Abstract
RegAnalyst is a user-friendly web interface that integrates MoPP (Motif Prediction Program), MyPatternFinder (pattern detection tool) and MycoRegDB (mycobacterial promoter and regulatory elements database). Since motif discovery is a challenging task, numerous tools have been developed over the past few years. Strikingly, the existing programs were not successful in detecting the known consensus in all mycobacterial (epitomizing degenerate) datasets even in the absence of noise and their performance was further reduced in the presence of noise. Consequently, MoPP, a de novo and greedy (for degeneracy) ‘inexact’ word-based tool that is tailored to enumerate significantly conserved degenerate oligonucleotide motifs was developed. Benchmarking on datasets from MycoRegDB and SCPD (http://rulai.cshl.edu/SCPD/) indicate that MoPP (i) consistently outperforms other motif discovery tools on highly degenerate as well as less degenerate datasets and (ii) successfully detects completely degenerate motifs (with no two instances of a pattern being exactly the same) even in the presence of noise. We have also developed another accessory program, MyPatternFinder, that scans a given sequence or genome to find exact or approximate matches to a query motif of any length identified by MoPP or any other user-defined motif. RegAnalyst will be a valuable tool for in silico analysis of regulatory networks and can be accessed at http://www.nii.ac.in/~deepak/RegAnalyst.
INTRODUCTION
Although transcriptional regulation is one of the most fundamental processes for all forms of life, it still remains an intriguing and challenging subject for biomedical research. Experimental endeavors towards understanding the regulation of genes are laborious, time-consuming and expensive but can be substantially accelerated with the use of in silico methods. Computational identification of transcription factor binding sites has proved to be extremely valuable for deciphering complex regulatory networks in functional genomic studies (1,2). Therefore, a variety of computational algorithms for identifying regulatory motifs from DNA sequences, with or without additional information, have been developed over the past few years (1–6). A motif can be represented as a word of length l that occurs in q sequences with k mismatches (7). Motif detection is acknowledged to be challenging, with various problems potentially requiring different algorithms or ensembles of different methods (8). Additionally, often a transcription factor recognizes a highly diversified (i.e. degenerate) set of elements that vary from each other at many positions (high k values). Such high degeneracy (as observed in mycobacteria) poses another obstacle in detecting motifs. A database of promoter and regulatory elements from various mycobacterial species, MycoRegDB, was created with the primary aim of addressing high levels of degeneracy. Surprisingly, the existing programs were not able to detect the obscured mycobacterial motifs very satisfactorily. Therefore, MoPP (Motif Prediction Program), an exhaustive motif discovery tool based on ‘inexact’ word detection was developed with a focus to detect highly degenerate regulatory elements. Analysis of various mycobacterial datasets from MycoRegDB unambiguously proves the ability of MoPP to identify degenerate motifs in the absence or presence of noise (i.e. background genomic sequences). Furthermore, limited tests suggest that MoPP may be useful in eukaryotes. We used MoPP to identify candidate binding sites in several well studied regulons of Saccharomyces cerevisiae. Our results indicate that MoPP outperforms other motif discovery programs on less degenerate datasets (such as those from yeast) as well.
Along with the growth of available genomic information (6,9), our knowledge of organism specific motifs such as promoters, Shine Dalgarno and regulatory sequences has increased (10–17). The ability to search genomic sequences to locate particular patterns in DNA is of considerable importance and also helps in designing primers with engineered restriction sites for use in molecular biology experiments. The program MyPatternFinder, which we describe here, is useful for detection of user-tailored motifs in DNA sequences. It uses an exact search method along with an alignment technique to find both exact and approximate copies (with/without indels). Its ability to detect copies with insertions and/or deletions (to any desired level) is unique.
We demonstrate the utility of MyPatternFinder, by successfully identifying and validating distinct motifs (such as promoters or hypoxia consensus sequences) in Mycobacterium tuberculosis which differ significantly from those present in other bacterial species, and detection of which proved to be difficult using existing tools. Bacterial persistence is a hallmark of tuberculosis and is thought to result from bacterial adaptation to the prevailing environment within tuberculous lesions and granulomas that are believed to be deficient in oxygen and/or nutrient supply (18). A whole genome microarray analysis revealed widespread changes in gene expression when M. tuberculosis was briefly subjected to in vitro hypoxic conditions (19). Among the genes that were induced was the two-component regulatory system devR-devS suggesting its possible role in mycobacterial latency. Recently, DevR (Rv3133c/DosR) was also reported to be a transcriptional regulator of the hypoxic response in M. tuberculosis (13). A hypoxia consensus motif (5′-TTSGGGACTWWAGTCCCSAA-3′) or a variant thereof was detected upstream of nearly all M. tuberculosis genes rapidly induced by hypoxia (12,13).
METHODS
MycoRegDB
Transcription start points (TSPs) and regulatory elements experimentally identified in various mycobacterial species [M. tuberculosis (strains H37Rv and CDC1551), M. bovis, M. leprae, M. smegmatis and M. avium subsp. paratuberculosis] were compiled.
MoPP
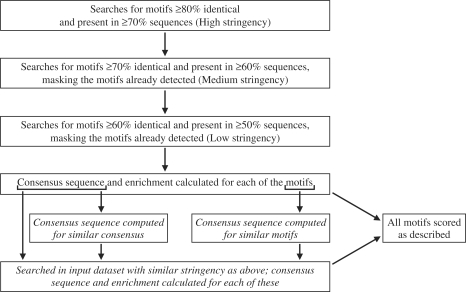
MoPP is an exhaustive motif discovery tool that is tailored to enumerate significantly conserved degenerate oligonucleotide patterns. Figure 1 shows the schematic representation of MoPP's algorithm. In the first step, MoPP identifies patterns that are overrepresented in the input dataset (FASTA format). By default, the program initially searches for motifs that are ≥80% identical and present in ≥70% of the sequences (high stringency). Subsequently, the stringency is reduced to detect motifs that are ≥70% identical and present in ≥60% of the sequences after masking out the motifs already found (medium stringency). Finally, the stringency is reduced to detect motifs that are ≥60% identical and present in ≥50% of the sequences after masking out the motifs already found (low stringency). Using advanced options, a user also has the freedom to specify the cut-offs for percent identity and percent sequences that should contain the motif. Consensus sequence (at each position a nucleotide or set of nucleotides present in ≥60% of the sequences is selected) and enrichment (ratio of copy number in input dataset to that in the non-coding regions of the genome) are then computed for each of the patterns.
Figure 1.
Schematic representation of MoPP's algorithm. The unparalleled ability of MoPP in detecting degenerate motifs is due to the steps indicated in italics.
In the second step, exact/degenerate matches to these consensus sequences are also searched for in the input dataset. Furthermore, for every group of similar patterns/consensus sequences identified in the first step, a consensus sequence is computed and searched for in the input dataset (as in the first step). All the patterns identified in the first and second steps are ranked on the basis of copy number (Rcp) and enrichment (Ren). The final score of each pattern is given by (1/Ravg) × 100, where Ravg = (Rcp + Ren)/2.
MoPP's performance was compared with other programs by motif level success rate score mSr, which is defined as the number of target motif groups Np that have at least one correctly predicted binding site divided by the total number of target motifs M [mSr = Np/M] (8). However, the programs YMF, PRISM and Oligo only report the detected motif, its statistical score(s) and/or count, but do not explicitly provide the binding sites or their locations. Therefore, while calculating mSr we have considered the detection of ‘motif’ instead of ‘binding site’ (on the presumption that if motif has been correctly detected then at least one binding site would definitely have been predicted correctly). Furthermore, to avoid any bias due to different number of motifs predicted by various programs, we have considered only the top five motifs for each program as suggested by Tompa et al. (20). The scalability issue, as to how the algorithm performance changes with the motif width and the sequence length, is also addressed (8). Therefore, yeast datasets for various motif lengths (6–10 bp) each with different margin sizes (extending on both sides of target motifs) of 50, 100, 200, 300, 400, 500 and 800 bp were generated and analyzed with MoPP by mSr as well as performance coefficient at binding site level [sPC] (8). The sPC score indicates whether predicted binding sites overlap with true binding sites (those that have ≥75% matches with the consensus) and is defined as, sPC = sTP/(sTP + sFP + sFN), where sTP is the number of predicted binding sites which overlaps with the true binding sites by at least 1 nucleotide, sFP is the number of predicted binding sites which have no overlaps with the true binding sites and sFN is the number of true binding sites that have no overlaps with any predicted binding sites.
In principle, MoPP has the capability to detect motifs of any length. However, by default, the program searches only for motif widths of 6–8 bp. In case 8-mer motifs are detected, user can repeat the search with longer motif width(s) of his interest. The algorithm also gives user the freedom to allow single/multiple hits of the motif in each input sequence.
MyPatternFinder
The algorithm that is used in MyPatternFinder is mentioned briefly as follows. In the first step (Option A), the input pattern of length N is aligned with the first N bases of the DNA sequence and percentage score is computed (1 for every match, 0 for a mismatch) in a sliding window with 1 base shift, along the entire sequence [the method was also incorporated in our previously developed program Spectral Repeat Finder (21)]. If indels are permitted (Option B), the input pattern of length N is aligned with the first M bases (M = N + number of mutations allowed by the user) of the DNA sequence using ClustalW (22) (with gap opening and gap extension penalties of 2.0 each); this allows for indels, and as before, a score can be computed in a sliding window. In the second step, windows where the percentage score exceeds a desired threshold are identified; if there are overlapping patterns, the one with the highest score is considered.
Flexibility has been incorporated into the MyPatternFinder algorithm, so that target patterns can be specified precisely, or with standard abbreviations B, D, H, V, K, M, W, R, S, Y and N if desired. However, in Option B no ambiguous bases can be specified in the query sequence since it is based on ClustalW. Query motifs can also be chosen from a list of available consensus sequences (these will constantly be updated). The first version of MyPatternFinder offers the choice of 34 distinct annotated DNA motifs [15 prokaryotic promoter elements (including 7 from mycobacteria), 4 eukaryotic promoter elements, 9 transcription factors and 6 response elements]. Searches can be carried out in various completely sequenced genomes (choice of >600 organisms is available at present and it will be kept up-to-date in future) and a detailed visualization of the patterns detected along with their positions is provided.
RESULTS AND DISCUSSION
MycoRegDB
MycoRegDB is currently the only available database of promoter/regulatory elements across various mycobacterial species. The first release of MycoRegDB (Supplementary Figure S1) contains 290 annotated DNA motifs (174 promoters and 116 transcription factor binding sites) described in 81 research papers. For each database entry, MycoRegDB gives a variety of information such as gene annotation, CDS positions, promoter/regulatory sequence (with TSP/binding site explicitly marked), TSP-CDS/Motif-CDS distance and hyperlinks to relevant reference(s). Wherever applicable, it also provides hyperlinks to gene information from TubercuList, BCGList and Leproma (http://genolist.pasteur.fr/). These resources are helpful for (i) retrieving DNA/protein sequences, (ii) knowing family classification of genes, and (iii) providing cross-references to UniProt, PDB, PFAM and COG databases. The MycoRegDB will be kept up-to-date in future releases.
Mycobacterial promoters are quite divergent
Among the 174 promoters in MycoRegDB, 118 are those for which the TSPs have been experimentally defined. Of these, for a large subset of 95 promoters the sigma factor(s) recognizing them is/are not known. A majority of these promoters are possibly regulated by the housekeeping sigma factor SigA (23). Alignment of the –10 and –35 regions revealed that there is only ∼60% conservation with the known SigA consensus in both these regions (Supplementary Figure S2). Only one of the –10 regions and six of the –35 regions showed perfect match to the –10 and –35 consensus, respectively. This indicates that there exists considerable degeneracy in mycobacterial promoters. Furthermore, our analysis does not seem to suggest that –35 regions are conserved to lesser extent in comparison to –10 regions (17). This discrepancy could possibly be due to accumulation of additional data over recent years. The remaining 23 promoters were divided into subsets on the basis of the involvement of a given sigma factor (SigC: 1, SigD: 6, SigF: 1, SigH: 10 and SigL: 5). Here again, the promoter elements were quite degenerate although less than SigA dataset (Supplementary Figure S3). However, it would be important to point out that these datasets are small in size and the level of degeneracy is expected to increase as more data get accumulated.
Evaluating MoPP and other motif prediction programs on mycobacterial datasets in absence of noise
The –10 and –35 regions from the SigA, SigD, SigH and SigL class (Supplementary Figures S2 and S3) were then used to evaluate MoPP, YMF (24), Oligo (25), MEME (26), PRISM (27) and SCOPE (28). MoPP was successful in detecting the known consensus in all eight datasets (Table 1). However, even in the absence of noise, the existing programs were not totally successful; MEME and Oligo, closely followed MoPP, with being successful in seven datasets (Table 1). The ensemble program SCOPE was able to detect the consensus in only five datasets. This enhanced ability of MoPP to detect highly degenerate motifs is because the algorithm (i) deduces the consensus sequences in three different ways, and (ii) allows imperfections not only in the initial step but also each time it searches for matches to the consensus in the input dataset (Figure 1).
Table 1.
Performance comparison of MoPP with five popular motif finders on mycobacterial datasets
| Regulon | Consensus | Size | MoPPa | YMF | PRISM | SCOPE | Oligo | MEME |
|---|---|---|---|---|---|---|---|---|
| MycoSigA-10 | TATAMT | 95 | TAYAVT (1)b | TATtrW (5) TATtAW (6) | tTAcAAT (3) | TANDVTgk (2) | TAgACT (1) TAcAAT (2) | TAgACT (1) |
| MycoSigA-35 | TTGACW | 95 | cTKGAC (1) cTBGAC (3) | TTGACW (6) | gnhWTGACW (1) | wyTTGMMW (1) | TTGACT (2) | TTGACT (1) |
| MycoSigA50bp | TATAMT | 95 | TATACT (2) TAKACT (3) | TAgWCW (14) | tTAcAAT (14) | ataTHDMAY (2)c | TAgACT (6) | TATtAT (11) |
| TTGACW | TRACTa (1) TaKACT (3) | TaGWCW (14) TWGACW (22) | TTGACT (4) | TTGACT (2) | ||||
| MycoSigD-10 | WNATGTd | 6 | gTTATG (1) gTTABG (4) | ACATaT (15) | ||||
| MycoSigD-35 | GTAACG | 6 | gGWAWC (3) | gGTAAC (2) | GTAACG (1) | GTAACG (1) | ||
| MycoSigH-10 | SGTTS | 10 | tCGTT (1) gCGKT (2) | SGTTar (21) | cGGTT (3) | cGGTT (3) | gCGTT (1) | cCGTT (2) |
| MycoSigH-35 | SGGAAC | 10 | GGGAAt (1) GGGAAY (2) | GGGAAC (1) GGGAAY (2) | CGGAA (2) | CGGAA (2) | GGGAAC (1) | GGGAAC (1) |
| MycoSigL-10 | CGTGTC | 5 | CGTGTC (1) | GTGTCa (5) | GTGTCa (1) CGTGTC (2) | GTGTCa (1) | ||
| MycoSigL-35 | TGAACC | 5 | tTGAAC (1) bTGAAC (2) | TGWACY (3) TGAACY (5) | TGAAC (1) | bTGAAC (1) | TGAACC (1) | cTGAAC (1) |
| mSr | 1.0 | 0.4 | 0.5 | 0.5 | 0.8 | 0.8 | ||
aWeeder could not be compared since the background file was not available.
bPattern is highlighted in bold if it matches the consensus with not more than one mismatch and ranks among top five. Number in parenthesis indicates rank of the pattern.
cNot considered a match since ≥80% of the matching residues are degenerate nucleotides or matching with degenerate nucleotides.
dAccording to MtbRegList (www.usherbrooke.ca/vers/MtbRegList).
Evaluating MoPP and other motif prediction programs on mycobacterial datasets in presence of noise
Input sequences for motif finding programs typically consist of motifs buried in noise. Therefore, to simulate real scenario, we made a dataset (MycoSigA50bp) by extracting 50 bp sequences upstream of TSPs (encompassing both –10 and –35 regions) for the SigA regulon. This formed an ideal dataset since it contained 95 genes with highly degenerate motifs. Here also, MoPP was successful in detecting both –10 and –35 consensus sequences (Table 1 and Figure 2). None of the other programs was able to detect the –10 consensus for which only a single perfect match occurred in the whole dataset. However, both MEME and Oligo were able to find a pattern TTGACT that matched with the –35 consensus since there existed six exact occurrences of this pattern in the dataset. This illustrates the ability of MoPP in detecting a completely degenerate motif (with not even two instances of exact match to the consensus) in the presence of noise.
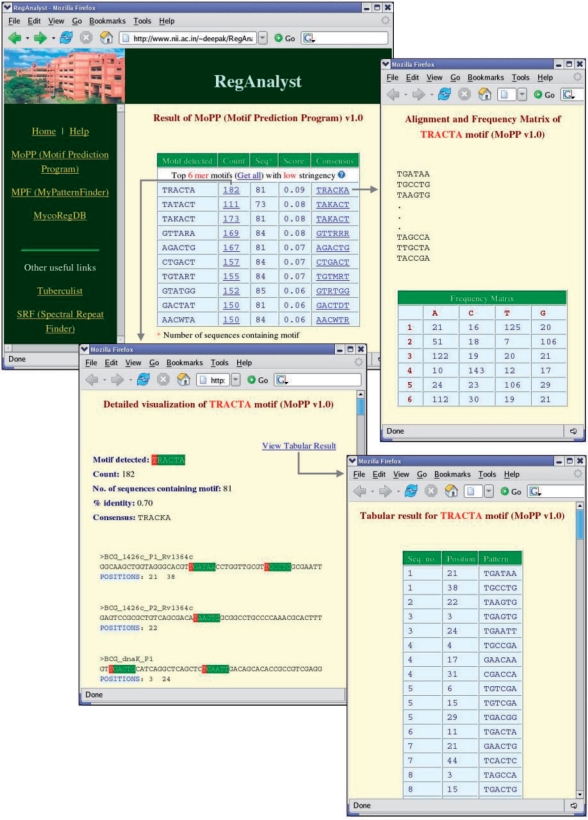
Figure 2.
A typical output of MoPP on analysis of a large (95 genes) and highly degenerate dataset, MycoSig50bp. MoPP successfully identified both –10 (motifs ranked 2 and 3) and –35 consensus (motifs ranked 1 and 3) sequences. For each of the detected motif, user can view (i) a colored display of patterns along with their positions (by clicking on the count link), (ii) a tabular output of patterns and their positions and (iii) alignment and frequency matrix of patterns (by clicking on the consensus sequence).
MoPP is not restricted to mycobacteria
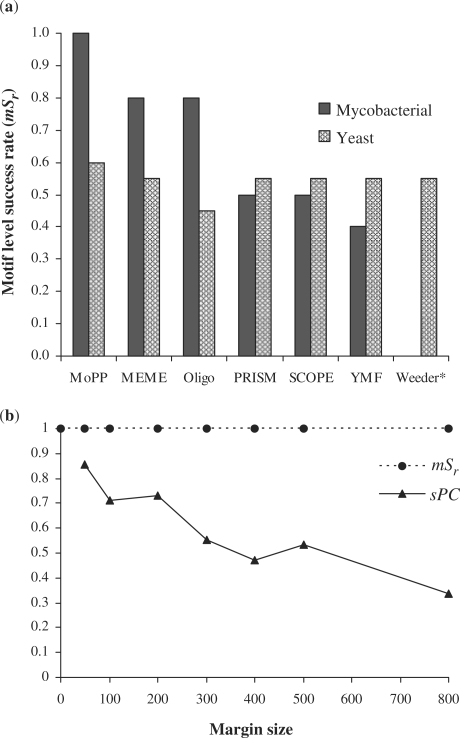
To demonstrate that MoPP algorithm is not organism specific, we compared MoPP against other programs on 20 well-characterized S. cerevisiae regulons (http://rulai.cshl.edu/SCPD/). MoPP was able to detect the known consensus in 12 of 20 regulons (Supplementary Table S1). Interestingly, MoPP (mSr = 0.60) outperformed all other programs including SCOPE (mSr = 0.55), which combines the output of three different programs. The overall comparison of MoPP with other tools across a total of 30 datasets derived from mycobacteria (with or without noise) and yeast also revealed that MoPP (mSr = 0.74) outperformed SCOPE (mSr = 0.54) (Figure 3a). MoPP was followed by MEME and Oligo which had an mSr of 0.64 and 0.57, respectively. However, it would be important to point out that the superior performance of MoPP was primarily because of its ability in detecting highly degenerate motifs present in mycobacterial datasets wherein it outperformed other programs by 20–60%.
Figure 3.
(a) Performance comparison of MoPP with other motif discovery tools on 30 datasets derived from mycobacteria and yeast. *Weeder could not be assessed on mycobacterial datasets since the background file was not available. (b) Scalability of MoPP in terms of motif level success rate (mSr) and performance coefficient at binding site level (sPC) with respect to the sequence length (margin size).
Furthermore, MoPP's motif level success rate (mSr) was not affected by sequence length and/or motif width since it is an exhaustive enumeration program (Figure 3b). These results are consistent with similar observations for MEME (8). It would also be important to mention that for each dataset (irrespective of the margin size) the motif (with ≥80% matches with the consensus) was correctly identified. The prediction accuracy at the binding site level (sPC) on yeast datasets (Figure 3b) was also higher (for all margin sizes) in comparison to those observed for other programs on Escherichia coli datasets (8).
Detecting known consensus sequences by MyPatternFinder
The consensus motif sequences of 7 of the 13 M. tuberculosis sigma factors (SigA, SigC, SigD, SigE, SigF, SigH and SigL) have been recently published (10,15). As representative examples, MyPatternFinder was used to search the exact consensus motifs of three sigma factors SigA, SigF and SigH (Table 2; complete details are available at http://www.nii.ac.in/~deepak/MyPattern/supl/sigma). No exact copy of the motif for the primary housekeeping sigma factor, SigA, was found and only four copies of the SigF motif could be located (15). This corroborates our observation that there exists considerable flexibility in promoter recognition and a search for promoter sequences must necessarily accommodate mismatches in sequence or spacing of the bipartite elements. We were indeed able to detect 20 copies of the SigA motif by allowing one mismatch with the consensus sequence, several of which were present upstream of various genes (Table 2). Some of these could possibly also be active in E. coli since they are almost identical to E. coli σ70 consensus promoter sequence; such comparisons with promoters of another organism(s) such as E. coli can help in predicting whether the organism(s) is a good candidate for studying these mycobacterial promoters (29). Another interesting finding was that out of the 150 exact copies of SigH motif identified, more than 80% were not present in the upstream region of genes but rather within the protein-coding regions.
Table 2.
Detection of exact sigma consensus sequences in the complete M. tuberculosis H37Rv genome by MyPatternFinder
| Sigma factor | Consensus sequence (Ref.) | Total number of hits | Genea,b | Distance from start codonc |
|---|---|---|---|---|
| SigA | TTGACW-N17-TATAMTd (15) | 0 | – | – |
| TTGACW-N16–21-TATAMT (15,17) | 0 | – | – | |
| TTGACW-N16–21-TATAMT (15,17) | 20e | Rv0068f | −84 | |
| Rv0305c (PPE) | −163 | |||
| Rvnr01 (16S rRNA) | −225 | |||
| Rv1403c | −84 | |||
| Rv2011cf | −50 | |||
| Rv2487c (PE_PGRS) | −288 | |||
| Rv2578cf | −35 | |||
| Rv3082c (virS)f | −44 | |||
| Rv3760 | −485 | |||
| SigF | GTTT-N17-GGGTAT (15) | 4 | Rv1248c (sucA) | −358 |
| Rv3287c (rsbW/usfX)g | −35 | |||
| Rv3349c | −264 | |||
| SigH | SGGAAC-N17–22-SGTTS (15) | 150 | Rv0384c (clpB)g | −72 |
| Rv0474 | −150 | |||
| Rv0563 (htpX) | −78 | |||
| Rv0569 | −475 | |||
| Rv1072 | −79 | |||
| Rv1535 | −93 | |||
| Rv1786 | −448 | |||
| Rv1792 | −112 | |||
| Rv1883c | −217 | |||
| Rv2018 | −182 | |||
| Rv2184c | −178 | |||
| Rv2308 | −34 | |||
| Rv2334 (cysK) | −364 | |||
| Rv2373c (dnaJ2) | −138 | |||
| Rv2466cg | −77 | |||
| Rv2525c | −345 | |||
| Rv2694c | −96 | |||
| Rv2745c | −66 | |||
| Rv2804c | −384 | |||
| Rv2839c (infB) | −313 | |||
| Rv3179 | −321 | |||
| Rv3482c | −248 | |||
| Rv3597c (lsr2) | −203 | |||
| Rv3832c | −481 | |||
| Rv3913 (trxB2)g | −66 |
aGene is reported only if the distance of consensus sequence is ≤500 bp upstream of the start codon and it has a non-coding upstream region of ≥25 bp.
bAccording to Cole et al. (36).
cLocation is relative to the translation start site as determined at http://genolist.pasteur.fr/TubercuList, except for Rv3287c (rsbW/usfX), where location is relative to transcription start site according to Beaucher et al. (37).
dW = A/T; M = A/C; S = G/C.
eBy allowing one mismatch in the consensus sequence.
fAlso predicted to be an E. coli σ70 promoter with one mismatch.
Using MyPatternFinder, we also searched for the hypoxia consensus motif (13) in the M. tuberculosis H37Rv genome. Complete details of the best 100 motifs identified are available at http://www.nii.ac.in/~deepak/MyPattern/supl/hypmotif. We were not only able to detect all the motifs reported by Park et al. (13), but also identified a number of additional motifs among which several were positioned upstream of coding regions (Table 3). Although most of these genes were not hypoxia responsive by microarray analysis (13), one of the genes, Rv3318 (sdhA), was repressed in hypoxia in M. tuberculosis H37Rv:ΔdosR (13) while another, Rv1039c (PPE15), was significantly induced within artificial granulomas in mice (30) substantiating our results. Further analysis revealed that a number of motifs (with significantly high scores) were present within protein-coding regions of genes, a majority of which were also not regulated by hypoxia. The possible significance of this observation is unclear at present.
Table 3.
Hypoxia responsive motifs present upstream of genesa
| Sequenceb | Scorec | Gened |
|---|---|---|
| ccGGGGAtgAAcGTCCCCgc | 11.8486 | Rv1039c (PPE15)e |
| TgCGGGACTAcAaTCCCGgg | 11.7186 | Rv1811 (mgtC) |
| ggCGGGACTATgGTCgCGAc | 11.414 | Rv1552 (frdA) |
| gTCGGGgCggTgGTCCCCgg | 11.2576 | Rv0345 |
| TTGGGGcCaTccGgCCCGgA | 11.195 | Rv0877 |
| aTaGtGACaTTcGaCCCGAA | 10.8046 | Rv3318 (sdhA)f |
| aTCGGGcCgAAcGTCaCGAt | 10.761 | Rv1824 |
| cTCGGGACaTTAcTtCCGtt | 10.7435 | Rv1881c (lppE) |
| caCGGGACgAgcaTCCCCAg | 10.7301 | Rv2194 (qcrC) |
| cTCGGGtgTgAgGTCCCatA | 10.6815 | Rv2221c (glnE) |
| gcCaGGACgTcgGgCCCGAg | 10.5356 | Rv1256c (cyp130) |
The utility of this server is also not limited to mycobacterial sequences: we screened for thyroid hormone response elements (TREs) which are regulatory sequences known to exist upstream of metallothionein genes (31). The metallothionein protein protects the cell against excess concentrations of heavy metals, by binding the metal and removing it from the cell. The gene is expressed at a basal level, but is induced to greater levels of expression by heavy metal ions (such as cadmium) or by glucocorticoids. The TRE has a binding site for transcription factor AP1 and this interaction is part of the mechanism for constitutive expression. Furthermore, this binding reaction is one of the mechanisms (not necessarily the only modality) by which phorbol esters such as TPA (an agent that promotes tumors) trigger a series of transcriptional changes. The TRE motif (TGACTCA) was identified, in 1–6 copies, upstream of various human metallothionein genes (MT1E, MT1K, MT2, MT3 and MT4) when the pattern was allowed to contain indels (Supplementary Figure S4; details are available at http://www.nii.ac.in/~deepak/MyPattern/supl/TRE). It is noteworthy to mention that motif discovery in datasets derived from large complex genomes pose certain additional challenges, and the speed and performance of the two algorithms (MoPP and MyPatternFinder) were not assessed on such datasets (e.g. genome-wide ChIP-chip or ChIP-seq datasets).
Validation of motifs detected
MyPatternFinder was used to detect matches to the various sigma consensus elements upstream of the experimentally determined TSPs in the Rv3134c-devR-devS operon (32). The P2Rv3134c promoter showed similarity to both M. tuberculosis SigA consensus as well as E. coli σ70 consensus. As predicted, the P2Rv3134c promoter was indeed found to be functional in both M. smegmatis [model for studying M. tuberculosis promoters since the transcriptional machinery is well conserved between the two organisms (33)] and E. coli (32) substantiating the results of MyPatternFinder.
Distant matches to the DevR consensus motif were also identified in the region encompassing the devR upstream region, Rv3134c coding sequence and Rv3134c upstream region (32). Although these low scoring Dev boxes did not show interaction with DevR (34), their comparison with various high scoring Dev boxes revealed the importance of C8 base in the consensus motif (35).
CONCLUSION
We have unambiguously proved the efficacy of MoPP (i) in prokaryotes and lower eukaryotes, (ii) in detecting motifs of various lengths, (iii) in detecting highly degenerate as well as less degenerate motifs, and (iv) in the presence of high noise (large sequence lengths). Similarly, the utility of MyPatternFinder has been shown (i) in prokaryotes and small eukaryotic sequences, (ii) in short sequences as well as complete less complex genomes, and (iii) for various consensus sequences (sigma factors, hypoxia motifs and TREs). Thus, both MoPP and MyPatternFinder work efficiently for smaller, less complex genomes and may also be useful for higher eukaryotes with larger, more complex genomes. The patterns detected using MyPatternFinder have been experimentally validated. The detection of conserved motifs (by MoPP) and user-defined patterns of interest (by MyPatternFinder) in genomic sequences should facilitate the understanding of gene expression and regulatory pathways in biological systems.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENT
D.S. is grateful to Dr R. Ramaswamy, Dr J. S. Tyagi, Dr G. P. S. Raghava and Dr Biju Issac for their valuable help during development of the MyPatternFinder program.
FUNDING
Research Fellowships from Department of Biotechnology (DBT), Government of India and Indian National Science Academy (INSA); core and BTIS project grants from DBT (to D.M.); Centre of Excellence by DBT (to A.S.). Funding for open access charge: Department of Biotechnology.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Wyrick JJ, Young RA. Deciphering gene expression regulatory networks. Curr. Opin. Genet. Dev. 2002;12:130–136. doi: 10.1016/s0959-437x(02)00277-0. [DOI] [PubMed] [Google Scholar]
- 2.Duret L, Bucher P. Searching for regulatory elements in human noncoding sequences. Curr. Opin. Struct. Biol. 1997;7:399–406. doi: 10.1016/s0959-440x(97)80058-9. [DOI] [PubMed] [Google Scholar]
- 3.Brazma A, Jonassen I, Vilo J, Ukkonen E. Predicting gene regulatory elements in silico on a genomic scale. Genome Res. 1998;8:1202–1215. doi: 10.1101/gr.8.11.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee N, Zhang MQ. Functional genomics as applied to mapping transcription regulatory networks. Curr. Opin. Microbiol. 2002;5:313–317. doi: 10.1016/s1369-5274(02)00322-3. [DOI] [PubMed] [Google Scholar]
- 5.Ohler U, Niemann H. Identification and analysis of eukaryotic promoters: recent computational approaches. Trends Genet. 2001;17:56–60. doi: 10.1016/s0168-9525(00)02174-0. [DOI] [PubMed] [Google Scholar]
- 6.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 7.D'Haeseleer P. What are DNA sequence motifs? Nat. Biotechnol. 2006;24:423–425. doi: 10.1038/nbt0406-423. [DOI] [PubMed] [Google Scholar]
- 8.Hu J, Li B, Kihara D. Limitations and potentials of current motif discovery algorithms. Nucleic Acids Res. 2005;33:4899–4913. doi: 10.1093/nar/gki791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kauer G, Blocker H. Applying signal theory to the analysis of biomolecules. Bioinformatics. 2003;19:2016–2021. doi: 10.1093/bioinformatics/btg273. [DOI] [PubMed] [Google Scholar]
- 10.Rodrigue S, Provvedi R, Jacques PE, Gaudreau L, Manganelli R. The σ factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 2006;30:926–941. doi: 10.1111/j.1574-6976.2006.00040.x. [DOI] [PubMed] [Google Scholar]
- 11.Camp E, Badhwar P, Mann GJ, Lardelli M. Expression analysis of a tyrosinase promoter sequence in zebrafish. Pigment Cell Res. 2003;16:117–126. doi: 10.1034/j.1600-0749.2003.00002.x. [DOI] [PubMed] [Google Scholar]
- 12.Florczyk MA, McCue LA, Purkayastha A, Currenti E, Wolin MJ, McDonough KA. A family of acr-coregulated Mycobacterium tuberculosis genes shares a common DNA motif and requires Rv3133c (dosR or devR) for expression. Infect. Immun. 2003;71:5332–5343. doi: 10.1128/IAI.71.9.5332-5343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park HD, Guinn KM, Harrell MI, Liao R, Voskuil MI, Tompa M, Schoolnik GK, Sherman DR. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 2003;48:833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puopolo KM, Madoff LC. Upstream short sequence repeats regulate expression of the alpha C protein of group B Streptococcus. Mol. Microbiol. 2003;50:977–991. doi: 10.1046/j.1365-2958.2003.03745.x. [DOI] [PubMed] [Google Scholar]
- 15.Manganelli R, Provvedi R, Rodrigue S, Beaucher J, Gaudreau L, Smith I. σ factors and global gene regulation in Mycobacterium tuberculosis. J. Bacteriol. 2004;186:895–902. doi: 10.1128/JB.186.4.895-902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J. The p53MH algorithm and its application in detecting p53-responsive genes. Proc. Natl Acad. Sci. USA. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez M, Smith I. In: Molecular genetics of Mycobacteria. Hatfull GF, Jacobs W.R. Jr, editors. Washington, DC: ASM Press; 2000. pp. 111–129. [Google Scholar]
- 18.Wayne LG, Sohaskey CD. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 19.Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding α-crystallin. Proc. Natl Acad. Sci. USA. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tompa M, Li N, Bailey TL, Church GM, De Moor B, Eskin E, Favorov AV, Frith MC, Fu Y, Kent WJ, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 21.Sharma D, Issac B, Raghava GP, Ramaswamy R. Spectral Repeat Finder (SRF): identification of repetitive sequences using Fourier transformation. Bioinformatics. 2004;20:1405–1412. doi: 10.1093/bioinformatics/bth103. [DOI] [PubMed] [Google Scholar]
- 22.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y, Coates AR. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J. Bacteriol. 1999;181:469–476. doi: 10.1128/jb.181.2.469-476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinha S, Tompa M. YMF: A program for discovery of novel transcription factor binding sites by statistical overrepresentation. Nucleic Acids Res. 2003;31:3586–3588. doi: 10.1093/nar/gkg618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Helden J, Andre B, Collado-Vides J. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 1998;281:827–842. doi: 10.1006/jmbi.1998.1947. [DOI] [PubMed] [Google Scholar]
- 26.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlson JM, Chakravarty A, Khetani RS, Gross RH. Bounded search for de novo identification of degenerate cis-regulatory elements. BMC Bioinformatics. 2006;7:254. doi: 10.1186/1471-2105-7-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlson JM, Chakravarty A, DeZiel CE, Gross RH. SCOPE: a web server for practical de novo motif discovery. Nucleic Acids Res. 2007;35:W259–W264. doi: 10.1093/nar/gkm310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verma A, Kinger AK, Tyagi JS. Functional analysis of transcription of the Mycobacterium tuberculosis 16S rDNA-encoding gene. Gene. 1994;148:113–118. doi: 10.1016/0378-1119(94)90243-7. [DOI] [PubMed] [Google Scholar]
- 30.Karakousis PC, Yoshimatsu T, Lamichhane G, Woolwine SC, Nuermberger EL, Grosset J, Bishai WR. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J. Exp. Med. 2004;200:647–657. doi: 10.1084/jem.20040646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewin B. Genes VI. New York: Oxford University Press; 1997. pp. 847–883. [Google Scholar]
- 32.Bagchi G, Chauhan S, Sharma D, Tyagi JS. Transcription and autoregulation of the Rv3134c-devR-devS operon of Mycobacterium tuberculosis. Microbiology. 2005;151:4045–4053. doi: 10.1099/mic.0.28333-0. [DOI] [PubMed] [Google Scholar]
- 33.Bashyam MD, Kaushal D, Dasgupta SK, Tyagi AK. A study of mycobacterial transcriptional apparatus: identification of novel features in promoter elements. J. Bacteriol. 1996;178:4847–4853. doi: 10.1128/jb.178.16.4847-4853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chauhan S, Tyagi JS. Cooperative binding of phosphorylated DevR to upstream sites is necessary and sufficient for activation of the Rv3134c-devRS operon in Mycobacterium tuberculosis: implication in the induction of DevR target genes. J. Bacteriol. 2008;190:4301–4312. doi: 10.1128/JB.01308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chauhan S, Tyagi JS. Interaction of DevR with multiple binding sites synergistically activates divergent transcription of narK2-Rv1738 genes in Mycobacterium tuberculosis. J. Bacteriol. 2008;190:5394–5403. doi: 10.1128/JB.00488-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry C.E., 3rd., et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 37.Beaucher J, Rodrigue S, Jacques PE, Smith I, Brzezinski R, Gaudreau L. Novel Mycobacterium tuberculosis anti-σ factor antagonists control σF activity by distinct mechanisms. Mol. Microbiol. 2002;45:1527–1540. doi: 10.1046/j.1365-2958.2002.03135.x. [DOI] [PubMed] [Google Scholar]
- 38.Manganelli R, Voskuil MI, Schoolnik GK, Dubnau E, Gomez M, Smith I. Role of the extracytoplasmic-function σ factor σH in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 2002;45:365–374. doi: 10.1046/j.1365-2958.2002.03005.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.