Abstract
Function prediction of proteins with computational sequence analysis requires the use of dozens of prediction tools with a bewildering range of input and output formats. Each of these tools focuses on a narrow aspect and researchers are having difficulty obtaining an integrated picture. ANNIE is the result of years of close interaction between computational biologists and computer scientists and automates an essential part of this sequence analytic process. It brings together over 20 function prediction algorithms that have proven sufficiently reliable and indispensable in daily sequence analytic work and are meant to give scientists a quick overview of possible functional assignments of sequence segments in the query proteins. The results are displayed in an integrated manner using an innovative AJAX-based sequence viewer. ANNIE is available online at: http://annie.bii.a-star.edu.sg. This website is free and open to all users and there is no login requirement.
INTRODUCTION
Advances in sequencing technology have taken the number of available sequences in databases to unprecedented levels (1). Unfortunately, the ability to determine the sequence of a particular gene has not been accompanied by an equally impressive gain in our ability to achieve insights into the biological function (including molecular and celullar) of these sequences. For example, the full genome sequence of the yeast Saccharomyces cerevisae became available in 1997 (2); nevertheless more than a decade later, of the 6000+ identified genes there are still over 1000 with uncharacterized function (3). In human, more than half of the genes are functionally characterized incompletely or not at all.
The classic route to functional characterization involving experimental methods from the genetic and biochemical toolbox-like specific knockouts, targeted mutations and a battery of biochemical assays is time consuming (depending on the model organism, it can take years) and costly. Therefore, there is a strong case for using in silico methods in a preliminary analysis for functional hypothesis generation to direct experimental planning in the laboratory.
There are literally hundreds of prediction algorithms described in the literature, although only some of those have a sensitivity and selectivity to be applicable for unsupervised function prediction of arbitrary query protein sequences (4). Each method concentrates on some specific structural or functional aspect of a sequence, e.g. the distribution of unstructured regions (5), its amino acid compositional particularities in sequence windows (6) or the existence of globular domains (7,8). The input formats, method of program invocation as well as the result presentation vary widely making it difficult to interconnect results and obtain an integrated picture of a possible functional assignment. Even when concentrating on a smaller set of reliable prediction methods, the results can still easily exceed several Megabytes of textual (ASCII-type) information, integration of which into an overall functional prediction can be a formidable task requiring days of work per sequence. The need for standardizing automated annotations as well as assessing their quality has been recognized by initiatives such as AFP (9).
There have been several attempts to address the interoperability problem (10–12). JAFA (13) is an example of an annotation meta-server that sends a query sequence to several function prediction servers and displays the overlap in Gene Ontology terms (14) as well as providing links to the original results. The ProFunc server (15) combines a range of methods for sequence analysis but requires the 3D structure of the query to be known in advance. There are also a number of databases that provide sequence annotations from various sources like UniProtKB (16) or Ensembl (17) as well as some services that predict a limited set of features for a given input sequence such as SMART (8), InterProScan (18,19) or TarO (20). It should be noted that, frequently, database annotations contain errors and, especially, function descriptions propagated by sequence similarity criteria might be dubious. Therefore, tools for de novo sequence annotation are important for reducing the dependence on potentially misleading or incomplete database comments (21,22).
ANNIE is unique in that it has been developed by a collaboration of sequence analysis as well as computer science experts. It provides over 20 of the most useful algorithms (Table 1) covering the first two steps of segment-based sequence analysis (23) that have proven indispensable in daily sequence analytic work for functional discovery (24–26). Of particular value is the inclusion of predictors for a number of post-translational modifications (27–32) as well as targeting signals (33,34) developed in-house.
Table 1.
Sequence analytic algorithms
| Algorithm | Description | Parameters |
|---|---|---|
| CAST (37) | Algorithm for low-complexity region (LCR) detection and selective masking | Threshold: 40 |
| IUPred (5) | Prediction method for recognizing ordered and intrinsically unstructured/disordered regions in proteins | Prediction type: long disorder |
| SAPS (6) | Statistical analysis of protein sequences with respect to amino acid composition and simple sequence motifs | n/a |
| SEG (38) | Prediction of low complexity regions | Three parameter sets: Window-size 12, Locut 2.2, Hicut 2.5 Window-size 25, Locut 3.0, Hicut 3.3 Window-size 45, Locut 3.4, Hicut 3.75 |
| Big-∏ (27–29) | Prediction of protein GPI lipid anchor cleavage sites | Taxon-specific learning set |
| NMT (30,31) | Prediction of N-terminal N-myristoylation of proteins | Taxon-specific parameter set |
| PrePS – FT (32) | Farnesylation prediction | n/a |
| PrePS – GGT1 (32) | Geranylgeranylation prediction | n/a |
| PrePS – GGT2 (32) | Rab geranylgeranylation Prediction | n/a |
| PeroPS/PTS1 (33,34) | Prediction of peroxisomal targeting signal 1 | Taxon-specific prediction function |
| DAS-TMfilter (39) | Prediction of transmembrane regions | Quality cutoff: 0.72 |
| HMMTOP (40) | Transmembrane topology prediction using Hidden Markov models | n/a |
| PHOBIUS (41) | Combined transmembrane topology and signal peptide predictor | n/a |
| TMHMM (42) | Transmembrane helix predictor | n/a |
| IMP-COIL (43) | Prediction of coiled-coil regions, modified implementation of the algorithm Lupas et al. by F. Eisenhaber | n/a |
| PROSITE (44) | Pattern search in the PROSITE database | n/a |
| PROSITE-Profile (44) | Profile search in the PROSITE database | n/a |
| HMMER (7) | Profile Hidden Markov Models | SMART (8) with e-value cutoff of 0.001 |
| IMPALA (45) | Tool to compare a query sequence against a library of position-specific scoring matrices | Wolf-library (e-value cutoff: 0.001) (46), Aravind-library (e-value cutoff: 1e-5) (47) |
| RPS-BLAST against CDD (48) | Reverse-position-specific BLAST against the Conserved Domain Database (CDD) | e-value cutoff: 0.001 |
The results of all algorithms are displayed in an integrated manner using a newly developed interactive sequence viewer as well as a number of views highlighting the distribution of features across sets of sequences. ANNIE enables scientists to gain a quick overview of possible functional assignments in protein sequence sets.
METHODS
Algorithms
Segment-based sequence analysis (23) starts with the assumption that proteins are chains of functional units which can be analyzed independently. The overall function arises from the synthesis of the functions predicted for each individual module.
The procedure first uses algorithms for the detection of nonglobular regions, which are segments with a compositional bias or repetitive patterns that often represent linker regions, fibrillar segments, flexible binding sites or points of post-translational modifications (35). The subsequent step is to run algorithms for the identification of known globular domains. These domains are conserved within groups of homologous proteins and are often associated with enzymatic or ligand-binding function. In the last step, it is assumed that the remaining parts of the sequence represent yet uncharacterized globular domains that need to be characterized within the homologous family concept. Iterative heuristic have to be applied to uncover weak links in sequence space and collect a family of protein sequence segments that contain yet unknown globular domains (36).
ANNIE provides a selection of algorithms covering the first two steps of this approach. Table 1 lists the algorithms which have been integrated together with a short description, references and the preselected runtime parameters. These parameters have been chosen so as to provide a reasonable compromise between the need to give a comprehensive and sensitive overview of sometimes weak signals and the ability of scientists not trained in sequence analysis to discard false positives. It should be noted that further relaxed parameterization might produce more prediction results; yet, their interpretation might require expert knowledge and experience. ANNIE is based on our extensive in-house sequence analytic pipeline ANNOTATOR, which is used to analyze proteomes and detect distant evolutionary relationships using computationally intensive iterative heuristics (36). The engine behind ANNIE has been in use for several years and has annotated millions of sequences. The online help pages contain a detailed description of each individual algorithm.
User-interface
There are two input methods allowing the user to either paste sequences in FASTA-format (a single sequence can also be pasted without a description line) or upload them from a corresponding FASTA-formatted file. There is currently a limit of 10 sequences per annotation run which might be increased in the future depending on actual usage patterns and the availability of compute server resources.
It is highly recommended to include taxonomic information in the classical NCBI square bracket notation at the end of the description line (e.g. [Homo sapiens]) in order for ANNIE to automatically choose the correct parameterization for predictors of post-translational modifications and targeting signals. Additionally, this will enable the user to view the taxonomic distribution of the uploaded sequence set.
The annotation process is started by pressing the corresponding ‘Annotate’ button. Requests are queued and, upon availability of resources, sent to a cluster of dedicated CPUs for execution of algorithms and parsing of output. The user will be directed to a page containing the current as well as past results. If an (optional) email address is provided, a message containing a link will be sent once all algorithms have completed. This gives the user access to past annotations for at least 72 h, after which they will be deleted.
There are a number of views that allow the user to look at different aspects of the annotation. Upon submission of an annotation request the user will normally click on the corresponding result folder and be presented with a view displaying the uploaded sequences with links to individual results. If a certain algorithm is still queued or running a special symbol will be displayed and the page reloads periodically until all algorithms have terminated (under average load this should take no more than 1 min).
Result view
Following the links for individual algorithms will display the corresponding result together with links to external resources where applicable (e.g. domain descriptions for HMMER). Each result also provides access for validation purposes to the ‘raw’ unparsed data generated by the executable.
Interactive sequence view
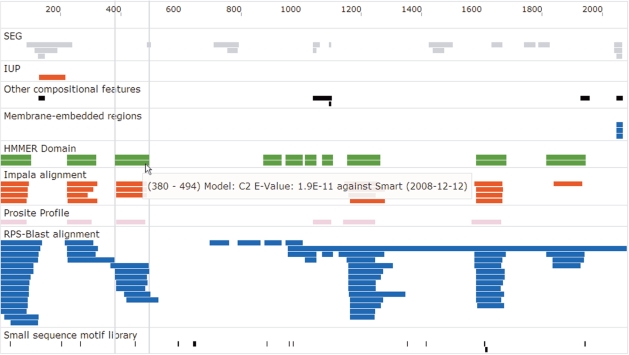
Clicking on the protein sequence symbol starts the interactive sequence view (Figure 1). The results of individual algorithms are displayed as rectangles projected onto the sequence ruler. Hovering over regions will display information specific to the result (e.g. e-values of globular domain model hits). Right-clicking on a region will allow examination of the particular feature in greater detail with algorithm-specific information as well as a compositional analysis of the sequence stretch.
Figure 1.
Interactive sequence view. This figure shows an exemplary interactive sequence view using the sequence of Dysferlin. The sequence features found by the various programs are organized in panes that coalesce findings with similar functional significance. The different color coding is just for the purpose of easing navigation.
Figure 1 displays the interactive sequence view of Dysferlin (49,50), a protein involved in a number of hereditary myopathies (it is provided as a sample sequence on the main page). The characteristic C2-domains (51) have been detected by a number of distinct tools (HMMER against Smart, IMPALA against Wolf-Library, PROSITE-Profile search, RPS-Blast against CDD) giving enhanced confidence to that particular finding. The detection of a C-terminal membrane-embedded region by three different methods also lends plausibility to the claim that Dysferlin is a transmembrane protein. It should be noted that there is a seventh C2-domain not shown in this view between residues 1338 and 1437 (the e-value = 0.025 is above the default threshold of 0.001),
Due to the AJAX-based technology of the viewer, zooming and panning is almost instantaneous, allowing fast and concise drill-down to a particular region. Additional feature-specific information can be obtained by right-clicking on a region. This will lead to a detailed compositional analysis of the sequence stretch and, were applicable, include alignment data as well as links to external resources.
Set view
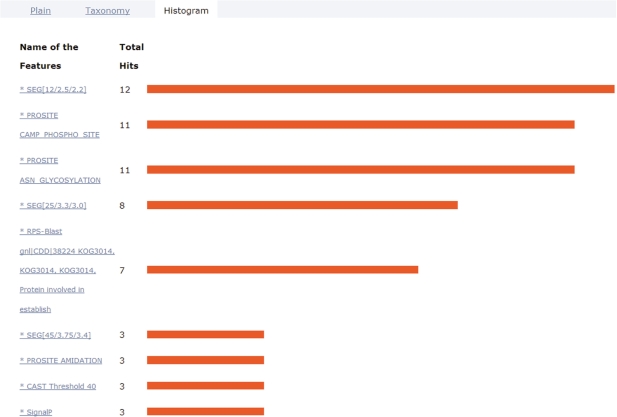
Uploading several sequences at once opens up the possibility to analyze the frequency of certain features within that set of sequences. ANNIE provides a special view called ‘Histogram’ (Figure 2). This view displays features found with diverse algorithms sorted by the number of occurrences. Clicking on the name of the feature will link to all the sequences in which it has been detected.
Figure 2.
Histogram view. This view shows the occurrence of sequence features in the sequence set under investigation. The features are sorted by their number of incidences in the set. Clicking on the link provided with the feature name will generate the sublist of sequences with this feature. In this example of Eco1-type proteins, the top four entries in the histogram are related to low-complexity regions as well as short motifs from PROSITE that are less reliable predictions. The fifth entry indicates the occurrence of the KOG3014 domain model that is characteristic for the Eco1-class of proteins necessary for the establishment of sister chromatid cohesion in mitosis.
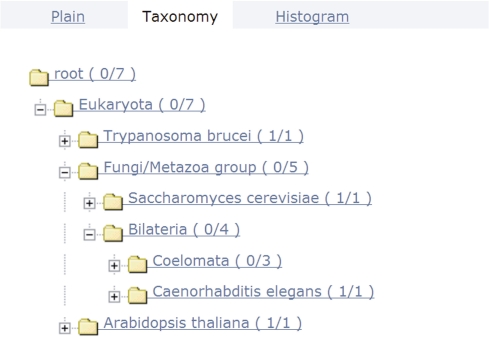
A third view called ‘Taxonomy’ (Figure 3) shows the taxonomic distribution of sequences within the set.
Figure 3.
Taxonomy view. The taxonomic distribution of the sequence set is displayed. The numbers in brackets refer to the number of sequences below a branch in the taxonomic tree and those assigned to a particular taxon. For the given Eco1 example set, this view shows that it contains one plant sequence (Arabidopsis thaliana) together with a trypanosome, one fungal sequence and four from Bilateria.
CONCLUSIONS AND OUTLOOK
We have presented ANNIE, a comprehensive de novo protein annotation system that integrates a large number of indispensable algorithms used in everyday sequence analytic work. The results of individual algorithms can be accessed separately or displayed together in an interactive AJAX-based sequence viewer. There are additional views for assessing the frequency of certain features across a set of sequences as well as revealing its taxonomic distribution.
New algorithms appearing in the literature are constantly being evaluated as to their potential contribution for function discovery and are eventually integrated. Future work will also see the inclusion of algorithms from the third step of segment-based sequence analysis if the necessary computational resources can be obtained.
FUNDING
Bioinformatics Insitute, A*Star, Singapore; Boehringer Ingelheim (2001-2007); and the Austrian Gen-AU BIN program (2004-2007) when the Eisenhaber group was located at the Research Institute of Molecular Pathology in Vienna (Austria). Funding for open access charge: Bioinformatics Institute, A*Star, Singapore.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank all colleagues that have contributed at various stages to the development of the software for protein sequence studies in the Eisenhaber group especially Georg Kraml, Miklos Kozlovszky, Florian Leitner, Georg Neuberger, Maria Novatchkova, Gernot Stocker, James Tay, Sun Tian and Wong Chee Hong.
REFERENCES
- 1.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cherry JM, Ball C, Weng S, Juvik G, Schmidt R, Adler C, Dunn B, Dwight S, Riles L, Mortimer RK, et al. Genetic and physical maps of Saccharomyces cerevisiae. Nature. 1997;387:67–73. [PMC free article] [PubMed] [Google Scholar]
- 3.Peña-Castillo L, Hughes TR. Why are there still over 1000 uncharacterized yeast genes? Genetics. 2007;176:7–14. doi: 10.1534/genetics.107.074468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponting CP. Issues in predicting protein function from sequence. Brief Bioinform. 2001;2:19–29. doi: 10.1093/bib/2.1.19. [DOI] [PubMed] [Google Scholar]
- 5.Dosztányi Z, Csizmók V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 6.Brendel V, Bucher P, Nourbakhsh IR, Blaisdell BE, Karlin S. Methods and algorithms for statistical analysis of protein sequences. Proc. Natl Acad. Sci. USA. 1992;89:2002–2006. doi: 10.1073/pnas.89.6.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 8.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodrigues APC, Grant BJ, Godzik A, Friedberg I. The 2006 automated function prediction meeting. BMC Bioinformatics. 2007;8 (Suppl. 4):S1–S4. doi: 10.1186/1471-2105-8-S4-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Letondal C. A web interface generator for molecular biology programs in Unix. Bioinformatics. 2001;17:73–82. doi: 10.1093/bioinformatics/17.1.73. [DOI] [PubMed] [Google Scholar]
- 11.Hull D, Wolstencroft K, Stevens R, Goble C, Pocock MR, Li P, Oinn T. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–W732. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilkinson MD, Links M. BioMOBY: an open source biological web services proposal. Brief Bioinform. 2002;3:331–341. doi: 10.1093/bib/3.4.331. [DOI] [PubMed] [Google Scholar]
- 13.Friedberg I, Harder T, Godzik A. JAFA: a protein function annotation meta-server. Nucleic Acids Res. 2006;34:W379–W381. doi: 10.1093/nar/gkl045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33:W89–W93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. UniProtKB/Swiss-Prot. Methods Mol. Biol. 2007;406:89–112. doi: 10.1007/978-1-59745-535-0_4. [DOI] [PubMed] [Google Scholar]
- 17.Hubbard TJP, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulder N, Apweiler R. InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol Biol. 2007;396:59–70. doi: 10.1007/978-1-59745-515-2_5. [DOI] [PubMed] [Google Scholar]
- 19.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Overton IM, van Niekerk CAJ, Carter LG, Dawson A, Martin DMA, Cameron S, McMahon SA, White MF, Hunter WN, Naismith JH, et al. TarO: a target optimisation system for structural biology. Nucleic Acids Res. 2008;36:W190–W196. doi: 10.1093/nar/gkn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilks WR, Audit B, De Angelis D, Tsoka S, Ouzounis CA. Modeling the percolation of annotation errors in a database of protein sequences. Bioinformatics. 2002;18:1641–1649. doi: 10.1093/bioinformatics/18.12.1641. [DOI] [PubMed] [Google Scholar]
- 22.Gilks WR, Audit B, de Angelis D, Tsoka S, Ouzounis CA. Percolation of annotation errors through hierarchically structured protein sequence databases. Math. Biosci. 2005;193:223–234. doi: 10.1016/j.mbs.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Eisenhaber F. Discovering Biomolecular Mechanisms with Computational Biology. Springer US; 2006. Prediction of protein function; pp. 39–54. [Google Scholar]
- 24.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 25.Ivanov D, Schleiffer A, Eisenhaber F, Mechtler K, Haering CH, Nasmyth K. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr. Biol. 2002;12:323–328. doi: 10.1016/s0960-9822(02)00681-4. [DOI] [PubMed] [Google Scholar]
- 26.Schleiffer A, Kaitna S, Maurer-Stroh S, Glotzer M, Nasmyth K, Eisenhaber F. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol. Cell. 2003;11:571–575. doi: 10.1016/s1097-2765(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 27.Eisenhaber B, Bork P, Eisenhaber F. Prediction of potential GPI-modification sites in proprotein sequences. J. Mol. Biol. 1999;292:741–758. doi: 10.1006/jmbi.1999.3069. [DOI] [PubMed] [Google Scholar]
- 28.Eisenhaber B, Wildpaner M, Schultz CJ, Borner GHH, Dupree P, Eisenhaber F. Glycosylphosphatidylinositol lipid anchoring of plant proteins. Sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiol. 2003;133:1691–1701. doi: 10.1104/pp.103.023580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eisenhaber B, Schneider G, Wildpaner M, Eisenhaber F. A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome-wide studies for Aspergillus nidulans, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae and Schizosaccharomyces pombe. J. Mol. Biol. 2004;337:243–253. doi: 10.1016/j.jmb.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 30.Maurer-Stroh S, Eisenhaber B, Eisenhaber F. N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. J. Mol. Biol. 2002;317:541–557. doi: 10.1006/jmbi.2002.5426. [DOI] [PubMed] [Google Scholar]
- 31.Maurer-Stroh S, Eisenhaber B, Eisenhaber F. N-terminal N-myristoylation of proteins: refinement of the sequence motif and its taxon-specific differences. J. Mol. Biol. 2002;317:523–540. doi: 10.1006/jmbi.2002.5425. [DOI] [PubMed] [Google Scholar]
- 32.Maurer-Stroh S, Eisenhaber F. Refinement and prediction of protein prenylation motifs. Genome Biol. 2005;6:R55. doi: 10.1186/gb-2005-6-6-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neuberger G, Maurer-Stroh S, Eisenhaber B, Hartig A, Eisenhaber F. Motif refinement of the peroxisomal targeting signal 1 and evaluation of taxon-specific differences. J. Mol. Biol. 2003;328:567–579. doi: 10.1016/s0022-2836(03)00318-8. [DOI] [PubMed] [Google Scholar]
- 34.Neuberger G, Maurer-Stroh S, Eisenhaber B, Hartig A, Eisenhaber F. Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence. J. Mol. Biol. 2003;328:581–592. doi: 10.1016/s0022-2836(03)00319-x. [DOI] [PubMed] [Google Scholar]
- 35.Eisenhaber B, Eisenhaber F. Posttranslational modifications and subcellular localization signals: indicators of sequence regions without inherent 3D structure? Curr. Protein Pept. Sci. 2007;8:197–203. doi: 10.2174/138920307780363424. [DOI] [PubMed] [Google Scholar]
- 36.Schneider G, Neuberger G, Wildpaner M, Tian S, Berezovsky I, Eisenhaber F. Application of a sensitive collection heuristic for very large protein families: evolutionary relationship between adipose triglyceride lipase (ATGL) and classic mammalian lipases. BMC Bioinformatics. 2006;7:164. doi: 10.1186/1471-2105-7-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Promponas VJ, Enright AJ, Tsoka S, Kreil DP, Leroy C, Hamodrakas S, Sander C, Ouzounis CA. CAST: an iterative algorithm for the complexity analysis of sequence tracts. Complexity analysis of sequence tracts. Bioinformatics. 2000;16:915–922. doi: 10.1093/bioinformatics/16.10.915. [DOI] [PubMed] [Google Scholar]
- 38.Wootton JC. Non-globular domains in protein sequences: automated segmentation using complexity measures. Comput. Chem. 1994;18:269–285. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
- 39.Cserzo M, Eisenhaber F, Eisenhaber B, Simon I. TM or not TM: transmembrane protein prediction with low false positive rate using DAS-TMfilter. Bioinformatics. 2004;20:136–137. doi: 10.1093/bioinformatics/btg394. [DOI] [PubMed] [Google Scholar]
- 40.Tusnády GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 41.Käll L, Krogh A, Sonnhammer ELL. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 42.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 43.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 44.Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJA. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schäffer AA, Wolf YI, Ponting CP, Koonin EV, Aravind L, Altschul SF. IMPALA: matching a protein sequence against a collection of PSI-BLAST-constructed position-specific score matrices. Bioinformatics. 1999;15:1000–1011. doi: 10.1093/bioinformatics/15.12.1000. [DOI] [PubMed] [Google Scholar]
- 46.Wolf YI, Brenner SE, Bash PA, Koonin EV. Distribution of protein folds in the three superkingdoms of life. Genome Res. 1999;9:17–26. [PubMed] [Google Scholar]
- 47.Chervitz SA, Aravind L, Sherlock G, Ball CA, Koonin EV, Dwight SS, Harris MA, Dolinski K, Mohr S, Smith T, et al. Comparison of the complete protein sets of worm and yeast: orthology and divergence. Science. 1998;282:2022–2028. doi: 10.1126/science.282.5396.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsuda C, Aoki M, Hayashi YK, Ho MF, Arahata K, Brown RH. Dysferlin is a surface membrane-associated protein that is absent in Miyoshi myopathy. Neurology. 1999;53:1119. doi: 10.1212/wnl.53.5.1119. [DOI] [PubMed] [Google Scholar]
- 50.Han R, Campbell KP. Dysferlin and muscle membrane repair. Curr. Opin. Cell Biol. 2007;19:409–416. doi: 10.1016/j.ceb.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nalefski EA, Falke JJ. The C2 domain calcium-binding motif: structural and functional diversity. Protein Sci. 1996;5:2375–2390. doi: 10.1002/pro.5560051201. [DOI] [PMC free article] [PubMed] [Google Scholar]