Abstract
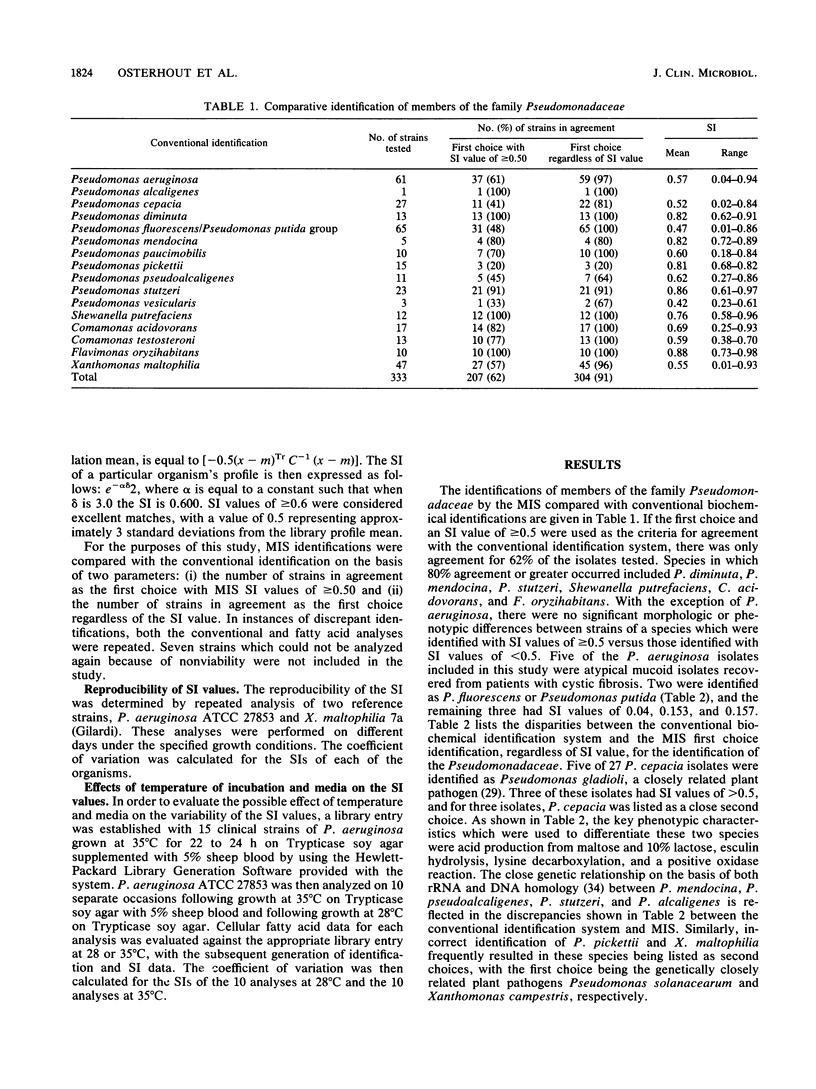
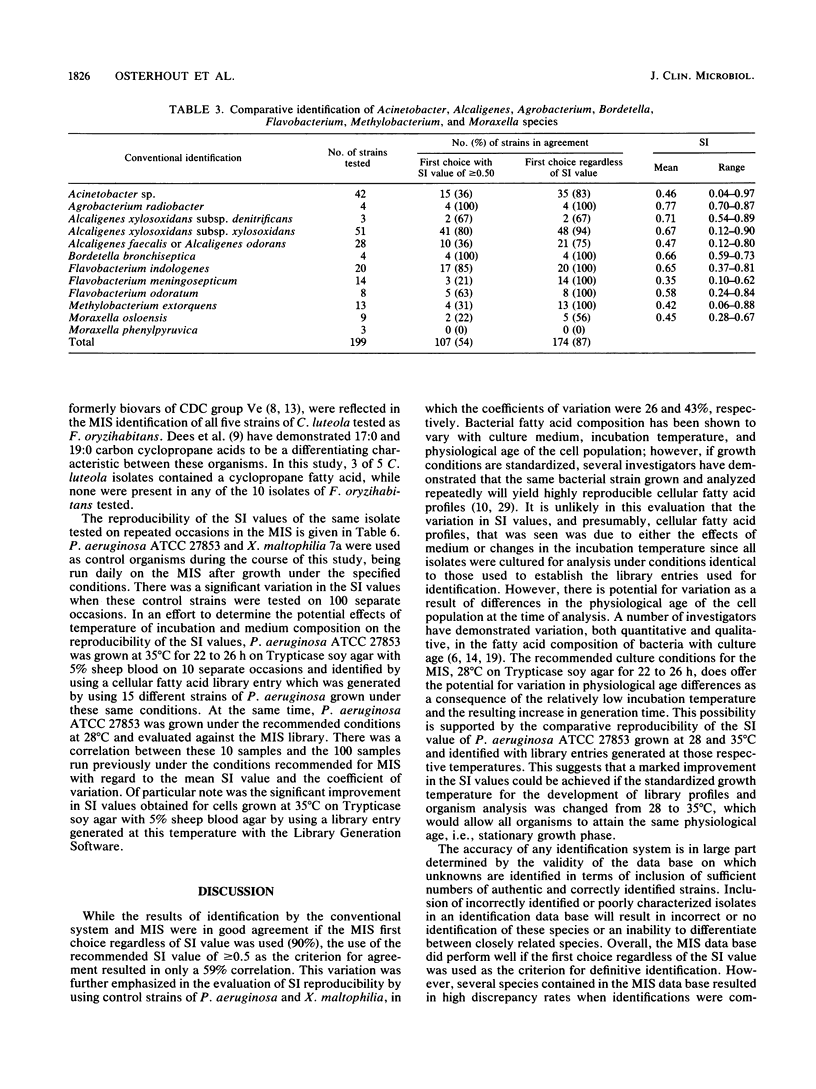
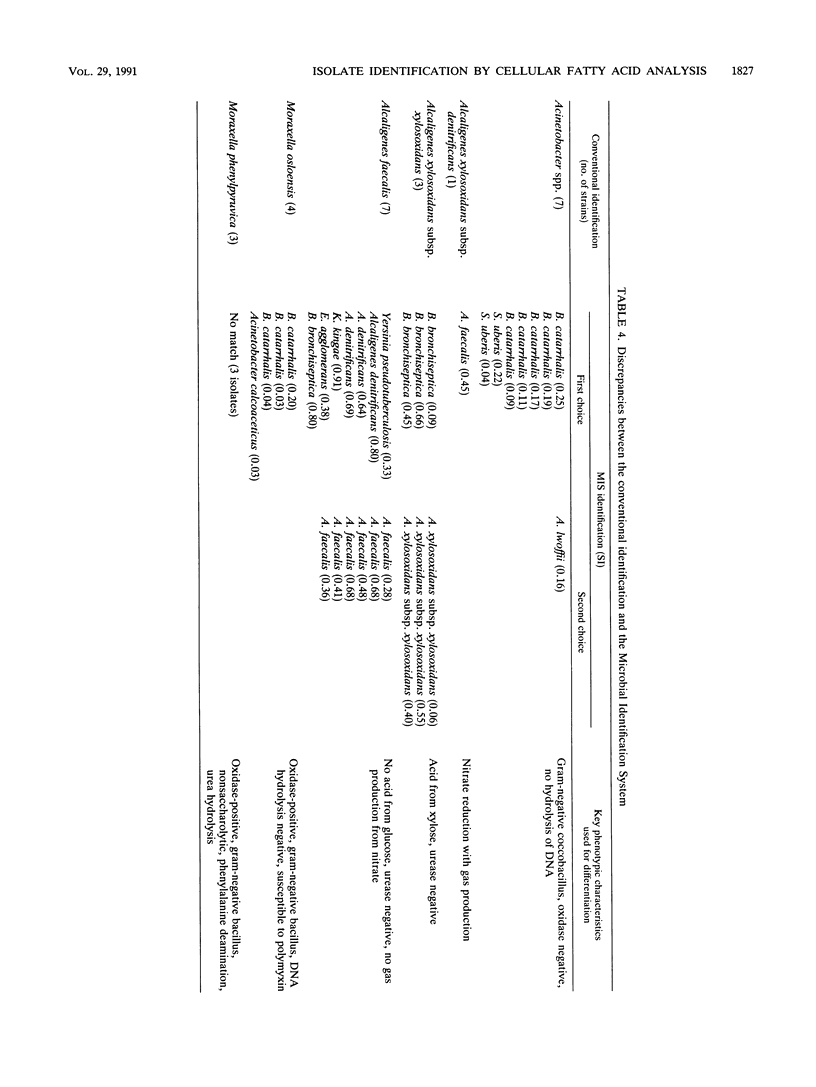
An automated cellular fatty acid (CFA) bacterial identification system, Microbial Identification System (MIS; Microbial ID, Newark, Del.), was compared with a conventional system for the identification of 573 strains of gram-negative nonfermentative bacteria. MIS identifications were based exclusively on the CFA composition following 22 to 26 h of growth at 28 degrees C on Trypticase soy agar. MIS identifications were listed with a confidence measurement (similarity index [SI]) on a scale of 0 to 1.0. A value of greater than or equal to 0.5 was considered a good match. The MIS correctly listed as the first choice 478 of 532 (90%) strains contained in the data base. However, only 314 (59%) had SI values of greater than or equal to 0.5. Of the 54 strains in which there was not agreement, 37 belonged to the genera Acinetobacter, Moraxella, or Alcaligenes or were Pseudomonas pickettii. Reproducibility studies suggest that SI variation is most likely a function of a difference in culture age at the time of analysis, which is due to the relatively low temperature and time of incubation. Other discrepancies were attributable to insufficiently characterized library entries or an inability to differentiate chemotaxonomically closely related species. The MIS, as the first automated CFA identification system, is an accurate, efficient, and relatively rapid method for the identification of gram-negative nonfermentative bacteria. The development of a CFA library with the media and incubation conditions routinely used for the isolation of clinical pathogens could further decrease the identification time and provide an increase in accuracy.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Appelbaum P. C., Stavitz J., Bentz M. S., von Kuster L. C. Four methods for identification of gram-negative nonfermenting rods: organisms more commonly encountered in clinical specimens. J Clin Microbiol. 1980 Aug;12(2):271–278. doi: 10.1128/jcm.12.2.271-278.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondz I., Olsen I. Multivariate analyses of cellular fatty acids in Bacteroides, Prevotella, Porphyromonas, Wolinella, and Campylobacter spp. J Clin Microbiol. 1991 Jan;29(1):183–189. doi: 10.1128/jcm.29.1.183-189.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christenson J. C., Welch D. F., Mukwaya G., Muszynski M. J., Weaver R. E., Brenner D. J. Recovery of Pseudomonas gladioli from respiratory tract specimens of patients with cystic fibrosis. J Clin Microbiol. 1989 Feb;27(2):270–273. doi: 10.1128/jcm.27.2.270-273.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen J., Phillips M. C., Shipley G. G. The effects of temperature on the composition and physical properties of the lipids of Pseudomonas fluorescens. Biochem J. 1971 Dec;125(3):733–742. doi: 10.1042/bj1250733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dees S. B., Moss C. W. Cellular fatty acids of Alcaligenes and Pseudomonas species isolated from clinical specimens. J Clin Microbiol. 1975 May;1(5):414–419. doi: 10.1128/jcm.1.5.414-419.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dees S. B., Moss C. W., Hollis D. G., Weaver R. E. Chemical characterization of Flavobacterium odoratum, Flavobacterium breve, and Flavobacterium-like groups IIe, IIh, and IIf. J Clin Microbiol. 1986 Feb;23(2):267–273. doi: 10.1128/jcm.23.2.267-273.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dees S. B., Moss C. W., Weaver R. E., Hollis D. Cellular fatty acid composition of Pseudomonas paucimobilis and groups IIk-2, Ve-1, and Ve-2. J Clin Microbiol. 1979 Aug;10(2):206–209. doi: 10.1128/jcm.10.2.206-209.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eerola E., Lehtonen O. P. Optimal data processing procedure for automatic bacterial identification by gas-liquid chromatography of cellular fatty acids. J Clin Microbiol. 1988 Sep;26(9):1745–1753. doi: 10.1128/jcm.26.9.1745-1753.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter M. I., Thirkell D. Variation in fatty acid composition of Sarcina flava membrane lipid with the age of the bacterial culture. J Gen Microbiol. 1971 Jan;65(1):115–118. doi: 10.1099/00221287-65-1-115. [DOI] [PubMed] [Google Scholar]
- Koestenblatt E. K., Larone D. H., Pavletich K. J. Comparison of the Oxi/Ferm and N/F systems for identification of infrequently encountered nonfermentative and oxidase-positive fermentative bacilli. J Clin Microbiol. 1982 Mar;15(3):384–390. doi: 10.1128/jcm.15.3.384-390.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämpfer P., Dott W. Evaluation of the Titertek-NF system for identification of gram-negative nonfermentative and oxidase-positive fermentative bacteria. J Clin Microbiol. 1989 Jun;27(6):1201–1205. doi: 10.1128/jcm.27.6.1201-1205.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe A. S., van der Reijden T. J. Evaluation of commercial test systems for the identification of nonfermenters. Eur J Clin Microbiol. 1984 Aug;3(4):301–305. doi: 10.1007/BF01977477. [DOI] [PubMed] [Google Scholar]
- Marr A. G., Ingraham J. L. EFFECT OF TEMPERATURE ON THE COMPOSITION OF FATTY ACIDS IN ESCHERICHIA COLI. J Bacteriol. 1962 Dec;84(6):1260–1267. doi: 10.1128/jb.84.6.1260-1267.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R., Siavoshi F., McDougal D. L. Comparison of Rapid NFT system and conventional methods for identification of nonsaccharolytic gram-negative bacteria. J Clin Microbiol. 1986 Dec;24(6):1089–1092. doi: 10.1128/jcm.24.6.1089-1092.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. T. Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxy acids. J Clin Microbiol. 1982 Sep;16(3):584–586. doi: 10.1128/jcm.16.3.584-586.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. J., Mawhinney H., Blackall P. J. Differentiation of Bordetella avium and related species by cellular fatty acid analysis. J Clin Microbiol. 1987 Jun;25(6):1059–1062. doi: 10.1128/jcm.25.6.1059-1062.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss C. W., Dees S. B. Cellular fatty acids and metabolic products of Pseudomonas species obtained from clinical specimens. J Clin Microbiol. 1976 Dec;4(6):492–502. doi: 10.1128/jcm.4.6.492-502.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss C. W., Dees S. B., Guerrant G. O. Gas-liquid chromatography of bacterial fatty acids with a fused-silica capillary column. J Clin Microbiol. 1980 Jul;12(1):127–130. doi: 10.1128/jcm.12.1.127-130.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss C. W. Gas-liquid chromatography as an analytical tool in microbiology. J Chromatogr. 1981 Jan 9;203:337–347. doi: 10.1016/s0021-9673(00)80305-2. [DOI] [PubMed] [Google Scholar]
- Moss C. W., Wallace P. L., Hollis D. G., Weaver R. E. Cultural and chemical characterization of CDC groups EO-2, M-5, and M-6, Moraxella (Moraxella) species, Oligella urethralis, Acinetobacter species, and Psychrobacter immobilis. J Clin Microbiol. 1988 Mar;26(3):484–492. doi: 10.1128/jcm.26.3.484-492.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukwaya G. M., Welch D. F. Subgrouping of Pseudomonas cepacia by cellular fatty acid composition. J Clin Microbiol. 1989 Dec;27(12):2640–2646. doi: 10.1128/jcm.27.12.2640-2646.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhofer T. R., Rowen J. W., Cunningham G. F. Characterization and identification of gram-negative, nonfermentative bacteria. J Clin Microbiol. 1977 Feb;5(2):208–220. doi: 10.1128/jcm.5.2.208-220.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhofer T. R. Use of the API 20E, Oxi/Ferm, and Minitek systems to identify nonfermentative and oxidase-positive fermentative bacteria: seven years of experience. Diagn Microbiol Infect Dis. 1983 Sep;1(3):241–256. doi: 10.1016/0732-8893(83)90024-x. [DOI] [PubMed] [Google Scholar]
- Otto L. A., Blachman U. Nonfermentative bacilli: evaluation of three systems for identification. J Clin Microbiol. 1979 Aug;10(2):147–154. doi: 10.1128/jcm.10.2.147-154.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett M. J., Greenwood J. R. Identification of oxidase-positive, glucose-negative, motile species of nonfermentative bacilli. J Clin Microbiol. 1986 May;23(5):920–923. doi: 10.1128/jcm.23.5.920-923.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoden D. L., Schable B., Smith P. B. Evaluation of PASCO MIC-ID system for identifying gram-negative bacilli. J Clin Microbiol. 1987 Dec;25(12):2363–2366. doi: 10.1128/jcm.25.12.2363-2366.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veys A., Callewaert W., Waelkens E., Van den Abbeele K. Application of gas-liquid chromatography to the routine identification of nonfermenting gram-negative bacteria in clinical specimens. J Clin Microbiol. 1989 Jul;27(7):1538–1542. doi: 10.1128/jcm.27.7.1538-1542.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace P. L., Hollis D. G., Weaver R. E., Moss C. W. Biochemical and chemical characterization of pink-pigmented oxidative bacteria. J Clin Microbiol. 1990 Apr;28(4):689–693. doi: 10.1128/jcm.28.4.689-693.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwood N. M., Blazevic D. J., Hofherr L. Comparison of the API 20E and Corning N/F systems for identification of nonfermentative gram-negative rods. J Clin Microbiol. 1979 Aug;10(2):175–179. doi: 10.1128/jcm.10.2.175-179.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]


