Abstract
Metabolomics is a newly emerging field of ‘omics’ research that is concerned with characterizing large numbers of metabolites using NMR, chromatography and mass spectrometry. It is frequently used in biomarker identification and the metabolic profiling of cells, tissues or organisms. The data processing challenges in metabolomics are quite unique and often require specialized (or expensive) data analysis software and a detailed knowledge of cheminformatics, bioinformatics and statistics. In an effort to simplify metabolomic data analysis while at the same time improving user accessibility, we have developed a freely accessible, easy-to-use web server for metabolomic data analysis called MetaboAnalyst. Fundamentally, MetaboAnalyst is a web-based metabolomic data processing tool not unlike many of today's web-based microarray analysis packages. It accepts a variety of input data (NMR peak lists, binned spectra, MS peak lists, compound/concentration data) in a wide variety of formats. It also offers a number of options for metabolomic data processing, data normalization, multivariate statistical analysis, graphing, metabolite identification and pathway mapping. In particular, MetaboAnalyst supports such techniques as: fold change analysis, t-tests, PCA, PLS-DA, hierarchical clustering and a number of more sophisticated statistical or machine learning methods. It also employs a large library of reference spectra to facilitate compound identification from most kinds of input spectra. MetaboAnalyst guides users through a step-by-step analysis pipeline using a variety of menus, information hyperlinks and check boxes. Upon completion, the server generates a detailed report describing each method used, embedded with graphical and tabular outputs. MetaboAnalyst is capable of handling most kinds of metabolomic data and was designed to perform most of the common kinds of metabolomic data analyses. MetaboAnalyst is accessible at http://www.metaboanalyst.ca
INTRODUCTION
Metabolomics is a field of ‘omics’ science that aims to study global metabolic changes in biological systems (1). It is largely based on the use of nuclear magnetic resonance (NMR) spectroscopy, gas chromatography mass spectrometry (GC–MS) and liquid chromatography mass spectrometry (LC–MS) to detect, identify and quantify small molecule metabolites from biological tissues and biofluids (2). Typically hundreds to thousands of compounds or spectral features can be seen in a typical high throughput metabolomic assay. While still a relatively new member of the ‘omics’ family, metabolomics is already finding applications in a wide variety of research fields including disease diagnosis (3), drug toxicity studies (4) and functional genomics (5). With the rapid growth of this field, there is an increasing demand for dedicated software tools that support the processing and analysis of metabolomic data from a variety of analytical platforms.
There are two major approaches to analyzing metabolomic data—chemometric approaches and quantitative approaches (2). With chemometric approaches the compounds are not initially identified—only their spectral patterns and intensities are recorded, statistically compared and used to identify the relevant spectral features that distinguish sample classes. Once these features have been identified, a variety of approaches may then used to identify the metabolites corresponding to the most important features. Chemometric approaches can be applied to data acquired by NMR, Fourier transform infrared spectroscopy (FTIR) and direct injection mass spectrometry (DIMS). In contrast to chemometric approaches, quantitative metabolomics (or targeted profiling) aims to formally identify and quantify all detectable metabolites from the spectra, prior to subsequent data analysis. Metabolites are identified and quantified by comparing the NMR, GC–MS or LC–MS spectrum of the biosample of interest to a set of authentic (isotopically labeled) standards or to a spectral reference library obtained from authentic standards. This approach requires that the compounds of interest to be known a priori. With the availability of several comprehensive metabolomic databases (6–9) and metabolome libraries, quantitative metabolomics is becoming increasingly common.
Whether the resulting data is chemometric data or quantitative data, what is not commonly realized by many metabolomics researchers is that metabolomics data share a great deal of similarity with microarray data. Both types of data matrices are large and feature rich. Likewise the objectives in microarray analysis are similar to those in metabolomic analysis. For example, both kinds of studies are aimed at identifying significant features associated with certain conditions (biomarker discovery) or for diagnosis (classification). Furthermore, both kinds of studies are often challenged with the problem of dealing with a limited sample size and a high-dimensional feature space. By using the robust data processing algorithms originally developed for microarray analysis and applying them to metabolomic analysis, we believed it could be possible to create a powerful suite of tools for metabolomic data processing. In particular, we chose to work with the resources available from the open source R-project (http://www.R-project.org) and the R-based Bioconductor project (10). These software repositories are widely considered to be the most complete collection of up-to-date statistical and machine learning algorithms for microarray data analysis. The algorithms are actively developed and conscientiously maintained by R-project team members and external contributors. In many respects they have become the reference tools for the microarray field. By combining these R-based analytical tools with metabolite identification and pathway mapping tools originally developed in our own laboratory (6,11), we have created MetaboAnalyst—a web-based server for processing, analyzing, visualizing and annotating high throughput metabolomic data.
In particular, MetaboAnalyst is able to process a wide variety of metabolomic data types including compound concentration tables (for quantitative metabolomics) as well as spectrally binned data, NMR/MS peak lists and GC/LC-MS spectra (NetCDF, mzXML, mzDATA—for chemometric metabolomics). It also provides a comprehensive list of analysis options for normalization, feature identification, dimensional reduction clustering and classification. Furthermore, MetaboAnalyst produces colorful graphical output and it supports a number of compound identification and pathway mapping tools for data annotation. Additional details about the server, its interface and its analytical features are provided in the following pages.
PROGRAM DESCRIPTION AND METHODS
MetaboAnalyst's web interface was developed using Java Server Faces (JSF) technology (http://java.sun.com/javaee/javaserverfaces). The backend statistical computing and visualization operations were carried out using functions from the R and Bioconductor packages. The integration between Java and R was established through the Rserve package (http://www.rforge.net/Rserve). Spectral matching and pathway identification software was developed in Java using the spectral libraries and pathway libraries developed for the Human Metabolome Project (6) and MetaboMiner (11).
JSF is a very powerful technology for developing Java-based web applications. It is designed to simplify the development of user interfaces for Java Enterprise Edition (Java EE) applications by automatic handling of low level HTTP requests and user input processing. JSF uses a component-based model for web development. Using the visual JSF web application tool offered by the NetBeans (http://www.netbeans.org) integrated development environment (IDE), components can be literally ‘painted’ on a virtual JSF page by dragging-and-dropping them from a palette of JSF component library. Event handlers can then be defined for each component the same way as for developing standalone Java graphic user interface (GUI) application. Finally, navigation rules are specified for each page from a central XML configuration file (faces-config.xml). User actions on a web interface will trigger an event whose return value determines which page is to be displayed subsequently based on the navigation rules specified for that page. This approach facilitates modular and flexible design, making web application development much simpler and faster.
MetaboAnalyst consists of several functional modules which will be discussed, in detail, later. These functions are carried out by several R scripts and Bioconductor function calls. Detailed information about the individual packages used and the R scripts can be downloaded from the MetaboAnalyst home page. When MetaboAnalyst is run, the executed R commands are recorded to a temporary text file. During the summary report generation, this R command history is examined and the last call for each analysis performed is re-evaluated using the R Sweave function which executes the R commands and writes text descriptions along with tabular and graphical results into a LaTeX file. Finally, the LaTeX file is converted into a PDF report describing the analysis, which is available to the user for download.
MetaboAnalyst is currently hosted on GlassFish (version 2 update release 2, https://glassfish.dev.java.net) installed on a Linux operating system (Fedora Core 9). The server is equipped with two Intel Pentium 4 processors (2.8 GHz each) and 4 GB of physical memory. The web application is platform independent and has been tested successfully under both Linux and Windows operating systems. R (version 2.8.0) is currently installed on the same machine with latest Bioconductor release 2.3 and Rserve 0.5–2.
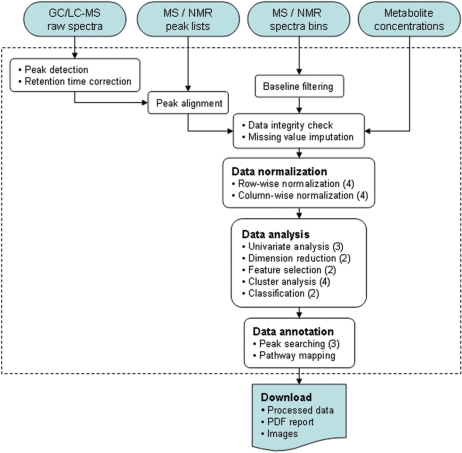
A diagram illustrating MetaboAnalyst's workflow is shown in Figure 1. MetaboAnalyst is not a ‘single-click’ analysis tool, but rather it is an on-line analysis pipeline similar in concept to several existing on-line microarray analysis tools such as GEPAS (12) and CARMAweb (13). It is primarily designed to allow users to conduct two-group discriminant analysis (i.e. control vs. non-control—the most common type of metabolomic analysis) for classification and ‘significant feature’ identification. MetaboAnalyst also supports both paired and unpaired data analyses. A typical MetaboAnalyst run consists of six steps: (i) data upload, (ii) processing, (iii) normalization, (iv) statistical analysis, (v) annotation and (vi) summary report download. Users are guided through these steps by MetaboAnalyst's intuitive interface and the navigation bar on the left panel of each page. Completed steps are indicated by a change in color. Certain downstream analyses may not be allowed depending on the context or type of analyses previously performed. Detailed descriptions, help files or helpful hints are either shown on the corresponding web pages or are provided as mouse-over pop-up balloons. This support is further enhanced by the availability of several step-by-step tutorials, sample data sets (NMR, GC/LC–MS, binned data, etc.), sample summary files and frequently asked questions (FAQs) available on MetaboAnalyst's web site.
Figure 1.
A diagram illustrating MetaboAnalyst's workflow and data processing options. Different data inputs are first transformed into compatible data matrices using several different processing methods. A variety of algorithms are implemented for data normalization, analysis, and annotation. The number of available options is shown inside the round brackets for each category. At the end of any given analysis, a comprehensive PDF report, the processed data, and high-resolution images are available for download.
Step 1: data upload
Users can begin a MetaboAnalyst analysis by pressing the ‘Click Here to Start’ link on the MetaboAnalyst's home page. This takes users to the data upload page. Because there is no widely-accepted standard format for reporting metabolomics experiments MetaboAnalyst has been designed to accept diverse data types including compound concentration tables (from quantitative metabolomic studies), binned spectral data, NMR or MS peak lists, as well as raw GC–MS and raw LC–MS spectra. For compound concentration or binned spectral data, MetaboAnalyst requires that they be uploaded as a comma separated values (CSV) table with class labels (control and abnormal, say) immediately following the sample names. For peak list data, MetaboAnalyst requires that they be uploaded as two zipped folders containing peak list files from the two respective groups. Each file should be a two or three-column CSV list indicating peak positions (chemical shift for NMR peaks, mass and/or retention time for MS peaks) and intensities, respectively. Examples of these formats and more detailed explanations of the formatting requirements are provided on the MetaboAnalyst home page. Vendor-specific, proprietary GC–MS or LC–MS spectra should be first converted to open exchange file formats (NetCDF, mzXML, mzDATA) and uploaded as two zipped folders corresponding to the two groups being analyzed. Detailed instructions on how to specify paired information (for paired data analysis) as well as examples for each data type are available through MetaboAnalyst's ‘Data Formats’ link on the home page.
Step 2: data processing and data integrity checking
Depending on the type of uploaded data, different processing strategies can be employed to convert the raw numbers into a data matrix suitable for downstream analysis. For compound concentration lists, the data can be used immediately after MetaboAnalyst's data integrity check. For binned spectral data, a linear filter is first applied in order to remove baseline noise. This is done because most data processing algorithms do not work properly with many near-zero values. For NMR and/or MS peak lists, MetaboAnalyst first groups the peaks across all samples based on their positions. For GC–MS and LC–MS spectra or total ion chromatograms, the program performs peak detection, peak grouping, and retention time correction sequentially using the popular XCMS package (14). Users can adjust the default parameters for each processing step.
Often there are large numbers of missing values in a typical quantitative metabolomics dataset (10–40% in our experience). Most of these missing values are due to various compounds in certain samples being below the instrument detection limits. To allow selected analyses to proceed (without divide-by-zero problems) these missing values are replaced by the half of the minimum value found in the dataset by default. We also implemented a variety of methods which enable users to manually or automatically perform missing value exclusion, missing value replacement, as well as missing value imputation by Probabilistic PCA (PPCA), Bayesian PCA (BPCA) and Singular Value Decomposition Imputation (SVDImpute) (15,16). In addition, as part of the data integrity check, MetaboAnalyst also verifies class labels and pair specification (if applicable) to make sure all the required information is present and consistent before proceeding to the next step.
Step 3: data normalization
At this stage, the uploaded data is compiled into a table in which each sample is formally represented by a row and each feature identifies a column. With the data structured in this format, two types of data normalization protocols—row-wise normalization and column-wise normalization—may be used. These are often applied sequentially to reduce systematic variance and to improve the performance for downstream statistical analysis. Row-wise normalization aims to normalize each sample (row) so that it is comparable to the other. Four commonly used metabolomic normalization methods have been implemented in MetaboAnalyst, including normalization to a constant sum, normalization to a reference sample (probabilistic quotient normalization) (17), normalization to a reference feature (creatinine or an internal standard) and sample-specific normalization (dry weight or tissue volume). In contrast to row-wise normalization, column-wise normalization aims to make each feature (column) more comparable in magnitude to the other. Four widely-used methods are offered in MetaboAnalyst—log transformation, auto-scaling, Pareto scaling and range scaling. Given the vast dynamic range of many features (compound concentration or ion abundance) in metabolomics data, normalization is highly recommended. The effects and utility of these different normalization strategies have been discussed in detail elsewhere (18) and are described further in MetaboAnalyst's online tutorials.
Step 4: data analysis
MetaboAnalyst's data analysis module is a collection of well-established statistical and machine learning algorithms that have been shown to be particularly robust for high-dimensional data analysis. These algorithms are organized into five analysis ‘paths’ for users to explore.
Univariate analysis path
Because of their simplicity and interpretability, univariate analyses are often first used to obtain an overview or rough ranking of potentially important features before applying more sophisticated analyses. Univariate analysis examines each variable separately and does not consider the effect of multiple comparisons. MetaboAnalyst's univariate analysis path supports three commonly used methods—fold-change analysis, t-tests and volcano plots. In a t-test one attempts to determine whether the means of two groups are distinct. Once a t-value is determined, a P-value can be calculated that can be used to determine whether this distinction is statistically significant. Both paired (same individuals measured before and after an intervention) and unpaired (individuals randomly assigned to two groups) analyses are supported. Volcano plots are used to compare the size of the fold change to the statistical significance level. The horizontal axis plots the fold change between the two groups (on a log scale), while the vertical axis represents the P-value for a t-test of differences between samples (on a negative log scale).
Chemometric analysis path
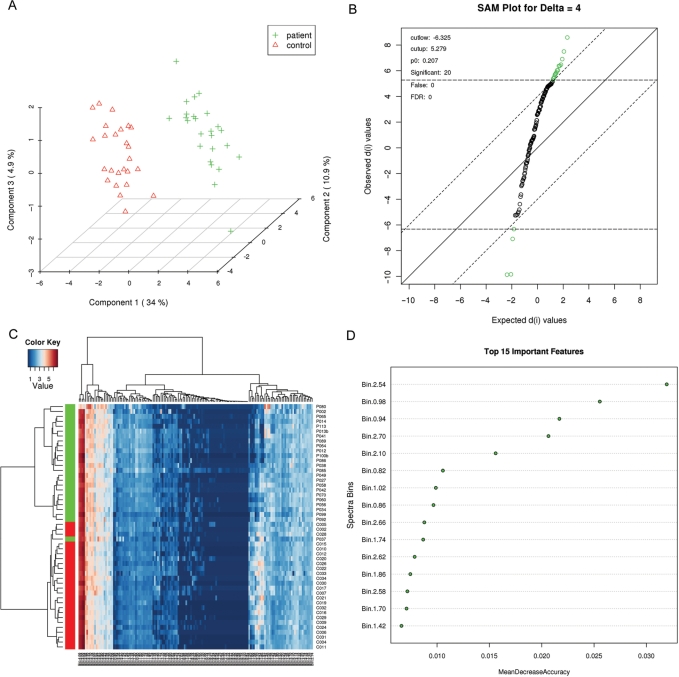
This analysis path offers the two most commonly used chemometric methods—principal component analysis (PCA) and partial-least squares discriminant analysis (PLS-DA). PCA is an unsupervised method aiming to find the directions of maximum variance in a data set (X) without referring to the class labels (Y). PLS-DA is a supervised method that uses multiple linear regression technique to find the direction of maximum covariance between a data set (X) and the class membership (Y). For both methods, the original variables are summarized into much fewer variables using their weighted averages. These new variables are called scores. The weighting profiles are called loadings. MetaboAnalyst provides various views commonly used for PCA and PLS-DA analysis. Users can specify each axis to view the patterns between different components. Both two-dimensional (2D) and three-dimensional (3D) views are implemented. A 3D PLS-DA score plot is shown in Figure 2A. As a supervised method, PLS-DA can perform both classification and feature selection. The algorithm uses cross-validation to select an optimal number of components for classification. Two feature importance measures are commonly used in PLS-DA. Variable importance in projection or VIP score is a weighted sum of squares of the PLS loadings. The weights are based on the amount of explained Y-variance in each dimension. The other importance measure is based on the weighted sum of PLS-regression coefficients. The weights are a function of the reduction of the sums of squares across the number of PLS components. Both importance measures are implemented in PLS-DA analysis for selecting important features. MetaboAnalyst's implementation of PLS-DA also supports several options for cross-validation including leave-one-out (LOOCV) and 10-fold cross validation. We also implemented PLS-DA permutation tests to help users determine the importance of class separation (19).
Figure 2.
Examples of some of the graphical output and analyses available from MetaboAnalyst. (A) PLS-DA class separation based on the top three components. (B) Significant features identified by SAM analysis. (C) Heat map generated from hierarchical clustering. (D) Features ranked by random forest. The binned NMR spectral data (test data #2) was used to generate these graphs.
Feature selection path
This analysis path provides two well-established methods widely used for identification of differentially expressed genes in microarray experiments—Significance Analysis of Microarrays (and Metabolites) (SAM) (20) and Empirical Bayesian Analysis of Microarrays (and Metabolites) (EBAM) (21). However, these methods are very general for identification of significant features in high-dimensional data and are not restricted to the analysis of microarray data. SAM is designed to address false discovery rate problems (FDR) when running multiple tests on high-dimensional data. It first assigns a significance score to each variable based on its change relative to the standard deviation of repeated measurements. Then it chooses variables with scores greater than an adjustable threshold and compares their relative difference to the distribution estimated by random permutations of the class labels. For each threshold, a certain proportion of the variables in the permutation set will be found to be significant by chance. The number is used to calculate the FDR. In this way SAM is able to perform permutation testing, something that is not done in MetaboAnalyst's t-tests. The EBAM algorithm is essentially a variation of the SAM method. The only difference is that EBAM uses a modified t-statistic in calculating its score. Typical SAM and EBAM plots are provided to assist users in choosing the best parameters and viewing the results. Tables containing numeric details are also available through hyperlinks in addition to these graphical presentations. A SAM plot is shown in Figure 2B.
Cluster analysis path
MetaboAnalyst's cluster analysis allows a closer interrogation of samples with similar abundance profiles. This path includes two major approaches of clustering analysis — hierarchical clustering and partitional clustering. Hierarchical (agglomerative) clustering begins with each sample considered as separate cluster and then proceeds to combine them until all samples belong to one cluster. A variety of dissimilarity measures (Euclidean distance, Pearson's correlation, and Spearman's rank correlation) and clustering methods (average linkage, complete linkage, single linkage and Ward's linkage) have been implemented in MetaboAnalyst. The result of hierarchical clustering is usually presented as a dendrogram or heat map, both of which are available in MetaboAnalyst. A heat map view is presented in Figure 2C using one of our test data sets. Partitional clustering attempts to directly decompose the data set into a user-specified number of disjoint clusters. Two widely used methods, k-means clustering and self-organizing map (SOM) have been implemented in MetaboAnalyst. k-Means clustering aims to create k clusters such that the sum of squares from points to the assigned cluster centers’ is minimized. SOM is an unsupervised neural network based around the concept of a grid of interconnected nodes, each of which contains a model. The model clusters begin as random values, but during the iterative training process, they are updated to represent different subsets of the training set. Users indicate the cluster number by specifying the expected dimension of the grid. The clusters from both k-means and SOM are presented as aggregated expression profiles in which samples in each cluster are plotted as line graphs on top of each other using their feature values.
Supervised classification path
Class prediction using metabolomics data is increasingly important in studies aiming for early diagnosis, prognosis or treatment outcomes. MetaboAnalyst offers three powerful supervised classification methods—PLS-DA, random forest (22) and support vector machine (SVM). These methods have proved to be robust for high-dimensional data and are widely used for other ‘omics’ data analysis. In addition, they can also help prioritize features that contribute significantly to the performance. PLS-DA based feature selection and classification was previously discussed in the chemometrics path. Random forest uses an ensemble of classification trees, each of which is grown by random feature selection from a bootstrap sample at each branch. Class prediction is based on the majority vote of the ensemble. During tree construction, about one-third of the instances are left out of the bootstrap sample. This data is then used as test sample to obtain an unbiased estimate of the classification (OOB) error. Variable importance is evaluated by measuring the increase of the OOB error when it is permuted. Figure 2D shows the important features ranked by random forest. The SVM classification algorithm aims to find a nonlinear decision function in the input space by mapping the data into a higher dimensional feature space and separating it by means of a maximum margin hyperplane (23). MetaboAnalyst's SVM analysis is done through recursive feature selection and sample classification using a linear kernel (24). Features are selected based on their relative contribution in the classification using cross validation error rates. The least important features are eliminated in the subsequent steps. This process creates a series of SVM models. The features used by the best model are considered to be important and are ranked by their frequencies of being selected in the model.
Step 5: Data annotation (peak search and pathway mapping)
A key step in placing statistically significant findings from chemometric analyses (as opposed to quantitative metabolomic analyses) into a biological context is to identify significantly altered compounds represented by certain spectral bins or certain clusters of spectral peaks. Once a user has identified lists of MS or NMR peaks that exhibit statistically significant changes, they may use one of several spectral comparison routines and spectral libraries to attempt to identify the compound(s) based on either lists of MS peaks (from MS or MS/MS data), GC-–MS peaks (from EI mass values and retention indices) or NMR peaks (from 1H, 13C or heteronuclear NMR spectra). These compound identification routines and spectral reference libraries were originally developed for the HMDB and for MetaboMiner (11). While not as comprehensive as some commercial libraries or commercial software, these freely available tools have been shown to be quite powerful in identifying many common compounds. Once compound information becomes available (via quantitative routes or via MetaboAnalyst's metabolite ID software), more insight can be obtained by which metabolic pathways are involved. Pathway mapping has been implemented in MetaboAnalyst using more than 70 pathway diagrams and metabolite libraries derived from the HMDB. Users simply type the names (or synonyms) of the metabolites identified and MetaboAnalyst provides the list of pathways in which these metabolites are found, along with hyperlinks to their pathway images. All results are linked to the HMDB where users can obtain more detailed information for each metabolite or pathway.
Step 6: summary report download
When users finish their analyses and click the download link, a comprehensive report will be generated containing a detailed description of each step performed embedded with graphical and tabular outputs. In addition, the processed numeric data, high-resolution images (PNG format), R scripts, as well as the R command history are also available for downloading. Users familiar with R can easily reproduce the results on their local machine after installation of R and the required packages. Users have the option of providing an email address (to which the summary report is sent) or simply downloading the compressed file that contains all the data (graphs, tables, etc.) produced during the analysis. A sample summary report is available for download from MetaboAnalyst's homepage. Raw data files are stored on the server in a temporary folder for up to 72 hrs and then deleted from the server using an automatic ‘cron’ job. After the user has analyzed their data and logs out (after providing their email address) they are sent a URL via email that will allow them to return to their analysis session. All images, figures and tables generated by a MetaboAnalyst session are downloadable and may be permanently stored on the user's own hard drive.
Tutorials and sample data sets
The inherent complexity of many data processing techniques combined with lack of familiarity that many users may have with some of the analytical approaches used by MetaboAnalyst led us to develop a number of tutorials and sample data sets. This was also done so that new users could become more familiar with MetaboAnalyst's expected inputs and outputs. Under the ‘Try our test data’ in the data upload window, users will find eight different data sets labeled as: (i) concentrations (a metabolite concentration table); (ii) NMR spectral bins; (iii) NMR peak lists; (iv) concentrations (paired, time series); (v) MS peak intensities; (vi) MS peak lists; (vii) LC-MS spectra in NetCDF format and (viii) GC-MS spectra in NetCDF format. Users may process these data by clicking on the radio button beside a given data set and pressing the Submit button. Alternately, these example data sets can be downloaded and subsequently ‘uploaded’ using the ‘Upload your data’ section. Once a test data set is submitted (or uploaded) the user may navigate through MetaboAnalyst in any way they choose.
MetaboAnalyst also has four step-by-step tutorials describing several analysis paths using a number of different data sets. These tutorials are available from both the homepage and from the data upload page. Tutorial #1 uses the Metabolite concentration list (data set #1). Tutorial #2 uses the Binned NMR spectra (data set #2), Tutorial #3 uses the paired concentration data (data set #4) and Tutorial #4 uses the LC-MS spectra in NetCDF format (data set #7). MetaboAnalyst also has ∼20 FAQs to complement the information found in the tutorials. These tutorials and FAQs will be updated frequently based on user feedback.
Comparison to other software and limitations
Many metabolomic analyses are currently done using local installations of commercial statistical software packages such as MatLab, MS-Excel, SigmPlot and SIMCA-P. SIMCA-P (Umetrics), in particular, is very widely used by the metabolomics community. While quite expensive, SIMCA-P offers excellent graphic capabilities and comprehensive analysis options for three multivariate methods (PCA, PLS/OPLS and SIMCA). MetaboAnalyst supports two of these multivariate methods (PCA and PLS) but it also offers many other methods (i.e. volcano plots, SAM, k-means, SOM, random forest, SVM) not found in SIMCA-P. While MetaboAnalyst does not have the graphical flexibility of SIMCA-P, it is designed to be more accessible (via the web), freely available, and easier to use. In addition, MetaboAnalyst provides its own metabolite and pathway identification tools—something that is not found in any dedicated statistical software package. However, MetaboAnalyst's dependence on the HMDB infrastructure means that its coverage of plant and microbial metabolism is somewhat incomplete.
To the best of our knowledge, the only other web application that offers a similar service to MetaboAnalyst is MeltDB (25). MeltDB is centered on MS-based metabolomics data storage, administration, analysis and annotation. Unfortunately, this server appears to have security certificate issues with a number of common browsers (Firefox, Netscape) and requires a user login and password to obtain access. According to the article, MeltDB appears to offer some of the features found in MetaboAnalyst such as t-tests, volcano plots, PCA and heat maps. However, these analyses are restricted to GC/LC-MS data only. MetaboAnalyst provides support for many more diverse data types, more advanced data analysis methods, more comprehensive data annotation tools as well as automated report generation utilities.
The current implementation of MetaboAnalyst primarily supports (i) biomarker discovery and (ii) two-group discrimination. We believe these kinds of analyses are most relevant to the widest range of metabolomics studies. Multiclass problems can always be converted into a series of two-class problems through pair-wise decomposition. Temporal studies (more than two time points) can be treated as a special case of multi-class problem and decomposed into a series of paired two-group analyses (see Tutorial #3 for an example of a time series analysis). We hope to add more functions to support simultaneous analysis of multiple time-points in the near future.
MetaboAnalyst makes extensive use of high-level data inputs (i.e. concentrations, peak lists) that requires users to perform some manual processing steps prior to uploading the data. The support for raw or partially processed GC-MS and LC-MS spectra is currently achieved through the XCMS package (14). However, software to handle unprocessed NMR spectra is not so readily available. Steps such as phasing, baseline correction, referencing, peak detection and deconvolution must be manually checked by an experienced analyst to ensure the integrity of the results. As a result, MetaboAnalyst does not accept raw NMR spectra. Likewise, MetaboAnalyst is not (yet) capable of handling or interpreting capillary electrophoretic (CE) data, FTIR data, coulometric electrode array (CEA) data or raw chromatographic (HPLC or UPLC) data. Certainly if the user community grows significantly in these areas, efforts will be made to accommodate these analytical platforms. Indeed, MetaboAnalyst's modular and flexible framework should facilitate future development efforts to keep up with this fast-changing field.
CONCLUSIONS
MetaboAnalyst is a comprehensive, web-based tool designed to facilitate high-throughput metabolomics studies. It accepts a variety of input data (NMR peak lists, binned NMR or MS spectra, MS peak lists, compound/concentration data) in a wide variety of formats. It also offers a number of options for metabolomic data processing, data normalization, multivariate statistical analysis, graphing, metabolite identification and pathway mapping. Through its intuitive interface and high quality graphics, users are presented with data overviews from different perspectives (i.e. PCA plots, heat maps), lists of candidate biomarkers identified by simple univariate analysis (i.e. volcano plots), as well as estimated classification performances by several powerful algorithms (i.e. random forest, SVM). Further biological insight can be gained by tapping into the HMDB using MetaboAnalyst's annotation tools. MetaboAnalyst's structured navigation, extensive documentation, as well as its comprehensive analysis reports should allow new users to analyze their data without significant training or without significant likelihood of statistical misadventure.
FUNDING
Alberta Ingenuity Fund (AIF), the Alberta Life Sciences Institute (ALSI), the Canadian Institutes for Health Research (CIHR) and Genome Alberta, a division of Genome Canada. Funding for open access charge: Canadian Institutes for Health Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fiehn O. Metabolomics—the link between genotypes and phenotypes. Plant Mol. Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- 2.Wishart DS. Quantitative metabolomics using NMR. Trends Anal. Chem. 2008;27:228–237. [Google Scholar]
- 3.Moolenaar SH, Engelke UFH, Wevers RA. Proton nuclear magnetic resonance spectroscopy of body fluids in the field of inborn errors of metabolism. Ann. Clin. Biochem. 2003;40:16–24. doi: 10.1258/000456303321016132. [DOI] [PubMed] [Google Scholar]
- 4.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Ann. Rev. Pharmacol. Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 5.Bino RJ, Hall RD, Fiehn O, Kopka J, Saito K, Draper J, Nikolau BJ, Mendes P, Roessner-Tunali U, Beale MH, et al. Potential of metabolomics as a functional genomics tool. Trends Plant Sci. 2004;9:418–425. doi: 10.1016/j.tplants.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, et al. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, Westler WM, Eghbalnia HR, Sussman MR, Markley JL. Metabolite identification via the Madison Metabolomics Consortium Database. Nat. Biotechnol. 2008;26:162–164. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
- 8.Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN—a metabolite mass spectral database. Therap. Drug Monitor. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 9.Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmuller E, Dormann P, Weckwerth W, Gibon Y, Stitt M, et al. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics. 2005;21:1635–1638. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- 10.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia J, Bjorndahl TC, Tang P, Wishart DS. MetaboMiner—semi-automated identification of metabolites from 2D NMR spectra of complex biofluids. BMC Bioinformatics. 2008;9:507. doi: 10.1186/1471-2105-9-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrero J, Al-Shahrour F, Diaz-Uriarte R, Mateos A, Vaquerizas JM, Santoyo J, Dopazo J. GEPAS: a web-based resource for microarray gene expression data analysis. Nucleic Acids Res. 2003;31:3461–3467. doi: 10.1093/nar/gkg591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rainer J, Sanchez-Cabo F, Stocker G, Sturn A, Trajanoski Z. CARMAweb: comprehensive R- and bioconductor-based web service for microarray data analysis. Nucleic Acids Res. 2006;34:W498–W503. doi: 10.1093/nar/gkl038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 15.Steinfath M, Groth D, Lisec J, Selbig J. Metabolite profile analysis: from raw data to regression and classification. Physiol. Plant. 2008;132:150–161. doi: 10.1111/j.1399-3054.2007.01006.x. [DOI] [PubMed] [Google Scholar]
- 16.Stacklies W, Redestig H, Scholz M, Walther D, Selbig J. pcaMethods—a bioconductor package providing PCA methods for incomplete data. Bioinformatics. 2007;23:1164–1167. doi: 10.1093/bioinformatics/btm069. [DOI] [PubMed] [Google Scholar]
- 17.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures: application in 1H NMR metabonomics. Anal. Chem. 2006;78:4281–4290. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- 18.van den Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics. 2006;7:142. doi: 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bijlsma S, Bobeldijk I, Verheij ER, Ramaker R, Kochhar S, Macdonald IA, van Ommen B, Smilde AK. Large-scale human metabolomics studies: a strategy for data (pre-) processing and validation. Anal. Chem. 2006;78:567–574. doi: 10.1021/ac051495j. [DOI] [PubMed] [Google Scholar]
- 20.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Efron B, Tibshirani R, Storey JD, Tusher V. Empirical Bayes analysis of a microarray experiment. J. Amer. Statist. Assoc. 2001;96:1151–1160. [Google Scholar]
- 22.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 23.Burges CJC. A tutorial on Support Vector Machines for pattern recognition. Data Mining and Knowledge Discovery. 1998;2:121–167. [Google Scholar]
- 24.Zhang X, Lu X, Shi Q, Xu XQ, Leung HC, Harris LN, Iglehart JD, Miron A, Liu JS, Wong WH. Recursive SVM feature selection and sample classification for mass-spectrometry and microarray data. BMC Bioinformatics. 2006;7:197. doi: 10.1186/1471-2105-7-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neuweger H, Albaum SP, Dondrup M, Persicke M, Watt T, Niehaus K, Stoye J, Goesmann A. MeltDB: a software platform for the analysis and integration of metabolomics experiment data. Bioinformatics. 2008;24:2726–2732. doi: 10.1093/bioinformatics/btn452. [DOI] [PubMed] [Google Scholar]