Abstract
Recovery from Friend Virus 3 (Rfv3) is a single autosomal gene encoding a resistance trait that influences retroviral neutralizing antibody responses and viremia. Despite extensive research for 30 years, the molecular identity of Rfv3 has remained elusive. Here we demonstrate that Rfv3 is encoded by Apobec3. Apobec3 maps to the same chromosome region as Rfv3 and has broad inhibitory activity against retroviruses including HIV. Not only did genetic inactivation of Apobec3 convert Rfv3-resistant mice to a susceptible phenotype, but Apobec3 was found to be naturally disabled by aberrant mRNA splicing in Rfv3-susceptible strains. The link between Apobec3 and neutralizing antibody responses highlights an Apobec3-dependent mechanism of host protection that might extend to HIV and other human retroviral infections.
The study of viral resistance factors has provided important insights into the evolutionary strategies of defense utilized by mammalian hosts (1–5). Recovery from Friend virus (FV) gene 3 (Rfv3) was first identified as a resistance trait in 1978 (6, 7) and later genetic studies showed that the phenotypes of decreased viremia and FV-specific neutralizing antibody responses segregated as a single gene (8). Since the generation of neutralizing antibodies is critical for recovery from FV infection (1, 9) and a desired but often unrealized outcome in various retroviral infections including HIV-1, we have focused our efforts on identifying the gene encoding Rfv3. The Rfv3 locus maps to a 0.83 centimorgan region of chromosome 15 (Fig. S1A) (10–12), which contains at least 61 annotated genes (table S1), one of which is murine Apobec3 (mA3), a member of a family of deoxycytidine deaminases with antiretroviral and anti-retroelement activity [as reviewed in (13)]. This fact, along with the presence of substantial polymorphism in mA3 (table S1), led us to consider mA3 as a prime candidate for Rfv3.
Since Rfv3 has no described in vitro phenotype, our investigation required the generation of mA3-deficient mice (14). First, an inactivated mA3 gene (Fig. S1B) was introduced into the Rfv3r/r C57BL/6 (B6) background to test its ability to act as a defective Rfv3 allele in matings with Rfv3s/s mice (Table 1). Since the Rfv3 resistance trait is dominant over susceptibility (7), Rfv3r/s F1 offspring should control viremia and mount effective neutralizing antibody responses. Conversely, if mA3 encodes Rfv3, then the gene from a mA3−/− parent will be null, and the resultant F1 offspring with an Rfv3−/s genotype are predicted to exhibit higher levels of viremia and weaker neutralizing antibody responses. To test this possibility, B6 × BALB/c F1 offspring were infected with FV and plasma viremia levels were measured. At 7 days post infection (dpi), the F1 mice containing an inactivated mA3 gene exhibited 15-fold higher levels of viremia than their congenic partners carrying the wild-type mA3 allele (Fig. 1A). These high viral loads in mA3− F1 mice were comparable to FV levels found in fully susceptible Rfv3s/s BALB/c parental mice. Thus, mA3 is a restriction factor contributing to the early control of FV infection in adult immunocompetent mice.
Table 1.
FV infection characteristics of various mouse strains used in this study.
| Type | Strain | General FV susceptibility |
Viremia | Rfv3 | Neutralizing Antibody |
H-2* | Cell-mediated immunity |
Fv2# | Splenomegaly induction |
|---|---|---|---|---|---|---|---|---|---|
| Wild-type | C57BL/6 (B6) | resistant | resistant | r/r | high | b/b | high | r/r | no |
| BALB/c | susceptible | chronic | s/s | low | d/d | very low | s/s | yes | |
| A.BY | susceptible | chronic | s/s | low | b/b | high | s/s | yes | |
| 129/Ola° | resistant | resistant | r/r | high | b/b | high | r/r | no | |
| F1 hybrids | B6 × BALB/c | susceptible | acute | r/s | high | b/d | low | r/s | yes |
| B6 × A.BY | susceptible | acute | r/s | high | b/b | high | r/s | yes |
H-2 is the murine major histocompatibility complex (MHC), which dictates cell-mediated immunity against FV (5,15).
Fv2 is a dominant FV susceptibility gene that facilitates splenomegaly induction through aberrant signaling in erythroblasts (4).
FV susceptibility data on 129/Ola were based on results from this study (see Suppl. Text and Fig. 2C). The cell-mediated immune response of this strain was inferred from its H-2 haplotype.
Fig. 1.
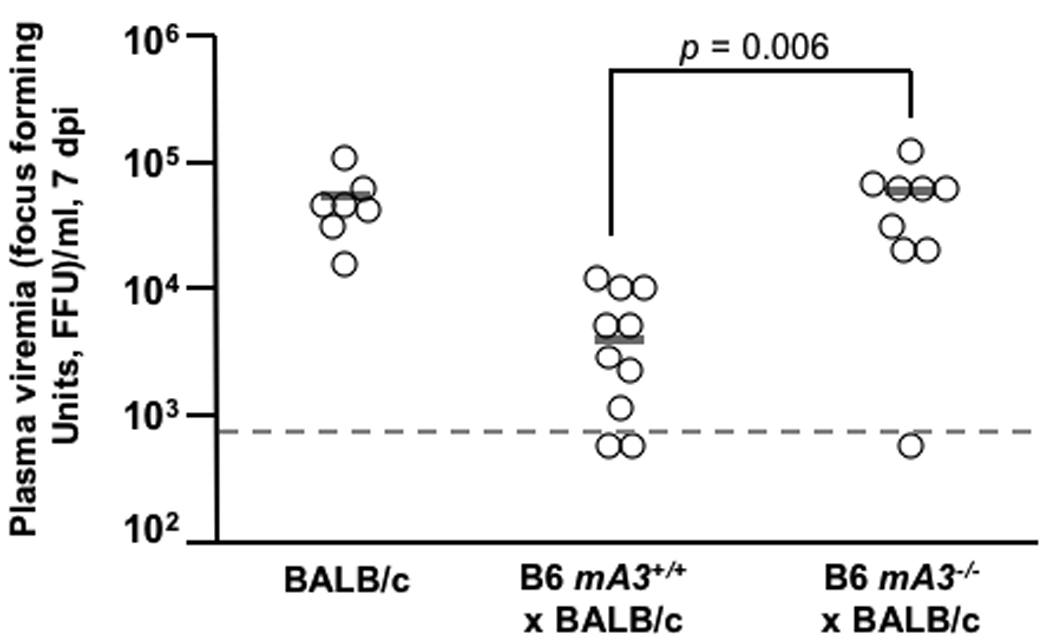
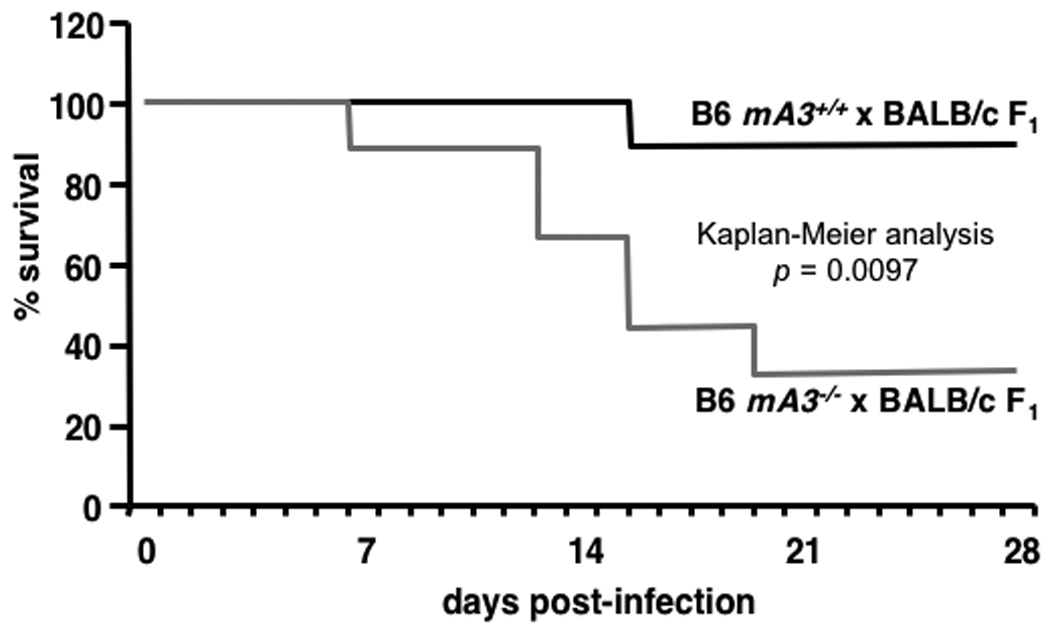
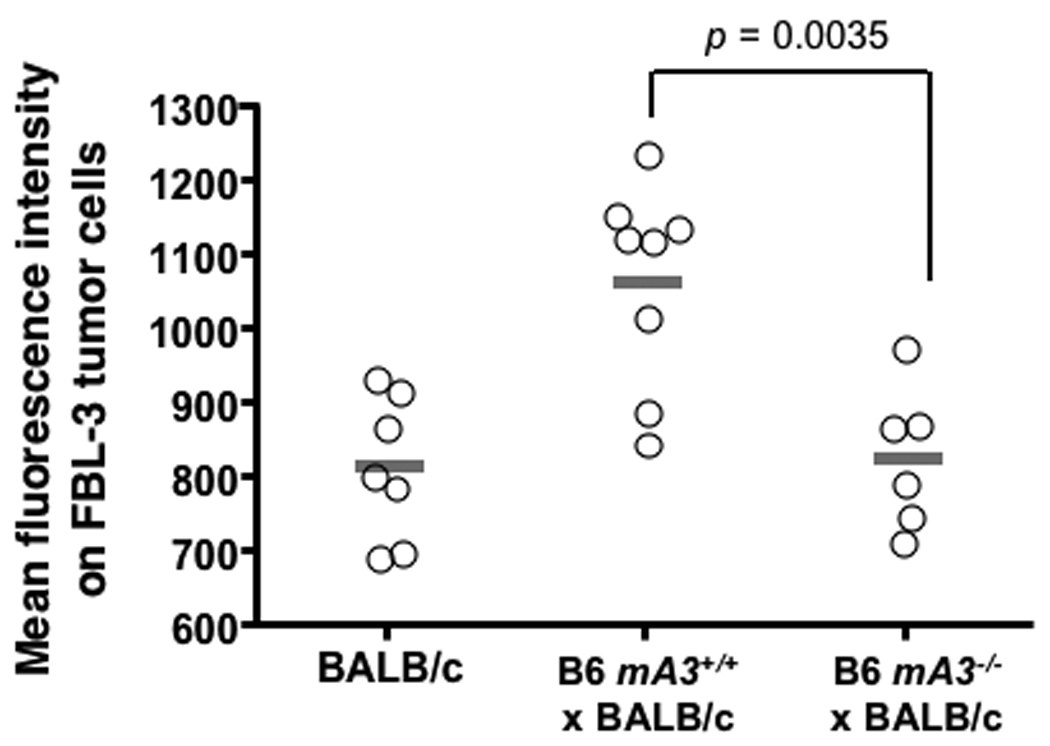
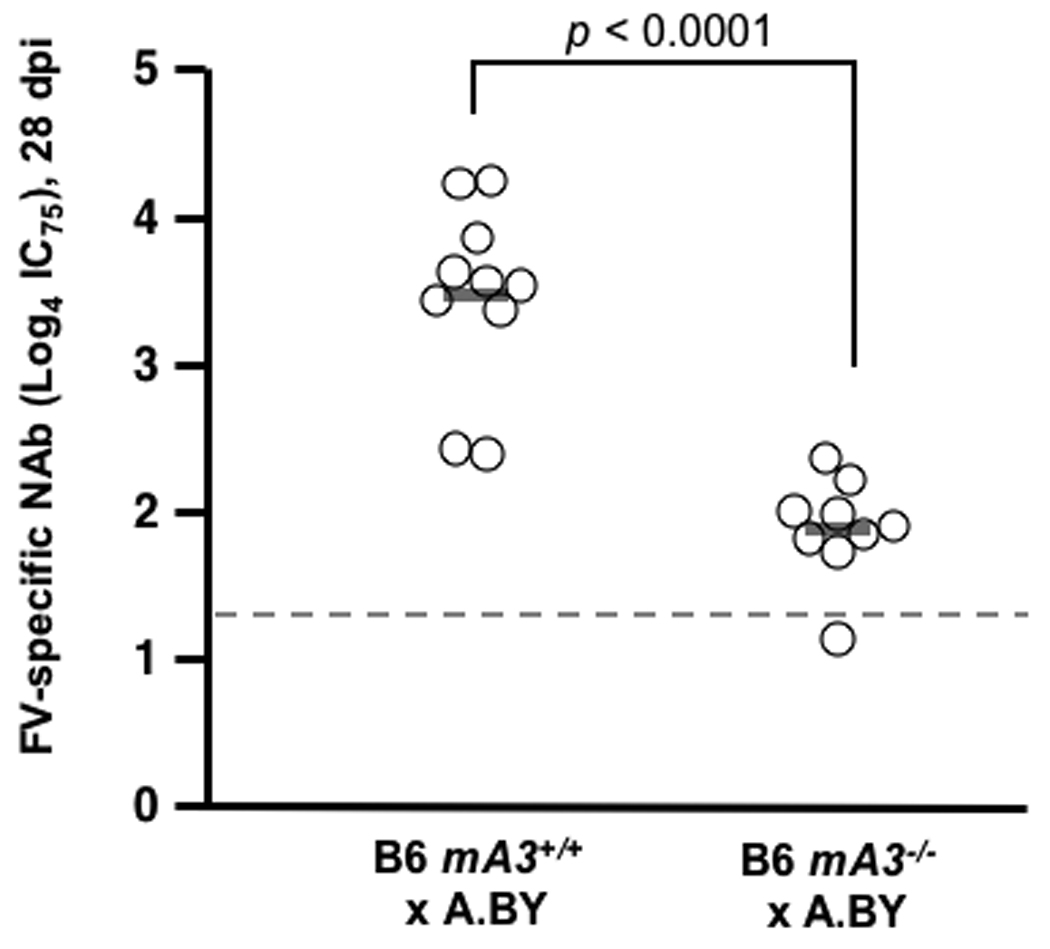
The Rfv3 genetic restriction is mediated by mA3 in vivo. (A–C) mA3 confers the Rfv3 phenotype in low-recovery H-2b/d mice. Congenic mA3+ and mA3− (B6 × BALB/c) F1 mice were infected with 140 spleen focus-forming units (SFFU) of FV. A, mA3 is required for viremic control at an early timepoint (7 dpi); B, mA3 inactivation compromises Rfv3-associated survival from FV-induced disease; C, mA3 influences FV-specific IgG production 14 dpi. FV binding IgG was measured by flow cytometry using FV antigen-expressing FBL-3 cells. (D) mA3 confers the Rfv3 phenotype in high-recovery H-2b/b mice. Congenic (A.BY × BALB/c)F1 mice were infected with 1400 SFFU of FV. Mean 28 dpi neutralizing antibody (NAb) titers (75% inhibitory concentration, IC75) are significantly lower in mA3 − F1 mice. Open circles indicate individual mice data, gray bars indicate means and dashed lines refer to the assay detection limit. Statistical analyses were performed with two-tailed Student’s t test.
Rfv3-mediated recovery from FV infection correlates strongly with FV-specific neutralizing antibody responses at 28 dpi (7). Thus, mA3+ and mA3− F1 congenic strains were monitored for up to one month following FV infection. However, F1 mice lacking the B6 mA3 allele suffered a markedly higher rate of FV-induced death (Fig. 1B) indicating that like Rfv3s susceptibility, mA3 inactivation compromised recovery from FV disease. Compared to mA3+ F1 mice, the three surviving mA3− F1 mice exhibited 14-fold higher mean viremia (Fig. S2A) and low FV-specific neutralizing antibody titers at 28 dpi (Fig. S2B), but the small number of surviving animals precluded obtaining statistically significant data. Therefore, separate cohorts of mice were studied for FV-specific antibodies at 14 dpi, prior to the steep decline in survival of mA3− F1 mice. The mA3− F1 mice exhibited significantly less FV binding antibody than mA3+ F1 mice and the low levels of FV antibodies in mA3− F1 mice proved comparable to levels detected in the parental Rfv3s/s BALB/c mice (Fig. 1C). These findings indicated that mA3 influenced FV-specific antibody responses.
To better assess FV-specific neutralizing antibody responses in mice expressing or lacking mA3, these studies were repeated in high-recovery B6 × A.BY F1 mice, which generally survive more than 1 month after FV infection due to protective cell-mediated immune responses associated with the H-2b/b haplotype (Table 1) (5, 15). Plasma samples obtained at 28 dpi revealed significantly lower FV-specific neutralizing antibody titers in mA3− F1 mice compared to mA3+ F1 mice (Fig. 1D). These findings confirmed that mA3 influenced FV-specific neutralizing antibody responses, and demonstrated that this effect operated independently of H-2, a known property of Rfv3 (7).
Purebred B6 mice are highly resistant to FV infection (Table 1), but their resistance can be overcome by inoculating aged mice with high doses of FV (16) or by using immunodeficient mice, including those that fail to produce specific antibodies (9). Genetic inactivation of mA3 in B6 mice might therefore recapitulate the Rfv3s susceptible phenotype without a requirement for outcrossing to susceptible strains. To test this possibility, >16 week-old B6 mA3+/+ and mA3−/− mice were infected with FV. Plasma viremia was 6.2-fold higher in mA3−/− mice than in mA3+/+ mice at 8 dpi (Fig. 2A). Furthermore, mA3−/− mice exhibited significantly lower neutralizing antibody titers than wild-type mice by 28 dpi (Fig. 2B). Thus, mA3 inactivation was sufficient both to enhance viremia and to diminish neutralizing antibody production even in the highly resistant B6 genetic background. These results were confirmed in a second highly resistant strain of mice, 129/Ola (Table 1, Supp. Text and Fig. 2C). Together, these findings demonstrate that genetic inactivation of mA3 recapitulates the Rfv3s phenotype and indicate that Rfv3 is encoded by mA3.
Fig. 2.
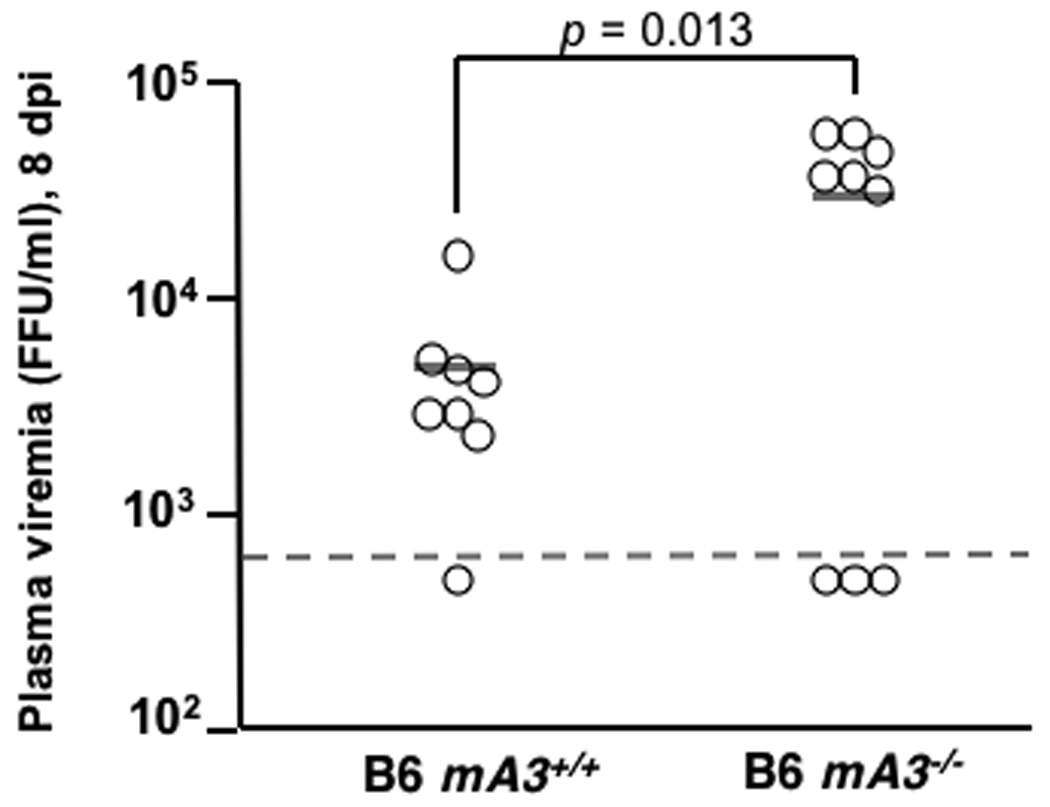
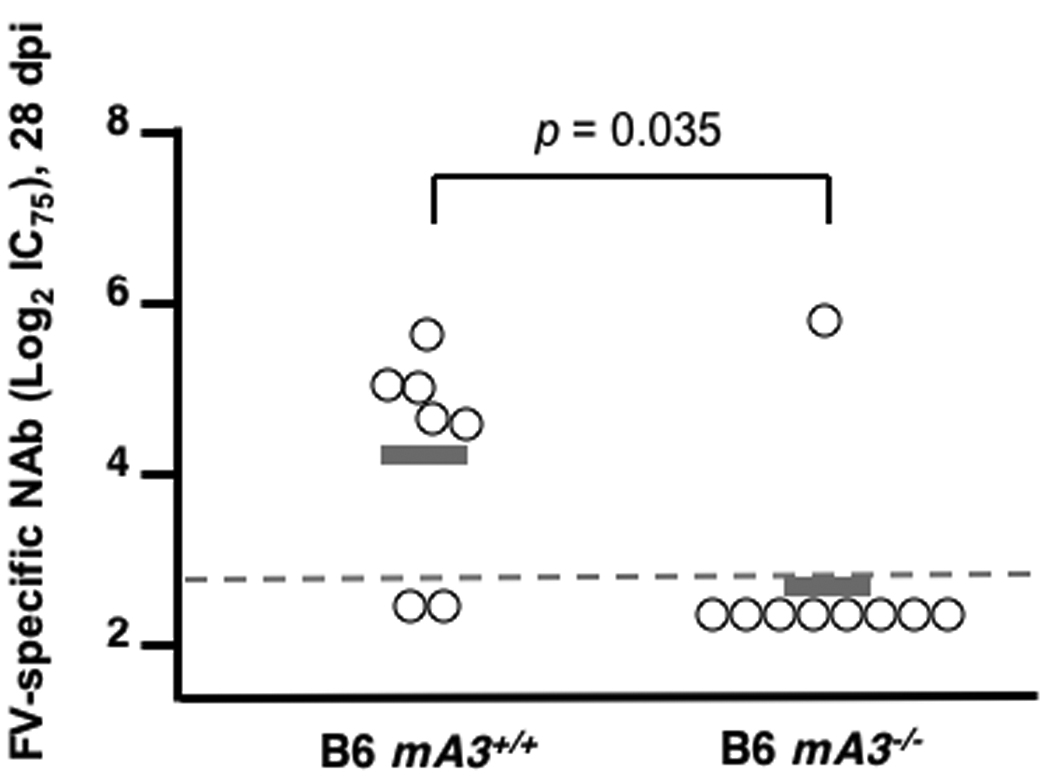
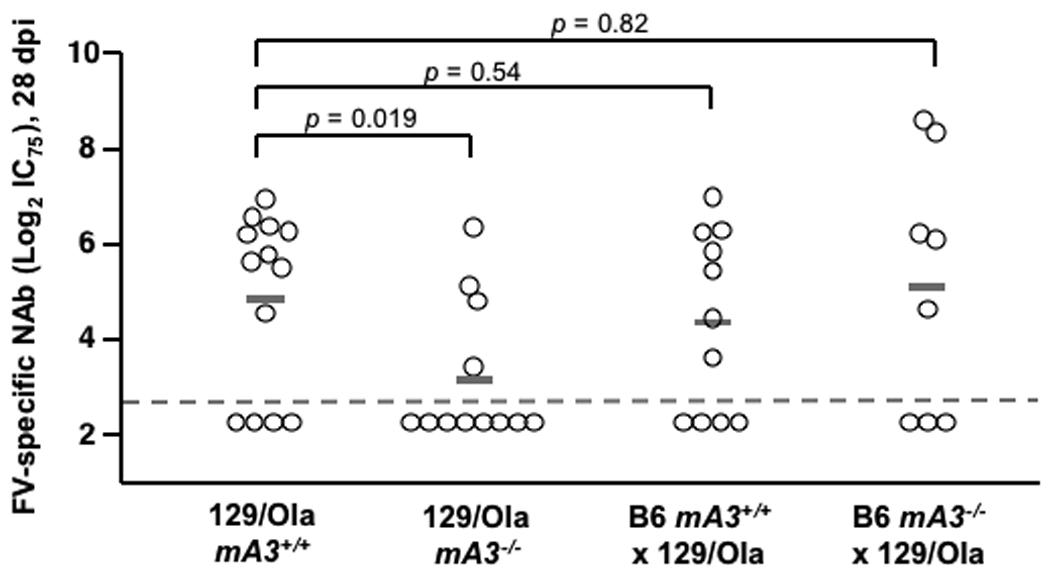
mA3 influences FV-specific neutralizing antibody responses in highly resistant (Fv2r/r) mice. (A–B) mA3+/+ and mA3−/− B6 mice (>16 weeks old) were infected with 5000 SFFU of FV. A, mA3 influences early viremic control at 7 dpi; B, mA3 is required for FV-specific neutralizing antibody production 28 dpi. (C) mA3 influences neutralizing antibody responses in 129/Ola mice. mA3+/+ and mA3−/− 129/Ola mice were crossed with mA3+/+ and mA3−/− B6 mice to generate F1 offspring. FV-specific neutralizing antibody titers (IC75) at 28 dpi with 5000 SFFU of FV are shown. Additional information is found in the Supplementary Text. Statistical analysis was performed using a two-tailed Student’s t test.
Both resistant and susceptible mouse strains contain the mA3 gene and express mA3 mRNA. Cloning of splenocyte mA3 mRNA from the Rfv3r/r 129/Ola strain revealed the predominant expression of a full-length mA3 transcript, while most mRNA transcripts from Rfv3r/r B6 mice lacked exon 5 sequences (Fig. S3 and Fig. S4) (17). mA3 transcripts from both Rfv3s/s strains BALB/c and A.BY were distinguished by the absence of exon 2 sequences (Fig. 3A and Fig. S4). Quantitative RT-PCR revealed similar levels of total mA3 mRNA in both the Rfv3r/r and Rfv3s/s strains but a 17-fold lower level of Exon2-containing transcripts in both Rfv3s/s mouse strains (Fig. 3B). Thus, the presence of an alternatively spliced mA3 mRNA lacking exon 2 correlated with the Rfv3s-susceptible phenotype.
Fig. 3.
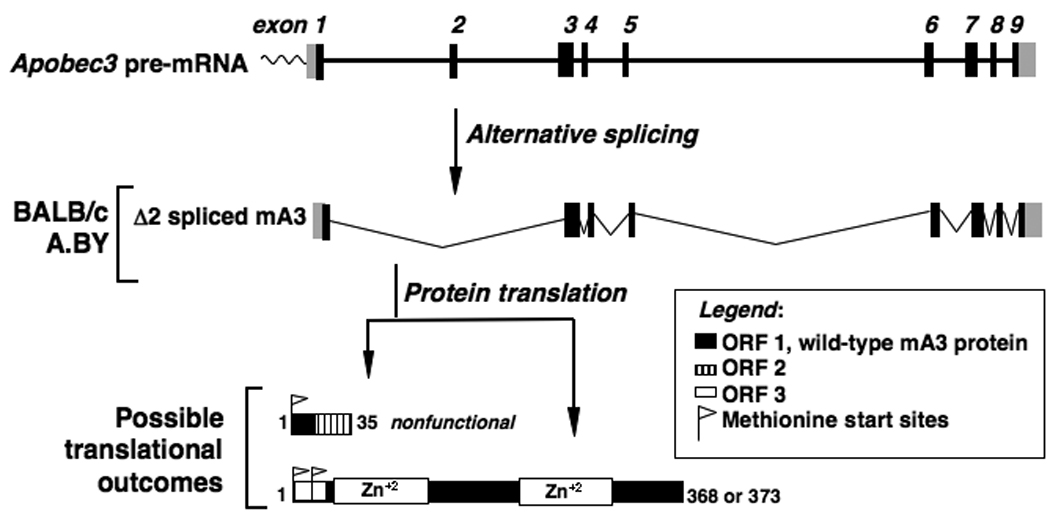
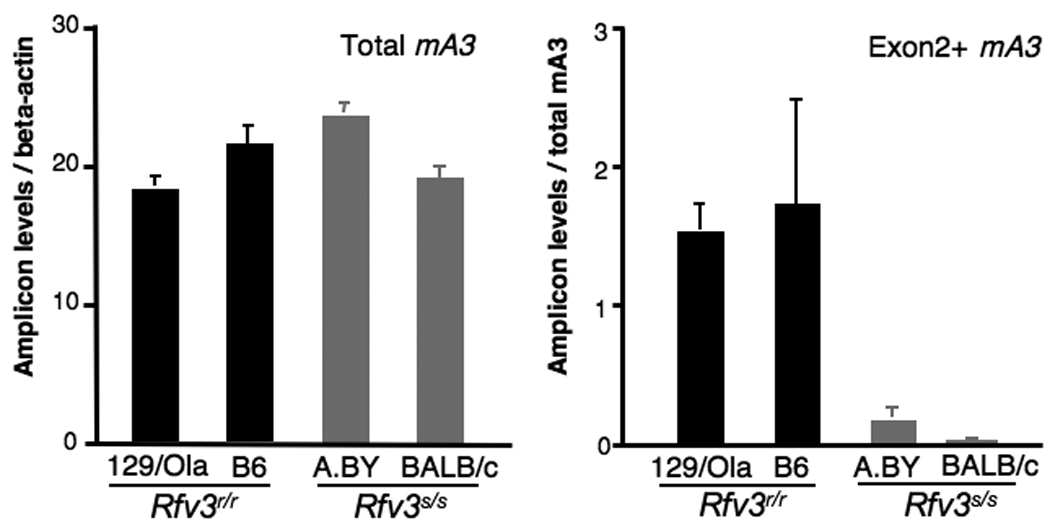
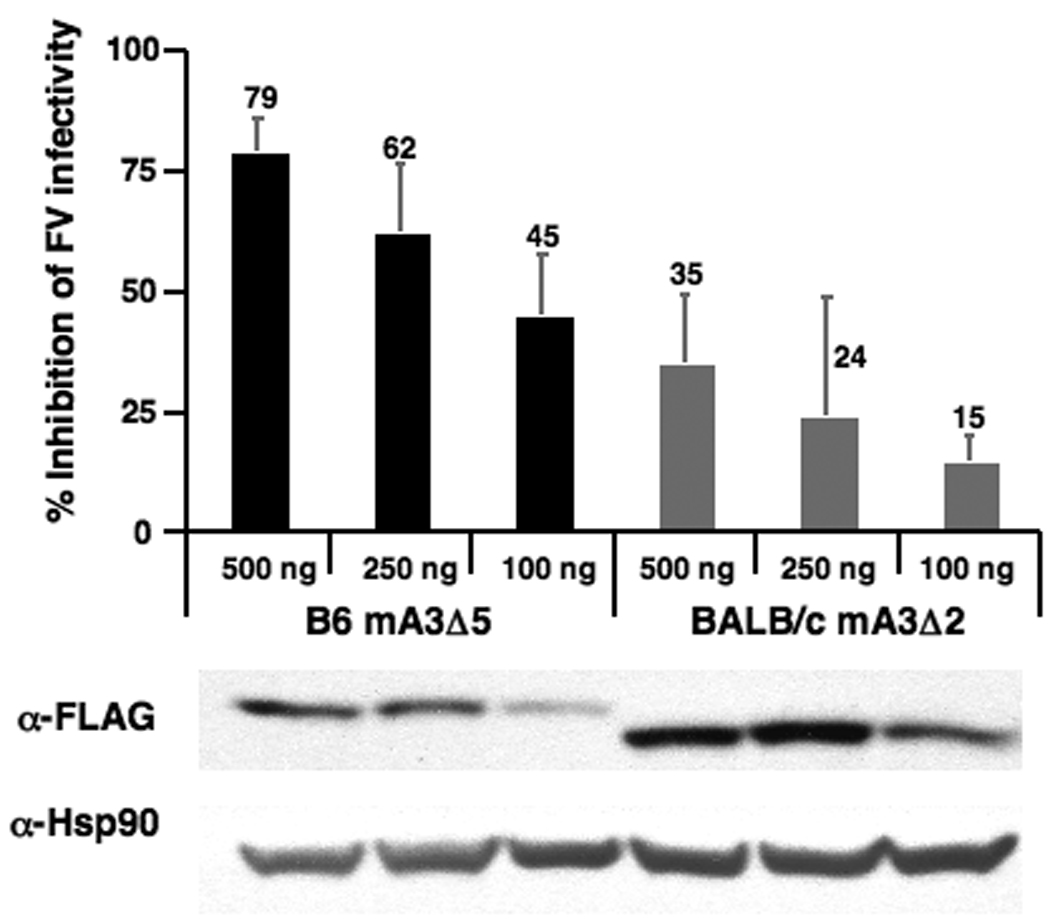
Molecular basis of Rfv3s susceptibility. (A) Aberrant mA3 exon 2 splicing in Rfv3s/s mice. If the wild-type mA3 open reading frame (ORF1) is used, frameshift-induced translational termination will result in a nonfunctional peptide. However, two start sites in an alternative reading frame (ORF3) may be utilized to translate a mutant mA3 protein with a novel N-terminus and a 56 amino acid deletion. (B) Decreased Exon2+, but not total mA3 transcripts, in Rfv3s/s mice. Quantitative RT-PCR of total (left) and Exon2+ (right) mA3 splenocyte transcripts in Rfv3r/r and Rfv3s/s mouse strains were performed. Amplification levels were normalized to beta-actin (left) or total mA3 (right). (C) Decreased antiviral activity of the mA3 Δexon2 relative to the mA3 Δexon5 spliceoform. An FV molecular clone was co-transfected in NIH3T3 cells with FLAG-tagged mA3 constructs. The infectivity of harvested virions were assayed in Mus dunni cells and normalized against reverse transcriptase activity. Co-transfections with FLAG vector alone were set at 0% inhibition (not shown). Expression of mA3 was assessed by anti-FLAG immunoblotting. Error bars correspond to SD from triplicate experiments.
Translational read-through of the mA3 mRNA lacking exon 2 predicts two possible outcomes: no mA3 protein expression, or a truncated mA3 protein (Fig. 3A and Fig. S5). We investigated the antiviral activity of this Δexon 2 mA3 protein by cloning it as a fusion with an N-terminal triple FLAG epitope tag. NIH3T3 cells were co-transfected with the expression construct and an FV molecular clone (pLRB302) (18) to test the infectivity of the resulting FV virions. For comparison, co-transfections were performed with a FLAG-tagged expression construct containing the mA3 cDNA from B6 mice lacking exon 5. When standardized for relative mA3 expression levels, the BALB/c Δexon 2 mA3 protein was at least 3 to 5-fold less potent at inhibiting FV infectivity than the B6 Δexon 5 mA3 protein (Fig. 3C). In control experiments, the full-length mA3 protein from 129/Ola also potently inhibited FV (Fig. S6). These data indicated that even if a truncated mA3 protein was expressed in Rfv3s/s mice, its antiviral activity would be significantly impaired.
The involvement of mA3 in the control of FV viremia before the onset of adaptive anti-FV immune responses confirms its stature as a bona fide innate immune factor in vivo. In addition, mA3 influences the development of virus-specific neutralizing antibody responses, perhaps by: (1) limiting the early viral antigenic load and evading a form of “high-zone tolerance” (19–21), or (2) by inhibiting early FV-induced injury of critical cell types, such as B cells and antigen-presenting cells, required for the development of FV-specific humoral immunity. However, mA3 is expressed in B cells and is evolutionarily related to activation-induced deaminase, an enzyme that controls somatic hypermutation and class-switch recombination in these cells (22). Thus, mA3 may also be involved in shaping the antibody repertoire.
The human Apobec3 family has been implicated in the control of HIV-1 infection, but HIV-1 encodes Vif, which thwarts the actions of Apobec3G (A3G) and Apobec3F (A3F) (23–26). Compromised A3G/A3F antiviral activity may therefore contribute to the generally poor neutralizing antibody response observed in HIV-1 infection (27). Vif antagonists, if and when they are available, may enhance the generation of effective humoral immune responses against HIV-1. Finally, studies exploring the apparent intrinsic resistance of individuals who are extensively exposed to HIV-1 yet remain uninfected, have genetically mapped this phenotype to chromosome 22q12–13 (12), a location distinguished by a tandem array of the seven human Apobec3 family members. Genome-wide studies of the entire human Apobec3 locus, with particular emphasis on functional differences induced by alternative splicing, are clearly merited to fully explore the potential contribution of this gene family to HIV resistance, neutralizing antibody production, and disease progression.
Supplementary Material
REFERENCES AND NOTES
- 1.Hasenkrug KJ, Chesebro B. Proc. Natl. Acad. Sci. USA. 1997;94:7811. doi: 10.1073/pnas.94.15.7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li W, Green WR. J. Virol. 2006;80:5777. doi: 10.1128/JVI.02711-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Best S, Le Tissier P, Towers G, Stoye JP. Nature. 1996;382:826. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 4.Persons DA, et al. Nat. Genet. 1999;23:159. doi: 10.1038/13787. [DOI] [PubMed] [Google Scholar]
- 5.Britt WJ, Chesebro B. J. Exp. Med. 1983;157:1736. doi: 10.1084/jem.157.6.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chesebro B, Wehrly K. In: Advances in Comparative Leukemia Research. Bentvelzen, editor. Elsevier: North-Holland Biomedical Press; 1978. pp. 69–73. [Google Scholar]
- 7.Chesebro B, Wehrly K. Proc. Natl. Acad. Sci. USA. 1979;76:425. doi: 10.1073/pnas.76.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doig D, Chesebro B. J. Exp. Med. 1979;150:10. doi: 10.1084/jem.150.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasenkrug KJ. J. Virol. 1999;73:6468. doi: 10.1128/jvi.73.8.6468-6473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasenkrug KJ, et al. J. Virol. 1995;69:2617. doi: 10.1128/jvi.69.4.2617-2620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Super HJ, et al. J. Virol. 1999;73:7848. doi: 10.1128/jvi.73.9.7848-7852.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanari Y, et al. AIDS. 2005;19:1015. doi: 10.1097/01.aids.0000174447.48003.dd. [DOI] [PubMed] [Google Scholar]
- 13.Chiu YL, Greene WC. Annu. Rev. Immunol. 2008;26:317. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 14.Materials and methods are found in the Supporting Online Material, http://___.
- 15.Chesebro B, Wehrly K. J. Exp. Med. 1976;143:73. doi: 10.1084/jem.143.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van der Gaag HC, Axelrad AA. Virology. 1990;177:837. doi: 10.1016/0042-6822(90)90561-5. [DOI] [PubMed] [Google Scholar]
- 17.Rulli SJ, Jr, et al. J. Virol. 2008;82:6566. doi: 10.1128/JVI.01357-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Portis JL, McAtee FJ, Kayman SC. J. Acquir. Immune Defic. Syndr. 1992;5:1272. doi: 10.1097/00126334-199212000-00011. [DOI] [PubMed] [Google Scholar]
- 19.Monroe JG, Lowy A, Granstein RD, Greene MI. Immunol. Rev. 1984;80:103. doi: 10.1111/j.1600-065x.1984.tb00497.x. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez-Fernandez A, Milstein C. Immunology. 1998;93:149. doi: 10.1046/j.1365-2567.1998.00423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim YT, Siskind GW. Clin. Exp. Immunol. 1974;17:329. [PMC free article] [PubMed] [Google Scholar]
- 22.Muramatsu M, et al. Cell. 2000;102:553. [Google Scholar]
- 23.Marin M, Rose KM, Kozak SL, Kabat D. Nat. Med. 2003;9:1398. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 24.Sheehy AM, Gaddis NC, Malim MH. Nat. Med. 2003;9:1404. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 25.Stopak K, de Noronha C, Yonemoto W, Greene WC. Mol. Cell. 2003;12:591. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 26.Zheng YH, et al. J. Virol. 2004;78:6073. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burton DR, et al. Nat. Immunol. 2004;5:233. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 28.We thank the Transgenic Core Laboratory, the Animal Facility and S. Espineda at the J. David Gladstone Institutes for technical assistance, L. Evans, J. Portis, R. Gallo, R. Locksley, and members of the Greene and Hasenkrug Laboratory for helpful discussions, and R. Givens, S. Cammack, G. Howard for manuscript preparation. This work was supported by the NIAID Division of Intramural Research at NIH to K.J.H and B.C., an NIH R01 AI065329 to W.C.G., and an NIH facility grant to the J. David Gladstone Institutes. Sequences are deposited in GenBank, with accession numbers EU707568-EU707571.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


