Abstract
The yeast spindle pole body (SPB), the functional equivalent of mammalian centrosome, duplicates in G1/S phase of the cell cycle and then becomes inserted into the nuclear envelope. Here we describe a link between SPB duplication and targeted translation control. When insertion of the newly formed SPB into the nuclear envelope fails, the SESA network comprising the GYF domain protein Smy2, the translation inhibitor Eap1, the mRNA-binding protein Scp160 and the Asc1 protein, specifically inhibits initiation of translation of POM34 mRNA that encodes an integral membrane protein of the nuclear pore complex, while having no impact on other mRNAs. In response to SESA, POM34 mRNA accumulates in the cytoplasm and is not targeted to the ER for cotranslational translocation of the protein. Reduced level of Pom34 is sufficient to restore viability of mutants with defects in SPB duplication. We suggest that the SESA network provides a mechanism by which cells can regulate the translation of specific mRNAs. This regulation is used to coordinate competing events in the nuclear envelope.
Keywords: Regulation of translation, spindle pole body, nuclear pore complex, Smy2, Eap1, Scp160
The yeast spindle pole body (SPB) is the functional equivalent of the mammalian centrosome (Jaspersen and Winey 2004; Jaspersen and Stearns 2008). Centrosomes and SPBs duplicate once per cell cycle, organize microtubules, and are associated with cell cycle regulators (Pereira and Schiebel 2001; Stegmeier and Amon 2004). In addition, both are connected with the nuclear envelope. The budding yeast SPB is embedded in the nuclear envelope in a similar way to the nuclear pore complexes (NPCs). This incorporation into the membrane differs from the centrosome of higher eukaryotes that is anchored to the nuclear envelope via a proteinaceous link (Jaspersen and Winey 2004; Tzur et al. 2006).
The yeast SPB duplicates in G1/S phase of the cell cycle by a template-based mechanism (Adams and Kilmartin 1999; Jaspersen and Winey 2004). An intermediate in this duplication process, the duplication plaque, develops on the cytoplasmic side of the nuclear envelope as an extension of the pre-existing SPB. The duplication plaque then becomes inserted into the nuclear envelope as a consequence of the action of the essential NDC1, MPS2, BBP1, and NBP1 genes (Winey et al. 1991, 1993; Adams and Kilmartin 1999; Schramm et al. 2000; Araki et al. 2006). A defect in these genes leads to a failure in SPB duplication such that the insertion of the duplication plaque into the nuclear envelope is either fully or partially compromised. The integral membrane protein Mps2 forms a tight complex with the cytoplasmic Bbp1. For SPB duplication the Mps2–Bbp1 complex cooperates with the Nbp1 protein, and the highly conserved integral membrane protein Ndc1 (Winey et al. 1993; Adams and Kilmartin 1999; Schramm et al. 2000; Araki et al. 2006; Mansfeld et al. 2006).
Ndc1 has an additional role in NPC biogenesis (Lau et al. 2004; Madrid et al. 2006; Mansfeld et al. 2006; Stavru et al. 2006). At the NPC, Ndc1 interacts with two other integral membrane proteins, Pom34 and Pom152 (Alber et al. 2007a,b; Onischenko et al. 2009). Ndc1, Pom34, and Pom152 have redundant functions in the biogenesis of NPCs (Madrid et al. 2006).
Curiously, NDC1 shows genetic interactions with EAP1 (Chial et al. 2000). In yeast, Eap1 and Caf20 are the two eIF4E-binding proteins that prevent formation of the eIF4E–eIF4G complex (Gingras et al. 1999), which is crucial for the recruitment of the 5′ end of mRNAs to the 40S ribosomal subunit during initiation of translation (Gingras et al. 1999). Eap1 and Caf20 inhibit general initiation of translation in response to stress conditions such as cadmium and diamides in the growth medium or the occurrence of membrane stress (Deloche et al. 2004; Mascarenhas et al. 2008).
SMY2 was discovered as a high dosage suppressor of the conditional lethal myo2-66 mutation (Lillie and Brown 1994). Based on bioinformatics and in vitro studies with the truncated protein, it was concluded that the GYF domain of Smy2 interacts with proteins containing PPG repeats such as Eap1 and pre-mRNA splicing factors (Kofler et al. 2005; Georgiev et al. 2007). In addition, Smy2 has GYF domain-independent functions at COPII vesicles (Higashio et al. 2008). Thus, the molecular role of Smy2 is largely not understood.
We now describe a novel translational initiation control mechanism that is exerted by the Smy2–Eap1–Scp160–Asc1 network of proteins, named SESA, to regulate the initiation of translation of POM34 mRNA, upon SPB duplication defects. The failure of the SPB to insert into the nuclear envelope triggers SESA to inhibit the translation initiation of POM34 mRNA. The concomitant reduction in the level of Pom34 protein restores SPB duplication and so ensures survival of cells that encounter difficulties in inserting their SPB into the nuclear envelope.
Results
A genetic link between the SMY2 gene and the SPB insertion machinery
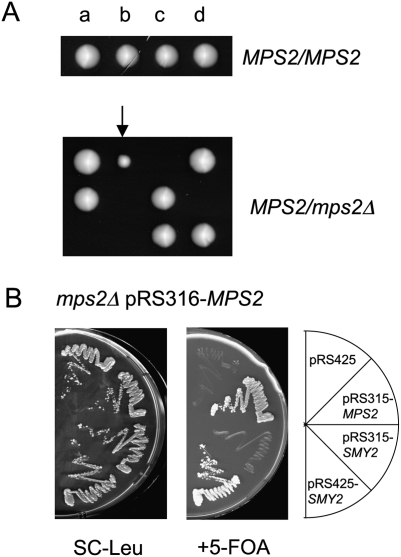
MPS2 encodes an essential, single membrane-spanning protein that inserts the newly formed SPB into the nuclear envelope (Winey et al. 1991; de la Cruz Munoz-Centeno et al. 1999; Schramm et al. 2000; Araki et al. 2006). Attempts to germinate mps2Δ spores from an mps2Δ/MPS2 parent frequently led to the appearance of slow-growing mps2Δ survivors indicating that a suppressor mutation can ensure survival of cells lacking MPS2 (Fig. 1A; arrow). A genetic screen for the wild-type genes that suppress the lethal growth defect of mps2Δ cells identified SMY2 as a high gene dosage suppressor (Fig. 1B, sector 4). Smy2 is an endoplasmic reticulum (ER)-associated, GYF domain protein whose function remains to be elucidated (Kofler et al. 2005; Higashio et al. 2008).
Figure 1.
Suppressor screen for the lethal phenotype of MPS2 deletion. (A) Spore analysis of MPS2/MPS2 and mps2Δ/MPS2 cells. a, b, c, and d indicate cells that developed from spores of one complete tetrad. The cells marked by an arrow did not contain the MPS2 gene. (B) SMY2 is a multicopy suppressor of the lethality of mps2Δ cells. mps2Δ pRS316-MPS2 cells were transformed with the indicated plasmids. Transformants were grown on SC-Leu or 5-FOA plates for 3 d at 23°C.
SMY2 functions through the translation inhibitor EAP1
The cellular function of SMY2 is obscured by reports on its interaction with proteins involved in translation control, pre-mRNA splicing, and protein secretion (Georgiev et al. 2007; Kofler et al. 2005; Higashio et al. 2008). To unravel the function of SMY2, we performed a synthetic lethal screen to reveal genes that become essential in the absence of SMY2 (Supplemental Fig. S1; Supplemental Material). The screen identified two groups of genes: one set that is involved in the regulation of protein translation, and the other in membrane function (Supplemental Fig. S1C). This data suggested that SMY2 may act as a functional bridge that connects these two processes.
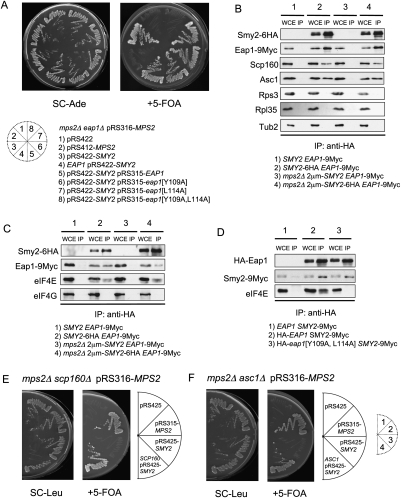
A link between Smy2 protein and control of protein translation was provided by the report that the GYF domain of Smy2 has the ability to interact with the PPG repeats of the translation inhibitor Eap1 (Kofler et al. 2005). To determine whether SMY2 functions through EAP1, we asked whether EAP1 becomes essential for viability of mps2Δ 2μm-SMY2 cells. Loss of EAP1 was lethal for the mps2Δ 2μm-SMY2 mutant cells (Fig. 2A, sector 3). In addition, the PPG mutant eap1[G624A] did not allow growth of mps2Δ cells (Supplemental Fig. S2C, row 8) and the GYF domain mutant smy2[Y234A] was no longer able to suppress the essential function of MPS2 (Supplemental Fig. S2A, sector 3). Thus, the interaction between the GYF element of Smy2 and the PPG domain of Eap1 is essential for the bypass of MPS2. In contrast, CAF20 that encodes the second eIF4E inhibitor (Ibrahimo et al. 2006), was not required for growth of mps2Δ 2μm-SMY2 cells (Supplemental Table 1). This data suggests that SMY2 functions through EAP1.
Figure 2.
EAP1, SCP160, and ASC1 are essential components of the SMY2 pathway. (A) Dependency of suppression on EAP1. mps2Δ eap1Δ pRS316-MPS2 cells were transformed with the listed plasmids and tested for growth on SC-Ade and 5-FOA plates. eap1[Y109A], eap1[L114A], and eap1[Y109A,L114A] code for Eap1 proteins with mutations that partially (eap1[Y109A], eap1[L114A]) or totally (eap1[Y109A,L114A]) disrupt binding to eIF4E (Fig. 2D; Ibrahimo et al. 2006). (B) Coimmunoprecipitation of Eap1, Scp160, and Asc1 by Smy2. Extracts from cells expressing SMY2 EAP1-9Myc or SMY2-6HA EAP1-9Myc were immunoprecipitated by anti-HA-coated magnetic beads and analyzed by immunoblotting with the indicated antibodies. (WCE) Whole-cell extract, 10% of the immunoprecipitation input; (IP) immunoprecipitation. (C) Coimmunoprecipitation of Eap1, eIF4E, and eIF4G by Smy2. Extracts from the yeast cells expressing SMY2 EAP1-9Myc or SMY2-6HA EAP1-9Myc were immunoprecipitated by anti-HA-coated magnetic beads and analyzed by immunoblotting with the indicated antibodies. Abbreviations as in B. (D) Coimmunoprecipitation of Smy2 and Eap1 and mutated Eap1 protein that fails to bind to eIF4E. Extracts from yeast cells with the indicated genotypes were immunoprecipitated by anti-HA-coated magnetic beads and analyzed by immunoblotting with anti-HA, anti-Myc, and anti-eIF4E antibodies. Abbreviations are as in B. (E,F) Dependency of suppression on SCP160 and ASC1. mps2Δ scp160Δ pRS316-MPS2 (E) or mps2Δ asc1Δ pRS316-MPS2 cells (F) were transformed with the indicated plasmids and tested for growth on SC-Leu and 5-FOA at 23°C.
Coimmunoprecipitation experiments were performed to demonstrate that Smy2 interacts with Eap1 in vivo (Fig. 2B). The Eap1–Smy2 interaction was equally observed in wild-type and mps2Δ 2μm-SMY2 cells (Fig. 2B), suggesting that the mps2Δ defect did not induce Smy2–Eap1 interaction. Moreover, the GYF domain of Smy2 was essential for the interaction, as Eap1 did not coimmunoprecipitate with the GYF domain-defective Smy2[Y234A] (Supplemental Fig. S2B). Together, this data suggests that Smy2 interacts in vivo with the translation inhibitor Eap1.
Eap1 allows bypass of MPS2 function by inhibiting translation factor eIF4E
Eap1 inhibits cap-dependent translation by competing with the initiation factor eIF4G (a subunit of the eIF4F cap-binding complex) for binding to eIF4E (Sonenberg and Gingras 1998). Because it was recently suggested that Eap1 may execute functions that are distinct from its role in the inhibition of translation (Chial et al. 2000), we asked whether EAP1 regulates translation or plays some other function in mps2Δ cells. Coimmunoprecipitation experiments showed that eIF4E was associated with Smy2 and Eap1 in both wild-type and mps2Δ 2μm-SMY2 cells, whereas eIF4G was excluded from this association (Fig. 2C). The latter was expected as Eap1 competes with eIF4G for the same site in eIF4E.
The interaction between Eap1 and eIF4E is impaired by the eap1[Y109A, L114A] mutations (Fig. 2D). This impairment of Eap1/eIF4E association relieves the inhibitory influence of Eap1 upon translation initiation (Ibrahimo et al. 2006). In this eap1[Y109A, L114A] background, SMY2 was no longer able to suppress the lethal phenotype of the mps2Δ mutation (Fig. 2A, sector 8), even though the Eap1[Y109A, L114A] protein still associated with Smy2 in coimmunoprecipitation assays (Fig. 2D). Thus, inhibition of translation factor eIF4E by Eap1 binding is crucial for the viability of mps2Δ cells.
SCP160, ASC1, and BFR1 become essential in cells lacking MPS2
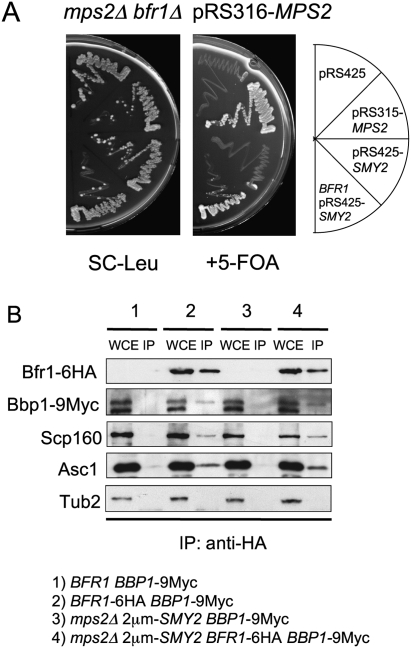
The proteins that regulate and cooperate with Eap1 in response to membrane defects are largely unknown (Deloche et al. 2004). With the mps2Δ 2μm-SMY2 cells in hand, we were able to test genes for function in conjunction with EAP1 and SMY2. We analyzed about 40 genes with roles in translation control, the UPR pathway (Sidrauski and Walter 1997), and nuclear envelope function (Supplemental Table 1) for their requirement in the viability of mps2Δ 2μm-SMY2 cells. This analysis identified the genes coding for the polysome-associated, mRNA-binding protein Scp160 that has been shown to exist in a complex with Eap1 (Mendelsohn et al. 2003), the Scp160-interacting protein Asc1 (essential for ER localization of Scp160) (Baum et al. 2004), and the brefeldin A resistance protein Bfr1 (a component of polyribosome-associated mRNP complexes) (Lang et al. 2001), as being essential in the mps2Δ 2μm-SMY2 background (Figs. 2E,F, 3A; Supplemental Table 1).
Figure 3.
BFR1 is essential for viability of cells lacking MPS2. (A) Dependency of suppression on BFR1. mps2Δ bfr1Δ pRS316-MPS2 cells were transformed with the indicated plasmids and tested for growth on SC-Leu and 5-FOA plates at 23°C. (B) Coimmunoprecipitation of Bbp1 by Bfr1. Extracts from the yeast cells with the indicated genotypes were subjected to immunoprecipitation with anti-HA-coated magnetic beads and analyzed by immunoblotting with the indicated antibodies. Abbreviations as in Figure 2B.
Coimmunoprecipitation experiments validated this genetic approach. Eap1, Scp160, and Asc1 were found to be in complex with Smy2 (Fig. 2B), and in addition, Scp160, Smy2, and Eap1 cofractionated in part in sucrose gradients (Supplemental Fig. S3). These interactions were not mediated by ribosomes, since the ribosomal proteins Rps3 (40 S ribosomal subunit) and Rpl35 (60 S ribosomal subunit) were not detected in the Smy2 immunoprecipitate (Fig. 2B). Interestingly, coimmunoprecipitation of Smy2 and Eap1 relied on the presence of the Scp160 protein but not on Asc1 (Supplemental Fig. S2D) and RNA, as RNaseA treatment only mildly affected the efficiency of coprecipitation (Fig. 4C). Thus, the Smy2–Eap1 interaction was stabilized by Scp160 but was not mediated by Asc1 and RNA.
Figure 4.
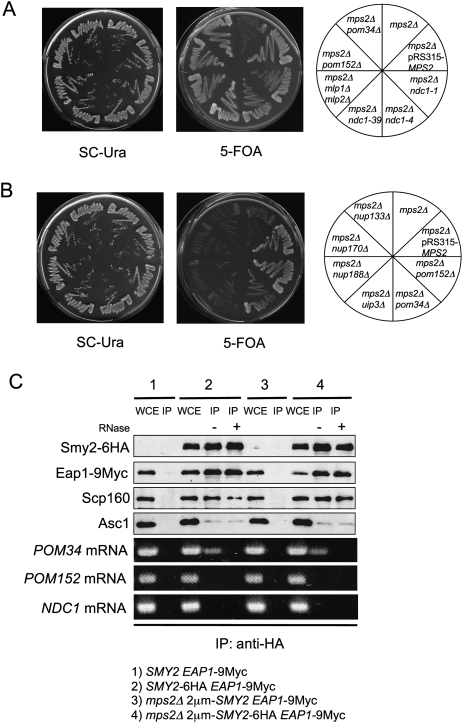
POM34 mRNA binds to SESA components. (A,B) The indicated yeast cells were tested for growth on SC-Ura and 5-FOA plates at 23°C. All strains harbored initially the pRS316-MPS2 plasmid. (C) Coimmunoprecipitation of POM34 mRNA together with Eap1, Scp160, and Asc1 by Smy2. Extracts from yeast cells were incubated with anti-HA-coated magnetic beads, with or without RNaseA treatment, and analyzed by immunoblotting with the indicated antibodies. RNA was isolated from the immunoprecipitates and analyzed by RT–PCR using primers specific to POM34, POM152, and NDC1. In this immunoprecipitation experiment the efficiency of Asc1 coimmunoprecipitation was reduced probably because of the longer incubation time due to RNaseA treatment (cf. Figs. 2 and 4C). Abbreviations are as in Figure 2B.
Bfr1 coimmunoprecipitated with the Scp160 and Asc1 proteins, but failed to coimmunoprecipitate with Smy2 (Fig. 3B; Supplemental Fig. S4A), which establishes a distinct set of interactions from the Smy2–Eap1–Scp160–Asc1 association (Fig. 2B). In addition, we confirmed that the interaction of Bfr1 with Scp160 and Asc1 was RNA-dependent (Supplemental Fig. S4B), as has been reported for the Bfr1–Scp160 interaction (Lang et al. 2001). Thus, Bfr1 concurs with Scp160 and Asc1 in a manner that is distinct from the association of these proteins with Smy2. Based on the common genetic and biochemical interactions, we suggest that the gene products of SMY2, EAP1, SCP160, and ASC1 closely cooperate in the suppression of MPS2 deletion. This network of genes was named SESA. Bfr1 may interact through an RNA link with SESA network components.
Interestingly, two hybrid interactions between the SPB component Bbp1, which is in complex with Mps2, and the Bfr1 protein have been reported previously (Xue et al. 1996; Schramm et al. 2000). Coimmunoprecipitation experiments showed complexes containing Bfr1 and Bbp1 (Fig. 3B) and this interaction was independent of RNA (Supplemental Fig. S4B). Thus, Bfr1 and the SPB protein Bbp1 are present in common complexes. This Bbp1–Bfr1 interaction may provide the functional link to couple SPB duplication and the SESA network.
POM34 mRNA is in a complex with Smy2
In previous studies, Eap1 was identified as a global inhibitor of the initiation of protein translation in response to conditions that induce membrane stress. This effect of Eap1 could be measured in sucrose gradients by the increase in monosomes with concomitant decrease in polysomes upon stress conditions (Deloche et al. 2004). Similar sucrose gradient analysis of the ratio between isolated polysomes and monosomes in wild-type and mps2Δ 2μm-SMY2 cells excluded the possibility of a global down-regulation of protein translation by Eap1 in mps2Δ 2μm-SMY2 cells (Supplemental Figs. S5 [cytoplasmic fraction], S6 [membrane-bound fraction]).
We therefore tested whether the interaction of the mRNA-binding protein Scp160 with Smy2 and Eap1 could restrict translation inhibition to a specific subset of mRNAs, which may bind to SESA through the known mRNA-binding protein Scp160 (Frey et al. 2001). Attempts to identify mRNAs that were clearly regulated by Scp160 on the level of translation were unsuccessful. We thus turned to a genetic approach to identify mRNA candidates that are bound to Scp160.
We reasoned that both translation inhibition of an mRNA by the SESA network and loss of the targeted gene could suppress the essential function of MPS2. A candidate approach was therefore adopted to screen for genes whose inactivation would suppress the lethality of mps2Δ cells. It has been previously reported that the SPB duplication defect of ndc1-1 cells is suppressed by the loss of the POM152 gene that encodes an integral membrane protein of the NPC (Chial et al. 1998). Thus, our suppression analysis was initially focused on NPC components. mps2Δ lethality was suppressed by deletion of POM34, POM152, the double deletion of MLP1 and MLP2, and conditional lethal NDC1 alleles (Fig. 4A). Ndc1 is an integral membrane protein that has overlapping functions with Pom152 and Pom34 in NPC biogenesis (Chial et al. 1998; Tcheperegine et al. 1999; Madrid et al. 2006). The myosin like Mlp1 and Mlp2 are associated with the nuclear side of the NPC and have diverse roles in the retention of nonspliced mRNAs, nuclear transport, and SPB duplication (Strambio-de-Castillia et al. 1999; Galy et al. 2004; Niepel et al. 2005). The mps2Δ suppression phenomenon was restricted to these NPC components. Deletion of other nucleoporins (e.g., NUP133, NUP170, NUP188) or other integral membrane protein components of the nuclear envelope (e.g., UIP3) did not overcome the lethality of mps2Δ (Fig. 4B; see Supplemental Table I for the full list of genes tested).
As outlined above, the SESA network may bind and down-regulate the translation of target mRNAs to suppress the essential requirement of MPS2. To test this notion, we immunoprecipitated Smy2 and then isolated the mRNAs that were precipitated in a complex with Smy2. RT–PCRs performed on these RNA samples revealed that POM34 mRNA associated with the SESA proteins, whereas POM152 and NDC1 mRNAs did not (Fig. 4C). Coimmunoprecipitation experiments performed in conjunction with UV cross-linking did not change these outcomes (data not shown). Other mRNAs tested and also found not to associate with Smy2 included MLP1, MLP2, SEC61, ADH1, and SIC1 (data not shown). We further established that the association of POM34 mRNA with Smy2 and Eap1 was dependent on the presence of Scp160 (Supplemental Fig. S7A,B). In contrast, disruption of SMY2 or EAP1 did not affect the association of POM34 mRNA with Scp160 (Supplemental Fig. S7C). Taken together, these data indicate that POM34 mRNA associates with the SESA network of proteins via an interaction with Scp160 protein.
SESA inhibits translation of POM34 mRNA
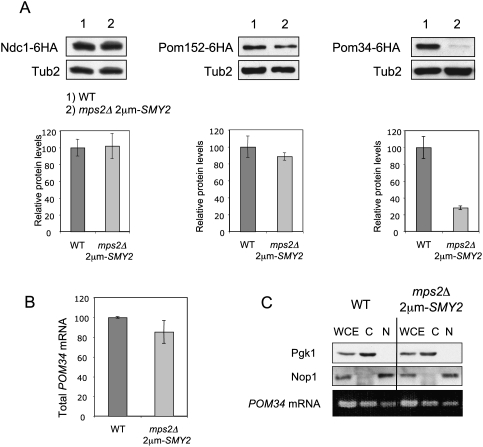
The presence of the translation initiation inhibitor Eap1 in the SESA network and the essential requirement of the Eap1–eIF4E interaction for mps2Δ suppression (Fig. 2A,D), prompted us to test whether SESA activity controls the translation of POM34 mRNA. A prediction of this hypothesis is that Pom34 protein levels would be lower in mps2Δ 2μm-SMY2 cells than in wild-type cells even though there would be no impact on POM34 mRNA levels. To address this point, we first quantified the protein levels of NPC components in mps2Δ 2μm-SMY2 and wild-type cells. Our analyses showed that Ndc1 levels were the same in both cell types, whereas Pom152 showed a modest decrease in mps2Δ 2μm-SMY2 cells (Fig. 5A). Mlp1, Mlp2, Nup133, Nup170, and Nup188 levels were also same in wild-type and mps2Δ 2μm-SMY2 cells (data not shown). In contrast, the level of Pom34 protein was significantly lower in mps2Δ 2μm-SMY2 cells than in wild type controls (Fig. 5A) even though the stability of the Pom34 protein assessed by the cycloheximide treatment was similar in both cell types (Supplemental Fig. S8). The modest decrease of Pom152 in mps2Δ 2μm-SMY2 cells may arise from the strongly reduced level of Pom34 that interacts with Pom152 (Alber et al. 2007a,b).
Figure 5.
Strongly reduced amount of Pom34 in mps2Δ 2μm-SMY2 cells. (A) Pom34 protein level is reduced in mps2Δ 2μm-SMY2 cells. Total cell extracts from yeast strains expressing NDC1-6HA, POM152-6HA, or POM34-6HA were analyzed by immunoblotting using anti-HA antibodies. Anti-Tub2 antibodies were used as loading control. The graphs underneath the immunoblots show the quantification of three independent experiments, normalized for the wild-type protein levels. Bars are standard deviations around the mean value. (B) POM34 mRNA levels are similar in wild-type and mps2Δ 2μm-SMY2 cells. Total RNA extracts from wild-type (WT) and mps2Δ 2μm-SMY2 cells were analyzed by quantitative RT–PCR using primers specific to POM34 mRNA. (C) mps2Δ 2μm-SMY2 cells do not show mRNA export defect. Wild-type and mps2Δ 2μm-SMY2 cells were fractionated into nuclear and cytoplasmic fractions and analyzed by immunoblotting with anti-Pgk1 and anti-Nop1 antibodies. RNA was isolated from the nuclear and cytoplasmic fractions and analyzed by RT–PCR using primers specific to POM34.
The data described above suggest that Pom34 levels are decreased in mps2Δ 2μm-SMY2 cells either because of a reduction in the efficiency with which POM34 mRNA is translated, or because nuclear export of this mRNA is impaired or, finally, that the stability of POM34 mRNA is reduced. Quantitative RT–PCR established that total levels of POM34 mRNA were not significantly affected in mps2Δ 2μm-SMY2 cells (Fig. 5B). Thus, we concluded that mRNA stability is probably not altered in mps2Δ 2μm-SMY2 cells. Furthermore, a comparison of the levels of POM34 mRNA in the cytoplasm of mps2Δ 2μm-SMY2 and wild-type cells revealed no differences (Fig. 5C), indicating that the export of POM34 mRNA was unperturbed by the mps2Δ 2μm-SMY2 genotype. We conclude that POM34 is likely regulated on the level of translation.
Membrane localization of POM34 mRNA requires initiation of translation
POM34 encodes an integral protein of the nuclear envelope (Miao et al. 2006). Its translation should therefore occur at the ER (Rapoport 1990). In order to pinpoint the step modulated by SESA control more precisely, we monitored the partitioning of POM34 mRNA between cytosolic and membrane-bound fractions of yeast cell extracts alongside an assessment of the abundance of Scp160, Sec61, and Pgk1 proteins and ribosomal subunits.
In both wild-type and mps2Δ 2μm-SMY2 cells, cytoplasmic Pgk1 was detected in the soluble cytoplasmic fraction and the integral membrane protein Sec61 (Deshaies et al. 1991) was found almost exclusively in the membrane fraction (Fig. 6A). Scp160 was enriched in the membrane-bound fraction, whereas the ribosomal subunit Rpl35 was more abundant in the cytoplasm. The latter distributions were as expected, since most ribosomes are present in cytoplasm while Scp160 is enriched with ribosomes at the ER (Supplemental Fig. S9A; Frey et al. 2001). Together, the cellular distributions of Scp160, Sec61, Pgk1, and Rpl35 proteins demonstrate that the fractionation into membrane and cytoplasmic fractions was successful.
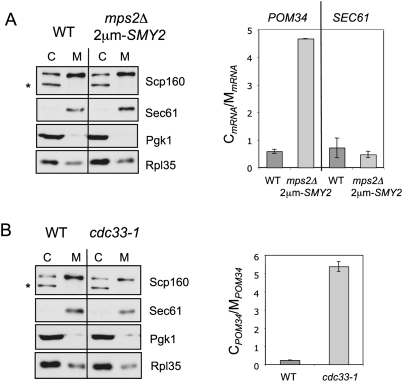
Figure 6.
POM34 mRNA accumulates in the cytoplasm in mps2Δ 2μm-SMY2 cells. (A) Total cell extracts from wild-type and mps2Δ 2μm-SMY2 cells were fractionated into cytosolic and membrane-bound fractions. Fractions were analyzed by immunoblotting using the indicated antibodies. The lower molecular weight band (asterisk) is a cytoplasmic degradation product of Scp160 (Frey et al. 2001). POM34 and SEC61 mRNA levels were determined by quantitative RT–PCR from three independent experiments. The graph shows the ratio of cytosolic POM34 mRNA levels to the membrane-bound POM34 mRNA levels. The bars indicate standard deviation of the results from the mean value. (C) Cytosolic fraction; (M) membrane-bound fraction. (B) Subcellular localization of POM34 mRNA in the translation initiation mutant cdc33-1. Cells were grown in YPAD at 23°C and were shifted for 2 h to 37°C. The cellular fractionation and analysis was performed as in A.
According to RT–PCR data, the mRNAs of SEC61 (Fig. 6A), NDC1, and POM152 (Supplemental Fig. S6), which all encode integral membrane proteins, were slightly enriched in the membrane-bound fraction in both wild-type and mps2Δ 2μm-SMY2 cells (Fig. 6A; Supplemental Fig. S6). In wild-type cells POM34 mRNA distribution between the membrane fraction and the cytoplasm was similar to SEC61 mRNA (Fig. 6A). Strikingly, however, in mps2Δ 2μm-SMY2 cells POM34 mRNA was mostly found in the cytoplasm, and only very little in the membrane fraction (Fig. 6A). Thus, the cellular localization of POM34 mRNA is significantly changed in mps2Δ 2μm-SMY2 cells.
Mislocalization of POM34 mRNA may arise from an inhibition of initiation of protein translation, which is required to target the mRNA in association with the nascent Pom34 protein to the ER membrane. If correct, a general inhibition of translation should result in the same cellular mislocalization of POM34 mRNA. To test this prediction, we analyzed POM34 mRNA localization in the conditional lethal translation initiation mutant cdc33-1, in which the function of yeast eIF4E is impaired (Brenner et al. 1988). POM34 mRNA had a predominately cytoplasmic distribution in cdc33-1 cells incubated at the restrictive temperature (Fig. 6B). The extent of the mislocalization of POM34 mRNA in cdc33-1 cells was only slightly higher than in mps2Δ 2μm-SMY2 cells (Fig. 6A). This establishes that inhibition of translation initiation is indeed sufficient to mislocalize POM34 mRNA to the cytoplasm.
We next asked whether translation elongation was also affected in mps2Δ 2μm-SMY2 cells. Separating the translating membrane-bound ribosomal subunits and polysomes via a sucrose gradient fractionation was used as experimental approach (Supplemental Fig. S6). Although less abundant in the membrane fraction, we observed no major difference in the relative POM34 mRNA distribution between membrane-bound polysomes and monosomes in mps2Δ 2μm-SMY2 cells with respect to wild-type cells (Supplemental Fig. S6C). Thus, our data do not suggest a defect in translation elongation for POM34 mRNA in mps2Δ 2μm-SMY2 cells. We also analyzed the cytoplasmic fraction of POM34 mRNA in a sucrose gradient (Supplemental Fig. S5). This analysis showed that the cytoplasmic POM34 mRNA, although more abundant in mps2Δ 2μm-SMY2 cells than in wild-type cells (Fig. 6A), was predominately associated with the 40S, 60S, and 80S fractions of ribosomes in both cell types (Supplemental Fig. S5). A polysomal cytoplasmic fraction was not observed, which is consistent with the notion that translation elongation of POM34 mRNA occurred at the ER. Thus, the main difference between wild type and mps2Δ 2μm-SMY2 cells is the strongly reduced translation initiation efficiency of POM34 mRNA in mps2Δ 2μm-SMY2 cells. This defect then leads to the accumulation of POM34 mRNA in the cytoplasm.
SPB duplication defects regulate Pom34 via the SESA network
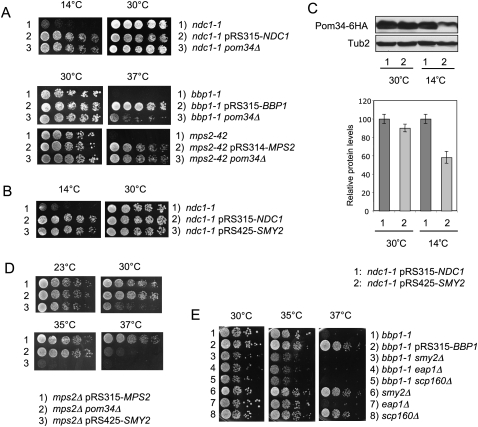
Our data indicate that the SESA network inhibits translation of the POM34 mRNA in mps2Δ 2μm-SMY2 cells. However, overexpression of SMY2 in otherwise wild-type cells was insufficient to down-regulate Pom34 (Supplemental Fig. S9B), suggesting that a defect in mps2Δ cells induces the regulation of POM34 mRNA. The implication is that defects in SPB duplication may activate SESA. In this respect it is important to note that the SESA network component EAP1 is essential for viability of the SPB duplication-defective ndc1-1 cells that have normal NPCs (Chial et al. 2000). In addition, as was the case for mps2Δ (Fig. 1), the lethal growth defect of ndc1-1 cells at 14°C was suppressed by the deletion of POM34 (Fig. 7A), or high gene dosage of SMY2 (Fig. 7B). Furthermore, Pom34 levels were reduced in ndc1-1 2μm-SMY2 cells (Fig. 7C). These similarities between the consequences of mutations in NDC1 and MPS2 support the notion that defects in SPB duplication down-regulate POM34 mRNA translation. Consistently, growth of the SPB duplication-defective bbp1-1, mps2-42, and ndc1-4 cells at the restrictive temperature was rescued by pom34Δ (Fig. 7A; Supplemental Fig. S10A). Although, high gene copy 2μm-SMY2 did not suppress the growth defect of these cells (data not shown), this is explained by the failure of SMY2 suppression at elevated temperatures (Fig. 7D).
Figure 7.
Genetic interactions between SESA and POM34 with genes involved in SPB duplication. (A) Deletion of POM34 suppresses ndc1-1, bbp1-1, and mps2-42. The listed yeast cells were tested for growth on YPAD plates at indicated temperatures. (B) SMY2 is a multicopy suppressor of the cold-sensitive phenotype of ndc1-1. ndc1-1 cells transformed with pRS315-NDC1 or pRS425-SMY2 were tested for growth on YPAD plates at 14°C and 30°C. (C) Pom34 protein level is reduced in ndc1-1 2μm-SMY2 cells. Total cell extracts from yeast strains expressing POM34-6HA were analyzed by immunoblotting using anti-HA antibodies. Anti-Tub2 antibodies were used as loading control. The graph shows the quantification of three independent experiments, normalized for the wild-type protein levels. Bars are standard deviations around the mean value. (D) mps2Δ 2μm-SMY2 and mps2Δ pom34Δ cells are temperature sensitive for growth. The listed yeast cells were tested for growth on YPAD plates at indicated temperatures. (E) Deletion of SESA components enhances bbp1-1 growth defects. The listed yeast cells were tested for growth on YPAD plates at the indicated temperatures.
To further demonstrate that SESA becomes important for survival of mutants defective in SPB duplication, we analyzed the consequence of SESA inactivation in bbp1-1 and ndc1-4 cells. Deletion of SESA network components enhanced the growth defect of bbp1-1 or ndc1-4 cells grown at elevated growth temperatures (Fig. 7E; Supplemental Fig. S10B), suggesting that SESA network is important under conditions where SMY2 is not overexpressed.
Down-regulation of POM34 restores SPB duplication of mps2 cells
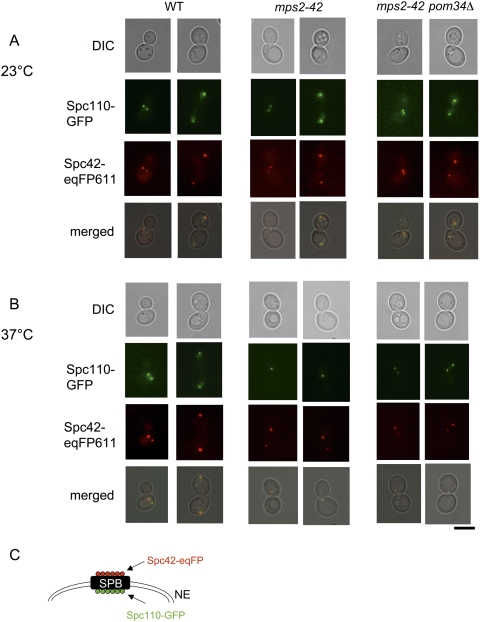
SPB duplication fails in the absence of BBP1, NDC1, and MPS2 function. In these mutant cells the duplication plaque is not inserted into the nuclear envelope. The nuclear side of the SPB with the component Spc110 is therefore not assembled, whereas the Spc42 becomes incorporated into the defective SPB (37°C: note mps2-42 cells with only one Spc110-GFP signal but two Spc42-eqFP611 signals) (Fig. 8B; Schramm et al. 2000). However, deletion of POM34 restored SPB duplication of mps2-42 cells at 37°C indicating the importance of Pom34 levels in SPB duplication mutants (Fig. 8B; mps2-42 pom34Δ cells at 37°C with two colocalizing Spc110-GFP and Spc42-eqFP611 signals). Furthermore, analysis of mps2Δ 2μm-SMY2 and mps2Δ pom34Δ cells at 23°C showed that the SPB duplicated in G1/S with similar kinetics as in wild-type cells (data not shown), and that the newly formed SPB carried the markers Ndc1-GFP, Bbp1-GFP, Nbp1-GFP, and Spc110-GFP (n = 150 cells) (Supplemental Figs. S11, S12). Thus, deletion or reduced translation of POM34 suppresses SPB duplication defects.
Figure 8.
SPBs are inserted into the nuclear envelope in mps2-42 pom34Δ cells. (A,B) Analysis of wild-type, mps2-42, and mps2-42 pom34Δ cells with SPC110-GFP SPC42-eqFP611 by fluorescence and phase contrast (DIC) microscopy at 23°C (A) and 37°C (B) (2 h). Note that in B Spc110-GFP is only associated with one of the two Spc42-eqFP611-marked SPBs of mps2-42 cells. This is the typical phenotype of cells with a defect in duplication plaque insertion (Schramm et al. 2000; Jaspersen and Winey 2004). Bars, 5 μm. (C) Shown is a cartoon of the SPB with the localization of Spc42 and Spc110 relative to the nuclear envelope (NE) (Adams and Kilmartin 1999).
Discussion
The initiation is the rate-limiting step in mRNA translation. Deregulating initiation by overexpression of the CAP-binding protein eIF4E leads to malignant transformation and therefore, not surprisingly, eIF4E is elevated in many human cancers (De Benedetti and Rhoads 1990; Lazaris-Karatzas et al. 1990). In addition, TOR signaling and stress situations including membrane defects inhibit global initiation of translation by regulating binding of proteins (4E-BPs) to the initiation factor eIF4E (Cosentino et al. 2000; Deloche et al. 2004; Matsuo et al. 2005; Ibrahimo et al. 2006).
In this study, we unraveled an unexpected link between the SPB duplication pathway and regulation of translation initiation of the POM34 mRNA. In response to SPB duplication defects, the SESA network, comprising of the known mRNA-binding protein Scp160 (Frey et al. 2001; Li et al. 2003, 2004; Baum et al. 2004), the ribosome-associated Asc1 (Baum et al. 2004; Gerbasi et al. 2004), the translation inhibitor Eap1 (Cosentino et al. 2000), and the protein Smy2 (Kofler et al. 2005), was identified as being responsible for the translation control of POM34 mRNA. We demonstrate that SESA inhibits translation of POM34 mRNA by binding of the 4E-BP Eap1 to the conserved translation initiation factor eIF4E. By showing that Smy2, Eap1, Scp160, and Asc1 physically and functionally interact, we provide a first understanding of how Eap1 is regulated on the molecular level. This regulation of POM34 mRNA by SESA is essential to ensure survival of cells with defects in SPB duplication.
SESA binds to POM34 mRNA
Based on the presence of the eIF4E inhibitor Eap1 in the SESA network, we expected to see a general inhibition of translation initiation in mps2Δ cells as is the case in cells exposed to membrane stress conditions (Deloche et al. 2004). Surprisingly, however, in mps2Δ 2μm-SMY2 cells translation of proteins was not inhibited on a global scale (Supplemental Figs. S5, S6) indicating that the SESA network regulates only a subset of mRNAs.
The mRNA-binding protein Scp160 could target mRNAs to SESA regulation (Frey et al. 2001; Baum et al. 2004; Li et al. 2004). A microarray analyses of mRNAs released from affinity isolated Scp160-containing complexes identified a limited set of mRNAs that bind to Scp160 (Li et al. 2003). We tested the two most prominent mRNAs identified by Li et al. (2003), the DHH1 and YOR338w transcripts, but did not find an enrichment of these mRNAs in anti-Smy2 immunoprecipitates nor any importance of the genes for survival of mps2Δ 2μm-SMY2 cells (B Sezen, unpubl.). Furthermore, a recent study identified mainly mRNAs coding for proteins of the cell wall, plasma membrane, ER and nucleolus in association with Scp160 (Hogan et al. 2008).
To identify mRNAs that are regulated by SESA in the context of MPS2 function, we turned to a genetic approach, which identified the POM34 mRNA (Fig. 4) as being associated with SESA components. In addition, Pom34 levels were down-regulated in mps2Δ cells that require SESA for viability (Fig. 5A). Together, this strongly supports the notion that POM34 mRNA is regulated by SESA.
SESA inhibits initiation of translation of POM34 mRNA
We suggest that SESA inhibits initiation of translation of POM34 mRNA (Fig. 9, step 4). This model is supported by a number of findings. First, Eap1 blocks the crucial binding of eIF4G to eIF4E (Fig. 2C), which is normally an essential step in translation initiation (Gingras et al. 1999). Second, the Eap1[Y109A, L114A] mutations, which impair the Eap1–eIF4E interaction (Fig. 2D; Ibrahimo et al. 2006), also abrogate the function of SESA in mps2Δ cells (Fig. 2A) indicating that Eap1–eIF4E binding is essential for the down-regulation of Pom34. Third, although the POM34 mRNA levels were unchanged in mps2Δ cells (Fig. 5B), Pom34 protein was strongly reduced in comparison with wild-type cells (Fig. 5A). A detailed analysis excluded POM34 mRNA nuclear export defects and altered Pom34 protein stability as factors that could decrease Pom34 protein levels in mps2Δ cells (Fig. 5C; Supplemental Fig. S8). However, analysis of the distribution of POM34 mRNA in mps2Δ cells clearly showed a shift toward the cytoplasmic fraction that has not initiated translation (Fig. 6A). A similar cytoplasmic shift of POM34 mRNA was observed in cells defective in the translation initiation factor eIF4E (Fig. 6B; Brenner et al. 1988), supporting the idea that SESA inhibits initiation of translation. Forth, conditions that inhibited POM34 translation did not affect translation of other mRNAs (NDC1, POM152, MLP1, MLP2, SEC61, ADH1, and SIC1). This implies that SESA is a specific inhibitor of translation of a subset of mRNAs. Smy2, Scp160, and Asc1 in the SESA network may confer Eap1 translation inhibition specificity toward a subset of mRNAs.
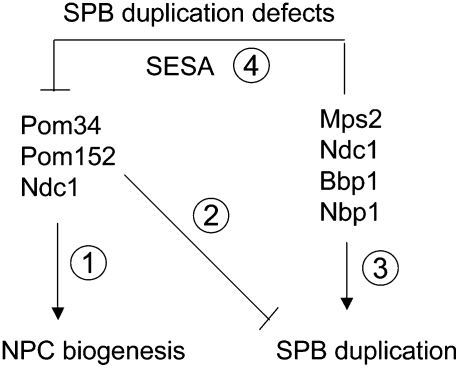
Figure 9.
Model for the function of SESA network. (Step 1) Pom34, Pom152, and Ndc1 form a complex that functions in NPC biogenesis (Chial et al. 1998; Madrid et al. 2006; Alber et al. 2007a,b; Onischenko et al. 2009). (Step 3) Ndc1 has a dual role and together with Mps2, Bbp1, and Nbp1 it also functions in SPB duplication (Winey et al. 1993; Araki et al. 2006). (Step 2) Deletion of POM34 or POM152 or mutations in ndc1 rescue SPB duplication defects (Fig. 4; Chial et al. 1998), suggesting an inhibitory role of the Pom34–Pom152–Ndc1 complex in SPB duplication. In response to SPB duplication defects, SESA down-regulates translation of POM34 (Figs. 4–7). (Step 4) This in turn rescues the defect and allows for SPB duplication (Fig. 8; Supplemental Fig. S12).
The Scp160 protein with its conserved KH RNA-binding domains (Frey et al. 2001) was deduced as the factor that binds the POM34 mRNA in the SESA network (Fig. 4C; Supplemental Fig. S7). Scp160 shows genetic and biochemical interactions with the Asc1 protein, which is a core component of the 40S ribosomal subunit and as such binds Scp160 to ribosomes (Baum et al. 2004; Nilsson et al. 2004). However, also a cytoplasmic pool of Asc1 exists (Brodersen and Nissen 2005) that may direct the SESA–POM34 mRNA complex to ribosomes. It is important to note that Asc1 functions as G-protein subunit coupled to glucose responsiveness in yeast (Zeller et al. 2007), and that the G protein α subunit Gpa1 transmits a signal through Scp160 (Guo et al. 2003). This raises the exciting possibility of a cross-talk between the SESA network and external stimuli.
Regulation of POM34 mRNA in response to SPB duplication defects
Mutants of NDC1 and MPS2 that failed to insert the SPB duplication plaque into the nuclear envelope showed genetic interactions with SESA components and, in addition, showed mislocalization of POM34 mRNA and reduced levels of Pom34 in comparison with wild-type cells. These dependencies suggest a mechanism by which SESA becomes active in response to defects in SPB duplication (Figs. 1, 7; Chial et al. 2000; Schramm et al. 2000). This model is consistent with the activation of mammalian 4E-BPs by external stimuli (Gingras et al. 1999) or of yeast Eap1 by membrane defects (Deloche et al. 2004). In addition, elevated gene levels of SMY2, may arise due to chromosome missegregation in mutants with SPB duplication defects, which are genetically unstable (Winey et al. 1991, 1993; Schramm et al. 2000; Araki et al. 2006). Increased gene dosage of SMY2 then likely makes the SESA network more sensitive to defects (Fig. 1).
The SESA network may be linked to the SPB by the Bfr1 protein and the interacting SPB component Bbp1 (Xue et al. 1996). Consistently, BFR1 was found to be essential in cells lacking MPS2 and the Bfr1 protein showed coimmunoprecipitation with Bbp1, Scp160, and Asc1 (Fig. 3). However, in contrast to SESA components, RNA mediated the interactions of Bfr1. Binding of Bfr1 to SESA-associated POM34 mRNA could activate the ability of Eap1 to inhibit the translation of this mRNA.
Why is the NPC component Pom34 important for SPB duplication? Pom34, Ndc1, and Pom152 belong to a group of functionally redundant and interacting integral membrane proteins. They are part of the membrane ring of the NPC and as such important for NPC biogenesis (Fig. 9, step 1; Wozniak et al. 1994; Chial et al. 1998; Madrid et al. 2006; Miao et al. 2006; Alber et al. 2007a,b; Onischenko et al. 2009). A functional link between SPB insertion and NPCs, which are frequently observed near duplicating SPBs, was already proposed (Adams and Kilmartin 1999; Jaspersen and Winey 2004). Curiously, deletion of either POM34 or POM152 or mutations in ndc1 suppress the essential function of MPS2 (Fig. 4A). Based on this observation we suggest that the Pom34–Pom152–Ndc1 complex inhibits SPB duplication (Fig. 9, step 2). The Pom34–Pom152–Ndc1 complex either directly binds to SPBs and inhibits its duplication or SPBs and NPCs compete for components of a common nuclear envelope insertion machinery. The dual function of Ndc1 in SPB duplication (Fig. 9, step 3) and NPC biogenesis (Fig. 9, step 1) supports the second competition model (Chial et al. 1998; Madrid et al. 2006). Defects in SPB duplication down-regulate expression of POM34 via SESA (Fig. 9, step 4). Reduced Pom34 levels then relieve the inhibitory function of the Pom34–Pom152–Ndc1 complex. Thus, depending on the conditions, the SESA network may promote either SPB duplication or NPC biogenesis.
This study unraveled a novel mechanism by which cells can regulate the translation of specific mRNAs. In contrast to the general translation inhibition in response to, for example, membrane stress (Deloche et al. 2004), regulation by the SESA pathway enables the cell to modify the proteome in a very specific way at the level of translation. Thus, in respect of specificity SESA regulation is similar to the translation control of mRNAs containing a cytoplasmic polyadenylation element (CPE) by Maskin or 4E-T (4E-BP) in Xenopus laevis. In this case the CPE-binding protein (CPEB) acts as the specific RNA-binding protein (Stebbins-Boaz et al. 1999; Minshall et al. 2007). Another such example is the translational repression of oskar mRNA in Drosophila by Cup (a 4E-BP) where Bruno acts as the mRNA-binding protein (Nakamura et al. 2004; Chekulaeva et al. 2006; for reviews, see Richter and Sonenberg 2005; Sonenberg and Hinnebusch 2009).
Materials and methods
Strain constructions and growth conditions
Gene deletions and epitope tagging of genes at their endogenous loci were performed using PCR-based methods (Janke et al. 2004). The strains and plasmids used in this study are listed in Supplemental Table 2. All yeast strains were derivatives of S228c with the exception of ndc1-4 and ndc1-39, which were derived from W303 and were compared with the corresponding wild type.
Typically cells were grown in yeast extract peptone glucose medium (YPD) at 23°C. For analysis of temperature-sensitive mutants they were shifted for 2 h to 37°C before analysis.
Construction of CDC33, SMY2, and EAP1 mutants
SMY2 and EAP1 with regulatory and coding regions were cloned into the LEU2-based yeast shuttle vector pRS425 and pRS315, respectively (Sikorski and Hieter 1989). Mutations in SMY2 and EAP1 were introduced by PCR-directed mutagenesis and confirmed by DNA sequencing. cdc33-1 with regulatory and coding regions were cloned into the LEU2-based yeast integration vector pRS305 (Sikorski and Hieter 1989) after amplification by PCR from the genomic DNA of a cdc33-1 strain (Brenner et al. 1988) and then inserted into our S288c strain background.
Antibodies and immunoblotting
Yeast extracts were prepared using alkaline lysis and TCA precipitation (Janke et al. 2004). To detect proteins by immunoblotting procedures, blocked membranes (Protean, Schleicher & Schuell) were incubated for 2 h at 20°C or overnight at 4°C with antibodies diluted in blocking buffer (PBS, 0.2% Tween 20, 5% dry milk powder) followed by peroxidase-conjugated secondary antibodies [Sigma] and detection with ECL (Roche Molecular Biochemicals). Anti-Scp160 (GST-Scp160), anti-Asc1 (NVIRVWQVMTAN-COOH), anti-Rps3 (VALISKKRKLVADC-CONH2), anti-Rpl35 (CPIRKYAIKV-COOH), anti-Sec61, anti-Clb2 (GST-Clb21–271), anti-Pds1 (GST-Pds11–173), and anti-Tub2 antibodies (yeast β-tubulin, GST-Tub2436–457) were prepared in rabbits or sheep against purified recombinant proteins or peptides (Frey et al. 2001; Pereira and Schiebel 2003; Baum et al. 2004). Monoclonal mouse anti-Myc (9E10) and anti-HA (12CA5) antibodies were from Roche Molecular Biochemicals. Anti-Pgk1 and anti-Nop1 antibodies were gifts from M. Knop. Anti-eIF4E and anti-eIF4G antibodies were gifts from M. Ashe.
Coimmunoprecipitation
Logarithmically growing cells (3 × 108) were disrupted with glass beads in 300-μL immunoprecipitation buffer (50 mM triethanolamine, 150 mM KCl, 5 mM EDTA, 5 mM EGTA) containing a protease inhibitor cocktail (Boehringer Mannheim), phenylmethylsulfonyl fluoride (PMSF) and benzamidine. Lysates were incubated with 1% Triton X-100 for 10 min at 4°C. Extracts were cleared by centrifugation (6000 rpm for 10 min at 4°C). After removing an aliquot that served as the input control, the resulting extract was incubated with monoclonal anti-HA antibody, 12CA5, coated magnetic beads (Dynal) for 2 h at 4°C. Beads were washed three times with immunoprecipitation buffer containing 0.1% Triton X-100.
mRNA coimmunoprecipitation
Protein–mRNA coimmunoprecipitations were performed as described (Munchow et al. 1999; Bohl et al. 2000; Long et al. 2000). In brief, 3 × 108 logarithmically growing cells were disrupted with glass beads in 200-μL breakage buffer BB (50 mM HEPES-KOH at pH 7.3, 50 mM potassium acetate, 2 mM magnesium acetate, 1% Triton X-100) containing a protease inhibitor cocktail (Boehringer Mannheim) and 0.5% BSA. Extracts were cleared by centrifugation (6000 rpm for 10 min). After removing an aliquot that served as input control, the resulting extract was incubated at 4°C with monoclonal anti-HA antibody, 12CA5, coated magnetic beads (Dynal). Beads were washed three times with BB lacking BSA. Pellets were extracted with phenol-chloroform, ethanol precipitated, resuspended in RQ1 DNase buffer, and treated with RQ1 DNase (Promega). The remaining RNA was extracted, precipitated, and resuspended in water. RT–PCR was performed with 1 μL RNA as a template using the Qiagen RT–PCR kit and the conditions suggested by the manufacturer. The number of amplification cycles was adjusted to avoid reaching a plateau during PCR. For amplification of POM34, POM152, SIC1, and ASH1 RNAs, we used 25 cycles, for amplification of NDC1, MLP1, and MLP2 RNAs we used 24 cycles, whereas 22 cycles were used for amplification of ADH1 RNA. The primers in Supplemental Table 3 were used for amplification.
RT-quantitative PCR
RNA was extracted from cells grown to logarithmic phase using Qiagen RNeasy kit following the manufacturer's protocol. A total of 4 μg of RNA was reverse transcribed using 7 μM oligo-dT and reverse transcriptase for 1 h at 37°C. The product was diluted 1:10 and used in the subsequent quantitative PCR reactions using POM34 and SEC61 primers in a Roche LightCycler using SYBR Green. Standard curves for each primer were generated using serial dilutions of yeast genomic DNA. Quantification of cDNA template concentrations was done using the standard curve for each primer.
Acknowledgments
We thank C. Mayer and I. Grummt for their help with quantitative PCR analyses. We thank K. Nasmyth, E. Hurt, and M. Winey for yeast strains; M. Ashe and M. Knop for plasmids and antibodies; P. Walter and K. Weis for plasmids; D. Kirkpatrick for the yeast genomic DNA library; and members of the Schiebel laboratory, I. Hagan, and G. Pereira for comments on the manuscript. We thank C. Schramm for the initial mps2Δ 2-μm screen performed at the Beatson Institute, Glasgow. The GRK1188 and SFB638 supported this work.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.524209.
Supplemental material is available at http://www.genesdev.org.
References
- Adams IR, Kilmartin JV. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae . J Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007a;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007b;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Araki Y, Lau CK, Maekawa H, Jaspersen SL, Giddings JTH, Schiebel E, Winey M. The Saccharomyces cerevisiae spindle pole body (SPB) component Nbp1p is required for SPB membrane insertion and interacts with the integral membrane proteins Ndc1p and Mps2p. Mol Biol Cell. 2006;17:1959–1970. doi: 10.1091/mbc.E05-07-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum S, Bittins M, Frey S, Seedorf M. Asc1p, a WD40-domain containing adaptor protein, is required for the interaction of the RNA-binding protein Scp160p with polysomes. Biochem J. 2004;380:823–830. doi: 10.1042/BJ20031962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohl F, Kruse C, Frank A, Ferring D, Jansen RP. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 2000;19:5514–5524. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner C, Nakayama N, Goebl M, Tanaka K, Toh-e A, Matsumoto K. CDC33 encodes mRNA cap-binding protein eIF-4E of Saccharomyces cerevisiae . Mol Cell Biol. 1988;8:3556–3559. doi: 10.1128/mcb.8.8.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen DE, Nissen P. The social life of ribosomal proteins. FEBS J. 2005;272:2098–2108. doi: 10.1111/j.1742-4658.2005.04651.x. [DOI] [PubMed] [Google Scholar]
- Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–533. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Chial HJ, Rout MP, Giddings TH, Winey M. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J Cell Biol. 1998;143:1789–1800. doi: 10.1083/jcb.143.7.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chial HJ, Stemm-Wolf AJ, McBratney S, Winey M. Yeast Eap1p, an eIF4E-associated protein, has a separate function involving genetic stability. Curr Biol. 2000;10:1519–1522. doi: 10.1016/s0960-9822(00)00829-0. [DOI] [PubMed] [Google Scholar]
- Cosentino GP, Schmelzle T, Haghighat A, Helliwell SB, Hall MN, Sonenberg N. Eap1p, a novel eukaryotic translation initiation factor 4E-associated protein in Saccharomyces cerevisiae . Mol Cell Biol. 2000;20:4604–4613. doi: 10.1128/mcb.20.13.4604-4613.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedetti A, Rhoads RE. Overexpression of eukaryotic protein synthesis initiation factor 4E in HeLa cells results in aberrant growth and morphology. Proc Natl Acad Sci. 1990;87:8212–8216. doi: 10.1073/pnas.87.21.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz Munoz-Centeno M, McBratney S, Monterrosa A, Byers B, Mann C, Winey M. Saccharomyces cerevisiae MPS2 encodes a membrane protein localized at the spindle pole body and the nuclear envelope. Mol Biol Cell. 1999;10:2393–2406. doi: 10.1091/mbc.10.7.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloche O, de la Cruz J, Kressler D, Doere M, Linder P. A membrane transport defect leads to a rapid attenuation of translation initiation in Saccharomyces cerevisiae . Mol Cell. 2004;13:357–366. doi: 10.1016/s1097-2765(04)00008-5. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Sanders SL, Feldheim DA, Schekman R. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature. 1991;349:806–808. doi: 10.1038/349806a0. [DOI] [PubMed] [Google Scholar]
- Frey S, Pool M, Seedorf M. Scp160p, an RNA-binding, polysome-associated protein, localizes to the endoplasmic reticulum of Saccharomyces cerevisiae in a microtubule-dependent manner. J Biol Chem. 2001;276:15905–15912. doi: 10.1074/jbc.M009430200. [DOI] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116:63–73. doi: 10.1016/s0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- Georgiev A, Sjostrom M, Wieslander A. Binding specificities of the GYF domains from two Saccharomyces cerevisiae paralogs. Protein Eng Des Sel. 2007;20:443–452. doi: 10.1093/protein/gzm041. [DOI] [PubMed] [Google Scholar]
- Gerbasi VR, Weaver CM, Hill S, Friedman DB, Link AJ. Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol Cell Biol. 2004;24:8276–8287. doi: 10.1128/MCB.24.18.8276-8287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Guo M, Aston C, Burchett SA, Dyke C, Fields S, Rajarao SJ, Uetz P, Wang Y, Young K, Dohlman HG. The yeast G protein α subunit Gpa1 transmits a signal through an RNA binding effector protein Scp160. Mol Cell. 2003;12:517–524. doi: 10.1016/s1097-2765(03)00307-1. [DOI] [PubMed] [Google Scholar]
- Higashio H, Sato K, Nakano A. Smy2p participates in COPII vesicle formation through the interaction with Sec23p/Sec24p subcomplex. Traffic. 2008;9:79–93. doi: 10.1111/j.1600-0854.2007.00668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:2297–2313. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahimo S, Holmes LE, Ashe MP. Regulation of translation initiation by the yeast eIF4E binding proteins is required for the pseudohyphal response. Yeast. 2006;23:1075–1088. doi: 10.1002/yea.1415. [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Stearns T. Exploring the pole: An EMBO conference on centrosomes and spindle pole bodies. Nat Cell Biol. 2008;10:1375–1378. doi: 10.1038/ncb1208-1375. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Winey M. The budding yeast spindle pole body: Structure, duplication, and function. Annu Rev Cell Dev Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Kofler M, Motzny K, Freund C. GYF domain proteomics reveals interaction sites in known and novel target proteins. Mol Cell Proteomics. 2005;4:1797–1811. doi: 10.1074/mcp.M500129-MCP200. [DOI] [PubMed] [Google Scholar]
- Lang BD, Li A, Black-Brewster HD, Fridovich-Keil JL. The brefeldin A resistance protein Bfr1p is a component of polyribosome-associated mRNP complexes in yeast. Nucleic Acids Res. 2001;29:2567–2574. doi: 10.1093/nar/29.12.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CK, Giddings THJ, Winey M. A novel allele of Saccharomyces cerevisiae NDC1 reveals a potential role for the spindle pole body component Ndc1p in nuclear pore assembly. Eukaryot Cell. 2004;3:447–458. doi: 10.1128/EC.3.2.447-458.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- Li AM, Watson A, Fridovich-Keil JL. Scp160p associates with specific mRNAs in yeast. Nucleic Acids Res. 2003;31:1830–1837. doi: 10.1093/nar/gkg284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AM, Vargas CA, Brykailo MA, Openo KK, Corbett AH, Fridovich-Keil JL. Both KH and non-KH domain sequences are required for polyribosome association of Scp160p in yeast. Nucleic Acids Res. 2004;32:4768–4775. doi: 10.1093/nar/gkh812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie SH, Brown SS. Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae . J Cell Biol. 1994;125:825–842. doi: 10.1083/jcb.125.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RM, Gu W, Lorimer E, Singer RH, Chartrand P. She2p is a novel RNA-binding protein that recruits the Myo4p–She3p complex to ASH1 mRNA. EMBO J. 2000;19:6592–6601. doi: 10.1093/emboj/19.23.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid AS, Mancuso J, Cande WZ, Weis K. The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. J Cell Biol. 2006;173:361–371. doi: 10.1083/jcb.200506199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld J, Guttinger S, Hawryluk-Gara LA, Pante N, Mall M, Galy V, Haselmann U, Muhlhausser P, Wozniak RW, Mattaj IW, et al. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol Cell. 2006;22:93–103. doi: 10.1016/j.molcel.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Mascarenhas C, Edwards-Ingram LC, Zeef L, Shenton D, Ashe MP, Grant CM. Gcn4 is required for the response to peroxide stress in the yeast Saccharomyces cerevisiae . Mol Biol Cell. 2008;19:2995–3007. doi: 10.1091/mbc.E07-11-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo R, Kubota H, Obata T, Kito K, Ota K, Kitazono T, Ibayashi S, Sasaki T, Iida M, Ito T. The yeast eIF4E-associated protein Eap1p attenuates GCN4 translation upon TOR-inactivation. FEBS Lett. 2005;579:2433–2438. doi: 10.1016/j.febslet.2005.03.043. [DOI] [PubMed] [Google Scholar]
- Mendelsohn BA, Li AM, Vargas CA, Riehman K, Watson A, Fridovich-Keil JL. Genetic and biochemical interactions between SCP160 and EAP1 in yeast. Nucleic Acids Res. 2003;31:5838–5847. doi: 10.1093/nar/gkg810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao M, Ryan KJ, Wente SR. The integral membrane protein Pom34p functionally links nucleoporin subcomplexes. Genetics. 2006;172:1441–1457. doi: 10.1534/genetics.105.052068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N, Reiter MH, Weil D, Standart N. CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J Biol Chem. 2007;282:37389–37401. doi: 10.1074/jbc.M704629200. [DOI] [PubMed] [Google Scholar]
- Munchow S, Sauter C, Jansen RP. Association of the class V myosin Myo4p with a localised messenger RNA in budding yeast depends on She proteins. J Cell Sci. 1999;112:1511–1518. doi: 10.1242/jcs.112.10.1511. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- Niepel M, Strambio-de-Castillia C, Fasalo J, Chait BT, Rout MP. The nuclear pore complex-associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. J Cell Biol. 2005;170:225–235. doi: 10.1083/jcb.200504140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, Sengupta J, Frank J, Nissen P. Regulation of eukaryotic translation by the RACK1 protein: A platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–1141. doi: 10.1038/sj.embor.7400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E, Stanton LH, Madrid AS, Kieselbach T, Weis K. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J Cell Biol. 2009;185:475–491. doi: 10.1083/jcb.200810030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Schiebel E. The role of the yeast spindle pole body and the mammalian centrosome in regulating late mitotic events. Curr Opin Cell Biol. 2001;13:762–769. doi: 10.1016/s0955-0674(00)00281-7. [DOI] [PubMed] [Google Scholar]
- Pereira G, Schiebel E. Separase regulates INCENP–Aurora B anaphase spindle function through Cdc14. Science. 2003;302:2120–2124. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- Rapoport TA. Protein transport across the ER membrane. Trends Biochem Sci. 1990;15:355–358. doi: 10.1016/0968-0004(90)90076-n. [DOI] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Schramm C, Elliott S, Shevchenko A, Shevchenko A, Schiebel E. The Bbp1p–Mps2p complex connects the SPB to the nuclear envelope and is essential for SPB duplication. EMBO J. 2000;19:421–433. doi: 10.1093/emboj/19.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Gingras AC. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr Opin Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavru F, Hulsmann BB, Spang A, Hartmann E, Cordes VC, Gorlich D. NDC1: A crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J Cell Biol. 2006;173:509–519. doi: 10.1083/jcb.200601001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins-Boaz B, Cao Q, de Moor CH, Mendez R, Richter JD. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol Cell. 1999;4:1017–1027. doi: 10.1016/s1097-2765(00)80230-0. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Amon A. Closing mitosis: The function of the Cdc14 phosphatase and its regulation. Annu Rev Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- Strambio-de-Castillia C, Blobel G, Rout MP. Proteins connecting the nuclear pore complex with the nuclear interior. J Cell Biol. 1999;144:839–855. doi: 10.1083/jcb.144.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcheperegine SE, Marelli M, Wozniak RW. Topology and functional domains of the yeast pore membrane protein Pom152p. J Biol Chem. 1999;274:5252–5258. doi: 10.1074/jbc.274.8.5252. [DOI] [PubMed] [Google Scholar]
- Tzur YB, Wilson KL, Gruenbaum Y. SUN-domain proteins: ‘Velcro’ that links the nucleoskeleton to the cytoskeleton. Nat Rev Mol Cell Biol. 2006;7:782–788. doi: 10.1038/nrm2003. [DOI] [PubMed] [Google Scholar]
- Winey M, Goetsch L, Baum P, Byers B. MPS1 and MPS2: Novel yeast genes defining distinct steps of spindle pole body duplication. J Cell Biol. 1991;114:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Hoyt MA, Chan C, Goetsch L, Botstein D, Byers B. NDC1: A nuclear periphery component required for yeast spindle pole body duplication. J Cell Biol. 1993;122:743–751. doi: 10.1083/jcb.122.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R, Blobel G, Rout MP. POM152 is an integral protein of the pore membrane domain of the yeast nuclear envelope. J Cell Biol. 1994;125:31–42. doi: 10.1083/jcb.125.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Shan X, Sinelnikov A, Melese T. Yeast mutants that produce a novel type of ascus containing asci instead of spores. Genetics. 1996;144:979–989. doi: 10.1093/genetics/144.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller CE, Parnell SC, Dohlman HG. The RACK1 ortholog Asc1 functions as a G-protein β subunit coupled to glucose responsiveness in yeast. J Biol Chem. 2007;282:25168–25176. doi: 10.1074/jbc.M702569200. [DOI] [PubMed] [Google Scholar]