Abstract
PCR amplification using COnsensus DEgenerate Hybrid Oligonucleotide Primers (CODEHOPs) has proven to be highly effective for identifying unknown pathogens and characterizing novel genes. We describe iCODEHOP; a new interactive web application that simplifies the process of designing and selecting CODEHOPs from multiply-aligned protein sequences. iCODEHOP intelligently guides the user through the degenerate primer design process including uploading sequences, creating a multiple alignment, deriving CODEHOPs and calculating their annealing temperatures. The user can quickly scan over an entire set of degenerate primers designed by the program to assess their relative quality and select individual primers for further analysis. The program displays phylogenetic information for input sequences and allows the user to easily design new primers from selected sequence sub-clades. It also allows the user to bias primer design to favor specific clades or sequences using sequence weights. iCODEHOP is freely available to all interested researchers at https://icodehop.cphi.washington.edu/i-codehop-context/Welcome.
INTRODUCTION
While databases containing nucleic and amino acid sequences continue to grow at an exponential rate, the data that they contain catalog only a fragment of the genetic content present in the millions of species on earth. A conservative estimate of global diversity anticipates some 3.5–10.5 million species (1) but, as of January 2009, only 200 000 species were represented in the organism taxonomy maintained by National Center for Biotechnology Information (2). Moreover, pathogen recombination events ensure that no sequence data repository could ever house a complete representation of nature's diversity. Yet, basic and clinical scientists require tools that help them characterize and study the structure, function and evolution of genes and proteins from widely disparate organisms. This article discusses such a tool; a web application called iCODEHOP that can design PCR primers capable of amplifying distantly related genes.
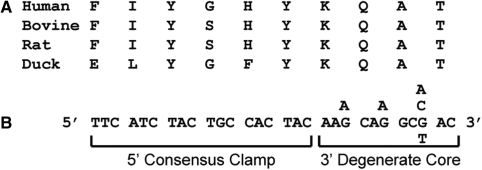
We have previously described a method for the prediction and use of COnsensus-DEgenerate Hybrid Oligonucleotide Primers (CODEHOPs) for the amplification of unknown genes (3). A CODEHOP is a hybrid primer consisting of a degenerate ‘core’ and non-degenerate ‘clamp’ region (Figure 1). The core region resides on the 3′-end of the primer and consists of the nucleotide sequences providing all possible codons for a highly conserved amino acid motif of three to four residues identified in a protein multiple alignment. The nondegenerate clamp region contains the most common nucleotide in each position of the codons for five to seven amino acid positions immediately adjacent to the conserved motif. This region is typically between 15 and 20 bases and its length can be adjusted by the user. The 3′-degenerate structure of CODEHOPs allows the PCR amplification to have a broad specificity for distantly related target gene templates, while the 5′-consensus clamp allows for a robust amplification from PCR product templates during the later cycles of the amplification reaction.
Figure 1.
A CODEHOP is a hybrid primer consisting of a 3′-degenerate ‘core’ and 5′-non-degenerate ‘clamp’ region. The DNA sequence of a CODEHOP and the amino acid multiple alignment it was derived from are shown.
The CODEHOP approach has been used to identify and characterize a number of novel genes and identify unknown pathogens. It has been especially useful in situations involving distantly related targets and limited template in complex mixtures (4–8). We previously developed a program to predict CODEHOPs from multiply aligned protein sequences and provided a Web interface for the scientific community (3,9). Several publications mention the successful application of CODEHOPs designed by this program (See: http://courses.washington.edu/bioinfo/CODEHOP/Codehop%20Genes.html) and log files indicate that this program has designed CODEHOPs for many hundreds of users since its release. However, its user interface was not optimal and its output was of limited use for primer selection and assay development. To address these issues, and provide new functions for primer design and selection, we have developed a new, interactive, web application called iCODEHOP.
WEB APPLICATION DESCRIPTION
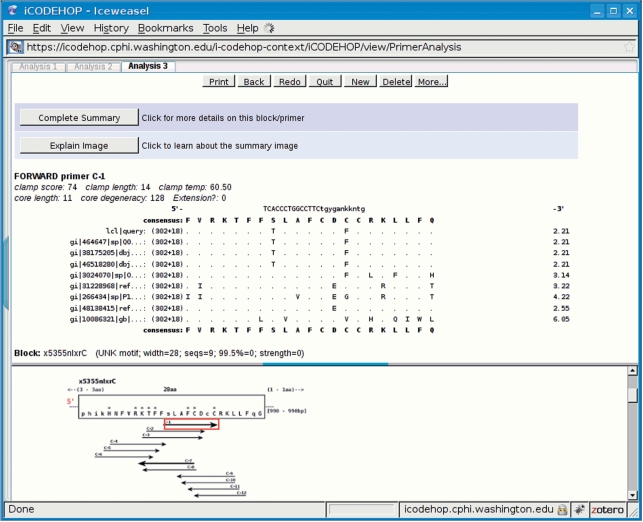
iCODEHOP is an interactive web application that intelligently guides users through the degenerate primer design process including uploading sequences, creating a gapped multiple alignment, converting the gapped alignment to an ungapped multiple alignment (a block), deriving CODEHOPs and calculating the range of annealing temperatures possible for degenerate primer pools. The input to iCODEHOP can be nonaligned protein sequences, CLUSTAL alignments, or an ungapped multiple alignment in BLOCKS format. Protein sequences can be specified by UniProt or GenBank accession number or by uploading a FASTA or GenBank formatted text file. The program's output includes a set of CODEHOPs and a variety of meta-data about each primer pool including its length, degeneracy and annealing temperature range. iCODEHOP enables users to quickly scan over an entire set of degenerate primers produced by the program to assess their relative quality and select individual degenerate primers for further analysis (Figures 2 and 3). The program also automatically generates and displays phylogenetic information for sub-selections of input sequences and allows users to easily design new degenerate primers or bias primer design towards sequences of their choice (Figure 4).
Figure 2.
A sample ‘CODEHOP summary page’ produced by iCODEHOP after creating CODEHOPs from a specific protein multiple alignment. The top pane of the split display shows summary details about a specific primer that a user has selected in the bottom pane using a mouse. The user can see more data on the selected primer, including all potential partner CODEHOPs, by clicking on the button titled ‘Complete Summary’.
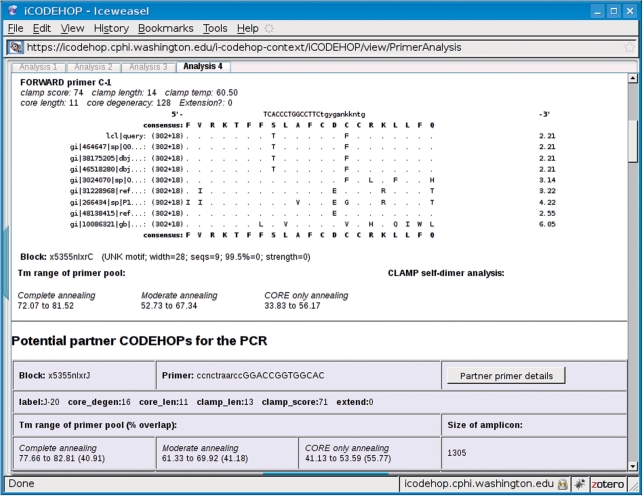
Figure 3.
The output of iCODEHOP after a user requests to view detailed information on a specific CODEHOP from the CODEHOP summary page shown in Figure 2. The program presents a number of details for the selected primer including its range of annealing temperatures at three levels of mismatch with the unknown target sequence, potential partner CODEHOPs and estimated amplicon length.
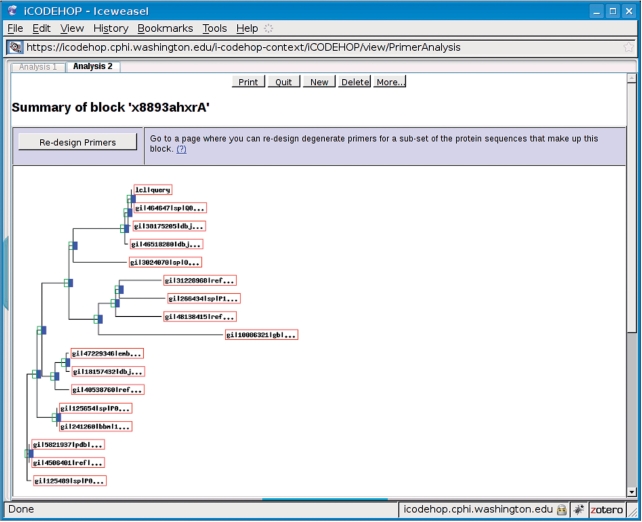
Figure 4.
In this figure, the user has chosen to view the details of a block for which no primer could be found by the program. The phylogram indicates that sequence homology falls into distinct clades along this block. The user can select which sequences to include in the next attempt to generate primers by selecting ‘Re-design Primers’.
The interested user can access the iCODEHOP web application for free and with no login requirement from the Internet Explorer (versions ≥ 7.0) and Firefox (versions ≥ 2.5) web browsers. A typical user would access the application intermittently to identify degenerate primers for a particular target gene family, carry out work in the lab using the primers, and then reuse the tool depending on the utility of the initial primer pair. Upon arriving at the application's home page for the first time, a user has the choice of running the iCODEHOP application from either an ‘anonymous’ or ‘named’ session. If the user selects to use a named session, the application will store data that the user creates while running the program for a short period of time (currently 72 h). This option ensures that a user can easily recover her/his data if something happens that disrupts the connection between the application and the user's web browser. Named sessions also facilitate remote collaboration by allowing multiple users at distinct locations to view or extend an analysis started by a single user. These features are not available should the user choose to run an anonymous session. Both session options provide the user with the option to download a file containing her/his data when they exit the program. This file can be reloaded into a new named session at a later time and enables the user to archive and share her/his analysis.
After a user starts a session, the iCODEHOP application initializes itself in a new window and provides the option to enter a degenerate primer design workflow or run other bioinformatics programs related to degenerate primer design. When a user selects to enter the degenerate primer design workflow, the program provides a form that (s)he can use to upload sequence data and choose which data to use for designing CODEHOPs. The user has a great deal of flexibility in selecting sequences and can choose:
one or more nonaligned sequences,
a single multiple alignment,
any combination of sequences from one or more multiple alignments,
any combination of nonaligned sequences and sequences from one or more multiple alignments and
any combination of blocks or block groups.
iCODEHOP advances the user down two different paths depending on the data that they select. If the user selects as input a single gapped multiple alignment or any number of nongapped multiple alignments (blocks), they are taken directly to a form that allows customization of CODEHOP design. Otherwise, iCODEHOP guides the user through a sub-workflow where (s)he creates a gapped multiple alignment using CLUSTALW (10) (iCODEHOP currently restricts the amount of amino acid sequence data that users can submit to its implementation of CLUSTALW to 20 000 residues). The program then attempts to identify blocks of highly conserved amino acids within the multiple alignment before taking the user to the CODEHOP design form.
At the CODEHOP design form, the user can change the default values for any of a large number of parameters that affect how CODEHOPs are designed. Here, we summarize the most interesting options; a complete description of all available options is available in the online iCODEHOP documentation. Users can:
Set the maximum degeneracy of each CODEHOP's 3′-core region.
Select from among more than 200 species-specific codon usage tables (11) to bias how the amino acid position-specific scoring matrix (PSSM) created for each block is back-translated to a DNA PSSM (12).
Adjust the approximate length of the 5′-clamp region of each CODEHOP by increasing or decreasing minimum annealing temperature (Tm) that each clamp region must satisfy.
Bias CODEHOP design to favor specific sequences in a protein family by adjusting the numerical weight assigned to each sequence when the program creates an amino acid PSSM from each block (This technique was used successfully by Rose et al. (3) to create CODEHOPs that amplified novel genes belonging to a species that was under represented in their protein multiple alignment).
When the user is ready to proceed, the program will then attempt to design CODEHOPs along each block of conserved amino acids that the program carved from the user's protein multiple alignment. iCODEHOP implements a revised version of the original CODEHOP design algorithm (3) that increases the maximum block width from 55 amino acid residues to 1000. This allows primer prediction to span the artificial block widths imposed by the original program. The new version also replaces the original method for calculating the Tm of clamp regions with a more recent nearest neighbor method based on experiments by SantaLucia and Hicks (13). Tests by our group and others have found that these changes produce primers that are nearly identical to experimentally verified primers produced by the old program.
Upon completion, the program presents a split window with one half showing a plot of the relative position of each CODEHOP along the block it was derived from and other half presenting details on specific blocks and CODEHOPs as the user mouses over them in the plot (Figure 2). Information present in this display includes the relative position along the block of the selected primer, the conservation at each amino acid position within the block, the primer's degeneracy, the length of the clamp and core regions of the primer, and the clamp's estimated annealing temperature.
The user can learn more about a specific primer by clicking on a button that links to a complete summary. A new tab will open within the application's window that shows the user detailed information on the selected primer and a brief summary of potential partner CODEHOPs for the PCR. Details on the selected primer include a phylogram of the block region that the primer was designed from and three different Tm range estimates (Figure 3). Each Tm range estimate represents a specific level of mismatch between primers in the degenerate pool and the unknown target. These include the case where (i) every primer in the pool perfectly complements the target, (ii) each primer in the pool matches the target perfectly over its core region but contains one mismatch every 3 nt over its clamp region and (iii) the primer is complementary to the target only over its core region. Summary information on each potential partner CODEHOP includes its sequence, position, degeneracy and a similar set of Tm range estimates as is shown for the selected primer. The program also shows, for each potential partner CODEHOP, the length of the amplicon that would be produced during a PCR experiment using it and the selected primer.
Support for problematic analysis
Under certain conditions, iCODEHOP is unable to design primers for a particular protein multiple alignment because conservation is too poor among the selected sequences for the program to find primers that satisfy degeneracy and clamp length constraints. In this case, the program supports two workflows that can ultimately lead to the successful design of CODEHOPs for the user's target gene family. In one workflow, the program presents the user with information regarding the phylogenetic relationships among the set of input sequences and allows the user to select a sub-clade for further analysis (Figure 4). This process is useful for eliminating sequences that prevent CODEHOP design due to a lack of conservation with other input sequences. It is also useful for identifying related clusters of input sequences that can be analyzed separately for CODEHOPs. In the other workflow, the user biases the sequence alignment that resulted in no primers to favor a specific sequence or sequence clade (for example, one that appears most closely related to the targeted gene family). The user accomplishes this by altering the numerical weight that was assigned to the chosen sequence(s) when the program created an amino acid PSSM from each block. Both methods are explained in detail in iCODEHOP's online documentation.
Evaluation
We conducted a formal usability study to evaluate the iCODEHOP user interface. The study was approved by the University of Washington's (UW) Institutional Review Board and involved three graduate students and one post-doctoral researcher from the UW Department of Pathobiology. We observed participants as they attempted to complete two focused tasks that differed in their difficulty using a ‘think aloud’ protocol (14). The first task required each participant to work her/his way through the basic primer design workflow implemented in iCODEHOP using a set of closely related sequences. The second task was similar, but required users to identify a specific sub-clade among the input sequences that would reduce the complexity of the sequence alignment, thus allowing the program to design primers for the chosen sub-clade. This study exposed several usability issues and led to refinements in the program's user interface. For example, each form that the program presents to the user was simplified and informational alerts were provided at each step of the primer design workflow. Subsequent users have found these changes to be very helpful. Further details on this study are available by request from the authors.
RELATED WORK
Although there are several other publicly available tools for designing degenerate PCR primers, iCODEHOP is the only one that aids in the design of primers with the consensus-degenerate hybrid format. It is also one of only a few systems that design primers from amino acid multiple-alignments. Several existing programs, including SCPrimer (15), Amplicon (16) and HYDEN (17) design degenerate primers exclusively from nucleic acid multiple alignments. This approach is very useful when nucleotide information is available and avoids the issue of back-translating amino acid sequences. However, it is sometimes not possible to identify conserved regions among the many nucleotide sequences that can encode a protein family of interest, even when conservation is discernible at the amino acid level.
Other researchers have proposed different strategies for designing degenerate primers from gapped amino acid sequence multiple alignments. Some methods, such as that proposed by Wie et al. (18), do not appear to have been validated at the laboratory bench. Alternate methods, such as that proposed by Pan et al. (19), have led to the PCR amplification of novel proteins but have not been implemented in any publicly available web application. To the best of our knowledge, GeneFisher (20,21) is the only other publicly available web application besides iCODEHOP that both designs degenerate primers from amino acid sequences and has produced degenerate primers that have been validated in laboratory experiments.
The original version of GeneFisher (20) identified potential degenerate primers by first creating a consensus sequence from a user-generated nucleotide multiple alignment then, excluding potential primers that did not meet a large set of customizable constraints. GeneFisher was replaced by an AJAX-enabled web application called GeneFisher2 (21) that adds the ability to design primers from back-translated amino acid sequences and no longer requires a user to create her/his own sequence multiple alignments. GeneFisher2 appears to allow degenerate positions at any position along the primers it creates. In contrast, the CODEHOPs produced by iCODEHOP have degenerate positions only within 11–12 bases of their 3′-end. The remaining positions of a CODEHOP are derived from the input amino acid multiple alignment by determining either the most common codons of the consensus amino acids or the codons with maximum weight in a DNA PSSM created from the alignment. As discussed above, this heuristic is known to be effective for designing degenerate primer for distantly related targets.
CONCLUSION
iCODEHOP is an enhanced, user-friendly, version of a program that predicts CODEHOPs from multiply aligned protein sequences. This project could result in more widespread use of a proven technique for identifying and amplifying new members of known gene families which could, in turn, increase the speed and accuracy of novel pathogen and gene discovery. Future efforts will focus on the development of metrics for choosing optimal CODEHOP primer pairs and the implementation of a process to utilize existing nucleic acid sequence information in CODEHOP primer design.
FUNDING
National Centers for Research Resources; National Institutes of Health (1R24RR021346). Funding for open access charge: National Centers for Research Resources - 1R24RR021346.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
iCODEHOP uses a web service provided by the European Bioinformatics Institute (22) to perform CLUSTAL alignments. The authors wish to thank Sherrilynne Fuller and Jim Wallace and the University of Washington Center for Public Health Informatics for hosting iCODEHOP. We also thank Steve and Jorja Henikoff for contributing source code from the original CODEHOP web application, Tobias Mann for contributing code from his Hyfi project that predicts annealing temperatures for nucleotide sequences and Nicholas Taylor for contributing the treeviewer program which iCODEHOP uses to produce interactive phylograms. Jeannette Staheli, Jonathan Ryan and Gregory Bruce contributed recommendations and feedback on the development of iCODEHOP and Sylvain Cibangu assisted with a usability study of the program conducted by coauthor Parmit Chilana.
REFERENCES
- 1.Alroy J. How many named species are valid? Proc. Natl Acad. Sci. USA. 2002;99:3706–3711. doi: 10.1073/pnas.062691099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wheeler DL, Chappey C, Lash AE, Leipe DD, Madden TL, Schuler GD, Tatusova TA, Rapp BA. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2000;28:10–14. doi: 10.1093/nar/28.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.VanDevanter DR, Warrener P, Bennett L, Schultz ER, Coulter S, Garber RL, Rose TM. Detection and analysis of diverse herpesviral species by consensus primer PCR. J. Clin. Microbiol. 1996;34:1666–1671. doi: 10.1128/jcm.34.7.1666-1671.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rose TM, Strand KB, Schultz ER, Schaefer G, Rankin G.W., Jr, Thouless ME, Tsai CC, Bosch ML. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J. Virol. 1997;71:4138–4144. doi: 10.1128/jvi.71.5.4138-4144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osterhaus AD, Pedersen N, van Amerongen G, Frankenhuis MT, Marthas M, Reay E, Rose TM, Pamungkas J, Bosch ML. Isolation and partial characterization of a lentivirus from talapoin monkeys (Myopithecus talapoin) Virology. 1999;260:116–124. doi: 10.1006/viro.1999.9794. [DOI] [PubMed] [Google Scholar]
- 7.Schultz ER, Rankin G.W., Jr, Blanc MP, Raden BW, Tsai CC, Rose TM. Characterization of two divergent lineages of macaque rhadinoviruses related to Kaposi's sarcoma-associated herpesvirus. J. Virol. 2000;74:4919–4928. doi: 10.1128/jvi.74.10.4919-4928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rose TM. CODEHOP-mediated PCR – a powerful technique for the identification and characterization of viral genomes. Virol. J. 2005;2 doi: 10.1186/1743-422X-2-20. 10.1186/1743-422X-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rose TM, Henikoff JG, Henikoff S. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 2003;31:3763–3766. doi: 10.1093/nar/gkg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from the international DNA sequence databases. Nucleic Acids Res. 1998;26:334. doi: 10.1093/nar/26.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henikoff S, Henikoff JG. Position-based sequence weights. J. Mol. Biol. 1994;243:574–578. doi: 10.1016/0022-2836(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 13.SantaLucia J, Jr, Hicks D. The thermodynamics of DNA structural motifs. Annu. Rev. Biophys. Biomol. Struct. 2004;33:415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen J. Usability Engineering. London, UK: Academic Press; 1993. [Google Scholar]
- 15.Jabado OJ, Palacios G, Kapoor V, Hui J, Renwick N, Zhai J, Briese T, Lipkin WI. Greene SCPrimer: a rapid comprehensive tool for designing degenerate primers from multiple sequence alignments. Nucleic Acids Res. 2006;34:6605–6611. doi: 10.1093/nar/gkl966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarman SN. Amplicon: software for designing PCR primers on aligned DNA sequences. Bioinformatics. 2004;20:1644–1645. doi: 10.1093/bioinformatics/bth121. [DOI] [PubMed] [Google Scholar]
- 17.Linhart C, Shamir R. The degenerate primer design problem: theory and applications. J. Comput. Biol. 2005;12:431–456. doi: 10.1089/cmb.2005.12.431. [DOI] [PubMed] [Google Scholar]
- 18.Wei X, Kuhn DN, Narasimhan G. Degenerate primer design via clustering. Proc. IEEE Comput. Soc. Bioinform. Conf. 2003;2:75–83. [PubMed] [Google Scholar]
- 19.Pan Z, Barry R, Lipkin A, Soloviev M. Selection strategy and the design of hybrid oligonucleotide primers for RACE-PCR: cloning a family of toxin-like sequences from Agelena orientalis. BMC Mol. Biol. 2007;8:32. doi: 10.1186/1471-2199-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giegerich R, Meyer F, Schleiermacher C. Proceedings of the Fourth International Conference on Intelligent Systems for Molecular Biology. Menlo Park, CA 94025: AAAI Press; 1996. GeneFisher-Software Support for the Detection of postulated genes; pp. 68–77. [PubMed] [Google Scholar]
- 21.Lamprecht AL, Margaria T, Steffen B, Sczyrba A, Hartmeier S, Giegerich R. GeneFisher-P: variations of GeneFisher as processes in Bio-jETI. BMC Bioinformatics. 2008;9(Suppl. 4):S13. doi: 10.1186/1471-2105-9-S4-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labarga A, Valentin F, Anderson M, Lopez R. Web services at the European bioinformatics institute. Nucleic Acids Res. 2007;35:W6–W11. doi: 10.1093/nar/gkm291. [DOI] [PMC free article] [PubMed] [Google Scholar]