SUMMARY
Binding of the HIV-1 envelope glycoprotein gp120 to the CCR5 coreceptor reduces constraints on the metastable transmembrane subunit gp41, thereby enabling gp41 refolding, fusion of viral and cellular membranes, and infection. We previously isolated adapted HIV-1JRCSF variants that more efficiently use mutant CCR5s, including CCR5(Δ18) lacking the important tyrosine sulfate-containing amino terminus. Effects of mutant CCR5 concentrations on HIV-1 infectivities were highly cooperative, implying that several may be required. However, because wild-type CCR5 efficiently mediates infections at trace concentrations that were difficult to accurately measure, analyses of its cooperativity were not feasible. New HIV-1JRCSF variants efficiently use CCR5(HHMH), a chimera containing murine extracellular loop 2. The adapted virus induces large syncytia in cells containing either wild-type or mutant CCR5s and has multiple gp120 mutations that occurred independently in CCR5(Δ18)-adapted virus. Accordingly, these variants interchangeably use CCR5(HHMH) or CCR5(Δ18). Additional analyses strongly support a novel energetic model for allosteric proteins, implying that the adaptive mutations reduce quaternary constraints holding gp41, thus lowering the activation energy barrier for membrane fusion without affecting bonds to specific CCR5 sites. In accordance with this mechanism, highly adapted HIV-1s require only one associated CCR5(HHMH), whereas poorly adapted viruses require several. However, because they are allosteric ensembles, complexes with additional coreceptors fuse more rapidly and efficiently than minimal ones. Similarly, wild-type HIV-1JRCSF is highly adapted to wild-type CCR5 and minimally requires one. The adaptive mutations cause resistances to diverse entry inhibitors and cluster appropriately in the gp120 trimer interface overlying gp41. We conclude that membrane fusion complexes are allosteric machines with an ensemble of compositions, and that HIV-1 adapts to entry limitations by gp120 mutations that reduce its allosteric hold on gp41. These results provide an important foundation for understanding the mechanisms that control membrane fusion and HIV-1’s facile adaptability.
Keywords: Human immunodeficiency virus, Membrane fusion, Allostery, CCR5, Viral adaptation
INTRODUCTION
HIV-1 envelope glycoproteins are trimers with two subunits, a surface subunit gp120 that binds to receptors and a transmembrane subunit gp41 that mediates membrane fusion 1; 2. In native virions, gp41 is in a metastable conformation and is constrained by the gp120 cap. Binding of gp120 to CD4 and then to a coreceptor (usually either CCR5 or CXCR4) reduces the constraint and enables gp41 to refold into a fusion-active conformation at an accelerated rate. After CD4/CCR5 binding, the gp41 trimers extend their amino termini in a harpoon-like manner to form a three-stranded coil (3SC) that embeds the hydrophobic amino terminal fusion peptides into the target cell membrane 1; 3. Membrane fusion is driven by folding of the 3SC into the more energetically stable six-helix bundle (6HB) as C-terminal helices from gp41 heptad repeat 2 pack in an antiparallel orientation into the grooves of N-terminal helices from heptad repeat 1 1; 4. This pulls the virus closer to the cell membrane and is irreversibly blocked by the 36 amino acid peptide enfuvirtide (T-20), which binds into the grooves of the 3SC 1. Sensitivity to T-20 is dictated by the 3SC half-life, which is influenced by CCR5 mutations and concentrations, and by sequences of gp120 and gp41 5; 6; 7. These results suggest that gp120 and CCR5 are present throughout gp41 refolding, in agreement with the idea that gp120 controls the magnitude of the activation energy barrier that constrains gp41 as well as the degree to which those constraints are reduced by gp120-CCR5 interactions. Optimal control by gp120 is important because premature gp41 refolding inactivates the virus and damages cells 8. Nevertheless, successful infection depends upon winning a race in which resolution of the 3SC coil with subsequent membrane fusion must occur before a competing inactivation or dissociation process eliminates the virus 6.
HIV-1 mutants resistant to small molecule CCR5 antagonists (SMCAs) have adaptive gp120 mutations in variable regions including V3 9; 10. Similarly, adaptations to other entry limitations and shifts to CXCR4 are principally determined by V3 and V3 also affects sensitivities to T-20 5; 11; 12. Although V3 interacts with coreceptors, the mechanisms by which this interaction modulates gp41 and controls infection are substantially unknown.
The tyrosine sulfate-containing amino terminus and extracellular loop 2 (ECL2) regions of CCR5 are most critical for its coreceptor activity 13; 14. Nevertheless, we isolated HIV-1JRCSF variants that efficiently use CCR5(Δ18) that lacks the amino terminus, and others that efficiently use CCR5(G163R), which has a partially inhibitory mutation in ECL2 12; 15. All adapted viruses had mutations in V3, and some also had single mutations in V2 and V4. Surprisingly, viruses adapted to CCR5(Δ18) efficiently used CCR5(G163R) but not the double mutant, implying that they can employ the CCR5 amino terminus but no longer depend on it. They accomplished this by infecting more rapidly as indicated by increased resistances to T-20.
During the latter investigations, we made panels of HeLa-CD4 cell clones that express wild-type or mutant CCR5s in discrete quantities 12; 16; 17. Wild-type CCR5 mediates HIV-1 entry efficiently at trace concentrations (e.g., ~6000/cell) that could not be accurately measured, and this prevented us from analyzing the precise relationship between cell surface CCR5 concentrations and HIV-1 infectivities. In contrast, the mutant coreceptors CCR5(Y14N) and CCR5(G163R) bound gp120-CD4 complexes weakly and mediated infections only at much higher concentrations that were easily measured. The relationships between cell surface concentrations of these mutant CCR5s and infectivities were sigmoidal in shape, indicating a high degree of cooperativity in their mediation of infections. Our analyses of this cooperativity for wild-type and adapted viral variants were consistent with the idea that infections require assembly of complexes with a uniform stoichiometry of 3–6 CCR5s 16. However, the viruses analyzed were not fully adapted, and there were indications in the data that the most adapted viruses might require fewer coreceptors. This evidence was compatible within experimental error with the generally accepted hypothesis that viral and cellular membrane fusion complexes are uniform assemblages with multiple subunits from each membrane that form a fusion pore 1; 18.
Here we extend these studies using a CCR5 mutant with a more severely damaged ECL2. Surprisingly, the adaptive mutations had been obtained independently in viruses adapted to CCR5(Δ18), which strongly suggested that they do not compensate for the entry limitation by increasing viral dependency on undamaged region(s) of CCR5. In addition, while the damaged CCR5s induce wild-type HIV-1JRCSF infections only weakly and in a highly cooperative manner, they induce infections of the adapted viruses much more efficiently at lower concentrations and with less cooperativity. These results imply that the adaptive mutations make the coreceptor-induced conformational changes more energetically favorable, thus enabling damaged CCR5s to mediate infections efficiently. Specific predictions of this mechanism, confirmed by our evidence, were as follows: (i) Because the adapted viruses are more easily triggered, they should more efficiently use damaged CCR5s regardless of the sites of damages and should also cause larger syncytia. (ii) The adaptive mutations should cluster in a region of gp120 important for constraining gp41. (iii) By enabling the conformational changes in the envelope trimers to occur more readily, the adaptive mutations should reduce the cooperativity of CCR5 binding and the numbers and concentrations of CCR5s required for infection 19; 20; 21; 22; 23. (iv) The adaptive mutations would be expected to reduce viral susceptibilities to entry inhibitors including SMCAs and T-20. Considered together, our analyses suggest that HIV-1 entry is limited energetically by an adjustable allosteric rheostat that is centered in the V3 region of the gp120 trimer interface overlying gp41, and that adaptive mutations in this region reduce the activation energy barrier that limits gp41 refolding, thus enabling infections to occur more efficiently.
RESULTS
Isolation and characterization of CCR5(HHMH)-adapted HIV-1JRCSF
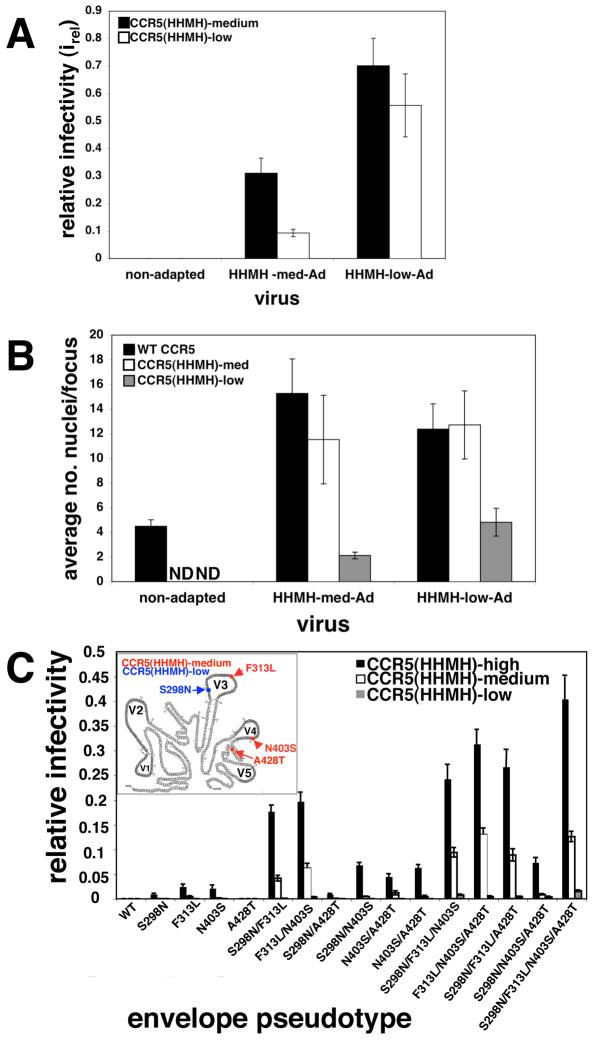
CCR5(HHMH), a chimera containing ECL2 from an NIH/Swiss mouse, was less active as a coreceptor for HIV-1JRCSF than other ECL2 mutants that we tested (see Fig S1 in the Supplementary material online). Indeed, titers of HIV-1JRCSF were extremely low in cells containing CCR5(HHMH) compared to our optimally susceptible reference cell clone JC.53 that contains a large amount of wild-type CCR5 (i.e., irel values were ~.003). Adapted viruses were isolated in a stepwise manner by first obtaining a variant that replicated efficiently in HeLa-CD4/CCR5(HHMH)-med cells that contain 1×105 CCR5(HHMH)/cell and then selecting for rapid replication in CCR5(HHMH)-low cells (2×104 CCR5(HHMH)/cell). The adapted viruses were unable to infect HeLa-CD4 cells that contain CXCR4 but lack CCR5 (results not shown), and they used CCR5(HHMH) almost as efficiently as they used wild-type CCR5 (Fig 1A). They also formed large syncytia in HeLa-CD4 cells that expressed either wild-type CCR5 or CCR5(HHMH)(Fig 1B), consistent with evidence described below that the adaptations facilitate membrane fusion and are not specific to the CCR5 mutation used for the selection. They were also highly sensitive to inactivation by soluble CD4, implying that their gp120-gp41-sCD4 complexes undergo irreversible conformational changes more readily than unadapted HIV-1JRCSF 12 (also, results not shown)
Fig 1.
Properties of CCR5(HHMH)-adapted viruses. (A) Infectivities of CCR5(HHMH)-adapted HIV-1JR-CSF. Viruses adapted to grow in HeLa-CD4 clones expressing medium or low amounts of CCR5(HHMH) were titered in these same cells and compared to wild-type HIV-1. Relative infectivities were obtained by dividing the titer obtained in CCR5(HHMH) cells by those obtained in JC.53 cells (N=5). Error bars are S.E.M. (B) Syncytia formation by unadapted and adapted viruses. The average numbers of nuclei per focus of infection were counted in cells with wild-type CCR5, HHMH-med, or HHMH-low. The data were obtained by examining 50–200 foci from a representative experiment. Error bars are S.E.M. N.D., (not done) due to the low titers on these cells. (C) Contributions of adaptive gp120 mutations to CCR5(HHMH) use. Viruses pseudotyped with single adaptive mutations or combinations thereof were used to infect HeLa-CD4 cells containing low, medium, or high amounts of CCR5(HHMH). Relative infectivity values (irel) were measured. The data represent the average of four independent experiments performed in duplicate. Error bars are S.E.M. Inset: Schematic showing locations of CCR5(HHMH)-adaptive mutations in JRCSF gp120. Variable regions are shown in gray (V1–V5), while branched structures denote glycosylation sites. Virus initially adapted in cells expressing medium amounts of CCR5(HHMH) had adaptive mutations in V3 (F313L), V4 (N403S) and C4 (A428T) (red). After further adaptation in cells a with low amount of CCR5(HHMH), the C4 A428T mutation disappeared and the V3 mutation S298N accumulated (blue).
Interestingly, there were only four functionally significant adaptive mutations, and they all occur in gp120 rather than gp41 (Fig 1C). Mutations that accumulated during selection in CCR5(HHMH)-med cells were F313L in V3, N403S in V4 that eliminates a site for N-linked glycosylation, and A428T in conserved region 4 (C4). Subsequent adaptation in CCR5(HHMH)-low cells resulted in retention of F313L and N403S, loss of A428T, and emergence of the V3 mutation S298N. Thus, the most adapted virus had only three mutations.
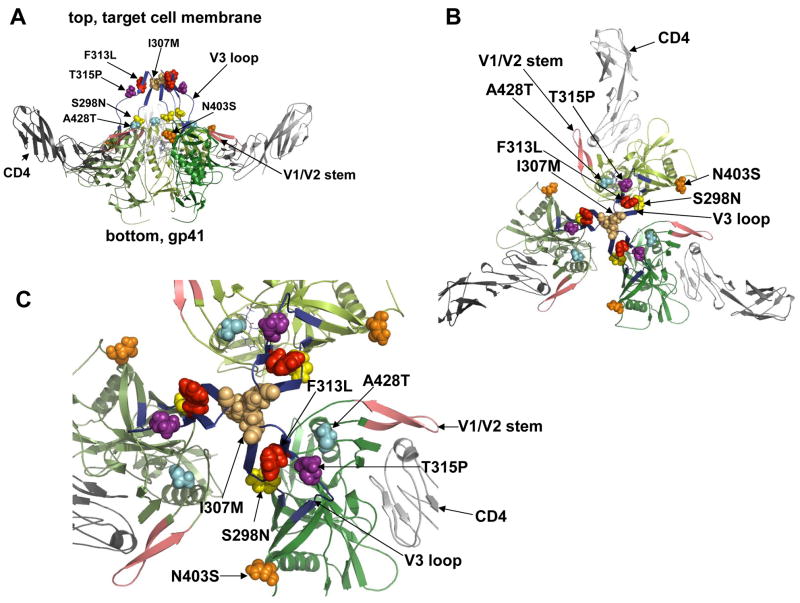
We examined the contributions to CCR5(HHMH) use of each gp120 mutation (Fig 1C). The mutations had additive effects in most combinations. More specifically, S298N, F313L and, N403S made major contributions to CCR5(HHMH) use, while A428T was very weak and contributed most significantly when F313L and/or N403S were present (Fig 1C). The mutant envelope glycoproteins were expressed, processed, and incorporated into virions to the same extents as the wild-type glycoprotein (Fig S2, which is in the Supplementary material online). Interestingly, these and other adaptive mutations described below all cluster in the gp120 trimer at the intersubunit interface that directly overlies gp41 (Fig 2).
Fig 2.
Structural modeling of the trimeric gp120/CD4 complex showing the adaptive mutations as space-filling objects. The structure of HIV-1 JR-FL gp120 core protein containing the third variable region (V3) was used to generate the trimeric model in accordance with previous evidence. Frame A, side view of the gp120 trimer (green shaded) with associated CD4s (in grey) and indicating positions of the adaptive mutations near the top and gp41 at bottom. Frame B shows a top view in low magnification, with adaptive mutations clustered at the gp120 interfaces overlying the central channel above gp41. Frame C shows the top view at a higher magnification. This clustering of adaptive mutations supports the hypothesis that they collaboratively control allosteric changes in quaternary structure that prevent gp41 refolding.
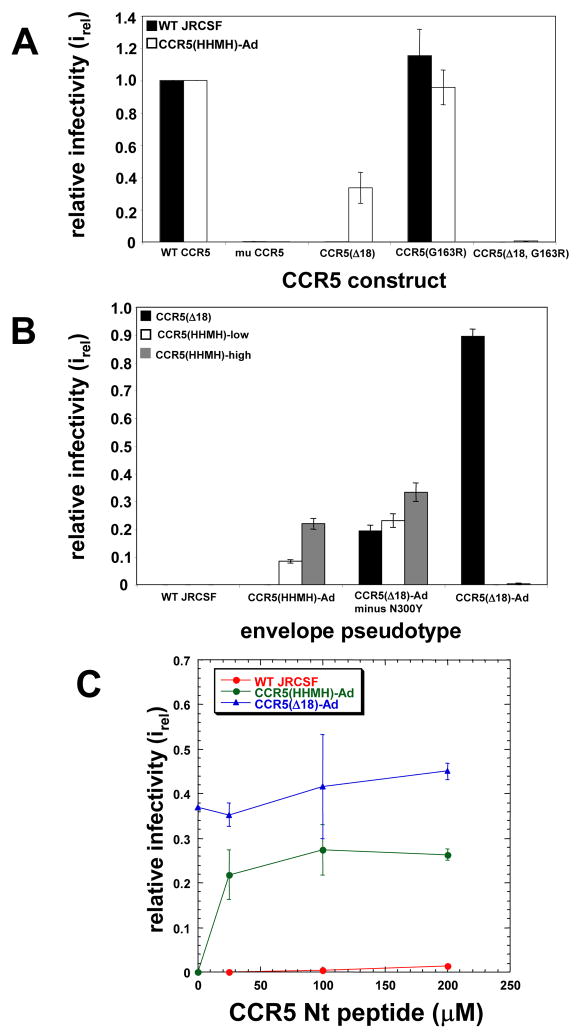
Surprisingly, the mutations S298N, F313L, and N403S that were present in the CCR5(HHMH)-low-adapted virus were previously found in HIV-1JRCSF variants selected for replication in HeLa-CD4/CCR5(Δ18) cells 12. This strongly suggested that these mutations facilitate use of CCR5(HHMH) and CCR5(Δ18) by a common mechanism. We emphasize, however, that the CCR5(Δ18)-adapted virus contains several additional adaptive mutations in gp120 V3 including N300Y, which enhances use of CCR5s with damaged amino termini but inhibits use of CCR5s with damaged ECL2 15. Therefore, while some mutations facilitate use of mutant CCR5s regardless of the site of damage, N300Y is more specific. Additionally, the CCR5(Δ18)-adapted virus contained I307M and T315P. In accordance with these considerations, the CCR5(HHMH)-low adapted virus used CCR5(Δ18) efficiently when it was coexpressed with CD4 in transiently transfected 293T cells, whereas wild-type HIV-1JRCSF was completely inactive (Fig 3A). Thus, although the CCR5(HHMH)2adapted virus contains only three of the gp120 mutations that contribute to CCR5(Δ18) usage, S298N, F313L, and N403S clearly enhance use of both CCR5(HHMH) and CCR5(Δ18). Whereas both wild-type HIV-1JRCSF and the CCR5(HHMH)-adapted virus used CCR5(G163R) efficiently in the conditions of this experiment, neither used murine CCR5 or a CCR5(Δ18;G163R) double mutant damaged in both its amino terminus and ECL2 (Fig 3A). In this context we note that attempts to use the CCR5(Δ18;HHMH) double mutant were uninformative because we could not detect it on cell surfaces. Consistent with these results, the CCR5(Δ18)-adapted virus that lacked N300Y used CCR5(HHMH) very efficiently (Fig 3B) implying that S298N, I307M, F313L, T315P, and N403S all facilitate use of both CCR5(HHMH) and CCR5(Δ18).
Fig 3.
Facile use of CCR5 domains by adapted HIV-1. (A) HIV-1 infections mediated by wild-type, mutant and murine CCR5s. Wild-type HIV-1JRCSF and the variant adapted to a low amount of CCR5(HHMH) were compared for abilities to infect 293T cells transiently expressing CD4 and murine CCR5 (mu CCR5), CCR5(Δ18), CCR5(G163R) or the double mutant CCR5(Δ18, G163R). The data are means of two independent experiments performed in triplicate. Error bars are S.E.M. (B) CCR5(Δ18) and CCR5(HHMH) use by HIV-gpt viruses containing adaptive gp120 mutations. Pseudotyped viruses were used to infect HeLa-CD4 cells expressing CCR5(Δ18) (2.7 × 104 molecules/cell), CCR5(HHMH)-low, or CCR5(HHMH)-high. Adaptive gp120 mutations in the virus pseudotypes were as follows: CCR5(HHMH)-Ad: S298N/F313L/N403S; CCR5(Δ18)-Ad minus N300Y: S298N/I307M/F313L/T315P/N403S; CCR5(Δ18)-Ad:S298N/N300Y/I307M/F313L/T315P/N403S. The data are from 2 independent experiments performed in duplicate. Error bars are S.E.M. (C) Infections mediated by CCR5(Δ18) plus sulfated N-terminal CCR5 peptide. HeLa-CD4 cells expressing 2.7 × 104 CCR5(Δ18) molecules/cell were infected in the presence of varying concentrations of CCR5 peptide (0, 25, 100, and 200 μM), and infectivities (irel) were measured relative to JC.53 cells. The replication competent CCR5(Δ18)-adapted, CCR5(HHMH)-adapted, and wild-type JRCSF (blue, green, and red curves, respectively) isolates were tested. The graph shows a representative experiment performed in duplicate. Error bars are the range.
Although the CCR5(HHMH)-adapted virus is less dependent on the CCR5 amino terminus than the wild-type virus, it counterintuitively uses the amino terminus much more efficiently when ECL2 is damaged (Fig 3A and B). This is substantiated in Fig 3C, which shows effects of a tyrosine sulfated amino terminal peptide on infection of HeLa-CD4/CCR5(Δ18) cells. The CCR5(HHMH)-adapted virus infected the cells efficiently when low concentrations of the peptide were present, whereas the wild-type virus was weakly infectious only when much larger concentrations were used. Single adaptive mutations also increased the ability of the virus to use the amino terminal peptide (results not shown). As expected, the CCR5(Δ18)-adapted virus was infectious in the absence of the peptide. Considered together, these results imply that the adaptive mutations do not increase gp120 binding to specific sites in the damaged CCR5s used for selection. Rather, they alter the virus so that it fuses more readily in a manner that is less dependent on any specific region of CCR5.
Role of allostery in the adaptive mechanism
(a) Effects of CCR5(HHMH) concentrations on viral infectivities
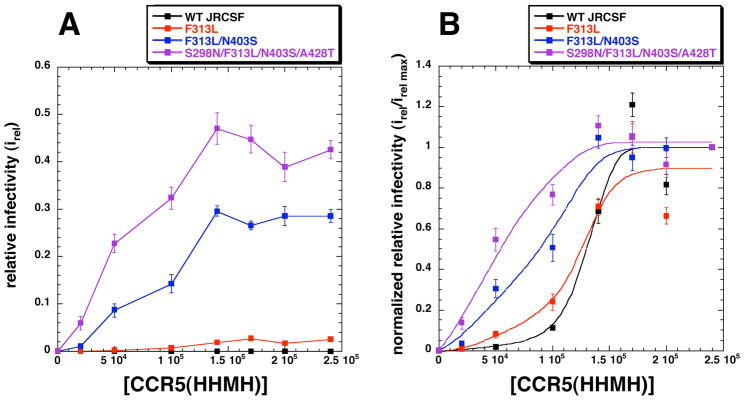
We analyzed infections using HeLa-CD4/CCR5(HHMH) clones that express discrete amounts of CCR5(HHMH). The wild-type and adapted mutant viruses were normalized to the same titers in the optimally susceptible HeLa-CD4/CCR5(wild-type) cell clone JC.53 and the relative titers were then measured in the CCR5(HHMH)-containing cells (Fig 4A). Although the wild-type and partially adapted viruses had low infectivities in these cell clones at all CCR5(HHMH) concentrations compared to the fully adapted virus, their titers were nevertheless highly significant and were accurately measured using less diluted virus samples. The curves, normalized relative to their maximum values, differ in positions on the CCR5(HHMH) concentration axis and in their shapes (Fig 4B). Specifically, the data for the highly adapted virus more closely resembles a simple saturation curve that extrapolates through the origin, whereas the data for the less infectious partially adapted and unadapted viruses have progressively more sigmoidal shapes suggestive of increasingly strong cooperative effects of CCR5(HHMH) concentrations. The differences in the curve shapes in Fig 4B and our other evidence (Figs 1–3) strongly imply that CCR5 activates gp120-gp41 trimers by a cooperative allosteric mechanism and that the adaptive mutations enable the allosteric transition to occur more readily. Thus, as proposed in the model of Monod et al 23 and supported by subsequent investigations 19; 20; 21, the above data suggest that quaternary interactions between gp120 subunits in the CD4-associated envelope trimers constrain the CCR5 binding sites and prevent gp41 refolding. Conversely, by enabling the gp120s in the trimers to adopt a less constrained conformation, the adaptive mutations would enhance gp120 affinities for damaged CCR5s regardless of the sites of damage and would simultaneously reduce the activation energy barrier that prevents gp41 refolding19; 20; 21. The clustering of adaptive mutations in the trimer interface between gp120 subunits (Fig 2) supports the idea that this interface forms a barrier that must be breached for infection to occur.
Fig 4.
Relationship of CCR5(HHMH) concentrations and viral infectivities. (A) Infectivities of wild-type and CCR5(HHMH)-adapted viruses in a HeLa-CD4 panel expressing distinct amounts of CCR5(HHMH). The infectivities of HIV-gpt viruses pseudotyped with wild-type envelopes or envelopes with the adaptive mutations designated in the figure were tested. Relative infectivities compared to JC.53 cells were plotted versus CCR5(HHMH) concentrations. The data represents the average of three independent experiments performed in triplicate. Error bars are S.E.M. (B) Normalization of the infectivity data in panel A to irel max values. The infectivity data for each virus was normalized to its maximum value, and the curves were drawn using Kaleidagraph (version 3.6) employing the least squares method. The infectivity data for wild-type virus with low titers on CCR5(HHMH) cells were easily measured using concentrated virus samples (average colony numbers on the highest expressing CCR5(HHMH) cell line ranged from 30 to 70 per well at a 1/5 virus dilution).
(b) Quantitative modeling of the allosteric mechanism
In order to better understand the HIV-1 entry mechanism, we derived an activation energy model of allostery that incorporates issues specifically relevant to virus infection (see Materials and Methods and Figs S3 and S4 in the Supplementary information online). In this context, it is notable that previous allosteric models including that of Monod et al 19; 20; 21; 22; 23 assume an equilibrium between two conformations of the allosteric protein, a predominating and functionally inhibited taut or constrained conformation that binds activating ligands relatively weakly, and a more relaxed functionally active conformation. Because activating ligands bind more strongly to the relaxed conformation, they shift the equilibrium in that direction. Although this concept is likely to be relevant to HIV-1 infection, the CCR5-induced conformation of the envelope trimer is unknown and the presumptive equilibria cannot be directly measured. Moreover, evidence in this field strongly suggests that the key CCR5-mediated process involves an irreversible breachment of gp120’s hold on gp41, and this breachment cannot be readily explained as an equilibrium between alternative conformations. Our model focuses on the activation energy barrier that constrains gp41 and indicates how the magnitude of this barrier would control the rate of irreversible gp41 conformational change. As in other allosteric models, we assume that the activating ligand forms an equilibrium assemblage with the protein of interest, in agreement with previous evidence concerning HIV-1 virion interactions with cell surface CD4 and CCR5 in our experimental system 6. In particular, the lag phase required for assembly of viral fusion complexes is very short compared to the time required for adsorbed virions to enter cells, and following this assembly phase the cohort of virions fuse in a stochastic manner consistent with an activation energy limitation rather than in a synchronous wave 6; 24 (see Discussion). Although our derivation would potentially be applicable to an allosteric protein with any number of subunits, we initially assumed a process involving trimers based on the HIV-1 envelope structure. We considered the possibility that the fusion complexes might contain several cooperating trimers, but we found as shown below that a model involving single trimers best fit our data. While this was somewhat surprising, it is compatible with other evidence 25, and we believe it is robustly supported by our data (see Discussion).
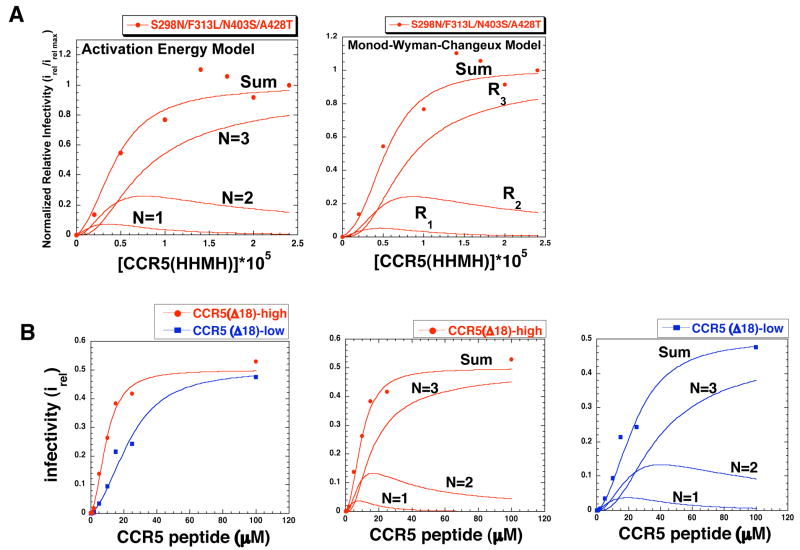
The infectivity data in Fig 4B were highly consistent with the hypothesis that gp120-gp41 trimers have three coreceptor binding sites and function by an allosteric mechanism in accordance with our model. We illustrate this for the most adapted virus in Fig 5A, which shows the close correspondence between the infectivity data and a theoretical curve based on equation 4 of our activation energy model. As deconvoluted in this analyses, the viral complexes with one or two CCR5(HHMH)s substantially contributed to the infections, especially at low CCR5(HHMH) concentrations. Even at high concentrations the complex with three CCR5(HHMH)s was responsible for only approximately 80% of the infectivity. Although it has a different theoretical basis, the allosteric model of Monod et al 23 also fit the data and gave similar estimates for the contributions to infectivity of viral complexes with different CCR5(HHMH) stoichiometries (Fig 5A). Thus, these models are quantitatively compatible with each other and with our data. However, the activation energy model is more useful for this analysis because the parameters quantitated by the curve fitting provide information concerning the effects of each CCR5(HHMH) addition on the gp120-imposed free energy barriers that limit infection (see Table 1). Thus, the viruses infect with different maximum efficiencies because the residual activation energy barriers that remain after formation of the saturated complexes (i.e., the values) differ. The wild-type gp120-gp41 trimer is highly constrained in the absence of a coreceptor as indicated its relatively large , and this large activation energy barrier is efficiently eliminated by binding wild-type CCR5 but not CCR5(HHMH). Consequently, wild-type HIV-1 efficiently uses wild-type CCR5 but inefficiently uses CCR5(HHMH). In contrast, the adapted viruses have lower indicating that they are less constrained in the absence of coreceptors and this weaker barrier is substantially overcome by binding even a single CCR5(HHMH), thus enabling infections to ensue. As expected for a cooperative process, our results suggest that the first CCR5(HHMH) that binds has to push against a larger constraint than subsequent CCR5(HHMH)s (i.e., f1<f2<f3 in equation 4). Consequently, this first CCR5(HHMH) has a relatively low binding affinity but makes a correspondingly large contribution to reducing the energy barrier. Analogously, the first oxygen that binds to hemoglobin has a relatively low affinity but makes a correspondingly large energetic contribution to switching the quaternary structure from a taut to a relaxed conformation 21. In this context, we note that the allosteric models were not useful for analyzing the wild-type and only slightly adapted viruses because their native conformations are very constraining and they consequently bind CCR5(HHMH) and undergo conformational changes needed for infection in a highly concerted manner without significant contributions by complexes that have only one or two CCR5(HHMH)s. These results suggest that the adaptive mutations shift the gp120-gp41 complex from a concerted form that requires three CCR5(HHMH)s toward a more flexible form that mediates infection when fewer CCR5(HHMH)s are bound. It should be understood that these activation energy estimates pertain only to the allosteric barriers imposed by gp120 rather than to the total free energy barrier required for membrane fusion (see “Implications of our activation energy measurements” in Supplementary information online). The conformational energy released during gp41 refolding makes a major energetic contribution to the fusion of the lipid bilayers 4.
Fig 5.
Role of allostery in HIV-1 infections. (A, left panel) Activation Energy Model. The activation energy model (Eq 4) was fit to the infectivity data (Fig 4) for the virus with S298N/F313L/N403S/A428T that is highly adapted to CCR5(HHMH). (A, right panel) Monod-Wyman-Changeux Model. The latter model also fit to the same infectivity data. Both models closely fit the data and gave similar estimates for the contributions to infectivity of the viral complexes with different CCR5(HHMH) stoichiometries, of 1 (N=1, R1), 2 (N=2, R2), or 3 (N=3, R3). (B) Infection triggered by a tyrosine sulfated N-terminal CCR5 peptide. The N-terminal CCR5 peptide induced infections of HeLa-CD4 cells expressing low (2.7 × 104) or high (6.6 × 104) amounts of CCR5(Δ18) by the virus highly adapted to use CCR5(HHMH)-low. (B,left panel) Fit of infectivity data (irel values) using the activation energy allosteric model equation 4. The irel max values for both cell clones were 0.5. (B, middle and right panels) Deconvolution analysis of the peptide triggering data for cells with high (middle panel) and low (right panel) CCR5(Δ18), showing how complexes with 1, 2, or 3 peptides contributed to infection. Parameters obtained by fitting the data using equation 4 are given in Table 1.
Table 1.
Effects of Wild-type and Mutant CCR5s on Activation Energy Barriers that Limit Infections of HIV-1 Isolates.
| Virus | CCR5 | Maximum Relative Infectivity(irel max)*1 |
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wild-type | wild-type | 1.0 | (>4.9)*3 | -- | -- | 0 | ||||
| Wild-type | HHMH | 0.00032 | (>4.9)*3 | -- | -- | 4.9 | ||||
| F313L | HHMH | 0.025 | -- | -- | -- | 2.3 | ||||
| F313L/N403S | HHMH | 0.286 | 4.0 | 1.2 | 0.77 | 0.77 | ||||
| Fully adapted (S298N/F313L/N403S/A428T) | HHMH | 0.43 | 2.8 | 1.0 | 0.59 | 0.53 | ||||
| Fully adapted | Δ18+ amino terminal peptide | 0.51 | 2.7 | 0.96 | 0.40 | 0.40 |
irel max is the maximum infectivity of each virus at saturating concentrations of the CCR5 used relative to the titer in JC.53 cells that express an optimally saturating amount of wild-type CCR5.
is the difference in kcal/mole between the activation free energy barrier that limits infection when the virus is saturated with the test CCR5 and the corresponding barrier when the virus is saturated with three wild-type CCR5s. This value was estimated from the irel max values for each virus-coreceptor combination as described in Experimental Procedures. Based on a previous estimate that the efficiency of infection by wild-type HIV-1JRCSF in JC.53 cells is ~0.2 6, we infer that the absolute for infection is ~ 1 kcal/mole larger than the relative values recorded in this column. For example, we estimate that there is a residual activation energy barrier of ~1 kcal/mole when the wild-type virus is saturated with wild-type CCR5 and ~5.9 kcal/mole when it is saturated with CCR5(HHMH). Similarly, , and are the free energy barriers in kcal/mole when the gp120-CD4 fusion complexes contain 0, 1, or 2 molecules of the test coreceptor. These values were estimated by fitting the data to equation 4 using Kaleidagraph, and using the resulting best-fit parameters in these estimates. Values for these sequential energy barriers were not obtained for the unadapted and poorly adapted viruses because they bind CCR5s in a highly concerted manner that was not amenable to allosteric analysis (see text). The f1, f2, f3, and k curve fitting values derived from use of equation 4 were as follows: 0.01, 0.5, 1.0, and 0.154 ×105/cell for the F313L/N403S virus using CCR5(HHMH), 0.05, 0.5, 0.9, and 0.154 ×105/cell for the fully adapted virus using CCR5(HHMH). The f1, f2, f3 values were 0.06, 0.40, and 1.0 for the fully adapted virus using the peptide plus CCR5(δ18). In that case, the apparent k values were 3.39μM and 8.13μM for the cells with high or lower amounts of CCR5(Δ18), respectively, consistent with the 2.4-fold difference predicted.
This is a minimum estimate of for the wild-type virus-CD4 complex lacking CCR5s. Since the complex saturated with CCR5(HHMH) has a residual activation energy of 4.9 kcal/mole, the unliganded complex must have a barrier at least that large.
As shown above, relatively low concentrations of the CCR5 amino terminal peptide strongly enhance infectivity of the CCR5(HHMH)-adapted virus in HeLa-CD4/CCR5(Δ 18)-high cells (Fig 3C). In contrast, much larger concentrations weakly stimulated wild-type HIV-1JRCSF infection, and the concentration dependency of this infectivity was reproducibly more sigmoidal in shape, implying a much stronger degree of peptide cooperativity. Since the concentration of the amino terminal peptide can be varied independently of CCR5(Δ 18), it provided a relatively stringent system to analyze our interpretations. Moreover, we had two clones of HeLa-CD4/CCR5(Δ 18) cells that express low versus higher amounts of CCR5(Δ 18) (i.e., 2.7 × 104 versus 6.6 × 104 CCR5(Δ 18)/cell, a difference of 2.4-fold), and we compared their abilities to mediate entry of the CCR5(HHMH)-adapted virus in the presence of different peptide concentrations. If our allosteric model is correct, the difference in CCR5(Δ 18) concentration should change the apparent microscopic dissociation constant k for the peptide by a factor of 2.4 without affecting other variables (i.e., the fi factors) in equation 4 (see Materials and Methods). The curves in Fig 5B left panel, which were based on this prediction, precisely fit the data points for both cell clones. Results in the adjacent panels deconvolute this analysis, indicating how complexes with one, two, or three peptides contributed to infections. Table 1 analyzes these results and shows how the peptide influenced affinities and residual activation energies of the viral entry complexes. Importantly, this close correspondence between our theoretical and experimental evidence strongly supports the idea that the viral fusion complexes contain one envelope trimer. If multiple trimers were required to form a fusion complex and/or if individual virions on cells simultaneously formed several independent fusion complexes, the precise correspondence between our calculated curves and these data would not have been expected.
Effects of adaptive mutations on entry inhibitor sensitivities
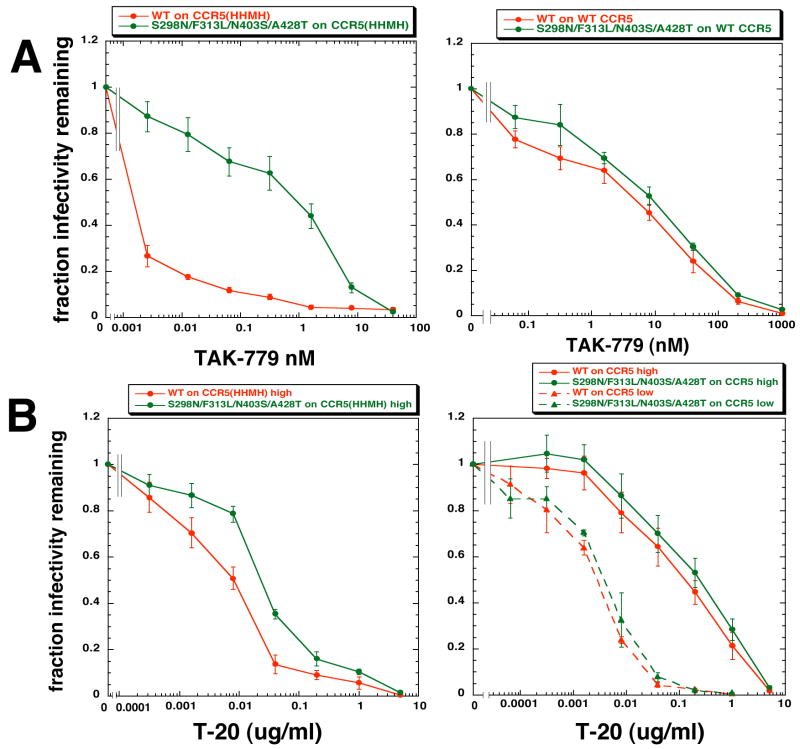
TAK-779 is a SMCA that binds to CCR5 irrespective of ECL2 mutations and prevents association of gp120 26; 27. TAK-779 sensitivities of infections mediated by CCR5(HHMH) are strikingly different for the wild-type and adapted variant HIV-1s, with the adapted virus being approximately 500-times more resistant (Fig 6A). In contrast, the viruses use wild-type CCR5 much more efficiently and similarly, with the displacement between the TAK-779 inhibition curves being only 3–4-fold. The IC50 values in the left versus right panels imply that the viruses bind more strongly to wild-type CCR5 than to CCR5(HHMH). As described above, the wild-type virus requires more associated CCR5(HHMH)s than the adapted virus, and this factor dramatically influences these TAK-779 sensitivities (see Fig S3 in Supplementary information online).
Fig 6.
TAK-779 and T-20 sensitivities of HIV-gpt viruses pseudotyped with envelopes containing wild-type gp120 or gp120 highly adapted to use CCR5(HHMH). (A) TAK-779 inhibition curves. Left: TAK-779 dose response assays performed using HeLa-CD4 cells expressing a high amount of CCR5(HHMH) (1.4 × 105 molecules/cell). Right: TAK-779 dose response assays using HeLa-CD4 cells expressing large amounts of wild-type CCR5 (1.9 × 105/cell). (B) T-20 inhibition curves. Left: T-20 dose response assays using HeLa-CD4 cells expressing high amounts of CCR5(HHMH). Right: T-20 dose response assays using HeLa-CD4 cells expressing high (1.9 × 105 molecules/cell) or low (6.0 × 103 molecules/cell) amounts of wild-type CCR5. Data are averages of 3 independent experiments performed in duplicate. Error bars are S.E.M.
The adaptative gp120 mutations reduce viral sensitivities to T-20 in the CCR5(HHMH)-containing cells by a factor of approximately five, implying that they accelerate the 3SC-to-6HB conformational change in gp41 (Fig 6B). Although the adapted virus is also more resistant than wild-type virus to T-20 in cells containing wild-type CCR5, the difference is smaller, confirming that the viruses use wild-type CCR5 more similarly than they use CCR5(HHMH). Moreover, the T-20 inhibition curves have significantly different IC50 values and shapes in cell clones that contain low or high amounts of wild-type CCR5. At low CCR5 concentrations, T-20 sensitivities are greater because each virus in the equilibrium ensemble spends only a fraction of its time associated with sufficient CCR5s to resolve its 3SC into a 6HB 6; 28. In this condition, the 3SC-to-6HB transition occurs slowly compared to the rate of CCR5 exchange between the complexes and the shape of the T-20 inhibition curve is therefore compatible with a uniform susceptibility of the viruses. In contrast, at high CCR5 concentrations the largest complexes fuse rapidly compared to the equilibration rate and these are relatively resistant to T-20. The shape difference in these inhibition curves is consistent with the idea that the fusion complexes are heterogeneous in composition. A similar analysis using cells with a small amount of CCR5(HHMH) was not feasible because the wild-type virus requires a large concentration of this coreceptor.
DISCUSSION
Our results, which are concordant with analyses of other multisubunit allosteric proteins 19; 20; 21, strongly support the simple idea that gp120 forms a restraining cap on gp41 that is relieved by association with CCR5, and that gp120-gp41 trimers function by a cooperative mechanism. An allosteric rheostat or regulator in gp120 that principally involves its V3 region and appropriately overlies gp41 (Fig 2) enables HIV-1 to dramatically alter its tropism for cells with different amounts of receptors (Figs 1, 3 and 4) and to overcome diverse entry limitations by a facile generic mechanism (Fig 6). The adaptive mutations cluster in a central channel above gp41 and at adjacent sites involved in gp120 subunit interactions, and our measurements strongly suggest that they reduce gp120’s hold on gp41 (Table 1). By folding back or loosening its grip, the V3 region would control a central passage for extension of the gp41 amino terminal 3SC coiled coil and for subsequent formation of the 6HB.
It has been widely assumed that type-1 membrane enveloped viruses assemble a uniform membrane fusion complex containing several envelope glycoprotein trimers and associated receptors and that similar compositional uniformity occurs in other membrane fusion systems 1; 18; 29. However, it is important to note that there is no agreement concerning the composition of any membrane fusion machine, and this uncertainty occurs even in the well-studied case of the low pH-triggered influenza virus hemagglutinin (HA) 30; 31; 32; 33. Indeed, this HA binds to sialic acids on diverse glycolipids and glycoproteins, and this receptor and microenvironmental heterogeneity can influence the fusion process 34; 35, perhaps by affecting the pKa values of critical HA residues or the requisite activation energies. Moreover, the influenza HA and rabies viral G proteins are commonly studied in complex artificial systems with erythrocytes or liposomes, and in these systems a process of hemifusion competes with full fusion, further complicating the analyses 34; 35; 36; 37. Recent evidence has implied that the viral glycoprotein complexes that mediate full fusion in these assays may differ in compositions or in microenvironments from those that cause only hemifusion 35; 36; 37. Thus, there is substantial heterogeneity in the fusion processes mediated by these viral glycoproteins. Although our data are compatible with the hypothesis that HIV-1 membrane fusion complexes have a uniform composition of envelope glycoprotein trimers (see below), our results suggest that these complexes differ substantially in their stoichiometries of the allosteric activator CCR5, and that these diverse fusion complexes function quantitatively as expected for a trimeric allosteric ensemble in equilibrium with CD4 and coreceptors (Fig 5 and Table 1). Accordingly, we infer that the HIV-1 fusion complexes contain a compositionally uniform central core of envelope glycoproteins that associate in physiologic conditions with a diverse equilibrium ensemble of activating allosteric ligands that strongly influence the overall process. Although we believe that similar mechanisms may regulate membrane fusion systems that are activated by other ligands such as protons or calcium ions, in these cases the ligands are likely to alter the membranes by multiple mechanisms, which would make the fusion complexes more difficult to analyze. Further investigations will be needed to evaluate these issues.
In contrast to previous allosteric models that consider only ligand binding and conformational equilibria20; 21; 22; 23, we believe that our model has important advantages for analyzing processes such as infections or signaling cascades that are irreversibly triggered by activating ligands. In particular, our model better explains infectivity differences between adapted and unadapted viruses (e.g., see Figs 4 and 5 and Table 1). Specifically, the adapted viruses have lower activation energy barriers that are substantially overcome by association with damaged CCR5s, whereas unadapted viruses are more constrained and they consequently bind damaged CCR5s less avidly in a manner that only partly reduces the barriers (Table 1). Because our model is based solely on chemical principles, the parameters quantified by the curve fitting (i.e., f1, f2, f3 and k in equation 4) provide important information about the energetics and mechanism of membrane fusion. In contrast, the parameters quantified using other allosteric models make predictions about quaternary structure equilibria that cannot easily explain irreversible processes and are often difficult to rigorously test 20; 21; 22; 23. We emphasize, however, that these models are complementary rather than mutually exclusive, and that structural (Fig 2) as well as energetic perspectives (Figs 4, 5 and Table 1) have contributed to our interpretations. Consequently, we consider our model an adjunct rather than an alternative to others.
We emphasize that our allosteric model contains several assumptions and that it provides only an initial framework for analyzing the infection process. Like previous allosteric models, we assume that the cell associated virus-CD4-CCR5 fusion complexes reach an equilibrium that depends on the concentrations and structures of its components. In strong support of this idea, in the conditions of our experiments the lag phase required for assembly of viral fusion complexes is very short compared to the time required for entry, and following this assembly phase the cohort of virions fuse in a stochastic manner consistent with an activation energy limitation rather than in a synchronous wave 6; 24. Although nonequilibrium processes including viral inactivation certainly occur during the time virions reside on the cell surfaces, our analyses suggest that this is not a confounding issue in the conditions of our experiments (see further discussion pertaining to Figs S3 and S4 in the Supplementary information online). In this context, it should be noted that assembly of fusion complexes often occurs relatively slowly in syncytial assays and in those systems cooperative processes also occur that may not be relevant to our experiments. For example, in syncytial assays, cells with viral glycoproteins adhere to receptor-bearing cells, and this causes the slow accretion and concentration of these components at the adherent surfaces with corresponding depletions elsewhere on the cells 38. Because these adhering surfaces are relatively large and form cooperatively and slowly, and because many fusion complexes accumulate in these regions, the assembly and fusion processes differ substantially from that occurring at the small virus-cell junctions. HIV-1 virions have only a small number of envelope trimers 39, and these cannot all contact the cells simultaneously. CD4 and CCR5 can diffuse rapidly in and out of these tiny contact sites, further suggesting that the viral fusion complexes are likely to reach equilibrium relatively quickly.
Our data implies that one diffusible CCR5 can mediate infections of highly adapted HIV-1s (Fig 5), but does not indicate whether that diffusible entity is a monomer or dimer40. Specifically, the infectivity versus coreceptor concentration curves for the CCR5(HHMH)-adapted virus in Figs 5A and B extrapolate through the origin and are only slightly sigmoidal in their shapes, strongly suggesting that fusion complexes containing one associated CCR5 are at least partially able to mediate infection. If more CCR5s were required, the curves would be much more sigmoidal implying a more cooperative process. Moreover, this interpretation of these curves is robust and is relatively independent of our modeling assumptions. While differences in dimerization efficiencies might conceivably occur in cell clones that express different amounts of CCR5s, this also could have only minor effects on our results or interpretations because the dramatic viral differences in CCR5 cooperativities, EC50s, and drug sensitivities that we detected were determined by gp120 mutations in assays that used the same cell clones (Figs 1 and 3–6).
A closely related matter concerns the envelope glycoprotein composition of the fusion site. Our mathematical analyses suggest that at least three CCR5s can contribute to entry, implying that membrane fusion complexes must contain at least three cooperating gp120s with accessible CCR5 binding sites. Although these could conceivably occur within a single glycoprotein trimer or in several juxtaposed trimers, our evidence discussed above suggests that highly adapted HIV-1s can infect to a significant degree when they are associated with only one CCR5 (Fig 5). Because one associated coreceptor would probably be incapable of simultaneously inducing conformational changes in several gp120-gp41 trimers, we conclude that entry of HIV-1JRCSF can be mediated in optimal conditions by one gp120-gp41 trimer, in agreement with a recent study of HIV-1YU2 25. These considerations strongly support the assumption in equation 4 of our allosteric model that the HIV-1 membrane fusion complex contains three coreceptor binding sites. In further agreement with this conclusion, equation 4 closely predicted the experimental data in Fig 5B.
Although wild-type HIV-1JRCSF uses CCR5(HHMH) inefficiently and requires the cooperative binding of several in order to infect cells, it is highly adapted to wild-type CCR5 and uses the latter very efficiently even at trace concentrations that are too low to accurately measure16. Moreover, our analyses using TAK-779 and T-20 imply that wild-type HIV-1JRCSF and the adapted virus use wild-type CCR5 with similar efficiencies (Fig 6). Since the adapted virus can infect cells when associated with one CCR5(HHMH) (Fig 5) and functions several times more efficiently when employing wild-type CCR5 (Figs 4, 6, and Table 1), we infer that it must also be able to mediate infections when associated with only one wild-type CCR5 and it follows from these considerations that this is also true for wild-type HIV-1JRCSF. Thus, although the wild-type virus requires the concerted binding of several cooperating CCR5(HHMH)s to weakly infect cells, these results suggest that it can infect cells to a significant extent when complexed with a single wild-type CCR5. Further investigations are needed to more directly analyze this important issue.
In summary, our results identify a novel mechanism by which a few mutations in gp120 enable HIV-1 to overcome diverse entry limitations. The adaptive mutations cluster on the gp120 subunit interfaces in a V3-containing region that forms a lid over gp41 and may be critical for transducing effects of CCR5 binding into other portions of the gp120-gp41 complex (Fig 2). HIV-1 variants resistant to SMCAs are also fusogenic and have adaptive mutations in the same gp120 regions that we have implicated in allosteric control 9; 10. Importantly, HIV-1s form an equilibrium ensemble of fusion-competent complexes with CD4 and CCR5 even when the virus and target cells are clonal (Fig 5), and this stoichiometric diversity can strongly influence entry inhibitor sensitivities (Fig 6 and Figs S3 and S4, which are in the Supplementary material online). Viruses that escape from inhibitors by allosteric adjustment enter cells more readily and are more fusogenic and infectious for cells that have low concentrations of CCR5. Interestingly, viruses with this constellation of properties are also relatively pathogenic and they accumulate in patients with advanced disease 41;42. Further understanding of these issues may be helpful for the design and evaluation of improved entry inhibitor and vaccine strategies for AIDS.
MATERIALS AND METHODS
Cells and viruses
HeLa-CD4 cells (HI-J) and derivative clones expressing wild-type or mutant CCR5s were propagated as described 12; 17; 43, as were HEK293T and COS7 cells16; 43. HeLa-CD4 clones expressing a chimeric CCR5 with a NIH/Swiss mouse ECL2 [CCR5(HHMH)] were generated by transduction of HI-J cells with the pSFF-CCR5(HHMH) retroviral vector 16; 44. Cell surface CCR5 antigen densities were measured by flow cytometry using the Dako Qifikit (Dako Corporation, Carpenteria, CA) according to the manufacturer’s instructions as previously described with additional standardization based on [125I]Mip-1β binding to cells with wild-type CCR516. Briefly, Qifikit calibration beads contain known concentrations of antigen and are incubated with the same goat anti-mouse FITC-conjugated secondary antibody as HeLa-CD4/CCR5 cells previously incubated with an anti-CCR5 monoclonal antibody. The 2D7 monoclonal antibody, which recognizes an epitope in ECL2, was used to determine coreceptor concentrations in cells expressing wild-type CCR5 or CCR5(Δ 18). Cells expressing CCR5(HHMH) lacks the 2D7 epitope, and in this case we used the CCR5 amino terminal specific monoclonal antibody CTC8 (R&D Systems, Minneapolis, MN), with normalization based on results using this antibody to assay cells with known amounts of wild-type CCR5. Propagation of virus produced from the HIV-1JR-CSF molecular clone, pYK-JRCSF, that was obtained from the NIH AIDS Research and Reference Reagent Program (ARRRP) Division of AIDS, NIAID, contributed by I. Chen and Y. Koyanagi, was described previously 15. Adapted isolates of HIV-1JR-CSF able to grow in HeLa-CD4 cell clones expressing small amounts of CCR5(G163R) or CCR5(Δ 18) have been described 12; 15. The CCR5(G163R)-adapted isolate was used to generate HeLa-CD4/CCR5(HHMH)-adapted viruses by forced passage of high titer virus on a cell line expressing 1.0 × 105 mutant CCR5s/cell. Infections were initiated by inoculating 5.0 × 105 – 1.0 ×106 cells seeded 24 hours previously in T-75 flasks with 2.0 × 106 ffu/ml CCR5(G163R)-adapted virus; cells were inoculated 2 times in an 8 h period, with the 3rd inoculation incubating overnight. The virus containing medium was passaged every 3 to 4 days onto fresh cultures of HeLa-CD4/CCR5(HHMH) cells. After 7 passages, syncytia became apparent. This CCR5(HHMH)-adapted virus population was also subjected to a more stringent selection in HeLa-CD4 cells that expressed a low concentration of CCR5(HHMH) (~2.0 × 104 molecules/cell). Syncytia in the cell cultures appeared after 6 passages. After syncytia formation became evident viruses were titered and dilute concentrations used to infect CCR5(HHMH) cells from which genomic DNA was harvested using the Easy-DNA reagents and protocol from Invitrogen (Carlsbad, CA).
Sequencing and construction of expression vectors for adapted envelope genes
Sequences were obtained from cloned PCR products generated from genomic DNA as previously described or from direct sequencing of PCR products 15. Gp120 mutations were introduced into pcDNA3.0-based envelope expression vectors using the QuickChange (Stratagene, La Jolla, CA) mutagenesis method according to the manufacturer’s instructions. The envelope genes were sequenced in their entirety to insure that only the desired mutations were introduced.
Construction, expression, and coreceptor activity of mutant CCR5s
Coreceptor constructs used in this study including the NIH/Swiss mouse CCR5, CCR5(Δ 18), CCR5(G163R), and CCR5(Δ 18, G163R) have been characterized and described previously 12; 14; 15; 16; 44.
Infectivity assays
Wild-type and CCR5(HHMH)-adapted replication competent HIV-1JR-CSF was titered using the focal infectivity assay as previously described 17; 45. Some infections with replication competent wild-type JR-CSF, CCR5(Δ 18) and CCR5(HHMH) adapted viruses were performed using CCR5(Δ 18) target cells in the presence of an amino-terminal CCR5 peptide. The 22 amino acid peptide MDYQVSSPIYDINYYTSEPSQK in which cysteine 20 was replaced by serine and tyrosines 10 and 17 (underlined) were sulfated was synthesized to 95% purity by the American Peptide Company (Sunnyvale, CA). The lyophilized peptide was resuspended in 100 mM sodium bicarbonate (pH 9.0) to concentration of 20 mg/ml (~7.2 × 10−3 M).
The contribution of adaptive mutations in gp120 to CCR5(HHMH) use was ascertained using HIV-gpt pseudotyped viruses. Infections of the HeLa-CD4/CCR5(HHMH) panel by pseudotyped HIV-gpt viruses with wild-type and adapted envelopes were used to investigate the effect of CCR5(HHMH) cell surface concentrations on titers and CCR5(HHMH) stoichiometries. Pseudotyped viruses were generated and used in infectivity assays as previously described 12; 43. Some infectivity assays using pseudotyped viruses were performed in the presence of the fusion inhibitor T-20 or the entry inhibitor TAK-77946. Both inhibitors were obtained from the ARRRP with Roche contributing the T-20 fusion inhibitor.
Structural modeling of the trimeric gp120/CD4 complex
The structure of HIV-1 JR-FL gp120 core protein containing the third variable region (V3) was used to generate the trimeric model [protein data base (PDB) accession code 2B4C]. The two membrane-distal domains of CD4 (D1D2) were superimposed onto the corresponding D1D2 domains (residues 1 to 168) of the structure of the four-domain extracellular region of CD4 (PDB accession code 1WIO). The superposition gave an RMS deviation of 1.1 Å for all atoms in residues 1 to 168. The superpositions, model rotations and translations were performed using Pymol (version 0.99), while energy minimization was performed using the CHARMM module (version c31b2) within InsightII (version 2005). The gp120 trimer was built using SymmDock 47, which uses an algorithm for prediction of complexes with Cn symmetry by geometry based docking. Since gp41 is a symmetric trimer in its isolated form and as a complex with gp120, the model of gp120 portion of the HIV-1 envelope glycoprotein was constrained to adopt a threefold-symmetry (n=3 in SymmDock). The residues involved in the trimer interface were those identified earlier by Kwong et al.48, using clustering analysis and solvent accessibility of residues that were conserved across all HIV-1 isolates 49. More specifically, the residues chosen for the constraining criteria were 102, 103, 113, 114, 204, 205, 207, 209, 211, 213, 214, 216, 221, 250, 297, 439, 491. When given the structure of the asymmetric unit of the multimer complex, SymmDock predicts the structure of the entire complex. The resulting trimer is a complex with rotational symmetry of order 3 about a symmetry axis and the rotation angle alpha is 120 degrees. The output of the method is a list of complexes that fulfill the cyclic symmetry constraints. Given these constraints, the SymmDock provided 14 plausible three-dimensional models for the gp120 portion of the trimeric HIV-1 envelope glycoprotein complex. These models were screened on the basis of the following conditions. (i) The surface of gp120 that is occluded in the trimer should maximize carbohydrate exclusion, (ii) be sterically compatible with binding of the 17b and X5 antibodies, and (iii) minimize the exposure of the conserved residues. The individual monomers were rotated 45° in the X-axis, −15° in the Y and Z axes. The models were then minimized using CharmM force fields 50 and visualized using Pymol, and were concordant with other evidence 39.
An allosteric-activation energy model for protein function
Refolding of gp41 is constrained by a gp120-imposed barrier. If gp120 is highly constraining, CCR5 will have to push harder in order to bind and in order to force gp120 to reduce its hold on gp41. Since gp41 refolding requires alleviation of the same allosteric constraints that are counteracted by CCR5 binding, these processes are coupled mechanistically and energetically. Because the result accurately simulates our data and is consistent with the trimer structure of gp120-gp41 complexes, we formulate this model assuming three binding sites for CCR5 in the viral fusion complexes although the derivation could apply to any number. Consistent with previous allosteric models, we also assume that the adsorbed virus associates with CD4 and CCR5 to form an equilibrium mixture of fusion complexes. Although many processes occur on the cell surfaces, we believe that this equilibrium assumption is valid for virus infections in the conditions of our experiments (see Discussion). Based on simple sequential binding equilibria and using microscopic dissociation constants and the binomial theorem as described elsewhere 19, it follows that
| (Eq 1.1) |
| (Eq 1.2) |
| (Eq 1.3) |
where Vi are concentrations of the viral complexes with i CCR5s and k1, k2, and k3 are the microscopic dissociation constants for the first, second, and third CCR5s that associate. For cooperative binding, k1>k2> k3, and we define them as k1=k/f1, k2=k/f2, and k3=k/f3 where fi are fractional terms that show how the allosteric constraints reduce the binding affinities of the CCR5 that is being used and k is the dissociation constant that would occur if the f value were 1.0. Defining α=[CCR5]/k, gives
| (Eq 2.1) |
| (Eq 2.2) |
| (Eq 2.3) |
| (Eq 2.4) |
where Pi is the probability that the fusion complex contains i CCR5s.
According to the transition state theories of Eyring and Kramer 21, the rate constant of a reaction is given by ν= βexp(−ΔG≠/RT), where ΔG≠ is the activation free energy barrier and β is a preexponential factor that differs for the Eyring and Kramer formulations but cancels out in our analyses. As described in the Supplementary information online, for HIV-1 infections in the conditions of our assays, the efficiency of infection of any virus complex is Ei=νιPi/kd, where kd is the uniform rate constant for inactivation of all virus complexes on the HeLa cell surfaces 6. When the concentration of the coreceptor is saturating, Emax=ν3/kd. We measure infectivities (irel values) relative to titers measured in optimally susceptible JC.53 cells that have a saturating excess of wild-type CCR5 17. The infectivity at saturating concentrations of the test coreceptor is . Therefore, by using the irel max values for different virus variants, we can estimate the differences in the activation energy barriers that limit infections of their saturated complexes (see Table 1), and this data supports our hypothesis that adaptation reduces the activation free energy barrier for infection. Therefore,
| (Eq 3) |
where the factors in equation 3 express the fact that the activation energy barrier that limits infection changes as the size of the complexes increase. This derivation relied on the relationships, , etc.
A substantial simplification in this derivation occurs because the fi factors described above derive from free energy costs that occur because the CCR5s have to force themselves into an allosterically constrained gp120 structure. However, because the pathway for refolding of gp41 requires the same changes in gp120 that are forced by CCR5 binding, these processes are energetically coupled. Consequently, the free energy cost for opposing the allosterically constrained gp120 structure in the binding reaction reduces to the same degree the free energy barrier that constrains gp41. Note that the affinity perturbations caused by the allosteric constraints can be written in the form fi=exp(−Δ Gi/RT)/exp(−Δ Gstandard/RT) and that the factors can conversely be written as fractional perturbations of dissociation constants. It follows that , and . Equation 3 can be rewritten as
| (Eq. 4) |
In some assays we varied the concentration of the amino terminal CCR5 peptide (P) and used cells that express constant amounts of CCR5(Δ 18). Accordingly, [CCR5] in the above equations was replaced by [CCR5(Δ 18)][P]. This made α=[CCR5(Δ 18)][P]/k=P/kapp, where kapp=k/[CCR5(Δ 18)] is an apparent dissociation constant of the peptide that is inversely proportional to the CCR5(Δ 18) concentration on the specific test cell clone. The data in Fig 5B was modeled using this modified form of Eq 4 based on the known CCR5(Δ 18) concentrations in the two CCR5(Δ 18)-containing cell clones.
The fi factors in this analysis refer only to constraints that are imposed on the CCR5 binding sites by allosteric interactions between the gp120s and that are alleviated by the wild-type or mutant CCR5 used in the assay. We assume that the free energy differences that distinguish the sequential binding steps result from changes in the allosteric protein rather than the ligand. However, because the constraints imposed on the ligand depend on stoichiometry, the ligands might differ somewhat in these sequential complexes. This would cause small differences between fi and terms in the derivation of equation 4. However, allosteric proteins have evolved to maximize the transfer of these effects between its subunits, and it has been assumed as a first approximation in all allosteric models that the ligands are energetically identical in sequential complexes. Graphical analyses and curve fitting employed Kaleidagraph version 3.6.2 (Synergy Software, Reading, PA). This process optimally fits the curves generated using Eq 4 to the experimental data, thereby obtaining estimates for the factors fi and α(and thus k) indicated in Table 1 legend.
Supplementary Material
Acknowledgments
This research was supported by NIH grant CA67358 to DK. Contributions were as follows: EJP and JPD planned and performed experiments. DK consulted about experiments, developed the activation energy model, and did the graphical analyses. US discussed experiments and performed molecular modeling. All authors helped with writing. We are especially grateful to Eric Barklis, Michael Chapman, Wolfhard Almers, and Meyer Jackson for expert critical reviews.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 2.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–8. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 3.He Y, Vassell R, Zaitseva M, Nguyen N, Yang Z, Weng Y, Weiss CD. Peptides trap the human immunodeficiency virus type 1 envelope glycoprotein fusion intermediate at two sites. Journal of Virology. 2003;77:1666–71. doi: 10.1128/JVI.77.3.1666-1671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melikyan GB, Markosyan RM, Hemmati H, Delmedico MK, Lambert DM, Cohen FS. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J Cell Biol. 2000;151:413–24. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derdeyn CA, Decker JM, Sfakianos JN, Zhang Z, O’Brien WA, Ratner L, Shaw GM, Hunter E. Sensitivity of human immunodeficiency virus type 1 to fusion inhibitors targeted to the gp41 first heptad repeat involves distinct regions of gp41 and Is consistently modulated by gp120 interactions with the coreceptor. Journal of Virology. 2001;75:8605–14. doi: 10.1128/JVI.75.18.8605-8614.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Platt EJ, Durnin JP, Kabat D. Kinetic factors control efficiencies of cell entry, efficacies of entry inhibitors, and mechanisms of adaptation of human immunodeficiency virus. J Virol. 2005;79:4347–56. doi: 10.1128/JVI.79.7.4347-4356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reeves JD, Lee FH, Miamidian JL, Jabara CB, Juntilla MM, Doms RW. Enfuvirtide resistance mutations: impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. Journal of Virology. 2005;79:4991–9. doi: 10.1128/JVI.79.8.4991-4999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madani N, Hubicki AM, Perdigoto AL, Springer M, Sodroski J. Inhibition of human immunodeficiency virus envelope glycoprotein- mediated single cell lysis by low-molecular-weight antagonists of viral entry. J Virol. 2007;81:532–8. doi: 10.1128/JVI.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhmann SE, Pugach P, Kunstman KJ, Taylor J, Stanfield RL, Snyder A, Strizki JM, Riley J, Baroudy BM, Wilson IA, Korber BT, Wolinsky SM, Moore JP. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. Journal of Virology. 2004;78:2790–807. doi: 10.1128/JVI.78.6.2790-2807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marozsan AJ, Kuhmann SE, Morgan T, Herrera C, Rivera-Troche E, Xu S, Baroudy BM, Strizki J, Moore JP. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D) Virology. 2005;338:182–99. doi: 10.1016/j.virol.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 11.Hartley O, Klasse PJ, Sattentau QJ, Moore JP. V3: HIV’s switch-hitter. AIDS Research and Human Retroviruses. 2005;21:171–89. doi: 10.1089/aid.2005.21.171. [DOI] [PubMed] [Google Scholar]
- 12.Platt EJ, Shea DM, Rose PP, Kabat D. Variants of human immunodeficiency virus type 1 that efficiently use CCR5 lacking the tyrosine-sulfated amino terminus have adaptive mutations in gp120, including loss of a functional N-glycan. J Virol. 2005;79:4357–68. doi: 10.1128/JVI.79.7.4357-4368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farzan M, Chung S, Li W, Vasilieva N, Wright PL, Schnitzler CE, Marchione RJ, Gerard C, Gerard NP, Sodroski J, Choe H. Tyrosine-sulfated peptides functionally reconstitute a CCR5 variant lacking a critical amino-terminal region. Journal of Biological Chemistry. 2002;277:40397–402. doi: 10.1074/jbc.M206784200. [DOI] [PubMed] [Google Scholar]
- 14.Siciliano SJ, Kuhmann SE, Weng Y, Madani N, Springer MS, Lineberger JE, Danzeisen R, Miller MD, Kavanaugh MP, DeMartino JA, Kabat D. A critical site in the core of the CCR5 chemokine receptor required for binding and infectivity of human immunodeficiency virus type 1. J Biol Chem. 1999;274:1905–13. doi: 10.1074/jbc.274.4.1905. [DOI] [PubMed] [Google Scholar]
- 15.Platt EJ, Kuhmann SE, Rose PP, Kabat D. Adaptive mutations in the V3 loop of gp120 enhance fusogenicity of human immunodeficiency virus type 1 and enable use of a CCR5 coreceptor that lacks the amino-terminal sulfated region. J Virol. 2001;75:12266–78. doi: 10.1128/JVI.75.24.12266-12278.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhmann SE, Platt EJ, Kozak SL, Kabat D. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J Virol. 2000;74:7005–15. doi: 10.1128/jvi.74.15.7005-7015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–64. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chernomordik LV, Kozlov MM. Protein-lipid interplay in fusion and fission of biological membranes. Annu Rev Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- 19.Cantor CRaSPR. Biophysical Chemistry, Part III: The Behavior of Biological Macromolecules. Vol. 3. W.H. Freeman and Company; New York: 1998. [Google Scholar]
- 20.Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308:1424–8. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 21.Jackson M. Molecular and Cellular Biophysics. Cambridge University Press; New York: 2006. [Google Scholar]
- 22.Koshland DE, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 23.Monod J, Wyman J, Changeux JP. On the Nature of Allosteric Transitions: A Plausible Model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 24.Raviv Y, Viard M, Bess J, Jr, Blumenthal R. Quantitative measurement of fusion of HIV-1 and SIV with cultured cells using photosensitized labeling. Virology. 2002;293:243–51. doi: 10.1006/viro.2001.1237. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Kurteva S, Ren X, Lee S, Sodroski J. Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. J Virol. 2005;79:12132–47. doi: 10.1128/JVI.79.19.12132-12147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seibert C, Ying W, Gavrilov S, Tsamis F, Kuhmann SE, Palani A, Tagat JR, Clader JW, McCombie SW, Baroudy BM, Smith SO, Dragic T, Moore JP, Sakmar TP. Interaction of small molecule inhibitors of HIV-1 entry with CCR5. Virology. 2006;349:41–54. doi: 10.1016/j.virol.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 27.Watson C, Jenkinson S, Kazmierski W, Kenakin T. The CCR5 receptor-based mechanism of action of 873140, a potent allosteric noncompetitive HIV entry inhibitor. Mol Pharmacol. 2005;67:1268–82. doi: 10.1124/mol.104.008565. [DOI] [PubMed] [Google Scholar]
- 28.Reeves JD, Gallo SA, Ahmad N, Miamidian JL, Harvey PE, Sharron M, Pohlmann S, Sfakianos JN, Derdeyn CA, Blumenthal R, Hunter E, Doms RW. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc Natl Acad Sci U S A. 2002;99:16249–54. doi: 10.1073/pnas.252469399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annual Review of Biochemistry. 2000;69:531–69. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 30.Bentz J, Mittal A. Architecture of the influenza hemagglutinin membrane fusion site. Biochim Biophys Acta. 2003;1614:24–35. doi: 10.1016/s0005-2736(03)00160-3. [DOI] [PubMed] [Google Scholar]
- 31.Danieli T, Pelletier SL, Henis YI, White JM. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. Journal of Cell Biology. 1996;133:559–69. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gunther-Ausborn S, Schoen P, Bartoldus I, Wilschut J, Stegmann T. Role of hemagglutinin surface density in the initial stages of influenza virus fusion: lack of evidence for cooperativity. J Virol. 2000;74:2714–20. doi: 10.1128/jvi.74.6.2714-2720.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imai M, Mizuno T, Kawasaki K. Membrane fusion by single influenza hemagglutinin trimers. Kinetic evidence from image analysis of hemagglutinin-reconstituted vesicles. J Biol Chem. 2006;281:12729–35. doi: 10.1074/jbc.M600902200. [DOI] [PubMed] [Google Scholar]
- 34.Mittal A, Bentz J. Comprehensive kinetic analysis of influenza hemagglutinin-mediated membrane fusion: role of sialate binding. Biophys J. 2001;81:1521–35. doi: 10.1016/S0006-3495(01)75806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhukovsky MA, Leikina E, Markovic I, Bailey AL, Chernomordik LV. Heterogeneity of early intermediates in cell-liposome fusion mediated by influenza hemagglutinin. Biophys J. 2006;91:3349–58. doi: 10.1529/biophysj.106.088005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mittal A, Leikina E, Chernomordik LV, Bentz J. Kinetically differentiating influenza hemagglutinin fusion and hemifusion machines. Biophys J. 2003;85:1713–24. doi: 10.1016/S0006-3495(03)74601-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roche S, Gaudin Y. Evidence that rabies virus forms different kinds of fusion machines with different pH thresholds for fusion. J Virol. 2004;78:8746–52. doi: 10.1128/JVI.78.16.8746-8752.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markovic I, Leikina E, Zhukovsky M, Zimmerberg J, Chernomordik LV. Synchronized activation and refolding of influenza hemagglutinin in multimeric fusion machines. J Cell Biol. 2001;155:833–44. doi: 10.1083/jcb.200103005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu P, Liu J, Bess J, Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–52. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 40.Chelli M, Alizon M. Rescue of HIV-1 receptor function through cooperation between different forms of the CCR5 chemokine receptor. Journal of Biological Chemistry. 2002;277:39388–96. doi: 10.1074/jbc.M205394200. [DOI] [PubMed] [Google Scholar]
- 41.Repits J, Oberg M, Esbjornsson J, Medstrand P, Karlsson A, Albert J, Fenyo EM, Jansson M. Selection of human immunodeficiency virus type 1 R5 variants with augmented replicative capacity and reduced sensitivity to entry inhibitors during severe immunodeficiency. J Gen Virol. 2005;86:2859–69. doi: 10.1099/vir.0.81111-0. [DOI] [PubMed] [Google Scholar]
- 42.Olivieri K, Scoggins RM, Bor YC, Matthews A, Mark D, Taylor JR, Jr, Chernauskas D, Hammarskjold ML, Rekosh D, Camerini D. The envelope gene is a cytopathic determinant of CCR5 tropic HIV-1. Virology. 2007;358:23–38. doi: 10.1016/j.virol.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 43.Platt EJ, Madani N, Kozak SL, Kabat D. Infectious properties of human immunodeficiency virus type 1 mutants with distinct affinities for the CD4 receptor. J Virol. 1997;71:883–90. doi: 10.1128/jvi.71.2.883-890.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuhmann SE, Platt EJ, Kozak SL, Kabat D. Polymorphisms in the CCR5 genes of African green monkeys and mice implicate specific amino acids in infections by simian and human immunodeficiency viruses. J Virol. 1997;71:8642–56. doi: 10.1128/jvi.71.11.8642-8656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chesebro B, Wehrly K. Development of a sensitive quantitative focal assay for human immunodeficiency virus infectivity. J Virol. 1988;62:3779–88. doi: 10.1128/jvi.62.10.3779-3788.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baba M, Nishimura O, Kanzaki N, Okamoto M, Sawada H, Iizawa Y, Shiraishi M, Aramaki Y, Okonogi K, Ogawa Y, Meguro K, Fujino M. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc Natl Acad Sci U S A. 1999;96:5698–703. doi: 10.1073/pnas.96.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–7. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwong PD, Wyatt R, Sattentau QJ, Sodroski J, Hendrickson WA. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. Journal of Virology. 2000;74:1961–72. doi: 10.1128/jvi.74.4.1961-1972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–59. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feliciangeli SF, Thomas L, Scott GK, Subbian E, Hung CH, Molloy SS, Jean F, Shinde U, Thomas G. Identification of a pH sensor in the furin propeptide that regulates enzyme activation. J Biol Chem. 2006;281:16108–16. doi: 10.1074/jbc.M600760200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.