Abstract
Carbon monoxide (CO) is a biologically active molecule produced in the body by the stress-inducible enzyme, heme oxygenase. We have previously shown that CO suppresses fibrosis in a murine bleomycin model. To investigate the mechanisms by which CO opposes fibrogenesis, we performed gene expression profiling of fibroblasts treated with transforming growth factor-β1 and CO. The most highly differentially expressed categories of genes included those related to muscular system development and the small proline-rich family of proteins. We confirmed in vitro, and in an in vivo bleomycin model of lung fibrosis, that CO suppresses α–smooth muscle actin expression and enhances small proline-rich protein-1a expression. We further show that these effects of CO depend upon signaling via the extracellular signal–regulated kinase pathway. Our results demonstrate novel transcriptional targets for CO and further elucidate the mechanism by which CO suppresses fibrosis.
Keywords: carbon monoxide, heme oxygenase-1, lung fibrosis, small proline-rich protein, α-smooth muscle actin
CLINICAL RELEVANCE
Although carbon monoxide (CO) has been shown to confer protection in the setting of lung fibrosis, the mechanism by which it does this remains poorly understood. This study points to potential mechanisms by which CO may inhibit fibrogenesis and introduces a new candidate molecule that may be involved in the pathogenesis of fibrosis: small proline-rich protein-1.
Previous studies from our laboratory indicate that matrix production by fibroblasts in vitro and in vivo is inhibited by exposure to a low concentration of carbon monoxide (CO) (1). CO is a biologically active molecule produced in the body via catabolism of heme by the stress-inducible enzyme, heme oxygenase. It has protective effects in the setting of lung injury from a number of different causes (2–5), including bleomycin-induced fibrosis (1). The mechanism for the suppression of fibrosis by CO is not known. To identify individual genes or pathways responsible for the effects of CO, we performed gene profiling of murine fibroblasts exposed to transforming growth factor (TGF)-β1 alone or TGF-β1 and CO for 24 hours. Among the functional categories of genes, those related to the ubiquitin proteasome pathway and to skeletal and muscular system development were most highly differentially expressed in the microarray. Genes in this second category are associated with the myofibroblast phenotype, and are known to be highly induced in fibroblasts by TGF-β1 (6). Fibroblasts expressing proteins involved in cell contraction, in particular α-smooth muscle actin (α-SMA), are responsible for the generation of contractile force and extracellular matrix deposition associated with wound healing (7).
Of the individual genes that were most highly differentially expressed, 3 of the top 10 most statistically significant genes belonged to the family of small proline-rich (Sprr) proteins. All three of these genes were modestly induced by TGF-β1, but were greatly upregulated by the addition of CO. Members of this family of genes was originally identified as markers of terminal squamous cell differentiation (8), acting as substrates of transglutaminase in the formation of the crosslinked envelope (9). Sprr genes comprise a four-member family (Sprr1–4) contained within a 170-kb region located on chromosome 1q21 (10). Given their biological annotation as mediators of keratinization, Sprr genes did not appear in any of the functional groupings of genes that were differentially expressed in the microarray. To date, there is no known function of Sprr family genes in fibroblasts or in the progression of fibrosis. However, Sprr genes have been identified as being among the most highly upregulated genes in the setting of injury or inflammation in the lung, intestine, biliary tree, skin, arterial intima, cardiomyocytes, and neurons (11–13). In the lung, cigarette smoke and vitamin A have been shown to induce epithelial expression of Sprr1a, and allergic inflammation in ovalbumin and aspergillus models has been shown to increase Sprr2a, Sprr2b, and Sprr1a expression (14). In these models of allergic inflammation, Sprr2 expression was shown to be increased in a broad spectrum of lung epithelial cells, as well as infiltrating mononuclear cells. The function of these proteins in the lung remains unknown. Recent reports suggest that Sprr1a provides protection against cardiomyocyte death in the setting of ischemia–reperfusion injury (12), and that Sprr2a promotes wound healing through enhanced epithelial cell migration (15). These reports suggest possible functions for Sprr proteins in the lung, but these have yet to be investigated.
MATERIALS AND METHODS
Animals
Male C57BL/6 mice were purchased from Jackson Laboratory (Bar Harbor, ME). All animals were housed in accordance with guidelines from the American Association for Laboratory Animal Care and Research Protocols, and were approved by the Animal Care and Use Committees of Pittsburgh University School of Medicine. Bleomycin (Bristol-Myers Squibb, Princeton, NJ) was administered intratracheally as previously described (1) at a dose of 0.075 units/mouse in 50 μl sterile saline.
CO Exposures
Mice or cells were exposed to compressed air with CO at a concentration of 250 ppm. CO exposure of mice began immediately after treatment with bleomycin, and exposure of cells began immediately after stimulation with TGF-β1. For cell culture experiments, the atmosphere included 5% CO2 for adjustment of pH. Details of CO administration have been previously described (3).
Cell Culture Experiments
Mouse lung fibroblasts were isolated from C57Bl/6 mice and were maintained and grown in Dulbecco's modified Eagle's medium containing 10% FBS and 1% pen-strep. TGF-β1 (R&D Systems, Minneapolis, MN) was reconstituted in sterile 4 mM HCL containing 1% BSA. Tricarbonyldichlororuthenium(II) dimer (CORM3) (Sigma, St. Louis, MO), PD098059, and SB203580 (Calbiochem, San Diego, CA) were dissolved in DMSO; control cells were treated with an equivalent volume of DMSO alone.
Microscopic Analysis of α-SMA Expression
At 7 days after the administration of bleomycin with or without the addition of CO, lungs were harvested and inflation fixed with 2% paraformaldehyde in PBS and immersed in the same fixative for 1 hour. The lungs were then placed in 30% sucrose in PBS overnight, and flash frozen in liquid nitrogen–cooled isopentane. Lung cryosections (5 μm) were cut and washed sequentially in PBS, then in PBS containing 0.5% BSA and 0.15% glycine. The sections were washed and incubated in a 1:1,000 dilution of monoclonal cy3–conjugated anti-mouse α-SMA antibody (American Research Products, Inc., Belmont, MA), with anti-mouse vimentin (Santa Cruz Biotechnology, Santa Cruz, CA) or anti-mouse pro–surfactant protein C (Chemicon, Atlanta, GA) for 1 hour. After three washes in BSA, the sections were stained with a 1:3,000 dilution of cy3-conjugated AffinPure Goat Anti-Rabbit IgG (H+L; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) for 1 hour, washed and stained with Hoechst dye (Sigma) for 30 seconds, and mounted in Gelvatol (Monsanto, St. Louis, MO). Images were collected with an Olympus Provis microscope (Olympus, Tokyo, Japan) as well as an Olympus Floview scanning confocal microscope.
Cell Extracts and Western Blot Analysis
Cellular protein extracts were collected and electrophoresed under denaturing conditions as previously described (1). α-SMA and Sprr1a were detected using rabbit polyclonal antibodies according to the manufacturer's suggestions (Santa Cruz Biotechnology and Abcam [Cambridge, MA], respectively). The imaging and analysis of the blots were performed as previously described (1).
α-SMA Promoter Constructs and Luciferase Assay
The α-SMA construct was kindly provided by Dr. S.H. Phan (Department of Pathology, University of Michigan Medical School, Ann Arbor, MI). Details have been previously published (16, 17). Transfection of fibroblasts was performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. pRL-TK control vector (Promega, Madison, WI) was used to normalize transfection efficiencies. Luciferase activity was assessed using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. The construct was tested in triplicate, and the data shown represent three to four independent experiments.
DNA Microarray Hybridization and Analysis
Fibroblasts treated with TGF-β1 alone for 24 hours were compared with fibroblasts treated with both TGF-β1 and CO 250 ppm for 24 hours. Control groups included untreated fibroblasts and cells exposed to CO alone. Extracted RNA was used to generate labeled cRNA and then hybridized to Codelink mouse 20K arrays that were scanned and normalized as previously described (18). Statistical analyses were performed using Scoregene as previously described (19). For data mining and visualization, we used Genomica, Ingenuity Pathways Analysis (Ingenuity Systems, Redwood City, CA) (20) and Spotfire Decision Site 8.0 (Spotfire Inc., Göteborg, Sweden). To identify individual genes that best distinguish between TGF-β1–treated and CO plus TGF-β1–treated cells, we used the threshold number of misclassification (TNoM) score (21), which counts the number of classification errors that occur between compared groups for each gene of the dataset, as well as the Student's t test and significance analysis of microarrays (SAM) (22).
Statistical Analysis
Experiments were repeated at least three times, and statistical evaluation was performed using an unpaired Student's t test. In all cases, two-tailed P values less than 0.05 were considered statistically significant.
RESULTS
Expression Profiling in Response to TGF-β1 and CO
To examine the effect of CO on fibroblast behavior in the setting of TGF-β1 stimulation, gene expression was compared between fibroblasts stimulated with TGF-β1 alone and those treated with both TGF-β1 and CO for a period of 24 hours. The most significantly enriched canonical pathway was the protein ubiquitination pathway, and the most significantly enriched functional categories included skeletal and muscular system development and function, cell migration, and cell death. Although most of the statistically significant genes were included in the functional categories listed above, strikingly, three of the most affected genes belonged to the small proline-rich family of proteins (Sprr1a, Sprr2a, and Sprr3). This protein family is not included in any of the functional categories or canonical pathways listed, as the gene ontology (GO) annotation biological process for these genes includes epidermis development and keratinization. These genes are therefore discussed separately from the brief description of each functional group of genes provided subsequently here.
The Protein Ubiquitination Pathway
The ubiquitin proteasome pathway, which is responsible for degradation of the majority of intracellular proteins, had the lowest log P value in the microarray analysis. Ubiquitin, which marks proteins for degradation, is affected by CO treatment, as are components of the 26S proteasome. All three enzymatic components required to link chains of ubiquitin to proteins destined for degradation (E1 [ubiquitin-activating enzyme], E2 [ubiquitin-carrier or conjugating proteins], E3 [ubiquitin-protein ligase]) have members demonstrating decreased transcription in response to CO. There is little known about the effects of CO on protein degradation. A recent study examining the effect of CO on endothelial cell apoptosis reported inclusion bodies with ubiquitin-decorated material, indicating altered proteolytic processing in response to CO (23). The functional consequence of this effect is unknown.
Genes Related to Skeletal and Muscular System Development and Function
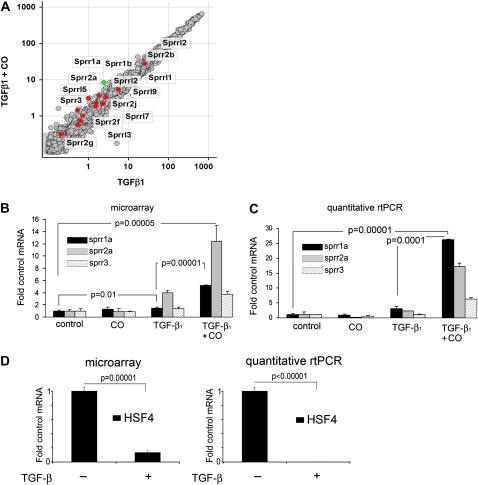
With the exception of EGR1 (5, 24), the genes in this functional category were not previously known to be regulated by CO, although the majority are known to be TGF-β1 responsive. For example, cytoskeletal genes involved in cell contraction were upregulated by TGF-β1, as might be expected during transition to a myofibroblast phenotype (ACTA2, TAGLN, ACTG2, TNNT3); expression of these genes was inhibited by CO. Similarly, the expressions of genes known to regulate myogenesis changed in response to both TGF-β1 and CO, including RRAS2, WNT10B, USMG1, and DACH2. One of the genes in this category most profoundly suppressed by TGF-β1 was the transcriptional regulator, heat shock factor (HSF)-4 (10-fold decrease, P = 0.00001); the addition of CO antagonized the effect of TGF-β1, partially restoring the expression of HSF4. We confirmed these findings by real-time PCR (shown in Figure 1). Known HSF4 target genes, fibroblast growth factor 7 and heat shock proteins 68 were also differentially expressed in our array in response to TGF-β1 and CO. This finding is notable as the first report of regulation of HSF4 by TGF-β1 and as a potential link between the heat shock response and fibrosis. HSF4 is expressed primarily in the adult brain and lung, and has been implicated, along with TGF-β1, in cataract formation (25).
Figure 1.
Small proline-rich (Sprr) family proteins are highly up-regulated in fibroblasts by the addition of carbon monoxide (CO) to transforming growth factor (TGF)-β1 stimulation. (A) The scatter plot shows Sprr family genes highlighted in red (Sprr1a is shown in green). The relative expression of Sprr1a, Sprr2a, and Sprr3 mRNA in response to TGF-β1 treatment and TGF-β1 plus CO treatment are shown by microarray analysis (B) and real-time PCR (C). (D) Expression of heat shock factor 4 (HSF4) mRNA in response to TGF-β1 treatment assessed by microarray analysis and real-time PCR.
Genes Related to Cell Movement
There is little known about the effects of CO on cell movement, although recent reports suggest that CO is capable of down-regulating leukocyte adhesion and migration (26, 27). The genes in this category overlap with the functional classification for tumor cell invasion, and the majority of these were previously known to be TGF-β1 responsive. In addition to regulating tumor invasion, TGF-β1 is thought to provide a stimulus for fibroblast migration in the setting of wound healing (28, 29). None of the genes in this functional classification was known to be responsive to CO, with the exception of EGR1, as described above. The genes affected by CO contribute to cell migration by a variety of mechanisms, such as cytoskeletal organization (e.g., ZYX, TAGLN, CRK, and NEDD9), proteolysis (e.g., SERPINA3, PLAUR, MMP9, and MMP13), cell adhesion (CD97), and transcriptional regulation (RUNX3 and EGR1).
Genes Related to Cell Death
Both TGF-β1 and CO have previously been implicated in regulation of apoptosis (30–34). Although genes in this functional category regulate apoptosis by a variety of mechanisms, this grouping includes p53 homolog p63 and a number of genes in the p53 network (WWOX, EGR1, HSPA1B, and POLB). p53 Tumor suppressor is a transcription factor that coordinates cellular responses to DNA damage and stress, initiating cell cycle arrest or triggering apoptosis. CO has previously been shown to induce growth arrest via modulation of expression of the p53 target gene, p21 (1, 35, 36), and the antiapoptotic action of CO in vascular smooth muscle cells has previously been associated with suppression of p53 expression (37). Our current findings support a role for p53 in CO-mediated suppression of apoptosis.
Small Proline-Rich Proteins
Of the individual genes that were most responsive to CO, three with the most statistically significantly up-regulated genes in response to CO belong to the family of Sprr proteins (Figure 1A). Sprr proteins were originally identified as markers for the terminal squamous cell differentiation and protein precursors of mammalian cornified envelope (9). This family of proteins is also expressed in nonsquamous cells, although their biological function in these tissues is not known. Several studies have shown that expression of Sprr1a and -2a is increased in response to cellular stress (11, 12, 38). We confirmed by real-time PCR that Sprr1a, Sprr2a, and Sprr3 mRNA are increased in fibroblasts stimulated concomitantly with TGF-β1 and CO (Figure 1C).
Effect of CO on α-SMA Expression In Vitro and In Vivo
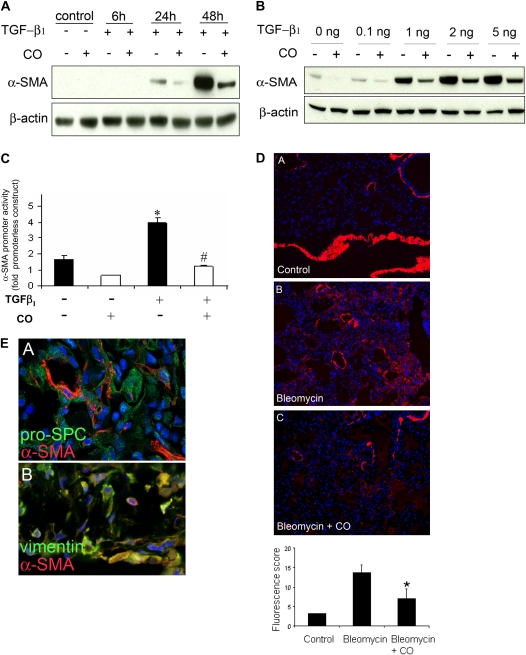
Given that genes related to skeletal and muscular system development were differentially expressed in the microarray experiment, we examined the effect of CO on fibroblast expression of α-SMA. Fibroblasts express α-SMA in response to TGF-β1 stimulation, and this change is associated with a profibrotic myofibroblast phenotype (39). We hypothesized that CO might inhibit lung fibrosis in part by suppressing the development of myofibroblasts from fibroblasts. To examine this question in vitro, cultured fibroblasts were stimulated with TGF-β1 (5 ng/ml) and then exposed to CO or control air. Cell lysate was harvested after 6, 24, and 48 hours for Western blot analysis of α-SMA expression. As illustrated in Figure 2A, TGF-β1 induced a time-dependent increase in α-SMA expression, and this was suppressed by treatment with CO. The same experiment was repeated using a TGF-β1 dose range of 0.5–5 ng/ml (Figure 2B); CO suppressed α-SMA expression throughout this dose range. CO had a similar effect on TGF-β1–induced α-SMA promoter activity, as shown in Figure 2C. To test whether the suppressive effect of CO on α-SMA expression occurs in vivo, bleomycin-treated mice were exposed to inhaled CO (250 ppm) or room air for 1 week, and lungs were harvested for α-SMA staining. As shown in Figure 2D (image A), mice that were untreated expressed little lung parenchymal α-SMA. After bleomycin treatment, α-SMA staining in the lung increased as expected (image B). Addition of inhaled CO to bleomycin treatment resulted in less α-SMA staining than bleomycin alone (image C). Quantitation of immunofluorescence for each group is shown in the Table 1. To address the question of which cell type was expressing α-SMA, costaining was performed using vimentin, which is primarily expressed in fibroblasts, and pro–surfactant protein C, which labels type-II pneumocytes. The greatest amount of costaining occurred with α-SMA and vimentin (Figure 2E).
Figure 2.
CO suppresses α–smooth muscle actin (α-SMA) expression in vitro and in vivo. Fibroblasts treated with TGF-β1 (5 ng/ml) were subsequently exposed to CO (250 ppm) or control incubator air. (A) Cells were analyzed by Western blotting for α-SMA expression at the time points indicated. (B) Effect of varying doses of TGF-β1. (C) The effect of CO on TGF-β1–induced activation of the α-SMA promoter was examined using a rat wild-type promoter construct with a luciferase reporter. *P < 0.05 compared with control; #P <0.05 compared with TGF-β1–treated. (D) The same suppressive effect of CO occurs in vivo; bleomycin-treated mice were exposed to inhaled CO (250 ppm) or room air for 1 week, and lungs were harvested for α-SMA staining. Image A is of untreated lung, image B is of bleomycin-treated lung, and image C is of bleomycin and CO–treated lung. Red stain represents α-SMA expression; immunofluorescence is quantitated in the accompanying graph (bleomycin-treated animals, n = 4–9; untreated control animals, n = 1). Quantitation excluded staining in airways and blood vessels to exclude smooth muscle cells. *P < 0.05 compared with bleomycin-treated wild type. (E) Costaining of α-SMA and cell markers vimentin and pro-pro–surfactant protein C. The majority of costaining (yellow) occurs in α-SMA and vimentin-stained cells shown in the lower image.
TABLE 1.
FOUR HIGHEST-SCORING CATEGORIES OF GENES GROUPED ACCORDING TO BIOLOGICAL FUNCTION
| Functional Category | Range P Values | Log P Value | Genes |
|---|---|---|---|
| Ubiquitin proteasome pathway | 1 × 10−4 to 2 × 10−2 | −4.0 | AMFR, BIRCH4, PSMB3, PSMB5, PSMB8, PSMD1, PSMD3, PSMD8, PSMD14, SMURF1, TCEB1, UBD, UBE2A, UBE2J2, USP1, USP5, USP26, USP38, USP9X, VDP, VHL |
| Skeletal and muscular system development and function | 1 × 10−4 to 4 × 10−2 | −3.8 | MMP9, MMP13, SMURF1, ANKH, MEOX1, PHC1, RPS6KA3, TP73L, SOX2, FHL2, ACTA2, TNNT2, TNNT3, TMOD4, WNT10B, EGR1, PLAUR, HSF4, PDLIM7, SPNB2, DACH2, RRAS2, USMG1, MTM1, TAGLN, ACTG2 |
| Cell movement | 2 × 10−4 to 4 × 10−2 | −3.7 | PLAUR, TAGLN, CD97, MMP9, CRK, EGR1, ANGPTL4, APBB2, BDNF, CXCL13, DUSP1, FHL2, NEDD9, PENK, PPBP, SERPINA3, SEMA3C, ZYX, SH2D2A, CXCR6, PRKD1, TP73L, ALOX5AP |
| Cell death | 2 × 10−4 to 4 × 10−2 | −3.6 | MMP9, MMP13, BDNF, PPBP, PLAUR, TUB, RUNX3, ATP1A1, MME, HSPA1B, EGR1, WWOX, TRP63, STC2, AKT3, POLB, TLR3, HDAC2 |
Effect of CO on Sprr1a Expression In Vitro and In Vivo
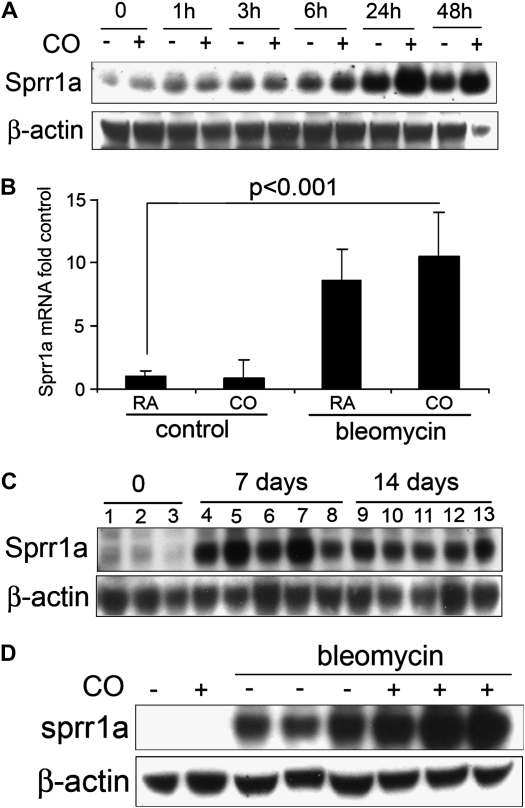
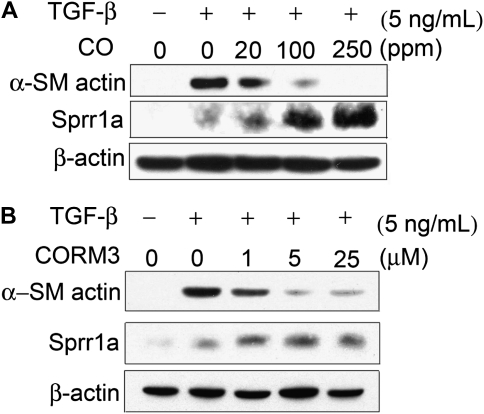
As Sprr1a was the most significantly differentially expressed gene in the microarray experiment, and, as this molecule had previously been shown to confer protection against cellular stress in a manner similar to CO (12), we chose to focus on this member of the Sprr family in our further studies. We confirmed by Western blotting that Sprr1a protein is increased in fibroblasts exposed to TGF-β1, and that the increase is further enhanced by CO exposure (Figure 3A). In mice treated with intratracheal bleomycin and inhaled CO, the expression of Sprr1a mRNA was significantly increased after 7 days (Figure 3B). The maximal expression of Sprr1a protein in whole-lung homogenate occurred at 7 days after bleomycin treatment, and persisted until 14 days (Figure 3C); the increase at 14 days was enhanced by inhaled CO (Figure 3D). The effect of CO on both α-SMA and Sprr1a expression in vitro was dose dependent (Figure 4A). The effect of exogenously administered CO gas was also recapitulated through use of a soluble CO donor molecule, CORM3 (Figure 4B).
Figure 3.
TGF-β1 and CO increase Sprr1a expression in vitro and in vivo. (A) Western blot of mouse lung fibroblasts stimulated with TGF-β1 (2 ng/ml) and exposed to CO or control incubator air. (B) Sprr1a mRNA expression increases in whole-lung homogenate in response to bleomycin and CO (7 d). (C) Sprr1a protein expression is increased in whole-lung homogenate 7 and 14 days after bleomycin exposure. (D) Sprr1a protein expression is further increased in whole-lung homogenate of lungs of mice exposed to CO.
Figure 4.
CO and the CO donor molecule, CORM3, alter Sprr1a and α-SMA expression in a dose-dependent fashion. (A) Treatment of fibroblasts in vitro with CO results in a dose-dependent suppression of α-SMA expression and increase in Sprr1a expression. (B) Treatment with the CO donor molecule, CORM3, has a similar effect.
The Effect of CO on α-SMA and Sprr1a Expression Depends upon Signaling Via Mitogen-Activated Protein Kinase Pathways
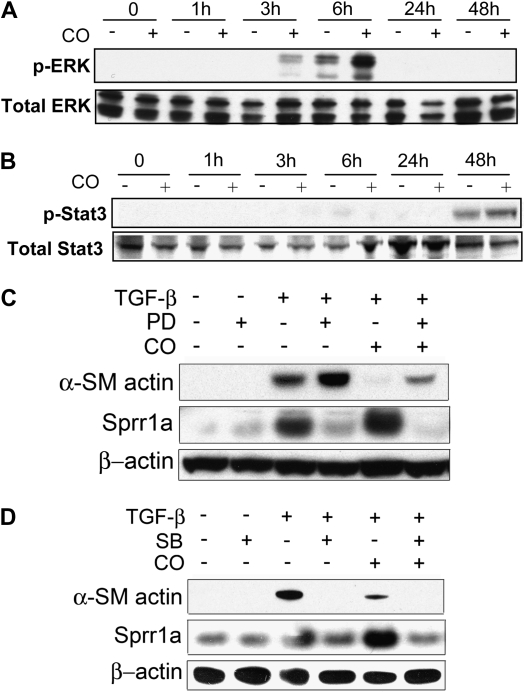
Sprr1a gene activation has previously been shown to depend upon extracellular signal–regulated kinase (ERK) activation downstream of gp130 (12). Because CO has been shown, in some systems, to activate Stat3 (2, 40), which is also downstream of gp130, we tested the effect of combined TGF-β1 and CO stimulation on both ERK and Stat3 activation in fibroblasts. As shown in Figure 5A, TGF-β1 stimulation led to a small increase in ERK phosphorylation at 6 hours; this effect was greatly enhanced by the addition of CO. Stat3 phosphorylation, on the other hand, was not increased by CO (Figure 5B). The mitogen-activated protein kinase kinase 1 (MEK1) inhibitor, PD098059, increased α-SMA expression in response to TGF-β1 and diminished the inhibitory effect of CO. Inhibition of the ERK pathway with PD098059 also abrogated the effect of CO on Sprr1a expression, in this case preventing the enhanced expression of Sprr1a (Figure 5C). Given previously reported involvement of the p38 pathway in CO signaling (35, 41, 42), we also tested the effect of chemical inhibition of the p38 pathway with SB203580. Interestingly, SB203580 led to a complete abrogation of α-SMA expression (Figure 5D). SB203580 inhibited Sprr1a expression in a manner similar to PD098059.
Figure 5.
Effect of extracellular signal–regulated kinase (ERK) and p38 inhibition on Sprr1a and α-SMA expression in the setting of CO treatment. (A) Fibroblasts were stimulated with TGF-β1 with or without CO, and cell lysate was analyzed by Western blotting for ERK phosphorylation. CO exposure enhanced ERK phosphorylation at 3- and 6-hour time points. (B) Stat3 phosphorylation was not significantly affected by CO exposure. (C) Inhibition of mitogen-activated protein/ERK1 kinase led to a loss of the effect of CO on both Sprr1a and α-SMA expression. (D) p38 Inhibition decreased both Sprr1a and α-SMA expression.
DISCUSSION
Although the myofibroblast is thought to play a central role in the development of pulmonary fibrosis (39, 43–45), factors affecting the emergence of this fibroblast phenotype are still poorly understood. We have previously shown that CO inhibits the development of lung fibrosis in a murine bleomycin model (1), and we postulated that this effect of CO may be due to its ability to prevent the emergence of myofibroblasts in the lung. We used microarray to screen for genes or pathways that might contribute to the mechanism of action of CO, and made some expected and unexpected observations. The inhibition by CO of expression of genes related to skeletal and muscular system development and function conformed to our hypothesis that CO was capable of modulating myofibroblast differentiation (8). The decrease in expression of α-SMA with CO treatment in bleomycin-treated mouse lung or TGF-β1–treated fibroblasts correlates with the decrease in collagen synthesis observed with CO treatment (1). Myofibroblasts are important sources of collagen production (39), and it is therefore reasonable to postulate that CO may affect matrix deposition by inhibiting myofibroblast activation.
An unexpected finding was that, among the individual genes most affected by CO treatment in conjunction with TGF-β1, the Sprr family proteins were strikingly up-regulated. These genes were not included in any of the functional groupings of genes, and they have no known function in fibrosis. Sprr proteins were originally identified as markers of terminal squamous cell differentiation (8). Although no role has been identified for Sprr proteins in fibroblasts, recent studies have shown that Sprr genes are induced in response to stress injury (12, 13, 46). When we looked for them in the AsthmaMap database (47), we found that Sprr1 and Sprr2 were both changed in animal models of asthma. Finkelman and colleagues (38) demonstrated in a model of murine asthma that IL-13 reproducibly induced the expression of three Sprr family members in the lung. In a manner similar to CO, up-regulation of Sprr proteins appears to contribute to cell protection (12). The protective effects of heme oxygenase and its catabolic byproduct, CO, have been well documented in a number of disease models, including transplantation, ischemia–reperfusion, vascular injury, and acute lung injury (2, 4, 36). Part of the mechanism for the protection afforded by CO includes inhibition of apoptosis, which can be modulated via the MAP kinase pathways (33, 34, 41, 48, 49), and there is evidence to suggest that Sprr proteins similarly inhibit apoptosis. Sprr proteins are substrates of transglutaminase in the formation of the crosslinked envelope (50), and transglutaminase appears to be involved in stabilizing apoptotic cells (51–53). Furthermore, ectopic expression of Sprr1a has been shown to prevent the disruption of myofibrils in cardiomyocytes, suggesting that stabilization of the actin cytoskeleton may contribute to this protective effect (12). Our microarray data indicate that one of the most differentially expressed categories of genes in response to CO treatment is that related to cell death; most of these are down-regulated, suggesting that CO may be antiapoptotic in this setting.
We have shown, in our model of fibroblast stimulation with TGF-β1, that the addition of CO significantly increases ERK pathway activation. The ERK pathway appears to play a role in the effect of CO on both α-SMA expression and Sprr1a expression, as shown in the experiments using the MEK1 inhibitor, PD098059. The ERK pathway inhibitor, PD098059, significantly inhibited Sprr1a induction in response to TGF-β1 and CO. This finding is consistent with previous studies in cardiomyocytes and human conducting airway epithelial cells, where PD098059 also blocked Sprr1 gene induction (12, 54). The effect of ERK pathway activation on α-SMA expression in fibroblasts is less well established. Absence of focal adhesion kinase-1 (FAK) and decreased ERK phosphorylation has been associated with increased α-SMA and stress fiber expression. On the other hand, incubation of rat kidney fibroblasts with TGF-β1, followed by PD98059 and connective tissue growth factor has been shown to abolish the increase in α-SMA attributable to connective tissue growth factor (55). Our findings suggest that loss of ERK activity enhances TGF-β1–induced α-SMA expression and prevents inhibition by CO. This implicates ERK signaling in CO effects on both α-SMA and Sprr1a expression. The involvement of the p38 pathway appears to be less clear. Inhibition of p38 had a similar effect on Sprr1a expression as inhibition of ERK activity; this is in keeping with the multiple reports of Sprr1a induction under conditions of stress (12, 11, 14, 50). However, the effect on α-SMA expression was different for ERK and p38 inhibition; whereas ERK inhibition reversed the suppressive effect of CO and enhanced α-SMA expression, p38 inhibition potently decreased α-SMA expression under all conditions. Although relatively little is known about the relationship between p38 and α-SMA expression, several recent reports do implicate the p38 pathway in myofibroblast differentiation. For instance, the neuronal adaptor protein, Unc119, has been shown to signal via p38 pathway to regulate myofibroblast differentiation (56), and renal tubular cells treated with p38 inhibitor express less α-SMA in response to TGF-β1 (57). The mechanism for this is not known, although p38 has been shown to alter nuclear trafficking of myocardin-related transcription factor, a serum response factor cofactor involved in α-SMA transcription, during the process of epithelial–mesenchymal transition (58).
In summary, these results provide the first evidence of a role for CO in the emergence of myofibroblasts, providing a potential mechanism for the antifibrotic effects of CO observed in a bleomycin model of lung fibrosis. These studies further demonstrate induction of expression of Sprr family proteins by TGF-β1 and CO in vitro and in vivo. Although the role of Sprr proteins in fibrosis remains to be defined, future studies should focus upon the role of Sprr1a in epithelial cells, where it may confer protection against cell death. We have shown that activation of the ERK signaling pathway is required for both of these effects of CO, lending new insight into the mechanism by which CO opposes fibrogenesis.
Acknowledgments
The authors thank Dr. Sem Phan for generously providing the α-smooth muscle actin promoter construct and Dr. S.M. Strittmatter for sharing the rabbit polyclonal anti-mouse SPRR1A antibody. They also thank Emeka Ifedigbo for his expert assistance with animal work.
This work was supported by National Institutes of Health grant 1RO1HL087122-01A1 (D.M.), the Veterans Administration, and the Parker B. Francis Family Foundation.
Originally Published in Press as DOI: 10.1165/rcmb.2007-0401OC on December 18, 2008
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Zhou Z, Song R, Fattman CL, Greenhill S, Alber S, Oury TD, Choi AM, Morse D. Carbon monoxide suppresses bleomycin-induced lung fibrosis. Am J Pathol 2005;166:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X, Shan P, Otterbein LE, Alam J, Flavell RA, Davis RJ, Choi AM, Lee PJ. Carbon monoxide inhibition of apoptosis during ischemia–reperfusion lung injury is dependent on the p38 mitogen-activated protein kinase pathway and involves caspase 3. J Biol Chem 2003;278:1248–1258. [DOI] [PubMed] [Google Scholar]
- 3.Otterbein LE, Mantell LL, Choi AM. Carbon monoxide provides protection against hyperoxic lung injury. Am J Physiol 1999;276:L688–L694. [DOI] [PubMed] [Google Scholar]
- 4.Song R, Kubo M, Morse D, Zhou Z, Zhang X, Dauber JH, Fabisiak J, Alber SM, Watkins SC, Zuckerbraun BS, et al. Carbon monoxide induces cytoprotection in rat orthotopic lung transplantation via anti-inflammatory and anti-apoptotic effects. Am J Pathol 2003;163:231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujita T, Toda K, Karimova A, Yan SF, Naka Y, Yet SF, Pinsky DJ. Paradoxical rescue from ischemic lung injury by inhaled carbon monoxide driven by derepression of fibrinolysis. Nat Med 2001;7:598–604. [DOI] [PubMed] [Google Scholar]
- 6.Chambers RC, Leoni P, Kaminski N, Laurent GJ, Heller RA. Global expression profiling of fibroblast responses to transforming growth factor-beta1 reveals the induction of inhibitor of differentiation-1 and provides evidence of smooth muscle cell phenotypic switching. Am J Pathol 2003;162:533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton-Piallat ML, Gabbiani G. The myofibroblast: one function, multiple origins. Am J Pathol 2007;170:1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kartasova T, van de Putte P. Isolation, characterization, and UV-stimulated expression of two families of genes encoding polypeptides of related structure in human epidermal keratinocytes. Mol Cell Biol 1988;8:2195–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baden HP, Kubilus J, Phillips SB, Kvedar JC, Tahan SR. A new class of soluble basic protein precursors of the cornified envelope of mammalian epidermis. Biochim Biophys Acta 1987;925:63–73. [DOI] [PubMed] [Google Scholar]
- 10.Cabral A, Voskamp P, Cleton-Jansen AM, South A, Nizetic D, Backendorf C. Structural organization and regulation of the small proline-rich family of cornified envelope precursors suggest a role in adaptive barrier function. J Biol Chem 2001;276:19231–19237. [DOI] [PubMed] [Google Scholar]
- 11.Tesfaigzi J, Th'ng J, Hotchkiss JA, Harkema JR, Wright PS. A small proline-rich protein, SPRR1, is upregulated early during tobacco smoke-induced squamous metaplasia in rat nasal epithelia. Am J Respir Cell Mol Biol 1996;14:478–486. [DOI] [PubMed] [Google Scholar]
- 12.Pradervand S, Yasukawa H, Muller OG, Kjekshus H, Nakamura T, St Amand TR, Yajima T, Matsumura K, Duplain H, Iwatate M, et al. Small proline-rich protein 1A is a gp130 pathway- and stress-inducible cardioprotective protein. EMBO J 2004;23:4517–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mueller A, O'Rourke J, Grimm J, Guillemin K, Dixon MF, Lee A, Falkow S. Distinct gene expression profiles characterize the histopathological stages of disease in Helicobacter-induced mucosa-associated lymphoid tissue lymphoma. Proc Natl Acad Sci USA 2003;100:1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmermann N, Doepker MP, Witte DP, Stringer KF, Fulkerson PC, Pope SM, Brandt EB, Mishra A, King NE, Nikolaidis NM, et al. Expression and regulation of small proline-rich protein 2 in allergic inflammation. Am J Respir Cell Mol Biol 2005;32:428–435. [DOI] [PubMed] [Google Scholar]
- 15.Demetris AJ, Specht S, Nozaki I, Lunz JG 3rd, Stolz DB, Murase N, Wu T. Small proline-rich proteins (SPRR) function as SH3 domain ligands, increase resistance to injury and are associated with epithelial–mesenchymal transition (EMT) in cholangiocytes. J Hepatol 2008;48:276–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu B, Wu Z, Jin H, Hashimoto N, Liu T, Phan SH. CCAAT/enhancer-binding protein beta isoforms and the regulation of alpha-smooth muscle actin gene expression by IL-1 beta. J Immunol 2004;173:4661–4668. [DOI] [PubMed] [Google Scholar]
- 17.Hu B, Wu Z, Phan SH. Smad3 mediates transforming growth factor-β–induced α–smooth muscle actin expression. Am J Respir Cell Mol Biol 2003;29:397–404. [DOI] [PubMed] [Google Scholar]
- 18.Pardo A, Gibson K, Cisneros J, Richards TJ, Yang Y, Becerril C, Yousem S, Herrera I, Ruiz V, Selman M, et al. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med 2005;2:e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dave NB, Kaminski N. Analysis of microarray experiments for pulmonary fibrosis. Methods Mol Med 2005;117:333–358. [DOI] [PubMed] [Google Scholar]
- 20.Kaminski N, Friedman N. Practical approaches to analyzing results of microarray experiments. Am J Respir Cell Mol Biol 2002;27:125–132. [DOI] [PubMed] [Google Scholar]
- 21.Ben-Dor A, Bruhn L, Friedman N, Nachman I, Schummer M, Yakhini Z. Tissue classification with gene expression profiles. J Comput Biol 2000;7:559–583. [DOI] [PubMed] [Google Scholar]
- 22.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001;98:5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thom SR, Fisher D, Xu YA, Notarfrancesco K, Ischiropoulos H. Adaptive responses and apoptosis in endothelial cells exposed to carbon monoxide. Proc Natl Acad Sci USA 2000;97:1305–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishra S, Fujita T, Lama VN, Nam D, Liao H, Okada M, Minamoto K, Yoshikawa Y, Harada H, Pinsky DJ. Carbon monoxide rescues ischemic lungs by interrupting MAPK-driven expression of early growth response 1 gene and its downstream target genes. Proc Natl Acad Sci USA 2006;103:5191–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujimoto M, Izu H, Seki K, Fukuda K, Nishida T, Yamada S, Kato K, Yonemura S, Inouye S, Nakai A. HSF4 is required for normal cell growth and differentiation during mouse lens development. EMBO J 2004;23:4297–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freitas A, Alves-Filho JC, Secco DD, Neto AF, Ferreira SH, Barja-Fidalgo C, Cunha FQ. Heme oxygenase/carbon monoxide–biliverdin pathway down regulates neutrophil rolling, adhesion and migration in acute inflammation. Br J Pharmacol 2006;149:345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urquhart P, Rosignoli G, Cooper D, Motterlini R, Perretti M. Carbon monoxide–releasing molecules modulate leukocyte–endothelial interactions under flow. J Pharmacol Exp Ther 2007;321:656–662. [DOI] [PubMed] [Google Scholar]
- 28.Singer AJ, Clark RA. Cutaneous wound healing. N Engl J Med 1999;341:738–746. [DOI] [PubMed] [Google Scholar]
- 29.Postlethwaite AE, Keski-Oja J, Moses HL, Kang AH. Stimulation of the chemotactic migration of human fibroblasts by transforming growth factor beta. J Exp Med 1987;165:251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oberhammer FA, Pavelka M, Sharma S, Tiefenbacher R, Purchio AF, Bursch W, Schulte-Hermann R. Induction of apoptosis in cultured hepatocytes and in regressing liver by transforming growth factor beta 1. Proc Natl Acad Sci USA 1992;89:5408–5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rotello RJ, Lieberman RC, Purchio AF, Gerschenson LE. Coordinated regulation of apoptosis and cell proliferation by transforming growth factor beta 1 in cultured uterine epithelial cells. Proc Natl Acad Sci USA 1991;88:3412–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brouard S, Berberat PO, Tobiasch E, Seldon MP, Bach FH, Soares MP. Heme oxygenase-1-derived carbon monoxide requires the activation of transcription factor NF-kappa B to protect endothelial cells from tumor necrosis factor-alpha–mediated apoptosis. J Biol Chem 2002;277:17950–17961. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Shan P, Alam J, Fu XY, Lee PJ. Carbon monoxide differentially modulates STAT1 and STAT3 and inhibits apoptosis via a phosphatidylinositol 3-kinase/Akt and p38 kinase–dependent STAT3 pathway during anoxia-reoxygenation injury. J Biol Chem 2005;280:8714–8721. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Wang Y, Kim HP, Nakahira K, Ryter SW, Choi AM. Carbon monoxide protects against hyperoxia-induced endothelial cell apoptosis by inhibiting reactive oxygen species formation. J Biol Chem 2007;282:1718–1726. [DOI] [PubMed] [Google Scholar]
- 35.Kim HP, Wang X, Nakao A, Kim SI, Murase N, Choi ME, Ryter SW, Choi AM. Caveolin-1 expression by means of p38beta mitogen-activated protein kinase mediates the antiproliferative effect of carbon monoxide. Proc Natl Acad Sci USA 2005;102:11319–11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otterbein LE, Zuckerbraun BS, Haga M, Liu F, Song R, Usheva A, Stachulak C, Bodyak N, Smith RN, Csizmadia E, et al. Carbon monoxide suppresses arteriosclerotic lesions associated with chronic graft rejection and with balloon injury. Nat Med 2003;9:183–190. [DOI] [PubMed] [Google Scholar]
- 37.Liu XM, Chapman GB, Peyton KJ, Schafer AI, Durante W. Carbon monoxide inhibits apoptosis in vascular smooth muscle cells. Cardiovasc Res 2002;55:396–405. [DOI] [PubMed] [Google Scholar]
- 38.Finkelman FD, Yang M, Perkins C, Schleifer K, Sproles A, Santeliz J, Bernstein JA, Rothenberg ME, Morris SC, Wills-Karp M. Suppressive effect of IL-4 on IL-13–induced genes in mouse lung. J Immunol 2005;174:4630–4638. [DOI] [PubMed] [Google Scholar]
- 39.Phan SH. The myofibroblast in pulmonary fibrosis. Chest 2002;122:286S–289S. [DOI] [PubMed] [Google Scholar]
- 40.Zhang X, Shan P, Jiang G, Zhang SS, Otterbein LE, Fu XY, Lee PJ. Endothelial STAT3 is essential for the protective effects of HO-1 in oxidant-induced lung injury. FASEB J 2006;20:2156–2158. [DOI] [PubMed] [Google Scholar]
- 41.Brouard S, Otterbein LE, Anrather J, Tobiasch E, Bach FH, Choi AM, Soares MP. Carbon monoxide generated by heme oxygenase 1 suppresses endothelial cell apoptosis. J Exp Med 2000;192:1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, Shan P, Alam J, Davis RJ, Flavell RA, Lee PJ. Carbon monoxide modulates Fas/Fas ligand, caspases, and Bcl-2 family proteins via the p38alpha mitogen-activated protein kinase pathway during ischemia–reperfusion lung injury. J Biol Chem 2003;278:22061–22070. [DOI] [PubMed] [Google Scholar]
- 43.Kuhn C, McDonald JA. The roles of the myofibroblast in idiopathic pulmonary fibrosis:ultrastructural and immunohistochemical features of sites of active extracellular matrix synthesis. Am J Pathol 1991;138:1257–1265. [PMC free article] [PubMed] [Google Scholar]
- 44.Phan SH, Zhang K, Zhang HY, Gharaee-Kermani M. The myofibroblast as an inflammatory cell in pulmonary fibrosis. Curr Top Pathol 1999;93:173–182. [DOI] [PubMed] [Google Scholar]
- 45.Roy SG, Nozaki Y, Phan SH. Regulation of alpha-smooth muscle actin gene expression in myofibroblast differentiation from rat lung fibroblasts. Int J Biochem Cell Biol 2001;33:723–734. [DOI] [PubMed] [Google Scholar]
- 46.Bonilla IE, Tanabe K, Strittmatter SM. Small proline-rich repeat protein 1A is expressed by axotomized neurons and promotes axonal outgrowth. J Neurosci 2002;22:1303–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Selman M, Pardo A, Barrera L, Estrada A, Watson SR, Wilson K, Aziz N, Kaminski N, Zlotnik A. Gene expression profiles distinguish idiopathic pulmonary fibrosis from hypersensitivity pneumonitis. Am J Respir Crit Care Med 2006;173:188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stec DE, Bishop C, Rimoldi JM, Poreddy SR, Vera T, Salahudeen AK. Carbon monoxide (CO) protects renal tubular epithelial cells against cold-rewarm apoptosis. Ren Fail 2007;29:543–548. [DOI] [PubMed] [Google Scholar]
- 49.Bainbridge SA, Belkacemi L, Dickinson M, Graham CH, Smith GN. Carbon monoxide inhibits hypoxia/reoxygenation-induced apoptosis and secondary necrosis in syncytiotrophoblast. Am J Pathol 2006;169:774–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tesfaigzi J, Carlson DM. Expression, regulation, and function of the SPR family of proteins: a review. Cell Biochem Biophys 1999;30:243–265. [DOI] [PubMed] [Google Scholar]
- 51.Piredda L, Amendola A, Colizzi V, Davies PJA, Farrace MG, Fraziano M, Gentile V, Uray I, Piacentini M, Fesus L. Lack of ‘tissue’ transglutaminase protein cross-linking leads to leakage of macromolecules from dying cells: relationship to development of autoimmunity in MRLIpr/Ipr mice. Cell Death Differ 1997;4:463–472. [DOI] [PubMed] [Google Scholar]
- 52.Nanda N, Iismaa SE, Owens WA, Husain A, Mackay F, Graham RM. Targeted inactivation of Gh/tissue transglutaminase II. J Biol Chem 2001;276:20673–20678. [DOI] [PubMed] [Google Scholar]
- 53.Antonyak MA, Jansen JM, Miller AM, Ly TK, Endo M, Cerione RA. Two isoforms of tissue transglutaminase mediate opposing cellular fates. Proc Natl Acad Sci USA 2006;103:18609–18614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deng J, Chen Y, Wu R. Induction of cell cornification and enhanced squamous-cell marker SPRR1 gene expression by phorbol ester are regulated by different signaling pathways in human conducting airway epithelial cells. Am J Respir Cell Mol Biol 2000;22:597–603. [DOI] [PubMed] [Google Scholar]
- 55.Yang M, Huang H, Li J, Li D, Wang H. Tyrosine phosphorylation of the LDL receptor-related protein (LRP) and activation of the ERK pathway are required for connective tissue growth factor to potentiate myofibroblast differentiation. FASEB J 2004;18:1920–1921. [DOI] [PubMed] [Google Scholar]
- 56.Vepachedu R, Gorska MM, Singhania N, Cosgrove GP, Brown KK, Alam R. Unc119 regulates myofibroblast differentiation through the activation of Fyn and the p38 MAPK pathway. J Immunol 2007;179:682–690. [DOI] [PubMed] [Google Scholar]
- 57.Sebe A, Leivonen SK, Fintha A, Masszi A, Rosivall L, Kähäri VM, Mucsi I. Transforming growth factor-beta–induced alpha–smooth muscle cell actin expression in renal proximal tubular cells is regulated by p38beta mitogen-activated protein kinase, extracellular signal–regulated protein kinase1,2 and the Smad signalling during epithelial–myofibroblast transdifferentiation. Nephrol Dial Transplant 2008;23:1537–1545. [DOI] [PubMed] [Google Scholar]
- 58.Sebe A, Masszi A, Zulys M, Yeung T, Speight P, Rotstein OD, Nakano H, Mucsi I, Szaszi K, Kapus A. Rac, PAK and p38 regulate cell contact–dependent nuclear translocation of myocardin-related transcription factor. FEBS Lett 2008;582:291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]