Abstract
GABA cell dysfunction in both schizophrenia (SZ) and bipolar disorder (BD) involves decreased GAD67 expression, although this change involves fundamentally different networks of genes in the 2 disorders. One gene that is common to these 2 networks is cyclin D2, a key component of cell cycle regulation that shows increased expression in SZ, but decreased expression in BD. Because of the importance of cell cycle regulation in maintaining functional differentiation and DNA repair, the current study has examined the genes involved in the G1 and G2 checkpoints to generate new hypotheses regarding the regulation of the GABA cell phenotype in the hippocampus of SZ and BD. The results have demonstrated significant changes in cell cycle regulation in both SZ and BD and these changes include the transcriptional complex (TC) that controls the expression of E2F/DP-1 target genes critical for progression to G2/M. The methyl-CpG binding domain protein (MBD4) that is pivotal for DNA repair, is significantly up-regulated in the stratum oriens (SO) of CA3/2 and CA1 in SZs and BDs. However, other genes associated with the TC, and the G1 and G2 checkpoints, show complex changes in expression in the SO of CA3/2 and CA1 of both SZs and BDS. Overall, the patterns of expression observed have suggested that the regulation of functional differentiation and/or genomic integrity of hippocampal GABA cells varies according to diagnosis and their location within the trisynaptic pathway.
Keywords: cyclin D2, DNA polymerase, G2 checkpoint, nicotinic receptors, p53
Schizophrenia (SZ) and bipolar disorder (BD) involve striking decreases in GAD67 expression (see Table S1) in the hippocampus, particularly in stratum oriens of sector CA3/2 (1), where preferential abnormalities have also been found in other postmortem studies (2). Network association analyses have suggested that GAD67 expression may be linked to cyclin D2 expression and has raised the possibility that the regulation of cell differentiation (1) and DNA damage (3) may play a role in GABA cell dysfunction in SZ and BD. Consistent with this idea, functional clusters of genes (refer to Table S2) showing the most robust changes in gene expression include neurogenesis, cell cycle regulation and the DNA damage response, particularly in stratum oriens of CA3/2 and CA1 (4). To learn more about the regulation of cell cycle in hippocampal GABA cells in SZ and BD, a post hoc analysis has assessed whether there is evidence for changes in the expression of genes involved in the G1 and G2 checkpoints, critical elements in the maintenance of terminal differentiation and DNA repair in postmitotic neurons (5). It was quite surprising to see that there were so many genes involved in the regulation of cell proliferation and cell identity or loss, showing significant changes in what are assumed to be terminally differentiated neurons in the adult hippocampus of both SZs and BDs.
The molecular events related to cell cycle regulation vary according to established canonical pathways that include not only cyclin D2, but also cyclin E; both form complexes with the cyclin dependent kinases, CDK4/6 and CDK2, respectively. The cyclin E/CDK2 complex and CDC42 (6) are particularly important for cell cycle progression because they can cause G1 arrest to fail and allow a cell to move into the (S) phase where DNA replication is stimulated (7). The S phase then progresses toward the G2 checkpoint where 3 possible outcomes are possible: (i) the formation of the cyclin B/CDC2 complex and entry into a mitotic division; (ii) the successful repair of damaged DNA and the survival of a functionally differentiated cell; or (iii) the inability to repair an over-whelming amount of DNA damage and entry into the apoptotic cascade (8). The G2 checkpoint, arguably the most critical stage, is mediated by p53, a sequence-specific transcription factor that can promote genomic integrity or cell death, depending on the degree to which DNA has been damaged (9). When p53 forms a complex with the replication protein (RPA3), DNA repair can occur (10) in a threshold-dependent manner (11). Other genes, such as BRCA1, a tumor suppressor that activates the G2 checkpoint genes (12) and a variety of DNA polymerases (see Table S3), play central roles in the regulation of the DNA repair response.
The progression of G1 toward the G2 checkpoint depends on many other genes, particularly those associated with a transcriptional complex that regulates the expression of cyclin E and other critical genes. The latter include E2F, a positive regulator of transcription (13) and EP300, a histone acetyltransferase that releases promoter activity via chromatin remodeling (14). Another key element is the retinoblastoma protein (Rb), which, by forming a complex with the tyrosine kinase ABL (15), suppresses the transcriptional complex (16). When E2F forms a protranscriptional complex with DP-1 (17) and Rb is phosphorylated by a CDK, the transcriptional complex can be switched to the “ON.” state This switch promotes the expression of key genes, such as E2F and cyclin E, that can further promote cell cycle progression. The transcriptional repressor activation of methyl-CpG binding protein 4 (MBD4) depends on histone deacetylase (HDAC) (18). When DNA is damaged, MBD4 forms a complex with the DNA mismatch-repair protein (19) and has the ability to repress the expression of E2F and other target genes. The response to DNA damage is determined by the expression of many different DNA polymerase genes (see Table S3).
Additionally, several different growth factors, such as FGF, drive the proliferation of neuronal precursor cells during development (20). For example, TGFβ inhibits this progression via the repressive effect of SMAD3 on CDC25A (21) and neuregulin I plays a role in the growth and differentiation of neurons (22) and glia (23). Finally, nicotinic receptors have also been associated with both the survival of differentiated neurons or cytotoxic changes in undifferentiated cells (24). Interestingly, nicotine treatment is associated with a failure of G1 arrest that involves a dysregulation of Rb and E2F activity (25) and over-expression of the alpha 7 nicotinic subunit has been found to prevent G1 arrest (26).
This was undertaken to generate hypotheses regarding the regulation of cell cycle regulation and the DNA damage response in SZ and BD. As discussed below, a remarkable number of the genes involved in cell division and cell identity showed significant expression changes in cells that are assumed to be terminally differentiated neurons in the adult hippocampus of both SZs and BDs. These findings suggest that cell cycle regulation in GABA cells may be circuitry-based and vary according to their location and integration within the trisynaptic pathway (see Fig. S1). Additionally, the results suggest that there may be fundamental differences in the status of cell differentiation and genomic integrity in GABAergic interneurons in SZs versus BDs.
Results
Samples of the strata oriens (SO), pyramidale (SP) and radiatum (SR) of sectors CA3/2 and CA1 of the hippocampus were obtained from normal controls (CONs), SZs and BDs (Table S4). The samples were collected by laser microdissection (LMD) of cresyl violet-stained cryostat sections as described in ref. 1. Although the percentage of present calls in the SP and SR were similar to those obtained for the SO, the data for these layers were not included in this report because there were relatively few individual genes that showed significant changes in expression and the composite probability, Pc, for the functional clusters of genes study did not reach the level (Pc ≤10−10) needed for inclusion in the post hoc analyses (27) (refer to Table S2). For these reasons, the data reported here are exclusively from the SO where the most significant changes in gene expression were observed.
Consistent with conventional microscopic criteria (28), interneurons in the SO showed a distinctive cresyl violet staining of their cytoplasm and light-staining, diffuse chromatin material in their nuclei. Glial cells typically showed no cytoplasmic staining, but large amounts of heterochromatic material in their nuclei. This fundamental difference between neurons and glia suggests that the concentrations of cytoplasmic RNA in interneurons is much higher than in glial cells. Additionally, in situ hybridization demonstrated that the antisense RNA associated with a broad array of genes, including, but not limited to GAD67, GAD65, HDAC1, DAXX, PAX5, and Runx2 (see Fig. S2 for HDAC1, DAXX, and PAX5), was abundantly localized over neuronal cell bodies in the SO, whereas little or no autoradiographic signals were detected over glia.
Functional Clusters of Genes.
The results of the GenMapp analyses demonstrated that there were 3 clusters of genes with particularly significant changes in expression and these included: neurogenesis, cell cycle, and the DNA damage response (Table S2 and Table S5).
Stratum Oriens of CA2/3.
Schizophrenia.
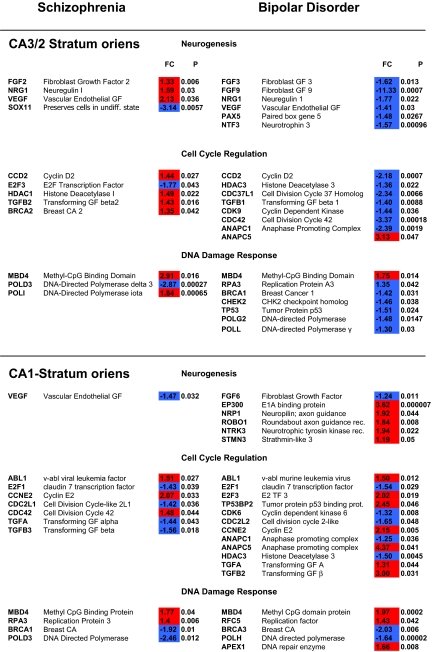
In SZs (Table 1 and Fig. 1), TGFβ2, neuregulin I, FGF2, VEGF, the nicotinic α4 receptor subunit (Table S6), and cyclin D2 were all up-regulated, suggesting that the phase of this GABA cell population may be arrested in early G1. For the transcriptional complex, MBD4, HDAC1 and its corepressor DAXX were also up-regulated, changes that could shut-down the transcription complex (29) associated with cell cycle progression. E2F showed decreased expression, providing further support for the possibility that the transcriptional complex is modulated in the “OFF” state and could result in reduced expression of target genes, such as cyclins A and E, which drive G1 toward the G2 checkpoint (30). Cyclin D2 showed a significant increase of expression and a similar finding has been observed in a recent postmortem study of the cingulate region in SZs (31, 32). Very few changes in the expression of genes specific for the G2/M phase were observed. p53 and CHK2 show normal expression in this group, which suggests that the G2 checkpoint is capable of inhibiting progression toward mitosis in SZs (12). Additionally, some DNA repair may be occurring at this locus I SZs, because BRCA1 and DNA polymerase iota (POLI) (see Table S3) were both up-regulated. The latter change could be associated with inaccurate copying of templates that result in point mutations on chromosomes of GABA cells in CA3/2-SO in SZ. In contrast, a decrease in the expression of POLD (Table 1 and Table S3), was also observed in the SZs (33).
Table 1.
Target Genes in Neurogenesis, Cell Cycle and DNA Damage Response of Schizophrenics and Bipolars
The data shown represent the fold changes (FC) and probability of significance (P) for individual genes within the neurogenesis, cell cycle regulation and DNA damage response clusters that are related to the maintenance of G1 arrest and/or its progression toward the G2 checkpoint in the stratum oriens of sectors CA3/2 and CA1 of subjects with schizophrenia and bipolar disorder. The red and blue fill patterns for FCs indicates increased or decreased expression, respectively, for the various genes. There are a total of 54 different genes listed, but many (42.4%) appear in both groups and/or in CA3/2 and/or CA1. For example, TGFβs, NRG1, VEGF, CCD2, ANAPC1 and ANAPC5, ABL1, E2F1 and E2F3, BRCA1, MBD4, POLD3, and RPA3 all appear in more than one locus or diagnostic group. MBD4 is unique because it showed a significant up-regulation in CA3/2 and CA1 of both SZs and BDs.
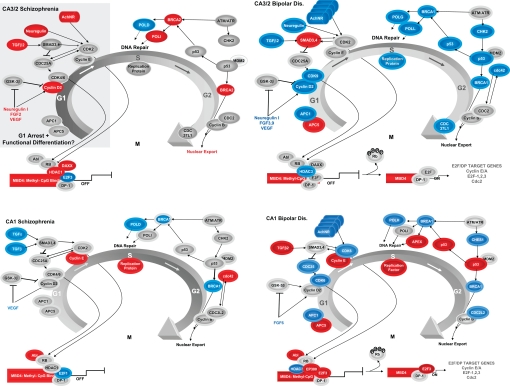
Fig. 1.
The regulation of G1/S and G2/M phase in GABA cells of the stratum oriens (SO) in CA3/2 and CA1 of schizophrenics (SZs) (Left) and bipolars (BDs) (Right). In each of the diagrams, the G1/S and G2/M phases of cell cycle and their associated genes. Genes showing increased, decreased, and no change in expression are indicated in red, blue, and gray, respectively. CDKs in G1 (arrows) are capable of phosphorylating Rb and turning the transcriptional complex “ON.” MBD4 is significantly up-regulated in the SO of CA3/2 and CA1 of both groups, suggesting that DNA repair may be commonly occurring in hippocampal GABA neurons at these loci in SZs and BDs. Other genes involved in the maintenance of G1 arrest and progression to G2 show many different expression patterns at each locus of the 2 groups. Overall, the data suggest that the transcriptional complex may be in the “OFF” state in SZs and in the “ON” state in BDs. Many different DNA polymerases (see Table S3) show significant changes in expression at the different loci of both SZs and BDs, suggesting that the cell cycle apparatus in these GABAergic interneurons may be primarily involved in the repair of damaged DNA.
Bipolar disorder.
In BDs, the regulation of the G1 and G2 checkpoint mechanisms in CA3/2 seemed to be quite different from that seen in SZs (Table 1 and Fig. 1). TGFβ1, neuregulin I, FGF3 and 9, neurotrophin 3, VEGF, cyclin D2, CDK9 and HDAC3 were all down-regulated. Four different nicotinic receptor subunits, including the alpha 7 isoform, showed a significant decrease in expression at this locus in BDs (Table S6). Significant decreases in the expression of ANAPC1 (APC1), the anaphase promoting complex 1 (34) were also observed in BDs. A decrease of APC1 activity can potentially contribute to a failure of G1 arrest and functional differentiation (35). However, another isoform ANAPC5 (APC5) was up-regulated at this locus in BDs, suggesting that these 2 changes could compensate one another, if both changes are present in the same GABA cells. The increased expression of SMAD3 (1), a key component of the TGFβ signaling pathway, could help to promote neuronal differentiation (36) and further suppress the down-regulated cyclin D2/CDK9 complex.
As in the SZ group, the expression of MBD4 was also significantly increased at this locus in BDs. In the setting of normal levels of the expression of E2F and the phosphorylation of Rb by CDK2, the transcriptional complex may be capable of increasing the expression of E2F/DP-1 target genes and could theoretically promote progression toward the G2 checkpoint. However, another key gene, CDC42, was also down-regulated.
Other G2 checkpoint genes, such as p53, CHK2 and BRCA1 were also down-regulated. Of these, p53 may be particularly important at the CA3/2-SO locus in BDs, because it is capable of promoting excisional repair (37). Accordingly, the decrease of p53 expression observed at this locus in BDs may contribute to the repair of single nucleotide mutations (9). Consistent with this, DNA polymerase lambda (POLL) and gamma (POLG) (refer to Table S3) both showed decreased expression at this locus in BDs (refer to Table 1). The replication protein (RPA3), which interacts directly with p53 in the setting of DNA damage (11), was also significantly decreased in expression.
SO-CA1.
Schizophrenia.
In the stratum oriens of CA1, there was an increased expression of MBD4 and ABL1, but a decreased expression of E2F, suggesting that the transcriptional complex at this locus, like its counterpart in CA3/2, may also be modulated in the “OFF” state in SZs (Table 1 and Fig. 1). Unlike CA3/2, however, TGFβ2 and VEGF both showed decreased expression, without any associated change in the expression of cyclin D2. Noteworthy was the finding of a significant increase in the expression of cyclin E and the RPA3 replication protein. Although these latter changes are consistent with the possibility that G1 arrest might have been failing in SZs, there appears to be a dissociation between these latter changes and those in the transcriptional complex, which may be modulated in the “OFF” state. POLD, another DNA polymerase (refer to Table S3), and BRCA1, were both decreased in expression, whereas CHK2 and p53 both showed normal expression. As shown in Table 1, SOX11, which ensures the expression of neuronal traits (38), showed a robust decrease in expression (FC = −3.14; P = 0.0057) at this locus of SZs.
Bipolar disorder.
The most striking changes in the transcriptional complex in CA1-SO of BDs was and increased expression of E2F3 (2.02-fold; P = 0.019) and EP300 (3.6-fold; P = 0.000007). These robust changes in E2F and EP300 could ensure that the transcriptional complex is regulated in the “ON” state at this locus in BDs. Consistent with this, cyclin E (2.15-fold; P = 0.005) also showed a significant increase of expression, a change that would further promote progression toward the G2 checkpoint. Additionally, CDC2L2, APC1 and CDK6 were also down-regulated. The expression of p53, the replication factor (RFC5) and the DNA polymerase APEX1 (refer to Table S3) were all increased. Another polymerase, POLH (Table S3), however, was down-regulated. CHES1and BRCA1 were also down-regulated. As with the SO in sector CA3/2 of BDs, 4 different nicotinic receptor subunits showed a significant decrease in expression in CA1 (see Table S6) and included the α1, α2, β3 and γ subunits. Significant increases in the expression of neuropilin (NRP1) (39), roundabout (ROBO1) (40), and strathmin 3 (STMN3) (41), genes that are believed to play a role in the plasticity of axons and dendrites, were also observed (Table 1)
SR and SP of CA2/3 and CA1.
There were very few changes in the regulation of genes associated with neurogenesis and cell cycle in the SR and SP of either sector in the SZ or BD groups.
Effects of Psychotropic Medications.
As shown in Fig. S3A and Fig. S3B for the genes included in the cell cycle and DNA damage response clusters, respectively, the SZ subjects were broken down into those receiving low (< 500 mgs per day) or high (>500 mgs per day) dose antipsychotic medications (APDs) during the year before death. The SZs showed many more genes with significant differences in the low dose group, whereas BDs had many more significant genes in the high dose group. Assessments of the expression patterns for individual genes with GenMapp functional clusters did not show significant overlap in the 2 groups. The SZ and BD groups showed a high degree of correspondence in the medication regimens, which suggests that psychotropic agents do not explain the changes in gene expression observed, particularly because the respective profiles were fundamentally different in the 2 groups.
Discussion
This study demonstrates that genes associated with the cell cycle apparatus and the transcriptional complex that regulates it show complex expression changes that vary according to sector, layer, and psychiatric diagnosis. These results support the hypothesis that the regulation of cell cycle in hippocampal GABA cells involves a complex interplay among transcription factors that regulate promoter activity associated with E2F/DP-1 target genes and their association with the G1 and G2 checkpoints. Additionally, a variety of growth factors, trophins, and nicotinic receptor subunits also help to determine whether G1 arrest is maintained or whether it progresses toward the G2 checkpoint. Particularly noteworthy is the significant increase in the expression of MBD4 in GABA cells in both CA3/2 and CA1 of SZs and BDs. This gene encodes a glycosylase that prefers substrates in which a G:T mismatch is present in the context of methylated or unmethylated CpG sites (42); an increase in its expression could indicate that the transcriptional complex may be suppressed. However, other genes associated with the regulation of this complex (e.g., HDAC, E2F, and ABL) probably help to determine whether the “ON” state may be achieved and whether progression from the G1 to the G2 checkpoint may occur in terminally differentiated GABA cells in the adult hippocampus. Generally speaking, the data presented here point to the transcriptional complex in GABA cells of the SO as playing a central role in the regulation of cell cycle progression and/or DNA repair in GABA cells of the SO in SZ and BD.
Taking together the expression changes in the transcriptional complex with those observed for cell cycle, it appears that GABA cells in the SO of CA3/2 of SZs may be more likely to suppress CpG islands of the transcriptional complex and help to maintain G1 arrest and functional differentiation. The analogous population of interneurons in CA3/2 of BDs showed widespread decreases in the expression of genes required for progression to the G2 checkpoint and DNA repair. The decreased expression of p53 and 2 different DNA polymerases at this locus may indicate that DNA repair is inhibited at this locus in BDs. In CA1-SO of BDs, however, the increased expression of p53, RFC5 and APEX1 suggest the opposite; GABA cells at this locus may be attempting to repair DNA damage associated with oxidative stress (43). Consistent with this possibility, GABAergic cells at this locus in BDs may be in a hypermetabolic state (see Fig. S1 Lower), as indicated by the very pronounced increase in the expression of genes involved in glycolysis, the Kreb cycle and the electron transport chain (1).
In summary, the results reported here suggest that genes associated with cell cycle regulation and DNA repair in hippocampal GABA cells show significant changes in expression that vary not only according to diagnosis, but also their integration within the trisynaptic pathway. As shown in Fig. S1, the patterns of connectivity are quite different within each locus of this circuit and this, in turn, may influence the regulation of gene expression in GABA cells (1). For example, in the SO of CA3/2, inputs from the basolateral amygdala and septal nuclei may help to establish gene expression changes that are unique to this locus (for a review of the connectivity, see ref. 4). The extrinsic and intrinsic afferent fiber systems that modulate the activity of GABA cells within the trisynaptic pathway probably contribute significantly to the maintenance of functional differentiation and genomic integrity in these terminally differentiated interneurons (27).
Overall, the interpretation of the expression changes reported here is based on studies of “simple” in vivo and in vitro systems in which cell proliferation or death can readily occur (44). In subjects with SZ or BD, however, the respective molecular endophenotypes within GABA cells of the adult hippocampus are probably much more complex than has heretofore been appreciated. In Alzheimer's disease, neurons that are at risk for degeneration are also at risk of reinitiating a cell cycle process that involves the expression of cell cycle proteins and DNA replication (44). Although failure of cell cycle regulation may be a root cause of several neurodegenerative disorders, it might also be a final common pathway for brain diseases, such as bipolar disorder.
Methods
Subjects.
Participating in the study were 7 normal controls (CONs), 7 schizophrenic subjects, and 7 bipolar subjects (BD) from the Harvard Brain Tissue Resource Center at McLean Hospital matched for age, postmortem interval (PMI), hemisphere, gender, and tissue pH (Table S4). The procedures for retrospective diagnoses and neuropathological evaluations are described in refs. 1 and 27.
Tissue Preparation and RNA Extraction.
A total of 7 frozen tissue sections (8 μm) were cut from each block on a Microm HM 560 CryoStar cryostat, mounted on LEICA Frame Slides with a PET-membrane (1.4 μm), and fixed in Streck tissue fixative (STF) (Streck Laboratories). The frame slides were mounted on a LEICA AS LMD apparatus and tissue samples from SO, SP, and SR of CA2/3 and CA1 were microdissected. Each vial into which the laser dissected specimens fell by gravity contained a small volume of a lysis/denaturing solution containing an RNase inhibitor. RNA extraction was undertaken with a Qiagen Rneasy micro kit yielding ≈20 to 30 ng of RNA. RNA quality was assessed using an Agilent 2100 bioanalyzer. Fifteen micrograms of biotinylated target RNA were fragmented and individually hybridized to the HU-133A arrays (Affymetrix). The microarrays were then stained with 2 rounds of streptavidin-phycoerythrin (Molecular Probes) and one round of biotinylated anti-streptavidin antibody (Vector Laboratories), and scanned twice. RNA quality was assessed using tissue pH, the 18S/28S ratio and the percentage of present calls for each case (see Table S4).
Data Analyses.
The DNA Chip Analyzer (dChip) Version 1.3 software package (32) was used to evaluate the percentage of present calls and the significance of differences between the normal controls vs. the SZs or BDs. The variance obtained with the perfect match model of dChip (R2 = 0.001) was considerably lower than that seen with any of the other models and was used throughout the first stage analysis of the microarray data. Biologically relevant clusters of genes were identified by using GenMapp algorithms (www.genmapp.org). A metric, called the composite probability, Pc, was computed for each GenMapp biopathway or cluster. The P value for inclusion of individual genes in each GenMapp cluster was set at P ≤ 0.05. Genes meeting this criterion were multiplied by one another (i.e., P1, P2 … Pi) for each pathway. This resulting number was divided by the ratio of the number of genes meeting the inclusionary criterion (ni) and the number of genes potentially showing significant changes (nt) in each pathway. The total number of GenMapp pathways was represented as Np. The following equation was used:
This equation provides 2 separate corrections for multiple comparisons by multiplying by both Np and nt. The α-level of significance for each GenMapp biopathway or cluster is established by examining the distribution of genes that met criteria for inclusion in the analysis and the final P value is equal to the value obtained with the equation described above. GenMapp pathways with Pc = 5 × 10−10 or lower were considered to be significant. The latter number was based on the number of genes per biological cluster showing significance and the robustness of the P values for individual gene within the clusters.
Validation Studies.
The method used for qRT-PCR described in ref. 1. As shown in Fig. S4, the genes studied covered a broad range of transmitter and receptor subunit isoforms, ion channels and transcription factors including GAD67, GAD65, GRIA1, GRIK1, GRIK2, HCN3, HCN4, KCNJ3, KCNJ6, HDAC1, LEF1, Runx2, and PAX5 (Fig. S4). The SO of CA3/2 and CA1 showed the most pronounced changes in expression when compared with SR and SP of these 2 sectors (see also Table S2). The target genes also showed changes in the same direction as those observed with the microarray analyses. One exception is GAD67 in SR of CA3/2 where expression was significantly decreased in the microarrays for the SZs, but showed no change with qRT-PCR. In addition to qRT-PCR validations, in situ hybridization was used to evaluate whether the expression of mRNA for HDAC1, DAXX, PAX5 (see Fig. S2), Runx2, GRIK1 and GRIK2 occurred in primarily interneurons versus glial cells.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grants MH42261, MH/NS 31862, and MH077175 and the William P. and Henry B. Test Endowment.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903066106/DCSupplemental.
References
- 1.Benes FM, et al. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci USA. 2007;104:10164–10169. doi: 10.1073/pnas.0703806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benes FM. and S. Berretta, GABAergic interneurons: Implications for understanding schizophrenia and bipolar disorder. Neuropsychopharmacology. 2001;25:1–27. doi: 10.1016/S0893-133X(01)00225-1. [DOI] [PubMed] [Google Scholar]
- 3.Buttner N, et al. DNA fragmentation is increased in non-GABAergic neurons in bipolar disorder but not in schizophrenia. Schizophr Res. 2007;93(1–3):33–41. doi: 10.1016/j.schres.2007.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benes FM, et al. Circuitry-based gene expression profiles in GABA cells of the trisynaptic pathway in schizophrenics versus bipolars. Proc Natl Acad Sci USA. 2008;105:20935–20940. doi: 10.1073/pnas.0810153105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sancar A, et al. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 6.Chou MM, Masuda-Robens JM, Gupta ML. Cdc42 promotes G1 progression through p70 S6 kinase-mediated induction of cyclin E expression. J Biol Chem. 2003;278:35241–35247. doi: 10.1074/jbc.M305246200. [DOI] [PubMed] [Google Scholar]
- 7.Cannon JD, et al. Granulosa cell expression of G1/S phase cyclins and cyclin-dependent kinases in PMSG-induced follicle growth. Mol Cell Endocrinol. 2007;264(1–2):6–15. doi: 10.1016/j.mce.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 8.Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 9.Helton ES, Chen X. p53 modulation of the DNA damage response. J Cell Biochem. 2007;100:883–896. doi: 10.1002/jcb.21091. [DOI] [PubMed] [Google Scholar]
- 10.Abramova NA, et al. Interaction between replication protein A and p53 is disrupted after UV damage in a DNA repair-dependent manner. Proc Natl Acad Sci USA. 1997;94:7186–7191. doi: 10.1073/pnas.94.14.7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bochkareva E, et al. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci USA. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dasika GK, et al. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene. 1999;18:7883–7899. doi: 10.1038/sj.onc.1203283. [DOI] [PubMed] [Google Scholar]
- 13.Reimer D, et al. Expression of the E2F family of transcription factors and its clinical relevance in ovarian cancer. Ann N Y Acad Sci. 2006;1091:270–281. doi: 10.1196/annals.1378.073. [DOI] [PubMed] [Google Scholar]
- 14.Mertens C, Roeder RG. Different functional modes of p300 in activation of RNA polymerase III transcription from chromatin templates. Mol Cell Biol. 2008;28:5764–5776. doi: 10.1128/MCB.01262-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Knudsen ES, Wang JY. Cells arrested in G1 by the v-Abl tyrosine kinase do not express cyclin A despite the hyperphosphorylation of RB. J Biol Chem. 1996;271:19637–19640. doi: 10.1074/jbc.271.33.19637. [DOI] [PubMed] [Google Scholar]
- 16.Riley DJ, Lee EY, Lee WH. The retinoblastoma protein: More than a tumor suppressor. Annu Rev Cell Biol. 1994;10:1–29. doi: 10.1146/annurev.cb.10.110194.000245. [DOI] [PubMed] [Google Scholar]
- 17.Ishida H, et al. Identification and characterization of novel isoforms of human DP-1: DP-1α regulates the transcriptional activity of E2F1 as well as cell cycle progression in a dominant-negative manner. J Biol Chem. 2005;280:24642–24648. doi: 10.1074/jbc.M500189200. [DOI] [PubMed] [Google Scholar]
- 18.Kondo E, et al. The thymine DNA glycosylase MBD4 represses transcription and is associated with methylated p16(INK4a) and hMLH1 genes. Mol Cell Biol. 2005;25:4388–4396. doi: 10.1128/MCB.25.11.4388-4396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ballestar E, Wolffe AP. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur J Biochem. 2001;268:1–6. doi: 10.1046/j.1432-1327.2001.01869.x. [DOI] [PubMed] [Google Scholar]
- 20.Li B, DiCicco-Bloom E. Basic fibroblast growth factor exhibits dual and rapid regulation of cyclin D1 and p27 to stimulate proliferation of rat cerebral cortical precursors. Dev Neurosci. 2004;26(2–4):197–207. doi: 10.1159/000082137. [DOI] [PubMed] [Google Scholar]
- 21.Ewen ME, et al. TGF beta inhibition of Cdk4 synthesis is linked to cell cycle arrest. Cell. 1993;74:1009–1020. doi: 10.1016/0092-8674(93)90723-4. [DOI] [PubMed] [Google Scholar]
- 22.Murphy SP, Bielby-Clarke K. Neuregulin signaling in neurons depends on ErbB4 interaction with PSD-95. Brain Res. 2008;1207:32–35. doi: 10.1016/j.brainres.2008.02.063. [DOI] [PubMed] [Google Scholar]
- 23.Birchmeier C, Nave KA. Neuregulin-1, a key axonal signal that drives Schwann cell growth and differentiation. Glia. 2008;56:1491–1497. doi: 10.1002/glia.20753. [DOI] [PubMed] [Google Scholar]
- 24.Berger F, Gage FH, Vijayaraghavan S. Nicotinic receptor-induced apoptotic cell death of hippocampal progenitor cells. J Neurosci. 1998;18:6871–6881. doi: 10.1523/JNEUROSCI.18-17-06871.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu M, Guo J, Chen CY. Long-term exposure to nicotine, via ras pathway, induces cyclin D1 to stimulate G1 cell cycle transition. J Biol Chem. 2005;280:6369–6379. doi: 10.1074/jbc.M408947200. [DOI] [PubMed] [Google Scholar]
- 26.Utsugisawa K, et al. Overexpression of alpha7 nicotinic acetylcholine receptor prevents G1-arrest and DNA fragmentation in PC12 cells after hypoxia. J Neurochem. 2002;81:497–505. doi: 10.1046/j.1471-4159.2002.00823.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhu X, Raina A.K., Smith M.A. Cell cycle events in neurons. Proliferation or death? Am J Pathol. 1999;155:327–329. doi: 10.1016/S0002-9440(10)65127-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benes FM, Davidson J., Bird E.D. Quantitative cytoarchitectural studies of the cerebral cortex of schizophrenics. Arch Gen Psych. 1986;43:31–35. doi: 10.1001/archpsyc.1986.01800010033004. [DOI] [PubMed] [Google Scholar]
- 29.Ecsedy JA, Michaelson JS, Leder P. Homeodomain-interacting protein kinase 1 modulates Daxx localization, phosphorylation, and transcriptional activity. Mol Cell Biol. 2003;23:950–960. doi: 10.1128/MCB.23.3.950-960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robertson KD, et al. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 31.Katsel P, et al. Abnormal indices of cell cycle activity in schizophrenia and their potential association with oligodendrocytes. Neuropsychopharmacology. 2008;33:2993–3009. doi: 10.1038/npp.2008.19. [DOI] [PubMed] [Google Scholar]
- 32.Li CX, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belyakova NV, et al. Proof-reading 3′→5′ exonucleases isolated from rat liver nuclei. Eur J Biochem. 1993;217:493–500. doi: 10.1111/j.1432-1033.1993.tb18269.x. [DOI] [PubMed] [Google Scholar]
- 34.Wu G, et al. The anaphase-promoting complex coordinates initiation of lens differentiation. Mol Biol Cell. 2007;18:1018–1029. doi: 10.1091/mbc.E06-09-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castro A, et al. The anaphase-promoting complex: A key factor in the regulation of cell cycle. Oncogene. 2005;24:314–325. doi: 10.1038/sj.onc.1207973. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Campmany L, Marti E. The TGFbeta intracellular effector Smad3 regulates neuronal differentiation and cell fate specification in the developing spinal cord. Development. 2007;134:65–75. doi: 10.1242/dev.02702. [DOI] [PubMed] [Google Scholar]
- 37.Lilling G, et al. p53-associated 3′→5′ exonuclease activity in nuclear and cytoplasmic compartments of cells. Oncogene. 2003;22:233–245. doi: 10.1038/sj.onc.1206111. [DOI] [PubMed] [Google Scholar]
- 38.Bergsland M, et al. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006;20:3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwarz Q, et al. Neuropilin 1 and 2 control cranial gangliogenesis and axon guidance through neural crest cells. Development. 2008;135:1605–1613. doi: 10.1242/dev.015412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez-Bendito G, et al. Robo1 and Robo2 cooperate to control the guidance of major axonal tracts in the mammalian forebrain. J Neurosci. 2007;27:3395–3407. doi: 10.1523/JNEUROSCI.4605-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poulain FE, Sobel A. The “SCG10-LIke Protein” SCLIP is a novel regulator of axonal branching in hippocampal neurons, unlike SCG10. Mol Cell Neurosci. 2007;34:137–146. doi: 10.1016/j.mcn.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 42.Bellacosa A. Role of MED1 (MBD4) Gene in DNA repair and human cancer. J Cell Physiol. 2001;187:137–144. doi: 10.1002/jcp.1064. [DOI] [PubMed] [Google Scholar]
- 43.Saitoh T, et al. Enhancement of OGG1 protein AP lyase activity by increase of APEX protein. Mutat Res. 2001;486:31–40. doi: 10.1016/s0921-8777(01)00078-7. [DOI] [PubMed] [Google Scholar]
- 44.Herrup K, Yang Y. Cell cycle regulation in the postmitotic neuron: Oxymoron or new biology? Nat Rev Neurosci. 2007;8:368–378. doi: 10.1038/nrn2124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.