Abstract
ADAR1 (adenosine deaminase acting on RNA-1) edits adenosines in nuclear transcripts of nervous tissue and is also required at the fetal liver stage of the developing mouse embryo. Here we show by inducible gene disruption in mice that ADAR1 was essential for maintenance of both fetal and adult hematopoietic stem cells (HSCs). Loss of ADAR1 in HSCs led to global upregulation of type-I and type-II interferon-inducible transcripts and rapid apoptosis. Our findings identify ADAR1 as an essential regulator of HSC maintenance and suppressor of interferon signaling that may protect the organism from the deleterious effects of interferon activation associated with numerous pathological processes, including chronic inflammation, autoimmune disorders and cancer.
Keywords: RNA editing, Liver, Bone Marrow, Stem Cells, Cytokines
Adenosine-to-inosine (A-to-I) editing of primary transcripts in the nucleus recodes exonic information and can lead to structural and functional changes in the encoded protein1. Recent studies revealing widespread A-to-I editing of Alu repeat-containing mRNAs2-4 and low-level modification of microRNA precursors5-7 suggest a broader role of A-to-I editing in posttranscriptional gene regulation. Two sequence-related genes encode candidate enzymes for A-to-I editing, termed ADAR1 and ADAR2, each endowed with regions for binding double-stranded RNA (dsRNA) and an enzyme domain distantly related to bacterial cytidine deaminase1. While site-selective A-to-I editing of primary transcripts in the central nervous system is an established in vivo function of ADAR1 (refs. 8,9) and ADAR2 (ref. 10), no function has been delineated for ADAR3 (RED2), a brain-specific protein with sequence similarity to the other two ADARs but without detectable editing activity on synthetic dsRNA or known ADAR substrates11.
ADAR1 exhibits several features that distinguish it from the other two, more closely related ADAR proteins. These features comprise two putative Z-DNA-binding domains12, a third dsRNA-binding region, more widespread expression13, 14 and transcription originating from at least two promoters15-17. One promoter directs type-I and type-II interferon (IFN) inducible transcripts encoding the full-length ADAR1 protein (p150). The other promoter provides for constitutive expression of an amino-terminally truncated ADAR1 protein (p110). Interestingly, p110 localizes to the nucleus, consistent with a role in pre-mRNA editing, whereas the p150 isoform is found in both the nucleus and cytoplasm18. The existence of an interferon-inducible ADAR1 isoform, together with the finding that certain viral RNAs are subject to A-to-I editing, has led to speculation that ADAR1 plays a role in interferon-mediated immune responses to viral infection1, 19.
Disruption of Adar, the gene that encodes ADAR1, in mice leads to death at embryonic days (E) 11.5−12.5 in association with liver disintegration and defects in hematopoiesis8, 9, 20. These prior findings are consistent with cell-intrinsic or secondary roles for ADAR1 in liver development and/or hematopoiesis. Here, we report that ADAR1 was essential for the in vivo maintenance of the hematopoietic stem and/or immature progenitor compartment in both fetal liver and adult bone marrow. Moreover, we identified ADAR1 as a suppressor of interferon (IFN) signaling in hematopoietic stem and progenitor cells in vivo.
RESULTS
Phenotypic HSCs are present in Adar−/− fetal liver
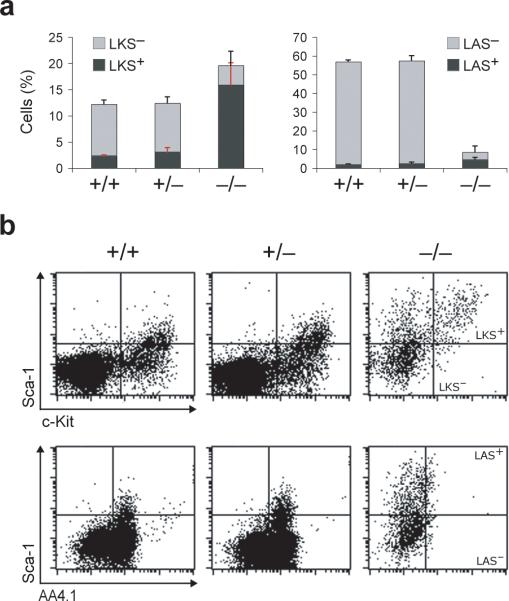
To assess the requirement, if any, for ADAR1 in the emergence of HSCs, or their migration from sites of production to the fetal liver (FL)21, we determined the immunophenotype and frequency of HSCs in E11.25 Adar−/− FL. We readily detected Lin− c-Kit+ Sca-1+ (LKS+) and Lin− AA4.1+ Sca-1+ (LAS+) cells, representing two HSC enriched populations in FL22, 23. Indeed, their frequencies were appreciably higher in Adar−/− FL as compared with wild-type and Adar+/− controls (Fig. 1). However, the absolute number of HSCs in Adar−/− FL was comparable to that in controls, due to reduced FL cellularity (not shown). In contrast, both the frequencies (Fig. 1a) and absolute numbers (not shown) of Lin− c-Kit+ Sca-1− (LKS−) and Lin− AA4.1+ Sca-1− (LAS−) progenitors were markedly decreased. Of note, Adar−/− cells displayed an increased intensity of Sca-1 surface expression, whereas the intensity of c-Kit surface staining was slightly reduced as compared with Adar+/− and wild-type cells (Fig. 1b). Detection of Lin− CD150+ CD48− CD244− cells representing a third HSC enriched population24, 25 confirmed the presence of phenotypic HSCs in E11.0 Adar−/− FL (Supplementary Fig. 1 online). We conclude that ADAR1 is dispensable for the emergence of phenotypic HSCs and their migration to the FL.
Fig. 1. ADAR1 is dispensable for the emergence of phenotypic HSCs and MPPs in the fetal liver.
(a) Graphs represent the frequencies of live, lineage depleted (Lin−) cells from E11.25 FL analyzed for c-Kit and Sca-1 (LKS) or AA4.1 and Sca-1 (LAS) expression LKS+ and LAS+ HSCs (dark grey bars), and LKS− and LAS− progenitors (light grey bars). Data are expressed as mean ± s.d. (3−5 tissues for each genotype). Statistical significance was determined by unpaired t test with p-values ranging from <0.0001 to 0.0328 (p<0.05 is considered statistically significant by this test). (b) Representative flow cytometry profiles of live Lin− c-Kit+ Sca-1+ (LKS+), Lin− c-Kit+ Sca-1− (LKS−), Lin− AA4.1+ Sca-1+ (LAS+), and Lin− AA4.1+ Sca-1− (LAS−).
ADAR1 is required in fetal liver derived HSCs
We next pursued possible roles for ADAR1 in the function of HSCs through inducible disruption of a conditional ADAR1 allele8. To rule out interference with ADAR1 expression of the pgk-neo gene present in a previously generated Adarf7−9 allele (now termed Adarfn), we derived a pgk-neo deleted version (Adarf) by Cre recombinase (Cre)-mediated excision in vivo (Supplementary Fig. 2 online). Adarf/f as well as Adarf/− mice that combine an Adarf allele with our previously generated AdarΔ2−13 null allele8 (now termed Adar−) were viable and appeared healthy, whereas both germ-line deleted AdarΔ/Δ and AdarΔ/− mice recapitulated the embryonic lethal phenotype of Adar−/− mice, as expected.
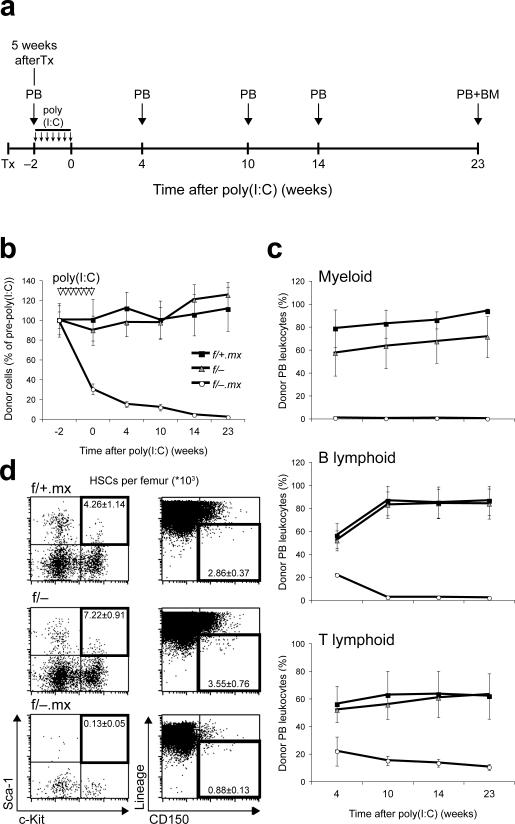
We interbred Adarf/− mice (Supplementary Fig. 2 online) with mice harboring the Mx1-Cre transgene26 that upon administration of the interferon inducer polyI-polyC (poly(I:C)) elicits efficient gene deletion throughout the hematopoietic system27, 28. Untreated Adarf/− Mx1-Cre mice were born at Mendelian frequency but, unexpectedly, died shortly after birth, possibly as a consequence of Adar deletion resulting from low-level Cre induction by endogenous interferon (Supplementary Fig. 3 online)28. However, Adarf/− Mx1-Cre embryos appeared largely intact at E14.5, despite a modest reduction in total FL cellularity (not shown). To circumvent the unanticipated early death of untreated Adarf/− Mx1-Cre mice, we performed transplant studies using FL derived hematopoietic cells (Fig. 2a). We distinguished donor and recipient cells in reconstituted wild-type animals by employing the CD45 congenic system27, 28 in combination with Adar allele-specific PCR. Five weeks after transplantation, Adarf/− Mx1-Cre as well as Adarf/− and Adarf/+ Mx1-Cre control FL cells had efficiently engrafted recipients and contributed multi-lineage peripheral blood cells (Fig. 2b and Supplementary Fig. 4 online). Mx1-Cre expression was then induced in the transplanted, FL-derived cells by administration of poly(I:C) (AdarΔ/− Mx1-Cre). Strikingly, two weeks after the first dose of poly(I:C), the contribution of Adarf/− Mx1-Cre FL-derived cells to peripheral blood leukocytes was reduced to one-fourth that of controls (Fig. 2b and Supplementary Fig. 4 online). Contribution of AdarΔ/− Mx1-Cre cells to peripheral blood cells, including myeloid, B lymphoid, and T lymphoid lineages, progressively declined and was negligible by 23 weeks after poly(I:C) treatment; the contribution of Adarf/− and Adarf/+ Mx1-Cre control cells was stable throughout the experiment (Fig. 2c). Flow cytometry of bone marrow cells confirmed the loss of Adarf/− Mx1-Cre contribution to multiple lineages observed in peripheral blood and revealed very few, if any, donor-derived LKS+ HSCs (Fig. 2d). Similar results were obtained when donor-derived HSCs were identified based on the combinatorial expression of SLAM cell surface receptors24 (Fig. 2d). Thus, the observed deficiency of HSCs was not due to an alteration of their immunophenotype in response to ADAR1 loss. These data demonstrate a cell-autonomous requirement for ADAR1 in FL-derived HSCs. We found no remarkable changes in the expression of genes associated with HSC maintenance in Adar−/− as compared with control Lin− CD150+ CD48− CD244− FL HSCs (Supplementary Fig. 5 online), suggestive of a novel mechanism through which ADAR1 regulates HSCs and immature hematopoietic progenitors.
Fig. 2. Near-complete loss of fetal liver contribution to adult bone marrow hematopoiesis after induced ADAR1 deficiency.
(a) Schematic diagram of experimental design. Peripheral blood (PB) and bone marrow (BM) were analyzed at indicated time points. PolyI-polyC (poly(I:C)) was administered in seven doses (small vertical arrows) over a two-week period beginning five weeks after transplantation (Tx) (time point −2 weeks). The day of the last poly(I:C) dose is designated 0 weeks after poly(I:C).
(b) Contribution of FL derived, CD45.2 expressing (donor) cells to PB leukocytes at indicated time points. (c) Donor contribution to peripheral myeloid (Mac-1+/Gr-1+), B-lymphoid (B220+), and T-lymphoid (CD4+/CD8+) lineages. (d) Representative flow cytometry profiles and absolute numbers per femur of donor derived LKS+ cells (left panel, black frames) and Lineage-negative CD150+CD48− (L150+) cells (right panel, black frames) in recipient bone marrow at 23 weeks after poly(I:C) treatment. Absolute HSC numbers per femur: 4,260 ± 114 LKS+ and 2,860 ± 370 L150+ cells (f/+.mx); 7,220 ± 910 LKS+ and 3,550 ± 760 L150+ cells (f/−); 130 ± 50 LKS+ and 880 ± 130 L150+ cells (f/−.mx). All data presented in (b-d) are expressed as mean ± s.e.m. (4−5 animals in each group).
ADAR1 is essential in adult-bone-marrow HSCs
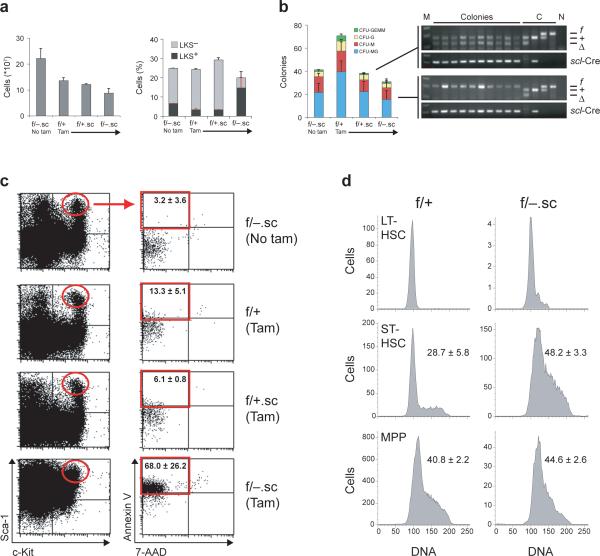
To study the in vivo consequences of ADAR1 loss in adult HSCs and to exclude potential confounding effects of interferon induced by poly(I:C) in the context of ADAR1 deficiency, we interbred Adarf/− mice with a tamoxifen-inducible SCL-Cre-ERT transgenic line that mediates deletion of floxed alleles in HSCs29. In absence of tamoxifen, Adarf/− SCL-Cre-ERT (hereafter referred to as Adarf/−.sc) and control mice appeared healthy, and no detectable excision of the Adarf allele was observed in peripheral blood (Supplementary Fig. 6 online). Upon induction of Adarf excision, we observed quantitative ADAR1 deletion, as determined by genomic PCR, in peripheral blood cells of Adarf/+.sc mice. In contrast, peripheral blood cells of Adarf/−.sc mice exhibited only modest excision, a finding most consistent with in vivo selection against ADAR1-deficient cells (Supplementary Fig. 6). Bone marrow analysis of Adarf/−.sc mice revealed markedly reduced cellularity as compared with control mice (Fig. 3a). Consistent with our findings in Adar−/− FL (Fig. 1), the frequency of LKS+ enriched HSCs in Adarf/−.sc bone marrow was appreciably increased, whereas the frequency of LKS− progenitors was markedly decreased as compared with induced and uninduced controls (Fig. 3a). Hematopoietic progenitors from unfractionated Adarf/−.sc bone marrow yielded only colonies with the unexcised Adarf allele, whereas all colonies derived from Adarf/+.sc bone marrow contained the excised allele (Fig. 3b). This finding confirms strong selection against ADAR1-deficient hematopoietic cells, as was observed in the peripheral blood.
Fig. 3. Induced deletion of ADAR1 in HSCs of adult mice leads to hyperproliferation and apoptosis.
(a) Absolute numbers of total bone marrow cells (grey bars), and frequencies of HSCs (LKS+) and progenitors (LKS−) in induced (Tam) and uninduced (No tam) adult mice. (b) Numbers of methylcellulose colonies grown from 12,500 bone-marrow cells of induced (Tam) and uninduced (No tam) Adar.sc mice, and representative Adar allele and scl-Cre transgene specific genomic PCR analysis of individual colonies from Adarf/+.sc (upper panel) and Adarf/−.sc (lower panel) bone marrow. DNA size markers (M) and no-template controls (N) are indicated. (c) Representative flow cytometry profiles of bone-marrow LKS+ HSCs (red ellipse) analyzed for apoptotic and necrotic cell death by Annexin V / 7-amino-actinomycin D (7-AAD) staining. Frequencies of apoptotic LKS+ HSCs (red frame): 3.2 ± 3.6 % (f/−, no tam); 13.3 ± 5.1 % (f/+, tam); 6.1 ± 0.8 % (f/+.sc, tam); 68.0 ± 26.2 % (f/− .sc, tam). (d) Representative cell cycle analysis of LKS+CD34lo long-term (LT-) HSCs, LKS+CD34hi short-term (ST-) HSCs, and LKS− multipotent progenitors (MPP). Numbers represent the frequency of cells in S-phase. All data presented in (a-d) are expressed as mean ± s.d. (3 mice per experimental group and 2 mice in each control group).
Maintenance of HSCs requires coordination of complex pathways that involve control of self-renewal, differentiation and apoptosis21, 30. To gain insight into the events underlying the loss of ADAR1-deficient cells, we quantified the frequency of apoptotic cells within HSC enriched LKS+ and progenitor populations in the bone marrow of Adarf/−.sc and control mice. The numbers of apoptotic cells within resting LKS+ and LKS− populations with characteristic low expression of the integrin Mac-1 (CD11b)31 in the bone marrow of induced Adarf/−.sc mice were only slightly higher than in controls (Supplementary Fig. 7 online). By contrast, 68 % of cycling (Mac-1-expressing) HSCs in Adarf/−.sc bone marrow, as compared with 3−13 % in controls, were found to be apoptotic based on AnnexinV staining (Fig. 3c). These observations are most consistent with a requirement for ADAR1 in cycling HSCs even though the data do not exclude the possibility that Mac-1-expressing immature progenitors32 contribute to the apoptotic population. Interestingly, the frequency of apoptotic cells was positively correlated with the surface expression of Sca-1; that is, the number of apoptotic cells was highest within the c-Kit+ Sca-1hi population (not shown). The few detectable LKS− progenitors in the bone marrow of Adarf/−.sc mice did not display increased apoptosis, consistent with our finding that all in vitro colony forming activity of hematopoietic progenitors in Adarf/−.sc bone marrow was derived from cells that contained the unexcised Adarf/−.sc allele (Fig. 3b). In support of a requirement for ADAR1 in cycling HSCs, cell cycle analysis of Adarf/−.sc as compared with Adarf/+.sc bone marrow LKS+ subsets and LKS− progenitors revealed a marked increase in the number of S-phase cells within the LKS+ CD34hi short-term (ST-) HSC but not in the LKS− progenitor population (Fig. 3d), perhaps reflecting activation in response to pancytopenia (Supplementary Table 1 online). Notably, the number of LKS+ CD34lo long-term (LT-) HSCs was markedly reduced in Adarf/−.sc relative to control bone marrow (Supplementary Fig. 8 online and Fig. 3d), indicating exhaustion of the LKS+ CD34lo population upon activation of the stem cell compartment.
Dependence on ADAR1 is not a general property of blood cells or stem cells, as we found ADAR1 to be dispensable in the myeloid lineage (J.C.H., C.R.W., S.H.O., unpublished) and for embryonic stem (ES) cell self-renewal, embryoid body formation in vitro (J.C.H., unpublished), and contribution to adult tissues of all three germ layers in chimeric mice, with the exception of hematopoietic tissues and liver8. Conversely, disruption of the Adar gene in the erythroid lineage causes loss of erythroblasts in the FL (J.C.H. and S.H.O., unpublished), demonstrating that the requirement for ADAR1 is not restricted to uncommitted hematopoietic progenitors.
ADAR1 deficiency elicits a global interferon response
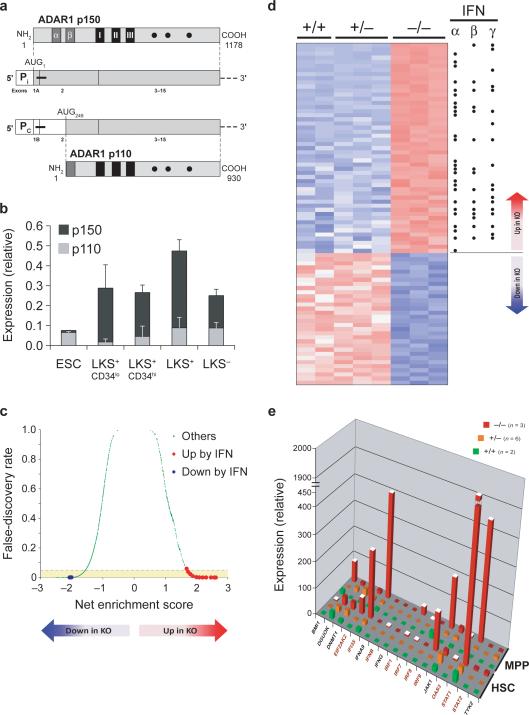
To approach the molecular mechanisms underlying the requirement for ADAR1 in HSCs, we determined the relative expression of the constitutively expressed ADAR1 p110 and the interferon-induced ADAR1 p150 transcripts (Fig. 4a) in sorted LKS populations from wild-type mice by real-time PCR (Fig. 4b). p150 transcripts were much more abundant than p110 transcripts in all LKS populations. Consistent with the hypothesis that p150 may be the critical isoform in HSCs, ES cells, in which ADAR1 is dispensable, expressed almost exclusively the p110 transcript (Fig. 4b). Given the predominant expression of the interferon-induced p150 transcript, we considered the possibility that the requirement for ADAR1 involves pathways activated by interferon in HSCs.
Fig. 4. ADAR1 deficiency in HSCs leads to a global upregulation of interferon-inducible transcripts.
(a) Schematic drawing of two murine ADAR1 mRNA and protein species. ADAR1 p150 protein is encoded by an mRNA transcribed from an interferon-inducible promoter (Pi), and ADAR1 p110 protein is encoded by an mRNA transcribed from a constitutively active promoter (PC). Translation start sites (AUG) within transcripts are indicated. White boxes, 5’-untranslated region (UTR); thick black horizontal lines, regions amplified by real-time PCR, broken horizontal lines, 3’-UTR; dark-grey boxes, putative Z-DNA binding domains (α and β); black boxes, RNA binding domains (I-III); closed circles, conserved residues within deaminase domain. (b) Real-time PCR analysis of ADAR1 mRNA expression in ES cells (ESC), long-term (LKS+CD34lo) and short-term (LKS+CD34hi) HSCs, total HSCs (LKS+), and multipotent progenitors (LKS−) using oligonucleotides specific for the IFN-inducible ADAR1 p150 (dark grey bars) and the constitutive ADAR1 p110 (light grey bars) transcripts. ADAR1 mRNA expression was normalized to YWHAZ (GenBank™/EBI accession number 22631) mRNA expression using the ΔΔCt method. Data are expressed as mean ± s.d. (c) Gene set enrichment analysis of genome-wide transcriptome expression data from ADAR1−/− (KO) relative to control HSCs. Among 1247 curated gene sets (represented by green dots), those that were annotated as upregulated by interferon signaling are colored in red, whereas those downregulated by interferon signaling are represented by blue dots. Positive net enrichment scores denote gene sets that were upregulated in KO cells. Negative scores reflect downregulated gene sets. The yellow area contains significantly altered gene sets with a false-discovery rate (FDR) of <0.05. (d) Heat map of the most significantly deregulated genes (FDR<0.1), ranked by t test score. Red color reflects high expression, and blue color indicates low expression. Black dots label genes known to be inducible by interferons (IFN) α, β, or γ. (e) Validation of gene array data by quantitative PCR. The graph shows mRNA expression of representative genes normalized to YWHAZ mRNA expression in lineage-depleted CD150+CD48−CD244− (HSC) and CD150−CD48+ (MPP) cells from early-E11 wild-type (+/+), Adar+/− (+/−), and ADAR1-deficient (−/−) FL. Data are expressed as mean ± s.e.m. (2−6 samples per group). White column tops indicate statistically significant differences (p value < 0.03) between Adar−/− and control cells, as determined by unpaired t test. Names of known interferon-inducible genes are in red color.
Sca-1 is expressed from an interferon-inducible gene33. We reasoned that the unexpected death of uninduced Adarf/− Mx1-Cre pups and the increased surface expression of Sca-1 on Adar−/− relative to wild-type and Adar+/− LKS+ cells (Fig. 1b) might reflect activation of the interferon pathway in response to ADAR1 loss. We tested this hypothesis by performing genome-wide transcriptome analysis in Adar−/− and control FL HSCs purified based on SLAM marker expression25 (Supplementary Figs. 1 and 9 online). Gene Set Enrichment Analysis (GSEA)34, a powerful computational method that determines statistically significant differences between gene sets obtained from microarray data, revealed strong correlation between ADAR1 deficiency and gene expression signatures of interferon-treated or virus-infected cells (Fig. 4d, Supplementary Fig. 10 and Table 2 online). At the single-gene level, Adar−/− Lin−CD150+ CD48− CD244− HSCs displayed a global up-regulation of transcripts inducible either by type I, type II, or both types of interferons (Fig. 4d, and Supplementary Table 3 online). We validated this finding for a subset of genes by quantitative PCR analysis of independently derived Lin−CD150+ CD48− CD244− FL HSCs and Lin− CD150+ CD48− progenitors (Fig. 4e). Genes significantly induced (up to 300 fold) by ADAR1 deficiency encompass the signal transducers and activators of transcription STAT1 and STAT2, the interferon regulatory factors IRF1, IRF7, and IRF9, the GTPases Mx1 and Mx2, the RNA-activated protein kinase PKR (EIF2AK2), the 2',5'-oligoadenylate synthetases OAS1−3, the ubiquitin-like modifiers Isg15 and Isg20, the interferon-induced proteins with tetratricopeptide repeats Ifit1−3 and ADAR1. The fact that most of the Affymetrix probes for Adar detect exons 12−15 permitted the analysis of Adar gene activity in Adar−/− cells lacking exons 2−13 of the Adar gene (Supplementary Fig. 2). We observed a similar induction of ADAR1 expression in erythroid cells lacking exons 7−9 of the Adar gene (not shown). As was observed in HSCs, gene expression profiling of ADAR1-deficient FL erythroid cells (J.C.H. and S.H.O., unpublished) revealed a global upregulation of interferon-inducible transcripts, strongly suggesting that the critical function of ADAR1 as a suppressor of interferon signaling was conserved in HSCs and erythroid cells. Notably, ADAR1 deficiency in macrophages and neutrophils did not affect the transcript abundance of STAT1, one of the most significantly upregulated genes in Adar−/− HSCs and erythroid cells, consistent with our finding that ADAR1 is dispensable in the myeloid lineage (J.C.H and S.H.O, unpublished). These findings hint at cell-specific differences in how ADAR1 may participate in regulation of interferon signaling.
Given that ADAR1 is a dsRNA binding and processing protein and ADAR1 deficiency might lead to an interferon response induced by intracellular dsRNA resulting from an unknown ADAR1-dependent mechanism we examined our expression data set for transcripts that have been shown experimentally to be inducible by dsRNA35. Among these, only transcripts known to be inducible by IFN were significantly upregulated in ADAR1-deficient as compared with wild-type and Adar+/− control HSCs (Supplementary Fig. 11a online). Likewise, among transcripts encoding known dsRNA-binding proteins, only those expressed from IFN-inducible genes, such as PKR (EIF2AK2), IFIH1 (RIG-1) and TLR3, were upregulated in ADAR1-deficient relative to control HSCs (Supplementary Fig. 11b online). The dsRNA binding protein NF90 has been shown previously to interact with ADAR136. Interestingly, the Adar−/− signature correlated with profiles obtained from cells that (over)express a carboxyterminal variant of NF90 protein37 (Supplementary Fig. 12 and Table 2 online), perhaps suggesting a functional link of NF90 and ADAR1.
To examine if the interferon response to ADAR1 deletion was unique to hematopoietic cells, we used quantitative PCR to analyze the expression of interferon-inducible transcripts in early E11 Adar−/− embryos that had their FL removed, termed ΔFL embryos (Supplementary Fig. 13a online). Similar to our observations in HSCs and erythroid cells, we found a significant upregulation of interferon-inducible transcripts in the ΔFL Adar−/− embryos as compared with control embryos. Interferon ELISAs performed on these embryos revealed >13 fold increased IFN-α and >80 fold increased IFN-β protein concentrations in the extracellular fluid of Adar−/− embryos relative to control embryos, whereas IFN-γ was not detectable in either group (Supplementary Fig. 13b). Notably, elevated concentrations of IFN-α and IFN-β protein in the extracellular fluid were not a general feature of embryos that exhibit a lethal phenotype associated with defective FL hematopoiesis, as demonstrated by absence of detectable interferon protein expression in the extracellular fluid of dying Gata2−/− embryos (data not shown). Gata2 deficiency causes loss of immature hematopoietic progenitors in the FL and embryonic death by ∼E11 (ref. 38). Thus, ADAR1 deficiency leads to a widespread failure to appropriately regulate an interferon response within the embryo that may be caused or enhanced by paracrine activities of type-I interferons secreted into the extracellular fluid from FL HSCs and erythroid cells upon activation of their interferon system.
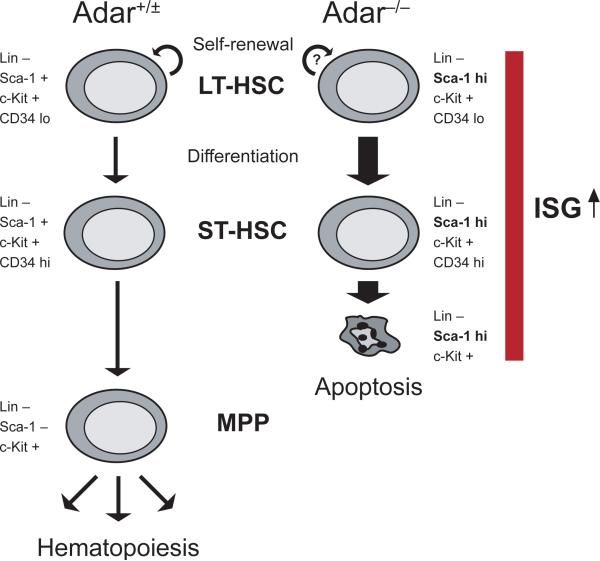
Here we identify ADAR1 as a regulator of HSC maintenance and suggest a model in which ADAR1 is dispensable for emergence of long-term reconstituting HSCs and, perhaps, their self-renewal activity. However, ADAR1 becomes essential for the survival of HSCs as they progress to the multipotent progenitor stage, a process that is associated with downregulation of Sca-1 surface expression (Fig. 5).
Fig. 5. Suggested model for the role of ADAR1 in HSCs.
Vertical arrows mark the transition from long-term (LT-) HSC via short-term (ST-) HSC to multipotent progenitor (MPP) as defined by lineage marker (Lin), cKit (K), Sca-1 (S), and CD34 surface expression. Semi-circled arrows indicate self-renewal. Both ADAR1-deficient (Adar−/−) HSCs and MPPs exhibit a global upregulation of transcripts encoding interferon-stimulated genes (ISGs), including Sca-1, as compared with wild-type and Adar+/− (Adar+/±)cells. Differentiation of lineage-negative, cKit-positive, Sca-1-positive, CD34-negative/low (LKS+ CD34lo) LT-HSCs via ST-HSCs (LKS+ CD34hi) and MPPs (LKS− CD34hi) is accompanied by an increase in CD34 and a decrease in Sca-1 surface expression. Perhaps as a consequence of the interferon pathway activation in absence of ADAR1, LKS+ HSCs fail to downregulate Sca-1 and undergo rapid apoptosis as they progress to the LKS−progenitor stage of hematopoiesis.
DISCUSSION
In addition to our finding that ADAR1 is essential for maintenance of hematopoiesis in the fetal liver and adult bone marrow, we identified ADAR1 as an essential suppressor of interferon signaling. Stringent control of this pathway, which is involved in numerous physiological processes, including inflammation and the innate immune response, is required to prevent the deleterious effects of excessive interferon activation. Importantly, defective interferon signaling has been associated with various pathological conditions, such as chronic inflammation, autoimmune disorders and cancer. The loss of HSCs and immature progenitors in the absence of ADAR1 was most likely a consequence of failure to hold interferon activation in check.
Conversion of adenosine to inosine in nuclear transcripts is the only established function of ADAR1. It is tempting, therefore, to speculate that ADAR1 regulates interferon signaling by editing critical, currently unknown transcripts. However, given the complexity of the interferon pathway that is activated by numerous stimuli it is instructive to consider alternative possibilities. First, as ADAR1 is a dsRNA binding protein and the innate immune response to dsRNA involves activation of type-I interferons35, we interrogated our expression data for genes induced by dsRNA. Indeed, a gene set obtained from cells treated with dsRNA correlated with the Adar−/− signature in that interferon-induced genes are significantly upregulated. ADAR1 deficiency enhanced the expression of transcripts encoding the interferon-inducible dsRNA binding proteins PKR (EIF2AK2), IFIH1 (RIG-1) and TLR3, whereas the expression of transcripts for other dsRNA binding proteins was not appreciably altered. This observation may suggest that immunoreactive dsRNA resulting from failure of an unknown ADAR1-dependent process contributes to the interferon response induced by ADAR1 deficiency. Second, ADAR1 has been shown previously to interact with the dsRNA binding protein NF90 via dsRNA and enhance NF90-mediated expression of genes, including the IFN-β gene, in an RNA-editing independent fashion36. Interestingly, the Adar−/− signature correlates with profiles obtained from cells that (over)express a carboxyterminal variant of NF90 protein37, raising the possibility that ADAR1 deficiency disrupts ADAR1-containing protein complexes involved in the regulation of interferon-induced gene expression. Third, a recent study suggests a role for ADAR1 in a cell-intrinsic DNA-sensing mechanism that involves the putative Z-DNA binding protein DAI, a sensor for cytosolic DNA39. Consistent with a possible role for ADAR1 in intracellular DNA sensing pathways, DAI transcript abundance was increased 15-fold in Adar−/− HSCs as compared with control HSCs (not shown). Finally, another recent study suggests that NF-κB-mediated transcriptional activation of the human IFNB gene upon virus infection involves the use of NF-κB binding sites embedded in an Alu repeat40. Interestingly, widespread A-to-I editing of Alu repeat-containing mRNAs has been described in humans2-4. Thus, ADAR1 may regulate the expression of interferon-inducible genes by targeting regulatory elements embedded in Alu or Alu-like repeats.
Future work will be required to elucidate whether ADAR1 plays a direct role in the regulation of interferon signaling, for example, by editing critical transcripts of protein-coding or regulatory RNA genes, or if activation of the interferon pathway by ADAR1 deficiency represents a cellular response to immunoreactive nucleic acid that may result from failure of an unknown ADAR1-dependent mechanism. Independent of the underlying mechanism, our study identifies ADAR1 as essential in vivo suppressor of interferon-induced gene expression.
Our finding that ADAR1 was essential during the activated state of a rare cell population comprising < 0.05 % of total bone marrow may explain, in part, why intensive efforts over the past years have failed to identify critical targets of ADAR1. Without substantial enrichment of specific, rare cells, the search for low abundance or edited transcripts may prove unsuccessful. Identifying the targets of ADAR1, which may include protein and/or non-protein coding transcripts, or possibly interacting proteins36, will be critical in a fuller understanding of the unanticipated requirements for ADAR1 in the maintenance of hematopoiesis and suppression of interferon signaling.
METHODS
Experimental mice
All mice were backcrossed at least six times to the C57BL/6 strain from a C57BL/6-Sv129 hybrid background. Adarf (floxed) mice were derived from crosses of ADAR1fn mice8 with several transgenic Cre recombinase (Cre) expressing mouse lines and selected by genomic PCR for recombination events that had excised the pgk-neor gene from the ADAR1fn allele without deleting the floxed ADAR1 segment (Supplementary Fig. 2). Pups that transmitted the Adarf allele through the germ line were identified by PCR after crossing out the Cre transgene. Mx1-Cre26 and SCL-Cre-ER(T)29 transgenic mice have been previously described. Experiments were performed with the approval of the respective Animal Ethics Committees (Dana-Farber Cancer Institute and Children's Hospital Boston).
Genotyping
All mice and tissues were genotyped by genomic PCR. ADAR1 mutant mice and tissues were genotyped as described previously8. For genotyping of Mx1-Cre mice, we designed Mx1-Cre-specific oligonucleotides (5'-CAATATCGATTTCCCAACCTCAG-3' and 5'-CTTGCACCATGCCGCCCACGAC-3'). The identity of the amplicon was confirmed by DNA sequencing. SCL-Cre-ER(T) mice were genotyped as described previously29.
Flow cytometry
Cells were processed and analyzed on a FACSCalibur or sorted on a FACSAria special cytometer as described previously41, 42. Lineage depletion, preceded by Ficoll-Paque Plus (GE Healthcare Life Sciences) density centrifugation, was performed with biotin-conjugated antibodies and fluorochrome-conjugated Streptavidin, or with magnetic beads as described by the manufacturer (Dynal) using affinity purified rat anti-mouse lineage antibodies (eBioscience). The lineage depletion cocktail included antibodies to CD4 (clone: GK1.5), CD5 (53−7.3), CD8a (53−6.7), B220 (RA3−6B2), Gr-1 (RB6−8C5), Mac-1 (M1/70), and TER-119 (TER-119). For recovery of fetal liver or activated bone marrow HSCs, Mac-1 antibody was omitted from the cocktail. Lineage-depleted FL HSCs and MPPs were analyzed using cKit (2B8) and Sca-1 (E13−161.7) or Sca-1 and C1qRp (AA4.1) antibodies, or sorted using antibodies to the SLAM proteins CD150 (TC15−12F12.2), CD48 (HM48−1), and CD244 (eBio244F4).
Long-term reconstitution
Fetal livers dissected from live E14.5 embryos were genotyped by genomic PCR, and single cell suspensions were prepared. Live cells, identified by Trypan Blue dye (Invitrogen) exclusion, were counted in a Neubauer hemocytometer. Equivalent numbers of cells from n = 2−3 fetal livers of each genotype were pooled, and 3 × 106 live cells in 0.2 ml sterile PBS were non-competitively transplanted into irradiated (2 × 5.25 Gy, 3 h apart) CD45.1 recipients. Cre expression was induced by intraperitoneal injections of poly(I:C) (Sigma) as described previously28, 41, 42. Donor cell contribution to peripheral blood was monitored by donor-specific flow cytometry analysis and genomic PCR.
Tamoxifen administration
12−20 week old mice were injected intraperitoneally with 2 mg tamoxifen in 0.2 ml corn oil per day and mouse or fed a standard mouse chow supplemented with tamoxifen at 1g/kg diet (Research Diets).
Methylcellulose culture
12,500 bone marrow cells were cultured in duplicate in Methocult M3434 (Stem Cell Technologies), and colonies were scored by microscopy. For single-colony PCR, individual colonies were picked, washed in PBS, and genomic DNA was isolated with the QIAamp DNA Blood Mini Kit as directed by the manufacturer (Qiagen).
Apoptosis
Lineage-depleted bone marrow cells were stained with fluorochrome-conjugated c-Kit (clone: 2B8) and Sca-1 (E13−161.7) antibodies, and AnnexinV protein in combination with 1 μg/ml 7-AAD (Molecular Probes) followed by analysis on a FACSCalibur flow cytometer (BD Biosciences) as described previously42.
Cell-cycle analysis
Live, lineage-depleted bone marrow cells were stained with fluorochrome-conjugated antibodies to c-Kit (clone: 2B8), Sca-1 (E13−161.7), and CD34 (RAM34), processed for cell-cycle analysis as described previously43, and 3 × 105 cells were analyzed on a FACSAria flow cytometer equipped with an UV laser (BD Biosciences). Data analysis was with FlowJo software (Tree Star, Inc.).
Gene expression profiling
Live CD150+CD48−CD244− HSCs (500−1500 cells per fetal liver) were isolated by flow cytometry from individual, viable early-E11 embryos. Total RNA was extracted with the microRNA kit (Qiagen), reverse-transcribed and amplified with the WT-Ovation™ Pico RNA Amplification System. Single-stranded cDNA amplification product was purified with the DNA Clean & Concentrator™ kit (Zymo Research, Inc.) and labeled with the FL-Ovation™ cDNA Biotin Module V2 (NuGEN Technologies, Inc.). Transcriptome analysis was performed by using GeneChip® Mouse Genome 430 2.0 Arrays (Affymetrix, Inc.) at the Broad Institute. Data were normalized by the RMA algorithm using the GenePattern software (http://www.broad.mit.edu/cancer/software/genepattern). Gene set enrichment analysis (GSEA, http://www.broad.mit.edu/gsea/) was then performed with the curated gene set collection C2 in MSigDB for distinction of Adar−/− versus Adar+/+ and Adar+/− HSCs. Gene sets were filtered to eliminate those with fewer than 15 or more than 800 genes, resulting in a total of 1247 gene sets. The dataset was row-normalized before subjecting to GSEA using signal-to-noise measurement and 30,000 permutations on the genes. For hierarchical clustering, expression data were filtered to eliminate genes with maximum expression values of less than 100 in all samples, with a standard deviation over mean of less than 0.5, and with present call percentage of less than 20% of samples. Data were then log2 transformed and clustered using Pearson correlation measurement and average linkage.
Real-time PCR
Live HSCs and progenitors were isolated from the femora, tibiae, and ilia of 10 male C57BL/6 mice (12 weeks old) or liver from viable, individual early E11 embryos by flow cytometry. Total RNA was extracted with the microRNA kit (Qiagen), and random-hexamer primed cDNA synthesis was performed with the Superscript III system (Invitrogen) as recommended. For samples containing less than 5,000 cells, total RNA was reverse-transcribed and amplified with the WT-Ovation™ Pico RNA Amplification System (NuGEN Technologies) as described above. Real-time PCR was performed in duplicate by using iQ SYBR Green Supermix in combination with the iQ5 Real-Time PCR Detection System (Bio-Rad). ADAR1-specific oligonucleotides for real-time PCR were designed based on sequence information obtained by 5'-RACE analysis of total RNA isolated from ES cells and murine embryonic fibroblasts with TRIZOL reagent (Invitrogen). Oligonucleotide sequences are listed in Supplementary Table 4 online. mRNA expression was normalized to YWHAZ, Cyclophilin A, or GAPDH mRNA expression using the ΔΔct method.
Interferon ELISA
Cell suspensions from live early-E11 embryos were prepared in ice-cold phosphate-buffered saline, centrifuged at moderate speed, and interferon protein concentrations in the cell-free supernatant were quantified with IFN-α, -β and -γ ELISA kits (R&D Systems) as directed by the manufacturer. Data were acquired on a Benchmark Plus microplate spectrophotometer and analyzed with MicroplateManager III software (Bio-Rad).
Statistics
Statistical significance of differences in the mean ± s.d. or mean ± s.e.m. was calculated with an unpaired t test, unless otherwise indicated.
Accession codes
The microarray data have been submitted to the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under the series accession number GSE12772.
Supplementary Material
Acknowledgments
We thank M. Higuchi and P. H. Seeburg for providing ADAR1 mutant mice, K. Rajewsky for providing Mx1-Cre mice, and J. Goethert for providing Scl-Cre-ERT mice; the DFCI and Children's Animal Facility Staff for care of experimental animals; J. Daley and S. Lazo-Kallanian of the DFCI HemNeo Flow facility for assistance with cell sorting. We appreciate the technical assistance of J. Shea. We thank T.R. Golub for his support in gene expression analysis. We are grateful to M. Higuchi, A. Athanasiadis, S. Maas, and L. Purton for critical comments on the manuscript. C.R.W. is a Special Fellow of the Leukemia & Lymphoma Society, and S.H.O. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Note: Supplementary information is available on the Nature Immunology website.
The authors declare no competing financial interests.
REFERENCES
- 1.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annual review of biochemistry. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS biology. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim DD, et al. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome research. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levanon EY, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nature biotechnology. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 5.Kawahara Y, et al. Science. Vol. 315. New York, N.Y: 2007. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. pp. 1137–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA. Vol. 10. New York, N.Y: 2004. RNA editing of a miRNA precursor. pp. 1174–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang W, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nature structural & molecular biology. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartner JC, et al. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. The Journal of biological chemistry. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, et al. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. The Journal of biological chemistry. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 10.Higuchi M, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 11.Melcher T, et al. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. The Journal of biological chemistry. 1996;271:31795–31798. doi: 10.1074/jbc.271.50.31795. [DOI] [PubMed] [Google Scholar]
- 12.Herbert A, et al. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:8421–8426. doi: 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Connell MA, et al. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Molecular and cellular biology. 1995;15:1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.George CX, Das S, Samuel CE. Organization of the mouse RNA-specific adenosine deaminase Adar1 gene 5'-region and demonstration of STAT1-independent, STAT2-dependent transcriptional activation by interferon. Virology. 2008 doi: 10.1016/j.virol.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.George CX, Samuel CE. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:4621–4626. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.George CX, Wagner MV, Samuel CE. Expression of interferon-inducible RNA adenosine deaminase ADAR1 during pathogen infection and mouse embryo development involves tissue-selective promoter utilization and alternative splicing. The Journal of biological chemistry. 2005;280:15020–15028. doi: 10.1074/jbc.M500476200. [DOI] [PubMed] [Google Scholar]
- 18.Eckmann CR, Neunteufl A, Pfaffstetter L, Jantsch MF. The human but not the Xenopus RNA-editing enzyme ADAR1 has an atypical nuclear localization signal and displays the characteristics of a shuttling protein. Molecular biology of the cell. 2001;12:1911–1924. doi: 10.1091/mbc.12.7.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samuel CE. Antiviral actions of interferons. Clinical microbiology reviews. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, Khillan J, Gadue P, Nishikura K. Science. Vol. 290. New York, N.Y: 2000. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. pp. 1765–1768. [DOI] [PubMed] [Google Scholar]
- 21.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordan CT, McKearn JP, Lemischka IR. Cellular and developmental properties of fetal hematopoietic stem cells. Cell. 1990;61:953–963. doi: 10.1016/0092-8674(90)90061-i. [DOI] [PubMed] [Google Scholar]
- 23.Okada S, et al. In vivo and in vitro stem cell function of c-kit- and Sca-1-positive murine hematopoietic cells. Blood. 1992;80:3044–3050. [PubMed] [Google Scholar]
- 24.Kiel MJ, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 25.Kim I, He S, Yilmaz OH, Kiel MJ, Morrison SJ. Enhanced purification of fetal liver hematopoietic stem cells using SLAM family receptors. Blood. 2006;108:737–744. doi: 10.1182/blood-2005-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Science. Vol. 269. New York, N.Y: 1995. Inducible gene targeting in mice. pp. 1427–1429. [DOI] [PubMed] [Google Scholar]
- 27.Hock H, et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431:1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 28.Mikkola HK, et al. Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature. 2003;421:547–551. doi: 10.1038/nature01345. [DOI] [PubMed] [Google Scholar]
- 29.Gothert JR, et al. Genetically tagging endothelial cells in vivo: bone marrow-derived cells do not contribute to tumor endothelium. Blood. 2004;104:1769–1777. doi: 10.1182/blood-2003-11-3952. [DOI] [PubMed] [Google Scholar]
- 30.Lessard J, Faubert A, Sauvageau G. Genetic programs regulating HSC specification, maintenance and expansion. Oncogene. 2004;23:7199–7209. doi: 10.1038/sj.onc.1207940. [DOI] [PubMed] [Google Scholar]
- 31.Randall TD, Weissman IL. Phenotypic and functional changes induced at the clonal level in hematopoietic stem cells after 5-fluorouracil treatment. Blood. 1997;89:3596–3606. [PubMed] [Google Scholar]
- 32.Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 33.Sinclair A, Daly B, Dzierzak E. The Ly-6E.1 (Sca-1) gene requires a 3' chromatin-dependent region for high-level gamma-interferon-induced hematopoietic cell expression. Blood. 1996;87:2750–2761. [PubMed] [Google Scholar]
- 34.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geiss G, et al. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. The Journal of biological chemistry. 2001;276:30178–30182. doi: 10.1074/jbc.c100137200. [DOI] [PubMed] [Google Scholar]
- 36.Nie Y, Ding L, Kao PN, Braun R, Yang JH. ADAR1 interacts with NF90 through double-stranded RNA and regulates NF90-mediated gene expression independently of RNA editing. Molecular and cellular biology. 2005;25:6956–6963. doi: 10.1128/MCB.25.16.6956-6963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krasnoselskaya-Riz I, et al. Nuclear factor 90 mediates activation of the cellular antiviral expression cascade. AIDS research and human retroviruses. 2002;18:591–604. doi: 10.1089/088922202753747941. [DOI] [PubMed] [Google Scholar]
- 38.Tsai FY, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, et al. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Apostolou E, Thanos D. Virus Infection Induces NF-kappaB-Dependent Interchromosomal Associations Mediating Monoallelic IFN-beta Gene Expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 41.Walkley CR, Orkin SH. Rb is dispensable for self-renewal and multilineage differentiation of adult hematopoietic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:9057–9062. doi: 10.1073/pnas.0603389103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walkley CR, Shea JM, Sims NA, Purton LE, Orkin SH. Rb regulates interactions between hematopoietic stem cells and their bone marrow microenvironment. Cell. 2007;129:1081–1095. doi: 10.1016/j.cell.2007.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gordon KM, Duckett L, Daul B, Petrie HT. A simple method for detecting up to five immunofluorescent parameters together with DNA staining for cell cycle or viability on a benchtop flow cytometer. Journal of immunological methods. 2003;275:113–121. doi: 10.1016/s0022-1759(03)00009-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.