Abstract
Aim
To define the Y-chromosomal genetic structure in a sample of Atayal men from Taiwan.
Methods
Buccal swab samples were collected from 170 unrelated healthy male volunteers from Taiwanese aboriginal Atayal population. Genomic DNA was extracted and 17 Y chromosome-specific short tandem repeat loci (DYS456, DYS389I, DYS390, DYS389II, DYS458, DYS19, DYS385a/b, DYS393, DYS391, DYS439, DYS635, DYS392, Y GATA H4, DYS437, DYS438, and DYS448) were analyzed using the AmpFlSTR Yfiler Polymerase Chain Reaction Amplification Kit.
Results
A total of 99 different haplotypes were identified, 69 (69.7%) of which were unique. Total haplotype diversity was 0.9887. The most common haplotype was shared by 9 individuals in the study sample. Gene diversities ranged from 0.0574 for DYS438 to 0.6749 for DYS456.
Conclusion
Our results will help provide the molecular genetic evidence for human settlement of the Pacific.
The Y chromosome is inherited only from father to son and remains generally unchanged over generations except for gradual accumulation of mutations. This unique mode of inheritance and the absence of recombination with the non-recombining portion of the X chromosome during meiosis lead to the maintenance of polymorphisms inherited through men of the same paternal lineage. This property can be exploited for forensic purposes and paternity testing. The polymorphisms of Y-chromosomal short tandem repeat (Y-STR) loci are a powerful tool for identification and confirmation of shared paternity and the relatedness of half-brothers or grandfather and grandson (1).
The population of Taiwan in 2008 was about 23 million, comprising 68.9% of Holo, 15.0% of Hakka, 14.0% of the mainlander population, and only 2.1% of indigenous population. There are 13 indigenous tribes with the total population of 488 700: Amis, Paiwan, Atayal, Bunun, Truku, Rukai, Puyuma, Tsou, Saisiyat, Yami, Kavalan, Thao, and Sakizaya. The population of the Atayal tribe amounts to 82 200. Traditionally, the Atayal tribe resides in the mid-northern mountain regions of Taiwan (Figure 1), including Wulai Township of Taipei County, Fuhsing Township of Taoyuan County, the Jianshih and Wufeng Townships of Hsinchu County, the Nanjuang and Taian Townships of Miaoli County, Heping Township of Taichung County, Renai Township of Nantou County, the Datong and Nanao Townships of Yilan County, and the Sioulin, Wanrung, and Juoshi Townships of Hualien County (2).
Figure 1.
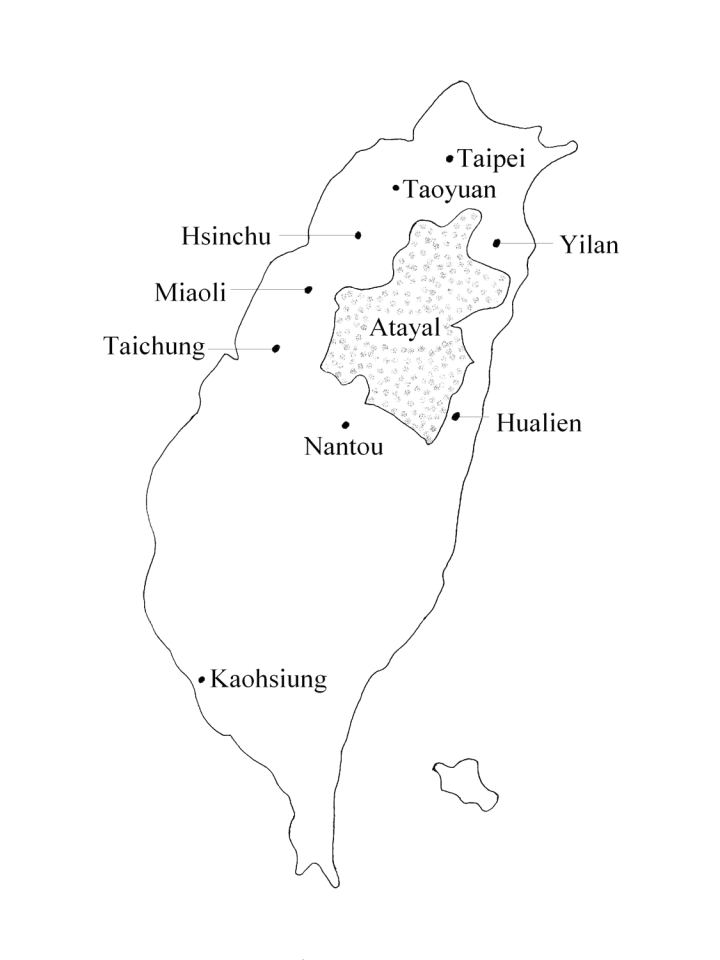
Map of regions in Taiwan where the Atayal population resides (adapted from http://www.apc.gov.tw/).
The aim of this study was to analyze 17 Y-STR loci in the Taiwanese Atayal population in order to investigate the genetic relationship between the Atayal tribe and the neighboring populations. The results obtained from this study could provide molecular genetic evidence for human settlement of the Pacific.
Materials and methods
Population
Buccal swab samples were collected from 170 unrelated male aboriginal Atayal volunteers residing in the mid-northern regions of Taiwan, who provided written informed consent. DNA was purified from the buccal swab samples using a Genomic DNA Extraction Kit (Yeastern company, Taipei, Taiwan, ROC); DNA concentrations were determined using the 7300 Real Time PCR System and the Quantifiler Y Human Male DNA Quantification Kit (Applied Biosystems company, Foster City, CA, USA).
DNA analysis
Target DNA (0.1 ng/μL) was amplified in a thermal cycler GeneAmp PCR System 9700 (Applied Biosystems) using the AmpFlSTR® Yfiler PCR Amplification Kit (Applied Biosystems) according to the manufacturers’ instructions. Single-tube polymerase chain reaction (PCR) was used for co-amplification of multiple STR loci, and 4-color detection of the resulting length-variant STR alleles allowed typing of 17 Y chromosome markers: DYS456, DYS389I, DYS390, DYS389II, DYS458, DYS19, DYS385a/b, DYS393, DYS391, DYS439, DYS635, DYS392, Y GATA H4, DYS437, DYS438, and DYS448. These include a 9-marker European minimal haplotype (minHt, the haplotype set of DYS19, DYS385a/b, DYS389I, DYS389II, DYS390, DYS391, DYS392, and DYS393) and an 11-marker Scientific Working Group on DNA Analysis Methods (SWGDAM) core set (minHt plus DYS438 and DYS439). PCR thermocycling parameters were the following: polymerase initial hot start at 95°C for 11 minutes; 30 cycles of denaturation 94°C for 1 minute, annealing at 61°C for 1 minute, and extension at 72°C for 1 minute; final extension was performed at 60°C for 80 minutes. PCR reactions were carried out in a final volume of 25 μL – 10 μL of target DNA, 5 μL of primer set, 9.2 μL of PCR reaction mix, and 0.8 μL of DNA polymerase. Electrophoresis of the PCR products, together with GeneScan-500 Internal Lane Size Standard (LIZ-500) for size determination, was performed on the ABI Prism® 3100 Avant genetic analyzer (Applied Biosystems) to separate Y-STR alleles. Data were analyzed using GeneMapper, version 3.1, software (Applied Biosystems) in conjunction with the Yfiler Allelic Ladder. Alleles were named as recommended by the DNA Commission of the International Society of Forensic Genetics (3).
Statistical analysis
Allele frequencies were determined by direct counting. Gene diversity and haplotype diversity were calculated according to Nei (4). Formulas applied were: gene diversity = 1-Σ Pi2, where Pi is the frequency of the i-th allele, and haplotype diversity = n(1-Σ fi2)/(n-1), where n is the sample size and fi is the frequency of the i-th haplotype. The standard error for haplotype diversity estimate was calculated according to the following equation (4): SE = {2[Σfi3-(Σfi2)2]/n}1/2. Comparisons between 2 populations were made by the analysis of molecular variance (AMOVA) test performed by Arlequin software, version 3.11 (5).
Results
Y-STR loci allele frequencies in 170 Atayal men are shown in Table 1. The most common alleles of the 17 Y-chromosomal STR loci were DYS456*17, DYS389I*13, DYS390*23, DYS389II*29, DYS458*15, DYS19*15, DYS385a/b*13/13, DYS393*13, DYS391*10, DYS439*12, DYS635*22, DYS392*14, Y GATA H4*12, DYS437*14, DYS438*10, and DYS448*18. Gene diversity values of these loci ranged from 0.0574 (DYS438) to 0.6749 (DYS456). Allele frequencies above 0.9 were found in 6 loci (DYS390, DYS393, DYS392, DYS437, DYS438, and DYS448). The gene diversities of the 6 loci were all below 0.16.
Table 1.
Allele frequencies and gene diversities of 17 Y-chromosomal short tandem repeat loci in 170 unrelated Atayal men from Taiwan
| Allele | Locus |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DYS456 | DYS389I | DYS390 | DYS389II | DYS458 | DYS19 | DYS393 | DYS391 | DYS439 | DYS635 | DYS392 | Y GATA H4 | DYS437 | DYS438 | DYS448 | Genotype | DYS385a/b | |
| 10 | 0.747 | 0.006 | 0.971 | 11,11 | 0.006 | ||||||||||||
| 11 | 0.006 | 0.253 | 0.053 | 0.006 | 0.171 | 0.024 | 12,13 | 0.035 | |||||||||
| 12 | 0.135 | 0.035 | 0.882 | 0.012 | 0.594 | 0.006 | 12,14 | 0.006 | |||||||||
| 13 | 0.806 | 0.947 | 0.059 | 0.035 | 0.229 | 12,19 | 0.006 | ||||||||||
| 14 | 0.018 | 0.053 | 0.006 | 0.024 | 0.018 | 0.006 | 0.947 | 0.965 | 12,20 | 0.006 | |||||||
| 15 | 0.065 | 0.665 | 0.818 | 0.035 | 13,13 | 0.629 | |||||||||||
| 16 | 0.118 | 0.253 | 0.141 | 13,14 | 0.265 | ||||||||||||
| 17 | 0.406 | 0.029 | 0.018 | 0.029 | 13,15 | 0.006 | |||||||||||
| 18 | 0.376 | 0.029 | 0.918 | 13,18 | 0.012 | ||||||||||||
| 19 | 0.018 | 0.018 | 0.012 | 0.024 | 14,14 | 0.006 | |||||||||||
| 20 | 0.029 | 0.024 | 14,20 | 0.006 | |||||||||||||
| 21 | 0.182 | 0.006 | 15,18 | 0.012 | |||||||||||||
| 22 | 0.012 | 0.629 | 15,19 | 0.006 | |||||||||||||
| 23 | 0.935 | 0.129 | |||||||||||||||
| 24 | 0.024 | 0.018 | |||||||||||||||
| 25 | 0.029 | ||||||||||||||||
| 26 | 0.006 | ||||||||||||||||
| 27 | 0.024 | ||||||||||||||||
| 28 | 0.129 | ||||||||||||||||
| 29 | 0.424 | ||||||||||||||||
| 30 | 0.394 | ||||||||||||||||
| 31 | 0.024 | ||||||||||||||||
| Gene diversity | 0.6749 | 0.3294 | 0.1237 | 0.6474 | 0.4921 | 0.3107 | 0.1015 | 0.3779 | 0.2152 | 0.5525 | 0.1017 | 0.5653 | 0.0681 | 0.0574 | 0.1559 | 0.5320 | |
A total of 99 haplotypes were identified in the Atayal population (Table 2), 69 of which were unique and 30 were present in 2 or more individuals. The 13, 7, 3, 3, 3, and 1 haplotypes were found in 2, 3, 4, 5, 6, and 9 individuals, respectively. Haplotype diversity was 0.9887 ± 0.0014 and the discrimination capacity was 0.5824 (99/170). The most frequent haplotype was H79, which was shared by 9 individuals. Alleles were 18 for DYS456, 13 for DYS389I, 23 for DYS390, 30 for DYS389II, 15 for DYS458, 15 for DYS19, 13-13 for DYS385a/b, 13 for DYS393, 10 for DYS391, 12 for DYS439, 22 for DYS635, 14 for DYS392, 11 for Y GATA H4, 14 for DYS437, 10 for DYS438, and 18 for DYS448.
Table 2.
Haplotypes observed of 17 Y-chromosomal short tandem repeat loci in 170 unrelated Atayal men from Taiwan
| Haplotype | n* | f† | Locus |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DYS456 | DYS389I | DYS390 | DYS389II | DYS458 | DYS19 | DYS385a/b | DYS393 | DYS391 | DYS439 | DYS635 | DYS392 | Y GATA H4 | DYS437 | DYS438 | DYS448 | |||
| H01 | 1 | 0.006 | 14 | 12 | 25 | 28 | 15 | 16 | 12,19 | 12 | 10 | 12 | 20 | 13 | 12 | 15 | 10 | 19 |
| H02 | 1 | 0.006 | 14 | 12 | 25 | 28 | 17 | 16 | 12,20 | 12 | 10 | 11 | 21 | 14 | 12 | 14 | 10 | 19 |
| H03 | 1 | 0.006 | 14 | 13 | 24 | 29 | 16 | 16 | 12,14 | 13 | 11 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H04 | 1 | 0.006 | 15 | 11 | 25 | 29 | 19 | 16 | 13,18 | 13 | 11 | 11 | 22 | 13 | 11 | 14 | 10 | 18 |
| H05 | 1 | 0.006 | 15 | 12 | 22 | 27 | 18 | 16 | 13,13 | 13 | 10 | 11 | 19 | 14 | 11 | 14 | 10 | 18 |
| H06 | 1 | 0.006 | 15 | 12 | 23 | 27 | 18 | 16 | 13,14 | 13 | 10 | 12 | 21 | 14 | 12 | 14 | 10 | 17 |
| H07 | 1 | 0.006 | 15 | 12 | 23 | 28 | 18 | 14 | 15,18 | 12 | 10 | 12 | 20 | 14 | 12 | 15 | 12 | 20 |
| H08 | 1 | 0.006 | 15 | 12 | 23 | 28 | 19 | 14 | 15,18 | 12 | 10 | 12 | 20 | 14 | 12 | 15 | 11 | 20 |
| H09 | 1 | 0.006 | 15 | 12 | 23 | 29 | 17 | 14 | 15,19 | 12 | 10 | 12 | 20 | 14 | 12 | 15 | 11 | 20 |
| H10 | 1 | 0.006 | 15 | 12 | 24 | 26 | 18 | 14 | 13,18 | 13 | 10 | 13 | 21 | 14 | 12 | 15 | 11 | 20 |
| H11 | 1 | 0.006 | 15 | 12 | 24 | 28 | 16 | 16 | 13,15 | 14 | 11 | 12 | 21 | 13 | 12 | 14 | 10 | 19 |
| H12 | 1 | 0.006 | 15 | 12 | 24 | 29 | 16 | 15 | 13,13 | 13 | 10 | 11 | 20 | 14 | 12 | 14 | 10 | 18 |
| H13 | 1 | 0.006 | 15 | 13 | 23 | 30 | 15 | 15 | 11,11 | 14 | 10 | 11 | 22 | 11 | 11 | 14 | 10 | 21 |
| H14 | 1 | 0.006 | 15 | 14 | 23 | 30 | 15 | 15 | 13,13 | 13 | 10 | 12 | 21 | 14 | 12 | 14 | 10 | 18 |
| H15 | 1 | 0.006 | 16 | 12 | 23 | 27 | 18 | 16 | 13,14 | 13 | 10 | 12 | 21 | 14 | 12 | 14 | 10 | 17 |
| H16 | 1 | 0.006 | 16 | 12 | 23 | 27 | 19 | 16 | 13,14 | 13 | 10 | 12 | 21 | 14 | 12 | 14 | 10 | 17 |
| H17 | 1 | 0.006 | 16 | 12 | 25 | 29 | 17 | 15 | 12,13 | 12 | 11 | 12 | 19 | 12 | 12 | 15 | 10 | 17 |
| H18 | 1 | 0.006 | 16 | 12 | 25 | 29 | 17 | 15 | 14,20 | 14 | 10 | 11 | 23 | 13 | 10 | 14 | 10 | 18 |
| H19 | 2 | 0.012 | 16 | 13 | 23 | 29 | 15 | 15 | 13,13 | 13 | 10 | 12 | 22 | 14 | 13 | 14 | 10 | 18 |
| H20 | 3 | 0.018 | 16 | 13 | 23 | 29 | 15 | 15 | 13,13 | 13 | 10 | 12 | 23 | 14 | 13 | 14 | 10 | 18 |
| H21 | 1 | 0.006 | 16 | 13 | 23 | 29 | 15 | 15 | 12,13 | 13 | 10 | 12 | 22 | 14 | 13 | 14 | 10 | 18 |
| H22 | 1 | 0.006 | 16 | 13 | 23 | 29 | 15 | 17 | 13,13 | 13 | 10 | 12 | 21 | 14 | 11 | 14 | 10 | 18 |
| H23 | 6 | 0.035 | 16 | 13 | 23 | 29 | 16 | 15 | 13,14 | 13 | 10 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H24 | 2 | 0.012 | 16 | 13 | 23 | 29 | 16 | 16 | 13,14 | 13 | 10 | 12 | 22 | 14 | 11 | 14 | 10 | 18 |
| H25 | 1 | 0.006 | 16 | 13 | 23 | 30 | 15 | 15 | 13,14 | 13 | 11 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H26 | 1 | 0.006 | 17 | 12 | 23 | 28 | 15 | 15 | 13,13 | 13 | 10 | 13 | 21 | 14 | 12 | 14 | 10 | 18 |
| H27 | 5 | 0.029 | 17 | 13 | 23 | 28 | 16 | 15 | 13,14 | 13 | 10 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H28 | 1 | 0.006 | 17 | 13 | 23 | 28 | 16 | 15 | 14,14 | 13 | 10 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H29 | 1 | 0.006 | 17 | 13 | 23 | 28 | 16 | 16 | 13,13 | 13 | 10 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H30 | 1 | 0.006 | 17 | 13 | 23 | 29 | 15 | 15 | 13,13 | 13 | 10 | 12 | 22 | 14 | 13 | 14 | 10 | 17 |
| H31 | 2 | 0.012 | 17 | 13 | 23 | 29 | 15 | 15 | 12,13 | 13 | 10 | 12 | 22 | 14 | 13 | 14 | 10 | 18 |
| H32 | 6 | 0.035 | 17 | 13 | 23 | 29 | 15 | 15 | 13,13 | 13 | 10 | 12 | 22 | 14 | 13 | 14 | 10 | 18 |
| H33 | 6 | 0.035 | 17 | 13 | 23 | 29 | 15 | 15 | 13,13 | 13 | 10 | 12 | 23 | 14 | 13 | 14 | 10 | 18 |
| H34 | 1 | 0.006 | 17 | 13 | 23 | 29 | 15 | 15 | 13,13 | 13 | 11 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H35 | 2 | 0.012 | 17 | 13 | 23 | 29 | 15 | 17 | 13,13 | 13 | 10 | 12 | 22 | 14 | 13 | 14 | 10 | 18 |
| H36 | 1 | 0.006 | 17 | 13 | 23 | 29 | 16 | 15 | 12,13 | 13 | 10 | 12 | 22 | 14 | 13 | 14 | 10 | 18 |
| H37 | 5 | 0.029 | 17 | 13 | 23 | 29 | 16 | 15 | 13,13 | 13 | 10 | 12 | 22 | 14 | 13 | 14 | 10 | 18 |
| H38 | 1 | 0.006 | 17 | 13 | 23 | 29 | 16 | 15 | 13,14 | 13 | 10 | 11 | 21 | 14 | 12 | 14 | 10 | 18 |
| H39 | 1 | 0.006 | 17 | 13 | 23 | 29 | 16 | 15 | 13,14 | 13 | 10 | 11 | 23 | 14 | 12 | 14 | 10 | 18 |
| H40 | 2 | 0.012 | 17 | 13 | 23 | 29 | 16 | 15 | 13,14 | 13 | 10 | 12 | 21 | 14 | 12 | 14 | 10 | 18 |
| H41 | 3 | 0.018 | 17 | 13 | 23 | 29 | 16 | 15 | 13,14 | 13 | 10 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H42 | 2 | 0.012 | 17 | 13 | 23 | 29 | 16 | 16 | 13,14 | 13 | 10 | 12 | 23 | 14 | 11 | 14 | 10 | 18 |
| H43 | 3 | 0.018 | 17 | 13 | 23 | 29 | 16 | 16 | 13,14 | 13 | 10 | 12 | 24 | 14 | 11 | 14 | 10 | 18 |
| H44 | 1 | 0.006 | 17 | 13 | 23 | 29 | 17 | 15 | 13,14 | 13 | 10 | 12 | 21 | 14 | 12 | 14 | 10 | 18 |
| H45 | 1 | 0.006 | 17 | 13 | 23 | 30 | 15 | 15 | 13,13 | 13 | 10 | 12 | 21 | 14 | 12 | 14 | 10 | 18 |
| H46 | 1 | 0.006 | 17 | 13 | 23 | 30 | 15 | 15 | 13,13 | 13 | 10 | 12 | 21 | 14 | 13 | 14 | 10 | 18 |
| H47 | 2 | 0.012 | 17 | 13 | 23 | 30 | 15 | 15 | 13,13 | 13 | 10 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H48 | 4 | 0.024 | 17 | 13 | 23 | 30 | 15 | 15 | 13,13 | 13 | 10 | 12 | 23 | 14 | 13 | 14 | 10 | 18 |
| H49 | 1 | 0.006 | 17 | 13 | 23 | 30 | 15 | 15 | 13,13 | 13 | 11 | 12 | 22 | 14 | 11 | 14 | 10 | 18 |
| H50 | 5 | 0.029 | 17 | 13 | 23 | 30 | 15 | 15 | 13,13 | 13 | 11 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H51 | 1 | 0.006 | 17 | 13 | 23 | 30 | 15 | 15 | 13,14 | 13 | 11 | 13 | 22 | 14 | 12 | 14 | 10 | 18 |
| H52 | 1 | 0.006 | 17 | 13 | 23 | 30 | 15 | 16 | 13,13 | 13 | 11 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H53 | 1 | 0.006 | 17 | 13 | 23 | 30 | 16 | 15 | 13,13 | 13 | 11 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H54 | 2 | 0.012 | 17 | 14 | 23 | 30 | 15 | 15 | 13,13 | 13 | 10 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H55 | 1 | 0.006 | 17 | 14 | 23 | 30 | 16 | 15 | 13,13 | 13 | 10 | 12 | 22 | 14 | 12 | 14 | 10 |
18 |
| H56 | 2 | 0.012 | 17 | 14 | 23 | 30 | 16 | 15 | 13,13 | 13 | 10 | 12 | 22 | 14 | 13 | 14 | 10 | 18 |
| H57 | 1 | 0.006 | 17 | 14 | 23 | 31 | 15 | 15 | 13,13 | 13 | 10 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H58 | 1 | 0.006 | 17 | 14 | 23 | 31 | 16 | 15 | 13,13 | 13 | 10 | 12 | 22 | 13 | 13 | 14 | 10 | 18 |
| H59 | 1 | 0.006 | 18 | 12 | 22 | 28 | 15 | 15 | 13,13 | 13 | 10 | 13 | 22 | 14 | 12 | 14 | 10 | 18 |
| H60 | 1 | 0.006 | 18 | 12 | 23 | 28 | 15 | 15 | 13,13 | 13 | 10 | 12 | 21 | 14 | 12 | 14 | 10 | 18 |
| H61 | 2 | 0.012 | 18 | 12 | 23 | 28 | 15 | 15 | 13,13 | 13 | 10 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H62 | 1 | 0.006 | 18 | 12 | 23 | 28 | 15 | 15 | 13,13 | 13 | 10 | 13 | 21 | 14 | 12 | 14 | 10 | 18 |
| H63 | 1 | 0.006 | 18 | 12 | 23 | 28 | 15 | 15 | 13,13 | 13 | 10 | 13 | 22 | 14 | 12 | 14 | 10 | 18 |
| H64 | 1 | 0.006 | 18 | 12 | 23 | 29 | 15 | 16 | 13,13 | 13 | 10 | 13 | 22 | 14 | 12 | 14 | 10 | 18 |
| H65 | 1 | 0.006 | 18 | 12 | 23 | 29 | 15 | 16 | 13,14 | 13 | 10 | 13 | 22 | 14 | 12 | 14 | 10 | 18 |
| H66 | 1 | 0.006 | 18 | 13 | 23 | 28 | 15 | 15 | 13,13 | 13 | 10 | 13 | 22 | 14 | 13 | 14 | 10 | 18 |
| H67 | 1 | 0.006 | 18 | 13 | 23 | 28 | 15 | 15 | 13,13 | 13 | 10 | 14 | 22 | 14 | 13 | 14 | 10 | 18 |
| H68 | 1 | 0.006 | 18 | 13 | 23 | 28 | 16 | 15 | 13,14 | 13 | 10 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H69 | 1 | 0.006 | 18 | 13 | 23 | 29 | 15 | 15 | 13,13 | 13 | 10 | 12 | 22 | 14 | 11 | 14 | 10 | 18 |
| H70 | 1 | 0.006 | 18 | 13 | 23 | 29 | 15 | 15 | 13,13 | 13 | 10 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H71 | 3 | 0.018 | 18 | 13 | 23 | 29 | 15 | 15 | 13,13 | 13 | 11 | 12 | 21 | 14 | 12 | 14 | 10 | 18 |
| H72 | 1 | 0.006 | 18 | 13 | 23 | 29 | 15 | 15 | 13,13 | 13 | 11 | 12 | 22 | 14 | 12 | 14 | 10 | 19 |
| H73 | 1 | 0.006 | 18 | 13 | 23 | 29 | 15 | 15 | 13,13 | 13 | 11 | 12 | 23 | 14 | 12 | 14 | 11 | 18 |
| H74 | 3 | 0.018 | 18 | 13 | 23 | 29 | 15 | 15 | 13,14 | 13 | 11 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H75 | 2 | 0.012 | 18 | 13 | 23 | 29 | 15 | 16 | 13,13 | 13 | 10 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H76 | 1 | 0.006 | 18 | 13 | 23 | 30 | 14 | 15 | 13,13 | 13 | 11 | 12 | 21 | 14 | 12 | 14 | 10 | 18 |
| H77 | 1 | 0.006 | 18 | 13 | 23 | 30 | 15 | 15 | 13,13 | 13 | 10 | 12 | 21 | 14 | 11 | 14 | 10 | 18 |
| H78 | 4 | 0.024 | 18 | 13 | 23 | 30 | 15 | 15 | 13,13 | 13 | 10 | 12 | 21 | 14 | 12 | 14 | 10 | 18 |
| H79 | 9 | 0.053 | 18 | 13 | 23 | 30 | 15 | 15 | 13,13 | 13 | 10 | 12 | 22 | 14 | 11 | 14 | 10 | 18 |
| H80 | 1 | 0.006 | 18 | 13 | 23 | 30 | 15 | 15 | 13,13 | 13 | 10 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H81 | 1 | 0.006 | 18 | 13 | 23 | 30 | 15 | 15 | 13,13 | 13 | 11 | 11 | 22 | 14 | 12 | 14 | 10 | 18 |
| H82 | 3 | 0.018 | 18 | 13 | 23 | 30 | 15 | 15 | 13,13 | 13 | 11 | 12 | 21 | 14 | 12 | 14 | 10 | 18 |
| H83 | 1 | 0.006 | 18 | 13 | 23 | 30 | 15 | 15 | 13,13 | 13 | 11 | 12 | 22 | 12 | 12 | 14 | 10 | 18 |
| H84 | 1 | 0.006 | 18 | 13 | 23 | 30 | 15 | 15 | 13,13 | 13 | 11 | 12 | 22 | 13 | 12 | 14 | 10 | 18 |
| H85 | 2 | 0.012 | 18 | 13 | 23 | 30 | 15 | 15 | 13,13 | 13 | 11 | 12 | 22 | 14 | 11 | 14 | 10 | 18 |
| H86 | 4 | 0.024 | 18 | 13 | 23 | 30 | 15 | 15 | 13,13 | 13 | 11 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H87 | 1 | 0.006 | 18 | 13 | 23 | 30 | 15 | 15 | 12,13 | 13 | 10 | 12 | 22 | 14 | 11 | 14 | 10 | 18 |
| H88 | 1 | 0.006 | 18 | 13 | 23 | 30 | 15 | 15 | 13,14 | 13 | 10 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H89 | 2 | 0.012 | 18 | 13 | 23 | 30 | 15 | 15 | 13,14 | 13 | 10 | 12 | 23 | 14 | 12 | 14 | 10 | 18 |
| H90 | 1 | 0.006 | 18 | 13 | 23 | 30 | 15 | 15 | 13,14 | 13 | 11 | 12 | 21 | 14 | 12 | 14 | 10 | 18 |
| H91 | 1 | 0.006 | 18 | 13 | 23 | 30 | 15 | 15 | 13,14 | 13 | 11 | 12 | 22 | 14 | 11 | 14 | 10 | 18 |
| H92 | 3 | 0.018 | 18 | 13 | 23 | 30 | 15 | 15 | 13,14 | 13 | 11 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H93 | 1 | 0.006 | 18 | 13 | 23 | 30 | 15 | 16 | 13,13 | 13 | 10 | 12 | 22 | 14 | 11 | 14 | 10 | 18 |
| H94 | 1 | 0.006 | 18 | 13 | 23 | 30 | 15 | 16 | 13,13 | 13 | 10 | 12 | 23 | 14 | 12 | 14 | 10 | 18 |
| H95 | 1 | 0.006 | 18 | 13 | 23 | 31 | 15 | 15 | 13,13 | 13 | 10 | 12 | 22 | 14 | 11 | 14 | 10 | 18 |
| H96 | 1 | 0.006 | 18 | 14 | 23 | 30 | 16 | 15 | 13,13 | 13 | 10 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
| H97 | 1 | 0.006 | 19 | 13 | 23 | 30 | 15 | 15 | 13,13 | 13 | 10 | 12 | 23 | 14 | 12 | 14 | 10 | 18 |
| H98 | 1 | 0.006 | 19 | 13 | 23 | 30 | 15 | 15 | 13,13 | 13 | 11 | 13 | 21 | 14 | 12 | 14 | 10 | 18 |
| H99 | 1 | 0.006 | 19 | 13 | 23 | 31 | 15 | 15 | 13,14 | 13 | 11 | 12 | 22 | 14 | 12 | 14 | 10 | 18 |
*Number of individuals.
†Haplotype frequency.
These haplotype data were compared with the Y Chromosome Haplotype Reference Database (a worldwide population sample of 26 586 haplotypes in a set of 235 populations for the haplotype of an 11-marker SWGDAM core set, http://www.yhrd.org). This comparison found only 10 matches (Table 3), while 89 (89.9%) haplotypes were not previously recorded.
Table 3.
Y-chromosomal short tandem repeat haplotypes of the Atayal found 10 matches of an 11-marker Scientific Working Group on DNA Analysis Methods core set at Y chromosome haplotype reference database
| Haplotype of DYS19, 385, 389I, 389II, 390, 391, 392, 393, 438, and 439 | Match with worldwide* | Match with Asian† | Match with European‡ | Haplotype No. of Atayal |
|---|---|---|---|---|
| 14-15,18-12-28-23-10-14-12-11-12 | 14 | 13 | 1 | H02 |
| 14-15,19-12-29-23-10-14-12-11-12 | 2 | 1 | 1 | H04 |
| 15-13,13-12-28-23-10-14-13-10-12 | 8 | 8 | H12, H13 | |
| 15-13,13-12-29-24-10-14-13-10-11 | 1 | 1 | H17 | |
| 15-13,13-13-30-23-11-14-13-10-11 | 1 | 1 | H43 | |
| 15-13,14-13-29-23-10-14-13-10-11 | 1 | 1 | H62, H63 | |
| 16-12,19-12-28-25-10-13-12-10-12 | 9 | 9 | H80 | |
| 16-13,13-12-29-23-10-14-13-10-13 | 1 | 1 | H83 |
*Worldwide population sample of 26 586 haplotypes in a set of 235 populations.
†Asian population sample of 8334 haplotypes in a set of 72 populations.
‡European population sample of 11 213 haplotypes in a set of 100 populations.
Discussion
The gene diversity of DYS385a/b is usually highest among Y-chromosomal STR loci (6-11) but in this study the diversity of DYS456 (0.6749), DYS389II (0.6474), DYS635 (0.5525), and Y GATA H4 (0.5653) was higher than that of DYS385a/b (0.5320). The discrimination capacity of the Atayal population (0.5824) was lower than that of neighboring populations, shown by other studies on individuals that had some degree of relatedness (6-10). Such findings might result from the fact that the Atayal reside in mountains and rarely interact with other peoples in Taiwan.
The native language of the Atayal belongs to the Formosan subfamily of Austronesian languages (12,13). Besides linguistic studies, genetic analyses have also confirmed that Taiwan’s aboriginal populations are close relatives of the Polynesian peoples, because of the first colonizations of the Polynesian islands from Taiwan (14). Forster et al (15) typed 30 aboriginal Taiwanese, including the Amis, Atayal, Bunun, and Paiwan for 5 Y-STRs (DYS19, DXYS156-Y, DYS391, DYS392, and DYS393) and sequenced the DYS390 locus; these profiles were compared with aboriginal Australians and Papuans. The aboriginal Taiwanese had the highest frequency at DYS390*23 (0.67) among 15 populations. Hagelberg et al (16) and Hurles (17) analyzed 27 and 39 Taiwanese aboriginals from 4 tribes (Ami, Atayal, Bunun, and Paiwan) for polymorphisms at DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, and DYS393 in order to find the genetic evidence for human settlement of the Pacific and to investigate the origins of the peoples speaking Austronesian languages. Hagelberg found that Taiwanese aboriginals were genetically closer to the populations of China, Java, Tolai, and Trobriands than to the populations of western Samoa, Papua New Guinea highlands, and Roro. Hurles found that among 11 populations Taiwan had the most similar population diversity to Papua New Guinea, based on the weighted mean intralineage mean pairwise difference. Kayser et al (14,18) investigated the Melanesian origin of the Polynesian peoples by typing 10 or 6 Atayal individuals and other globally dispersed human populations for Y chromosomal microsatellite polymorphisms at DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, and DYS393. They found Polynesian ancestors originated from Asia/Taiwan but did not move rapidly through Melanesia; rather, they interacted with and mixed extensively with Melanesians, leaving behind their genes and incorporating many Melanesian genes before colonizing the Pacific. Su et al (19) screened 24 Atayal for 19 Y-chromosome biallelic loci and these were compared with Polynesian peoples: markers studied were M3 (C→T mutation), M5 (A→G mutation), M7 (C→G mutation), M9 (C→G mutation), M15 (9-bp insertion), M17 (1-bp deletion), DYS287 (YAP), M45 (G→A mutation), M50 (T→C mutation), M88 (A→G mutation), M89 (C→T mutation), M95 (C→T mutation), M103 (C→T mutation), M110 (T→C mutation), M111 (4-bp deletion), M119 (A→C mutation), M120 (T→C mutation), M122 (T→C mutation), and M134 (1 bp deletion). They found the Y-chromosome data did not favor Taiwanese origin of Polynesians. Capelli et al (20) screened 50 Atayal for 9 Y-chromosomal haplogroups (B, C, D, E, F, G, H, I, and L), 49 of which fell into haplogroup H and 1 into haplogroup L and found that the haplogroups of the Atayal were not shared by the populations of Irian Jaya and Madang of Melanesia. More recently, Li et al (21) used fluorescent-labeled primers to type 7 microsatellite markers (STR) on the Y-chromosome (DYS19, DYS388, DYS389I, DYS390, DYS391, DYS392, and DYS393) in 22 Atayal individuals and compared their haplotypes with western Austronesians and Daic populations, showing that Taiwan aborigines likely originated from the Daic populations based on their paternal lineages.
While previous studies of the Atayal people have analyzed fewer than 50 individuals and DNA typing has included HLA, mtDNA, Y-SNPs, and Y-STRs, the present study analyzed Y-STR polymorphisms in 170 male Atayal individuals. These data can be of considerable utility in ethnological studies and geographical genetics.
The Atayal reside in mid-northern mountain regions of Taiwan with little intercrossing with other Taiwanese peoples. The genetic structure of the Atayal has therefore been largely conserved, and over half of 17 Y-STR loci typed contained high frequency alleles. At the DYS391 and DYS437 loci, only 2 alleles were recorded with little polymorphic variation.
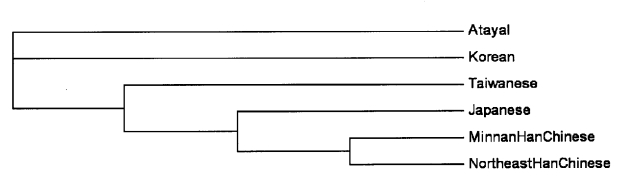
The present data on Y-chromosome haplotypes in the Atayal and the data on minHt of Y-STR loci of Chinese population in Taiwan (monomorphic [TCTG]3 at DYS389I and DYS389II including a repeat +3 compared with the available data) (6), Minnan Han Chinese (7), northeast Han Chinese (8), Korean (9), and Japanese (10) were analyzed using the Arlequin software package to determine the molecular variance between these populations. RST values were 0.318, 0.478, 0.448, 0.265, and 0.362, respectively, all of which were significant (P < 0.001). Neighbor-joining tree of the 6 populations (22,23) is shown as Figure 2.
Figure 2.
Neighbor-joining tree constructed from Slatkin's RST genetic distances based on the allelic frequencies of the minimal haplotype (minHt, the haplotype set of DYS19, DYS385a/b, DYS389I, DYS389II, DYS390, DYS391, DYS392, and DYS393) of Y-chromosomal short tandem repeat loci in 6 Asian populations.
A Y-STR haplotype database for Atayal population could be of great utility in determining the geographic origin of male individuals. In addition, because the Y chromosome haplotype has remained generally unchanged over the generations, this database can be used for determining the origin of different closely-related populations and contribute to better understaning of geographic effects and population migration.
Acknowledgments
We thank the County Governments of Taoyuan, Hsinchu, and Nantou, the Public Health Bureaus of Taoyuan, Hsinchu, and Nantou, the Township Health Centers of Fuhsing, Jianshih, and Renai, and Taipei County Indigenous People Home Cared Cooperation for their assistance in collecting samples. This study was supported by the National Science Council of the Republic of China, grant No. 95-1301-36-00-00-00-00-14.
References
- 1.Pestoni C, Cal ML, Lareu MV, Rodriguez-Calvo MS, Carracedo A. Y chromosome STR haplotypes: genetic and sequencing data of the Galician population (NW Spain). Int J Legal Med. 1999;112:15–21. doi: 10.1007/s004140050191. [DOI] [PubMed] [Google Scholar]
- 2.Department of Statistics. Monthly Bulletin of Interior Statistics. 2007-08. Available from: http://www.moi.gov.tw/stat/english/monthly.asp Accessed: July 9, 2008.
- 3.Gusmao L, Butler JM, Carracedo A, Gill P, Kayser M, Mayr WR, et al. DNA Commission of the International Society of Forensic Genetics (ISFG): an update of the recommendations on the use of Y-STRs in forensic analysis. Forensic Sci Int. 2006;157:187–97. doi: 10.1016/j.forsciint.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Nei M. Molecular evolutionary genetics. New York: Columbia University; 1987. [Google Scholar]
- 5.Excoffier L, Laval G, Schneider S. Arlequin ver. 3.11: an integrated software package for population genetics data analysis. Berne: Computational and Molecular Population Genetics Laboratory, Institute of Zoology, University of Berne; 2007. [PMC free article] [PubMed] [Google Scholar]
- 6.Wu FC, Pu CE. Multiplex DNA typing of short tandem repeat loci on Y chromosome of Chinese population in Taiwan. Forensic Sci Int. 2001;120:213–22. doi: 10.1016/S0379-0738(01)00388-7. [DOI] [PubMed] [Google Scholar]
- 7.Hu SP. Genetic polymorphism of 12 Y-chromosomal STR loci in the Minnan Han Chinese in Southeast China. Forensic Sci Int. 2006;159:77–82. doi: 10.1016/j.forsciint.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 8.Yang B, Gu M, Wang G, Li X, Liu Y, Yang W. Population data for 11 Y-chromosome STRs in northeast China Han. Forensic Sci Int. 2006;164:65–71. doi: 10.1016/j.forsciint.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 9.Park MJ, Lee HY, Yoo JE, Chung U, Lee SY, Shin KJ. Forensic evaluation and haplotypes of 19 Y-chromosomal STR loci in Koreans. Forensic Sci Int. 2005;152:133–47. doi: 10.1016/j.forsciint.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 10.Hashiyada M, Nagashima T, Itakura Y, Sakai J, Kanawaku Y, Kanetake J, et al. 12 Y-chromosomal STR haplotypes in Japanese. Forensic Sci Int. 2006;158:204–12. doi: 10.1016/j.forsciint.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 11.Ljubkovic J, Stipisic A, Sutlovic D, Definis-Gojanovic M, Bucan K, Andelinovic S. Y-chromosomal short tandem repeat haplotypes in southern Croatian male population defined by 17 loci. Croat Med J. 2008;49:201–6. doi: 10.3325/cmj.2008.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellwood P. The Austronesian dispersal and the origin of languages. Sci Am. 1991;265:88–93. [Google Scholar]
- 13.Chu JY, Huang W, Kuang SQ, Wang JM, Xu JJ, Chu ZT, et al. Genetic relationship of populations in China. Proc Natl Acad Sci U S A. 1998;95:11763–8. doi: 10.1073/pnas.95.20.11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kayser M, Brauer S, Weiss G, Underhill PA, Roewer L, Schiefenhovel W, et al. Melanesian origin of Polynesian Y chromosomes. Curr Biol. 2000;10:1237–46. doi: 10.1016/S0960-9822(00)00734-X. [DOI] [PubMed] [Google Scholar]
- 15.Forster P, Kayser M, Meyer E, Roewer L, Pfeiffer H, Benkmann H, et al. Phylogenetic resolution of complex mutational features at Y-STR DYS390 in aboriginal Australians and Papuans. Mol Biol Evol. 1998;15:1108–14. doi: 10.1093/oxfordjournals.molbev.a026018. [DOI] [PubMed] [Google Scholar]
- 16.Hagelberg E, Kayser M, Nagy M, Roewer L, Zimdahl H, Krawczak M, et al. Molecular genetic evidence for the human settlement of the Pacific: analysis of mitochondrial DNA, Y chromosome and HLA markers. Philos Trans R Soc Lond B Biol Sci. 1999;354:141–52. doi: 10.1098/rstb.1999.0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurles ME, Nicholson J, Bosch E, Renfrew C, Sykes BC, Jobling MA. Y chromosomal evidence for the origins of oceanic-speaking peoples. Genetics. 2002;160:289–303. doi: 10.1093/genetics/160.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kayser M, Krawczak M, Excoffier L, Dieltjes P, Corach D, Pascali V, et al. An extensive analysis of Y-chromosomal microsatellite haplotypes in globally dispersed human populations. Am J Hum Genet. 2001;68:990–1018. doi: 10.1086/319510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su B, Jin L, Underhill P, Martinson J, Saha N, McGarvey ST, et al. Polynesian origins: insights from the Y chromosome. Proc Natl Acad Sci U S A. 2000;97:8225–8. doi: 10.1073/pnas.97.15.8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capelli C, Wilson JF, Richards M, Stumpf MP, Gratrix F, Oppenheimer S, et al. A predominantly indigenous paternal heritage for the Austronesian-speaking peoples of insular Southeast Asia and Oceania. Am J Hum Genet. 2001;68:432–43. doi: 10.1086/318205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Wen B, Chen SJ, Su B, Pramoonjago P, Liu Y, et al. Paternal genetic affinity between Western Austronesians and Daic populations. BMC Evol Biol. 2008;8:146. doi: 10.1186/1471-2148-8-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J Hered. 2001;92:371–3. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- 23.Xia X. Data analysis in molecular biology and evolution. Boston: Kluwer Academic publishers; 2000. [Google Scholar]