SUMMARY
Drosophila Dscam encodes a vast family of immunoglobulin (Ig)-containing proteins that exhibit isoformspecific homophilic binding. This diversity is essential for cell recognition events required for wiring the brain. Each isoform binds to itself but rarely to other isoforms. Specificity is determined by “matching” of three variable Ig domains within an ∼220 kD ectodomain. Here, we present the structure of the homophilic binding region of Dscam, comprising the eight N-terminal Ig domains (Dscam1−8). Dscam1−8 forms a symmetric homodimer of S-shaped molecules. This conformation, comprising two reverse turns, allows each pair of the three variable domains to “match” in an antiparallel fashion. Structural, genetic, and biochemical studies demonstrate that, in addition to variable domain “matching,” intramolecular interactions between constant domains promote homophilic binding. These studies provide insight into how “matching” at all three pairs of variable domains in Dscam mediates isoform-specific recognition.
INTRODUCTION
Molecular recognition regulates cellular interactions in many biological contexts. So that complex intercellular interactions can be carried out, a repertoire of recognition proteins has evolved through diversification of conserved domains. The prime example is the immunoglobulin (Ig) domain, which provides an evolutionarily conserved scaffold found in many recognition proteins (called Ig superfamily [IgSF] proteins). These proteins regulate diverse cellular processes, including morphogenesis, growth, differentiation, neuronal wiring, and the immune response. Molecular diversification within IgSFs, such as antibodies and T cell receptors, gives rise to vast repertoires of structurally related proteins that exhibit distinct recognition specificities. Recent studies have shown that molecular diversification of Dscam (Down syndrome cell adhesion molecule) IgSF proteins in Drosophila is essential for wiring the fly brain (Chen et al., 2006; Hattori et al., 2007). The molecular recognition provided by Dscam diversity plays a crucial role in regulating cellular interactions necessary for neural circuit assembly (Hughes et al., 2007; Matthews et al., 2007; Soba et al., 2007; Wojtowicz et al., 2004; Zhan et al., 2004).
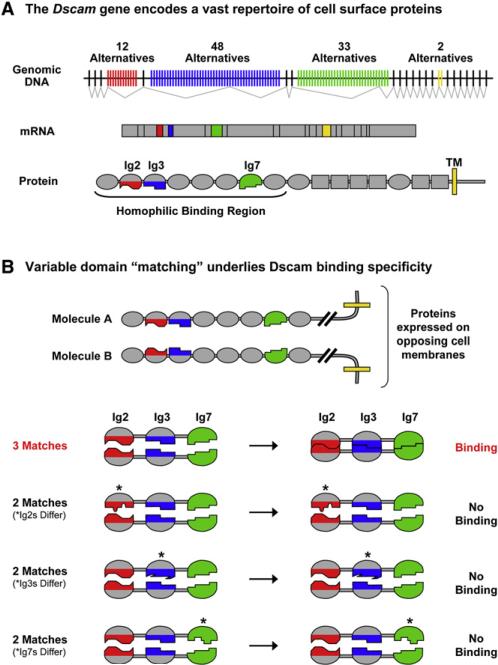
The key to Dscam function is the ability of isoforms to distinguish between each other with high fidelity. These isoforms share a common domain structure and contain variable amino acid sequences within three Ig domains (Schmucker et al., 2000)(Figure 1A). The variable domains mediate isoform-specific homophilic binding of Dscam proteins (Wojtowicz et al., 2004). Structural and biochemical studies have provided some clues to the molecular basis of this binding specificity. The three variable domains (i.e., Ig2, Ig3, and Ig7) engage in self-binding or “matching” in a modular fashion; Ig2 in one molecule matches Ig2 in an opposing molecule, Ig3 matches Ig3, and Ig7 matches Ig7 (Wojtowicz et al., 2004, 2007; Meijers et al., 2007)(Figure 1B). Only identical opposing domains (with rare exceptions) match each other. The self-binding properties of identical domains demonstrated that the Dscam gene potentially gives rise to 18,048 (i.e., 12 Ig2s × 47 Ig3s × 32 Ig7s) isoform-specific homophilic binding proteins (Wojtowicz et al., 2007). And, because matching at all three variable domains is required for binding to occur between opposing molecules, binding is largely restricted to isoforms sharing identity at all three variable domains. Indeed, in all but a small fraction of cases, no binding above the threshold of the assay, rather than weaker binding, is observed between isoforms that differ at only one of the three variable domains, resulting in “all-or-none” binding.
Figure 1. Dscam Gives Rise to a Vast Family of Isoform-Specific Homophilic Binding Proteins.
(A) Dscam encodes cell surface molecules comprising immunoglobulin (Ig) domains (ovals), fibronectin type III domains (rectangles), a transmembrane domain (yellow line), and a cytoplasmic tail. The Dscam gene contains four variable exon blocks. Isoforms contain one variant from each block. These encode the N-terminal halves of Ig2 (red; 12 alternatives) and Ig3 (blue; 48 alternatives), all of Ig7 (green; 33 alternatives), and the transmembrane domain (yellow; two alternatives). Dscam proteins engage in isoform-specific homophilic binding. Biochemical studies led to the proposal that 18,048 of the 19,008 ectodomains encoded by the locus mediate isoform-specific homophilic binding (Wojtowicz et al., 2007). The minimal region of the ectodomain required to support homophilic binding includes the N-terminal eight Ig domains (Dscam1−8) (Wojtowicz et al., 2004).
(B) Schematic illustration of the principles underlying Dscam isoform-specific homophilic binding. Top: The homophilic binding regions of Dscam proteins expressed on opposing cell surfaces. Bottom: Only the Ig2, Ig3, and Ig7 variable domains in two opposing isoforms are shown. “Matching” (as represented by the same shape) at all three variable domains is required for protein binding (with rare exceptions).
Recent studies have demonstrated that each of the three variable domains engages in matching via a two-fold symmetric interface. A dimer present in the crystal structure of the first four Ig domains of Dscam demonstrated that the Ig2 and Ig3 interfaces comprise residues along one strand within the Ig domain (Meijers et al., 2007), whereas biochemical and molecular modeling studies argued that the Ig7 interface is formed by multiple strands comprising one face of the Ig domain (Wojtowicz et al., 2007). That these self-binding interface regions reside at the interface of many, if not all, variants of each domain and that these regions are sufficient to confer self-binding specificity was demonstrated through biochemical studies of many chimeric variable Ig domains (Wojtowicz et al., 2007).
Despite these advances, the molecular basis of the curious “all-or-none” binding specificity of Dscam isoforms remained an enigma. For instance, why must all three pairs of variable domains match for binding to occur? And, furthermore, how is self-binding at each variable domain maintained in such a highly specific manner? Binding studies have revealed that a single interface residue difference between one pair of opposing variable domains is sufficient to prevent binding between isoforms even when they are identical at the other two variable domain pairs (Wojtowicz et al., 2007). These binding properties present a remarkable example of binding specificity and raise the question of how such small differences at only one of three binding domains have such a dramatic effect on the overall binding of Dscam proteins. The crystal structure of the homophilic binding region of Dscam comprising the N-terminal eight Ig domains (Dscam1−8) and additional modeling studies described herein illuminate the molecular basis of this “all-or-none” Dscam binding specificity.
RESULTS
Structure Determination
A segment comprising the N-terminal eight Ig domains of Dscam that we term Dscam1−8 was chosen for structural studies because it contains all three variable domains and is sufficient to specify homophilic binding, whereas fragments containing the N-terminal six Ig domains or fewer do not support homophilic binding (Wojtowicz et al., 2004). The crystal structure of Dscam1−8 containing variable domains Ig2.1, Ig3.30, and Ig7.30 was solved by molecular replacement using a structure of the first four N-terminal Ig domains (Dscam1−4; PDB ID 2V5M) (Meijers et al., 2007) as a search model. The Dscam1−4 structure used for molecular replacement contains the same Ig2 variable domain but a different Ig3 variable domain (i.e., Ig3.34) that differs in 23 out of 40 variable residues. The remaining Ig domains (i.e., Ig5−Ig8) could be visualized in difference Fourier maps and were manually positioned. Three Dscam1−8 molecules were found in the asymmetric unit. Molecules A and B pair together in a non-crystallographic dimer, and molecule C pairs with a copy of itself about the two-fold crystallographic b axis, offering two independent views of the Dscam dimer. All residues are modeled in molecule A, but Ig8 is disordered in molecules B and C. The final model has been refined at 4.2 Å resolution to a crystallographic R factor of 28.0% and Rfree of 32.7% (Table S1 available online).
The Serpentine Fold of the Dscam Molecule
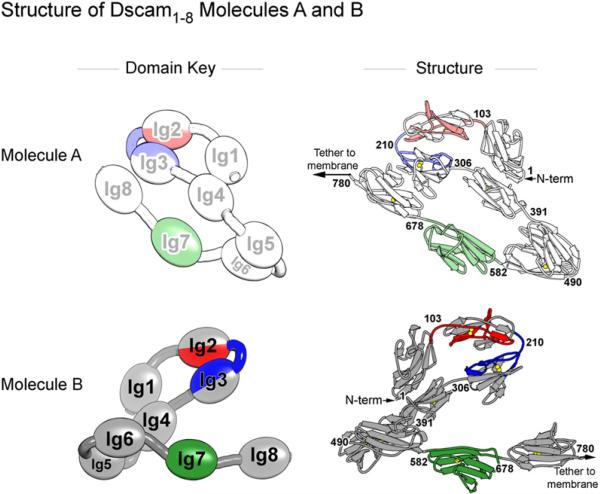
The Ig domains of Dscam1−8 trace out a serpentine, or “S,” shape (Figure 2). Domains Ig1–Ig4 form a rigid structure comprising the top half of the “S,” a shape corresponding to the previously described horseshoe configuration of two Dscam1−4 isoforms (one containing Ig2.1 and Ig3.34 [Dscam1−4 (1.34)] and the other containing Ig2.9 and Ig3.9 [Dscam1−4 (9.9)]) (Meijers et al., 2007), hemolin1−4 (Su et al., 1998), and axonin-11−4 / TAG-11−4 (Freigang et al., 2000; Mörtl et al., 2007). Rigidity within the horseshoe is maintained by extensive interfaces between Ig1 and Ig4 (1351 Å2 surface area involving strands A and G of Ig1 and strands F and G of Ig4) and between Ig2 and Ig3 (1416 Å2 surface area involving strands B, E, and D of Ig2 and Ig3). Indeed, these interfaces are so rigid that the root mean square deviation (RMSD) for 380 out of a total 388 αC's is less than 1.3 Å for all pairwise comparisons of eight Dscam structures: molecules A, B, and C of Dscam1−8, three independent views of Dscam1−4 (1.34) (PDB ID 2V5M and 2V5R [chains A and B]), and two independent views of Dscam1−4 (9.9) (PDB ID 2V5S, chains A and B). This similarity is observed despite only 40%−50% amino acid sequence identity between variable domains in these structures.
Figure 2. Crystal Structure of Dscam1−8 Molecules A and B.
Dscam1−8 forms a dimer (see Figure 3) containing molecules A and B, shown separately. Left: Schematics illustrate orientation of Ig domains within each molecule. Molecules A and B adopt an S shape. Molecule B is flipped 180 degrees relative to molecule A, revealing the back face. The domains are colored as in Figure 1. Molecule A is represented as a lighter version of these colors. Right: Crystal structures of molecules A and B shown in cartoon. Ig8 in molecule B is modeled with coordinates from model A. Sulphur atoms in disulfide bonds are drawn as spheres. Numbers on the structures indicate amino acid residues at Ig domain boundaries. N terminus is indicated. The remaining eight C-terminal domains of Dscam (not present in the crystal structure) would presumably lead to the two opposing cells (left for molecule A and right for molecule B). Domains Ig1–Ig4 comprise the “horseshoe” in the top half of the “S.” Domains Ig5–Ig8 comprise the bottom half of the “S.”
Domains Ig5–Ig8 form a reverse turn in the bottom half of the “S.” There are no large interfaces between any pair of domains in Ig5–Ig8, suggesting flexibility in their hinges (Table S2). Only the Ig5:Ig6 interface has any appreciable surface area, 842 Å2. Other intramolecular domain-domain interfaces are less than 443 Å2. Indeed, hinge flexibility is evident in the comparison of the three molecules in the asymmetric unit: the hinge angle between Ig4 and Ig5 varies up to 33°; the hinge angle between Ig6 and Ig7 varies up to 13° (Figure S1); and the hinge angle between Ig7 and Ig8 is so variable that Ig8 is disordered in molecules B and C (Table S3). Notably, the Ig5-Ig6 hinge is the only domain-domain hinge in the bottom half of the “S” that shows a complete lack of flexibility (variation less than 0.5°), in keeping with its relatively larger interface.
Quaternary Fold of the Dscam Dimer
Two S-shaped Dscam1−8 molecules associate in a two-fold symmetric dimer (Figure 3). The dimer interface buries 4500 Å2 of surface area, 80% of which is contributed by the three pairs of variable domains, Ig2-Ig2, Ig3-Ig3, and Ig7-Ig7. The three variable domains assume positions in the “S” curve roughly where the staff in a dollar sign (“$”) crosses the “S.” That is, the three variable domains are positioned in a line. Parallel to this line, the two-fold symmetry axis passes between the two S-shaped Dscam molecules marking the center of the Ig2-Ig2, Ig3-Ig3, and Ig7-Ig7 interfaces. The Ig2-Ig2 and Ig3-Ig3 contacts reside in the top half of the “S” flanking the horseshoe bend and lie in close proximity to each other (18 Å apart). The Ig7-Ig7 contact resides in the bottom half of the “S” and is separated from the Ig3-Ig3 contact by a variable distance of ∼32−45 Å through the flexibly linked Ig4-Ig5 and Ig6-Ig7 hinges (Figure S1). This flexibility leads to high RMS deviations when the two dimers in their entirety are superimposed (RMSD 6.1 Å over 670 αC pairs). But, taken individually, the Ig2-Ig2, Ig3-Ig3, and Ig7-Ig7 interfaces superimpose very well (RMSD 0.2 Å over 210 αC pairs, 0.1 Å over 190 αC pairs, and 0.6 Å over 188 αC pairs, respectively).
Figure 3. Structure of Dscam1−8 Dimer.
View of the Dscam1−8 dimer from the front (A), side (B), bottom (C), and top (D). The dimer forms a two-fold symmetric double “S.” Schematics of the eyes in (A) domain key show the perspective illustrated in (B)–(D). Left: Schematics illustrate the orientation of the eight Ig domains within each molecule. Right: Structures of the dimer in stereo view. Ig8 in molecule B is modeled with coordinates from model A. Coloring is as in Figure 2. Front view (A) shows view of dimer perpendicular to the two-fold symmetry axis. Note that the intermolecular contacts between molecules A and B occur between opposing Ig2-Ig2, Ig3-Ig3, and Ig7-Ig7 domains. Black ovals (C and D) indicate the symmetry axis.
Structural comparison of the Ig1–Ig4 horseshoe dimer of Dscam1−8 with Dscam1−4 (1.34) (Meijers et al., 2007) suggests that the homophilic dimer pairing geometry reported here is conserved among different Dscam isoforms. The three independent views of the Ig1–Ig4 dimer in Dscam1−8 can be superimposed on the dimer of Dscam1−4 (1.34) with a RMSD of 1.5 Å for 686 αC pairs (there are 389 αCs in each Ig1–Ig4 segment or 778 αCs per dimer) (Figure S2). A single exception, the dimer of Dscam1−4 (9.9) (Meijers et al., 2007), differs by a 36° rotation and a 7 Å shift in registry of interacting β strands at the Ig2-Ig2 interface. This incongruity is further reflected in the RMSD of 13.4 Å for the superimposition of the Ig1–Ig4 dimers formed in Dscam1−8 and Dscam1−4 (9.9) (760 αC pairs). The difference could represent an alternate mode of Dscam1−4 dimerization; however, multiple lines of evidence suggest that the dimer interface observed in the Dscam1−4 (9.9) structure is instead a crystallization artifact (see Figure S2 and below).
The number of constant residues involved in the Dscam1−8 dimer interface is minimal, consistent with the homophilic binding properties of Dscam isoforms and the lack of heterophilic binding between isoforms. A few dimer contacts (395 Å2 of surface area) in the top half of the Dscam1−8 “S” involve constant residues of Ig1 and variable residues from Ig2 (Figure S3). These contacts position the Ig1 domains such that they abut either side of the Ig2-Ig2 interface, possibly to guide the proper registration between opposing Ig2 domains by inhibiting sliding across the relatively flat interface. There are also some dimer contacts between variable regions of Ig2 and Ig3 (about 630 Å2).
The intramolecular interface between domains Ig5 and Ig6 also plays an important role in dimer formation. The Ig5:Ig6 interface stabilizes a sharp bend between the domains that is required for bringing the variable domains into alignment for homophilic pairing. We discuss this intramolecular interface as well as the intermolecular variable domain interfaces separately in the following sections.
Ig2-Ig2 and Ig3-Ig3 Composite Interface
Intramolecular interactions between Ig1:Ig4 and Ig2:Ig3 within the Ig1–Ig4 horseshoe rigidly constrain the variable Ig2 and Ig3 domains within the molecule. As such, the Ig2-Ig2 and Ig3-Ig3 self-binding interfaces comprise two halves of a composite interface (Figures 4A and 4B). These findings are in agreement with the Dscam1−4 crystal structure (Meijers et al., 2007). We discuss each half of the Ig2-Ig2/Ig3-Ig3 composite interface separately in this section.
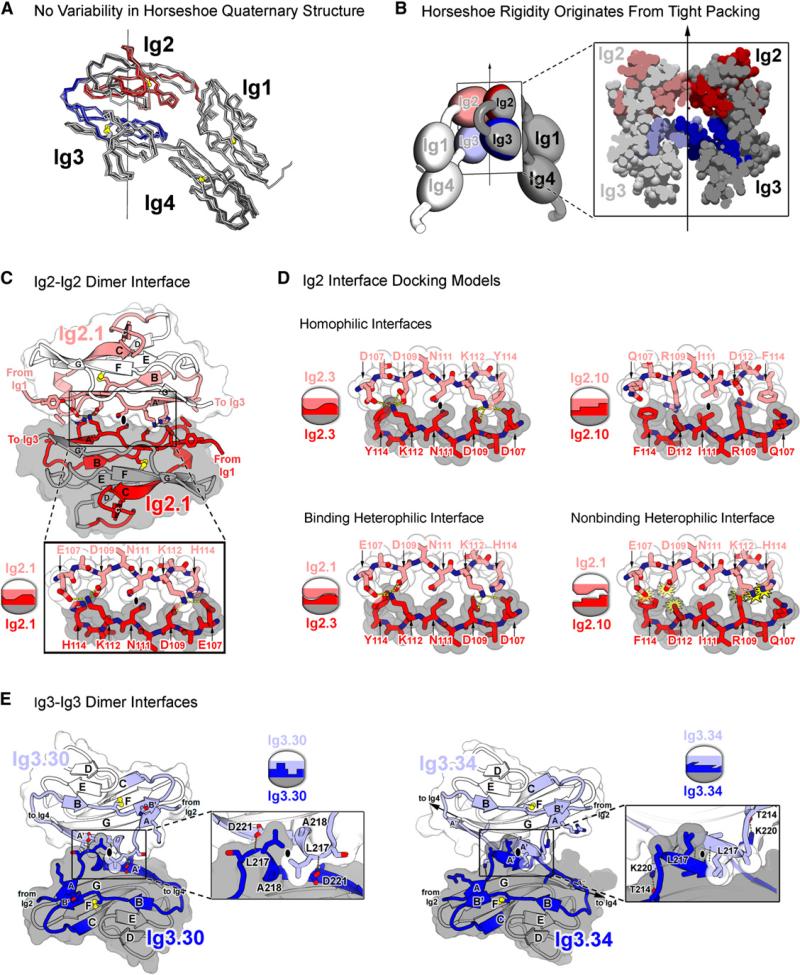
Figure 4. Ig2 and Ig3 Self-Binding Variable Domains Form a Composite Interface.
(A) Superimposition of horseshoe domains from eight different Dscam structures reveals lack of variability in domain-domain interactions, suggesting conformational rigidity in Ig1–Ig4. Structures superimposed (listed from dark to light) are the following: Dscam1−8, molecules A, B, and C; Dscam1−4 (1.34) chains A and B (PDB ID 2V5M and 2V5R); and Dscam1−4 (9.9) chains A and B (PDB ID 2V5S). RMS deviations in αCs are under 1.3 Å for all pairwise comparisons. Red and blue colors represent variable regions of Ig2 and Ig3, respectively.
(B) Rigidity of Ig1–Ig4 horseshoe originates from tight intramolecular packing of Ig2 and Ig3 within each molecule as illustrated via cross-section of the Ig1–Ig4 dimer. Light and dark shading distinguishes molecules in the dimer.
(C) Structure of Ig2.1-Ig2.1 interface in Dscam1−8 dimer. The Ig2-Ig2 interface is viewed down the two-fold symmetry axis. Molecule A is in lighter colors. The Ig2.1-Ig2.1 interface is two-fold symmetric and formed between identical segments along the A and A′ strands (residues 107−114). Opposing N111 residues pack against one another at the symmetry axis (black oval). Left and right networks flank the symmetry axis. K112 forms hydrogen bonds (H bonds) with E107 and D109. D109 additionally forms H bonds with H114. Intermolecular H bonds are drawn with yellow dashed lines. The Ig2.1 interface was previously described (Meijers et al., 2007).
(D) Ig2 interface docking models. Binding properties based on previous binding studies (Wojtowicz et al., 2007). Coloring is as in (A). Schematics illustrate presence or absence of electrostatic and shape complementarity at interface. Top: Ig2.3-Ig2.3 and the Ig2.10-Ig2.10 homophilic interfaces are shown. Opposing N111 and I111 residues, respectively, pack against one another at the symmetry axis (black oval). Flanking left and right networks comprise unique networks that exhibit electrostatic and shape complementarity. Bottom left: Ig2.1-Ig2.3 binding heterophilic interface is shown. These isoforms engage in heterophilic binding, albeit at levels lower than the homophilic binding of each. Lower levels of binding may be attributed to the left network, which is bulkier than its wild-type counterpart (D107-Y114 versus E107-H114), and may introduce steric constraint and to the right network, which is less bulky than its wild-type counterpart (D107-H114 versus D107-Y114) and may destabilize the H bonds. Bottom right: Ig2.1-Ig2.10 nonbinding heterophilic interface is shown. Electrostatic and shape noncomplementarity (yellow starbursts) are observed at the left and right networks. The left network contains three negatively charged residues (E107, D109, and D112). The right network contains steric clash between positively charged residues (K112 and R109).
(E) Structure of Ig3-Ig3 interfaces in Dscam1−8 and Dscam1−4 (1.34) dimers. View of Ig3-Ig3 interface down the two-fold symmetry axis (black ovals) is shown. Ig3.30-Ig3.30 (left) and Ig3.34-Ig3.34 (right; [Meijers et al., 2007]) dimer interfaces are shown. Molecule A is in lighter colors. Interface is formed between the identical transition segment between the A and A′ strands in opposing domains. In Ig3.34, this region forms a single turn of a helix; in Ig3.30, it forms a β strand. Both interfaces exhibit electrostatic and shape complementarity. Given the differences in structure, it is not surprising that these domains do not bind to each other (Wojtowicz et al., 2007).
The Ig2-Ig2 interface is located at the edge of Ig2's β sandwich fold and covers 1100 Å2 of surface area (Figure 4C). The Ig2 variant in Dscam1−8 was present in the Dscam1−4 (1.34) structure, and, as such, the interface is the same as that described by Meijers et al. (2007). It involves primarily polar residues (residues 105−116) in strands A and A′ and, to a lesser extent, the ends of strands B (residues 128−130), and G (residues 197 and 204). The primary dimer contacts between the A and A′ strands occur along an eight-residue segment (residues 107−114) oriented in an antiparallel fashion between molecules, giving rise to a two-fold symmetric interface comprising a central residue flanked by hydrogen bond networks. That this interface represents the interface formed during homophilic binding is supported by previous biochemical swapping experiments that demonstrated that this eight-residue segment is sufficient to confer the binding specificity of all 12 Ig2 variants (Wojtowicz et al., 2007).
To investigate how a unique binding specificity arises from each of the 12 Ig2 interface segments, we generated docking models for each variant using Rosetta (Figure 4D and Figures S4 and S5). All variable Ig2 interface segments adopt the same configuration observed in the crystal structures of Ig2.1, containing a central residue capable of packing against itself by two-fold rotational symmetry flanked by residues that accommodate the formation of a unique network. It is important to note that the Ig2.9 modeled interface differs from the interface observed in the crystal structure of Ig2.9 (Meijers et al., 2007) (see above and Figure S2). The salient feature of the 12 Ig2 interface models is that they all exhibit electrostatic and shape complementarity. The interfaces of the four Ig2 pairs that exhibit heterophilic binding and several pairs that do not were also modeled (binding properties based on Wojtowicz et al., 2007). In the four heterophilic cases, the interface segments exhibit electrostatic and shape complementarity (albeit to a lesser degree than the complementary observed between identical pairs). In all cases where binding does not occur between Ig2 variants, electrostatic and/or shape noncomplementarity is observed. These models suggest that binding specificity at the Ig2 interface is dependent upon both electrostatic and shape complementarity and that the Ig2.1 interface represents the self-binding interface of all 12 variable Ig2 domains.
The Ig3-Ig3 interface is also located at the edge of the Ig β sandwich (Figure 4E) but buries approximately half of the surface area of the Ig2-Ig2 interface (550 Å2). The Ig3-Ig3 interface contains mostly hydrophobic residues in a single turn of helix between the A and A′ strands (residues 216−222). This Ig3 interface structure differs locally from the Dscam1−4 (1.34) structure described previously (Meijers et al., 2007) because of the different Ig3 variant in Dscam1−8. That this segment resides at the interface formed during homophilic binding is supported by previous biochemical swapping experiments demonstrating that this segment is sufficient to confer the binding specificity of all Ig3 variants tested (Wojtowicz et al., 2007).
Ig7-Ig7 Interface
Ig7 domains interact in an antiparallel fashion through the face of one sheet in the β sandwich. Specifically, the interface comprises residues on strands A (residues 585−592), B (residues 600−609), E (residues 635−639), and D (residues 641−647) (Figure 5A). The directions of the strands A, B, E, and D are very nearly antiparallel to the directions of the equivalent strands across the two-fold symmetric interface. Thus, strand A interacts with strand A across the interface, strand B interacts with strand B, etc. The contacts formed between Ig7 domains are more extensive (approximately 1300 Å2 surface area buried) than those formed by the self-binding Ig2 and the self-binding Ig3 contacts but less than the composite Ig2-Ig2/Ig3-Ig3 interface (i.e., 1659 Å). The Ig7.30 interface in Dscam1−8 agrees well with the structure proposed through our previous biochemical and molecular modeling studies using other Ig7 variants (Wojtowicz et al., 2007). The Dscam1−8 Ig7.30 dimer can be superimposed on a docking model of Ig7.25 with an RMSD of 2.1 Å for 180/190 αC pairs despite a 62% difference in residues (i.e., 59/95) (Figure S6). Although a variety of Ig7 variants were used in the biochemical and modeling analyses, all biochemically characterized interface residues reside at the Ig7.30-Ig7.30 interface. These findings strongly suggest that the Ig7-Ig7 interface observed in Dscam1−8 can be generalized to most, if not all, Ig7 variants.
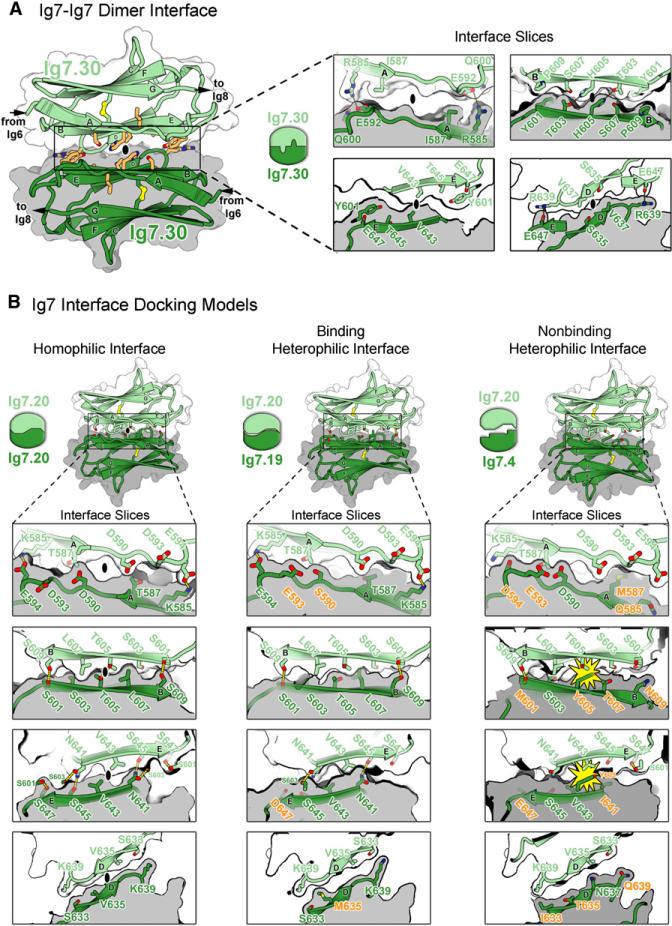
Figure 5. Ig7 Self-Binding Variable Domain Interfaces.
(A) Structure of the Ig7.30-Ig7.30 interface in the Dscam1−8 dimer. Left: View of Ig7-Ig7 interface down the two-fold symmetry axis (black oval). Molecule A is in lighter colors. The Ig7.30-Ig7.30 interface is two-fold symmetric and formed between identical segments along the ABED strands comprising one face of the Ig domain. The two domains are aligned so that their A, B, E, and D strands are nearly antiparallel. The residues represented in sticks (light orange) have been shown to affect binding specificity (Wojtowicz et al., 2007). Right: Slices of the Ig7.30-Ig7.30 interface taken from four different depths illustrate surface complementarity between subunits. Intermolecular H bonds are drawn as dashed yellow lines.
(B) Ig7 docking models. Binding properties are based on binding studies (Wojtowicz et al., 2007). Left: Ig7.20-Ig7.20 homophilic interface exhibits complementarity across all of the ABED strands. Middle: Despite several residue differences (orange), the Ig7.20-Ig7.19 heterophilic binding interface exhibits complementarity across all of the ABED strands. Lower levels of binding between Ig7.20-Ig7.19 as compared with the self-binding exhibited by each suggest these residue differences decrease the complementarity each exhibits with itself. Right: Multiple residue differences (orange) at the Ig7.20-Ig7.4 heterophilic nonbinding interface lead to electrostatic and shape noncomplementarity (yellow starbursts) between residues on the B and E strands. Given this noncomplementary, it is not surprising these domains do not bind to each other.
To investigate how a unique binding specificity arises from each of the 32 self-binding Ig7 variants (Ig7.33 doesn't exhibit self-binding), we generated docking models of ten Ig7 variants using Rosetta (Figure 5B, Figure S7, and data not shown). The docking models, along with the structure of Ig7.30, demonstrate that the Ig7 interface exhibits electrostatic and shape complementarity across the ABED interface strands. In cases where binding does not occur between Ig7 variants, electrostatic and/or shape noncomplementarity is observed. These models suggest that binding specificity at the Ig7 interface is dependent upon both electrostatic and shape complementarity and that the Ig7.30 interface represents the self-binding interface of many, if not all, variable Ig7 domains.
Ig5:Ig6 Intramolecular Interface
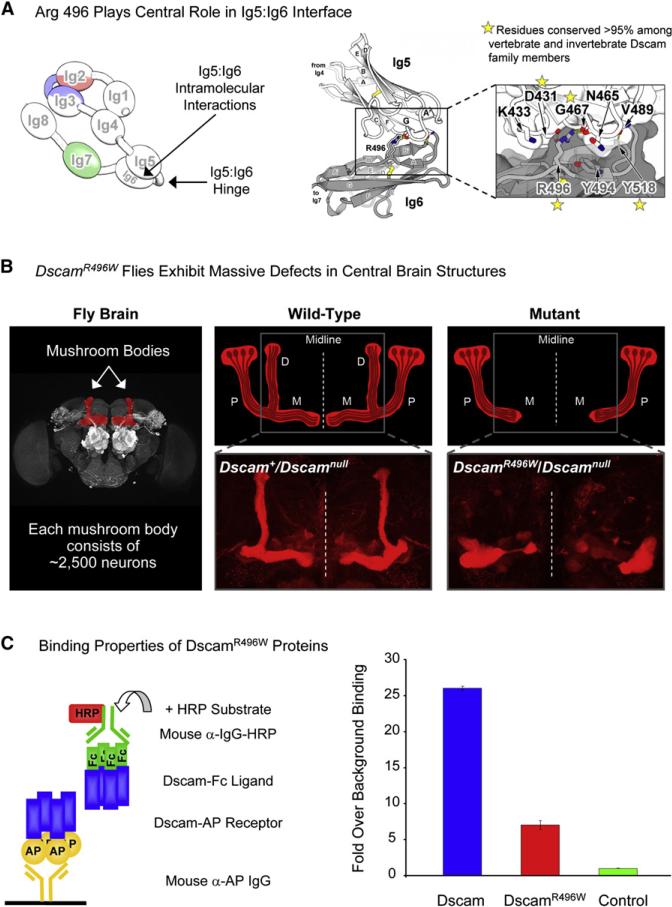
A sharp bend between Ig5 and Ig6 forms the bottom curve of the “S.” This bend allows more extensive intramolecular contacts to form between Ig5 and Ig6 than are observed between other tandem domains within the molecule (842 Å2 relative to ∼400 Å2) (see Table S2). Residue R496 located on the A strand of Ig6 plays a central role in the Ig5:Ig6 interface by forming hydrogen bonds with the backbone oxygen atoms of N465 and G467 in Ig5 (Figure 6A and Figure S8). Another hydrogen bond is formed between the backbone oxygen of V489 in Ig5 and the backbone nitrogen of Y518 in Ig6. There are also many van der Waals contacts between Ig5 and Ig6. That the Ig5:Ig6 interface plays a critical role in the formation of the homophilic dimer is supported by sequence conservation, genetic studies, and biochemical analysis. We discuss each of these below.
Figure 6. Intramolecular Ig5:Ig6 Interactions Are Important for Homophilic Binding In Vitro and Dscam Function In Vivo.
(A) Left: Schematic of molecule A. Right: The Ig5:Ig6 intramolecular interface. R496 in Ig6 makes significant contributions to the Ig5:Ig6 interface by forming H bonds with the backbone oxygen atoms of N465 and G467 in Ig5. Sequence alignment of 94 vertebrate and invertebrate Dscam proteins (see Figure S9) revealed that R496 and other residues are highly conserved.
(B) Flies containing the DscamR496W mutation exhibit phenotypes in neuronal patterning. Genotypes are as indicated. Left: Mushroom bodies within the central brain are highlighted in red. Upper middle and right: Schematics of mutant and wild-type. Four neurons within each mushroom body are shown. The cell body (oval) of each neuron resides in the Calyx. During development, axons branch at the base of the peduncle (P), giving rise to two sister neurites. Homophilic binding between identical sets of Dscam isoforms expressed on each sister neurite leads to repulsive interactions between sister neurites. Repulsion causes one sister neurite to extend into the dorsal lobe (D) and the other to extend into the medial lobe (M). Because sister branches share few, if any, isoforms with branches of other neurons, they selectively repel one another, leading to sister branch segregation with high fidelity. Gray box outlines region stained in lower panels. Lower middle and right: Axons in the D and M lobes are highlighted by anti-FasII staining. In DscamR496W flies, the D lobe is missing and the M lobe fails to extend to the midline (dotted line).
(C) DscamR496W mutant protein shows reduced homophilic binding. Left: Schematic illustration of ELISA-based binding assay utilized to assess homophilic binding is shown. Dscam ectodomain (blue) fused to alkaline phosphatase (Dscam-AP) is captured on a 96-well plate with anti-AP antibody. Dscam ectodomain fused to the Fc region of IgG (Dscam-Fc) is added and binding between Dscam-AP and Dscam-Fc is detected with anti-Fc antibody conjugated with horseradish peroxidase (HRP). Right: Homophilic binding levels of Dscam and DscamR496W protein are shown. The isoform used for binding is the same isoform used for crystallization studies of Dscam1−8. Binding was quantified by measurement of HRP enzymatic activity. A control isoform differing at all three variable domains was used to provide a value for background binding. Data are represented as average ± SD.
Alignment of Dscam1−8 protein sequences from 94 Dscam genes (including invertebrate Dscam, Dscam2, Dscam3, Dscam4 and vertebrate DSCAM and DSCAM-L) reveals that residues involved in the Ig5:Ig6 intramolecular interface are highly conserved. These include R496 (99% conserved; Aplysia californica contains K496), G467, Y518, and D431 (>95% conserved) and the composition of the hinge region (G491, L492, and P493), which facilitates the sharp bend between Ig5 and Ig6 (Figure S9).
In a genetic screen for recessive lethal alleles at the Dscam locus, we identified a missense mutation that retains normal protein levels (data not shown) and behaves as a loss-of-function allele. Remarkably, this allele contains a single point mutation resulting in the change R496W (i.e., DscamR496W). Similar to protein null Dscam mutations, DscamR496W flies exhibit gross phenotypes in a central brain structure called the mushroom body (Figure 6B). During development of the mushroom body, which comprises ∼2,500 neurons, each neuron grows in a nerve tract from the Calyx to the base of the peduncle, where it branches, giving rise to two sister neurites. One sister neurite extends within a nerve tract into the dorsal lobe and the other extends within a nerve tract into the medial lobe. In DscamR496W flies, the dorsal lobe nerve tract does not form and the medial lobe fails to extend to the midline. These phenotypes are 100% penetrant and exhibit similar but slightly weaker expressivity than Dscamnull.
Because DscamR496W flies exhibit a loss-of-function phenotype in vivo, we sought to assess whether this phenotype reflects a loss of homophilic binding. We tested the ability of DscamR496W mutant proteins to engage in homophilic binding in vitro using an enzyme-linked immunosorbent assay (ELISA)-based binding assay (Wojtowicz et al., 2007). DscamR496W mutant proteins exhibit an ∼70% reduction in homophilic binding (Figure 6C). It is likely that this reduction results from a disruption of the hydrogen bonds formed between R496 and the backbone carbonyl oxygen atoms of N465 and G467, leading to destabilization of the Ig5:Ig6 interface (Figure 6A).
In summary, these structural, genetic, and biochemical studies, combined with sequence conservation at the Ig5:Ig6 interface, indicate that the intramolecular Ig5:Ig6 interface plays an important role in formation of the S-shaped homophilic dimer.
DISCUSSION
The array of cell recognition proteins encoded by the Drosophila Dscam locus plays a crucial role in the formation of neuronal circuits. To function as unique identification tags in the developing nervous system, each of the possible 18,048 Dscam isoforms (Wojtowicz et al., 2007) must recognize itself and exclude all other isoforms. The Dscam1−8 structure provides insight into how this recognition is achieved.
The S Shape and Variable Domain Complementarity Underlie Dscam Binding Specificity
The vast majority of intermolecular dimer contacts in the Dscam1−8 crystal structure are formed by the two-fold symmetric interfaces of the three variable domains. Conceptually, achieving two-fold symmetry at all three variable domains would appear to pose an architectural challenge. That is, a reverse turn is required after each variable domain: one between Ig2 and Ig3 and one between Ig3 and Ig7. Dscam's S shape does this exactly, presenting an elegant solution to the problem of arranging two-fold symmetric dimer interfaces at all three variable domains. High sequence conservation of the Ig2-Ig3 and Ig5-Ig6 reverse turn hinges among invertebrate and vertebrate Dscam family members (see Figure S9) suggests that the double S-shape structure represents a stereotyped Dscam homophilic binding conformation.
Specificity at each variable domain interface arises from favorable electrostatic and shape complementarity, as demonstrated by the variable domain interfaces in the Dscam1−4(1.34) (Meijers et al., 2007) and Dscam1−8 crystal structures as well as our extensive modeling studies for Ig2 and Ig7. Although identical pairs fit together, nonidentical pairs (with rare exceptions) do not fit. Nonidentical pairs are destabilized by steric overlap, electrostatic repulsion, and poor shape complementarity between mismatched side chains across the heterophilic interface. Thus, noncomplementarity between nonidentical domains creates an inhibitory energy barrier that prevents the unfavorable interface from forming. For Ig3, this barrier may be further enhanced by structural differences (α-helical versus β strand) found at the Ig3 interface (Figure 4E).
In summary, the double “S” structure allows matching at all three variable domain interfaces, which fit together like pairs of children's building blocks.
Structural Insight into Variable Domain Modularity
Previous biochemical studies demonstrated that variable domain self-binding occurs in a modular fashion: thus, any combination of Ig2, Ig3, and Ig7 self-binding domains gives rise to a protein that exhibits isoform-specific homophilic binding (Wojtowicz et al., 2004, 2007). Modularity requires that variable sequences within a domain are interchangeable and compatible with alternate sequences in other variable domains. As such, structural changes due to sequence variation at the dimer interface of one pair of variable domains should not strain or disrupt the dimer interface at the other pairs of variable domains in the homodimer. The structure of the Dscam1−8 dimer reveals that this modularity is achieved in two distinct ways.
Modularity of Ig7 is ensured through a flexible region (linkers connecting Ig4 and Ig5, and Ig6 and Ig7) separating it from the Ig2 and Ig3 modules. Therefore, structural differences that arise from the presence of different Ig7 variants are unlikely to propagate to Ig2 and Ig3. Additionally, a comparison of Ig7 in the crystal structure and previous Ig7 modeling experiments (Wojtowicz et al., 2007) suggests that variation in Ig7 dimer geometry is small, perhaps less than 2 Å RMSD. In summary, the dimer interfaces of different Ig7 variants, though different in detail, are unlikely to stray far from the orientation described here, and these differences are unlikely to affect the Ig2 and Ig3 modules.
The Ig2 and Ig3 self-binding modules also function independently of each other, even though they are both part of the same rigid Ig1–Ig4 horseshoe and form a composite interface. Although no flexible linker insulates Ig2 from Ig3, sequence variations at the Ig2 and Ig3 interface segments cause only small, localized changes so that a constant distance and orientation between binding modules can be maintained, regardless of which combination of variants is encoded. This is evident in a comparison of Dscam1−8 and Dscam1−4 (1.34) (Meijers et al., 2007), where sequence variation between the different Ig3 domains in these structures results in structural differences at the Ig3-Ig3 interface but the structure of the Ig2-Ig2 interface is unperturbed between isoforms. In summary, dimer interfaces of different Ig2 and Ig3 variants, though different in detail, are unlikely to stray far from the intermolecular distance and orientation described here, so that the Ig2 and Ig3 binding modules may function in a modular way while also comprising a composite interface within the rigid Ig1–Ig4 horseshoe.
Together, these findings argue that any combination of Ig2, Ig3, and Ig7 variants is compatible with the double “S” scaffold of the Dscam1−8 homophilic dimer.
Structural Insight into “All-or-None” Binding Specificity
The structure of the Dscam1−8 dimer argues that both inter-molecular and intramolecular interactions are key to the “all-or-none” nature of Dscam binding specificity. As described above, the Ig2 and Ig3 domains function as distinct self-binding modules. However, matching at one domain is likely disrupted when the other does not match. This is due to intramolecular interactions between Ig2 and Ig3 that torsionally constrain these variable domains. As such, the Ig2-Ig2 and Ig3-Ig3 contacts function as two halves of a composite interface. When both halves match, they do not affect one another and, hence, function as modular units. However, when one half does not match, the strain of noncomplementarity is predicted to propagate strain to the other half of the interface, even if it is complementary. In this way, tight coupling between Ig2 and Ig3 further destabilizes a heterophilic complex by communicating an asymmetric pairing at Ig2 to Ig3, and vice versa. In the case of Ig7, the interface comprises multiple strands within one sheet and is, therefore, internally constrained. A single residue mismatch along one interface strand between opposing Ig7 variants will cause a shift in the interface. And, because the strands are rigidly coupled within the sheet, this shifts and, hence, destabilizes the entire Ig7 interface.
That intramolecular Ig5:Ig6 interactions are crucial for homophilic dimer formation is supported by genetic and biochemical data and conservation of multiple residues at this interface. Mutation of an evolutionarily conserved residue at the Ig5:Ig6 interface leads to a dramatic reduction in homophilic binding in vitro and wiring defects in vivo. These data suggest that matching at all three variable domains, although essential, is not sufficient to support robust binding. We propose that the free energy provided by the Ig5:Ig6 constant domain interaction is required to surpass the loss in entropy incurred by matching at all three variable domains.
A Structure-Based Model for Dscam Function In Vivo
Genetic studies have demonstrated that homophilic binding activates intracellular signaling and that this is dependent upon sequences in the cytoplasmic domain (Matthews et al., 2007). Therefore, the signal of homophilic binding is communicated to the cytoplasmic domain. This raises the intriguing possibility that the signal of homophilic binding is, somehow, communicated to the cytoplasmic domain by the double S-shape structure. We speculate that the double S shape may form during homophilic binding and trigger cytoplasmic signaling (Figure 7). Consistent with this notion, the S shape of each molecule in the Dscam1−8 dimer reveals a marked difference in shape from those observed in electron micrographs of monomeric Dscam1−8 molecules (Meijers et al., 2007). These studies demonstrated that although Ig1–Ig4 adopts a compact horseshoe configuration, the rest of the molecule (i.e., Ig5–Ig8) adopts multiple conformations, suggesting that in the absence of homophilic binding, the region comprising Ig5–Ig8 may be relatively unstructured. Although additional studies are required to establish that the Ig5–Ig8 region is indeed unstructured in monomers in solution, the lack of structure described by Meijers et al. suggests that the C-terminal half of Dscam1−8 may undergo a marked conformational change upon formation of the double S-shape dimer. Further studies will be necessary to assess whether a large-scale conformational change of Dscam occurs during homophilic binding and whether such a change affects the signaling events that underlie Dscam function in vivo.
Figure 7. Proposed Model for Transition of Dscam Monomers into S-Shaped Structures.
Top: Electron micrographs suggest that Dscam monomers exhibit the Ig1–Ig4 horseshoe, whereas the remainder of the protein exhibits various conformations (Meijers et al., 2007). This lack of a precise topology is incompatible with dimer formation, as suggested previously (Schmucker, 2007). Bottom: We propose that matching at all three variable domains and Ig5:Ig6 intramolecular interactions that facilitate a sharp bend between Ig5 and Ig6 lead to the formation of the double “S” upon homophilic binding. This proposed conformational change upon homophilic binding may play a role in transducing the signal of homophilic binding to the cytoplasmic domain (orange strarburst), which initiates repulsive signaling leading to downregulation of the homophilic binding complex and cytoskeletal rearrangements, causing neurites A and B to be repelled from one another.
EXPERIMENTAL PROCEDURES
Protein Production
Secreted unlabeled and selenomethionine-labeled Dscam1−8 proteins containing variable domains Ig2.1, Ig3.30, and Ig7.30 and a C-terminal 6xHis tag were produced in insect cells by the Protein Expression Center at the CalTech Beckman Institute. Dscam1−8 was purified via the 6xHis tag with cobalt resin (BD Biosciences Clonetech), dialyzed into 20 mM Tris, 100 mM NaCl and 2.5% (+/−)-2-methyl-2,4-pentanediol and concentrated to 35 mg/ml. Mass spectrometry analysis revealed two major peaks at 93.8 and 94.0 kD (unlabeled) and one major peak at 92.41 kD and minor peaks at 92.14, 92.27, and 92.55 kD (labeled).
Crystal Preparation
The Mosquito crystallization robot (TTP LabTech) was used to screen commercially available crystallization buffer kits. Crystals were optimized by the hanging-drop vapor-diffusion method. Reservoir solution used for crystallization contained 1.28 M ammonium sulfate and 0.2 M 2-(N-morpholino)ethanesulfonic acid (MES) (pH 6.0). Rectangular block-shaped crystals appeared within 3 days and reached full size after ∼1 month. Crystals were quickly cry-oprotected in a 65% reservoir and 35% glycerol solution and immediately flash frozen in a nitrogen cryostream (100 K). Crystals belonged to the space group I222 with unit cell dimensions: a = 118.6 Å, b = 177.9 Å, c = 434.3 Å. With three molecules in the asymmetric unit, the solvent content of the crystals was large (71% by volume).
Data Collection
X-ray data were collected on the Se-met-labeled crystal at 100K at the Advanced Light Source, beamline 8.2.2 with ADSC Quantum 315 3×3 CCD array. Three-hundred and sixty 1.0° oscillation frames were collected in inverse-beam mode at a wavelength of 0.9797 Å to maximize the anomalous signal from the Se atoms. Each exposure lasted 8 s. Data reduction and scaling were performed with DENZO/SCALEPACK (Otwinowski and Minor, 1997). Diffraction was anisotropic, extending to 3.9 Å in the b* and c* directions but only to 4.2 Å in the a* direction. Data were truncated to 4.2 Å resolution.
Structure Determination
An initial set of phases for the Dscam1−8 structure was obtained by molecular replacement using Dscam1−4 (PDB ID 2V5M) as a search model for Ig1–Ig4. Two chains (A and B) were readily located with PHASER at 5.0 Å resolution (McCoy et al., 2007) then refined using rigid-body refinement with REFMAC5 (Murshudov et al., 1997). Two features immediately validated the molecular replacement solution: first, the relative position and orientation of Ig1–Ig4 recapitulated the dimer interface observed in the crystal packing of 2V5M; second, an anomalous difference Fourier map calculated with phases of the molecular replacement model was of sufficient quality to produce numerous 7 σ peaks, indicating the position of the Se atoms. Many of these overlapped the Met sulfurs of the constant regions of Ig1–Ig4 in the model. The remaining peaks were used to aid in threading the registry of Ig5–Ig8. The gross features of Ig5–Ig8 could be visualized in difference Fourier maps. Homology models of Ig5, Ig6, Ig7, and Ig8 were computed by SWISS-MODEL (Schwede et al., 2003) and were individually positioned into the appropriate density, manually. A third Dscam molecule (chain C) was also found in difference density; it formed a crystallographic two-fold symmetric dimer.
The first refinement steps were performed with CNS (Brünger et al., 1998), using simulated annealing and conjugate gradient algorithms and the aid of a hydrogen bond potential function (Fabiola et al., 2002). Strong noncrystallographic symmetry restraints were used throughout. After each refinement step, the model was visually inspected in Coot (Emsley and Cowtan, 2004), with both 2Fo-Fc and Fo-Fc difference maps. Later rounds of refinement were performed with REFMAC5 (Murshudov et al., 1997) to benefit from TLS parameterization of domain disorder (Winn et al., 2003). The model was validated with the following structure-validation tools: PROCHECK (Laskowski et al., 1993), ERRAT (Colovos and Yeates, 1993), and VERIFY3D (Lüthy et al., 1992). All structure figures were prepared with PyMol (http://www.pymol.org).
Molecular Modeling
Ig2 and Ig7 variable domain docking models were generated as described previously (Wojtowicz et al., 2007).
Genetics and Immunohistochemistry
DscamR496W was isolated in a screen for recessive lethal alleles at the locus induced by ethylmethane sulfonate. Pupal brains from DscamR496W and Dscam23 null were dissected, and immunostaining was carried out essentially as described previously (Hummel et al., 2003).
Binding Assay
DscamR496W binding was assessed with the ELISA-based binding assay as previously described (Wojtowicz et al., 2007). The R496W mutation was introduced into Dscam containing Ig2.1, Ig3.30, and Ig7.30 with the QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's protocol.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Inderjit Nangiana, Jost Vielmetter, and the late Peter Snow of the CalTech Protein Expression Facility for production of Dscam1−8 protein, Julian Whittelegge for mass spectrometry analysis, Jeff Abramson for use of the Mosquito crystallization robot, Brent Graveley for providing annotated fly Dscam genomic sequences, and Srivatsan Raman for molecular models. We thank Daisuke Hattori for sequencing and initial characterization of DscamR496W and for help with genetics and immunohistochemistry and Aljoscha Nern for comments on the manuscript. We also thank Duilio Cascio and the staff at the Advanced Light Source, beamline 8.2.2, for assistance in collecting X-ray diffraction data. This work was supported by a National Institutes of Health (NIH) predoctoral training grant (United States Public Health Service National Research Service Award GM07185, W.M.W.), The Knut and Alice Wallenberg Foundation (I.A.), The American Leukemia and Lymphoma Society Fellowship (B.Q.), and the NIH. D.B., S.L.Z., and D.E. are investigators of the Howard Hughes Medical Institute.
Footnotes
ACCESSION NUMBERS
Coordinates and structure factor amplitudes of the Dscam1−8 structure (isoform 1.30.30) have been deposited in the Protein Data Bank under accession number 3DMK.
SUPPLEMENTAL DATA
Supplemental Data include Supplemental Experimental Procedures, nine figures, and four tables and can be found with this article online at http://www.cell.com/cgi/content/full/134/6/1007/DC1/.
REFERENCES
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chen BE, Kondo M, Garnier A, Watson FL, Puettmann-Holgado R, Lamar DR, Schmucker D. The molecular diversity of Dscam is functionally required for neuronal wiring specificity in Drosophila. Cell. 2006;125:607–620. doi: 10.1016/j.cell.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Colovos C, Yeates TO. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fabiola F, Bertram R, Korostelev A, Chapman MS. An improved hydrogen bond potential: Impact on medium resolution protein structures. Protein Sci. 2002;11:1415–1423. doi: 10.1110/ps.4890102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freigang J, Proba K, Leder L, Diederichs K, Sonderegger P, Welte W. The crystal structure of the ligand binding module of axonin-1/TAG-1 suggests a zipper mechanism for neural cell adhesion. Cell. 2000;101:425–433. doi: 10.1016/s0092-8674(00)80852-1. [DOI] [PubMed] [Google Scholar]
- Hattori D, Demir E, Kim HW, Viragh E, Zipursky SL, Dickson BJ. Dscam diversity is essential for neuronal wiring and self-recognition. Nature. 2007;449:223–227. doi: 10.1038/nature06099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Bortnick R, Tsubouchi A, Baumer P, Kondo M, Uemura T, Schmucker D. Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron. 2007;54:417–427. doi: 10.1016/j.neuron.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel T, Vasconcelos ML, Clemens JC, Fishilevich Y, Vosshall LB, Zipursky SL. Axonal targeting of olfactory receptor neurons in Drosophila is controlled by dscam. Neuron. 2003;37:221–231. doi: 10.1016/s0896-6273(02)01183-2. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- Lüthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, Grueber WB. Dendrite sef-avoidance is controlled by Dscam. Cell. 2007;129:593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijers R, Puettmann-Holgado R, Skiniotis G, Liu JH, Walz T, Wang JH, Schmucker D. Structural basis of Dscam isoform specificity. Nature. 2007;449:487–491. doi: 10.1038/nature06147. [DOI] [PubMed] [Google Scholar]
- Mörtl M, Sonderegger P, Diederichs K, Welte W. The crystal structure of the ligand-binding module of human TAG-1 suggests a new mode of homophilic interaction. Protein Science. 2007;16:2174–2183. doi: 10.1110/ps.072802707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macro-molecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Schmucker D. Molecular diversity of Dscam: Recognition of molecular identity in neuronal wiring. Nat. Neurosci. 2007;8:915–920. doi: 10.1038/nrn2256. [DOI] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soba P, Zhu S, Emoto K, Younger S, Yang SJ, Yu HH, Lee T, Jan LY, Jan YN. Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron. 2007;54:403–416. doi: 10.1016/j.neuron.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su XD, Gastinel LN, Vaughn DE, Faye I, Poon P, Bjorkman PJ. Crystal structure of hemolin: A horseshoe shape with implications for homophilic adhesion. Science. 1998;281:991–995. doi: 10.1126/science.281.5379.991. [DOI] [PubMed] [Google Scholar]
- Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtowicz WM, Wu W, Andre I, Qian B, Baker D, Zipursky SL. A vast repertoire of Dscam binding specificities arises from modular interactions of variable Ig domains. Cell. 2007;130:1134–1145. doi: 10.1016/j.cell.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan XL, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, Vasconcelos ML, Chess A, Zipursky SL. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 2004;43:673–686. doi: 10.1016/j.neuron.2004.07.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.