Abstract
The cohesin-dockerin interaction in Clostridium thermocellum cellulosome mediates the tight binding of cellulolytic enzymes to the cellulosome-integrating protein CipA. Here, this interaction was used to study the effect of different cellulose-binding domains (CBDs) on the enzymatic activity of C. thermocellum endoglucanase CelD (1,4-β-d endoglucanase, EC3.2.1.4) toward various cellulosic substrates. The seventh cohesin domain of CipA was fused to CBDs originating from the Trichoderma reesei cellobiohydrolases I and II (CBDCBH1 and CBDCBH2) (1,4-β-d glucan-cellobiohydrolase, EC3.2.1.91), from the Cellulomonas fimi xylanase/exoglucanase Cex (CBDCex) (β-1,4-d glucanase, EC3.2.1.8), and from C. thermocellum CipA (CBDCipA). The CBD-cohesin hybrids interacted with the dockerin domain of CelD, leading to the formation of CelD-CBD complexes. Each of the CBDs increased the fraction of cellulose accessible to hydrolysis by CelD in the order CBDCBH1 < CBDCBH2 ≈ CBDCex < CBDCipA. In all cases, the extent of hydrolysis was limited by the disappearance of sites accessible to CelD. Addition of a batch of fresh cellulose after completion of the reaction resulted in a new burst of activity, proving the reversible binding of the intact complexes despite the apparent binding irreversibility of some CBDs. Furthermore, burst of activity also was observed upon adding new batches of CelD–CBD complexes that contained a CBD differing from the first one. This complementation between different CBDs suggests that the sites made available for hydrolysis by each of the CBDs are at least partially nonoverlapping. The only exception was CBDCipA, whose sites appeared to overlap all of the other sites.
All potent cellulolytic bacteria and fungi produce a battery of cellulases, which act synergistically to solubilize crystalline cellulose (1–3). Cellulolytic systems can be associated into multienzymatic complexes (called cellulosomes) or unassociated as individual enzymes. In both cases, enzymes have a modular structure. The unassociated enzymes consist generally of a catalytic domain responsible for the hydrolysis reaction and of a cellulose-binding domain (CBD) mediating binding of the enzymes to the substrate. The two domains are joined by a linker peptide, which must be sufficiently long and flexible to allow efficient orientation and operation of both domains (4–6). The cellulosomal enzymes are bound noncovalently to the cellulosome-integrating protein, which carries a CBD.
Cellulases have been traditionally divided into exoglucanases and endoglucanases. Exoglucanases, or cellobiohydrolases (1,4-β-d-glucan cellobiohydrolase, EC3.2.1.91), release cellobiose units mainly from the chain ends and degrade preferentially crystalline cellulose in a processive manner. Endoglucanases (1,4-β-d-glucan glucanohydrolase, E.C. 3.2.1.4), like Clostridium thermocellum endoglucanases D (CelD), on the other hand, are thought to act more randomly along the cellulose chain and be more active on amorphous cellulose. In addition, CBDs contribute significantly to the activity of cellulases against native cellulose. This was shown in several cases by comparing the activity of bifunctional holoenzymes with core enzymes containing the catalytic domain only or by grafting a CBD onto cellulases originally consisting of a single catalytic domain (for a review see ref. 3).
So far, more than 180 different CBDs have been identified and classified into 13 families according to their amino acid sequence similarities (7). Most of the reported CBDs belong to families I, II, and III. Family I CBDs are compact polypeptides of 32–36 residues, which are found only in fungi. The CBDs of families II and III are much larger and contain 90–100 and 130–172 residues, respectively. They are specific for bacterial enzymes. Besides different structures, CBDs also have quite diverse properties. In terms of substrate binding, they have different affinities and different specificities, with some binding to crystalline cellulose whereas others are restricted to the amorphous substrate. Furthermore, in some cases the binding of isolated CBDs follows a simple thermodynamic equilibrium (8) whereas in other cases it does not and appears irreversible (9, 10). In contrast to family I CBDs, family II CBDs have been reported to enhance the physical disruption of cellulose fibers and to release small particles from cotton fibers (11, 12). However, few studies have compared the effect of different CBDs in stimulating the activity of a given cellulase (12, 13).
In this work, we compared the effects of four CBDs representative of the three main CBD families on the cellulolytic activity of Clostridium thermocellum endoglucanase CelD. CBDCBH1 and CBDCBH2, derived from Trichoderma reesei cellobiohydrolases I and II, belong to family I. They are located at the C terminus and N terminus, respectively, of the corresponding enzymes. Isolated CBDCBH1 binds reversibly to crystalline cellulose (8), whereas CBDCBH2 appears to bind irreversibly (10). The CBD of the exoglucanase/xylanase (Cex) (β-1,4-glucanase, EC3.2.1.8) (CBDCex) from Cellulomonas fimi belongs to family II and its binding to crystalline cellulose is apparently irreversible (9, 14, 15). The CBD of the cellulosome-integrating protein CipA (CBDCipA) from C. thermocellum belongs to family III (16) and binds to crystalline cellulose in a reversible manner (17). The tertiary structures of those four CBDs have been solved, and they all exhibit a flat face carrying the aromatic residues involved in the interaction with the crystalline substrate (16, 18–20).
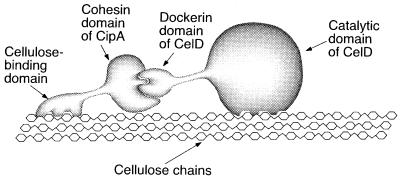
To provide CelD with different CBDs, we used the noncovalent interaction responsible for the assembly of the multienzyme cellulase complex (cellulosome) of C. thermocellum (21). Like other cellulosome components, CelD contains a noncatalytic dockerin domain, which can bind to any of the nine complementary receptor domains (cohesin domains) borne by the cellulosome integrating protein CipA. Thus, each of the CBDs to be tested was fused to the seventh cohesin domain of CipA, and each of the resulting hybrid polypeptides was allowed to interact with the dockerin domain of CelD (Fig. 1). The CelD–CBD complexes then were compared for their capacity to degrade cellulose. We analyzed to which extent they promoted binding of CelD to the substrate, increased the percentage of substrate degraded, and were able to move onto freshly added substrate. We also determined whether mixtures containing two different CelD–CBD complexes were able to degrade a higher percentage of the substrate than either of the complexes by itself. The results suggest that all four enzyme complexes can bind in a reversible mode and also that these four CBDs can bind to different areas on crystalline cellulose, thus affecting the ability of the catalytic domain to solubilize the crystalline substrate.
Figure 1.
Diagram showing the formation of an active, noncovalent complex between CelD and a chimeric polypeptide comprising a CBD fused to a cohesin domain.
Materials and Methods
Construction of Recombinant Proteins.
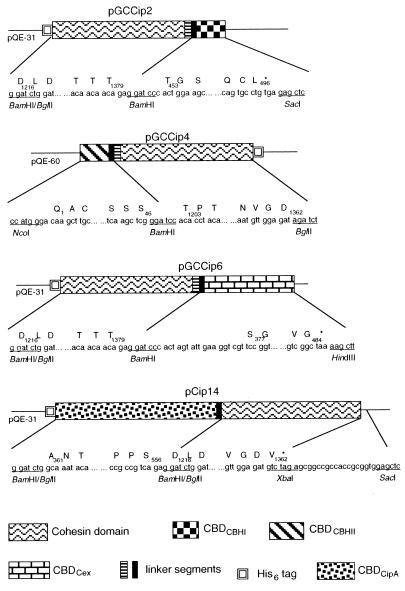
Four constructs encoding the CBD-cohesin hybrids used in this study are shown in Fig. 2. The plasmids pCip14 and pCT672, encoding the CBDCipA-cohesin hybrid and CelD, respectively, were already available (22). For the remaining constructs, DNA segments encoding protein domains to be combined were amplified by PCR, using primers containing appropriate restriction sites, so that they could be fused and inserted as desired in expression vectors. The segment encoding the seventh cohesin domain of CipA, plus a 16-residue linker segment, was cloned from pCip3 (22). The sequence encoding CBDCBH1 with a six-residue linker was cloned from pEMF5 (23). The segment encoding CBDCBH2 and six residues from the adjacent linker was carried by pTTc9 (24). The segment encoding CBDCex and seven residues from the adjacent linker was cloned from pOxscFv-CBD (25). Expression vectors were plasmids of the pQE series (Qiagen, Chatsworth, CA), enabling fusion of the polypeptides with a His6 tag. All DNA manipulations were done according to standard procedures (26). Cloned PCR fragments were verified by sequencing. Escherichia coli XL1 blue (Stratagene) was used as a host strain during the cloning steps. The final constructs were transferred to E. coli BL21 (pREP4) (22) for production of each polypeptide.
Figure 2.
Schematic representation of the pGCip2, pGCCip4, and pGCCip6 plasmids harboring CBDCBH1-cohesin, CBDCBH2-cohesin, and CBDCex-cohesin hybrids, respectively. The segments encoding the different modules are represented by boxes of different patterns. The plain boxes contained by the different modules represent the adjacent linker of each domain. The numbers under the cohesin, CBDCipA, CBDCBH1, CBDCBH2, and CBDCex boxes refer to the amino acid sequence of CipA, CBHI, CBHII, and Cex, respectively. The GenBank accession numbers for the sequences of the inserts borne by pGCCip2, pGCCip4, pGCCip6, and pCip14 are AF283514, AF283515, AF283516, and AF283517, respectively.
Protein Expression and Purification.
One-liter cultures of BL21 (pREP4) harboring pGGip2, pGCCip4, pGCCip6, and pCip14 were grown at 30°C to an OD600 of 0.4–0.6 in LB medium (27) containing 50 μg/ml carbenicillin and 10 μg/ml kanamycin. Isopropyl β-d-thiogalactoside was added to a final concentration of 1 mM, and the cultures were further incubated for 5 h at 30°C. The cells were harvested by centrifugation and resuspended in 40 ml of 50 mM Tris⋅HCl, pH 7.0 containing 1 mg DNase I, 5 mg lysozyme, and two tablets of protease inhibitor mixture (Complete Mini, Boehringer Manheim). After incubating overnight at 4°C, the cell suspension was frozen and thawed four times from −80°C to 22°C and centrifuged at 20,000 × g for 30 min. The supernatant was collected and mixed end over end for 1 h at 22°C with 3 g of CBinD 100 Resin (Novagen, 70120). The cellulose resin was sedimented and washed twice with 50 ml of 50 mM Tris⋅HCl, pH 7.0. Bound proteins were eluted with 100% ethylene glycol. The eluted fraction then was diluted 1/30 into 40 ml of Tris⋅HCl/0.7 M NaCl/20 mM imidazole, pH 8.0 (buffer A), loaded onto a 5-ml Cu2+-loaded Chelatin Sephadex (HR 10/10, Amersham Pharmacia), washed with buffer A, and eluted with a linear gradient of imidazole (20–500 mM) in buffer A. Fractions containing the purified protein were concentrated by ultrafiltration, and the buffer was exchanged to 50 mM Tris⋅HCl, pH 7.0 on a Econopac 10 DG desalting column (Bio-Rad). The intact 68-kDa form of CelD was purified by Ni2+-nitrilotriacetic acid affinity chromatography and ion exchange chromatography on Sepharose Q Fast Flow (Amersham Pharmacia) from inclusion bodies produced in E. coli BL21(pREP4) (pCT672) (M. Matuschek, and P. B., unpublished work). Purity of the protein was checked on SDS/PAGE (27). Protein concentrations were determined on the basis of A280 values. Extinction coefficient calculated from the content in aromatic residues (28) were: 120,600 M−1⋅cm−1 for CelD,, 10,840 M−1⋅cm−1 for cohesin-CBDCBH1, 21,180 M−1⋅cm−1 for CBDCBH2-cohesin, 33,930 M−1⋅cm−1 for cohesin-CBDCex and 40,600 M−1⋅cm−1 for CBDCipA-cohesin.
Complex Formation.
Complexes between CelD and the different CBD-cohesin hybrids were formed by incubating proteins together overnight at 4°C in 50 mM Tris⋅HCl, pH 7.0 containing 10 mM CaCl2, with molar ratios of CelD/CBD-cohesin ranging from 0.1 to 5. Complexes were analyzed on native 10% PAGE, using the Laemmli buffer system (27) without SDS nor βmercaptoethanol.
Activity Measurements.
Activity against Avicel, phosphoric acid-swollen cellulose (29), or bacterial microcrystalline cellulose (BMCC) (30) was determined at 45°C by incubating CelD or preformed CelD–CBD complexes at a final concentration of 80 nM with 3.6 g/liter substrate suspended in 2 ml of 50 mM Mes buffer, pH 6.0 containing 10 mM CaCl2. After suitable time intervals, aliquots were filtered, and the reaction was stopped by adding 0.5 vol of 100 mM glycine, pH 11.0 in 90% ethanol. Soluble reducing sugars were assayed with p-hydroxybenzoic acid hydrazide by using d-glucose as a standard (31). The error range was approximately 5–12% of the given value on the basis of two independent experiments. Soluble sugars produced during the hydrolysis of BMCC by CelD or CelD–CBD complexes were identified by HPLC on a Carbopac PA-1 column (Dionex) by using cellooligosaccharides from dimer to hexamer (Seikagaku, Kogyo, Tokyo) as standards. Insoluble reducing sugars present in the cellulose fraction were assayed by the dinitrosalicylate method (23), using glucose as a standard. The hydrolysis of 5 mM of p-nitrophenyl-β-d-cellobioside was assayed by monitoring the release of p-nitrophenol at 60°C in 50 mM Mes buffer, pH 6.0 containing 10 mM CaCl2 (32).
Adsorption of Proteins to Cellulose.
The adsorption of CelD and the CelD–CBD complexes to cellulose during hydrolysis was followed by measuring the intrinsic fluorescence of tryptophanyl residues using a RF 5000 spectrofluorophotometer (Shimadzu) (emission 280 nm/bandwidth 3 mm, excitation 350 nm/bandwidth 20 mm) (33). The bound fraction was estimated by deducting the fluorescence intensity of the soluble fraction from the total fluorescence of the proteins in absence of cellulose.
Results
Cellulolytic Activity of CelD and CelD–CBD Complexes.
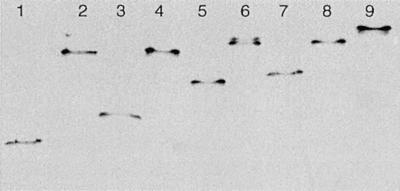
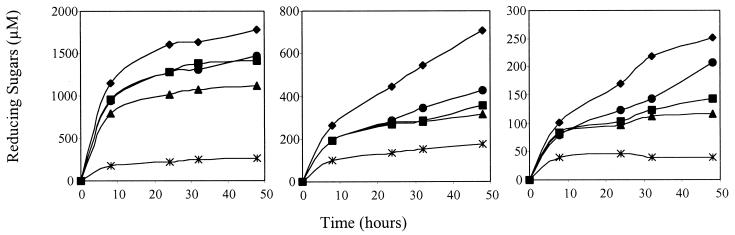
CBD-cohesin fusion proteins were purified by cellulose-based affinity chromatography, ensuring functionality of the CBDs. Nondenaturing gel electrophoresis indicated that all CBD-cohesin hybrids formed stable complexes with CelD (Fig. 3). Titration of CelD with increasing amounts of CBD-cohesin was complete at a molar ratio of 1:1. No dissociation was observed after incubating the complexes for 5 days at 60°C in the buffer used for activity assays. The activity of CelD against p-nitrophenyl-β-d-cellobioside remained the same after complex formation with any of the CBD-cohesin hybrids and was stable for at least 50 h at 45°C in the buffer used for activity assays (data not shown). Fig. 4 shows that CelD and the various CelD-CBD complexes digested insoluble forms of cellulose to clearly different extents. For all insoluble substrates studied, the trend was similar. CelD alone had the least activity. Among the different CBDs, CBDCBH1 was less active than the others in stimulating CelD activity. CBDCBH2 and CBDCex had an intermediate, rather similar, effect except on BMCC, for which CBDCex was more active. For all substrates, the strongest stimulation was observed with CBDCipA.
Figure 3.
Nondenaturing gel electrophoresis of purified CBD-cohesin hybrids and the CelD–CBD complexes. Complexes were formed as indicated in Materials and Methods using 90 pmol of each partner. Lane 1: CBDCBH1-cohesin; lane 2: CelD–CBDCBH1 complex; lane 3: CBDCBH2-cohesin; lane 4: CelD–CBDCBH2 complex; lane 5: CBDCex-cohesin; lane 6: CelD–CBDCex complex; lane 7: CBDCipA-cohesin; lane 8: CelD–CBDCipA complex; lane 9: CelD alone.
Figure 4.
Stimulation of CelD activity by the different CBDs on insoluble substrate at pH 6, 45°C. Production of reducing sugars was followed on (Left) amorphous cellulose, (Center) AVICEL, or (Right) BMCC. The protein complexes used in this study were: CelD–CBDCBH1 (▴), CelD–CBDCBH2 (■), CelD–CBDCex (●), and CelD–CBDCipA (⧫). CelD (*) was used as a control. Details of the assay are given in Materials and Methods.
Stimulation of CelD activity depended on the CelD/CBD-cohesin ratio, with no significant increase past the equivalence point for molar ratios up to 5 (data not shown). This finding suggests that stimulation of CelD activity strictly depends on the formation of complexes and that free CBD-cohesin hybrids do not contribute to the reaction. Indeed, prior incubation of BMCC for 50 h at 45°C of cellulose with any of the CBD-cohesin hybrids failed to increase the release of reducing sugars by CelD when EGTA was added to inhibit the cohesin-dockerin interaction by chelating Ca2+. Likewise, adding an excess of any of the free CBD-cohesin hybrids after 50 h of hydrolysis by the CelD–CBDCBHI complex failed to enhance the reaction (data not shown). This indicates furthermore that CelD remained complexed with the CBDCBHI-cohesin hybrid and did not form new complexes with a more potent CBD.
The soluble products from the BMCC hydrolysis were analyzed by HPLC after 8 and 24 h. In all cases, glucose and cellobiose were the only soluble sugars detected, and cellobiose was the major product. The production of insoluble sugars associated with crystalline cellulose followed the same trend as the release of soluble sugars. Under the condition of Fig. 3, after hydrolysis of BMCC by CelD for 24 h, the concentration of insoluble glucose equivalents reached 250 μM. The concentrations of insoluble sugars measured after hydrolysis under the same conditions by CelD–CBDCBH1, CelD–CBDCBH2, CelD–CBDCex, and CelD–CBDCipA were 380, 490, 480, and 540 μM, respectively. The calculated ratios of the soluble sugars to insoluble sugars ranged from 0.2 to 0.3 and were thus not significantly different for CelD and CelD–CBD complexes.
The amount of protein adsorbed to cellulose during the hydrolysis of BMCC was determined by tryptophan fluorescence. The results show that after 8 h of incubation 85% of CelD and 100% of each CelD–CBD complex was bound to the crystalline substrate (BMCC) and remained bound for the whole duration of the experiment (50 h).
Exhaustion of Substrate Sites Available for Hydrolysis.
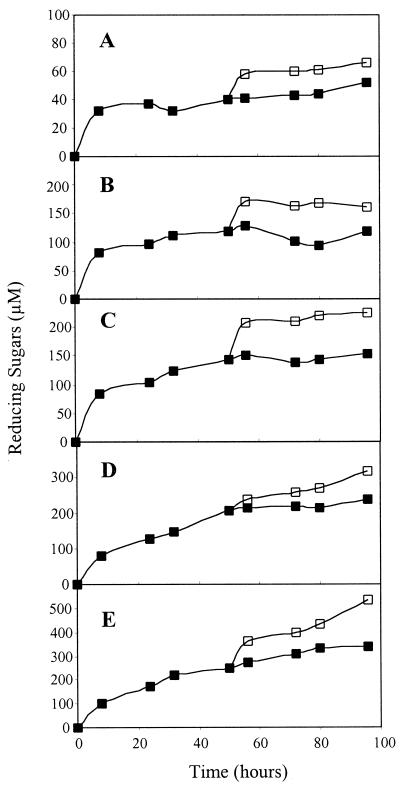
Cellulose hydrolysis by CelD or CelD–CBD complexes stopped after only a small fraction of the substrate had been degraded (up to 2.5% of BMCC), despite the stability of the complexes in the conditions of the assay. This limited hydxolysis could be caused by product inhibition of the enzyme or by exhaustion of the most easily degraded fraction of the substrate. In the case of BMCC, product accumulation was too low to cause significant inhibition (22). The addition of a fresh batch of BMCC to CelD or CelD–CBD complexes resulted in a new burst of activity, indicating that early termination of BMCC hydrolysis was caused by the limiting number of accessible substrate sites (Fig. 5). The increase of reducing sugars concentration after addition of fresh substrate was at least 2.5-fold higher for CelD–CBD complexes than for CelD alone. This finding suggests that all four intact complexes could detach from BMCC and were able to bind to the new batch of substrate.
Figure 5.
Addition of 3.6 mg of fresh BMCC (□) or the equivalent volume of buffer (■) after 50 h of hydrolysis performed with CelD (A), CelD–CBDCBH1 (B), CelD–CBDCBH2 (C), CelD–CBDCex (D), or CelD–CBDCipA (E).
Complementation of CBDs During Hydrolysis.
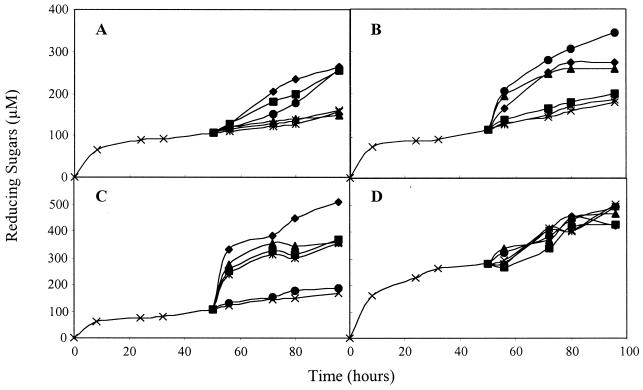
The different amounts of reducing sugars released by the different CelD–CBD complexes suggested that the substrate sites made available for hydrolysis might not be identical for all CBDs. This hypothesis was tested by incubating BMCC for 50 h with each of the CelD–CBD complexes before adding fresh batches of complexes containing either the same or different CBDs. With all complexes, addition of a second batch of the same complex failed to enhance the release of reducing sugars over that observed with buffer alone (Fig. 6). This finding confirms that, for a given CelD–CBD complex, the exhaustion of the accessible sites is the limiting factor for hydrolysis and not the inactivation of the protein. However, with CelD-CBDCBH1, CelD-CBDCBH2, and CelD-CBDCex the release of reducing sugars was boosted when a second batch of complex containing a different CBD was added. Hydrolysis in the presence of CelD–CBDCipA could not be boosted by adding any of the four complexes. Adding CelD alone in the second batch failed to increase the release of reducing sugars, except in the case of CelD–CBDCex. These results suggest that the substrate sites made accessible by the different CBDs are at least partially nonoverlapping in the case of CBDCBH1, CBDCBH2, and CBDCex. The sites made accessible by CBDCipA seem to include those revealed by the three other CBDs. All sites available to CelD alone are also available to CelD–CBDCBH1, CelD–CBDCBH2, and CelD–CBDCipA, but not to CelD–CBDCex.
Figure 6.
Complementation of different CBDs during cellulose hydrolysis. After 50 h of hydrolysis (×) by CelD–CBDCBH1 (A), CelD–CBDCBH2 (B), CelD–CBDCex (C), and CelD–CBDCipA (D); buffer (×), CelD (*), CelD–CBDCBH1 (▴), CelD–CBDCBH2 (■), CelD–CBDCex (●), and CelD–CBDCipA (⧫) were added to the reaction mixture, and the hydrolysis was continued for another 50 h.
Discussion
The hydrolysis of cellulose by the endoglucanase CelD is limited by the availability of substrate sites accessible to the enzyme (22). The results reported here show that different CBDs increase the range of available sites to different extents. Furthermore, the sites targeted for hydrolysis by different CBDs can be different, implying that each CBD possesses its own specificity for the heterogeneous sites present on the surface of BMCC. Consequently, mixtures of complexes containing the same catalytic domain but different CBDs may have access to a more extended range of cleavable sites than each complex by itself.
Previous studies have shown that the substrate preference and activity of a given cellulase depends on the type of CBD. Coutinho et al. (13) replaced the original family II CBD of C. fimi endoglucanase CenA by the family IV CBD of endoglucanase CenC. The resulting chimeric protein was less active than the native protein on crystalline cellulose, as might be expected from the fact that the type IV CBD of CenC preferentially binds to amorphous cellulose. Tomme et al. (12) exchanged the original family II CBD of C. fimi exoglucanase/xylanase Cex for the family I CBD of T. reesei cellobiohydrolase I. They observed the strongest difference for the activity on BMCC, for which Cex carrying its own CBD was 2.4 times more active than the hybrid carrying CBDCBH1. Our data show the same trend (Fig. 4), but the difference is less pronounced and, contrary to the previous study, it is seen mostly with acid-swollen cellulose rather than with BMCC. Because CelD and Cex are enzymes with different substrate preference, it is not unexpected that they should be influenced in a different manner by the nature of the CBD. In contrast to these studies, however, Srisodsuk et al. (34) found no significant change in the activity of CBHI upon replacing the original CBD of the enzyme with the CBD of T. reesei endoglucanase I, despite the fact that the two CBDs had different affinities for the substrate.
It has been claimed that the efficiency of cellulases is directly related to their affinity for the substrate (35, 36). However, enhanced adsorption to cellulose can hardly explain the increase in BMCC hydrolysis, because 85% of CelD was able to bind to the substrate by itself, and 100% when it was associated with any of the CBD-cohesin hybrids. Clearly, bulk adsorption of the enzyme does not accurately reflect the fraction that is actually poised to hydrolyze the substrate.
Furthermore, there was no obvious correlation between the efficiency of the CBD-cohesin hybrids in promoting cellulose hydrolysis and the reversible or apparently irreversible character of cellulose binding reported for each of the CBDs. Hydrolysis was less extensive when CelD was complexed with CBDCBH1, which binds reversibly, than with CBDCex or CBDCBH2, which bind with apparent irreversibility. However, the most efficient CBD was CBDCipA, which binds reversibly. At any rate, our data show that the binding of the CelD–CBD complexes can be reversible, regardless of the type of CBD, because the complexes were able to migrate and bind to freshly added substrate. Thus, the CBDs may behave differently when associated with a catalytic domain hydrolyzing the substrate thus changing the microenvironment of the enzyme. Indeed, the binding of whole cellobiohydrolase II containing a catalytic core covalently linked to the CBD is partially reversible during hydrolysis (37). This reversibility is expected because the enzyme behaves as a renewable catalyst.
None of the free CBDs affected the activity of CelD or CelD–CBD complexes. Therefore, physical disruption of the substrate probably played a minor role in the degradation process, at least as long as CBDs were not linked to CelD. This finding appears to contrast with previous observations indicating that CBDs by themselves might have a disrupting action on native cellulose, which must be because of different experimental conditions. Din et al. (11) reported that the family II CBD of C. fimi endoglucanase CenA was able to stimulate the hydrolysis of cotton and ramie fibers when added together with the catalytic core of CenA. Pagès et al. (38) observed a partial synergism on colloidal Avicel between a CBD-cohesin polypeptide (mini-CipC1) of C. cellulolyticum and a truncated endoglucanase (CelA3), which was no longer able to associate with the CBD-cohesin hybrid. However, synergism required the addition of a 30-fold excess of mini-CipC1 relative to the endoglucanase.
Because CBDs should prevent desorption of CelD, they might force the enzyme to progress continuously along the cellulose surface and proceed with a higher processivity. However, processivity was similar for CelD and CelD–CBD complexes, as shown by the small variation of ratios between soluble and insoluble reducing sugars produced in the reaction. Thus, changes in the cleavage pattern of CelD are unlikely to be responsible for the different potencies of the CBDs.
Of all studied CBD-cohesin hybrids, CBDCipA was the most efficient. It promoted the largest extent of substrate hydrolysis and was as efficient by itself as when combined with any of the other CBDs. A versatile CBD may be needed for the cellulosome, because CBDCipA has to enhance the activity of the whole set of cellulolytic enzymes making up the complex. In contrast, the CBDs of unassociated cellulases, such as CBHI or CBHII, may be more specialized and direct each catalytic domain toward its preferred type of substrate. In this respect, it could be of interest to characterize the structure of the regions corresponding to the substrate sites whose hydrolysis is preferentially enhanced by defined CBDs.
Acknowledgments
This work was supported by a fellowship of the Ministère Français de l'Enseignement Supérieur et de la Recherche and by a grant from the Ella and Georg Ehrnrooth Foundation.
Abbreviations
- CBD
cellulose-binding domain
- CBHI and CBHII
Trichoderma reesei cellobiohydrolase I and II, respectively
- CelD
Clostridium thermocellum endoglucanase D
- Cex
Cellulomonas fimi xylanase/exoglucanase
- CipA
Clostridium thermocellum cellulosome-integrating protein
- BMCC
bacterial microcrystalline cellulose
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.160216697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.160216697
References
- 1.Béguin P, Aubert J P. FEMS Rev. 1994;13:25–58. doi: 10.1111/j.1574-6976.1994.tb00033.x. [DOI] [PubMed] [Google Scholar]
- 2.Henrissat B. Cellulose. 1994;1:169–196. [Google Scholar]
- 3.Tomme P, Warren R A J, Gilkes N R. Adv Microb Physiol. 1995;37:1–81. doi: 10.1016/s0065-2911(08)60143-5. [DOI] [PubMed] [Google Scholar]
- 4.Ferreira L M A, Durrant A J, Hall J, Hazelwood G P, Gilbert H J. Biochem J. 1990;269:261–264. doi: 10.1042/bj2690261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen H, Schmuck M, Pilz I, Gilkes N R, Kilburn D G, Miller R C J, Warren R A J. J Biol Chem. 1991;266:11335–11340. [PubMed] [Google Scholar]
- 6.Srisodsuk M, Reinikainen T, Penttila M, Teeri T T. J Biol Chem. 1993;268:20756–20761. [PubMed] [Google Scholar]
- 7.Tomme P, Warren A J, Miller R C J, Kilburn D G, Gilkes N R. In: Enzymatic Degradation of Insoluble Carbohydrates. Saddler J N, Penner M H, editors. Vol. 618. San Diego: ACS; 1995. pp. 145–163. [Google Scholar]
- 8.Linder M, Teeri T T. Proc Natl Acad Sci USA. 1996;93:12251–12255. doi: 10.1073/pnas.93.22.12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jervis E J, Haynes C A, Kilburn D G. J Biol Chem. 1997;272:24016–24023. doi: 10.1074/jbc.272.38.24016. [DOI] [PubMed] [Google Scholar]
- 10.Carrard G, Linder M. Eur J Biochem. 1999;262:637–643. doi: 10.1046/j.1432-1327.1999.00455.x. [DOI] [PubMed] [Google Scholar]
- 11.Din N, Gilkes N R, Tekant B, Miller R C J, Warren R A J, Kilburn D G. Bio/Technology. 1991;9:1096–1099. [Google Scholar]
- 12.Tomme P, Driver D P, Amandoron E A, Miller R C J, Antony R, Warren J, Kilburn D G. J Bacteriol. 1995;177:4356–4363. doi: 10.1128/jb.177.15.4356-4363.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coutinho J B, Gilkes N R, Kilburn D G, Warren R A J, Miller R C J. FEMS Microbiol Lett. 1993;113:211–218. [Google Scholar]
- 14.Ong E, Gilkes N R, Miller R C, Warren R A J, Kilburn D G. Biotechnol Bioeng. 1993;42:401–409. doi: 10.1002/bit.260420402. [DOI] [PubMed] [Google Scholar]
- 15.Bray M R, Johnson P E, Gilkes N R, Mcintosh L P, Kilburn D G, Warren R A J. Protein Sci. 1996;5:2311–2318. doi: 10.1002/pro.5560051117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tormo J, Chirino A J, Morag E, Bayer E A, Shoham Y, Steitz T A. EMBO J. 1996;15:5739–5751. [PMC free article] [PubMed] [Google Scholar]
- 17.Yaron S, Bayer E A, Morag E, Lamed R, Shoham Y. In: Genetics, Biochemistry and Ecology of Cellulose Degradation: Mie Bioforum 98. Ohmiya K, Hayashi K, Sakka K, Kobayashi Y, Karita S, Kimura T, editors. Suzuka, Japan: Uni; 1998. pp. 45–46. [Google Scholar]
- 18.Kraulis P, Clore G, Nilges M, Jones T A, Pettersson G, Knowles J, Gronenborn A. Biochemistry. 1989;28:7241–7257. doi: 10.1021/bi00444a016. [DOI] [PubMed] [Google Scholar]
- 19.Hoffrén A M, Teeri T T, Teleman O. Protein Eng. 1995;8:443–450. doi: 10.1093/protein/8.5.443. [DOI] [PubMed] [Google Scholar]
- 20.Xu G Y, Ong E, Gilkes N R, Kilburn D G, Muhandiram D R, Harris-Brandts M, Carver J P, Kay L E, Harvey T S. Biochemistry. 1995;34:6993–7009. [PubMed] [Google Scholar]
- 21.Béguin P, Lemaire M. Crit Rev Biochem Mol Biol. 1996;31:201–236. doi: 10.3109/10409239609106584. [DOI] [PubMed] [Google Scholar]
- 22.Kataeva I, Guglielmi G, Béguin P. Biochem J. 1997;326:617–624. doi: 10.1042/bj3260617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ståhlberg J, Divne C, Koivula A, Piens K, Claeyssens M, Teeri T T, Jones T A. J Mol Biol. 1996;264:337–349. doi: 10.1006/jmbi.1996.0644. [DOI] [PubMed] [Google Scholar]
- 24.Koivula A, Reinikainen T, Ruohonen L, Valkeajärvi A, Claeyssens M, Teleman O, Kleywegt G J, Szardenings M, Rouvinen J, Jones T A, Teeri T T. Protein Eng. 1996;9:691–699. doi: 10.1093/protein/9.8.691. [DOI] [PubMed] [Google Scholar]
- 25.Reinikainen T, Takkinen K, Teeri T T. Enzyme Microbiol Technol. 1997;20:143–149. [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 27.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Pace N C, Vajdos F, Fee L, Grimsley G, Gray T. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood T M. Methods Enzymol. 1988;160:19–25. [Google Scholar]
- 30.Gilkes N R, Jervis E, Henrissat B, Tekant B, Miller R C, Warren R A J, Kilburn D G. J Biol Chem. 1992;267:6743–6749. [PubMed] [Google Scholar]
- 31.Lever M. Anal Biochem. 1972;47:273–279. doi: 10.1016/0003-2697(72)90301-6. [DOI] [PubMed] [Google Scholar]
- 32.Chauvaux S, Béguin P, Aubert J P, Bhat K M, Gow L A, Wood T M, Bairoch A. Biochem J. 1990;265:261–265. doi: 10.1042/bj2650261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinikainen T, Ruohonen L, Nevanen T, Laaksonen L, Jones T A, Knowles J K C, Teeri T T. Proteins. 1989;14:475–482. doi: 10.1002/prot.340140408. [DOI] [PubMed] [Google Scholar]
- 34.Srisodsuk M, Lehtio J, Linder M, Margolles-Clark E, Reinikainen T, Teeri T T. J Biotechnol. 1997;57:49–57. doi: 10.1016/s0168-1656(97)00088-6. [DOI] [PubMed] [Google Scholar]
- 35.Klyosov A A. Biochemistry. 1986;25:540–542. [Google Scholar]
- 36.Klyosov A A. Biochemistry. 1990;29:10577–10585. doi: 10.1021/bi00499a001. [DOI] [PubMed] [Google Scholar]
- 37.Palonen H, Tenkanen M, Linder M. Appl Environ Microbiol. 1999;65:5229–5233. doi: 10.1128/aem.65.12.5229-5233.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pagès S, Gal L, Belaich A, Gaudin C, Tardif C, Belaich J P. J Bacteriol. 1997;179:2810–2816. doi: 10.1128/jb.179.9.2810-2816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]