Abstract
Dynein is a minus-end–directed microtubule motor important for mitotic spindle positioning. In budding yeast, dynein activity is restricted to anaphase when the nucleus enters the bud neck, yet the nature of the underlying regulatory mechanism is not known. Here, the microtubule-associated protein She1p is identified as a novel regulator of dynein activity. In she1Δ cells, dynein is activated throughout the cell cycle, resulting in aberrant spindle movements that misposition the spindle. We also found that dynactin, a cofactor essential for dynein motor function, is a dynamic complex whose recruitment to astral microtubules (aMTs) increases dramatically during anaphase. Interestingly, loss of She1p eliminates the cell-cycle regulation of dynactin recruitment and permits enhanced dynactin accumulation on aMTs throughout the cell cycle. Furthermore, localization of the dynactin complex to aMTs requires dynein, suggesting that dynactin is recruited to aMTs via interaction with dynein and not the microtubule itself. Lastly, we present evidence supporting the existence of an incomplete dynactin subcomplex localized at the SPB, and a complete complex that is loaded onto aMTs from the cytoplasm. We propose that She1p restricts dynein-dependent spindle positioning to anaphase by inhibiting the association of dynein with the complete dynactin complex.
INTRODUCTION
Proper positioning of the mitotic spindle is essential for successful cell division and requires precise coordination of motor protein activity with the cell cycle. In Saccharomyces cervisiae the spindle is assembled in the mother cell and must be oriented perpendicular to the bud neck, the future site of cell division, and inserted across the bud neck before chromosome segregation occurs (Yeh et al., 2000). Two partially redundant pathways move the spindle by generating pulling forces on astral microtubules (aMTs) that emanate from the cytoplasmic face of the spindle pole bodies (SPB) (Shaw et al., 1997; Adames and Cooper, 2000). The first pathway utilizes the adenomatous polyposis coli-related protein Kar9p and a type V myosin to orient the spindle and position it adjacent to the bud neck before anaphase (Hwang et al., 2003; Kusch et al., 2003; Lee et al., 2003). The second pathway utilizes dynein and its cofactors Bik1p, a CLIP-170 homolog; Kip2p, a kinesin-related protein; and the dynactin complex to position the spindle across the bud neck at anaphase onset (Adames and Cooper, 2000; Lee et al., 2003; Carvalho et al., 2004).
In budding yeast, dynein activity is both spatially and temporally regulated to achieve correct spindle positioning within the dividing cell. Before mitosis, dynein is asymmetrically loaded onto the daughter-bound SPB and its associated aMTs (Grava et al., 2006; Segal and Bloom, 2001). This localization ensures unidirectional spindle movement toward the bud and is governed by Cdk1, spindle pole components, and bud neck kinases (Grava et al., 2006). Furthermore, analysis of dynein mutants showed that (1) dynein is necessary for establishing proper spindle position during, but not before, anaphase; and (2) cortical MT-sliding events mediated by dynein motor activity occur only during anaphase (Adames and Cooper, 2000). These data indicate that dynein activity is strictly limited to anaphase.
Dynein activity appears to be cell cycle–regulated in other organisms as well. In the C. elegans one-cell-stage embryo, overall dynein-dependent spindle displacement and oscillation increase dramatically during metaphase and climax in anaphase. Modeling studies attribute this change in activity to a gradual increase in dynein motor processivity (Pecreaux et al., 2006), but no molecular mechanism has been revealed. In fact, how dynein-dependent spindle positioning is temporally regulated during the cell cycle has not been determined in any organism.
MATERIALS AND METHODS
Yeast Strains and Media
The yeast strains used in this study are listed in the Supplementary Material, Table S2. The she1Δ null mutant was generated by PCR product-mediated gene deletion (Longtine et al., 1998). Other null mutants originated from the Research Genetics Collection. Jnm1-3HA and Jnm1-tdTomato were generated by inserting PCR-amplified 3HA::HIS3 or tdTomato::KanMX cassettes at the 3′ end of Jnm1 at its endogenous locus (Longtine et al., 1998, Shaner et al., 2004). For construction of 3GFP-tagged strains, ≈500-bp fragments of the DYN1, JNM1, ARP1, LDB18, and NIP100 open reading frames were subcloned into the BamHI site of pYS47 (Wong et al., 2007). A (GlyAla)3 linker was inserted between each gene and the 3GFP sequence. Each resulting plasmid was linearized and transformed into a haploid strain to integrate the fusion construct at the endogenous locus. The resulting strains showed no visible defects in spindle positioning. Furthermore, the DYN1-3GFP, JNM1-3GFP, LDB18-3GFP, and NIP100-3GFP alleles showed no synthetic growth defects in combination with a kar9Δ allele. However, the ARP1-3GFP kar9Δ strain was slightly sick. Thus, the DYN1-3GFP, JNM1-3GFP, LDB18-3GFP, and NIP100-3GFP fusions were completely functional, whereas the ARP1-3GFP fusion was partially functional.
We found that the localization of Jnm1-3GFP, Nip100-3GFP, and Arp1-3GFP to MTs was impaired in haploid cells expressing mCherry-Tub1 (a generous gift from E. Schiebel (ZMBH, Heidelberg, Germany); Khmelinskii et al., 2007) in combination with one copy of Tub1p. MT-associated localization of dynactin was restored when we increased expression of wild-type alpha tubulin. Therefore, we performed all of our dynactin visualization experiments using diploid strains heterozygous for the mCherry-TUB1 allele integrated at the URA3 locus and homozygous for wild-type TUB1. (mCherry-TUB1::URA3/ura3-52; TUB1/TUB1).
Fluorescence Microscopy
Images were obtained using an Olympus IX-71 microscope, 100× NA 1.4 objective and an Orca-ER camera (Hamamatsu, Hamamatsu City, Japan). Two-color images were obtained by sequential switching between RFP and GFP filter sets. For GFP-Tub1 time-lapse microscopy of pre-anaphase spindle movement, images were collected at 10-s intervals with 300-ms exposures. For time-lapse microscopy of spindle elongation, images were collected at 90-s intervals for 55 min. Each image represents a maximum intensity projection from a Z-stack containing 6 to 9 planes 0.2 μm apart. To determine the foci/cytoplasmic ratio of Arp1-3GFP and Nip100-3GFP fluorescence, fluorescence intensity of a GFP spot (referred to as a “focus”), the adjacent cytoplasm, and background were measured using a 5 × 5 pixel area. Background intensity was subtracted from the GFP spot and cytoplasm intensity values before the ratio was calculated. All image processing was performed using Metamorph.
Photobleaching
Photobleaching was performed using a Zeiss AxioObserver fluorescence microscope equipped with a spinning-disk confocal head (Solamere Technology Group, Salt Lake City, UT) and a Cascade II EMCCD camera (Photometrics, Tucson, AZ). Location of laser ablation was controlled using a MicroPoint system (Photonic Instruments, St. Charles, IL). Areas to be photobleached were subjected to two iterations of three pulses from a UV laser.
Immunoblotting
To detect Jnm1-3GFP, Arp1-3GFP, and Nip100-3GFP, the upper half of the membrane (containing proteins >75 kDa) was probed with 1:2500 anti-GFP (Torrey Pines Biolabs, East Orange, NJ) and 1:5000 anti-rabbit HRP-conjugated antibody (GE Healthcare, Piscataway, NJ). To detect Pgk1, the lower half of the membrane (containing proteins <75 kDa) was probed with 1:10,000 anti-Pgk1 (Molecular Probes, Eugene, OR) and 1:10,000 anti-mouse HRP-conjugated antibody (GE Healthcare). To detect Jnm1-3HA, the membrane was probed with 1:1000 rabbit anti-HA (Covance, HA.11). To detect Jnm1p, the membrane was probed with 1:1000 rabbit anti-Jnm1p serum (a gift from Kelly Tatchell, Louisiana State University Health Sciences Center, Shreveport, LA).
Immunoprecipitation
Immunoprecipitation was performed essentially as described previously (Moore et al., 2008) with a few minor exceptions. Lysates were incubated with rabbit anti-GFP antibody for one hour and then passed over protein A sepharose beads (Amersham Biosciences, Piscataway, NJ) to purify Nip100-3GFP. The column was washed with lysis buffer and eluted with SDS loading buffer.
Online Supplemental Material
Supplemental Figure S1 shows that loss of She1p can cause unequal distribution of spindle poles at the end of mitosis. Supplemental Figure S2 shows localization of dynein in wild-type and she1Δ cells. Supplemental Figure S3 shows expression levels of Jnm1-3GFP, Nip100-3GFP, and Arp1-3GFP. Supplemental Table S1 details lengths of aMTs and their dynamics in wild-type and she1Δ cells. Supplemental Table S2 lists the strains used in this study. Supplemental Movies S1-S5 show either wild-type or she1Δ cells that express GFP-Tub1.
RESULTS
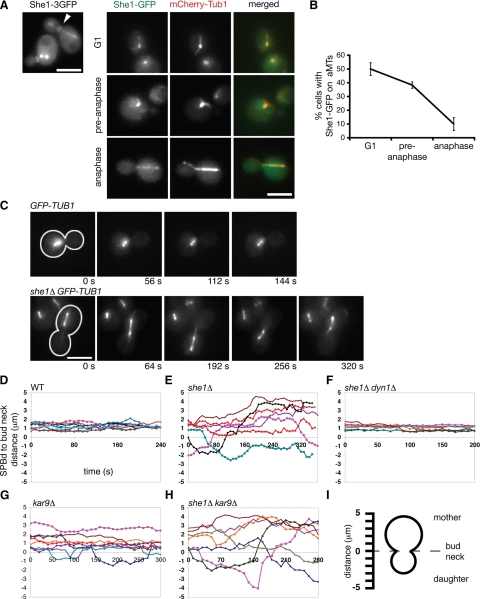
We set out to uncover determinants of dynein activity by first identifying proteins important for spindle positioning in budding yeast. We reasoned that She1p could influence spindle positioning based on its reported localization: in addition to appearing along the mitotic spindle, a 3GFP-tagged version of She1p localizes to the bud neck (Figure 1A; Wong et al., 2007), a structure that establishes polarity in the cell and serves as a landmark for spindle position and cell division (Grava et al., 2006). Further analysis revealed that She1-GFP localizes along aMTs in a cell cycle–dependent manner. Greater than 40% of G1 and pre-anaphase cells displayed aMT-localized She1-GFP, whereas <10% of anaphase cells displayed aMT-localized She1-GFP (Figure 1, A and B).
Figure 1.
She1p is a microtubule and bud neck–associated protein required to inhibit dynein activity before anaphase. (A) She1-3GFP localizes along the entire length of the mitotic spindle and at the bud neck (arrowhead). She1-GFP predominately localizes along astral microtubules in G1 and pre-anaphase cells, but not in anaphase cells. (B) Quantification of She1-GFP localization to aMTs. (C) Time-lapse images of pre-anaphase cells expressing GFP-Tub1. Cells were arrested with hydroxyurea to prevent entry into mitosis. Cell shape is outlined in white. Scale bar, 5 μm. (D through H) Graphs plotting the distances between the daughter-bound SPB (SPBd) and the bud neck over time for cells expressing GFP-Tub1. Each line represents the spindle position for an individual cell. Comparison of spindle position between wild-type (n = 9; D), she1Δ (n = 7; E), she1Δ dyn1Δ (n = 7; F), kar9Δ (n = 8; G), and she1Δ kar9Δ cells (n = 7; H). The addition of the dyn1Δ mutation, but not the kar9Δ mutation, eliminates spindle transiting seen in she1Δ cells. (I) Diagram depicting the measurements represented in D through H. Positive distances indicate that the SPBd was in the mother cell, whereas negative distances indicate that the SPBd was in the daughter cell.
To test directly whether She1p plays a role in orienting the spindle, we monitored pre-anaphase spindle movements in wild-type and she1Δ mutant cells expressing GFP-Tub1 (α tubulin) fusion protein. Spindle movement was quantified by measuring the distance between the daughter-bound spindle pole body (dSPB) and the bud neck over time (see Figure 1I for schematic). In wild-type cells, spindles stayed relatively fixed near the bud neck in the mother cell and were oriented perpendicular to the plane of division (Figure 1, C and D, and Supplemental Movie S1). However, in she1Δ cells, spindles exhibited dramatic movements. These motile spindles were lead by long aMTs that appeared to glide around the cell cortex, reminiscent of dynein-directed cortical aMT sliding normally seen during anaphase. Additionally, ≈29% of these spindles also traveled back and forth between the mother and daughter cells (Figure 1, C and E, and Supplemental Movie S2). To test whether dynein activity was responsible for these dramatic spindle movements, we monitored spindle position in cells lacking both She1p and the dynein heavy chain subunit (Dyn1). Indeed, pre-anaphase spindles in the she1Δ dyn1Δ mutant resembled wild-type spindles, lacking any dramatic movement (Figure 1F). We also noticed that she1Δ pre-anaphase spindles were longer than wild-type pre-anaphase spindles. Depletion of dynein in she1Δ cells restored pre-anaphase spindle length to normal (she1Δ : 1.75 ± 0.4 μm, n = 32; wild-type: 1.34 ± 0.3 μm, n = 19; she1Δ dyn1Δ : 1.28 ± 0.3 μm, n = 15), suggesting that ectopic dynein activity can “stretch” the spindle. Because dynein ordinarily is inactive before anaphase in budding yeast, these results suggest that She1p represses dynein activity until anaphase.
Premature spindle migration between the mother and the daughter cell has been reported in kar9Δ cells, raising the possibility that She1p could be an activator of the Kar9 pathway (Yeh et al., 2000). We found that spindle movements in kar9Δ cells (Figure 1G) were not as dramatic as those in she1Δ cells (Figure 1E), and that addition of the she1Δ mutation enhanced the spindle movements seen in kar9Δ cells (Figure 1H; mean maximum displacement = 2.93 ± 1 μm [she1Δ, n = 7], 1.66 ± 0.8 μm [kar9Δ, n = 8], 3.63 ± 0.6 μm [she1Δ kar9Δ, n = 7]). Also, we observed no synthetic interactions between the she1Δ and dyn1Δ alleles (unpublished data). Traditionally, null mutations in all known Kar9p pathway components show synthetic lethal/sick interactions with the dyn1Δ mutation (Lee et al., 2003; Grava et al., 2006). These results support the notion that She1p is not an activator of the Kar9p pathway but rather a repressor of the dynein pathway.
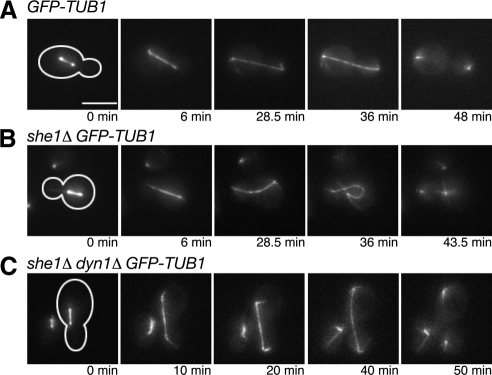
The results above indicate that She1p inhibits dynein activity before anaphase. We next asked whether She1p also negatively regulates dynein after anaphase. Once the cell completes anaphase, it must suppress dynein activity to permit proper spindle orientation in the next cell cycle. The absence of aMT-sliding on the cell cortex from late anaphase onward suggests that dynein suppression occurs before the next cell cycle begins (Adames and Cooper, 2000). If She1p is necessary to inactivate dynein during late anaphase, then cortical aMT sliding events may be observed during that time in she1Δ cells. To test this hypothesis, we monitored GFP-Tub1–expressing cells undergoing anaphase. In all wild-type cells, the spindle entered the bud, elongated until the spindle poles reached the ends of the dividing cell, and summarily disassembled (n = 7; Figure 2A and Supplemental Movie S3). In no case did cortical MT sliding occur once the spindle poles reached the cortex. In contrast, in 16 of 19 she1Δ cells, the spindle elongated properly but was then pulled around the cortex, likely by aMTs sliding along the cortex. These forces consequently bent the spindle to create a distinctive curled morphology (Figure 2B and Supplemental Movie S4). Although the presence of aMT-sliding suggests that spindle curling is caused by dynein activity, it is still possible that spindle curling is a side effect of spindle overextension. However, in all she1Δ dyn1Δ cells observed, late anaphase spindles remained straight throughout extension, implicating ectopic dynein activity as the cause of spindle curling in she1Δ cells (n = 7; Figure 2C). These results, combined with the observation that kar9Δ cells do not display spindle curling (unpublished data), further support the conclusion that She1p is an inhibitor of dynein and not an activator of the Kar9p pathway.
Figure 2.
She1p is required to repress dynein activity at the end of anaphase. (A through C) Time-lapse images of wild-type, she1Δ, and she1Δ dyn1Δ anaphase cells expressing GFP-Tub1. Cell shape is outlined in white. Scale bar, 5 μm. (A) The wild-type spindle remains straight throughout anaphase spindle elongation. (B) Without She1p, the spindle poles move around the cortex of the cell and create a distinctive curled morphology. Note the long cortical astral MT emanating from the motile spindle pole in the mother cell. (C) Addition of the dyn1Δ mutation eliminated the spindle curling seen in she1Δ cells.
Occasionally, during spindle curling in she1Δ cells, an aMT pulled one spindle pole far enough to penetrate the other cell, resulting in the formation of one cell with two spindle poles and one without any after cytokinesis (2 of 16 events; Supplemental Figure S1A and Supplemental Movie S5). Because chromosomes stay very closely attached to the spindle pole throughout the entire cell cycle in yeast, we suspected that spindle curling could cause unequal distribution of chromosomes. We tested this possibility by arresting haploid yeast in G1 and visualizing fluorescently marked chromosome III (Chr III). Wild-type cells possessed only one Chr III (99% with one GFP “dot”), whereas she1Δ cells frequently possessed two Chr IIIs (18% with two GFP dots; Supplemental Figure S1B). Further analysis revealed that she1Δ cells correctly segregated Chr III in early anaphase, indicating that the unequal distribution of Chr III seen in G1 cells was not a result of chromosome nondisjunction (unpublished data). These data suggest that She1p is required to inactivate dynein at the end of anaphase to ensure equal distribution of spindle poles and their associated chromosomes between the mother and daughter cell.
We next addressed how She1p affects dynein activity. The prevailing model for dynein function proposes that dynein is loaded onto aMTs, targeted to the plus ends, and then off-loaded to the cortex. Once anchored at the cortex, dynein uses its minus-end-directed motor activity to pull the attached aMT and the connected spindle toward the site of dynein anchorage (Lee et al., 2003; Li et al., 2005). Because She1p localizes along MTs, we first asked if She1p affects dynein activity indirectly by modifying aMT dynamics. However, there was no significant difference in aMT length, growth rate, and shrinkage rate between she1Δ and wild-type cells (Supplemental Table S1). Next, we tested whether She1p interferes with dynein loading onto aMTs or recruitment to aMT plus ends. Dynein is found on aMTs throughout the cell cycle, and, although its localization to aMT plus ends does increase as the cell enters mitosis, at least 50% of all cells retain dynein at aMT plus ends regardless of cell cycle stage (Sheeman et al., 2003). Therefore, it is unlikely that loading of dynein onto aMTs or its recruitment to the plus end contributes to the cell cycle–dependent regulation of its activity. Nevertheless, we studied the localization of dynein in wild-type and she1Δ cells using a 3GFP-tagged version of the dynein heavy chain (Dyn1-3GFP). Consistent with previous reports (Lee et al., 2003; Grava et al., 2006), we observed Dyn1-3GFP at SPBs, aMT plus ends, and the cell cortex in wild-type cells. This localization was unchanged in she1Δ cells (Supplemental Figure S2A). We also found that the percentage of cells with dynein localized to aMT plus ends increased slightly as the cells entered anaphase. Only a small, statistically insignificant change in the amount of plus end-localized dynein was observed in she1Δ cells (G1: 67.9 ± 8.9% [wt] vs. 85.2 ± 5.2% [she1Δ] P = 0.14; pre-anaphase: 84.5 ± 6.3% versus 87.3 ± 0.4% P = 0.59; anaphase: 90.3 ± 2% versus 92.3 ± 10.9% P = 0.82; Supplemental Figure S2B). Loss of She1p induces premature dynein activity without affecting dynein localization in pre-anaphase cells, suggesting that She1p inhibits dynein activity by a mechanism other than restricting its loading onto aMTs or recruitment to aMT plus ends.
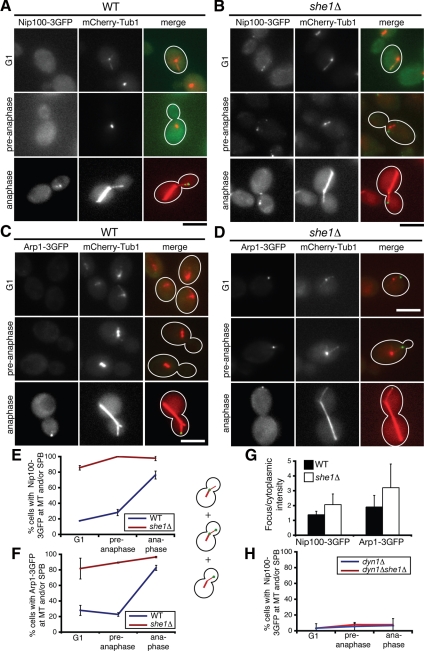
Another possibility is that She1p regulates a known enhancer of dynein motor function. One such candidate is the multi-subunit dynactin complex, which is essential for dynein activity but dispensable for dynein recruitment to aMTs in yeast (Schroer, 2004; Sheeman et al., 2003; Moore et al., 2008). We tested whether She1p regulates dynactin function by monitoring the localization of four prominent dynactin subunits: the p150glued ortholog Nip100p, the actin-related protein Arp1p, the dynamitin ortholog Jnm1p, and the p24 ortholog Ldb18p. For visualization, we tagged the endogenous copies of each protein with 3GFP at the C terminus. Haploid cells expressing the Jnm1-3GFP, Ldb18-3GFP, and Nip100-3GFP fusions displayed normal spindle positioning and were viable when Kar9p was depleted, indicating that the fusion proteins were functional. However, the Arp1-3GFP fusion was partially functional (see Materials and Methods). In addition, we coexpressed each 3GFP fusion protein with the microtubule marker mCherry-Tub1. We noticed that reducing the ratio of modified tubulin to wild-type tubulin enhanced dynactin localization to concentrated foci. Hence, we studied dynactin localization in homozygous TUB1/TUB1 diploid cells with only one copy of the mCherry-TUB1 allele integrated at the URA3 locus (see Materials and Methods). Consistent with previous reports (Moore et al., 2008), in wild-type cells, all four dynactin subunits were found at aMT plus ends, near SPBs, and the cell cortex (Figures 3 and 4).
Figure 3.
She1p affects the cell cycle–dependent localization of the dynactin components Nip100 (p150glued) and Arp1. (A and B) Colocalization of Nip100-3GFP and mCherry-Tub1 in (A) wild-type and (B) she1Δ G1, pre-anaphase, or anaphase cells, as indicated. Nip100-3GFP foci are largely absent until anaphase in wild-type cells (A) but are present throughout the cell cycle in she1Δ cells (B). (C and D) Colocalization of Arp1-3GFP and mCherry-Tub1 in (C) wild-type and (D) she1Δ G1, pre-anaphase, or anaphase cells, as indicated. Localization of Arp1-3GFP is similar to localization of Nip100-3GFP. Scale bars, 5 μm. (E and F) The percentage of cells with Nip100-3GFP (E) or Arp1-3GFP (F) foci at aMTs or SPBs as a function of cell cycle stage (n >45). (G) Ratio of the fluorescence intensity of Nip100-3GFP and Arp1-3GFP localized in foci versus the cytoplasm in wild-type and she1Δ cells (n = 23 to 30). (H) The percentage of dyn1Δ or dyn1Δ she1Δ cells with Nip100-3GFP localized to aMTs or SPBs (n = 90). Dyn1 is required for Nip100-3GFP localization to aMTs. All error bars represent SD.
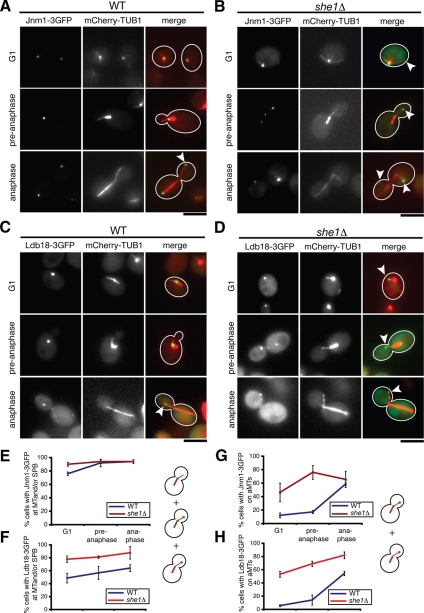
Figure 4.
She1p affects the cell cycle–dependent recruitment of Jnm1p (dynamitin) and Ldb18p (p24) to astral MTs. (A and B) Colocalization of Jnm1-3GFP and mCherry-Tub1 in (A) wild-type and (B) she1Δ G1, pre-anaphase, or anaphase cells, as indicated. (C and D) Colocalization of Ldb18-3GFP and mCherry-Tub1 in (C) wild-type and (D) she1Δ G1, pre-anaphase, or anaphase cells, as indicated. Jnm1-3GFP and Ldb18-3GFP foci primarily localize to SPBs in G1 and pre-anaphase cells and then appear on aMTs during anaphase. White arrowheads denote Jnm1-3GFP or Ldb18-3GFP localized at an astral MT plus end. Loss of She1p permits enhanced association of Jnm1-3GFP and Ldb18-3GFP with aMTs throughout the cell cycle. Scale bars, 5 μm. (E and F) The percentage of cells with Jnm1-3GFP (E) or Ldb18-3GFP (F) foci localized to aMTs or SPBs as a function of cell cycle stage. (G and H) Percentage of cells with Jnm1-3GFP (G) or Ldb18-3GFP (H) present on astral MTs (n >50). Error bars represent SD.
We found that the localization of Nip100-3GFP and Arp1-3GFP varied dramatically with the cell cycle. Nip100-3GFP and Arp1-3GFP were largely absent from SPBs or aMTs until anaphase: both proteins were found on SPBs or aMTs in ≈25% of G1 and pre-anaphase cells and in ≈80% of anaphase cells, representing an ≈3.2-fold increase (Figure 3, A, C, E, and F). When they did appear, Nip100-3GFP and Arp1-3GFP predominately localized to plus ends (unpublished data; Moore et al., 2008). However, in >80% of she1Δ cells, Nip100-3GFP and Arp1-3GFP appeared on aMTs regardless of cell cycle stage (Figure 3, B, D, E, and F), suggesting that She1p governs the cell cycle–dependent recruitment of Nip100p and Arp1p to aMTs. We also considered the possibility that the cell cycle–dependent appearance and disappearance of Nip100p and Arp1p is regulated by changes in protein degradation or expression. However, we found that the presence of She1p had no detectable impact on Nip100p or Arp1p protein levels in asynchronous, G1, and pre-anaphase cells (Supplemental Figure S3A and unpublished data). Yet, the ratio of Nip100-3GFP and Arp1-3GFP fluorescence detected in MT-associated foci versus the cytoplasm was ≈50% higher in anaphase she1Δ cells than in anaphase wild-type cells (Figure 3G), supporting the idea that more Nip100p and Arp1p are recruited to aMTs from the cytoplasmic pool in the absence of She1p. We then asked whether dynactin recruitment to aMTs occurs via direct binding to the microtubule or indirectly through dynein. We observed that Nip100-3GFP localization to SPBs and aMTs was almost entirely eliminated in both dyn1Δ and dyn1Δ she1Δ cells, suggesting that dynactin is recruited to aMTs through its interaction with dynein (Figure 3H). This result is consistent with the report that a mutant version of p150glued lacking its MT-binding domain still localizes to MTs in Drosophila melanogaster S2 cells (Kim et al., 2007). Overall, these data suggest that She1p precludes the interaction of dynein and dynactin before anaphase.
Interestingly, the behavior of Jnm1p and Ldb18p differed from the behavior of the other dynactin subunits. Unlike Nip100-3GFP and Arp1-3GFP, Jnm1-3GFP and Ldb18-3GFP almost always localized in foci associated with SPBs or aMTs throughout the cell cycle in wild-type cells (Figure 4, A, C, E, and F). In G1 and pre-anaphase cells, the majority of Jnm1-3GFP and Ldb18-3GFP foci localized at or very near SPBs (Figure 4, A and C, and unpublished data). However, similar to Arp1-3GFP and Nip100-3GFP, the frequency of Jnm1-3GFP and Ldb18-3GFP on aMTs increased ≈3.4-fold as cells entered anaphase (Figure 4, A, C, G, and H). Again, loss of She1p eliminated the dramatic cell cycle-dependent change in Jnm1-3GFP and Ldb18-3GFP localization and enhanced the presence of both subunits on aMTs (Figure 4, B, D, G, and H). These results suggest that She1p inhibits Jnm1p and Ldb18p recruitment to aMTs until anaphase.
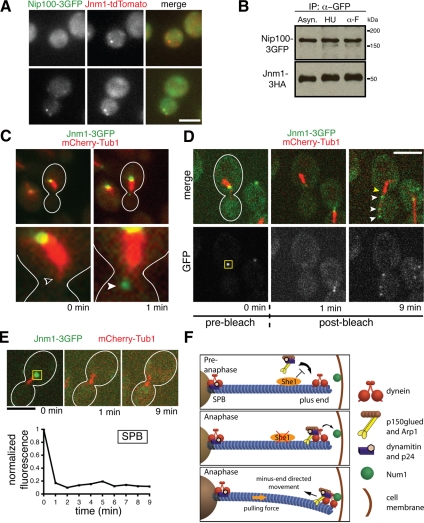
The difference in localization patterns of Jnm1p, Ldb18p, Arp1p, and Nip100p suggests that the dynactin complex could exist in subcomplexes. Indeed, only ≈60% of Jnm1-tdTomato foci colocalized with Nip100-3GFP foci in asynchronous wild-type cells (n = 114; Figure 5A). The presence of dynactin subcomplexes is further supported by previous cosedimentation and coimmunopurification experiments that identified large pools of Jnm1p and Ldb18p unassociated with Nip100p and Arp1p (Moore et al., 2008; Amaro et al., 2008). Given these findings and our observations that Jnm1p and Ldb18p localize to SPBs throughout the cell cycle but appear on aMTs only during anaphase, it is possible that the complete dynactin complex is assembled during anaphase from two spatially segregated subcomplexes: (1) an SPB-localized subcomplex that contains Jnm1p and Ldb18p, and (2) a cytoplasmic subcomplex that contains Nip100p and Arp1p. However, roughly equal amounts of Jnm1-3HA coimmunoprecipitated with Nip100-3GFP in G1, pre-anaphase, and asynchronous wild-type cells, suggesting that a complete version of the dynactin complex exists throughout the cell cycle (Figure 5B). In addition, time-lapse microscopy following a cell entering anaphase showed that Jnm1p suddenly appeared at the aMT plus end, rather than being transported there from the SPB (Figure 5C). Furthermore, after photobleaching SPB-localized Jnm1-3GFP signal, we observed an increase in GFP fluorescence on the aMT but no recovery at the SPB (Figure 5D), ruling out the possibility that aMT-localized Jnm1-3GFP foci result from transport and concentration of undetectable amounts of SPB-localized Jnm1-3GFP. The fact that we never observed recovery of fluorescence at the SPB indicates that the Jnm1p localized there comprises a static structure (Figure 5E). Therefore, the Jnm1p that appeared on the aMT was recruited from the cytoplasm, most likely in complex with the other dynactin subunits. In total, these results suggest that the dynactin complex comprises complete and incomplete varieties, and that She1p specifically hinders recruitment of the complete version to aMTs until anaphase.
Figure 5.
Dynactin exists in complete and incomplete complexes that are spatially distinct. (A) Approximately 60% of Jnm1-tdtomato colocalizes with Nip100-3GFP in asynchronous cells (n = 114). (B) Immunoprecipitation of Nip100-3GFP from asynchronous (Asyn.), hydroxyurea-arrested (HU), and alpha factor–arrested (α-F) cells. Essentially equal amounts of Jnm1-3HA copurify with Nip100-3GFP at different cell cycle stages, indicating that a complete dynactin complex exists throughout the cell cycle. (C) Before anaphase, Jnm1-3GFP localizes to the SPB but is absent from the aMT plus end (unfilled arrowhead). Near the anaphase transition, Jnm1-3GFP suddenly appeared at the plus end (filled arrowhead), rather than being transported from the SPB. Anaphase onset was indicated by rapid elongation of the mitotic spindle shortly after Jnm1-3GFP appeared on the aMT (not shown). (D) Jnm1-3GFP fluorescence at the SPB was photobleached in pre-anaphase cells. The yellow box represents the area that was photobleached at t = 0.5 min. Appearance of GFP foci (white arrowheads) occurred along the aMT as the cell entered anaphase. There was no recovery of GFP fluorescence at the SPB (yellow arrowhead). (E) The GFP fluorescence at the SPB in a different cell also did not recover after photobleaching, indicating that SPB-localized dynactin subcomplexes are very static. The yellow box represents the area that was photobleached at t = 0.5 min. Scale bars, 5 μm. (F) A model for cell cycle control of dynein activation. She1p precludes stable association of the complete dynactin complex with dynein until anaphase.
DISCUSSION
The majority of the work detailing mitotic spindle positioning has focused on activation of the Kar9p and dynein pathways. Less studied, but equally important, is the issue of how these pathways are silenced to prevent unnecessary spindle movement. In this study, we explored the mechanisms that restrict dynein-dependent spindle positioning to anaphase in budding yeast. We obtained strong genetic evidence that She1p is an inhibitor of dynein activity in vivo. Also, we found that the dynactin complex is a dynamic entity whose localization is coordinated with the cell cycle. She1p regulates the localization of dynactin, preventing the complete complex from associating with aMTs, via dynein, until anaphase.
Connection between She1p and the Dynactin Complex
Our results indicate that She1p hinders the stable interaction between dynein and dynactin by a mechanism that remains to be determined. Unfortunately, we cannot predict She1p function based on amino acid sequence, as we were unable to identify any instructive homology to previously characterized proteins. We have also purified She1p from yeast and analyzed copurifying proteins by mass spectrometry; however, no previously known dynein pathway components were identified (unpublished data). Nevertheless, our data support the conclusion that She1p specifically affects dynactin. Loss of She1p dramatically enhances dynactin recruitment to aMTs. The frequency of G1 and pre-anaphase cells displaying aMT-associated dynactin is ≈4 times higher for she1Δ cells than for wild-type cells (Figures 3 and 4). This effect is not an indirect consequence of alterations in aMT dynamics or length, because She1p has no significant impact on these parameters. Also, it is unlikely that the change in dynactin localization is attributable to enhanced function of other MT-bound proteins needed for dynein activity. Dynein plus-end localization requires proper functioning of NudEL, CLIP170, and LIS1 (Li et al., 2005). Because She1p has little impact on dynein recruitment to aMTs or tracking to the plus ends, She1p likely does not affect the activity of NudEL, CLIP170, and LIS1.
Currently, we suspect that She1p may impact dynein-dynactin interaction indirectly through a phosphoryation-dependent mechanism. Overexpression of the protein phosphatase I cofactor Bud14p in budding yeast causes dynein-dependent spindle transiting in pre-anaphase cells (Knaus et al., 2005). This phenotype is similar to the one that we have observed in she1Δ cells. In addition, Vaughan et al. (2001) reported that phosphorylation of the dynein IC (DIC) disrupts binding of p150glued to dynein. These studies both argue for dephosphorylation of DIC as a means to regulate dynein activity. It is enticing to speculate that She1p inhibits dynactin association with dynein by encouraging DIC phosphorylation. However, it is equally possible that other factors, including She1p itself, are phospho-targets.
Possibility of Dynactin Subcomplexes
Rate-zonal sedimentation experiments (Amaro et al., 2008; Moore et al., 2008) revealed pools of p24 (Ldb18p) and dynamitin (Jnm1p) that did not sediment with the other dynactin subunits, raising the possibility that dynactin subunits could form subcomplexes with discrete functions. These studies, however, did not report any major differences in dynactin subunit localization. It is possible that these studies reached this conclusion because their analyses focused on anaphase cells, when the localization of all dynactin subunits to aMTs is most prominent. In our study, we examined cells at different cell cycle stages and noticed that dynamitin and p24 shared a localization pattern that differed from the p150glued and Arp1 localization pattern. Although all four subunits appeared on aMTs primarily during anaphase, dynamitin and p24 localized to SPBs throughout the cell cycle. Thus, the SPB-localized dynamitin and p24 subunits could comprise a subcomplex distinct from the aMT-localized complete dynactin complex. This possibility is consistent with findings from Moore et al. (2008) that dynamitin and p24 still localize to SPBs, but not aMTs, in the absence of p150glued.
Because of the sensitivity of dynactin to modification of its subunits, especially dynamitin, which disrupts the complex when overexpressed (Schroer, 2004), it is possible that the SPB localization of dynamitin and p24 is an artifact caused by addition of the 3GFP tag. However, both Jnm1-3GFP and Ldb18-3GFP are functional because they rescue the nuclear migration defect characteristic of jnm1Δ and ldb1Δ null mutants (see Materials and Methods). In addition, we verified that the 3GFP did not alter expression or stability of Jnm1p (Supplemental Material, Figure 3B). Finally, the localization data corroborates the biochemical analysis of native Jnm1p and Ldb18p performed by Amaro et al. (2008) and Moore et al. (2008).
Cell Cycle Regulation of She1p Activity
Our data suggest that She1p activity is also regulated in a cell cycle stage–dependent manner. Activation of the anaphase promoting complex/cyclosome (APC/c) triggers movement of the anaphase spindle into the bud (Ross and Cohen-Fix, 2004). However, it is unlikely that She1p activity is silenced through APC/c-mediated degradation because She1p protein levels remain unchanged throughout anaphase (Figure 1A and unpublished data). Instead, considering that She1-GFP localization along aMTs drastically diminishes during anaphase, it is likely that She1p activity is regulated through its controlled loading and removal from aMTs. In the future, it will be worthwhile to uncover the factors that determine She1p localization.
Model for Cell Cycle Regulation of Dynein
Based on our work and previous studies, we propose that cell cycle–regulated association between dynein and dynactin controls dynein-driven spindle positioning (Figure 5F). In our model, there exist at least two versions of dynactin: an incomplete version located at the SPB containing at least dynamitin (Jnm1p) and p24 (Ldb18p), and a complete version located in the cytoplasm containing at least dynamitin, p24, Arp1, and p150glued (Nip100p). Before anaphase, She1p inhibits association of the complete dynactin complex with dynein, thus rendering dynein inactive. During anaphase, removal of She1p from the aMT permits binding of the complete dynactin complex with dynein, which then stimulates dynein motor activity and primes it for off-loading onto the cell cortex. As a result, dynein pulls the spindle into the bud only during anaphase.
Coordination of spindle positioning with the cell cycle requires precise control of microtubule-bound motor proteins like dynein. This work provides a possible explanation for how dynein becomes active strictly during anaphase in budding yeast. Given that the proteins and mechanisms involved in dynein-dependent spindle positioning are evolutionarily conserved throughout eukaryotes (Gomes et al., 2005), we expect our findings to be generally applicable.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Schiebel for providing the mCherry-TUB1 plasmid, Roger Tsien (University of California, San Diego) for providing the td-tomato cassette, Kelly Tatchell (Louisiana State University Health Sciences Center) for providing Jnm1p antibody, and C. Boone (University of Toronto) for sharing unpublished data. We also thank Voytek Okgreglak, Ross Rounsevell, Yidi Sun, and Randall Tyers for experimental help and suggestions. This work was supported by NIH grant GM 47842 to G.B. and a National Science Foundation Graduate Research Fellowship to J.B.W.
Abbreviations used:
- MT
microtubule
- aMT
astral microtubule
- SPB
spindle pole body
- Chr III
chromosome III
- td-tomato
tandem dimer tomato.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-03-0186) on April 29, 2009.
REFERENCES
- Adames N. R., Cooper J. A. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro I. A., Costanzo M., Boone C., Huffaker T. C. The Saccharomyces cerevisiae homolog of p24 is essential for maintaining the association of p150Glued with the dynactin complex. Genetics. 2008;178:703–709. doi: 10.1534/genetics.107.079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Gupta M. L., Jr., Hoyt M. A., Pellman D. Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev. Cell. 2004;6:815–829. doi: 10.1016/j.devcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Gomes E. R., Jani S., Gundersen G. G. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Grava S., Schaerer F., Faty M., Philippsen P., Barral Y. Asymmetric recruitment of dynein to spindle poles and microtubules promotes proper spindle orientation in yeast. Dev. Cell. 2006;10:425–439. doi: 10.1016/j.devcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Hwang E., Kusch J., Barral Y., Huffaker T. C. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J. Cell Biol. 2003;161:483–488. doi: 10.1083/jcb.200302030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., Lawrence C., Roostalu J., Schiebel E. Cdc14-regulated midzone assembly controls anaphase B. J. Cell Biol. 2007;177:981–993. doi: 10.1083/jcb.200702145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Ling S. C., Rogers G. C., Kural C., Selvin P. R., Rogers S. L., Gelfand V. I. Microtubule binding by dynactin is required for microtubule organization but not cargo transport. J. Cell Biol. 2007;176:641–651. doi: 10.1083/jcb.200608128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus M., Cameroni E., Pedruzzi I., Tatchell K., De Virgilio C., Peter M. The Bud14p-Glc7p complex functions as a cortical regulator of dynein in budding yeast. EMBO J. 2005;24:3000–3011. doi: 10.1038/sj.emboj.7600783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch J., Liakopoulos D., Barral Y. Spindle asymmetry: a compass for the cell. Trends Cell Biol. 2003;13:562–569. doi: 10.1016/j.tcb.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Lee W. L., Oberle J. R., Cooper J. A. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J. Cell Biol. 2003;160:355–364. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lee W. L., Cooper J. A. NudEL targets dynein to microtubule ends through LIS1. Nat. Cell Biol. 2005;7:686–690. doi: 10.1038/ncb1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A, Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Moore J. K., Li J., Cooper J. A. Dynactin function in mitotic spindle positioning. Traffic. 2008;9:510–527. doi: 10.1111/j.1600-0854.2008.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecreaux J., Röper J. C., Kruse K., Jülicher F., Hyman A. A., Grill S. W., Howard J. Spindle oscillations during asymmetric cell division require a threshold number of active cortical force generators. Curr. Biol. 2006;16:2111–2122. doi: 10.1016/j.cub.2006.09.030. [DOI] [PubMed] [Google Scholar]
- Ross K. E., Cohen-Fix O. A role for the FEAR pathway in nuclear positioning during anaphase. Dev. Cell. 2004;6:729–735. doi: 10.1016/s1534-5807(04)00128-5. [DOI] [PubMed] [Google Scholar]
- Schroer T. A. DYNACTIN. Annu. Rev. Cell. Dev. Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- Segal M., Bloom K. Control of spindle polarity and orientation in Saccharomyces cerevisiae. Trends Cell Biol. 2001;11:160–166. doi: 10.1016/s0962-8924(01)01954-7. [DOI] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp.red fluorescent protein. Nat. Biotech. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shaw S. L., Yeh E., Maddox P., Salmon E. D., Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J. Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman B., Carvalho P., Sagot I., Geiser J., Kho D., Hoyt M. A., Pellman D. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr. Biol. 2003;13:364–372. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- Vaughan P. S., Leszyk J. D., Vaughan K. T. Cytoplasmic dynein intermediate chain phosphorylation regulates binding to dynactin. J. Biol. Chem. 2001;276:26171–26179. doi: 10.1074/jbc.M102649200. [DOI] [PubMed] [Google Scholar]
- Wong J., Nakajima Y., Westermann S., Shang C., Kang J. S., Goodner C., Houshmand P., Fields S., Chan C. S., Drubin D., Barnes G., Hazbun T. A protein interaction map of the mitotic spindle. Mol. Biol. Cell. 2007;18:3800–3809. doi: 10.1091/mbc.E07-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Yang C., Chin E., Maddox P., Salmon E. D., Lew D. J., Bloom K. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol. Biol. Cell. 2000;11:3949–3961. doi: 10.1091/mbc.11.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.