Abstract
Primers4clades is an easy-to-use web server that implements a fully automatic PCR primer design pipeline for cross-species amplification of novel sequences from metagenomic DNA, or from uncharacterized organisms, belonging to user-specified phylogenetic clades or taxa. The server takes a set of non-aligned protein coding genes, with or without introns, aligns them and computes a neighbor-joining tree, which is displayed on screen for easy selection of species or sequence clusters to design lineage-specific PCR primers. Primers4clades implements an extended CODEHOP primer design strategy based on both DNA and protein multiple sequence alignments. It evaluates several thermodynamic properties of the oligonucleotide pairs, and computes the phylogenetic information content of the predicted amplicon sets from Shimodaira–Hasegawa-like branch support values of maximum likelihood phylogenies. A non-redundant set of primer formulations is returned, ranked according to their thermodynamic properties. An amplicon distribution map provides a convenient overview of the coverage of the target locus. Altogether these features greatly help the user in making an informed choice between alternative primer pair formulations. Primers4clades is available at two mirror sites: http://maya.ccg.unam.mx/primers4clades/and http://floresta.eead.csic.es/primers4clades/. Three demo data sets and a comprehensive documentation/tutorial page are provided for easy testing of the server's capabilities and interface.
INTRODUCTION
Polymerase chain reaction (PCR) remains the most widely used technology to gain molecular markers for molecular ecology and systematics studies. With the ongoing accumulation of fully sequenced genomes in public sequence databases, these research areas, including metagenomics, are increasingly focusing on the analysis of protein coding genes and sequences (CDSs) to understand ecological, metabolic and evolutionary processes in nature (1–3). This trend is reflected in the huge interest of studying the diversity and expression patterns of ‘functional genes’ in the environment, such as antibiotic resistance and virulence genes (4,5), photosynthesis (6) or nitrogen fixation genes (7), to mention a few. Furthermore, multilocus sequence analysis (MLSA) and typing (MLST) of protein-coding genes are the new standards in molecular systematics (8–10) and molecular epidemiology (11,12).
However, it still remains a major challenge to design optimal PCR primers to specifically amplify CDSs from target lineages directly from environmental DNA samples or from novel organisms. Here we introduce primers4clades, a publicly available and easy-to-use web server that uses phylogenetic trees for the targeted design of PCR primers for the above mentioned purposes. Our empirical validation studies have proven its utility to study diversity of protein-coding genes in complex metagenomic DNA samples, as well as from previously uncharacterized microorganisms.
COMPARISON WITH RELATED WEB TOOLS
Primers4clades implements an extended and fully automated CODEHOP (Consensus Degenerate Hybrid Oligonucleotide Primer) design strategy (9,10), based on both DNA and protein multiple sequence alignments of protein-coding sequences. The original CODEHOP (http://blocks.fhcrc.org/codehop.html) and the very recent iCODEHOP (https://icodehop.cphi.washington.edu/i-codehop-context/Welcome Analysis) servers both rely only on protein sequences and use a single codon usage table (CUT) out of a limited choice of CUTs to derive the primer formulations. Primers4clades automatically uses the CUTs for all species identified in the input data set for which a CUT is available at the Codon Usage Database (13), as well as an alignment-specific CUT computed on the fly. Furthermore, none of these servers allows the user to specify a desired amplicon size range, which is a convenient filter implemented in primers4clades.
The PrimaClade (14), Greene SCPrimer (15), PriFi(16) and GeneFisher-P (17) servers take any DNA multiple sequence alignment as input and implement different strategies to identify PCR primer-binding sites and degenerate primer formulations. The QPRIMER web server (18) generates ‘universal’ primers for conserved regions of vertebrate genomes, whereas the Muplex server (19) provides a service for the design of primers for multiplex PCR.
How does primers4clades fit in this context? To our knowledge, it is the only freely available web-based tool that uses phylogenetic trees to interactively target the search for oligonucleotide formulations to particular sequence clusters (Figure 1). A very useful feature of primers4clades is that it returns a non-redundant set of primer pair formulations, ranked according to their thermodynamic properties. Many of the related web servers return highly redundant oligonucleotide formulations for long and conserved sequence alignments. Primers4clades checks that the resulting amplicon sets for the primer pairs do not overlap more than 80%, ensuring a high coverage of the target locus, but filtering excessive redundancy, as shown on the amplicon distribution maps (Figure 2). Furthermore, the phylogenetic information content of the aligned amplicon sets each primer pair would theoretically amplify, given the input sequences, is also computed, which is a unique and valuable feature of primers4clades. Together, these features are very useful to make an informed choice among alternative, non-redundant primer pair formulations, considering both the thermodynamic properties of the primers and the phylogenetic information content of the expected amplicon sets.
Figure 1.
First output generated by primers4clades run in the interactive ‘cluster sequences’ mode using the provided alpha-proteobacterial atpD demo data set. (A) Provides a summary of the user-specified run parameters and alignment statistics for the input data set. (B) Shows the labeled NJ tree computed from the alignment of the input dataset. (C) Shows the cluster selection panel based on the labels shown on the NJ tree. Hitting the re-cluster button parses the alignment to use only the selected sequences. Hitting the get primers button starts the primer design and evaluation steps.
Figure 2.
Output summary of the primer design and evaluation steps. (A) Shows the positions of the different amplicon sets mapped on the first sequence of the input alignment at the protein level. The nonredundant codon usage tables used for primer design are shown. All primer pair formulations and their thermodynamic properties can be downloaded using the TAB link. (B) The CODEHOP (bold) and the three codon-based primer formulations returned by the system (only the forward formulation is shown), aligned with the underlying codons. An ‘!’ sign denotes positions corrected by the system based on the codon alignment. (C) Output summary for the (forward) primer thermodynamic and phylogenetic quality evaluations.
INPUT DATA AND THEIR PROCESSING BY THE PRIMERS4CLADES PIPELINE
Implementation, input data processing and run modes
Primers4clades was mainly written in Perl and uses several Bioperl modules (20) along with the open source software cited below to perform different computations.
The input for the server is a set of homologous protein-coding genes in FASTA format, which may be aligned or not, with or without introns. The server excises introns if their coordinates are indicated in the FASTA header (see the server's documentation and the fungal alpha-tubulins demo data set), collapses redundant sequences to haplotypes, translates the CDSs with user-selected translation tables and aligns them using Muscle (21). The alignment step is skipped if the server detects that the uploaded DNA sequences are previously aligned. The protein alignment is projected on the underlying DNA sequences to compute the corresponding codon alignment, along with maximum likelihood (ML) distance matrices from the protein (WAG+G) or the codon (HKY85+G) alignments, using Tree-Puzzle (22). The latter are used to compute and display a neighbor-joining (NJ) tree with ‘neighbor’ from the PHYLIP package (23). If the server is run in the ‘advanced’ and interactive ‘cluster sequences’ mode, the user can select a clade from the displayed NJ tree to target the primer design towards its sequence members (Figure 1). In the default, non-interactive ‘get primers’ run mode, all the uploaded sequences will be considered to compute the primer formulations.
THE EXTENDED CODEHOP ALGORITHM IMPLEMENTED IN PRIMERS4CLADES
Primers4clades implements an extended CODEHOP primer design strategy based on both DNA and protein multiple sequence alignments of the CDSs. The CODEHOP algorithm (24,25) is based on the identification of highly conserved regions within protein BLOCKS (26) and the use of a particular CUT and position specific scoring matrix to derive the CODEHOP formulation. The extensions included in primers4clades comprise: (i) the automatic evaluation of a non-redundant set of codon usage tables (nrCUTs) for all organisms recognised in the input file FASTA header, as well as the computation of an alignment-specific CUT (Figure 2A, server's documentation/tutorial page). (ii) In addition to the CODEHOP formulations derived from the nrCUTs, the server computes what we call a corrected CODEHOP in which the degeneracy level is corrected considering the target codon alignment. (iii) The server also computes a so-called relaxed corrected CODEHOP which has an extended degenerate region as compared to the corrected CODEHOP in case that the latter has a degeneracy level <24. (iv) A fourth, fully degenerated oligonucleotide formulation is also computed based on the codon alignment. (v) A comprehensive set of thermodynamic parameters is calculated for each oligonucleotide pair. (vi) The coordinates of each CODEHOP in a primer pair are used to extract the reference in-silico amplicon set out of the original protein and codon alignments for the evaluation of their phylogenetic information content and to display them on the amplicon distribution map, as shown in Figure 2 and explained below.
Return of sorted, non-redundant primer formulations and their interactive refinement
The first useful result displayed by the server is an amplicon distribution map, showing the positions of each theoretical amplicon set with respect to the first protein sequence translated from the original input data set (Figure 2A). As mentioned above, the primers4clades pipeline returns four alternative primer formulations, which are displayed on screen, aligned with the corresponding codon multiple sequence alignment, along with their degeneracy level and expected amplicon size for easy visual inspection of the results (Figure 2B and C; see the online documentation for detailed recommendations about which type of primer to choose in different scenarios). Additionally, the phylogenetic information content parameter computed for each of the predicted aligned amplicons is also displayed on screen (Figure 2C; more details in the server's documentation page).
The thermodynamic parameters of oligonucleotides and primer pairs (max. and min. Tm of the pool of degenerate primers found, their max and min hairpin loop formation-, cross-hybridization- and self-priming potential) are computed using functions from Amplicon (27). Relatively relaxed cut-off values are defined for these parameters (see Table 2 of the online documentation). If any of them is worse than the specified cut-off values, then a quality warning is signaled and displayed on screen. An arbitrary quality scale is defined based on these cut-off values, which decreases from 100% (no warnings) downwards (Figure 2A and C). A tab-formatted file containing all the computed thermodynamic properties for each primer pair can be downloaded from the server (Figure 2A).
The evaluation of the phylogenetic information content relies on computing the mean and median Shimodaira–Hasegawa-like branch support values of ML phylogenies estimated by PhyML (28,29), as described previously (10), either at the DNA or protein levels, under user-specified substitution models or matrices. This parameter essentially describes the level of resolution achieved by the tree computed from the current amplicon set alignment, ranging from 1 (best resolution) to 0. These ML trees can be visualized online and downloaded, along with the alignments of each amplicon set (Figure 2C).
In the ‘advanced’ interactive ‘cluster sequences’ mode, after a first set of oligonucleotides has been found, the user can further refine primer formulations by selecting particular sequences to be excluded by clicking on check boxes displayed along the reference NJ tree (see the online documentation).
In summary, a non-redundant set of primer pairs is returned to the user, sorted according to the ‘thermodynamic quality’ score, excluding pairs inferred from the same CUT that overlap more than 20% of the amplicon sequence, and filtered by the user-specified length range of the amplicons (Figure 2).
GENOME-SCALE BENCHMARK ANALYSIS OF PRIMERS4CLADES PERFORMANCE
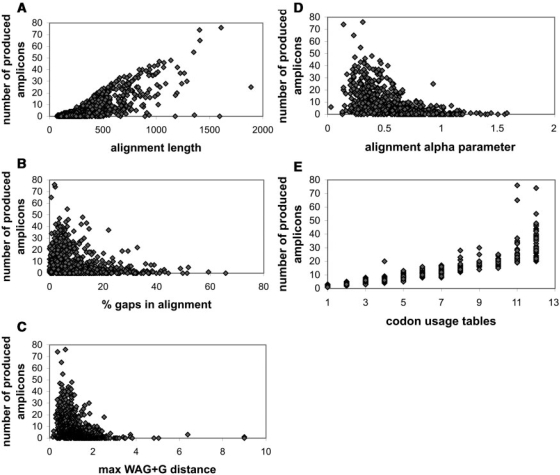
It is important to acknowledge key observations and parameters that affect the value of results generated by primers4clades. In order to identify those parameters and their critical cut-off values, we performed a genome-wide benchmark analysis using 983 orthologous gene families shared by 19 fully sequenced rhizobial genomes listed on the tree shown in Figure 8 of the server's documentation page, and identified as detailed therein. For this analysis we specifically tested the influence of the following parameters on the numbers of predicted primer pairs per locus: (i) protein-alignment length. (ii) Percentage of gaps in the alignment. (iii) Maximum WAG+G ML distance between pairs of sequences in a gene family multiple sequence alignment (at the protein level). (iv) Among site rate variation in the protein alignment, measured as a function of the alpha (shape) parameter of the gamma distribution, estimated under ML using the WAG+G model with 8 discrete rate categories. (v) Number of codon tables used per alignment.
The results of these analyses are summarized in Figure 3A–E, which demonstrate that the number of predicted primer pairs per locus increases linearly with the alignment length (Figure 3A) and with the number of codon usage tables (Figure 3E) analyzed, whereas a linear decrease in predicted primer pairs per locus is observed with an increasing percentage of gapped sites (Figure 3B). Interestingly, it was found that for alignments containing sequences with a WAG+G ML distance >2.5 (Figure 3C) the primers4clades pipeline will have a very low chance of finding suitable primer-binding sites. It is also noteworthy that an among-site rate variation level accommodated by an alpha value in the range of 0.3–0.6 is optimal (Figure 3D).
Figure 3.
Genome-scale benchmark analysis to test the influence of diverse alignment parameters on the numbers of predicted primer pairs obtained per locus. The analyses were performed on a set of 983 orthologous gene family alignments (at the protein level) for 19 Rhizobiales genomes. (A) Protein alignment length. (B) Percentage of gaps in the alignment. (C) Maximum WAG + G ML distance between pairs of sequences in a gene family multiple sequence alignment. (D) Among site rate variation in the protein alignment, measured as a function of the alpha (shape) parameter of the gamma distribution, estimated under ML using the WAG + G model with eight discrete rate categories. (E) Number of codon tables used per alignment.
EXPERIMENTAL VALIDATION EXAMPLES
As experimental validation examples we show the efficiency of our system to selectively amplify rpoB sequence fragments from environmental mycobacteria using as template metagenomic DNA extracted from three contrasting tropical and temperate soils, described in the online documentation page along with the primer formulations and details of the library construction procedure. Ten clones from each library were randomly chosen for sequencing. All sequences belonged to Actinobacteria, and over 90% of them clustered within the Mycobacterium clade as judged from a ML gene tree inferred from the sample and reference sequences downloaded from the Integrated Microbial Genomes site, and shown in Figure 11 of the server's documentation page. Furthermore, the environmental Mycobacterium rpoB sequences clustered within both the fast- and slow-growing clades of mycobacteria, demonstrating the utility of the primers4clades primer design pipeline to develop clade-specific oligonucleotides for metagenomic and microbial ecology studies. Large scale sequencing and analysis of the libraries will be reported elsewhere. We also show the amplification results of dnaE, fusA, lon, pheS and rpoB fragments from a diverse world-wide collection of 28 Bradyrhizobium strains (10) for which these loci had not previously been studied in a molecular phylogenetic context.Figure 12 and Table 3 of the documentation/tutorial page show the amplification results and the primer formulations with associated thermodynamic parameters, respectively. Figure 13 shows a Bayesian phylogeny estimated from the five new molecular markers using partition-specific best-fitting substitution models. The high overall tree resolution (most bipartitions have a posterior probability = 1) reflects the high phylogenetic information content of the markers, even though they are relatively short amplicons, as shown in Table 3 of the online documentation.
CONCLUSIONS AND FUTURE DEVELOPMENT
Primers4clades is currently the only publicly available server that integrates alternative primer-design strategies with phylogenetic trees to interactively target the search for oligonucleotide formulations to specific sequence clusters, and to evaluate the phylogenetic information content of the new molecular markers. These attributes make of primers4clades a novel and useful tool for the targeted design and informed selection of PCR primers for metagenomic and diversity studies, as demonstrated by our experimental validation studies. The development of the tool is now coupled to its recent implementation in a phylogenomics analysis pipeline to construct an interactive primer database for phylogenetic clades at different taxonomic and phylogenetic depths. The graphical interface, analysis options and parameter evaluation procedures will be improved, extended and refined.
FUNDING
DGAPA/PAPIIT-UNAM (IN201806-2); CONACyT-Mexico (P1-60071); Consejo Superior de Investigaciones Científicas (200720I038). Funding for open access charge: Universidad Nacional Autónoma de México.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to thank Dr David Romero (CCG-UNAM) for his valuable comments on the manuscript. Romualdo Zayas-Laguna and Víctor del Moral are acknowledged for their technical help.
REFERENCES
- 1.Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, Delong EF. Microbial community gene expression in ocean surface waters. Proc. Natl Acad. Sci. USA. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth's biogeochemical cycles. Science. 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 3.Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science. 2008;320:1081–1085. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- 4.Castiglioni S, Pomati F, Miller K, Burns BP, Zuccato E, Calamari D, Neilan BA. Novel homologs of the multiple resistance regulator marA in antibiotic-contaminated environments. Water Res. 2008;42:4271–4280. doi: 10.1016/j.watres.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Manning SD, Motiwala AS, Springman AC, Qi W, Lacher DW, Ouellette LM, Mladonicky JM, Somsel P, Rudrik JT, Dietrich SE, et al. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl Acad. Sci. USA. 2008;105:4868–4873. doi: 10.1073/pnas.0710834105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yutin N, Suzuki MT, Beja O. Novel primers reveal wider diversity among marine aerobic anoxygenic phototrophs. Appl. Environ. Microbiol. 2005;71:8958–8962. doi: 10.1128/AEM.71.12.8958-8962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zehr JP, Jenkins BD, Short SM, Steward GF. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 2003;5:539–554. doi: 10.1046/j.1462-2920.2003.00451.x. [DOI] [PubMed] [Google Scholar]
- 8.Edwards SV. Is a new and general theory of molecular systematics emerging? Evolution. 2009;63:1–19. doi: 10.1111/j.1558-5646.2008.00549.x. [DOI] [PubMed] [Google Scholar]
- 9.Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, Stackebrandt E, Van de Peer Y, Vandamme P, Thompson FL, et al. Opinion: re-evaluating prokaryotic species. Nat. Rev. Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- 10.Vinuesa P, Rojas-Jimenez K, Contreras-Moreira B, Mahna SK, Prasad BN, Moe H, Selvaraju SB, Thierfelder H, Werner D. Multilocus sequence analysis for assessment of the biogeography and evolutionary genetics of four Bradyrhizobium species that nodulate soybeans on the Asiatic continent. Appl. Environ. Microbiol. 2008;74:6987–6996. doi: 10.1128/AEM.00875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metzker ML, Mindell DP, Liu XM, Ptak RG, Gibbs RA, Hillis DM. Molecular evidence of HIV-1 transmission in a criminal case. Proc. Natl Acad. Sci. USA. 2002;99:14292–14297. doi: 10.1073/pnas.222522599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maiden MC. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 2006;60:561–588. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gadberry MD, Malcomber ST, Doust AN, Kellogg EA. Primaclade–a flexible tool to find conserved PCR primers across multiple species. Bioinformatics. 2005;21:1263–1264. doi: 10.1093/bioinformatics/bti134. [DOI] [PubMed] [Google Scholar]
- 15.Jabado OJ, Palacios G, Kapoor V, Hui J, Renwick N, Zhai J, Briese T, Lipkin WI. Greene SCPrimer: a rapid comprehensive tool for designing degenerate primers from multiple sequence alignments. Nucleic Acids Res. 2006;34:6605–6611. doi: 10.1093/nar/gkl966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fredslund J, Schauser L, Madsen LH, Sandal N, Stougaard J. PriFi: using a multiple alignment of related sequences to find primers for amplification of homologs. Nucleic Acids Res. 2005;33:W516–W520. doi: 10.1093/nar/gki425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamprecht AL, Margaria T, Steffen B, Sczyrba A, Hartmeier S, Giegerich R. GeneFisher-P: variations of GeneFisher as processes in Bio-jETI. BMC Bioinformatics. 2008;9(Suppl. 4):S13. doi: 10.1186/1471-2105-9-S4-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim N, Lee C. QPRIMER: a quick web-based application for designing conserved PCR primers from multigenome alignments. Bioinformatics. 2007;23:2331–2333. doi: 10.1093/bioinformatics/btm343. [DOI] [PubMed] [Google Scholar]
- 19.Rachlin J, Ding C, Cantor C, Kasif S. MuPlex: multi-objective multiplex PCR assay design. Nucleic Acids Res. 2005;33:W544–W547. doi: 10.1093/nar/gki377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al. The Bioperl toolkit: perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 23.Felsenstein J. Phylogeny Inference Package v. 3.6 Distributed by the author. University of Washington: Department of Genetics; 2004. PHYLIP. Seattle. [Google Scholar]
- 24.Rose TM, Henikoff JG, Henikoff S. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 2003;31:3763–3766. doi: 10.1093/nar/gkg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henikoff JG, Henikoff S, Pietrokovski S. New features of the blocks database servers. Nucleic Acids Res. 1999;27:226–228. doi: 10.1093/nar/27.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarman SN. Amplicon: software for designing PCR primers on aligned DNA sequences. Bioinformatics. 2004;20:1644–1645. doi: 10.1093/bioinformatics/bth121. [DOI] [PubMed] [Google Scholar]
- 28.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 2006;55:539–352. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 29.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]