Abstract
Detection of structural motif of residues in protein structures allows identification of structural or functional similarity between proteins. In the field of protein engineering, structural motif identification is essential to select protein scaffolds on which a motif of residues can be transferred to design a new protein with a given function. We describe here the RASMOT-3D PRO webserver (http://biodev.extra.cea.fr/rasmot3d/) that performs a systematic search in 3D structures of protein for a set of residues exhibiting a particular topology. Comparison is based on Cα and Cβ atoms in two steps: inter-atomic distances and RMSD. RASMOT-3D PRO takes in input a PDB file containing the 3D coordinates of the searched motif and provides an interactive list of identified protein structures exhibiting residues of similar topology as the motif searched. Each solution can be graphically examined on the website. The topological search can be conducted in structures described in PDB files uploaded by the user or in those deposited in the PDB. This characteristic as well as the possibility to reject scaffolds sterically incompatible with the target, makes RASMOT-3D PRO a unique webtool in the field of protein engineering.
INTRODUCTION
Structural genomics projects have led to the exponential growth of the number of protein structures deposited in the Protein Data Bank (1), creating an urgent need for efficient bioinformatics tools to extract the considerable amount of information contained in this database. A large number of developed methods are based on global 3D structures similarities. These fold comparison methods often do not allow identifying similarities among functionally significant residues such as metal-binding sites, catalytic sites of enzymes or ‘hot spot’ residues (2) involved in protein–protein interactions. Indeed, proteins with the same fold or even homologous proteins can exhibit a variety of biochemical functions (3). Conversely, proteins with different folds can perform the same function with the same set of residues and a similar mechanism (4). Specific methods should then address this particular problem of identifying functional motif similarity among proteins of different folds. These methods have been used for instance to identify specific enzymatic activity (5), to design proteins ligands (6–8) and new enzymes (9–11).
Several webservers currently give access to 3D motifs based methods for protein structures analysis. MultiBind (12) recognizes 3D-binding patterns common to several protein structures submitted. The KFC server (13) predicts binding hot spots at a particular protein–protein interface. MegaMotifBase (14) provides a compilation of structural motifs identified in protein families that may permit the user to assign a particular protein to one of these families. Other webservers search for known motifs in protein structures. PAR-3D (15) uses 3D motifs to identify several different classes of proteases or metal-binding sites in a submitted protein structure. Superimpose (16) allows searching for a specific 3D motif in protein structure databases. The SPASM server (17) allows identification of 3D motif in a PDB derived database. However, none of these webservers are specifically dedicated to the identification of protein scaffolds to transfer residues for protein ligand design. Such a website would ideally include an extensive search in all structures deposited in the PDB including the many conformers deposited in each file of NMR structure. Another essential characteristic of 3D search methods dedicated to protein ligand design is to take into account the steric aspects of the interaction of the protein scaffold with the considered target. These characteristics could permit to extensively take part of the topological information contained in the PDB and to identify very rapidly good protein scaffold candidates.
Here we describe RASMOT-3D PRO, a webserver that permits to search in protein structures for residues exhibiting similar topology as a user-defined reference 3D motif. It can then be useful in the fields of function identification and protein design. It can also be used to identify any type of 3D specific arrangement of residues such as super-secondary structures or small domains. The webserver is freely accessible at http://biodev.extra.cea.fr/rasmot3d/.
IMPLEMENTATION
RASMOT-3D searches in protein structures for sets of residues in a topology similar to the motif given in input (hereby called reference motif). Each protein structure file is examined independently. Comparison is based on Cα and Cβ atoms exclusively and can be divided into two sequential steps: inter-atomic distances comparison and root mean square deviation (RMSD). We consider for illustration a reference motif R composed of n residues {r1, r2, …, rn} and an examined protein P of N residues {p1, p2, p3, …, pN).
(i) Inter-atomic distances comparison step is described in the following paragraph and a corresponding scheme is provided in Supplementary Data S1. The initial step consists in calculating the 2n(n−1) inter-atomic distances between all Cα and Cβ atoms of the residues composing the reference motif. Then, examined protein residues are combined sequentially, trying to form sets of residues S, composed of n residues {s1, s2, …, sn}, with Cα and Cβ inter-atomic distances similar to those calculated in the reference motif. Starting from two residues s1 = pi and s2 = pj, considered as equivalent to r1 and r2, distances between Cα and Cβ atoms of these two residues s1 and s2 are calculated. If one of these distances differs by more than the threshold (delta-dist) from the corresponding distance calculated in the (r1, r2) pair, residue pj in position s2 is rejected and a pj+1 is tested. Conversely, if all these distances differ by less than delta-dist, residue s3 = pk is added and distances between Cα and Cβ atoms of s3 with Cα and Cβ atoms of s1 and s2 are calculated and compared to the corresponding distances characterizing (r1, r2, r3) in the reference motif. Residues are added this way to the set. When a set of n residues {s1, s2, …, sn} satisfies all the inter-atomic distance restraints, the second topological filter RMSD is applied [see (ii)]. Then the following set (with a new residue in position sn) is tested. This method, which allows pruning of the search tree as early as possible, is applied until all combinations of examined protein residues have been tested.
(ii) Root mean square deviation (RMSD) filter is calculated on sets of residues that satisfy all inter-atomic distance restraints [see (i)]. Each of these set of residues S {s1, s2, …, sn} are superimposed onto the reference motif R {r1, r2, …, rn} by root mean square fitting of the Cα and Cβ atoms. Its coordinates are rotated and translated to minimize the RMSD on the Cα and Cβ atoms of S relative to R. After superimposition, the resulting RMSD value is compared to the threshold set in input (RMSD-max). The tested set of residues S {s1, s2, …, sn} is rejected if the RMSD is larger than this threshold.
(iii) In addition, an optional steric filter was implemented. When searching for a binding or a catalytic site, it can be useful to select only scaffolds allowing residues topologically equivalent to the reference motif to interact with a specific target T. Indeed, it is not uncommon that scaffolds selected via identification of a motif S, topologically similar to the reference motif R, possesses structural elements that preclude the binding to T due to steric hindrance. To address this specific problem, we implemented a steric score calculated for each protein P exhibiting a set of residues S that satisfies the RMSD criterion. When one has in hand, a structure of the reference protein containing motif R in interaction with the target T, the motif S in the identified protein scaffold P is superimposed on the reference motif R. Then, the inter-atomic distances are calculated between all the atoms of protein P and target T. If the distance between an atom of P and an atom of T is lower than the sum of their radii, the score is increased by a value taking into account the interpenetration distance and the distance of the atom of P to the main chain of this protein. This allows giving less weight to the steric clash involving side-chain atoms than those involving the main chain ones. Finally, if this score is larger than a threshold, the corresponding set of residues S is rejected. This threshold has been set empirically to allow for minor interpenetration. This is justified because protein and target are treated as rigid bodies by the program.
From the above description, it can be seen that the search method implemented in RASMOT-3D PRO shares some similarities with the SPASM program (18) but presents specific features dedicated to the identification of protein scaffolds onto which transfer functional motifs. One central feature of RASMOT-3D PRO is that the type of each residue in the selected motif can be different than the corresponding residue in the reference motif. This is possible without bias because the search is based on the Cα and Cβ atoms. The method treats protein chains in a single PDB file as independent structures. For NMR derived structures, all the models are evaluated. For each identified set of residues, only the model with the lowest RMSD and satisfying the steric criteria is presented in the results.
USING THE RASMOT-3D PRO WEBSERVER
Submitting a query
The RASMOT-3D PRO only requires the reference motif residues coordinates uploaded as a PDB file to be launched. Several other parameters are available but are optional or set to default values. They are divided into four subgroups:
Examined protein files
In this part, the user can determine the PDB files containing the coordinates of the proteins in which to search for the motif. Two options are available. The user can upload its own PDB files (up to 10) or search into one of the four NCBI non-redundant PDB chain sets (http://www.ncbi.nlm.nih.gov/Structure/VAST/nrpdb.html) obtained by clustering using four different sequence-similarity cutoffs (P-values of 10−7, 10−40, 10−80 and 100% identity).
Selection parameters
These parameters permit to set the threshold values described in the previous section: the maximal deviation for inter-atomic distances (delta-dist) and the maximal RMSD. They are set to default values but can be changed by the user. In addition, two pre-filters are available. (a) Motif search can be restricted to residues identical or with similar physical properties than their equivalent in the reference motif. (b) Examined scaffolds can be restricted to proteins with length size within defined limits.
Steric filter
If needed, the user can upload the coordinates in PDB format of a target positioned relative to the reference motif to eliminate identified scaffolds that make important steric clashes with this target. The steric score threshold is set to a fixed value determined empirically to give acceptable results.
Personal information
Before submitting, the user can optionally provide an e-mail address where a link to the results will be sent when the run is completed.
Choosing parameters
Calculation can take from few seconds for uploaded structures search to several hours for non-redundant PDB search. For the latter case, delta-dist thresholds must be chosen with caution. Large values for these parameters will increase exponentially the number of sets of residues to fit on the reference motif. Therefore, the computational time will increase dramatically. As the number of solutions reported is limited to the 250 lowest RMSD, there is no advantage to choose large delta-dist value. Thus, we suggest the users of the RASMOT-3D PRO to start their search with default parameters and to increase progressively the threshold if needed.
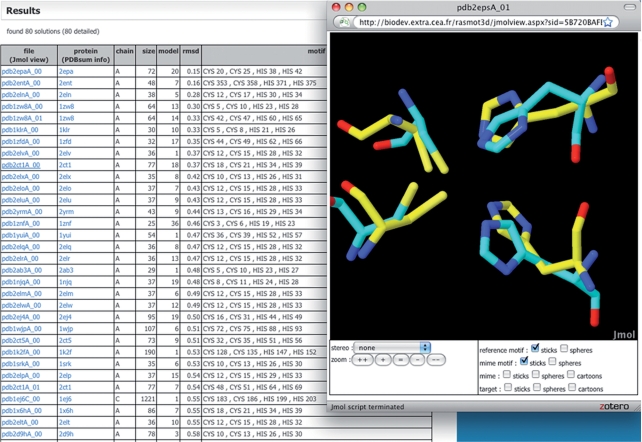
Viewing the results
Figure 1 shows an example of RASMOT-3D PRO results page. Solutions are sorted according to the motif RMSD. Only the 250 firsts scaffolds are displayed. For each solution, the data reported are: the PDB file name of the protein containing the set of residues of similar topology, the chain id, the size of this chain, the best model id for NMR derived structures, the RMSD, and the identity of the residues in the set identified. For known PDB file names, a link to the PDBsum (19) is provided. Finally, the scaffolds identified can be examined with the Jmol interactive online molecular viewer (http://www.jmol.org) without any plugin installation. Clicking on the name of the solution in the results table opens a window with the online molecular viewer. Opening it in a separate window allows simultaneous examination of several solutions that can therefore be easily compared. The reference motif given in input, the superimposed set of residues identified in the particular PDB file and the target, if provided, can be visualized. Reference motif is colored in cyan, identified residues and scaffold in yellow and target in grey. The user can choose which molecule or motif to display and select different representation modes.
Figure 1.
RASMOT-3D PRO output example: results for Cys-Cys-His-His zinc finger motif search into the non-redundant PDB chain set (P-value of 10–7).
When the search is conducted on one of the four NCBI non-redundant PDB chain sets, the online results pages are accessible via the URL sent by e-mail during 24 h. An archive can be downloaded from the server. It contains: (i) a file with the parameters used, (ii) a results file with one solution per line and fields separated by tabulations that can be easily imported in a spreadsheet program, and for each solution (iii) a PDB file containing the coordinates of the scaffold and (iv) a PyMol (http://www.pymol.org) visualization script file.
CASE STUDIES/DISCUSSION
Ligand design is still a considerable goal in biology with obvious applications in basic sciences, diagnosis and therapeutics, but it remains a challenging task. RASMOT-3D PRO was initially elaborated to identify platforms to transfer a functional motif by systematic examination of the structures deposited in the PDB. In a previous work (7) we used this approach to design a Kv1.2 potassium channel blocker and we obtained several micromolar blockers for this channel. With a similar method, using Cα and Cβ inter-atomic distances, RMSD and steric filtering, Liu and coworkers designed the pleckstrin homology domain PLCδ1-PH to bind the human erythropoietin receptor by grafting the key interacting residues of the human erythropoietin (8). These works clearly demonstrated the value of the approach to design protein ligands. However, one conclusion of our previous work (7) was that the success of the method depends on the number of identified scaffolds. Indeed, after topological in silico scaffold selection, several steps must be overcome. In particular, the designed molecule must be produced, folded and purified that, in some case, can be a very difficult or even an impossible task. Other very impressive works in the field of computational design of enzymes relying on the selection of scaffolds showed that, despite the sophistication of the model used, only a fraction of the designed enzymes displayed a significant activity (20). Consequently, in computational design methods relying on the identification of scaffolds, it is essential to analyze extensively the PDB to return a diversity of protein scaffolds thereby increasing the chance of success. As an illustration of the capacity of RASMOT-3D PRO to identify protein scaffolds by systematic examination of the PDB, we considered the work of Vita and coworkers that engineered a mini-protein binding the HIV-1 gp120 by transfer of a group of CD4-binding residues onto scyllatoxin (21). At the time of this work, the selection of the scaffold was made without the help of any bioinformatics means, but on a visual basis. Scyllatoxin was selected because it presented a β-hairpin motif similar to the CD4-binding region. We used RASMOT-3D PRO to search for scaffolds possessing a β-hairpin similar to that formed by residues 38–47 in CD4 making no major steric clash with the HIV-1 gp120 once the motifs are superimposed. We used the most non-redundant pdb chain set with P-value of 10–7, delta-dist of 1.5 Å and RMSD of 1.0 Å. CD4–gp120 complex coordinates were taken from PDB file 1g9n. We restricted the search to proteins smaller than CD4 (less than 180 residues). This search returned 23 proteins of different size and scaffold among which several scyllatoxin analogs (Table 1). We also identified scaffolds that better reproduce the R59 critical residue topology, as illustrated in Figure 2, which could be used as well to design mimetic protein of the CD4. A second example of RASMOT-3D PRO use illustrates its ability to identify proteins sharing a similar function relying on the presence of a conserved functional motif but located in very different structural contexts. We considered serine endopeptidases that include proteins with different folds (22), which are all characterized by the serine/histidine/aspartate catalytic triad. We then searched for this three residues motif, using target steric filtering and default parameters (in the most non-redundant protein structures database, with delta-dist and RMSD threshold both set to 0.8 Å). Coordinates of target and motif were extracted from beta-trypsin/BPTI complex described in PDB file 2PTC. We found 47 solutions, all of them being serine proteases from different organisms, with different folds (23). Figure 3 displays one example of two serine proteases identified by RASMOT-3D PRO, possessing the Ser/His/Asp motif supported by completely different architectures. RASMOT-3D PRO is then able to identify proteins sharing similar function on the basis of common 3D functional motif. These two examples show that the webserver RASMOT-3D PRO might give a very useful contribution in scaffold-based protein engineering and in protein function assignment.
Table 1.
Sorted solutions of the CD4 β-hairpin motif search in the non-redundant pdb chain set with RASMOT-3D PRO
| Name | Size | Description | RMSD | |
|---|---|---|---|---|
| 1 | 1cdy | 178 | CD4 mutant G47S | 0.52 |
| 2 | 2z59 | 109 | adrm1 | 0.55 |
| 3 | 1kla | 112 | tgf-b1 growth factor | 0.55 |
| 4 | 1mm0 | 36 | termicin antimicrobial peptide | 0.63 |
| 5 | 1v5r | 97 | gas2 domain of growth arrest protein 2 | 0.70 |
| 6 | 1ne5 | 42 | Herg specific scorpion toxin cnerg1 | 0.70 |
| 7 | 1sis | 35 | scorpion insectotoxin i5a | 0.70 |
| 8 | 2oox | 93 | transferase | 0.71 |
| 9 | 2hgc | 78 | unknown function | 0.71 |
| 10 | 3ca7 | 50 | EGF domain of Spitz | 0.72 |
| 11 | 1pnh | 31 | PO5-NH2 | 0.72 |
| 12 | 2k1n | 55 | abrB | 0.73 |
| 13 | 1rpy | 85 | dimeric sh2-signaling protein | 0.74 |
| 14 | 2ea9 | 103 | unknown function | 0.75 |
| 15 | 1du9 | 28 | BMP02 scorpion toxin | 0.77 |
| 16 | 2dir | 98 | THUMP domain RNA-binding protein | 0.79 |
| 17 | 2jna | 104 | unknown function | 0.79 |
| 18 | 2jtv | 65 | unknown function | 0.81 |
| 19 | 2qhd | 122 | ecarpholin | 0.84 |
| 20 | 1a96 | 150 | Xprtase | 0.87 |
| 21 | 2k5l | 81 | unknown function | 0.96 |
| 22 | 2k6z | 120 | unknown function | 0.96 |
| 23 | 1quz | 34 | scorpion toxin hstx1 | 1.00 |
The structure representative of the cluster of the scyllatoxin in the non-redundant pdb chain set is represented in italic.
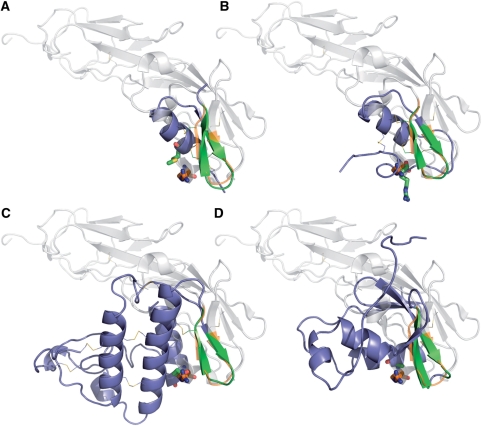
Figure 2.
Comparison of the superimposition of the CD4 with the scyllatoxin and three scaffolds identified by RASMOT-3D PRO. CD4 is colored in light grey with beta-hairpin motif and R59 in orange. The mimetic scaffolds are colored in blue with beta-hairpin motif and R59 equivalent residue in green. (A) scyllatoxin (1scy) identified by Vita et al. (21), (B) Cnerg1 (1ne5) another scorpion toxin, (C) ecarpholin (2 qhd) and (D) gas domain (1v5r) with an unrelated fold.
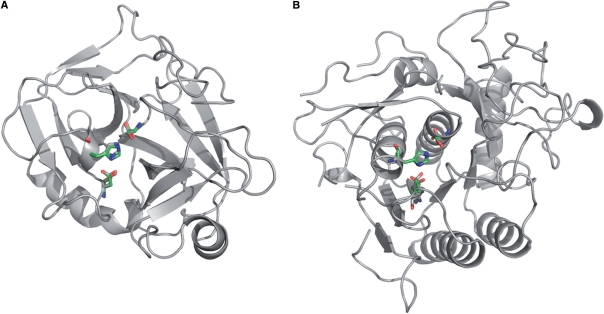
Figure 3.
Comparison of two different serine protease folds obtained by searching the Ser-His-Asp catalytic motif with RASMOT-3D PRO: (A) trypsin (1os8) and (B) sphericase. Reference motif is represented in green, identified scaffold in grey.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Commissariat à l'Energie Atomique, France.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 3.Todd AE, Orengo CA, Thornton JM. Evolution of protein function, from a structural perspective. Curr. Opin. Chem. Biol. 1999;3:548–556. doi: 10.1016/s1367-5931(99)00007-1. [DOI] [PubMed] [Google Scholar]
- 4.Hegyi H, Gerstein M. The relationship between protein structure and function: a comprehensive survey with application to the yeast genome. J. Mol. Biol. 1999;288:147–164. doi: 10.1006/jmbi.1999.2661. [DOI] [PubMed] [Google Scholar]
- 5.Torrance JW, Bartlett GJ, Porter CT, Thornton JM. Using a library of structural templates to recognize catalytic sites and explore their evolution in homologous families. J. Mol. Biol. 2005;347:565–581. doi: 10.1016/j.jmb.2005.01.044. [DOI] [PubMed] [Google Scholar]
- 6.Looger LL, Dwyer MA, Smith JJ, Hellinga HW. Computational design of receptor and sensor proteins with novel functions. 2003;423:185–190. doi: 10.1038/nature01556. [DOI] [PubMed] [Google Scholar]
- 7.Magis C, Gasparini D, Lecoq A, Le Du MH, Stura E, Charbonnier JB, Mourier G, Boulain JC, Pardo L, Caruana A, et al. Structure-based secondary structure-independent approach to design protein ligands: application to the design of Kv1.2 potassium channel blockers. J. Am. Chem. Soc. 2006;128:16190–16205. doi: 10.1021/ja0646491. [DOI] [PubMed] [Google Scholar]
- 8.Liu S, Liu S, Zhu X, Liang H, Cao A, Chang Z, Lai L. Nonnatural protein-protein interaction-pair design by key residues grafting. Proc. Natl Acad. Sci. USA. 2007;104:5330–5335. doi: 10.1073/pnas.0606198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellinga HW, Richards FM. Construction of new ligand binding sites in proteins of known structure. I. Computer-aided modeling of sites with pre-defined geometry. J. Mol. Biol. 1991;222:763–785. doi: 10.1016/0022-2836(91)90510-d. [DOI] [PubMed] [Google Scholar]
- 10.Zanghellini A, Jiang L, Wollacott AM, Cheng G, Meiler J, Althoff EA, Röthlisberger D, Baker D. New algorithms and an in silico benchmark for computational enzyme design. Protein Sci. 2006;15:2785–2794. doi: 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang L, Althoff EA, Clemente FR, Doyle L, Röthlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, 3rd, et al. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shulman-Peleg A, Shatsky M, Nussinov R, Wolfson HJ. MultiBind and MAPPIS: webservers for multiple alignment of protein 3D-binding sites and their interactions. Nucleic Acids Res. 2008;36:W260–W264. doi: 10.1093/nar/gkn185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darnell SJ, LeGault L, Mitchell JC. KFC Server: interactive forecasting of protein interaction hot spots. Nucleic Acids Res. 2008;36:W265–W269. doi: 10.1093/nar/gkn346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pugalenthi G, Suganthan PN, Sowdhamini R, Chakrabarti S. MegaMotifBase: a database of structural motifs in protein families and superfamilies. Nucleic Acids Res. 2008;36:D218–D221. doi: 10.1093/nar/gkm794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goyal K, Mohanty D, Mande SC. PAR-3D: a server to predict protein active site residues. Nucleic Acids Res. 2007;35:W503–W505. doi: 10.1093/nar/gkm252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bauer RA, Bourne PE, Formella A, Frömmel C, Gille C, Goede A, Guerler A, Hoppe A, Knapp EW, Pöschel T, et al. Superimpose: a 3D structural superposition server. Nucleic Acids Res. 2008;36:W47–W54. doi: 10.1093/nar/gkn285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madsen D, Kleywegt GT. Interactive motif and fold recognition in protein structures. J. Appl. Cryst. 2002;35:137–139. [Google Scholar]
- 18.Kleywegt GT. Recognition of spatial motifs in protein structures. J. Mol. Biol. 1999;285:1887–1897. doi: 10.1006/jmbi.1998.2393. [DOI] [PubMed] [Google Scholar]
- 19.Laskowski RA. PDBsum new things. Nucleic Acids Res. 2009;37:D355–D359. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Röthlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 21.Vita C, Drakopoulou E, Vizzavona J, Rochette S, Martin L, Ménez A, Roumestand C, Yang YS, Ylisastigui L, Benjouad A, et al. Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein. Proc. Natl Acad. Sci. USA. 1999;96:13091–13096. doi: 10.1073/pnas.96.23.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 23.Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2008;36:D320–D325. doi: 10.1093/nar/gkm954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.