from lymphoblastoid cell lines of 1064 individuals from the HGDP-CEPH Diversity Panel (http://www.cephb.fr/HGDP-CEPH-Panel/) (42). Samples from 15 known duplicate and atypical individuals were excluded (43). Genotyping was performed using Sequenom iPLEX assays. SNP identifiers, coordinates and genotyping success rates are documented in Supplementary Material, Table S4. Following quality control for missing genotypes, a set of 974 individuals were selected for further analysis (average genotyping success rate 96.7%; rs3211938 success rate 99.8%).
Statistical analysis.
Phased haplotypes and missing data were inferred using PHASE (version 2.1) (44,45). Genotype data from each population was phased on its own. EHH calculations were performed using a web-based calculator (http://ihg2.helmholtz-muenchen.de/cgi-bin/mueller/webehh.pl). REHH was assessed as described previously (15,46). REHH was assessed both sides of the core SNP (5′ and 3′). The average REHH values for the two markers closest to and either side of 0.25 cM were used for the LRH test [5′, rs704871 (0.267 cM) and rs2944398 (0.225 cM); 3′, rs10487878 (0.236 cM) and rs4731861 (0.325 cM)].
Genetic association studies
Phenotype definition.
All cases were children admitted to hospital with evidence of P. falciparum on blood film and clinical features of severe malaria (47,48). We used a Blantyre coma score of ≤2 as a criterion of CM (because of the limitations of the available data, we used ≤3 in Ghanaian cases) and <5 g/dl or packed cell volume <15% as a criterion of SA. Some individuals had both CM and SA. Of the severe malaria cases that were not CM or SA by these criteria, most had lesser degrees of coma (Blantyre coma score 3) or anaemia (haemoglobin 5–6 g/dl), or other complications such as respiratory distress. In Gambia, Malawi and Kenya, controls were cord blood samples obtained from birth clinics in the same locations as the cases. Ghanaian controls were community controls matched for age, ethnic group and location of origin.
Subjects.
Patient samples were collected during ongoing epidemiological studies of severe malaria at the Royal Victoria Hospital, Banjul, The Gambia; the Queen Elizabeth Central Hospital, Blantyre, Malawi; Kilifi District Hospital, Kilifi, Kenya; and Navrongo War Memorial Hospital, Ghana. All DNA samples were collected and genotyped following approval from the relevant research ethics committees and informed consent from participants. See Supplementary Material, Table S5, for demographic details of patients and controls.
Power calculations.
Power calculations were performed using the Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/) (49). For a type I error rate of 0.05, the Gambian case–control and family studies had 85.7 and 98.7% power, respectively. This is based on an allelic OR of 1.3 and a high-risk allele frequency of 0.25 (the average minor allele frequency of the SNPs screened in the Gambians). With lower allele frequencies (0.1, 0.05 and 0.01), the case–control studies (each roughly 700 cases and 700 controls) had lower power, 59.1, 36 and 11.4%, respectively. The additional family studies (approximately 250 trios) each had ∼25.8, 16.0 and 7.2% power. The power of the combined case–control study (2945 cases and 2736 controls) was 99.3, 89.6 and 31.5%. The combined family study (1747 trios) had 93.3, 71.6 and 21.3% power.
SNP selection.
SNP data from HapMap release 21a (January 2007, http://www.hapmap.org) within 10 kb of the CD36 gene was downloaded and assessed using HAPLOVIEW (version 3.32) (50). Polymorphic SNPs (minor allele frequency >5%) were identified in the HapMap Yoruba. Tagging SNPs were defined using the TAGGER algorithm (51). Sequenom iPLEX genotyping assays were designed for 69 tagging SNPs and three additional variants, T1264G, G1439C and rs3173798 (located at a conserved splice site). Following quality control, 70 SNPs were accepted for association analysis. This final set of SNPs tagged 116 of 133 (87%) polymorphic markers (>5% frequency) across CD36 (including 10 kb up- and downstream), using pairwise tagging and an r2 >0.8 (mean r2 = 0.97), in the HapMap Yoruba.
Sample preparation and genotyping.
Genomic DNA samples used in the association analysis underwent whole-genome amplification through either primer extension pre-amplification (52) or multiple displacement amplification (53), before Sequenom iPLEX genotyping. Genotype data underwent quality control for missing data (<10%), Hardy–Weinberg equilibrium (P > 0.001) and Mendelian error rate. Genotype counts and quality control measures are documented in Supplementary Material, Table S6.
Statistical analysis.
Case–control association analysis was performed by logistic regression using covariates of ethnic group, gender and haemoglobin S (HbS) status. Sequenom genotyping for the HbS variant was performed for all samples as described previously (54). Family-based association analysis was performed using a case–pseudo-control approach and conditional logistic regression based on parental genotypes. Trios were drawn from a larger pool of samples assessed for relationship misspecification by Mendelian error rates. We utilized the R statistical application (version 2.6.0) along with the dgc.genetics library (http://www-gene.cimr.cam.ac.uk/clayton/software/) and the SNPassoc package (55). In both family- and population-based studies, we tested each marker in allelic (or ‘multiplicative’), dominant, recessive and overdominant (or ‘heterozygote advantage’) genetic models and with three phenotypes (CM, SA and all severe cases). Pooling across case–control and family studies was performed using the UNPHASED application (version 3.0.12) (http://www.mrc-bsu.cam.ac.uk/personal/frank/software/unphased/) (56). Ethnic origin was a significant confounder and was retained as a covariate in the UNPHASED analysis.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by the Wellcome Trust (GR074586AIA) through a Clinical Research Training Fellowship to A.E.F. and UK Medical Research Council funding (to D.P.K.). T.N.W. was funded by the Wellcome Trust, MalariaGEN and the European Union Network 6 BioMalPar Consortium. The work of the Ghanaian study was supported by funding from the National Institute of Allergy and Infectious Diseases/National Institutes of Health (contract number AI95363) to the Noguchi Memorial Institute for Medical Research. Funding to pay the Open Access charge was provided by the Wellcome Trust.
ACKNOWLEDGEMENTS
We would like to thank the MalariaGEN consortium for providing access to the Illumina 650Y Gambian parent–offspring trio data. This manuscript is published with the permission of the director of the Kenya Medical Research Institute. The views expressed here are the personal ones of the authors and do not purport to represent those of the United States Department of the Navy.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Newbold C., Craig A., Kyes S., Rowe A., Fernandez-Reyes D., Fagan T. Cytoadherence, pathogenesis and the infected red cell surface in Plasmodium falciparum. Int. J. Parasitol. 1999;29:927–937. doi: 10.1016/s0020-7519(99)00049-1. [DOI] [PubMed] [Google Scholar]
- 2.Heddini A., Pettersson F., Kai O., Shafi J., Obiero J., Chen Q., Barragan A., Wahlgren M., Marsh K. Fresh isolates from children with severe Plasmodium falciparum malaria bind to multiple receptors. Infect. Immun. 2001;69:5849–5856. doi: 10.1128/IAI.69.9.5849-5856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rogerson S.J., Tembenu R., Dobano C., Plitt S., Taylor T.E., Molyneux M.E. Cytoadherence characteristics of Plasmodium falciparum-infected erythrocytes from Malawian children with severe and uncomplicated malaria. Am. J. Trop. Med. Hyg. 1999;61:467–472. doi: 10.4269/ajtmh.1999.61.467. [DOI] [PubMed] [Google Scholar]
- 4.Udomsangpetch R., Reinhardt P.H., Schollaardt T., Elliott J.F., Kubes P., Ho M. Promiscuity of clinical Plasmodium falciparum isolates for multiple adhesion molecules under flow conditions. J. Immunol. 1997;158:4358–4364. [PubMed] [Google Scholar]
- 5.Baruch D.I., Gormely J.A., Ma C., Howard R.J., Pasloske B.L. Plasmodium falciparum erythrocyte membrane protein 1 is a parasitized erythrocyte receptor for adherence to CD36, thrombospondin, and intercellular adhesion molecule 1. Proc. Natl Acad. Sci. USA. 1996;93:3497–3502. doi: 10.1073/pnas.93.8.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Febbraio M., Hajjar D.P., Silverstein R.L. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J. Clin. Invest. 2001;108:785–791. doi: 10.1172/JCI14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Urban B.C., Willcox N., Roberts D.J. A role for CD36 in the regulation of dendritic cell function. Proc. Natl Acad. Sci., USA. 2001;98:8750–8755. doi: 10.1073/pnas.151028698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pain A., Ferguson D.J., Kai O., Urban B.C., Lowe B., Marsh K., Roberts D.J. Platelet-mediated clumping of Plasmodium falciparum-infected erythrocytes is a common adhesive phenotype and is associated with severe malaria. Proc. Natl Acad. Sci. USA. 2001;98:1805–1810. doi: 10.1073/pnas.98.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikeda H., Mitani T., Ohnuma M., Haga H., Ohtzuka S., Kato T., Nakase T., Sekiguchi S. A new platelet-specific antigen, Naka, involved in the refractoriness of HLA-matched platelet transfusion. Vox Sang. 1989;57:213–217. doi: 10.1111/j.1423-0410.1989.tb00826.x. [DOI] [PubMed] [Google Scholar]
- 10.Urwijitaroon Y., Barusrux S., Romphruk A., Puapairoj C. Frequency of human platelet antigens among blood donors in northeastern Thailand. Transfusion. 1995;35:868–870. doi: 10.1046/j.1537-2995.1995.351096026370.x. [DOI] [PubMed] [Google Scholar]
- 11.Curtis B.R., Aster R.H. Incidence of the Nak(a)-negative platelet phenotype in African Americans is similar to that of Asians. Transfusion. 1996;36:331–334. doi: 10.1046/j.1537-2995.1996.36496226147.x. [DOI] [PubMed] [Google Scholar]
- 12.Aitman T.J., Cooper L.D., Norsworthy P.J., Wahid F.N., Gray J.K., Curtis B.R., McKeigue P.M., Kwiatkowski D., Greenwood B.M., Snow R.W., et al. Malaria susceptibility and CD36 mutation. Nature. 2000;405:1015–1016. doi: 10.1038/35016636. [DOI] [PubMed] [Google Scholar]
- 13.Yanai H., Chiba H., Fujiwara H., Morimoto M., Abe K., Yoshida S., Takahashi Y., Fuda H., Hui S.P., Akita H., et al. Phenotype–genotype correlation in CD36 deficiency types I and II. Thromb. Haemost. 2000;84:436–441. [PubMed] [Google Scholar]
- 14.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabeti P.C., Reich D.E., Higgins J.M., Levine H.Z., Richter D.J., Schaffner S.F., Gabriel S.B., Platko J.V., Patterson N.J., McDonald G.J., et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 16.Sabeti P.C., Schaffner S.F., Fry B., Lohmueller J., Varilly P., Shamovsky O., Palma A., Mikkelsen T.S., Altshuler D., Lander E.S. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 17.Pain A., Urban B.C., Kai O., Casals-Pascual C., Shafi J., Marsh K., Roberts D.J. A non-sense mutation in Cd36 gene is associated with protection from severe malaria. Lancet. 2001;357:1502–1503. doi: 10.1016/S0140-6736(00)04662-6. [DOI] [PubMed] [Google Scholar]
- 18.Amodu O.K., Gbadegesin R.A., Ralph S.A., Adeyemo A.A., Brenchley P.E., Ayoola O.O., Orimadegun A.E., Akinsola A.K., Olumese P.E., Omotade O.O. Plasmodium falciparum malaria in south-west Nigerian children: is the polymorphism of ICAM-1 and E-selectin genes contributing to the clinical severity of malaria? Acta Trop. 2005;95:248–255. doi: 10.1016/j.actatropica.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 19.Ayodo G., Price A.L., Keinan A., Ajwang A., Otieno M.F., Orago A.S., Patterson N., Reich D. Combining evidence of natural selection with association analysis increases power to detect malaria-resistance variants. Am. J. Hum. Genet. 2007;81:234–242. doi: 10.1086/519221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bamshad M.J., Wooding S., Watkins W.S., Ostler C.T., Batzer M.A., Jorde L.B. Human population genetic structure and inference of group membership. Am. J. Hum. Genet. 2003;72:578–589. doi: 10.1086/368061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qamar R., Ayub Q., Mohyuddin A., Helgason A., Mazhar K., Mansoor A., Zerjal T., Tyler-Smith C., Mehdi S.Q. Y-chromosomal DNA variation in Pakistan. Am. J. Hum. Genet. 2002;70:1107–1124. doi: 10.1086/339929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fry A.E., Trafford C.J., Kimber M.A., Chan M.S., Rockett K.A., Kwiatkowski D.P. Haplotype homozygosity and derived alleles in the human genome. Am. J. Hum. Genet. 2006;78:1053–1059. doi: 10.1086/504160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asch A.S., Barnwell J., Silverstein R.L., Nachman R.L. Isolation of the thrombospondin membrane receptor. J. Clin. Invest. 1987;79:1054–1061. doi: 10.1172/JCI112918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lepretre F., Linton K.J., Lacquemant C., Vatin V., Samson C., Dina C., Chikri M., Ali S., Scherer P., Seron K., et al. Genetic study of the CD36 gene in a French diabetic population. Diabetes Metab. 2004;30:459–463. doi: 10.1016/s1262-3636(07)70143-x. [DOI] [PubMed] [Google Scholar]
- 25.Love-Gregory L., Sherva R., Sun L., Wasson J., Schappe T., Doria A., Rao D.C., Hunt S.C., Klein S., Neuman R.J., et al. Variants in the CD36 gene associate with the metabolic syndrome and high-density lipoprotein cholesterol. Hum. Mol. Genet. 2008;17:1695–1704. doi: 10.1093/hmg/ddn060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savill J., Hogg N., Haslett C. Macrophage vitronectin receptor, CD36, and thrombospondin cooperate in recognition of neutrophils undergoing programmed cell death. Chest. 1991;99:6S–7S. doi: 10.1378/chest.99.3_supplement.6s-a. [DOI] [PubMed] [Google Scholar]
- 27.Dawson D.W., Pearce S.F., Zhong R., Silverstein R.L., Frazier W.A., Bouck N.P. CD36 mediates the in vitro inhibitory effects of thrombospondin-1 on endothelial cells. J. Cell Biol. 1997;138:707–717. doi: 10.1083/jcb.138.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laugerette F., Passilly-Degrace P., Patris B., Niot I., Febbraio M., Montmayeur J.P., Besnard P. CD36 involvement in orosensory detection of dietary lipids, spontaneous fat preference, and digestive secretions. J. Clin. Invest. 2005;115:3177–3184. doi: 10.1172/JCI25299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Curtis B.R., Ali S., Glazier A.M., Ebert D.D., Aitman T.J., Aster R.H. Isoimmunization against CD36 (glycoprotein IV): description of four cases of neonatal isoimmune thrombocytopenia and brief review of the literature. Transfusion. 2002;42:1173–1179. doi: 10.1046/j.1537-2995.2002.00176.x. [DOI] [PubMed] [Google Scholar]
- 30.Yanai H., Watanabe I., Ishii K., Morimoto M., Fujiwara H., Yoshida S., Hui S.P., Matsuno K., Chiba H. Attenuated aerobic exercise capacity in CD36 deficiency. J. Med. Genet. 2007;44:445–447. doi: 10.1136/jmg.2007.050070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGilvray I.D., Serghides L., Kapus A., Rotstein O.D., Kain K.C. Nonopsonic monocyte/macrophage phagocytosis of Plasmodium falciparum-parasitized erythrocytes: a role for CD36 in malarial clearance. Blood. 2000;96:3231–3240. [PubMed] [Google Scholar]
- 32.Smith T.G., Serghides L., Patel S.N., Febbraio M., Silverstein R.L., Kain K.C. CD36-mediated nonopsonic phagocytosis of erythrocytes infected with stage I and IIA gametocytes of Plasmodium falciparum. Infect. Immun. 2003;71:393–400. doi: 10.1128/IAI.71.1.393-400.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Omi K., Ohashi J., Patarapotikul J., Hananantachai H., Naka I., Looareesuwan S., Tokunaga K. CD36 polymorphism is associated with protection from cerebral malaria. Am. J. Hum. Genet. 2003;72:364–374. doi: 10.1086/346091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newbold C., Warn P., Black G., Berendt A., Craig A., Snow B., Msobo M., Peshu N., Marsh K. Receptor-specific adhesion and clinical disease in Plasmodium falciparum. Am. J. Trop. Med. Hyg. 1997;57:389–398. doi: 10.4269/ajtmh.1997.57.389. [DOI] [PubMed] [Google Scholar]
- 35.Traore B., Muanza K., Looareesuwan S., Supavej S., Khusmith S., Danis M., Viriyavejakul P., Gay F. Cytoadherence characteristics of Plasmodium falciparum isolates in Thailand using an in vitro human lung endothelial cells model. Am. J. Trop. Med. Hyg. 2000;62:38–44. doi: 10.4269/ajtmh.2000.62.38. [DOI] [PubMed] [Google Scholar]
- 36.Turner G.D., Morrison H., Jones M., Davis T.M., Looareesuwan S., Buley I.D., Gatter K.C., Newbold C.I., Pukritayakamee S., Nagachinta B., et al. An immunohistochemical study of the pathology of fatal malaria. Evidence for widespread endothelial activation and a potential role for intercellular adhesion molecule-1 in cerebral sequestration. Am. J. Pathol. 1994;145:1057–1069. [PMC free article] [PubMed] [Google Scholar]
- 37.Silamut K., Phu N.H., Whitty C., Turner G.D., Louwrier K., Mai N.T., Simpson J.A., Hien T.T., White N.J. A quantitative analysis of the microvascular sequestration of malaria parasites in the human brain. Am. J. Pathol. 1999;155:395–410. doi: 10.1016/S0002-9440(10)65136-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kashiwagi H., Tomiyama Y., Honda S., Kosugi S., Shiraga M., Nagao N., Sekiguchi S., Kanayama Y., Kurata Y., Matsuzawa Y. Molecular basis of CD36 deficiency. Evidence that a 478C–>T substitution (proline90–>serine) in CD36 cDNA accounts for CD36 deficiency. J. Clin. Invest. 1995;95:1040–1046. doi: 10.1172/JCI117749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kashiwagi H., Tomiyama Y., Nozaki S., Kiyoi T., Tadokoro S., Matsumoto K., Honda S., Kosugi S., Kurata Y., Matsuzawa Y. Analyses of genetic abnormalities in type I CD36 deficiency in Japan: identification and cell biological characterization of two novel mutations that cause CD36 deficiency in man. Hum. Genet. 2001;108:459–466. doi: 10.1007/s004390100525. [DOI] [PubMed] [Google Scholar]
- 40.Longmate J.A. Complexity and power in case–control association studies. Am. J. Hum. Genet. 2001;68:1229–1237. doi: 10.1086/320106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slager S.L., Huang J., Vieland V.J. Effect of allelic heterogeneity on the power of the transmission disequilibrium test. Genet. Epidemiol. 2000;18:143–156. doi: 10.1002/(SICI)1098-2272(200002)18:2<143::AID-GEPI4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 42.Cann H.M., de Toma C., Cazes L., Legrand M.F., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 43.Rosenberg N.A. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann. Hum. Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- 44.Stephens M., Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stephens M., Smith N.J., Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sabeti P.C., Walsh E., Schaffner S.F., Varilly P., Fry B., Hutcheson H.B., Cullen M., Mikkelsen T.S., Roy J., Patterson N., et al. The case for selection at CCR5-Delta32. PLoS Biol. 2005;3:e378. doi: 10.1371/journal.pbio.0030378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Division of Control of Tropical Diseases, World Health Organization. Severe and complicated malaria. Trans. R. Soc. Trop. Med. Hyg. 1990;84(Suppl. 2):1–65. [PubMed] [Google Scholar]
- 48.Marsh K., Forster D., Waruiru C., Mwangi I., Winstanley M., Marsh V., Newton C., Winstanley P., Warn P., Peshu N., et al. Indicators of life-threatening malaria in African children. N. Engl. J. Med. 1995;332:1399–1404. doi: 10.1056/NEJM199505253322102. [DOI] [PubMed] [Google Scholar]
- 49.Purcell S., Cherny S.S., Sham P.C. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 50.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 51.de Bakker P.I., Yelensky R., Pe'er I., Gabriel S.B., Daly M.J., Altshuler D. Efficiency and power in genetic association studies. Nat. Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L., Cui X., Schmitt K., Hubert R., Navidi W., Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc. Natl Acad. Sci. USA. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonzalez J.M., Portillo M.C., Saiz-Jimenez C. Multiple displacement amplification as a pre-polymerase chain reaction (pre-PCR) to process difficult to amplify samples and low copy number sequences from natural environments. Environ. Microbiol. 2005;7:1024–1028. doi: 10.1111/j.1462-2920.2005.00779.x. [DOI] [PubMed] [Google Scholar]
- 54.Fry A.E., Griffiths M.J., Auburn S., Diakite M., Forton J.T., Green A., Richardson A., Wilson J., Jallow M., Sisay-Joof F., et al. Common variation in the ABO glycosyltransferase is associated with susceptibility to severe Plasmodium falciparum malaria. Hum. Mol. Genet. 2008;17:567–576. doi: 10.1093/hmg/ddm331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gonzalez J.R., Armengol L., Sole X., Guino E., Mercader J.M., Estivill X., Moreno V. SNPassoc: an R package to perform whole genome association studies. Bioinformatics. 2007;23:644–645. doi: 10.1093/bioinformatics/btm025. [DOI] [PubMed] [Google Scholar]
- 56.Dudbridge F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum. Hered. 2008;66:87–98. doi: 10.1159/000119108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.