Abstract
Neurotrophic factors such as nerve growth factor (NGF) promote a wide variety of responses in neurons, including differentiation, survival, plasticity, and repair. Such actions often require changes in gene expression. To identify the regulated genes and thereby to more fully understand the NGF mechanism, we carried out serial analysis of gene expression (SAGE) profiling of transcripts derived from rat PC12 cells before and after NGF-promoted neuronal differentiation. Multiple criteria supported the reliability of the profile. Approximately 157,000 SAGE tags were analyzed, representing at least 21,000 unique transcripts. Of these, nearly 800 were regulated by 6-fold or more in response to NGF. Approximately 150 of the regulated transcripts have been matched to named genes, the majority of which were not previously known to be NGF-responsive. Functional categorization of the regulated genes provides insight into the complex, integrated mechanism by which NGF promotes its multiple actions. It is anticipated that as genomic sequence information accrues the data derived here will continue to provide information about neurotrophic factor mechanisms.
Neurotrophins, exemplified by nerve growth factor (NGF), exert a variety of actions on their targets, including regulation of proliferation, differentiation, neurite growth, neurotransmission, plasticity, repair, and survival (1, 2). Good progress has been made in uncovering initial steps in the receptor-dependent signaling mechanism by which NGF and other neurotrophins work (3, 4). Beyond the initial signaling events, NGF promotes its actions by means of both transcription-independent and -dependent pathways (3, 5, 6). Understanding the mechanism and consequences of neurotrophin responses therefore requires a description of the genes that are subject to regulation by these factors.
Detection of neurotrophin-regulated genes necessitates cellular models that can be compared before and after factor exposure. Because many neurotrophin-responsive cells require the factors for survival, they are not optimally suited for such experiments. For this reason, a large percentage of NGF gene regulation studies have used the PC12 line of rat pheochromocytoma cells (7, 8). These do not require NGF in serum-containing media, but respond to NGF by changing their phenotype from that of proliferating chromaffin-like cells to that resembling nonproliferating, neurite-bearing sympathetic neurons. Application of a variety of approaches has identified on the order of 50 genes that respond to NGF (9–13). These include immediate early genes as well as those that are regulated relatively late in the differentiation process and that encode proteins with clear roles in neuronal function (1–4).
Despite such progress, present data suggest that many additional NGF-responsive genes remain to be identified. It has been estimated that 5–10% of the genes expressed in PC12 cells may be NGF-regulated (11, 12), which would suggest regulation of at least 1,000 transcripts. Detecting and identifying such transcripts is important because it provides key insight at the molecular level as to how trophic factors exert their many actions. Although the PC12 cell model is not without limitations, the extent and utility of information regarding NGF mechanisms and neuronal function that it has furnished strongly support its continued exploitation for defining transcriptional actions of neurotrophins.
To carry out a comprehensive analysis of NGF-dependent gene regulation in PC12 cells, we chose the SAGE (serial analysis of gene expression) technology first described by Velculescu et al. (14). Among the advantages of SAGE is that it has the potential to provide detection of all genes expressed in a given cell type, provides quantitative information about the relative expression of such genes, permits ready comparison of gene expression in the same cell type under different conditions (e.g., before and after NGF exposure), and yields sequence information that can be used to identify the detected genes. Thus far, SAGE methodology has proved itself to reliably detect expression of regulated and nonregulated genes in a variety of cell types (15–17). Here, we describe results garnered from comparative SAGE profiling of approximately 157,000 tags from PC12 cells before and after long-term exposure to NGF.
Materials and Methods
Cell Culture.
PC12 and PC12-derived cells were cultured as described (8) on 100-mm plastic dishes coated with rat tail collagen in complete medium (85% RPMI medium, 10% horse serum, and 5% FBS). For NGF treatment, cells were plated at approximately 20% confluency and maintained for 4 days in serum-free RPMI medium and 50 ng/ml recombinant human NGF (a kind gift of Genentech) and then for 5 days in complete medium with the same NGF concentration. The initial exposure to serum-free conditions ensured death of any NGF-nonresponsive cells, and the reintroduction of complete medium assured that both naïve and NGF-treated cultures were exposed to the same serum concentrations.
SAGE.
Total cellular RNA was isolated from naive and NGF-treated cells after two washes with ice-cold PBS and extraction with 1 ml of Tri-reagent per dish (Molecular Research Products, Cincinnati). Polyadenylated mRNA was isolated by oligo(dT) chromatography using the MessageMaker kit (Life Technologies, Grand Island, NY). Five micrograms of mRNA was used to construct each SAGE library, using SAGE protocol version 1.0c (14). In brief, mRNA transcripts were converted to cDNAs with biotinylated oligo(dT)18 as the primer (cDNA synthesis kit, Life Technologies). The cDNAs were digested with NlaIII, and the 3′ ends were bound to streptavidin-coated magnetic beads and then ligated to linkers 1 and 2. For the naïve cell library, we modified the protocol to use 1/5 the amount of the linkers to reduce subsequent nonspecific incorporation of partial linker sequences into concatemers. BsmF1 IIS then was used to release the tags, which then were blunted at their 3′ ends and ligated to form ditags, which then were amplified by PCR. The amplified ditags were isolated by gel electrophoresis and, after extraction, purified through a silica membrane column and redigested with NlaIII to remove the linkers. The glass-membrane purification step was used to enhance the reliability of digestion with NlaIII (18). The ditags were concatemerized at their NlaIII overhangs (14). Concatemers with minimum size of 500 bp were obtained by gel purification. These were ligated into cloning vector pcZERO plasmid (Invitrogen) and transformed into DH10B bacteria (Life Technologies) by electroporation. For both libraries, plasmids with concatemer inserts were cycle sequenced with Big Dye terminator chemistry and analyzed either on a 377 or 3700 Applied Biosystems DNA sequencer. The tags were extracted from the sequences and analyzed by using the sage 300 software package (14).
Matching Tags with Transcripts.
We considered only tags encountered two or more times for the two libraries combined. Tags were initially analyzed with the National Center for Biotechnology Information (NCBI) rat SAGE tag to gene mapping database (ftp://ncbi.nlm.nih.gov/pub/sage/map/Rn/Nla3), which matches possible 14-mer tags with known rat genes and expressed sequence tags (ESTs). With the use of sequences present in the NCBI unigene rat database, potential matches were further scrutinized to determine whether there was a match at the 15th base and to determine whether the matched sequence could be confirmed as being at the most 3′ end of a known rat transcript or EST. We considered only cases in which a clear poly(A) tail and/or polyadenylation signal was present on the transcript or EST. Matched ESTs were further analyzed by an advanced blast search for homology with known genes.
Northern Blot Analysis.
Ten micrograms of total RNA (isolated as described above) from each sample was analyzed as described (19). The blots were exposed and quantified with a Storm PhosphorImager (Amersham Pharmacia–Molecular Dynamics). Probes were kindly supplied by R. Dalla-Favera, Columbia University (human β-actin), R. Vallee, University of Massachusetts, Worcester (LIC-2; ref. 20), P. Cserjesi, Columbia University (MASH-1), and D. Park, University of Ottawa (CDK4). Full-length probes for rat cofilin and galectin-1 were generated by PCR using primers that flank their ORFs. Probes for 14-3-3-γ, ATFx, MAP4, PYBP2, BTF3, pleiotrophin, CD9, Phox2a, and Tsc-22 were rat ESTs produced by the University of Iowa rat genome project and purchased from Research Genetics, Huntsville, AL. All probes were sequenced to verify identity.
Western Blotting.
Western blots were carried out as described (19) with a mouse anti-cofilin antibody (1:400) and rabbit anti-ADF antiserum (1:5,000) kindly provided by J. Bamburg, Colorado State University, Fort Collins (21).
Results and Discussion
Generation and Analysis of PC12 Cell SAGE Tags.
The present study used matched cultures of PC12 cells (passage 27) harvested at comparable cell density (25–50% confluency) with or without 9 days of NGF exposure. This relatively long treatment was chosen because it elicits extensive neuritogenesis, cessation of proliferation, and acquisition of a neuronal phenotype. The cultures were used to construct appropriate libraries that were subjected to the SAGE protocol.
The present analysis considered (after exclusion of duplicate ditags, tags of less than 15 bases, mitochondrial genes, and repetitive elements) 74,880 15-mer tags from NGF-untreated (naïve) cells and 82,743 from NGF-treated cells. These represented 75,317 different transcripts for the two libraries combined. This number is unlikely to reflect extensive sequencing errors. Zhang et al. (16) reported a maximal sequencing error rate of 0.7% per base in their SAGE study; repeat analysis of the same concatemers in our study indicated an error rate per base of less than 0.2%. By comparison, single libraries analyzed by the Cancer Genome Anatomy Project (CGAP) SAGE consortium (as of January 2000) consisting of 50,000–100,000 tags reported up to 33,000 novel transcripts (http://www.ncbi.nlm.nih.gov/CGAP/). The higher level of transcripts detected here may in part reflect our use of 15-mer tags rather than the 14-mer tags analyzed by CGAP (see below), as well as apparent stimulation of low-abundance genes by NGF (see Table 2, which is published as supplemental material on the PNAS web site, www.pnas.org). Additionally, PC12 cells may express basal levels of many transcripts. This possibility is supported by the observation that even at the present level of analysis, 3/4 of the transcripts are represented by only a single tag.
If one considers transcripts represented by tags detected at least twice, then 16,199 were observed for the naïve cell library and 12,761 for the library of NGF-treated cells. For both libraries combined, 21,086 unique transcripts were represented two or more times. These data compare well with those of Velculescu et al. (17) who reported 23,580 unique genes expressed in brain libraries totaling 202,448 tags represented two or more times. Of the 21,000 unique transcripts here, approximately 2,400 (11%) were matched to known, named genes and an additional 3,500 (17%) to undefined rat ESTs, based on sequence information present in NCBI databases.
Although several major SAGE projects used 14-mer base tags (CATG+10 bp), we found that 15-mer tags (CATG+11 bp) are more reliable. The NCBI rat SAGE tag to gene mapping database, which matches possible 14-mer tags with known rat genes and ESTs, in a number of cases correctly matched the same tag with 3′ sequences representing four different transcripts. However, this ambiguity was in almost all cases resolved when the 15th base was added. As one example, four different 15-mer tags [CATGGTGTGGCACA (T)/(C)/(A)/(G)] were matched by the 14-mer database as ribosomal protein S13. Consideration of the 15th base led to an unambiguous match (CATGGTGTGGCACAG) with the published S13 sequence.
Reliability of Quantitative SAGE Analysis of PC12 Cell Transcripts.
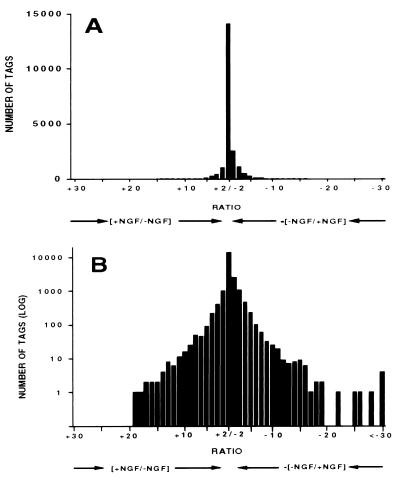
There are several criteria by which to judge the fidelity of our SAGE data to the status of transcripts in our cells. One is that housekeeping genes should not undergo major NGF regulation. As anticipated, four different EF-1 proteins (α, β, γ, and δ) showed little, if any, differential expression. Several additional examples of such nonresponsive genes are given in Table 3, which is published as supplemental data, including cytoplasmic β actin and alcohol dehydrogenase. Fig. 1 illustrates the distribution of fold-changes in tag number between naïve and NGF-treated cells for tags detected two or more times. This reveals that approximately 90% of the tags show less than a 3-fold difference in expression for the two libraries, which is another indication that most expressed genes are nonresponse to NGF. The 10% of NGF-responsive transcripts detected by our analysis is in line with previous estimates (11, 12).
Figure 1.
Distribution of NGF-promoted changes in expression of transcripts in PC12 cells as revealed by SAGE. Transcripts for which two or more tags were detected are included. Up-regulated transcripts are to the left, down-regulated transcripts to the right. Tag numbers are expressed either on a linear (A) or log (B) scale.
Another measure of the reliability of our analysis is that it should detect changes in expression of transcripts already known to be NGF responsive. Our data revealed 17 such transcripts previously reported as NGF-responsive in PC12 cells or other systems. In all cases, the changes in expression shown by the SAGE data were consistent with the changes previously reported (see supplemental Table 4).
Gene Regulation by NGF.
Approximately 10% of the 21,000 tags detected two or more times changed in abundance by 3-fold or greater in response to NGF. The data in Fig. 1 indicate that NGF-down-regulated transcripts exceed the number that are up-regulated. Fig. 1 further shows that the numbers of regulated transcripts diminished at greater levels of change so that approximately 3.8% (800) changed by 6-fold or greater whereas 149 (0.7%) changed by 10-fold or more. Of tags regulated by 6-fold or more, 150 were assignable (see Materials and Methods) to known, named genes and 238 to currently novel, unmatched rat ESTs. Approximately 130 of the identified genes have not been previously reported to our knowledge as targets for NGF regulation; these are listed in Table 1.
Table 1.
Named transcripts identified by SAGE as NGF-regulate in PC12 cells
| Tag | +/− NGF | Identity |
|---|---|---|
| Intracellular signaling | ||
| AATGTGAGTCA | 14:0 | 14-3-3 protein γ-subtype |
| GATTGTCTTGA | 12:0 | 25-kb FK506 binding protein (FKBP25) |
| AAGCCTTGCTG | 9:0 | Ash-m protein |
| TCCTGTGTCCT | 7:1 | Inositol trisphosphate receptor 3 (IP3R-3) |
| ATCAACACCGC | 100:27 | Guanine nucleotide BP Gsalpha/Galpha8 |
| ATAGCTGGGGC | 18:5 | MAP kinase kinase (MEK1) |
| TGCCCAATAAA | 0:15 | NIPK-death-induced kinase |
| TTCTGCCTCCA | 1:8 | Similar to novel Ser/Thr kinase PKL12/MPSK |
| TCTCACCCACT | 0:6 | Phosphoprotein HASPP28 |
| ACGAGCTTTAA | 1:6 | FK506-binding protein FKBP23 |
| Transcription factors/regulators | ||
| CCCTTCACCTC | 8:1 | Drl-associated polypeptide (DRAP1) |
| CGAAGTCAGGC | 8:1 | CIIDBP/HMG 1 protein homolog |
| TTCCCCACACA | 8:1 | Transcriptional regulator TSC-22 |
| CAAATAAGTTT | 7:0 | Transcriptional cofactor pirin |
| AGCCTGTTTTC | 6:1 | TFIID 30-kDa subunit (TAFII-30) |
| AGAACCTAGTC | 5:126 | ATFx/ATF5 |
| TGCTCCGTGTA | 1:9 | Supt5hp homolog |
| GAGAACATCAC | 0:6 | BTF3 |
| TGATCTTTTTG | 2:11 | ATF-4/CREB2 |
| AGAAGCGCAAG | 5:27 | Arix1/PHOX2A homeodomain protein |
| Proliferation/DNA synthesis | ||
| ACTGAGTGCTT | 11:1 | Deleted in oral cancer homolog/DOC1 |
| AAGGGTCCCCG | 8:0 | PRPPS-associated protein 39 |
| TTCCTGTGCCC | 6:0 | Anaphase-prom complex subunit 2 (APC2) |
| CAAACTGCATT | 0:9 | Cyclin-dependent kinase 4 (CDK4) |
| GGGCAGACAGG | 1:9 | Replication protein A p32 (RPA32) |
| CGCAAGAAGGT | 1:8 | DNA polymerase δ catalytic subunit |
| GAGCATTTAGT | 1:6 | DNA primase small subunit |
| RNA processing/splicing/stability | ||
| AAGCTGGTTTA | 9:0 | snRNP B |
| ATGAGGAACTT | 7:0 | U1 snRNP C (U1-C) |
| GTCGCTTCTGA | 1:9 | Similar to splicing factor U2AF p35 |
| GAAGAGTGTAA | 2:15 | Pyrimidine-rich tract binding protein |
| CACAACTGTGA | 0:7 | SR-related protein SRrp129 |
| GAAATGTAAGA | 2:12 | Poly(rC) binding protein 2 (PCBP-2) |
| TTGATCGAAGT | 0:6 | Poly(A) polymerase, isoform VI |
| GCCCGAAAGAT | 0:6 | Suppressor of white apricot homolog 2 |
| Protein folding/chaperones | ||
| AGCCTCCCTTG | 12:1 | C terminal of Hsp70-interacting protein |
| TAGAGCGTGCT | 11:0 | Nucleosome assembly protein 2/NAP2 |
| TGGGTTAGACC | 8:1 | Prefoldin subunit 1 |
| AGAAGTTCAGA | 15:2 | Heat shock protein 60 (HSP60) |
| GAGCGTTTTGG | 159:37 | Cyclophilin A |
| Microtubule function related | ||
| TGGAACCTTGC | 16:1 | Cytoplasm dynein light intermediate chain/LIC2 |
| GAGGAGGGGGA | 8:0 | α-6 tubulin |
| GTATGTGGATC | 11:2 | Homologous with MAP 1A/B light chain 3 |
| Actin cytoskeleton regulation | ||
| AGTTTGCTGAT | 38:3 | Transgelin-like protein |
| AGCTTTCCTGT | 9:1 | Cortactin B (brain isoform) |
| TGGGTTAGACC | 8:1 | Prefoldin subunit 1 |
| GCAATAAATGG | 6:0 | Actin binding protein drebrin A |
| TGTATAATCAG | 6:1 | Myosin regulatory light chain 2B |
| TTGCACCTTCT | 6:0 | ARP2/3 complex 34-kDa subunit |
| GAAGCAGGACC | 44:10 | Cofilin |
| Protein/vesicle trafficking | ||
| TCAGGCATTTT | 9:0 | Ras-related protein RAB-1B |
| TGTGAAGTAGC | 17:2 | ADP-ribosylation factor ARF1 |
| TGTGCAGTGAA | 17:2 | Signal peptidase 12-kDa subunit |
| GCACACTGTGT | 7:0 | Endophilin III/SH3p13 |
| TGGTGACTAAG | 7:1 | ADP-ribosylation factor BP GGA 2 |
| AGAATGAAGTT | 6:0 | MUM-2 |
| CTGTCTGACTC | 0:12 | β adaptin |
| ACCTTGCCCTC | 3:18 | Translocon-associated protein β subunit |
| Granules/synaptic vesicle components | ||
| CTAGACACCTG | 7:1 | Secretogranin III (SgIII) |
| TCCTGTGTCCT | 7:1 | IP3R-3 |
| Calcium binding proteins | ||
| CCCCCTGGATC | 24:3 | Calcyclin/S100A6 |
| TCATCTTTAAC | 1:7 | Calreticulin |
| ACGAGCTTTAA | 1:6 | FK506-binding protein FKBP23 |
| Ubiquitination/proteasome components | ||
| GTGCTGGACCT | 10:1 | Proteasome activator PA28 β |
| AGACGCCTGTG | 9:0 | 26S proteasome-associated PAD1 homolog |
| TTCCTGTGCCC | 6:0 | Anaphase-promoting complex subunit 2 |
| ACAAGTATACT | 6:1 | Proteasome ɛ-chain precursor |
| ATCAGTGGCTT | 21:4 | Proteasome β-type RN3 subunit |
| Intercellular/substrate interactions | ||
| GCGGCGGATGG | 209:15 | β-gal-binding lectin, Galectin 1 |
| AAATCCTTTCA | 11:0 | Pleiotrophin |
| GGTGTGCCAGG | 10:0 | Putative integral membrane receptor |
| ATTCTGTGCTG | 10:1 | Tetraspanin family member CD9 |
| ATCTAAGCCAG | 9:1 | Galectin 3 |
| TCTGTCCTGCT | 8:0 | Degen spermatocyte (des) homolog |
| CCCTTCCCTGC | 7:0 | Cortexin |
| CGCGCGCGCGC | 7:0 | BIT/MFP/SHPS-1/P84 |
| CTCTGACTTTA | 26:4 | OX47/basigin/neurothelin/M6/CE9 |
| AGCTCTACAGG | 6:0 | G10 protein homolog (edg2) |
| TGAGCCCAGCC | 6:0 | Ephrin type-B receptor 1 precursor |
| GAGTGGACTCT | 12:2 | Tetraspanin AD1/ME491/CD63 |
| GAGACCCTCAG | 6:1 | Brain sigma receptor |
| GCTACAGGGAG | 6:1 | Rod outer segment membrane protein 1/ROM1 |
| TACTTGTGTTC | 6:1 | IgG superfamily member gp65 |
| CTACAGTTCCT | 0:13 | Latrophilin/CIRL/CL1 |
| Neuroprotection | ||
| GTGGCCCACTT | 22:2 | Cyt c oxidase assembly protein COX17 |
| TATGCACAGGC | 8:1 | DNA-repair protein XRCC1 |
| GTCTAGGTCAC | 7:0 | Hepatitis B virus X interacting protein |
| GAGAGGCATTT | 6:0 | Glutathione S-transferase 8/GST8 |
| ACCGGCCTTAG | 11:2 | GST-microsomal form |
| Energy metabolism | ||
| GCATACGGCGC | 18:0 | Mitochondrial ATP synthase subunit e |
| CCAAGGAAAAC | 51:8 | Lactate dehydrogenase A |
| ATTAACTTGGT | 6:1 | Glutamate dehydrogenase |
| CGCTGAGGCCT | 6:1 | Hom 64-kDa ubiquinol-cyt c reductase |
| CTCTGTTTTCC | 6:0 | Similar to 14-kDa NDUFS5 |
| CATCCTTGATG | 11:2 | Brain creatine kinase B |
| TTCCAGCTGCT | 26:5 | Brain phosphoglycerate mutase B |
| TACTAGAAAAG | 1:7 | NADH-ubiquinol oxidoreductase NDUFS2 |
| Miscellaneous metabolism | ||
| TGCTCCTGTGA | 13:0 | β-hexosaminidase α-subunit |
| GCTGGAATTGA | 10:1 | Farnesyl pyrophosphate synthetase |
| GAGAAGAAGGA | 6:1 | Peroxisomal enoyl-CoA hydrat-like protein |
| ACCTTGTTGAT | 2:30 | D-dopachrome tautomerase |
| ACCTACAGGAT | 1:12 | Branched chain α-ketoacid dehydrogenase E1α |
| CTTGTGACAGG | 0:9 | Monoglyceride lipase |
| Ion pumps/transporters | ||
| TTCTAGCATAT | 11:0 | Na+, K+ −ATPase β-1 subunit |
| TCCCCCTGCTA | 6:0 | Organic cation transporter BOCT |
| TTGGTGAGGTA | 1:16 | LAT4 Na-independent neutral aa transporter |
| ATTCTCTGGAT | 1:7 | Sarco/endoplasmic reticulum Ca-ATPase |
| CTGGAGCTGGG | 1:8 | Mitochondria dicarboxylate carrier |
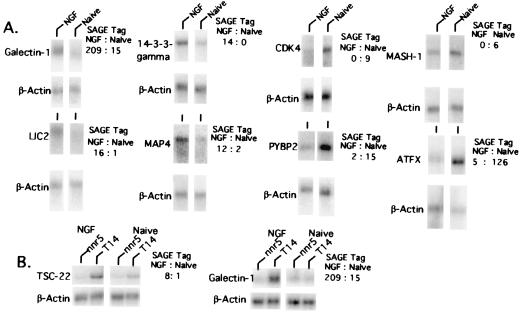
To confirm our analysis, a set of regulated transcripts was subjected to Northern blotting using RNA from naïve and NGF-treated cultures independent from those used to establish our SAGE libraries. Fourteen named genes and 13 genes corresponding to ESTs were probed by using β-actin (which SAGE revealed as nonresponsive) as a loading control. In all cases, the blots showed changes consistent with those revealed by SAGE. Several such blots for named genes are shown in Fig. 2. In one case our SAGE data were inconsistent with published findings. It was reported that the transcription factor MASH-1 is up-regulated by NGF in PC12 cells (22). In contrast, our SAGE data indicate a drop in expression (tags +NGF/−NGF, 0:6). This was confirmed by our Northern blot analysis (Fig. 2A) and would be the anticipated response in that MASH-1 influences early specification of autonomic lineage and is not expressed by postmitotic sympathetic neurons (23). Also consistent with this, expression of the transcription factor Phox2a, which appears to be downstream of MASH-1 in specification of the sympathetic lineage (24), also is down-regulated by NGF in our SAGE analysis (tags +NGF/−NGF, 5:27).
Figure 2.
Northern blot analysis of NGF-promoted gene regulation in (A) PC12 cells and (B) PC12-cell-nnr5 derived cells with (T14) and without (nnr5) Trk expression. Total cellular RNA was isolated from cells treated without (naïve) or with NGF for 7–14 days (NGF). Ten micrograms of RNA was analyzed as described in Materials and Methods with the indicated probes. Blots were stripped and reprobed with β-actin to indicate relative loading. Ratios to side of blots indicate numbers of tags in PC12 cell SAGE analysis +/− NGF.
Two distinct NGF receptor types have been recognized; Trk is a receptor tyrosine kinase required for regulation of at least several NGF-stimulated genes (4, 19) and p75 is a tumor necrosis factor α receptor superfamily member that appears capable of evoking death and whose signaling mechanism is at best partially understood (1, 4). To carry out an initial analysis of Trk and p75 involvement in regulation of the genes described here, Northern blots were performed on RNA isolated from PC12nnr5 cells expressing only p75 receptors or from Trk-transfected PC12nnr5-T14 cells (19). For the six transcripts tested [TSC-22 (Fig. 2B), MAP4, galectin 1 (Fig. 2B), tetraspanin CD9, 14-3-3γ, and pleiotrophin], regulation by NGF was observed only for the cells expressing Trk. Thus, p75 alone is not sufficient to regulate the NGF-responsive genes we assessed to date, whereas the presence of Trk is necessary for this action.
Table 1 lists approximately 130 transcripts of named genes indicated by SAGE to undergo significant levels of long-term regulation by NGF in PC12 cell cultures. To facilitate evaluation of these changes from a biological perspective, the transcripts have been loosely grouped into functional categories. Although it is beyond the scope of this report to discuss each change in detail, a few points can be made about how these data begin to illuminate our understanding of how NGF-promoted transcriptional changes contribute to neuronal differentiation, plasticity, and function.
Cytoskeleton.
Prior studies have drawn attention to the importance of microtubules in NGF-promoted neurite outgrowth. Aside from their structural role, microtubules play a key role in supporting retro- and antero-grade neuronal transport. In this respect, the striking up-regulation of transcripts encoding LIC2, a cytoplasmic dynein light intermediate chain putatively involved in organelle transport (20), is especially interesting.
Dynamic regulation of the actin network, particularly at the growth cone, is another important element of neuronal differentiation and plasticity. SAGE reveals substantial up-regulation of transcripts encoding proteins involved in actin gelation (a transgelin-like protein, ref. 25); actin depolymerization (cofilin, ref. 26); formation and organization of actin filaments (ARP 2/3 34-kDa subunit, ref. 27); linkage of signaling pathways to f-actin reorganization (cortactin B, brain isoform, ref. 28); and alteration of actin organization in dendrites and growth cones (drebrin A, ref. 29). The diversity of the roles of these proteins underscores the complexity of the mechanisms by which NGF regulates actin function and points to several new candidates that may mediate NGF-promoted synaptic plasticity.
Because cofilin regulates neuronal actin polymerization (26) and NGF causes rapid cofilin dephosphorylation and relocalization (21), we carried out Western blotting with anticofilin in extracts of naïve and NGF-treated PC12 cells to determine whether the observed transcriptional changes led to changes in protein expression (see Fig. 3, which is published as supplemental material). This revealed an increase in cofilin protein expression similar to that for cofilin transcripts. In contrast, levels of the closely related actin depolymerizing factor (ADF) protein (26) were unchanged, thus indicating the selectivity of the actions of NGF on this class of molecules.
Regulation of Proliferation.
A striking response of PC12 cells to NGF is cessation of proliferation (7). Understanding how this occurs is relevant both to the neuroblast-neuron transition and the regulation of tumor cell growth. In this context, it is of interest that NGF up-regulates transcripts for a putative tumor suppressor (DOC1, ref. 30); for an enzyme that suppresses nucleotide synthesis (phosphoribosylpyrophosphate synthetase-associated protein, ref. 31): and for a subunit of a complex with ubiquitin-protein ligase activity that triggers degradation of proteins required for mitosis (anaphase-promoting complex subunit 2, ref. 32). In addition, there is substantial down-regulation of transcripts encoding the DNA replication protein RPA32 (33), the proliferating cell nuclear antigen-associated catalytic subunit of DNA polymerase delta (34), and the small subunit of DNA primase (35). These observations identify multiple genes whose regulation may participate in generating the neuronal postmitotic state. In addition to roles in mitosis, cell-cycle associated molecules have been implicated in apoptotic neuronal death evoked by loss of support by trophic factors such as NGF (36). Cdk4 and cyclin-dependent kinase p130-PITSLRE are two such examples considered as possible elements in the apoptotic mechanism (36, 37). These are down-regulated by NGF and hence would be expected to increase expression upon NGF deprivation, and thereby potentially contribute to the apoptotic mechanism.
Membrane and Vesicle Trafficking and Formation.
Axon generation entails significantly enhanced formation and trafficking of membranes and their components. Within this context, the up-regulation of transcripts for Rab-1B (38), ARF-binding protein GGA2 (39), MUM-2 (40), and the 12-kDa signal peptidase subunit (41) all appear to be functionally significant. NGF-promoted neuronal differentiation in PC12 cells also includes formation of synaptic vesicles (7). The up-regulated transcripts for ARF1 (42) and endophilin III (43) presumably contribute to vesicle genesis and recycling, respectively, whereas those for secretogranin III (44) and IP3R-3 (45) would provide vesicular components.
Transcription Factors.
A number of NGF-regulated genes were found to encode transcriptional regulators, which suggests that such factors themselves may play important roles in mediating NGF's actions on gene expression. Among those that are up-regulated, TSC-22 may be especially notable in that this evolutionarily conserved transcriptional repressor also is induced by other growth inhibitory molecules including transforming growth factor type β (46). TSC-22 is expressed in many developing mammalian tissues including sympathetic neurons (47) and in the fly, its homologue shortsighted/bunched, plays a required role in development of the nervous system (48). The up-regulated CIIDBP, a high mobility group I protein (HMGI) homologue, is also a potentially significant mediator of NGF actions in that HMGI proteins appear to play important roles in coordinating proliferation and differentiation during development (49). Of those transcription factors that are down-regulated, ATFx/ATF5 is reported to suppress cAMP-regulated gene expression (50), indicating that NGF may effectively increase neuronal responsiveness to cAMP and other effectors that use CREB-dependent signaling.
14-3-3 Family.
Aside from revealing transcript levels for diverse proteins involved in a common function, SAGE provides information about relative expression and regulation of transcripts within protein families. One example is the 14-3-3 family of highly homologous proteins that bind to and affect the distribution, availability, and/or activity of other proteins, thereby regulating a variety of enzymatic activities and signaling pathways (51). The differences in regulation and activities of the seven major mammalian 14-3-3 isoforms are not well understood. Six isoforms were detected in PC12 cells by SAGE (see supplemental Table 5); all are highly expressed in brain (52, 53). Transcripts for the γ form were highly up-regulated in response to NGF (14:0); η, the most abundant form, was up-regulated by about 3-fold as was the less abundant ζ isoform. In contrast, the β and ɛ isoforms underwent little or no regulation whereas the θ form was significantly down-regulated. These observations indicate that even closely related isoforms are differentially regulated by NGF and that they may play specific roles in the NGF response.
NGF and Glutathione S-Transferases (GSTs).
Protection of neurons from oxidative and toxic stresses is an important action of NGF (54). GST, which comprises a multigene family as well as microsomal and mitochondrial forms, is among the major enzymes involved in such protection, and it is reported that NGF up-regulates GST activity in brain (55). SAGE reveals seven distinct GST transcripts in PC12 cells (see supplemental Table 6) of which three (GSTb, GST YB3, and microsomal GST) appear to be significantly up-regulated. These observations not only illuminate a potential mechanism by which NGF confers neuroprotection, but also illustrate selective actions of the factor on a group of genetically and functionally related molecules.
Concluding Remarks.
Our aims in undertaking the present analysis have been several-fold. One has been to generate a database of the genes expressed in naïve and neuronally differentiated PC12 cells. This cell line has been and continues to be an extensively used model for a variety of aspects of neuroendocrine/neuronal behavior and function, and such information should enhance its utility in this regard. The similarity of PC12 cells to chromaffin cells and sympathetic neurons also should provide useful information regarding genes potentially expressed in these and other neuronal/neuroendocrine cell types. Approximately 2,400 transcripts for named genes have been identified in the cells to date and this number should grow significantly as additional genomic information enters public databases. A second aim has been to provide a comprehensive analysis of genes that undergo long-term regulation in response to NGF and thereby to gain insight about the molecular basis of neurotrophic factor actions. Aside from confirming known NGF responsive genes, we identified approximately 130 additional named genes that appear to undergo significant levels of regulation by NGF. Even this fragmentary list of regulated, known genes has begun to yield insights about the NGF mechanism and it can be anticipated that its size and impact will grow as a consequence of advancing genomic information. The next steps will be to test the functional consequences of manipulating the expression of these genes in neuronal systems and to determine temporal and causal relationships between the regulated genes. Further analyses also will be required to identify genes whose regulation by NGF is transient in nature. A final goal has been to provide information for discovery of new neuronal genes, particularly those subject to NGF regulation. The several hundred currently unmatched ESTs and roughly 500 unmatched tags regulated by more than 6-fold provide fertile ground in this respect.
Supplementary Material
Acknowledgments
We thank Claudine L. Bitel for outstanding technical assistance and Drs. R. Vallee, R. Dalla-Favera, P. Cserjesi, D. Park, and J. Bamburg for providing critical reagents. We are also grateful to Drs. A. Chalazonitis, N. Papadopoulos, R. Parsons, K. Kinzler, V. Velculescu, and L. Zhang for helpul advice. This work is supported in part by grants from the National Institute of Neurological Disorders and Stroke (L.A.G. and M.L.S.).
Abbreviations
- NGF
nerve growth factor
- SAGE
serial analysis of gene expression
- NCBI
National Center for Biotechnology Information
- EST
expressed sequence tag
- GST
glutathione S-transferase
References
- 1.Frade J M, Barde Y A. BioEssays. 1998;20:137–145. doi: 10.1002/(SICI)1521-1878(199802)20:2<137::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 2.Tessarollo L. Cytokine Growth Factor Rev. 1998;9:125–137. doi: 10.1016/s1359-6101(98)00003-3. [DOI] [PubMed] [Google Scholar]
- 3.Segal R A, Greenberg M E. Annu Rev Neurosci. 1996;19:463–489. doi: 10.1146/annurev.ne.19.030196.002335. [DOI] [PubMed] [Google Scholar]
- 4.Friedman W J, Greene L A. Exp Cell Res. 1999;253:131–142. doi: 10.1006/excr.1999.4705. [DOI] [PubMed] [Google Scholar]
- 5.Burstein D E, Greene L A. Proc Natl Acad Sci USA. 1978;75:6059–6063. doi: 10.1073/pnas.75.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greene L A. Trends Neurosci. 1984;7:91–94. [Google Scholar]
- 7.Greene L A, Tischler A S. Proc Natl Acad Sci USA. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greene L A, Farinelli S E, Cunningham M E, Park D S. In: Culturing Nerve Cells. 2nd Ed. Banker G, Goslin K, editors. Cambridge, MA: MIT Press; 1998. pp. 161–187. [Google Scholar]
- 9.Greenberg M E, Greene L A, Ziff E B. J Biol Chem. 1985;260:14101–14110. [PubMed] [Google Scholar]
- 10.Leonard D G, Ziff E B, Greene L A. Mol Cell Biol. 1987;7:3156–3167. doi: 10.1128/mcb.7.9.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee N H, Weinstock K G, Kirkness E F, Earle-Hughes J A, Fuldner R A, Marmaros S, Glodek A, Gocayne J D, Adams M D, Kerlavage A R, et al. Proc Natl Acad Sci USA. 1995;92:8303–8307. doi: 10.1073/pnas.92.18.8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayumi K, Yaoi T, Kawai J, Kojima S, Watanabe S, Suzuki H. Biochim Biophys Acta. 1998;1399:10–18. doi: 10.1016/s0167-4781(98)00081-5. [DOI] [PubMed] [Google Scholar]
- 13.Brown A J, Hutchings C, Burke J F, Mayne L V. Mol Cell Neurosci. 1999;13:119–130. doi: 10.1006/mcne.1999.0736. [DOI] [PubMed] [Google Scholar]
- 14.Velculescu V E, Zhang L, Vogelstein B, Kinzler K W. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 15.Velculescu V E, Zhang L, Zhou W, Vogelstein J, Basrai M A, Bassett D E, Jr, Hieter P, Vogelstein B, Kinzler K W. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Zhou W, Velculescu V E, Kern S E, Hruban R H, Hamilton S R, Vogelstein B, Kinzler K W. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 17.Velculescu V E, Madden S L, Zhang L, Lash A E, Yu J, Rago C, Lal A, Wang C J, Beaudry G A, Ciriello B P, et al. Nat Genet. 1999;23:387–388. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
- 18.Angelastro J M, Klimaschewski L, Vitolo O V. Nucleic Acids Res. 2000;28:e62. doi: 10.1093/nar/28.12.e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loeb D M, Greene L A. J Neurosci. 1993;13:2919–2929. doi: 10.1523/JNEUROSCI.13-07-02919.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes S M, Vaughan K T, Herskovits J S, Vallee R B. J Cell Sci. 1995;108:17–24. doi: 10.1242/jcs.108.1.17. [DOI] [PubMed] [Google Scholar]
- 21.Meberg P J, Ono S, Minamide L S, Takahashi M, Bamburg J R. Cell Motil Cytoskeleton. 1998;39:172–190. doi: 10.1002/(SICI)1097-0169(1998)39:2<172::AID-CM8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 22.Johnson J E, Birren S J, Anderson D J. Nature (London) 1990;346:858–861. doi: 10.1038/346858a0. [DOI] [PubMed] [Google Scholar]
- 23.Lo L C, Johnson J E, Wuenschell C W, Saito T, Anderson D J. Genes Dev. 1991;5:1524–1537. doi: 10.1101/gad.5.9.1524. [DOI] [PubMed] [Google Scholar]
- 24.Hirsch M R, Tiveron M C, Guillemot F, Brunet J F, Goridis C. Development (Cambridge, UK) 1998;125:599–608. doi: 10.1242/dev.125.4.599. [DOI] [PubMed] [Google Scholar]
- 25.Prinjha R K, Shapland C E, Hsuan J J, Totty N F, Mason I J, Lawson D. Cell Motil Cytoskeleton. 1994;28:243–255. doi: 10.1002/cm.970280307. [DOI] [PubMed] [Google Scholar]
- 26.Bamburg J R. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 27.Welch M D. Trends Cell Biol. 1999;9:423–427. doi: 10.1016/s0962-8924(99)01651-7. [DOI] [PubMed] [Google Scholar]
- 28.Ohoka Y, Takai Y. Genes Cells. 1998;3:603–612. doi: 10.1046/j.1365-2443.1998.00216.x. [DOI] [PubMed] [Google Scholar]
- 29.Shirao T. J Biochem (Tokyo) 1995;117:231–236. doi: 10.1093/jb/117.2.231. [DOI] [PubMed] [Google Scholar]
- 30.Tsuji T, Duh F M, Latif F, Popescu N C, Zimonjic D B, McBride J, Matsuo K, Ohyama H, Todd R, Nagata E, et al. J Biol Chem. 1998;273:6704–6709. doi: 10.1074/jbc.273.12.6704. [DOI] [PubMed] [Google Scholar]
- 31.Kita K, Ishizuka T, Ishijima S, Sonoda T, Tatibana M. J Biol Chem. 1994;269:8334–8340. [PubMed] [Google Scholar]
- 32.Zachariae W. Curr Opin Cell Biol. 1999;11:708–716. doi: 10.1016/s0955-0674(99)00041-1. [DOI] [PubMed] [Google Scholar]
- 33.Iftode C, Daniely Y, Borowiec J A. Crit Rev Biochem Mol Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- 34.Yuzhakov A, Kelman Z, Hurwitz J, O'Donnell M. EMBO J. 1999;18:6189–6199. doi: 10.1093/emboj/18.21.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foiani M, Lucchini G, Plevani P. Trends Biochem Sci. 1997;22:424–427. doi: 10.1016/s0968-0004(97)01109-2. [DOI] [PubMed] [Google Scholar]
- 36.Park D S, Levine B, Ferrari G, Greene L A. J Neurosci. 1997;17:8975–8983. doi: 10.1523/JNEUROSCI.17-23-08975.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lahti J M, Xiang J, Kidd V J. Prog Cell Cycle Res. 1995;1:329–338. doi: 10.1007/978-1-4615-1809-9_27. [DOI] [PubMed] [Google Scholar]
- 38.Dugan J M, deWit C, McConlogue L, Maltese W A. J Biol Chem. 1995;270:10982–10989. doi: 10.1074/jbc.270.18.10982. [DOI] [PubMed] [Google Scholar]
- 39.Hirst J, Lui W W Y, Bright N A, Totty N, Seaman M N J, Robinson M S. J Cell Biol. 2000;149:67–80. doi: 10.1083/jcb.149.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiari R, Foury F, De Plaen E, Baurain J F, Thonnard J, Coulie P G. Cancer Res. 1999;59:5785–5792. [PubMed] [Google Scholar]
- 41.Kalies K U, Hartmann E. J Biol Chem. 1996;271:3925–3929. doi: 10.1074/jbc.271.7.3925. [DOI] [PubMed] [Google Scholar]
- 42.Faundez V, Horng J T, Kelly R B. J Cell Biol. 1997;138:505–515. doi: 10.1083/jcb.138.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ringstad N, Gad H, Low P, Di Paolo G, Brodin L, Shupliakov O, De Camilli P. Neuron. 1999;24:143–154. doi: 10.1016/s0896-6273(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 44.Dopazo A, Lovenberg T W, Danielson P E, Ottiger H P, Sutcliffe J G. J Mol Neurosci. 1993;4:225–233. doi: 10.1007/BF02821554. [DOI] [PubMed] [Google Scholar]
- 45.Blondel O, Takeda J, Janssen H, Seino S, Bell G I. J Biol Chem. 1993;268:11356–11363. [PubMed] [Google Scholar]
- 46.Shibanuma M, Kuroki T, Nose K. J Biol Chem. 1992;267:10219–10224. [PubMed] [Google Scholar]
- 47.Dohrmann C E, Belaoussoff M, Raftery L A. Mech Dev. 1999;84:147–151. doi: 10.1016/s0925-4773(99)00055-6. [DOI] [PubMed] [Google Scholar]
- 48.Treisman J E, Lai Z C, Rubin G M. Development (Cambridge, UK) 1995;121:2835–2845. doi: 10.1242/dev.121.9.2835. [DOI] [PubMed] [Google Scholar]
- 49.Zhou X, Chada K. Keio J Med. 1998;47:73–77. doi: 10.2302/kjm.47.73. [DOI] [PubMed] [Google Scholar]
- 50.Nishizawa M, Nagata S. FEBS Lett. 1992;299:36–38. doi: 10.1016/0014-5793(92)80094-w. [DOI] [PubMed] [Google Scholar]
- 51.Skoulakis E M, Davis R L. Mol Neurobiol. 1998;16:269–284. doi: 10.1007/BF02741386. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe M, Isobe T, Ichimura T, Kuwano R, Takahashi Y, Kondo H. Brain Res Mol Brain Res. 1993;17:135–146. doi: 10.1016/0169-328x(93)90082-z. [DOI] [PubMed] [Google Scholar]
- 53.Martin H, Rostas J, Patel Y, Aitken A. J Neurochem. 1994;63:2259–2265. doi: 10.1046/j.1471-4159.1994.63062259.x. [DOI] [PubMed] [Google Scholar]
- 54.Semkova I, Krieglstein J. Brain Res Rev. 1999;30:176–188. doi: 10.1016/s0165-0173(99)00013-2. [DOI] [PubMed] [Google Scholar]
- 55.Guegan C, Ceballos-Picot I, Chevalier E, Nicole A, Onteniente B, Sola B. Neurobiol Dis. 1999;6:180–189. doi: 10.1006/nbdi.1999.0240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.