Abstract
Oligodendrocyte development is regulated by the interplay of repressors and activators in a complex transcriptional network. Here we report that two histone-modifying enzymes, HDAC1 and HDAC2, are required for oligodendrocyte formation. Genetic deletion of both HDAC1 and HDAC2 in oligodendrocyte lineage cells resulted in stabilization and nuclear translocation of β-catenin, which negatively regulates oligodendrocyte development by repressing Olig2 expression. We further identified an oligodendrocyte-restricted transcription factor TCF7L2/TCF4 as a bipartite co-effector of β-catenin for regulating oligodendrocyte differentiation. Targeted disruption of TCF7L2 in mice leads to severe defects in oligodendrocyte maturation, while expression of its dominant repressive form promotes precocious oligodendrocyte specification in developing chick neural tube. Transcriptional co-repressors HDAC1 and HDAC2 compete with β-catenin for TCF7L2 interaction to regulate downstream genes involved in oligodendrocyte differentiation. Hence, crosstalk between HDAC1/2 and the canonical Wnt signaling pathway mediated by TCF7L2 serves as a regulatory mechanism for oligodendrocyte differentiation.
Keywords: oligodendrocyte differentiation, Olig1, Olig2, Wnt, β-catenin, TCF/LEF factors, ID proteins, conditional mutagenesis, chromatin remodeling, epigenetic regulation
INTRODUCTION
Epigenetic chromatin remodeling events such as histone modification are essential for many biological processes, including neural cell fate specification and differentiation 1. The histone deacetylases (HDACs) that oppose the action of histone acetyltransferases, remove acetyl groups from histone lysine tails, resulting in chromatin compaction. There are four classes of HDACs that comprise a highly conserved enzyme family 2. HDACs can be recruited to multiprotein transcriptional complexes on the genome and serve as epigenetic co-repressors to facilitate inhibition of target gene transcription.
HDAC activity is shown to be required for oligodendrocyte maturation 3, 4. Using HDAC inhibitors, valproic acid and trichostatin A, oligodendrocyte differentiation is repressed in vitro in tissue culture studies and in vivo during animal development 3, 4. Since these HDAC inhibitors have significant affinities for multiple HDAC proteins, it is not clear which HDAC proteins are required for oligodendrocyte differentiation in vivo or how HDACs function in this process. Although morpholino-based knockdown of HDAC1 in zebrafishes leads to a loss of Olig2 5, an essential transcriptional regulator for oligodendrocyte development 6-8, it is not clear that the effect of knockdown is due to cell-autonomous or non-autonomous function of HDAC1. In addition, the molecular mechanism underlying the role of HDAC proteins in oligodendrocyte differentiation has yet to be fully understood.
Wnt signaling plays a critical role in embryonic patterning, tissue homeostasis and cell fate determination 9, 10. In the absence of Wnt signals, β-catenin is phosphorylated at several N-terminal residues, and subjected to ubiquitination and proteasome-mediated degradation. Wnt signaling activation inhibits the kinase activity of a destruction complex and therefore stabilizes β-catenin in the cytoplasm, which then translocates and accumulates in the nucleus. Nuclear β-catenin forms a bipartite transcriptional complex with a member of TCF (T-cell factor)/LEF (lymphoid enhancer family) transcription factor family. In the canonical Wnt/β-catenin pathway, the TCF factor (LEF1, TCF7/TCF1, TCF7L1/TCF3 or TCF7L2/TCF4) transduces Wnt/β-catenin signals to activate downstream target genes 9. This process requires the recruitment of multiple chromatin remodeling components including HDACs 11, 12. The Wnt signaling pathway appears to negatively regulate oligodendrocyte differentiation in vitro 13. Interestingly, Wnt signaling is constitutively activated in the retina of zebrafish HDAC1 homolog (Adh1) mutants, suggesting that HDAC function may antagonize Wnt signaling 14.
In this report, we explored the function of HDAC1 and HDAC2 in oligodendrocyte development. HDAC1 and HDAC2 proteins share 85% identity and have redundant functions during embryogenesis 15. We generated mice with mutations of both HDAC1 and HDAC2 specifically in the oligodendrocyte lineage and demonstrated a critical redundant role of HDAC1/2 for oligodendrocyte differentiation. Activation of the canonical Wnt/β-catenin pathway was shown to contribute to the inhibition of oligodendrocyte differentiation in HDAC1/2 double mutant mice. We further identified a Wnt/β-catenin effector TCF7L2 as an oligodendrocyte lineage-specific transcription factor and demonstrated that TCF7L2 is essential for oligodendrocyte differentiation. In addition, we showed that binding between β-catenin and HDAC proteins switches TCF7L2 from a repressor to an activator for oligodendrocyte differentiation. Collectively, our findings indicate that HDAC1/2 regulate oligodendrocyte differentiation, at least in part, by inhibiting Wnt signaling through disrupting β-catenin-TCF interactions.
RESULTS
HDAC1/2 are required for oligodendrocyte differentiation
To assess the role of class I HDACs in oligodendrocyte development, we selectively deleted HDAC1 and HDAC2 in the oligodendrocyte lineage by mating HDAC1/2 floxed mice with Olig1-Cre mice 16. When Olig1-Cre mice were intercrossed with a ROSA26-LacZ reporter strain 17, LacZ expression was initially detected in the precursor pMN domain for oligodendrocytes and motoneurons at E12.5 and persisted in oligodendrocytes at postnatal stages (Supplementary Fig. 1a,b). Consistent with these findings, ROSA-YFP reporter mice 18 crossed with Olig1-Cre mice showed co-expression of YFP with a differentiated oligodendrocyte marker CC1 and an OPC marker Pdgfrα (Supplementary Fig. 1c-j). Thus, Olig1-Cre activity will delete HDAC1/2 floxed alleles in oligodendrocytes and their progenitors at embryonic and postnatal stages, as well as in motor neuron progenitors.
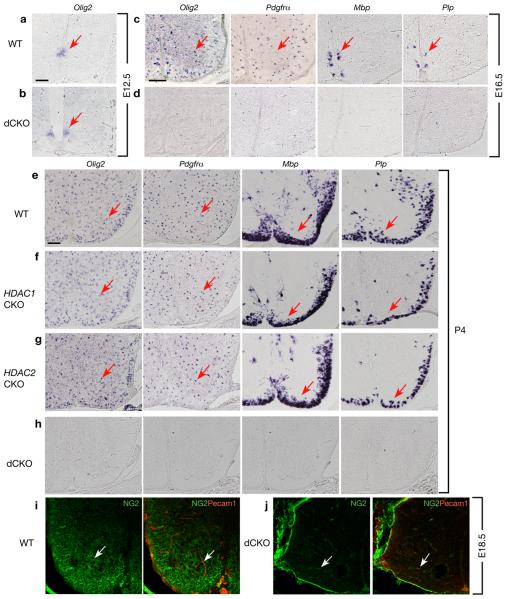
Mice with double deletion of HDAC1 and HDAC2 by Olig1-Cre (designated dCKO) were born in normal Mendelian ratios, however, they developed severe tremor, reminiscent of myelin deficient mice 19, and died around postnatal week 2. In dCKO animals at E12.5, Olig2 expression was maintained in the ventral ventricular zone of the spinal cord (Fig. 1a,b). At E16.5, expression of OPC markers Pdgfrα and Olig2 as well as mature oligodendrocyte markers myelin basic protein (Mbp) and Plp/DM20 were undetectable in dCKO mice (Fig. 1d vs. c). Even at the postnatal day P4, these markers were not observed in dCKO mice (Fig. 1h vs. e). In contrast, mice with a single deletion of HDAC1 (HDAC1CKO) or HDAC2 (HDAC2CKO) showed normal expression patterns of OPC and mature oligodendrocyte markers (Fig. 1f,g). These mice were also phenotypically identical to wild-type mice (data not shown). The deficit of OPCs was further confirmed with the expression of another OPC marker NG2 20. The NG2+ OPCs were essentially diminished in dCKO mutant spinal cords at E18.5 (Fig. 1j) compared to the control (Fig.1i).
Figure 1. HDAC1 and HDAC2 are required for oligodendrocyte development in the spinal cord.
In situ hybridizations of cross-sections of spinal cord from wild-type (WT) (a, c, e), HDAC1lox/lox; Olig1-Cre (HDAC1CKO; f), HDAC2 lox/lox; Olig1-Cre (HDAC2CKO; g), HDAC1/2lox/lox; Olig1-Cre (dCKO; b, d, h) at ages E12.5, E16.5, or P4 as indicated using probes for oligodendrocyte lineage markers Olig2, Pdgfrα, Plp/DM20, and Mbp. i-j, spinal cords from WT and dCKO at e18.5 were subject to double immunostaining with anti-NG2 (green) and anti-PECAM1 (red) antibodies. The anti-PECAM1 was used to distinguish OPCs from pericytes since NG2 labels both OPCs and pericytes within the spinal cord. Arrows indicate in situ labeled cells (a-h) and NG2+ or PECAM1+ cells (i-j), respectively. Scale bars in a-b, c-d and e-j, 100 μm.
Similar to the finding in spinal cord, dCKO mice displayed a complete absence of mature oligodendrocytes and their progenitors in the postnatal brain (Supplementary Fig. 2). The failure of OPC formation persisted into postnatal stages of dCKO mice, suggesting that HDAC1/2 are required for oligodendrocyte specification and differentiation.
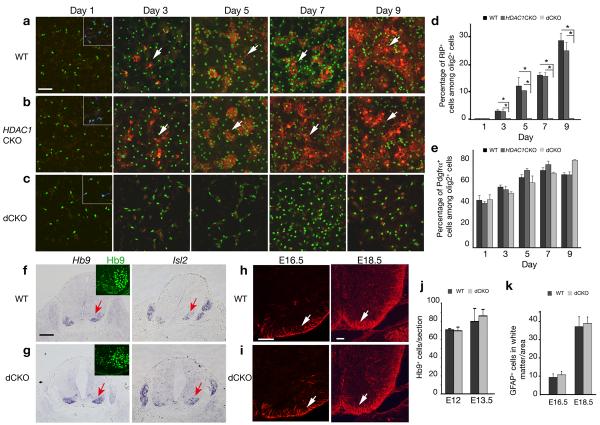
Oligodendrocyte progenitor culture from wild-type, HDAC1 or HDAC2 mutant embryonic cortices express a oligodendrocyte differentiation marker RIP beginning at day 3 after plating in oligodendrocyte differentiation medium (Fig. 2a,b, data not shown). However, RIP expression was not detected in cells isolated from dCKO mutant embryos (Fig. 2c,d). In contrast to the lack of OPC marker expression in the spinal cord of dCKO mice (Fig. 1), we observed Olig2+ and Pdgfrα+ OPCs under the in vitro condition and they maintained at the precursor stage without differentiation (Fig. 2c,e). These findings show that although OPCs can be formed from dCKO mutant embryos under certain cell culture conditions, these cells do not mature into differentiated oligodendrocytes.
Figure 2. HDAC1 and HDAC2 are essential for oligodendrocyte differentiation in vitro and is not required for motor neurons and astrocyte development.
a-c) Cortical progenitors from WT, HDAC1CKO; and dCKO embryos at E15.5 were cultured in oligodendrocyte differentiation media. Cells were immunostained antibodies to RIP, Olig2 and Pdgfrα at defined days as indicated. Pdgfrα expression (blue). was detected in control and dCKO culture shown as inserts in panels. d-e) Histographs depict the percentage of RIP+ (d) or Pdgfrα+ (e) cells among Olig2+ cells. Data are derived from experiments in parallel cultures of at least three age-matching littermates. *p<0.01, ANOVA in post hoc Newman-Keuls Multiple comparison test. f-g) In situ hybridization of transverse sections of ventral spinal cord from WT and dCKO at E12.5 using probes for motor neuron markers Hb9 and Isl2. Motor neurons in the ventral horn immunolabeled by an Hb9 (green) antibody were shown as inserts. h-i) Immunostaining of spinal cord of WT and dCKO at E16.5 and E18.5 using GFAP antibody. j) Quantification of Hb9+ cells per section was shown in the spinal cord of WT and dCKO at E12 and E13.5 (n=3). k) Quantification of GFAP+ cells was shown in the white matter of WT and dCKO spinal cords per unit area (0.1 mm2) at E16.5 and E18.5 (n=3). All values are presented as mean ± SD in the graphs. Arrows indicate immuno- or in situ labeling cells. Scale bar in a-c: 50 μm; in f-g: 200 μm, and in h-i; 100 μm.
Motor neuron and astrocyte formation in HDAC1/2 mutants
Motor neurons, which are derived from the Olig1-Cre+ progenitor domain in the spinal cord, were examined in wild-type and dCKO embryos using motor neuron markers Hb9 and Isl2 23. We did not detect any defects in the formation of somatic motor neurons in the developing dCKO spinal cord (Fig. 2g, j).
To determine whether astrocyte development is perturbed in dCKO mice, we examined expression of glial fibrillary acid protein (GFAP), an astrocyte marker. Similar levels of GFAP were observed in the spinal cord of both wild-type and dCKO mice at E16.5 and E18.5 (Fig. 2h,i,k). We did not observe significant apoptosis, as assessed by caspase-3-immunostaining and TUNEL staining, or alteration of cell proliferation and neurogenesis in the dCKO spinal cord (Supplementary Fig. 3; data not shown). In addition, we did not detect any defect in Schwann cell formation at all stages examined in the Olig1-Cre mediated dCKO mice (data not shown). Collectively, our data indicate that Olig1-Cre mediated oligodendrocyte-specific ablation of HDAC1/2 selectively disrupts oligodendrocyte differentiation but does not perturb neuron or astrocyte development.
Stabilization of β-catenin in the absence of HDAC1/2
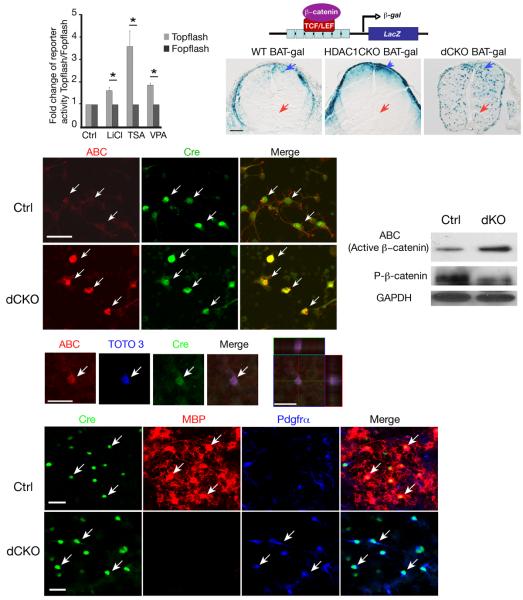
In zebrafish retina, HDAC1 was found to negatively regulate Wnt and Notch signaling 12, which has been shown to inhibit oligodendrocyte differentiation 13. Cortical progenitor cells isolated from dCKO embryos did not exhibit an increase of expression of Notch effectors Hes1 and Hes5 (data not shown). To examine whether inhibition of HDAC activity affects Wnt signal transduction, we transfected hippocampus-derived adult neural progenitor cells (HCN) 24 with a β-catenin/TCF luciferase reporter, TOPFLASH, carrying a β-catenin-responsive promoter 25. This reporter responded to the treatment of LiCl, an activator of Wnt signaling (Fig. 3a). Cells treated with an HDAC inhibitors (trichostatin A, TSA) and VPA (valproic acid) showed enhanced TOPFLASH reporter activity over the negative control FOPFLASH reporter (Fig. 3a). These data suggest that inhibition of HDAC activity enhances canonical Wnt signal transduction.
Figure 3. Activation of Wnt signaling by stabilizing β-catenin in HDAC1 and HDAC2 mutant progenitors.
a) HCN cells were transfected with TOPFLASH and FOPFLASH treated with LiCl and TSA (100 nM) and VPA (100 μM) for 48hr. Fold changes of the reporter activity of TOPFLASH relative to FOPFLASH were presented. Data are derived from three independent experiments with error bars representing mean±SD (*P<0.01, Student's twotailed t test). b) A schematic diagram is shown for a BAT-gal reporter line under the control of β-catenin/TCF. c-e) β-galactosidase (β-gal) expression in the E13.5 spinal cord is shown in WT;BAT-gal and HDAC1CKO;BAT-gal and dCKO;BAT-gal mice. Arrows indicate β-gal expressing cells. f-k) Immunostain of cortical progenitors from E15.5 HDAC1/2 flox/+;Olig1Cre (Ctrl) and dCKO embryos cultured in oligodendrocyte growth medium, using antibodies to a stable active form of β-catenin (ABC, red) and Cre (green). Arrows indicate Olig1-Cre+ cells and ABC expression (i-k,). l-p) Confocal imaging showed that ABC accumulation (arrow) in dCKO Cre+ cells in the nucleus visualized with TOTO3 nuclear staining dye. Orthogonal reconstructions of confocal images at the z-axis level were shown in side panels (p). q) Lysates of oligodendrocyte enriched culture from HDAC1/2 flox/+;Olig1Cre and dCKO embryos at E15.5 were subject to Western blot analysis for a stable form (ABC) or a phosphorylated form of β-catenin as indicated. r-y) Immunostaining of oligodendrocyte-enriched culture from HDAC1lox/+; Olig1-Cre (Ctrl; r-u) and dCKO embryos using antibodies to Cre (green), MBP (red), and Pdgfrα (blue). Arrows indicate Cre+ cells. Scale bars in c-e:100 μm; in f-o and r-v: 50 μm.
To further assess in vivo Wnt/β-catenin activation in Olig1-Cre mediated HDAC1/2 mutant animals, we bred a BAT-gal reporter line with HDAC1/2 floxed mice to generate dCKO carrying BAT-gal reporter. In the BAT-gal transgenic line, beta-galactosidase (β-gal) reporter is expressed under the control of multiple β-catenin/T cell factor responsive elements 26 (Fig. 3b). As compared to control mice at E13.5 (Fig. 3c,d), where β-gal activity is mainly detected in the dorsal spinal cord, which exhibits a high Wnt/β-catenin activity, Olig1-Cre-mediated HDAC1/2 deletion results in wide-spread β-gal reporter expression in unidentified cells throughout the cord (Fig. 3e). These observations suggest that HDAC 1 and 2 deletion could, in part, lead to Wnt signaling activation during normal oligodendrocyte development.
Cytoplasmic stabilization and nuclear localization of β-catenin are key events in canonical Wnt signaling to regulate target gene transcription. To examine β-catenin stabilization in dCKO mice, we isolated cortical progenitors from dCKO and its control embryos and cultured them in oligodendrocyte differentiation medium to promote oligodendrocyte formation 27. The cultures were immunostained with antibodies against the stabilized, active form of β-catenin (anti-Active-β-Catenin; ABC)28 and Olig1-Cre. In control Cre+ cells, β-catenin immunostaining displayed a weak and diffuse cytoplasmic membrane pattern with a minimal amount of cytoplasmic accumulation. No nuclear accumulation of β-catenin was present in Cre+ cells (Fig. 3f-h). In contrast, in the dCKO mutants, expression of the stable form of β-catenin in the Olig1-Cre+ cells appeared to accumulate in the cytoplasm, and we observed nuclear localization of β-catenin (Fig. 3i-k), which was confirmed by confocal analysis at the Z-axis level (Fig. 3l-p). Accumulation of the stable form of β-catenin in dCKO-derived cells was further confirmed with Western blot analysis, which showed an approximate 3.2-fold increase over the control determined by densitometry (Fig. 3q). Furthermore, phosphorylation of β-catenin in the culture derived from dCKO embryos was significantly reduced by approximately 4.5 folds (Fig. 3q). The decrease of phosphorylation-dependent degradation of β-catenin could result in a stabilized form of β-catenin, which could then enter the nucleus as a transcription coactivator for gene transcription28. Compared to control, Olig1-Cre+ cells from dCKO mutants failed to mature into MBP+ oligodendrocytes even in the culture medium containing factors that promote oligodendrocyte maturation such as thyroid hormone T3 and CNTF 21, 22 (Fig. 3v-y). They maintained as Pdgfrα+ OPCs. These observations suggest that the deletion of HDAC1/2 results in stabilization and nuclear localization of β-catenin, leading to Wnt signaling activation and oligodendrocyte differentiation failure.
Wnt signaling inhibits oligodendrocyte differentiation
To determine whether the activation of canonical Wnt signaling alone is sufficient to inhibit oligodendrocyte differentiation, we crossed the β-catenin gain-of-function allele Catnlox(ex3) mouse line 29 with the Olig1-Cre line, which excised exon 3 of the β-catenin allele, producing a shortened, stable form of β-catenin that lacks N-terminal phosphorylation and ubiquitination sites 29, which result in constitutive Wnt signaling activation in oligodendrocyte lineage cells.
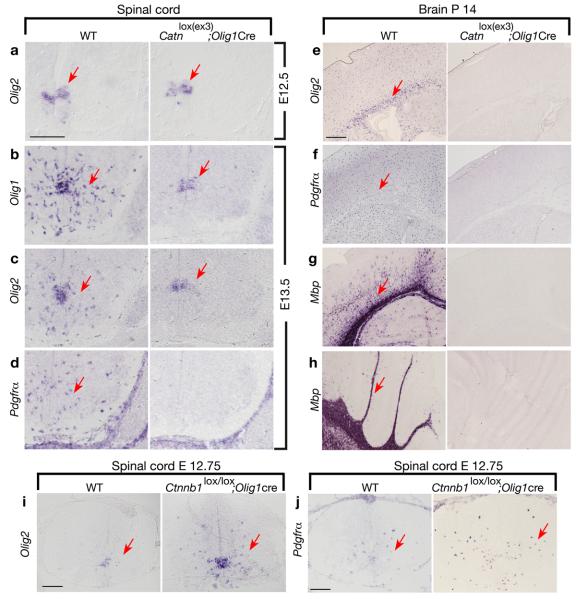
In the spinal cord of Catnlox(ex3); Olig1Cre embryos at E12.5, expression of an OPC marker Olig2 in the ventral ventricular zone was comparable to the control (Fig. 4a). At E13.5, expression of Olig1 and Olig2 decreased and maintained in a domain of the ventral ventricular zone (Fig. 4b,c). Strikingly, Pdgfrα expression was essentially undetectable (Fig. 4d). Thus, Olig2+ neuroepithelial cells in the mutant were unable to migrate and differentiate into OPCs and oligodendrocytes, and they maintained as progenitor cells in the ventral ventricular zone. We did not detect any alteration of motor neuron and astrocyte development or increase of cell death assayed with an active form of Caspase 3 and TUNEL in mutants at different stages (data not shown). Thus, activation of β-catenin mediated by Olig1-Cre inhibits differentiation of Olig2+ neuroepithelial cells into oligodendrocyte lineage cells in the developing spinal cord.
Figure 4. Activation of canonical Wnt signaling in oligodendrocyte lineage cells inhibits oligodendrocyte differentiation.
a-h) In situ hybridization of sections of spinal cord (a-d), forebrain (e-g) or cerebellum (h) taken from WT, Catnlox(ex3); Olig1Cre mice at indicated ages using probes to Olig2, Olig1, Pdgfrα, Plp, and Mbp. i-j) In situ hybridization of spinal cord sections of from WT and β-catenin KO (Ctnnb1lox/lox; Olig1Cre) mice at age E12.75 using probes to Olig2 and Pdgfrα as indicated. Arrows indicate the in situ labeled cells. Scale bars in a-d, e-h and i-j:100 μm.
Failure of oligodendrocyte formation persists throughout the postnatal stage in Catnlox(ex3); Olig1-Cre mutants. In the hindbrain at P4, expression of Olig2, Pdgfrα, and oligodendrocyte markers Plp and Mbp was not detected in mutant mice (Supplementary Fig.4). Even at P14, OPCs and mature oligodendrocytes were not detected in the forebrain or cerebellum (Fig. 4e-h). Thus, our data shows that constitutive activation of β-catenin in Olig1-expressing cells inhibits OPC specification, paralleling our observations in oligodendrocyte formation in mice lacking both HDAC1 and HDAC2.
Oligodendrocyte differentiation defects were also observed in the spinal cord of β-catenin gain-of-function mediated by oligodendrocyte-expressing Cre line, CNP-Cre 30. Since Cre expression directed by CNP promoter begins at or after OPCs form, Pdgfrα+ OPCs in this mutant were largely spared (Supplementary Fig. 5).
Conversely, we generated compound mice with Olig1-Cre mediated β-catenin loss-of-function alleles (Catnnb1lox/lox; Olig1-Cre). In the developing spinal cord of the β-catenin mutant animals at E12.75, we observed a pre-scheduled increase and dispersal of oligodendrocyte progenitor markers Olig2, a paralog of Olig1, and Pdgfrα, in contrast to their restricted expression in a domain of the ventral ventricular zone in controls (Fig.4i, j). Our observations indicate that Wnt/β-catenin signaling participates in and negatively regulates oligodendrocyte development.
To further confirm whether the stabilized form of β-catenin represses Olig2 expression in vitro, we transfected HCN cells with a constitutively active form of β-catenin (ΔN89β-catenin), which lacks N-terminal 89 amino acid phosphorylation motif preventing degradation by destruction complex, while preserving transcriptional activity 31. Four days following transfection in the presence of differentiation promoting factor IGF-1 24, control vector transfected HCN cells differentiated into RIP+ oligodendrocytes with multiple elaborated cellular processes (Supplementary Fig. 6), while ΔN89β-catenin expressing cells are confined to a progenitor cell morphology lacking RIP+ elaborated processes (Supplementary Fig. 6b). Olig2 expression was observed in all the control-vector transfected HCN cells in the presence of IGF-1 (Supplementary Fig. 6c), but its expression was reduced or absent in a significant percentage of ΔN89β-cateninexpressing cells (Supplementary Fig. 6d,e). This in vitro study is consistent with the observation of in vivo β-catenin gain-of-function alleles that the active form of β-catenin negatively regulates Olig2 expression and therefore oligodendrocyte differentiation.
TCF7L2 is an oligodendrocyte-specific β-catenin effector
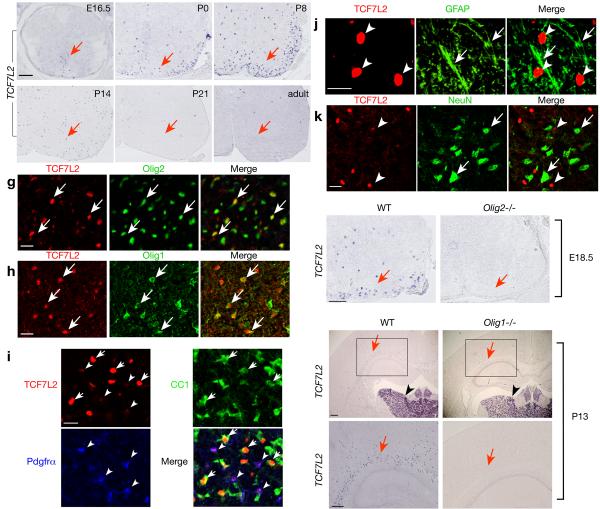
®-catenin, which does not bind to DNA, activates canonical Wnt signaling by forming a bipartite transcriptional activator with a member of the DNA-binding TCF/LEF transcription factor family 9. To determine which TCF transcription factor(s) may potentially regulate oligodendrocyte differentiation, we examined mRNA expression of all TCF/LEF factor family (LEF-1, TCF7, TCF7L1 and TCF7L2) in the developing spinal cord by in situ hybridization. In contrast to low or non-specific expression of LEF-1, TCF7, TCF7L1, the expression pattern of TCF7L2 is reminiscent of oligodendrocyte markers Zfp488, Plp and Mbp in the developing spinal cord (Fig. 5a-f)32. TCF7L2 expression appears mainly in the ventral spinal cord and increases primarily in the white matter at perinatal stages, while it is reduced in the adult spinal cord (Fig. 5f).
Figure 5. Identification of Wnt/β-catenin effector TCF7L2 as an oligodendrocyte-specific transcription factor.
a-f) In situ hybridization of transversed sections of spinal cord from WT at different ages as indicated using a probe to TCF7L2. g-k) Immunostain of P7 spinal cord using antibodies to TCF7L2, Olig2, Olig1, Pdgfrα, CC1, GFAP and NeuN as indicated. In panels g-i, arrows show co-labeling of TCF7L2 with Olig2 (g), Olig1 (h), CC1 (i). Arrowheads in i indicate Pdgfrα+/TCF7L2+ OPCs (arrowheads). TCF7L2 (arrowheads) was not detected in GFAP+ astrocytes (j, arrows) or NeuN+ neurons (k, arrows). l-q) In situ hybridization of spinal cord of WT and Olig2-/- mice at e18.5 or at forebrain of Olig1 null mice at P13 using a TCF7L2 probe. Arrow in l indicates TCF7L2 expressing cells in the lateral white matter, which is absent in Olig2 null animals (m). Arrows and arrowheads in n-q indicate TCF7L2 expression in the cerebral white matter region and neuronal populations in the thalamus, respectively. The boxed areas in n and o were showed in a high magnification in p and q, respectively. Scale bars in a-f, in g-k, 100 μm and l-q, 200 μm.
Using immunocytochemistry, we demonstrated that TCF7L2 co-localizes with oligodendrocyte lineage markers Olig2, Olig1, CC1 and Pdgfrα in the spinal cord at P7 (Fig. 5g-i). The expression of TCF7L2 detected in Pdgfrα+ cells is weaker (arrowheads in Fig.5i) as compared to strong expression of TCF7L2 in CC1+ differentiated oligodendrocytes (Fig. 5i, arrows). TCF7L2 was not observed in GFAP+ astrocytes or NeuN+ neurons (Fig. 5j,k). Examination of mice lacking Olig2 showed that TCF7L2 was undetectable in the mutant (Fig. 5l,m). Similarly, TCF7L2 expression was detected in the cerebral white matter of the wild-type forebrain at P13, but it was absent in Olig1 null mice (Fig. 5o,q vs. n,p). This result is consistent with our transcriptome profile analysis of Olig1 null mice 16, which showed that TCF7L2 is downregulated in Olig1 mutants (unpublished observation). Of note, TCF7L2 expression in neuronal populations of the thalamus region in the brain was not affected in Olig1 null mice (Fig. 5n, o; arrowheads). Thus, our data suggest that TCF7L2 is expressed in oligodendrocyte lineage cells.
TCF7L2 is critical for oligodendrocyte differentiation
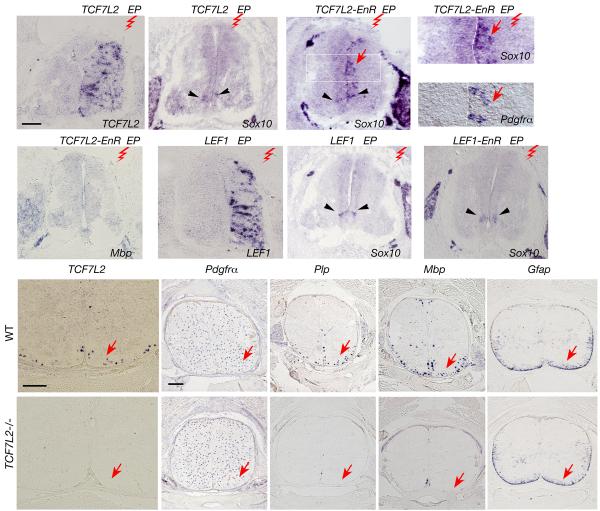
To examine the role of TCF7L2 in oligodendrocyte differentiation, we first carried out a gain-of-function study in ovo. A TCF7L2 expression vector was electroporated into the neural tube of E2.5 chick embryos and harvested at E5.5, when the differentiation of endogenous oligodendrocytes has not yet occurred 33. Expression of transgenes in the electroporated side was conformed by in situ hybridization (Fig. 6a, g; data not shown). Misexpression of TCF7L2 alone did not induce ectopic oligodendrocyte differentiation (Fig. 6b). This implies that TCF7L2 alone cannot activate transcription of oligodendroglial genes and may require additional cofactors 11. In contrast, misexpression of TCF7L2-EnR, which encodes a dominant-repressive form of TCF7L2 with its Cterminal fused to an Engrailed repressor domain (EnR) and a lack of β-catenin binding domain 34, was able to promote ectopic expression of a differentiated oligodendrocyte marker Sox10 and an OPC marker Pdgfrα (Fig. 6c-e). Ectopic expression of these early oligodendrocyte markers was mainly detected near the ventricular zone of the neural tube (Fig. 6c-e; arrows), although no mature oligodendrocytes were evident as determined by the lack of Mbp expression (Fig. 6f).
Figure 6. A dominant-repressive form of TCF7L2 promotes ectopic and precocious oligodendrocyte specification.
a-f) Expression vectors for TCF7L2 or TCF7L2-EnR were electroporated into the neural tube of E2.5 chick embryos and harvested at E5.5. The spinal cord sections were analyzed by in situ hybridization with probes to TCF7L2 (a), Sox10 (b-d), Pdgfrα (e) and Mbp (f) as indicated. Red zigzags indicate the electroporated side (EP). The boxed area in c is shown with a larger magnification in d. Arrows in c-e indicate ectopic expression of Sox10 and Pdgfrα detected on the electroporated side of chick neural tubes. g-i) In situ hybridization analysis in the chick neural tube electroporated with LEF1 (g, h) and LEF1-EnR (i) with probes to LEF1 (g) and Sox10 (h,i). Arrowheads in a-i indicate endogenous Sox10 expression. j-s) Expression of mRNA transcripts for TCF7L2 (j,o), Pdgfrα (k,p), Plp (l,q), and Mbp (m,r) and Gfap (n,s) was analyzed in situ on spinal cord sections of WT and TCF7L2 null animals at E17.5 as indicated by arrows. Scale bars in a-c and f-i: 100 μm; in j,o and k-s: 200 μm.
In contrast, ectopic expression of another TCF family member LEF1 and its dominant repressive form LEF1-EnR did not alter or promote ectopic expression of Sox10 and Pdgfrα (Fig. 6h,i; data not shown). These data suggest that a unique TCF7L2-mediated repressor interaction promotes oligodendroglial gene expression and de novo oligodendrocyte precursor formation. However, it is possible that other TCF factors could mediate the progression from neuroepithelial precursors to committed OPCs and that this process can be mimicked by TCF7L2-EnR.
To determine whether the oligodendrocyte-specific Wnt signaling effector TCF7L2 is required for oligodendrocyte development, we analyzed expression of the markers for mature oligodendrocytes and their precursors in the spinal cord of TCF7L2 null embryos, which die at birth 35. At E17.5 in TCF7L2 null embryos, expression of the OPC marker Pdgfrα is detected throughout the spinal cord and is comparable to that of wild-type littermates (Fig. 6k,p). In contrast, expression of myelin genes Plp and Mbp was undetectable compared to robust expression in the lateral white matter of wild-type spinal cords (Fig. 6q,r vs. l,m). In addition, expression of Gfap was not altered in the absence of TCF7L2 (Fig. 6s). These data suggest that the Wnt/β-catenin effector TCF7L2 is required for oligodendrocyte differentiation or maturation but not the formation of oligodendrocyte precursors.
HDAC1/2 suppress Wnt/β-catenin signaling activation
To examine the change in expression of myelin genes and differentiation inhibitors in response to Wnt signaling, we treated HCN cells with a Wnt signaling ligand Wnt3a in the presence of IGF-1 and analyzed gene expression by qRT-PCR analysis. Expression of both oligodendrocyte inhibitors ID2 and ID4 36, 37 was up-regulated after Wnt3A treatment. We did not observed a significant increase of Hes1 and Hes5, Notch signaling effectors for oligodendrocyte differentiation 38 (Fig. 7a). As expected, expression of myelin genes such as Mbp and Cnp was significantly reduced (Fig. 7a). Similar responses to Wnt3a treatment were obtained with primary OPCs isolated from rat neonates (Fig. 7a), consistent with a previous study that Wnt3a treatment inhibits OPC differentiation 13.
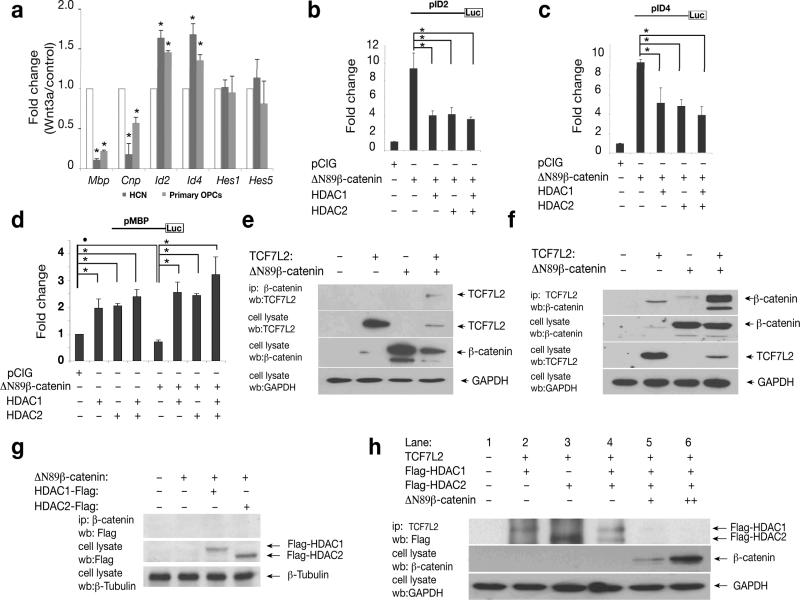
Figure 7. Competition between β-catenin and HDAC1/2 proteins for TCF7L2 interaction regulates expression of Wnt target genes.
a) HCN cells and primary rat OPCs were treated with Wnt signaling ligand Wnt3a (100ng/ml) for 48 hours. Expression of Mbp, Cnp, Id2/4, Hes1 and Hes5 were examined by qRT-PCR analysis. Gapdh served an internal control. b-d) HCN cells were transfected with vectors carrying ID2 (b) or ID4 (c) or MBP (d) promoter-driven luciferase reporters together with pCIG-△N89β-catenin and HDAC1, or HDAC2 or both HDAC1/2 as indicated. The luciferase activity of transfected cell lysates was measured 48 hr posttransfection. Values in a-d represent the average of three independent experiments. Error bars shown are the mean ± S.D. (•P<0.05, *P<0.01, ANOVA in post hoc Newman-Keuls Multiple comparison test). e-g) Expression vector encoding △N89β-catenin was cotransfected with TCF7L2 (e-f), or flag-tagged HDAC1 or HDAC2 (g) and individual controls. Co-immunoprecipitation with anti-△N89β-catenin (e) or anti-TCF7L2 (f) was performed from cell lysates 48 hr after transfection. h) TCF7L2 was co-transfected with expression vectors for △N89β-catenin and HDAC1 or/and HDAC2 as indicated. The TCF7L2 complex was immunoprecipitated with TCF7L2 antibody. Lane 2-4, in the absence of β-catenin, both HDAC1 and 2 were detected in the TCF7L2 complex. Lanes 5, in the presence of a low level of △N89β-catenin, flag-tagged HDAC 1 but not HDAC2 was detected in the TCF7L2 complex. Lane 6, in the presence of high level of β-catenin, neither HDAC1 nor HDAC2 was observed to be associated with the TCF7L2 complex.
ID2 is a direct target gene of β-catenin-TCF transactivation 39, 40 and negatively regulates oligodendrocyte differentiation 36. Mutation of TCF7L2 binding motif in an ID2 promoter abolishes the promoter activity activated by β-catenin in colon carcinoma cells 40. To determine the relationship of β-catenin and HDAC1/2 in regulating expression of Wnt target genes, we examined the ID2 promoter activity in the presence of β-catenin and/or HDAC1/2. Due to poor transfection efficiency of primary OPCs, we chose to use HCN for biochemical assays. The HCN cells have been established as precursor cells for studying oligodendrocyte differentiation 24. Over-expression of ΔN89β-catenin significantly transactivated the ID2 promoter-luciferase reporter (Fig. 7b). The activation of the ID2 promoter activity by ΔN89β-catenin was suppressed by co-transfection of either HDAC1 or HDAC2, or both HDAC1 and HDAC2 (Fig.7b). Similarly, activation of an ID4 promoter reporter 41 by active β-catenin was repressed by HDAC1 or HDAC2 or both (Fig. 7c).
We monitored the effect of HDACs and β-catenin signaling on myelin gene expression by co-expressing an Mbp promoter driven-luciferase reporter 42 with ΔN89β-catenin, HDAC1 and HDAC2. When co-transfected with the Mbp-reporter, HDAC1 or HDAC2 or both activated the Mbp promoter activity approximately 2-fold over control (Fig. 7d) whereas ΔN89β-catenin reduced the promoter activity (Fig. 7d). In contrast, when HDAC1 or HDAC2 or both were co-transfected with the active form of β-catenin, there was a statistically significant increase in Mbp promoter-driven reporter activity even in the presence of an active form of β-catenin (Fig. 7d). These data suggest that the inhibitory effect of β-catenin/TCF signaling on the myelin gene promoter can be reversed by overexpression of HDAC1 or HDAC2 or both.
Competing Interaction of β-catenin and HDACs with TCF7L2
To investigate the interaction between TCF7L2, β-catenin and HDAC1 or HDAC2, we co-transfected cDNA constructs encoding TCF7L2 and ΔN89β-catenin, as well as HDAC1 or HDAC2 and performed co-immunoprecipitation on cell lysates 48 hr after transfection. Consistent with β-catenin binding to a member in the LEF/TCF transcription factor family 11, we detected that ΔN89β-catenin was associated with TCF7L2 (Fig. 7e). Similarly, immunoprecipitation of TCF7L2 was able to pull down β-catenin in a complex (Fig. 7f). In contrast, no β-catenin was detected to associate with HDAC1 or HDAC2 in the co-immunoprecipitated complex (Fig. 7g).
Inhibition of β-catenin-driven transactivation by HDAC1/2 could result from either displacement of TCF7L2 binding to β-catenin by HDACs or the formation of a ternary complex (HDAC-TCF7L2-β-catenin) that has no transactivation potential. To distinguish between these possibilities, TCF7L2 was co-transfected with expression vectors for ΔN89β-catenin, HDAC1 or HDAC2. After transfection, the TCF7L2 complex was immunoprecipitated with TCF7L2 antibody. In the absence of ΔN89β-catenin, both HDAC1 and HDAC2 were detected in the same complex containing TCF7L2 by Western blot analysis (Fig. 7h). In the presence of a low level of ΔN89β-catenin, a small amount of Flag-tagged HDAC1 but not HDAC2 was detected in the co-immunoprecipitated complex (Fig. 7h). In the presence of a high level of ΔN89β-catenin, neither HDAC1 nor HDAC2 was associated with the TCF7L2 complex (Fig. 7h).
These data are consistent with a model (Supplementary Fig.7) whereby β-catenin competes with HDAC1 and/or HDAC2 to interact with TCF7L2 to inhibit oligodendrocyte differentiation. Conversely, dissociation and attenuation of TCF7L2-bound β-catenin by HDAC1/2 result in a template that is poised for oligodendrocyte differentiation.
DISCUSSION
Oligodendrogenesis occurs in a spatially and temporally specific manner and is regulated by interplay of multiple integrated genetic and epigenetic factors. In this report, we demonstrate that chromatin remodeling enzymes HDAC1/2 regulate oligodendrocyte specification and differentiation, at least in part, by inhibiting β-catenin/TCF7L2 complex formation.
Redundant roles of HDAC1/2 in oligodendrocyte development
HDACs have been implicated in oligodendrocyte development based on the ability of HDAC inhibitors to block oligodendrocyte differentiation 3, 4. However, it is important to determine whether the effects of HDAC inhibitors reflect the combined inhibition of multiple HDACs or specific functions of individual HDACs. A recent study with a morpholino-antisense knockdown approach in zebrafishes suggests that HDAC1 is essential for oligodendrocyte formation 5. In addition, siRNA mediated knockdown of either HDAC1 or HDAC2 is sufficient to block oligodendrocyte differentiation in vitro 43. We showed that mutant mice with oligodendrocyte lineage-specific ablation of HDAC1 or HDAC2 alone did not develop discernable defects in oligodendrocyte development, whereas simultaneous deletion of both HDAC1 and HDAC2 resulted in a complete loss of oligodendrocytes. The discrepancies with previous studies might be due to species difference e.g. mouse versus zebrafish and potential cross-targeting of HDAC1 or HDAC2 siRNA on both HDACs due to high sequence homology between HDAC1 and HDAC2.
The ablation of HDAC1/2 directed by Olig1-Cre does not affect formation of other neural cell types including motor neurons and astrocytes in the developing CNS. This observation most likely reflects a specific requirement of HDAC1/2 in oligodendrocyte formation rather than a general requirement for neural cell type specification.
The presence of OPCs in cortical progenitor culture from dCKO embryos is intriguing since no committed OPCs were detected in vivo, although HDAC1/2 ablation inhibits terminal differentiation of OPCs in vitro. Nonetheless, such altered differentiation potential of progenitor populations in culture has been observed in other cell types 44. In this vein, it is possible that extrinsic components in the culture medium, or the absence of inhibitory signals that are normally present in the CNS, enable OPC formation in vitro in the absence of HDAC1/2. Alternatively, OPCs were specified in cortical progenitor culture before Olig1-Cre mediated HDAC1/2 deletion.
β-catenin/TCF7L2 regulates oligodendrocyte differentiation
We show that HDAC1/2 ablation leads to Wnt signaling activation by stabilization and nuclear translocation of β-catenin. Constitutive activation of Wnt/β-catenin signaling inhibits oligodendrocyte differentiation by, at least partially, activating expression of differentiation inhibitors Id2/4, while repressing Olig2 and myelin gene expression. Interesting, neural progenitor cells with either Wnt3a treatment or HDAC1/2 deletion did not exhibit an increase of Notch signaling effectors, suggesting that the effect of Wnt signaling activation on oligodendrocyte differentiation is independent of the Notch pathway. Considering its strong inhibitory effects, the activation of canonical Wnt signaling may serve as a key inhibitory factor to block oligodendrocyte differentiation in the developing CNS and, perhaps, oligodendrocyte remyelination after demyelinating injury.
Through expression pattern analysis of TCF/LEF family members and transcriptome profiling analysis of wildtype and Olig1-/- mice, we identified a β-catenin co-effector TCF7L2 as an oligodendrocyte lineage-specific transcription factor. Overexpression of a dominant-repressive form of TCF7L2 but not LEF1 was observed to promote ectopic and premature OPC specification in the developing chick neural tube. This observation suggests that a unique association of TCF7L2 with transcriptional corepressor(s) would promote oligodendrocyte specification from neural progenitor cells.
The defect of oligodendrocyte maturation in TCF7L2 null mutants suggests that TCF7L2 is critical for oligodendrocyte differentiation. Thus, TCF7L2 serves a key DNA-binding component in the transcriptional complex to regulate oligodendrocyte development. The absence of apparent defects in oligodendrocyte precursor formation in TCF7L2 null mutants may reflect additional TCF factors that may utilize HDAC1/2 as a co-repressor to promote oligodendrocyte precursor specification from neural progenitor cells.
Convergence of β-catenin and HDAC1/2 on TCF7L2 interaction
How do HDAC1/2 suppress the Wnt signaling pathway? As HDAC proteins are recruited by transcriptional repressors such as Groucho/TLE-related factors 11, 45, it is possible that HDAC1/2 co-repressors switch off transcription of Wnt signaling-activated genes. Our data suggest that HDAC1/2 antagonize Wnt signaling to suppress expression of differentiation inhibitors IDs and permit or promote myelin gene expression. Alternatively, HDACs may affect other unidentified pathways besides competing with the inhibitory β-catenin pathway for oligodendrocyte differentiation.
The fact that a dominant-repressive form of TCF7L2 promotes ectopic precocious oligodendrocyte formation suggests that oligodendrocyte differentiation is mediated by TCF7L2/co-repressor complex. Our observations are consistent with that HDAC1/2 or their-associated co-repressors may directly compete with β-catenin to interact with TCF7L2. The competition of HDAC1/2 proteins with β-catenin converts TCF7L2 from a transcriptional repressor to an activator for oligodendrocyte differentiation, suggesting that TCF7L2 functions as a convergent regulator for oligodendrocyte differentiation. Such a direct competition model suggests that a balance or ratio between HDAC1/2 and β-catenin on TCF7L2 transcriptional activity may control the timing of oligodendrocyte differentiation and maturation (Supplementary Fig. 7).
Implications for human disease
HDACs have been implicated in a wide range of cellular processes and disease states based on the ability of HDAC inhibitors to improve various disease pathologies 46, 47. However, inhibition of HDACs, particularly HDAC1/2, could cause white matter injury and the potential cancer risk of Wnt signaling activation. Our studies and others 43 emphasize the need to develop specific HDAC inhibitors to avoid potential side effects in myelin regeneration. In addition, inhibition of canonical β-catenin mediated Wnt signaling in conjunction with augment of HDAC1/2 activity may offer a therapeutic approach to promote oligodendrocyte regeneration and myelin repair.
Methods
Tissue collection and RNA in situ hybridization
Brains and spinal cords from wild type and mutants at various embryonic and postnatal stages were harvested from ketamine/xylazine anesthetized animals, fixed in 4% paraformaldehyde at 4°C overnight, infused with 20% sucrose in PBS overnight, embedded in OCT and cryosectioned at 16 μm. Digoxigenin-labeled riboprobes were used to perform RNA in situ hybridization, as described previously 6, and the probes used were: murine Olig1, Olig2, Pdgfrα, Plp/Dm-20, Mbp, TCF7L2, LEF1 and Gfap and chick Pdgfrα Sox10 and Mbp. Detailed protocols are available upon request. Recombinant mouse Wnt-3a was purchased from R&D systems (Minneapolis, MN). In all cases of rodent embryonic litters analyzed, the day of detection of vaginal plug at noon was considered 0.5 dpc or E0.5.
Quantitative Real Time Polymerase Chain Reaction (QRT-PCR)
QRT-PCR was performed using the ABI Prism 7700 Sequence Detector System (Perkin-Elmer Applied Biosystems). QRT-PCR was performed as previously described 16. Primers used for expression analyses are available upon request.
Hippocampus-derived adult neural progenitor cells and oligodendroglial cell cultures
The hippocampus-derived adult neural progenitor cells (HCN cells) were originally isolated from adult (8-10 wks old) female Fisher 344 rats and have been characterized previously 24. They give rise to homogeneous populations of oligodendrocytes upon IGF-1 stimulation 24. HCN cells were cultured in N2-FGF media [DMEM:F12 with N2 supplement (Invitrogen) and basic fibroblast growth factor (20 ng/ml FGF-2)]. In the presence of 100 ng/ml IGF-1, the HCN cells express Olig2 (Supplementary Fig. 6) and can be induced into differentiated oligodendrocytes 24. Using the HCN cells for the reporter assay and biochemical studies has been established in previous studies including myelin gene promoter-driven reporter assay 24, 48.
Primary oligodendrocyte precursor cells (OPCs) were isolated from rat pups at P2 as previously described 27. For mouse primary oligodendroglial enriched cell culture, cortical precursors were isolated from E15.5 as described previously 27. The mouse cortical neural stem/progenitors were derived from control and HDAC mutant embryos were grown in the N2-bFGF growth medium for five days to enrich Olig2+ OPCs 49, then passaged by trypsinization and cultured in oligodendrocyte growth medium (N2 supplemented with bFGF and PDGFAA) and oligodendrocyte differentiation medium (N2 supplemented with T3 and CNTF) to promote oligodendrocyte differentiation as previously described 27. Most of neurons did not survive after precursor cell passaging while cultured in the oligodendrocyte growth medium (data not shown). Thus, oligodendrocyte precursors were enriched in serum-free oligodendrocyte growth medium 27.
Transient transfection, luciferase assay and immunohistochemistry
HCN progenitor cells were seeded and grown in Dulbecco's modified Eagle medium with N2 supplemented with IGF-1 (100ng/ml) before transfection with Amaxa according to the manufacturer's protocol and assayed 72-hour post-transfection for immunocytochemistry and qRT-PCR analysis. The immunohistochemical staining procedure was performed as described previously 16. Antibodies used: Olig1 and Olig2 (gift of Chuck Stiles), PDGFRα (BD Bioscience), CC1 (Oncogene research), NG2, PECAM1, NeuN and Glutamine synthetase (Chemicon), BrdU, GFAP (Sigma), Ki67 (Swant), anti-ABC, phosphorylated β-catenin, TCF7L2 and caspase-3 (Upstate). Monoclonal antibodies to Hb9 (MNR2) and RIP were obtained from the Developmental Studies Hybridoma Bank at the University of Iowa under the auspices of the National Institute of Child Health and Human Development. X-Gal staining for β-galactosidase activity was performed as previously described 6. Luciferase assay was performed by using Promega luciferase assay kit. In addition, the pRSV-renilla luciferase plasmid was included to control for variable transfection efficiencies between different experiments.
Co-immunoprecipitation and immunoblotting
HCN cells or Hek293 cells were transiently transfected with 10 μg each of TCF7L2, p△N89β-catenin and pFlag-HDAC1 or 2 in pCIG vector 50. Whole cell lysates were prepared 48 h after transfection by using 1× Passive lysis Buffer (Promega, Madison, WI) supplemented with a protease inhibitor cocktail (1:200, Sigma, St. Louis, MO). For immunoprecipitation, 600 μg of cell lysate proteins were incubated with 10μg anti-TCF7L2 or anti-β-catenin in immunoprecipitation (IP) assay as described previously 32. After Western blotting, proteins carrying the Flag epitope tag were detected with mouse anti-Flag mAb (Clontech Laboratories, Palo Alto, CA) by using chemiluminescence with the ECL kit (Pierce, Rockford, IL.) according to the instruction of the manufacturer.
Chick embryo in ovo electroporation
Chicken eggs were incubated at ~38 °C. Approximately 1 μl (1.5 μg/μl) of expression vectors (pCIG) carrying GFP, △N89β-Catenin, TCF7L2, LEF1, TCF7L2-EnR, LEF1-EnR were injected into a chicken embryo neural tube at stage HH 13-15 (E2.5) with the aid of Picospritzer III (Parker Hannifin, Cleveland, OH). The subsequent electroporation was performed by using a square wave electroporator (CUY21, BEX Co. LTP, Japan) with 5 pulses of electrical shock (25V, 50 msec for each pulse). Embryos were harvested 3 days after electroporation. At this stage (E5.5), none of oligodendrocyte markers are normally expressed in the neural tube. The green fluorescent segment of neural tube was dissected, fixed for 1 hour in 4% paraformaldehyde-PBS on ice and embedded in OCT for sectioning on a cryostat for in situ hybridization or immunohistochemistry. At least five embryos with expression of each transgene were analyzed and characterized.
Statistic analysis
Quantifications were performed from at least three experimental groups. Data are presented as mean ± SD in the graphs. p values are from Student's twotailed t test between control and experimental groups. For multiple comparisons, which were done using one-way ANOVA with post-test: Newman-Keuls multiple-comparison test. Animal use and studies were approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center at Dallas.
Supplementary Material
ACKNOWLEDGEMENTS
Authors would like to thank J. Johnson and B. Cregg for a critical reading of the manuscript and D. Rowitch for communicating their unpublished results. We thank W. Walker, A. Iavarone, Y. Yokota, K. Kim, R. Miskimins and J. Suh for ID2 and ID4 and Mbp promoter reporters, β-catenin and TCF expression vectors. This study was funded by grants from the US National Multiple Sclerosis Society (RG3978 and PP0144) and the US National Institutes of Health (NS050389 to QRL). QRL is a Harry Weaver Neuroscience Scholar and a Basil O'Connor Scholar.
REFERENCES
- 1.Hsieh J, Gage FH. Chromatin remodeling in neural development and plasticity. Curr Opin Cell Biol. 2005;17:664–671. doi: 10.1016/j.ceb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Bio. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunliffe VT, Casaccia-Bonnefil P. Histone deacetylase 1 is essential for oligodendrocyte specification in the zebrafish CNS. Mech Dev. 2006;123:24–30. doi: 10.1016/j.mod.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Lu QR, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 8.Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12:1157–1163. doi: 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- 9.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 10.Grigoryan T, Wend P, Klaus A, Birchmeier W. Deciphering the function of canonical Wnt signals in development and disease: conditional loss- and gain-of-function mutations of beta-catenin in mice. Genes Dev. 2008;22:2308–2341. doi: 10.1101/gad.1686208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurlstone A, Clevers H. T-cell factors: turn-ons and turn-offs. Embo J. 2002;21:2303–2311. doi: 10.1093/emboj/21.10.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Billin AN, Thirlwell H, Ayer DE. Beta-catenin-histone deacetylase interactions regulate the transition of LEF1 from a transcriptional repressor to an activator. Mol Cell Biol. 2000;20:6882–6890. doi: 10.1128/mcb.20.18.6882-6890.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimizu T, et al. Wnt signaling controls the timing of oligodendrocyte development in the spinal cord. Dev Biol. 2005;282:397–410. doi: 10.1016/j.ydbio.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi M, et al. Histone deacetylase 1 regulates retinal neurogenesis in zebrafish by suppressing Wnt and Notch signaling pathways. Development. 2005;132:3027–3043. doi: 10.1242/dev.01881. [DOI] [PubMed] [Google Scholar]
- 15.Montgomery RL, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xin M, et al. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci. 2005;25:1354–1365. doi: 10.1523/JNEUROSCI.3034-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 18.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nave KA. Neurological mouse mutants and the genes of myelin. J Neurosci Res. 1994;38:607–612. doi: 10.1002/jnr.490380602. [DOI] [PubMed] [Google Scholar]
- 20.Nishiyama A, Lin XH, Giese N, Heldin CH, Stallcup WB. Colocalization of NG2 proteoglycan and PDGF alpha-receptor on O2A progenitor cells in the developing rat brain. J Neurosci Res. 1996;43:299–314. doi: 10.1002/(SICI)1097-4547(19960201)43:3<299::AID-JNR5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 21.Barres BA, Lazar MA, Raff MC. A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development. 1994;120:1097–1108. doi: 10.1242/dev.120.5.1097. [DOI] [PubMed] [Google Scholar]
- 22.Stankoff B, et al. Ciliary neurotrophic factor (CNTF) enhances myelin formation: a novel role for CNTF and CNTF-related molecules. J Neurosci. 2002;22:9221–9227. doi: 10.1523/JNEUROSCI.22-21-09221.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arber S, et al. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23:659–674. doi: 10.1016/s0896-6273(01)80026-x. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh J, et al. IGF-I instructs multipotent adult neural progenitor cells to become oligodendrocytes. J Cell Biol. 2004;164:111–122. doi: 10.1083/jcb.200308101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korinek V, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 26.Maretto S, et al. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci U S A. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, et al. Isolation and culture of rat and mouse oligodendrocyte precursor cells. Nat Protoc. 2007;2:1044–1051. doi: 10.1038/nprot.2007.149. [DOI] [PubMed] [Google Scholar]
- 28.van Noort M, Meeldijk J, van der Zee R, Destree O, Clevers H. Wnt signaling controls the phosphorylation status of beta-catenin. J Biol Chem. 2002;277:17901–17905. doi: 10.1074/jbc.M111635200. [DOI] [PubMed] [Google Scholar]
- 29.Harada N, et al. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. Embo J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lappe-Siefke C, et al. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet. 2003;33:366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- 31.Munemitsu S, Albert I, Rubinfeld B, Polakis P. Deletion of an aminoterminal sequence beta-catenin in vivo and promotes hyperphosporylation of the adenomatous polyposis coli tumor suppressor protein. Mol Cell Biol. 1996;16:4088–4094. doi: 10.1128/mcb.16.8.4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang SZ, et al. An oligodendrocyte-specific zinc-finger transcription regulator cooperates with Olig2 to promote oligodendrocyte differentiation. Development. 2006;133:3389–3398. doi: 10.1242/dev.02522. [DOI] [PubMed] [Google Scholar]
- 33.Ono K, Bansal R, Payne J, Rutishauser U, Miller RH. Early development and dispersal of oligodendrocyte precursors in the embryonic chick spinal cord. Development. 1995;121:1743–1754. doi: 10.1242/dev.121.6.1743. [DOI] [PubMed] [Google Scholar]
- 34.Kardon G, Harfe BD, Tabin CJ. A Tcf4-positive mesodermal population provides a prepattern for vertebrate limb muscle patterning. Dev Cell. 2003;5:937–944. doi: 10.1016/s1534-5807(03)00360-5. [DOI] [PubMed] [Google Scholar]
- 35.Korinek V, et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 36.Wang S, Sdrulla A, Johnson JE, Yokota Y, Barres BA. A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron. 2001;29:603–614. doi: 10.1016/s0896-6273(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 37.Kondo T, Raff M. The Id4 HLH protein and the timing of oligodendrocyte differentiation. Embo J. 2000;19:1998–2007. doi: 10.1093/emboj/19.9.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S, et al. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron. 1998;21:63–75. doi: 10.1016/s0896-6273(00)80515-2. [DOI] [PubMed] [Google Scholar]
- 39.Memezawa A, et al. Id2 gene-targeted crosstalk between Wnt and retinoid signaling regulates proliferation in human keratinocytes. Oncogene. 2007;26:5038–5045. doi: 10.1038/sj.onc.1210320. [DOI] [PubMed] [Google Scholar]
- 40.Rockman SP, et al. Id2 is a target of the beta-catenin/T cell factor pathway in colon carcinoma. J Biol Chem. 2001;276:45113–45119. doi: 10.1074/jbc.M107742200. [DOI] [PubMed] [Google Scholar]
- 41.Yu L, et al. Global assessment of promoter methylation in a mouse model of cancer identifies ID4 as a putative tumor-suppressor gene in human leukemia. Nat Genet. 2005;37:265–274. doi: 10.1038/ng1521. [DOI] [PubMed] [Google Scholar]
- 42.Miskimins R, Srinivasan R, Marin-Husstege M, Miskimins WK, Casaccia-Bonnefil P. p27(Kip1) enhances myelin basic protein gene promoter activity. J Neurosci Res. 2002;67:100–105. doi: 10.1002/jnr.10080. [DOI] [PubMed] [Google Scholar]
- 43.Shen S, et al. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nat Neurosci. 2008 doi: 10.1038/nn.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hasty P, et al. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- 45.Brantjes H, Roose J, van De Wetering M, Clevers H. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 2001;29:1410–1419. doi: 10.1093/nar/29.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 47.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 48.Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116:779–793. doi: 10.1016/s0092-8674(04)00248-x. [DOI] [PubMed] [Google Scholar]
- 49.Gabay L, Lowell S, Rubin LL, Anderson DJ. Deregulation of dorsoventral patterning by FGF confers trilineage differentiation capacity on CNS stem cells in vitro. Neuron. 2003;40:485–499. doi: 10.1016/s0896-6273(03)00637-8. [DOI] [PubMed] [Google Scholar]
- 50.Megason SG, McMahon AP. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087–2098. doi: 10.1242/dev.129.9.2087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.