Abstract
CXXC finger protein 1 (Cfp1) is a regulator of both cytosine methylation and histone methylation. Murine embryonic stem (ES) cells lacking Cfp1 exhibit a decreased plating efficiency, decreased cytosine methylation, elevated global levels of histone H3-Lys4 trimethylation, and a failure to differentiate in vitro. Remarkably, transfection studies reveal that expression of either the amino half of Cfp1 (amino acids 1 to 367 [Cfp11-367]) or the carboxyl half of Cfp1 (Cfp1361-656) is sufficient to correct all of the defects observed with ES cells that lack Cfp1. However, a point mutation (C169A) that abolishes DNA-binding activity of Cfp1 ablates the rescue activity of the Cfp11-367 fragment, and a point mutation (C375A) that abolishes the interaction of Cfp1 with the Setd1 histone H3-Lys4 methyltransferase complexes ablates the rescue activity of the Cfp1361-656 fragment. Introduction of both the C169A and C375A point mutations ablates the rescue activity of the full-length Cfp1 protein. These results indicate that retention of either the Cfp1 DNA-binding domain or Setd1 interaction domain is required for Cfp1 rescue activity, and they illustrate the functional complexity of this critical epigenetic regulator. A model is presented for how epigenetic cross talk may explain the finding of redundant functional domains within Cfp1.
Epigenetics refers to heritable patterns of gene expression that occur without a change in the nucleotide sequence of DNA. Epigenetic modifications include DNA methylation and covalent modification of histone proteins (8, 20). Histones are subject to a variety of covalent modifications, including acetylation, phosphorylation, methylation, ubiquitination, sumoylation, and ADP-ribosylation (41). DNA methylation involves the addition of a methyl group to the carbon-5 position of the cytosine ring and is correlated with heterochromatin and gene repression (38, 50). Cytosine methylation plays essential roles in development, gametogenesis, X chromosome inactivation, genomic imprinting, and genome stability (3, 28), and dynamic changes in DNA methylation and histone modifications are necessary for in vitro differentiation of embryonic stem (ES) cells (18, 22, 45).
Cytosine methylation is catalyzed by DNA methyltransferase (Dnmt) enzymes in the context of CpG dinucleotides in mammalian cells (50). The maintenance methyltransferase Dnmt1 exhibits a preference for hemimethylated substrates and is primarily responsible for copying cytosine methylation patterns from the parental to daughter DNA molecules following semiconservative DNA replication (3, 36). In contrast, Dnmt3A and Dnmt3B are primarily responsible for the establishment of de novo cytosine methylation patterns during early embryonic development (3, 36, 63).
Histone methylation is carried out by a family of histone methyltransferases, many of which contain a catalytic SET [Su(var)3-9, Enhancer of Zeste, trithorax] domain (13). Methylation of histone H3 at Lys4 (H3-Lys4) is associated with transcriptionally active euchromatin. Saccharomyces cerevisiae expresses only a single H3-Lys4 methyltransferase, referred to as Set1, which associates with a complex known as COMPASS (complex proteins associated with Set1) (34). In contrast, mammalian cells contain numerous histone methyltransferases that exhibit specificity for histone H3-Lys4, including Setd1A, Setd1B, Mll1, Mll2, Mll3, Mll4, Smyd1, Smyd2, Smyd3, and Set7/9 (13, 15, 23, 25).
CXXC finger protein 1 (Cfp1), encoded by the CXXC1 gene, is an epigenetic regulator of both cytosine and histone methylation that physically interacts with both Dnmt1 (7) and with the Setd1 histone H3-Lys4 methyltransferase complexes (23, 25). Cfp1 contains several conserved protein domains, including two plant homeodomains (PHD). PHD finger domains are found in several dozen proteins involved in chromatin-mediated transcriptional control (17) and may be involved in the recognition of differentially modified histone tails (1, 44, 49). Cfp1 contains a cysteine-rich CXXC DNA-binding domain that exhibits binding affinity to DNA sequences containing unmethylated CpG dinucleotides (26, 59). Cfp1 also contains acidic, basic, and coiled-coil domains and a Set1 interaction domain (SID) that is required for interaction with the Setd1A and Setd1B histone H3-Lys4 methyltransferase complexes (7, 26, 59).
Cfp1 is crucial for vertebrate development, and disruption of the murine CXXC1 gene results in early embryonic lethality. Embryos that lack the CXXC1 gene (CXXC1−/−) fail to gastrulate and exhibit peri-implantation lethality around 4.5 to 6.5 days postcoitus (9). Depletion of Cfp1 in zebrafish embryos results in decreased global cytosine methylation and a failure of primitive hematopoiesis (64), and depletion of Cfp1 in the PLB-985 human myeloid cell line results in reduced survival and impaired myeloid cell differentiation (65). CXXC1−/− murine ES cells exhibit a variety of defects, including a ∼70% decrease in global cytosine methylation and an increase in histone H3-Lys4 methylation, which are indicative of reduced levels of heterochromatin. ES cells lacking Cfp1 also suffer from an inability to achieve in vitro differentiation (9). Reduced cytosine methylation in CXXC1−/− ES cells is a consequence of impaired maintenance cytosine methylation. In contrast, de novo cytosine methylation is normal in these cells (9). These data demonstrate that Cfp1 plays an important role in the regulation of cytosine and histone methylation and cellular differentiation. Importantly, the defects observed in CXXC1−/− ES cells are corrected upon introduction of a full-length Cfp1 expression vector, indicating that the CXXC1−/− ES cell phenotype is specifically due to the absence of Cfp1 and that epigenetic perturbations that occur in the absence of Cfp1 are reversible (9).
The ability of wild-type Cfp1 to rescue the defects observed in CXXC1−/− ES cells provides a convenient method to assess structure-function relationships of Cfp1. The purpose of this study was to determine the functional importance of various Cfp1 domains and properties. Unexpectedly, these studies reveal that expression of either the amino half of Cfp1 (amino acids [aa] 1 to 367 [Cfp11-367]) or the carboxyl half of Cfp1 (Cfp1361-656) is sufficient to correct the defects observed in CXXC1−/− ES cells and that Cfp1 contains redundant functional domains. Additional studies revealed that retention of either the DNA-binding domain or SID is required for Cfp1 function.
MATERIALS AND METHODS
Cell culture.
HEK-293 (human embryonic kidney) cells were grown in Dulbecco's modified Eagle's medium (Gibco BRL Life Technologies, Grand Island, NY) supplemented with 10% fetal calf serum (HyClone, Logan, UT). Isolation of CXXC1−/− murine ES cell lines was previously described (9). ES cells were cultured on 0.1% gelatin-coated tissue culture dishes in high-glucose Dulbecco's modified Eagle's medium (Gibco BRL Life Technologies) supplemented with 20% fetal bovine serum (Gibco BRL), 100 units/ml penicillin-streptomycin (Invitrogen, Carlsbad, CA), 2 mM l-glutamine (Invitrogen), 1% nonessential amino acids (Invitrogen), 0.2% leukemia inhibitory factor (LIF)-conditioned medium, 100 nM β-mercaptoethanol, 0.025% HEPES, pH 7.5 (Invitrogen), and 1% Hank's balanced salt solution (Invitrogen). In vitro differentiation of ES cells was performed using the hanging drop method in the absence of LIF, as previously described (9). Alkaline phosphatase activity was histochemically detected using an alkaline phosphatase leukocyte detection kit (Sigma-Aldrich) as previously described (9). To perform colony forming assays, 400 ES cells were plated, and colonies were allowed to form for 7 to 9 days; they were then fixed in methanol-acetic acid (3:1) and stained with 0.02% crystal violet and counted.
Plasmid construction and transfection of ES cells.
cDNA constructs which encode full-length FLAG epitope-tagged human Cfp1 (aa 1 to 656) or various Cfp1 fragments and mutations were subcloned into the pcDNA3.1/Hygro mammalian expression vector (Invitrogen). The amino-terminal bipartite nuclear localization signal of Cfp1 (aa 109 to 121) was inserted between the FLAG epitope and Cfp1 sequence for constructs containing Cfp1361-656 or Cfp1361-656 C375A. Amino acid substitutions within Cfp1 were carried out using a QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) per the manufacturer's protocol using primers as previously described (7, 26).
Twenty-five micrograms of each plasmid was linearized and electroporated into CXXC1−/− ES cells. Hygromycin B-containing medium was added after 24 h and replaced every other day for approximately 2 weeks before single colonies were isolated for expansion. For experiments, cells were grown in ES culture medium containing 50 μg/ml hygromycin B. Expression of FLAG-tagged Cfp1 fragments was verified by Western blot analysis using anti-FLAG (Sigma- Aldrich) or anti-Cfp1 (25) antibodies. For each construct, several independent clones that expressed FLAG-Cfp1 at a level exceeding 50% of the wild-type Cfp1 levels were selected for analysis.
Production of His6-tagged proteins and electrophoretic mobility shift assays (EMSAs).
Production of His6-tagged Cfp1 protein fragment was performed as previously described (26). Briefly, isopropyl-β-d-thiogalactopyranoside-induced Escherichia coli cells were sonicated on ice for 10 min. Following centrifugation at 15,000 rpm for 30 min, supernatants were loaded onto a His-Trap purification column (Amersham Pharmacia Biotech, GE Healthcare, United Kingdom) and eluted with 500 mM imidazole. For Western blot analysis, 0.5 μg of partially purified protein was subjected to 10 to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and proteins were transferred onto a nitrocellulose membrane (Amersham, GE Healthcare). The membrane was then incubated with anti-His6 tag monoclonal antibody (R&D Systems, Minneapolis, MN) followed by horseradish peroxidase-labeled anti-mouse antiserum and detected using an ECL detection kit (Amersham, GE Healthcare).
An EMSA was performed as previously described (26, 51), using 0.01 to 0.1 μg of partially purified Cfp1 protein fragment and an end-labeled double-stranded oligonucleotide probe containing a CpG dinucleotide (5′-CTATGCTTCTTCTTCCGGTGAGGAAATGAAAACAGCAG-3′).
Immunoprecipitation.
Nuclear extracts of HEK-293 cells expressing FLAG-Cfp1 proteins were prepared as previously described (23). Nuclear extracts were incubated with FLAG-immunoglobulin G-agarose slurry (Sigma-Aldrich) for 4 h and washed five times with extraction buffer containing 300 mM NaCl. Recovered proteins were analyzed by Western blotting as described below.
Analysis of cytosine methylation.
Global cytosine methylation was assessed utilizing a methyl acceptance assay as previously described (2, 9, 64). Briefly, 500 ng of genomic DNA was incubated with 2 μCi of S-adenosyl-l-[methyl- 3H]methionine (15 Ci/mmol; Amersham, GE Healthcare), and 3 units of SssI (CpG) methylase (New England Biolabs, Ipswich, MA) for 1 h at 30°C. In vitro methylated DNA was isolated by filtration through Whatman DE-81 ion exchange filters (Fisher Scientific, Pittsburgh, PA), and the incorporated radioactivity was measured by scintillation counting. Three independent methylation experiments were performed in duplicate for each ES clone analyzed.
To analyze cytosine methylation at intracisternal A particle (IAP) retroviral repeats, genomic DNA was digested with MspI or the methyl-sensitive isoschizomer HpaII (New England Biolabs), and Southern blot analysis was performed, as previously described (9). Membranes were probed with a 32P-labeled IAP repetitive element probe, generously provided by En Li (Novartis Institutes for Biomedical Research, Cambridge, MA).
Histone protein preparation.
Histones were isolated using a histone extraction protocol (Abcam Inc., Cambridge, MA). Briefly, ES cells were collected and resuspended in Triton extraction buffer ([TEB] phosphate-buffered saline containing 0.5% Triton X-100, 2 mM phenylmethylsulfonyl fluoride, 0.02% NaN3) on ice for 10 min with gentle agitation. After centrifugation, pellets were briefly washed with TEB and resuspended in 0.2 N hydrochloric acid (HCl), and histones were extracted overnight at 4°C. Acid-insoluble proteins were removed by centrifugation, and supernatant containing histones was collected. Histones were precipitated by adding 8 volumes of acetone and incubating the mixture at −20°C overnight. The histone pellets were then washed twice with acetone-100 mM HCl (10:1) and three times with acetone. Pellets were dried and resuspended in distilled water.
Western blot analysis.
Protein expression of Cfp1 and FLAG-tagged Cfp1 fragments was assessed following isolation of whole-cell protein extracts as previously described (23). Dnmt1 protein expression was assessed following isolation of nuclear extracts as previously described (23). Protein concentration was quantified using the Bradford method (6). In order to separate proteins, 60 μg of whole-cell protein or 100 μg of nuclear extract was subjected to electrophoresis on 7% (Cfp1), 4 to 12% (Dnmt1), or 12% (total histone H3 and trimethylated H3-Lys4) PAGEr-Gold precast Tris-glycine gradient gels (Lonza Group, Ltd., Switzerland) and transferred to nitrocellulose membranes (Amersham, GE Healthcare). The membranes were probed with primary antiserum for 1 h at room temperature or overnight at 4°C, washed, and incubated with an appropriate horseradish peroxidase-labeled secondary antibody diluted for 1 h. Membranes were washed, and signal was detected with an ECL kit (Amersham, GE Healthcare) and quantified by densitometry (Image J, NIH). The following antisera were used for immunoblotting: rabbit polyclonal Cfp1 (25); mouse monoclonal β-actin and mouse monoclonal FLAG (Sigma-Aldrich); rabbit polyclonal Dnmt1 (7); rabbit polyclonal trimethylated H3-Lys4 and total histone H3 (Abcam, Cambridge, MA); rabbit polyclonal Setd1A, Setd1B, Wdr82, and Wdr5 (25); or rabbit polyclonal Ash2 or Rbbp5 (Bethyl Laboratories, Montgomery, TX).
RT-PCR analysis.
Developmentally and lineage-specific mRNA in ES cells was detected by reverse transcription-PCR (RT-PCR) as previously described (23). The following primer pairs were used: Hprt, 5′-CACAGGACTAGAACACCTGC-3′ and 5′-GCTGGTGAAAAGGACCTCT-3′ (21); Brachyury, 5′-ATCAAGGAAGGCTTTAGCAAATGGG-3′ and 5′-GAACCTCGGATTCACATCGTGAGA-3′ (62); βH1, 5′-AGTCCCCATGGAGTCAAAGA-3′ and 5′-CTCAAGGAGACCTTTGCTCA-3′; c-fms, 5′-CTGAGTCAGAAGCCCTTCGACAAAG-3′ and 5′-CTTTGCCCCAGACCAAAGGCTGTAGC-3′ (33); gp-IIB, 5′-AGGCAGAGAAGACTCCGGTA-3′ and 5′-TACCGAATATCCCCGGTAAC-3′ (57); Gata-4, 5′-CACTATGGGCACAGCACAGCAGCTGG-3′ and 5′-TTGGAGCTGGCCTGCGATGTC-3′ (62); and Oct4, 5′-GGCGTTCTCTTTGGAAAGGTGTTC-3′ and 5′-CTCGAACCACATCCTTCTCT-3′ (generously provided by Rebecca Chan, Indiana University). The ratio of Oct4 to Hprt expression was determined by quantifying band intensities using densitometry.
Statistical analysis.
Statistical significance was assessed by a two-tailed Student t test with equal variance, compared to the data for CXXC1−/− ES cells expressing the vector control or full-length Cfp1 (Cfp11-656). A P value < 0.05 was interpreted as statistical significance.
RESULTS
Decreased plating efficiency of CXXC1−/− ES cells is rescued by redundant Cfp1 domains.
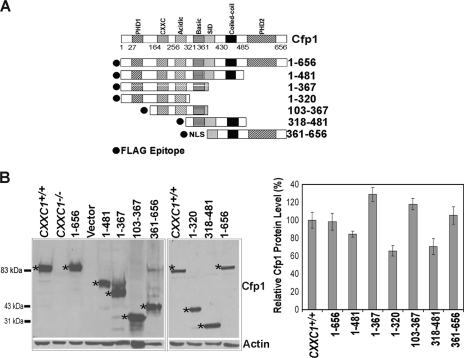
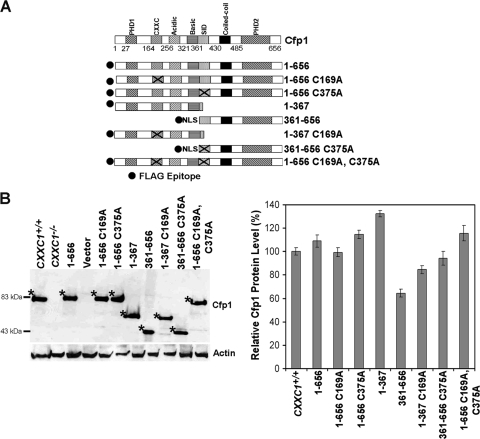
Various cDNA constructs encoding FLAG-tagged Cfp1 fragments (Fig. 1A) were electroporated into CXXC1−/− ES cells. Stably transfected clones were selected, and clones were screened for FLAG-Cfp1 protein expression by Western blot analysis using anti-Cfp1 antiserum (Fig. 1B). ES cells heterozygous for the disrupted CXXC1 allele (CXXC1+/−) express ∼50% of Cfp1 protein compared to wild-type (CXXC1+/+) ES cells and do not exhibit a phenotype (9). Consequently, clones that expressed at least 50% of the level of Cfp1 observed in CXXC1+/+ ES cells were selected for analysis (Fig. 1B).
FIG. 1.
Isolation of CXXC1−/− ES cell clones expressing Cfp1 fragments. (A) A schematic representation of each Cfp1 construct is shown. The filled circle at the N terminus of each construct represents the FLAG epitope, and NLS denotes a nuclear localization signal (24). (B) The level of Cfp1 expression in CXXC1+/+, CXXC1−/−, and CXXC1−/− ES cells expressing either full-length Cfp1 (1-656) or the indicated Cfp1 truncations (or carrying the empty expression vector) was assessed by Western analysis using antiserum directed against Cfp1 (25). A representative blot is presented. Asterisks denote the position of Cfp1 and each FLAG-Cfp1 protein. The blot was reprobed for β-actin as a loading control. The graph summarizes the results from at least three independent experiments, and error bars represent standard error.
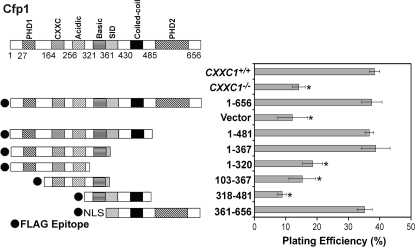
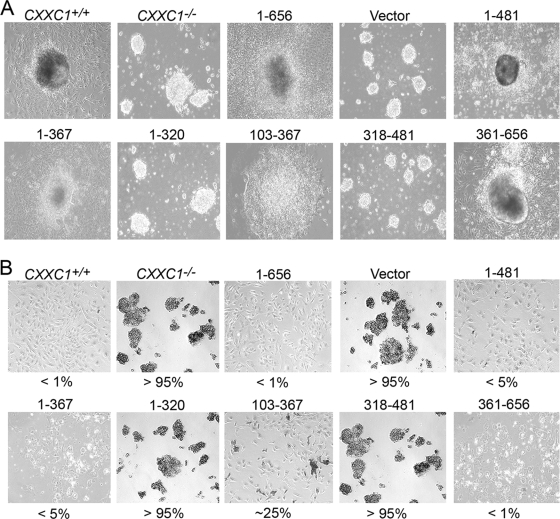
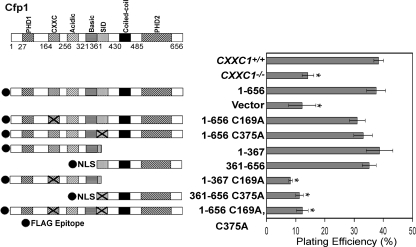
Plating efficiency is the percentage of seeded cells that give rise to cell colonies. Plating efficiency is approximately 38% for CXXC1+/+ ES cells and is significantly decreased (∼14%) in CXXC1−/− ES cells (Fig. 2). Thus, fewer ES cells survive and form a colony in the absence of Cfp1. This reduction in plating efficiency is rescued upon expression of full-length Cfp1 (Cfp11-656), indicating that decreased plating efficiency is specifically due to loss of Cfp1. CXXC1−/− ES cells expressing Cfp11-481 (containing the PHD1, CXXC, acidic, basic, and coiled-coil domains) exhibit a normal plating efficiency (Fig. 2), demonstrating that the PHD2 domain is not necessary for the rescue of plating efficiency. Surprisingly, expression of either the amino half of Cfp1 (Cfp11-367 containing the PHD1, CXXC, acidic, and basic domains) or the carboxyl half of Cfp1 (Cfp1361-656 containing the coiled-coil, SID, and PHD2 domains) is sufficient to rescue the plating efficiency defect observed in CXXC1−/− ES cells. However, removal of the basic domain or PHD1 domain from the N-terminal half of Cfp1 (Cfp11-320 or Cfp1103-367, respectively) or removal of the PHD2 domain from the C-terminal half of Cfp1 (Cfp1318-481) results in a failure to rescue the defect in plating efficiency. Thus, no single domain of Cfp1 is necessary to rescue the plating efficiency defect observed in CXXC1−/− ES cells.
FIG. 2.
Cfp1 contains redundant functional domains sufficient to rescue the deficiency in plating efficiency observed in CXXC1−/− ES cells. The graph summarizes the plating efficiency of CXXC1+/+, CXXC1−/−, and CXXC1−/− ES cells expressing full-length Cfp1 (1-656) or the indicated Cfp1 fragments (or carrying the empty expression vector). Plating efficiency was determined by counting the number of colonies formed after plating 400 cells. The graph summarizes data from at least three independent experiments performed in triplicate, and error bars represent standard error. Asterisks denote statistically significant (P < 0.05) differences compared to CXXC1−/− ES cells expressing Cfp11-656.
DNA-binding activity of Cfp1 is not required for appropriate cytosine methylation in ES cells.
ES cells lacking Cfp1 exhibit a ∼70% decrease in global and site-specific cytosine methylation (9). The cysteine-rich CXXC domain functions as the sole DNA-binding domain of Cfp1 (26). S. cerevisiae, Schizosaccharomyces pombe, and Caenorhabditis elegans lack cytosine methylation, and their respective Cfp1 homologues lack the CXXC domain. Therefore, the CXXC domain is expected to play a crucial role in the regulation of cytosine methylation. Consequently, experiments were performed to determine if the DNA-binding activity of Cfp1 plays an essential role in the regulation of cytosine methylation. Global cytosine methylation was analyzed using a DNA methyl acceptance assay. This assay utilizes an in vitro methylation reaction to quantify transfer of radiolabeled methyl groups from S-adenosyl-l-methionine onto genomic DNA sites that are not methylated (2). Therefore, the number of radiolabeled methyl groups incorporated into a DNA sample is inversely related to the native cytosine methylation status.
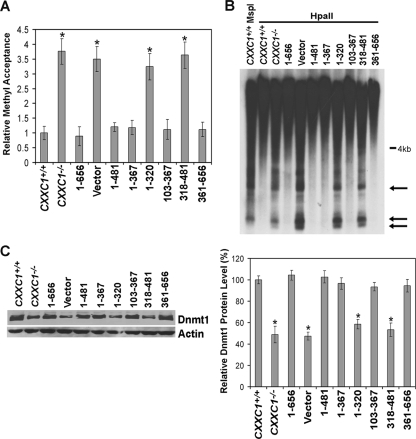
Consistent with a previous report (9), DNA isolated from CXXC1−/− ES cells (or ES cells carrying the empty expression vector) accepts approximately 3.5-fold more methyl groups than DNA derived from CXXC1+/+ ES cells, indicating a ∼70% decrease in global cytosine methylation in cells lacking Cfp1 (Fig. 3A). The reduced cytosine methylation observed in CXXC1−/− ES cells is rescued by expression of Cfp11-656 (the full-length protein) (Fig. 3A). Consistent with the rescue of plating efficiency, expression of Cfp11-481 in CXXC1−/− ES cells also rescues global cytosine methylation. In addition, expression of either the amino half of Cfp1 (Cfp11-367) or the carboxyl half of Cfp1 (Cfp1361-656) is sufficient to rescue the cytosine methylation defect (Fig. 3A). Therefore, Cfp1 DNA-binding activity is not essential to maintain appropriate cytosine methylation levels because the CXXC DNA-binding domain is absent from the Cfp1361-656 fragment. The basic domain is indispensable for rescue activity of Cfp11-367 because removal of the basic domain from the N-terminal Cfp11-320 fragment results in failure to rescue global cytosine methylation (Fig. 3A). In contrast to its inability to rescue plating efficiency, Cfp1103-367 does rescue global cytosine methylation, indicating that the PHD1 domain is not essential for appropriate cytosine methylation. Lastly, CXXC1−/− ES cells expressing Cfp1318-481 exhibit decreased cytosine methylation, indicating that the PHD2 domain is required for cytosine methylation rescue activity of the C-terminal Cfp1 fragment.
FIG. 3.
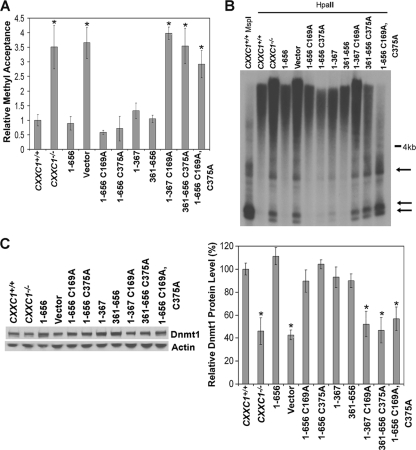
Redundant functional domains of Cfp1 are sufficient to rescue reduced cytosine methylation in CXXC1−/− ES cells. A. Global cytosine methylation levels were determined by methyl acceptance assay for genomic DNA isolated from CXXC1+/+, CXXC1−/−, and CXXC1−/− ES cells expressing full-length Cfp1 (1-656) or the indicated Cfp1 fragments (or carrying the empty expression vector). The graph represents the results from three independent experiments, and error bars represent standard error. Asterisks denote statistically significant (P < 0.05) differences compared to CXXC1−/− ES cells expressing Cfp11-656. (B) Genomic DNA was isolated from the indicated ES cell lines and digested with MspI or HpaII, and Southern blot analysis was performed using a radiolabeled probe specific for IAP retrovirus repetitive elements. Arrows indicate bands that demonstrate cytosine hypomethylation. (C) Western blot analysis was performed on protein extracts collected from the indicated ES cell lines, using antiserum directed against Dnmt1 (7). The blot was reprobed for β-actin as a loading control. The graph summarizes the results from at least three independent experiments, and error bars represent standard error. Asterisks denote statistically significant (P < 0.05) differences compared to CXXC1−/− ES cells expressing Cfp11-656.
Southern blot analysis was used to analyze cytosine methylation specifically at IAP retroviral repeats (Fig. 3B). IAP repeats are endogenous retroviral sequences present in multiple copies in the mouse genome and normally demonstrate a high level of cytosine methylation (43). The methylation status of the IAP repeats was investigated by Southern blot analysis following digestion of genomic DNA with the restriction enzyme MspI or its cytosine methylation-sensitive isoschizomer HpaII. Consistent with the rescue of global cytosine methylation (Fig. 3A), IAP fragments smaller than 4 kb were not observed following HpaII digestion of DNA isolated from CXXC1+/+ ES cells and CXXC1−/− ES cells expressing Cfp11-656, Cfp11-481, Cfp11-367, Cfp1103-367, or Cfp1361-656, indicating a high degree of cytosine methylation present at IAP repeats. In contrast, low-molecular-weight fragments were observed for DNA isolated from CXXC1−/− ES cells and CXXC1−/− ES cells expressing Cfp11-320 and Cfp1318-481(or carrying the empty vector), indicating reduced cytosine methylation at IAP repeats. Thus, either the amino (aa 1 to 367) or carboxyl (aa 361 to 656) fragment of Cfp1 is sufficient to rescue both global and site-specific cytosine methylation.
ES cells lacking Cfp1 exhibit a ∼50% decrease in Dnmt1 protein expression and maintenance DNA methyltransferase activity but demonstrate normal de novo DNA methyltransferase activity (9). Western blot analysis was performed to determine the expression level of Dnmt1 protein in CXXC1−/− ES cells expressing various Cfp1 fragments (Fig. 3C). CXXC1−/− ES cells expressing Cfp11-656, Cfp11-481, Cfp11-367, Cfp1103-367, or Cfp1361-656 express Dnmt1 protein levels similar to the level in CXXC1+/+ ES cells. In contrast, a significant decrease in Dnmt1 protein expression was observed in CXXC1−/− ES cells and CXXC1−/− ES cells expressing Cfp11-320 or Cfp1318-481 (or carrying the empty vector). These data reveal a strong correlation between rescue of Dnmt1 protein levels and restoration of normal levels of cytosine methylation.
Cfp1 contains redundant functional domains that are sufficient to restore appropriate histone methylation.
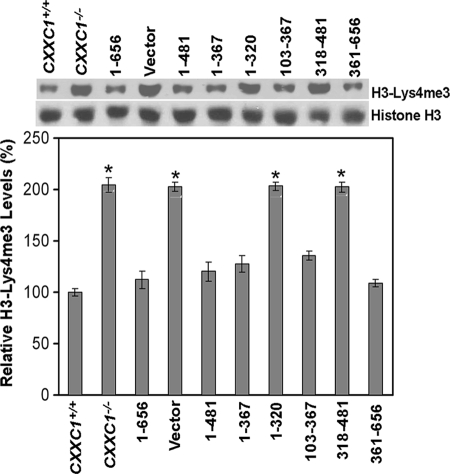
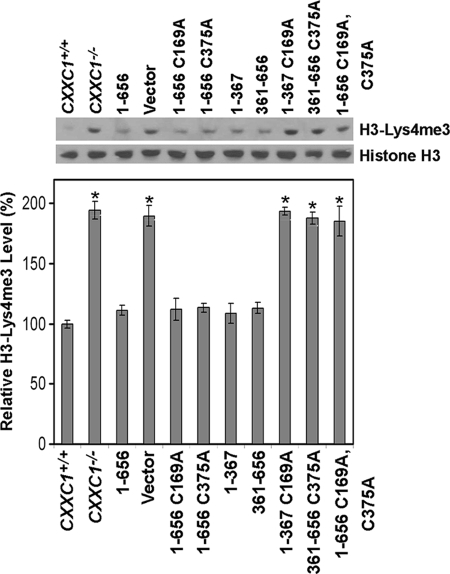
Global levels of histone H3-Lys4 methylation transiently decline once ES cells start to differentiate (22). However, following induction of in vitro differentiation, CXXC1−/− ES cells exhibit an inappropriate increase in histone H3-Lys4 trimethylation 6 days following the removal of LIF from the medium (23). Consequently, Cfp1 is postulated to restrict the activity of the Setd1A and Setd1B histone H3-Lys4 methyltransferase complexes. ES cells were cultured under conditions to promote differentiation, and histones were isolated after 6 days to assess the global levels of histone H3-Lys4 trimethylation. The level of histone H3-Lys4 trimethylation is significantly elevated (approximately twofold) in CXXC1−/− ES cells and cells carrying the empty expression vector (Fig. 4). However, global levels of histone H3-Lys4 trimethylation are reduced to normal levels in CXXC1−/− ES cells expressing Cfp11-656. As with the rescue of other aspects of the Cfp1-deficient phenotype, histone H3-Lys4 trimethylation is rescued upon expression of either the amino half of Cfp1 (Cfp11-367) or carboxyl half of Cfp1 (Cfp1361-656) (Fig. 4). Also, expression of Cfp11-481 or Cfp1103-367 restores normal histone H3-Lys4 trimethylation, indicating that the PHD1 and PHD2 domains are not required. However, the basic domain is required for histone methylation rescue activity of Cfp11-320, and the PHD2 domain is required for rescue activity of Cfp1318-481.
FIG. 4.
Redundant functional domains of Cfp1 are sufficient to restore appropriate levels of histone H3-Lys4 trimethylation in CXXC1−/− ES cells following induction of differentiation. ES cells were induced to differentiate for 6 days by the removal of LIF from the culture medium. Histone protein extracts were isolated from CXXC1+/+, CXXC1−/−, and CXXC1−/− ES cells expressing full-length Cfp1 (1-656) or the indicated Cfp1 fragments (or carrying the empty expression vector) and subjected to Western blot analysis using antiserum directed against histone H3-Lys4 trimethylation (H3-Lys4me3). The membrane was also probed with total histone H3 antiserum as a loading control. The graph represents average levels of global histone H3-Lys4 trimethylation from at least three independent experiments, and error bars represent standard error. Asterisks denote statistically significant (P < 0.05) increases in histone H3-Lys4 tri-methylation compared to CXXC1+/+ ES cells expressing Cfp11-656.
Cfp1 contains redundant functional domains that are sufficient to rescue in vitro differentiation.
ES cells lacking Cfp1 fail to differentiate in vitro following the removal of LIF from the growth medium, and this defect is rescued by introduction of a full-length Cfp1 cDNA expression vector (9). Experiments were performed to determine which functional domains of Cfp1 are necessary and sufficient to support in vitro differentiation of ES cells.
Following growth in medium lacking LIF, CXXC1+/+ and CXXC1−/− ES cells expressing either Cfp11-656, Cfp11-481, Cfp11-367, or Cfp1361-656 grow large cell aggregates (embryoid bodies) and form a prominent outgrowth characteristic of endoderm differentiation (Fig. 5A). CXXC1−/− ES cells expressing Cfp1103-367 exhibited a slight decrease in the amount of outgrowth present. In contrast, CXXC1−/− and CXXC1−/− ES cells expressing Cfp11-320 or Cfp1318-481 (or carrying the empty expression vector) produce cell aggregates that remain small and fail to produce an outgrowth (Fig. 5A), indicating an inability to achieve in vitro differentiation.
FIG. 5.
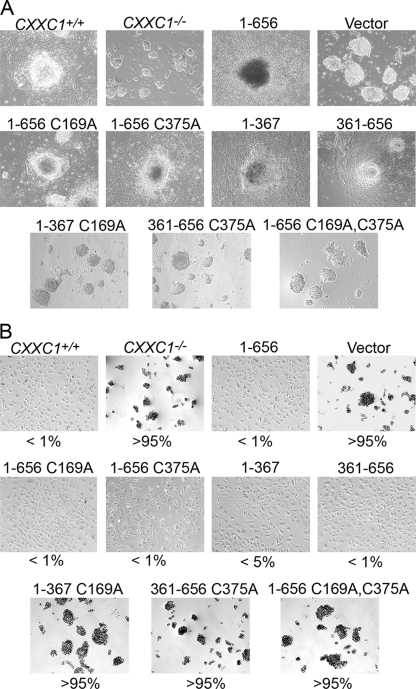
Cfp1 contains redundant functional domains that are sufficient to rescue in vitro differentiation. (A) Colony morphology was examined following induction of differentiation. CXXC1+/+, CXXC1−/−, and CXXC1−/− ES cells expressing full-length Cfp1 (1-656) or the indicated Cfp1 fragments (or carrying the empty expression vector) were cultured in the absence of LIF for 10 days. Magnification, ×10. (B) Cells were induced to differentiate as in panel A, harvested, disaggregated with trypsin, reseeded into gelatin-coated culture dishes, and stained for alkaline phosphatase activity. Numbers represent the percentage of cells that express alkaline phosphatase activity. At least 200 cells were scored for each clone. Magnification, ×10.
Alkaline phosphatase is a marker of pluripotency in ES cells that is downregulated upon cellular differentiation (61). Consistent with colony morphology results (Fig. 5A), CXXC1+/+ and CXXC1−/− ES cells expressing either Cfp11-656, Cfp11-481, Cfp11-367, or Cfp1361-656 demonstrate a very low number of cells (1 to 5%) that express alkaline phosphatase activity 10 days following removal of LIF, indicating that almost all of these cells differentiate (Fig. 5B). Approximately 25% of CXXC1−/− ES cells expressing Cfp1103-367 express alkaline phosphatase activity following induction of differentiation, indicating an intermediate degree of differentiation that is consistent with colony morphology. In contrast, CXXC1−/− and CXXC1−/− ES cells expressing Cfp11-320 or Cfp1318-481 (or carrying the empty expression vector) retain alkaline phosphatase activity following removal of LIF, indicating that these cells retain stem cell properties and fail to differentiate (Fig. 5B).
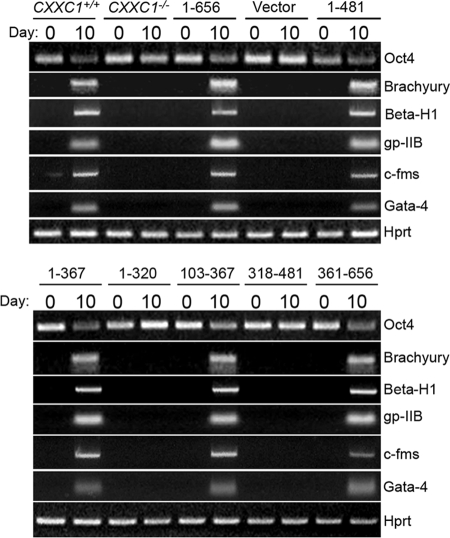
Oct4 is a transcription factor that is expressed in pluripotent ES and germ cells. It functions as a suppressor of differentiation and is a master regulator of pluripotency that controls lineage commitment (31, 35, 40). Oct4 becomes downregulated when ES cells are induced to differentiate. Oct4 mRNA expression was assessed by RT-PCR in undifferentiated ES cells (day 0) and 10 days following induction of differentiation. The ratio of Oct4 mRNA expression to Hprt was used to assess whether Oct4 mRNA expression changed upon removal of LIF. Similar to alkaline phosphatase activity, Oct4 mRNA expression is downregulated in CXXC1+/+ ES cells and CXXC1−/− ES cells expressing either Cfp11-656, Cfp11-481, Cfp11-367, Cfp1103-367, or Cfp1361-656, indicating the ability to differentiate (Fig. 6). In contrast, Oct4 mRNA expression persists in CXXC1−/− and CXXC1−/− ES cells expressing Cfp11-320 or Cfp1318-481 (or carrying the empty expression vector) following removal of LIF, indicating that these cells fail to differentiate (Fig. 6).
FIG. 6.
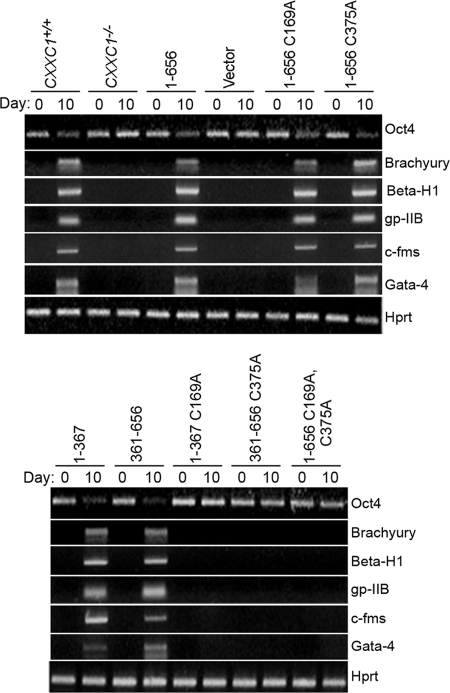
Identification of Cfp1 fragments that are sufficient to rescue in vitro differentiation. CXXC1+/+, CXXC1−/−, and CXXC1−/− ES cells expressing full-length Cfp1 (1-656) or various Cfp1 fragments (or carrying the empty expression vector) were cultured in the absence of LIF to induce differentiation. Cells were collected at day 0 (undifferentiated) and day 10 after induction of differentiation. Total RNA was isolated, and RT-PCR was performed to analyze the expression of Oct4, a marker of undifferentiated ES cells, along with the following lineage-specific markers: Brachyury (mesoderm), β-H1 (primitive erythroid), c-fms (myeloid), gp-IIB (megakaryocyte), Gata-4 (endoderm), and Hprt, a housekeeping gene that serves as a control for cDNA quantity and integrity.
CXXC1+/+ ES cells also induce expression of lineage- specific gene markers following removal of LIF and induction of differentiation, including Brachyury (mesoderm), β-H1 (primitive erythroid), gp-IIb (megakaryocyte), c-fms (myeloid), and Gata-4 (visceral/parietal endoderm). In contrast, ES cells lacking Cfp1 fail to induce expression of these differentiation markers upon removal of LIF (8) (Fig. 6). CXXC1−/− ES cells expressing either Cfp11-656, Cfp11-481, Cfp11-367, Cfp1103-367, or Cfp1361-656 induce expression of these lineage-specific markers 10 days following removal of LIF (Fig. 6). In contrast, CXXC1−/− and CXXC1−/− ES cells expressing Cfp11-320 or Cfp1318-481 (or carrying the empty expression vector) fail to induce expression of these markers (Fig. 6).
Taken together, these results are consistent with cytosine methylation and histone methylation rescue activity and indicate that expression of either the N-terminal (Cfp11-367) or the C-terminal (Cfp1361-656) half of Cfp1 in CXXC1−/− ES cells is sufficient to rescue in vitro differentiation. In addition, the rescue activity of Cfp1103-367 reveals that the PHD1 domain is not essential for cellular differentiation. However, the basic domain is required for the amino terminal rescue activity because removal of the basic domain (Cfp11-320) results in an inability to rescue differentiation. In addition, loss of the PHD2 domain from the C-terminal Cfp1318-481 fragment leads to a loss of differentiation rescue activity, demonstrating a requirement for the PHD2 domain.
Retention of either the DNA-binding domain or Setd1 association domain is required for Cfp1 rescue activity.
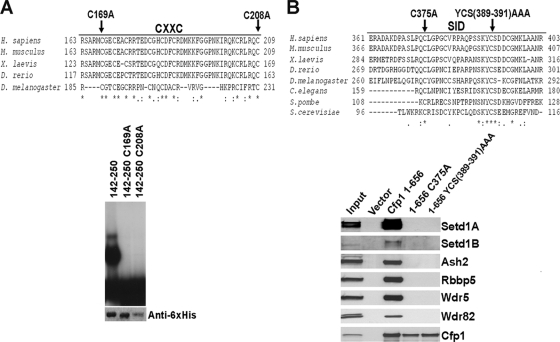
To further define the functional domains required for Cfp1-mediated rescue activity, point mutations that abolish the DNA-binding activity of Cfp1 or ablate the interaction of Cfp1 with the Setd1 histone H3-Lys4 methyltransferase complexes were analyzed. Protein alignment between species reveals conservation of the CXXC and SID domains of Cfp1 (Fig. 7). The C169A and C208A mutations were previously shown to abolish the DNA-binding activity of Cfp1 (26). EMSAs reveal that the C169A and C208A mutations abolish DNA-binding activity of Cfp1 to an oligonucleotide probe that contains an unmethylated CpG dinucleotide (Fig. 7A).
FIG. 7.
Mutations within the CXXC domain and SID of Cfp1 abolish DNA-binding activity or interaction of Cfp1 with the Setd1 complexes. (A) A multispecies alignment of the CXXC domain of Cfp1 is shown. Asterisks mark identical amino acid residues conserved between species, a colon indicates a conserved amino acid substitution, and period indicates a semiconserved amino acid substitution. The amino acids chosen for site-directed mutagenesis (C169 and C208) are indicated with arrows. As previously published (26), EMSA was performed using a CpG-oligonucleotide probe containing a nonmethylated CpG motif and wild-type or mutated Cfp1142-260 expressed as His6-tagged fusion proteins. The bottom panel presents Western blot analysis using anti-His6 antibody, demonstrating the expression and recovery of each recombinant protein. A portion of this figure was reprinted from reference 26 with permission from the publisher. (B) A multispecies alignment of the SID of Cfp1 is shown. The amino acids chosen for site-directed mutagenesis (C375 and YCS389-391AAA) are indicated with arrows. As previously published (7), FLAG immunoprecipitation was carried out using full-length wild-type or mutated Cfp1. Western blot analysis of immunoprecipitates was performed to determine interaction of Cfp1 with Setd1A, Setd1B, Ash2, Rbbp5, Wdr5, Wdr82. This figure is reprinted from reference 7 with permission from the publisher.
The C375A mutation and the mutations Y389A C390A S391A (YCS389-391AAA) within the SID were previously shown to ablate the interaction of Cfp1 with the Setd1A and Setd1B histone H3-Lys4 methyltransferase complexes (7). FLAG immunoprecipitation was carried out using wild-type or mutated (C375A or YCS389-391AAA) forms of full-length Cfp1. Western blot analysis demonstrates that the C375A and YCS389-391AAA Cfp1 mutations abolish Cfp1 interaction with the Setd1A and Setd1B proteins, along with the other protein components of the Setd1 complexes, including Ash2, Rbbp5, Wdr5, and Wdr82 (Fig. 7B).
Transfection experiments were performed to assess the functional significance of DNA-binding and Setd1 complex interaction domains of Cfp1. Various cDNA constructs encoding FLAG-tagged Cfp1 truncations and point mutations (Fig. 8A) were electroporated into CXXC1−/− ES cells. Stably transfected clones were isolated and analyzed for protein expression, and clones expressing at least 50% of the level of Cfp1 observed in CXXC1+/+ ES cells were selected for analysis (Fig. 8B).
FIG. 8.
Isolation of CXXC1−/− ES cell clones expressing Cfp1 fragments or point mutations. (A) A schematic of each Cfp1 construct is presented. The approximate location of the C169A and C375A mutations within the CXXC domain and SID, respectively, are denoted by X. (B) The level of Cfp1 expression in CXXC1+/+, CXXC1−/−, and CXXC1−/− ES cells expressing either full-length Cfp1 (1-656) or the indicated Cfp1 truncations or point mutations (or carrying the empty expression vector) was assessed by Western analysis using antiserum directed against Cfp1 (25). A representative blot is presented. Asterisks denote the position of each FLAG-Cfp1 protein. The blot was reprobed for β-actin as a loading control. The graph summarizes the results from at least three independent experiments, and error bars represent standard error.
CXXC1−/− ES cells expressing full-length Cfp1 that lacks DNA-binding activity (Cfp11-656 C169A) or full-length Cfp1 that fails to interact with the Setd1 histone H3-Lys4 methyltransferase complexes (Cfp11-656 C375A) exhibit normal plating efficiency (Fig. 9). This was expected given the finding that either half of Cfp1 is sufficient to provide rescue activity (Fig. 3). However, ablation of DNA-binding activity within the amino fragment of Cfp1 (Cfp11-367 C169A) or disruption of interaction with the Setd1 complexes within the carboxyl fragment of Cfp1 (Cfp1361-656 C375A) results in loss of rescue activity (Fig. 9). In addition, rescue of plating efficiency was not observed in CXXC1−/− ES cells expressing Cfp11-656 C169A C375A. This indicates that retention of either DNA-binding domain or SID of Cfp1 is required to rescue ES cell plating efficiency.
FIG. 9.
Retention of either the DNA-binding domain or SID of Cfp1 is required for rescue of plating efficiency of CXXC1−/− ES cells. The graph presents plating efficiency of CXXC1+/+, CXXC1−/−, and CXXC1−/− ES cells expressing full-length Cfp1 (1-656) or the indicated Cfp1 truncations or mutations (or carrying the empty expression vector). Plating efficiency was determined by counting the number of colonies formed after plating 400 cells. The graph summarizes data from at least three independent experiments performed in triplicate, and error bars represent standard error. Asterisks denote statistically significant (P < 0.05) differences compared to CXXC1−/− ES cells expressing Cfp11-656.
Genomic DNA isolated from CXXC1−/− ES cells expressing full-length Cfp1 lacking DNA-binding activity (Cfp11-656 C169A), or full-length Cfp1 that fails to interact with the Setd1 H3-Lys4 histone methyltransferase complexes (Cfp11-656 C375A) exhibit global cytosine methylation levels similar to CXXC1+/+ and CXXC1−/− ES cells expressing Cfp11-656 (Fig. 10A). In contrast, ablation of the DNA-binding activity in the amino terminal Cfp11-367 C169A or disruption of the Setd1 complex interactions of the carboxyl terminal Cfp1361-656 C375A results in loss of rescue activity (Fig. 10A). In addition, CXXC1−/− ES cells expressing Cfp11-656 C169A C375A also exhibit reduced cytosine methylation (Fig. 10A).
FIG. 10.
Retention of either the DNA-binding domain or SID of Cfp1 is required to rescue cytosine methylation. (A) Global cytosine methylation levels were determined by methyl acceptance assay for genomic DNA isolated from CXXC1+/+, CXXC1−/−, and CXXC1−/− ES cells expressing full-length Cfp1 (1-656) or the indicated Cfp1 truncations or mutations (or carrying the empty expression vector). The graph summarizes three independent experiments, and error bars represent standard error. Asterisks denote statistically significant (P < 0.05) differences compared to CXXC1−/− ES cells expressing Cfp11-656. (B) Genomic DNA was isolated from the indicated ES cell lines and digested with MspI or HpaII, and Southern blot analysis was performed using a radiolabeled probe specific for IAP retrovirus repetitive elements. Arrows indicate bands that demonstrate cytosine hypomethylation. (C) Western blot analysis was performed on protein extracts derived from the indicated ES cell lines, using antiserum directed against Dnmt1 (7) and β-actin as a loading control. The graph summarizes data from at least three independent experiments, and error bars represent standard error. The data for the CXXC1+/+, CXXC1−/−, Cfp11-656 (1-656), and vector lanes are reproduced from Fig. 3 to facilitate comparison of rescue activity. Asterisks denote statistically significant (P < 0.05) differences compared to CXXC1−/− ES cells expressing Cfp11-656.
Southern blot analysis of IAP repeats reveals a high level of cytosine methylation in DNA isolated from CXXC1+/+ and CXXC1−/− ES cells expressing Cfp11-656 C169A or Cfp11-656 C375A (Fig. 10B). In contrast, low-molecular-weight fragments (less than 4 kb), indicating decreased cytosine methylation, were observed at IAP repeats in DNA isolated from CXXC1−/− and CXXC1−/− ES cells expressing Cfp11-367 C169A, Cfp1361-656 C375A, or the doubly mutated Cfp11-656 C169A C375A) (Fig. 10B).
CXXC1+/+ ES cells and CXXC1−/− ES cells expressing Cfp11-656 C169A or Cfp11-656 C375A express Dnmt1 protein levels similar to the levels in of CXXC1+/+ ES cells and CXXC1−/− ES cells expressing Cfp11-656 (Fig. 10C). In contrast, decreased Dnmt1 protein expression is not rescued in CXXC1−/− ES cells expressing Cfp11-367 C169A, Cfp1361-656 C375A, or Cfp11-656 C169A C375A (Fig. 10C). Consistent with plating efficiency, these results indicate that retention of either the DNA-binding domain or SID of Cfp1 is required for appropriate Dnmt1 protein expression and global and site-specific cytosine methylation.
Neither DNA-binding activity (Cfp11-367 C169A) nor interaction with the Setd1 complexes (Cfp1361-656 C375A) is essential for full-length Cfp1 to restore normal histone H3-Lys4 trimethylation in CXXC1−/− ES cells following removal of LIF from the growth medium (Fig. 11). However, disruption of DNA-binding activity within Cfp11-367 C169A or disruption of interaction with the Setd1 complexes within Cfp1361-656 C375A results in a failure to rescue the inappropriately increased levels of histone H3-Lys4 trimethylation in CXXC1−/− ES cells following induction of differentiation (Fig. 11). In addition, a significant increase in the level of histone H3-Lys4 trimethylation is observed in CXXC1−/− ES cells expressing Cfp11-656 C169A C375A, indicating that retention of either the DNA-binding domain or Setd1 complex interaction domain is required to restore normal regulation of histone methylation.
FIG. 11.
Retention of either DNA-binding activity of Cfp1 or association of Cfp1 with the Setd1 complexes is required for appropriate histone H3-Lys4 trimethylation following induction of differentiation. ES cells were grown for 6 days following removal of LIF and induction of differentiation. Western blot analysis was performed on histone extracts isolated from CXXC1+/+, CXXC1−/−, and CXXC1−/− ES cells expressing full-length Cfp1 (1-656) or the indicated Cfp1 truncations or mutations (or carrying the empty expression vector), using antiserum directed against histone H3-Lys4 trimethylation and total histone H3 as a loading control. The graph represents relative levels of H3-Lys4 trimethylation from at least three independent experiments, and error bars represent standard error. Asterisks denote statistically significant (P < 0.05) differences compared to CXXC1−/− ES cells expressing Cfp11-656.
Upon culture in medium without LIF, CXXC1+/+ and CXXC1−/− ES cells expressing Cfp11-656, Cfp11-656 C169A, Cfp11-656 C375A, Cfp11-367, or Cfp1361-656 grow large cell aggregates and form a prominent embryoid body outgrowth characteristic of differentiation (Fig. 12A). In contrast, CXXC1−/− and CXXC1−/− ES cells expressing Cfp11-367 C169A, Cfp1361-656 C375A, or Cfp11-656 C169A C375A form small cell aggregates and fail to produce an outgrowth, indicating an inability to differentiate (Fig. 12A).
FIG. 12.
Retention of either the DNA-binding domain or SID of Cfp1 is required to rescue in vitro differentiation. (A) Colony morphology following induction of differentiation. CXXC1+/+, CXXC1−/−, and CXXC1−/− ES cells expressing full-length Cfp1 (1-656) or the indicated Cfp1 truncations or mutations (or carrying the empty expression vector) were cultured in the absence of LIF for 10 days. Magnification, ×10. (B) Cells were grown as in described for panel A and then disaggregated with trypsin, reseeded into gelatin-coated culture dishes, and stained for alkaline phosphatase activity using a leukocyte detection kit. Numbers represent percentages of cells positive for alkaline phosphatase activity. At least 200 cells were scored for each clone. Magnification, ×10.
Molecular markers were monitored to further assess the differentiation capacity of CXXC1−/− ES cells expressing various Cfp1 mutations. After 10 days of culture in the absence of LIF, CXXC1+/+ and CXXC1−/− ES cells expressing Cfp11-656, Cfp11-656 C169A, Cfp11-656 C375A, Cfp11-367, or Cfp1361-656 demonstrate a very small number of cells that express alkaline phosphatase activity, indicating that nearly all of these cells differentiate. In contrast, alkaline phosphatase activity persists in CXXC1−/− and CXXC1−/− ES cells expressing Cfp11-367 C169A, Cfp1361-656 C375A, or Cfp11-656 C169A C375A, indicating that these cells retain stem cell properties and fail to differentiate (Fig. 12B).
Similar to alkaline phosphatase activity, Oct4 mRNA expression is downregulated, and lineage-restricted differentiation markers are induced in CXXC1+/+ and CXXC1−/− ES cells expressing Cfp11-656, Cfp11-656 C169A, Cfp11-656 C375A, Cfp11-367, or Cfp1361-656, following growth in medium lacking LIF, indicating the ability of these cells to achieve in vitro differentiation (Fig. 13). In contrast, CXXC1−/− and CXXC1−/− ES cells expressing Cfp11-367 C169A, Cfp1361-656 C375A, or Cfp11-656 C169A C375A fail to repress Oct4 mRNA expression and fail to induce lineage-restricted differentiation markers, indicating that these cells fail to differentiate (Fig. 13).
FIG. 13.
Retention of either the DNA-binding domain or SID of Cfp1 is required to rescue in vitro differentiation of CXXC1−/− ES cells. CXXC1+/+, CXXC1−/−, and CXXC1−/− ES cells expressing full-length Cfp1 (1-656) or the indicated Cfp1 truncations or mutations (or carrying the empty expression vector) were cultured in the absence of LIF to induce differentiation. Cells were collected at day 0 (undifferentiated) and day 10 after induction of differentiation. Total RNA was isolated, and RT-PCR was performed to analyze the expression of Oct4, a marker of undifferentiated ES cells, along with the following lineage and developmentally specific markers: Brachyury (mesoderm), β-H1 (primitive erythroid), c-fms (myeloid), gp-IIB (megakaryocyte), Gata-4 (endoderm), and Hprt, a housekeeping gene that serves as a control for cDNA quantity and integrity.
Taken together, these results demonstrate that Cfp11-656 C169A and Cfp11-656 C375A exhibit rescue activity for a variety of defects observed in CXXC1−/− ES cells, including plating efficiency, appropriate regulation of cytosine methylation and histone methylation, and in vitro differentiation. However, ablation of the DNA-binding activity in Cfp11-367 C169A or disruption of the Setd1 complex interaction in Cfp1361-656 C375A results in loss of rescue activity. In addition, mutation of both the DNA-binding domain and the SID in Cfp11-656 C169A C375A ablates rescue activity. Similar findings were obtained when independent mutations that ablate Cfp1 DNA-binding activity (C208A) or Cfp1 interaction with the Setd1 complexes (YCS389-391AAA) were analyzed (data not shown). These findings indicate that retention of either the DNA-binding activity of Cfp1 or interaction of Cfp1 with the Setd1 histone H3-Lys4 methyltransferase complexes is required for Cfp1 function in ES cells.
DISCUSSION
The results of the structure-function studies reported here demonstrate that expression of either the amino half of Cfp1 (aa 1 to 367) or the carboxyl half of Cfp1 (aa 361 to 656) in CXXC1−/− ES cells is sufficient to rescue defects in ES cell plating efficiency, cytosine methylation, histone methylation, and in vitro differentiation. Analysis of mutations within the Cfp1 CXXC domain (C169A) and SID (C375A) reveals that retention of either DNA-binding or Setd1 histone H3-Lys4 methyltransferase interaction domains of Cfp1 is required for rescue function in ES cells. Thus, no single domain of Cfp1 is required for rescue activity, and Cfp1 contains redundant functional domains.
DNMT1−/− ES cells exhibit an ∼80 to 90% decrease in cytosine methylation and undergo apoptosis when induced to differentiate (3, 19). Therefore, the observed 50% reduction in Dnmt1 protein and 70% reduction in cytosine methylation may be causally related to the inability of CXXC1−/− ES cells to achieve in vitro differentiation. Rescue of normal levels of Dnmt1 protein in CXXC1−/− ES cells upon expression of various Cfp1 mutations correlates with rescue of cytosine methylation, histone methylation, and in vitro differentiation. However, DNMT1+/− ES cells express similarly reduced levels of Dnmt1 protein but retain normal levels of cytosine methylation (11, 29). Therefore, it is unlikely that decreased Dnmt1 protein in CXXC1−/− ES cells completely accounts for the observed decrease in cytosine methylation. Cfp1 may additionally be necessary for genomic targeting of Dnmt1 or regulating Dnmt1 enzymatic activity. Both amino and carboxyl fragments of Cfp1 interact with Dnmt1 in coimmunoprecipitation assays (7). Thus, Cfp1 may recruit Dnmt1 to unmethylated CpG motifs to initiate cytosine methylation and transcriptional silencing. Importantly, the C375A point mutation within the SID does not ablate the interaction between Cfp1361-656 and Dnmt1 (7), indicating that Cfp1 interacts independently with the DNA methyltransferase and the histone methyltransferase complexes. The finding that Cfp1361-656 C375A retains interaction with Dnmt1 but lacks rescue activity highlights the functional importance of the association of the Setd1A and Setd1B histone H3-Lys4 methyltransferase complexes with the C-terminal fragment of Cfp1.
Deletion of the PHD2 domain ablates the rescue activity of the carboxyl fragment of Cfp1. In addition, the PHD1 domain is specifically necessary for the amino terminal fragment of Cfp1 to rescue plating efficiency. Many PHD-containing proteins associate with chromatin and play a role in chromatin-mediated transcriptional control (4, 47). Recent studies have shown that PHD domains are binding modules for unmodified and methylated H3-Lys4 and methylated H3-Lys36 (30, 32, 39, 46, 48, 49). Interestingly, the PHD1 domain of Spp1, the yeast homologue of Cfp1, binds methylated histone H3-Lys4 (49). It remains to be determined whether the PHD domains of mammalian Cfp1 also bind to modified H3-Lys4 and whether they function in transcriptional regulation.
Removal of the basic domain from the Cfp11-320 results in ablation of rescue activity. Previous studies revealed that the basic domain (aa 318 to 367) is required for efficient subnuclear targeting of Cfp1 to nuclear speckles, association with the nuclear matrix, and transcriptional trans-activation activity (24). The studies reported here reveal that, in the context of the amino-terminal fragment, the Cfp1 basic domain is additionally required for appropriate cytosine methylation, histone methylation, plating efficiency, and in vitro differentiation.
Retention of either the DNA-binding (CXXC) domain or SID of Cfp1 is essential for rescue of ES cell cytosine methylation, histone methylation, plating efficiency, and differentiation. CXXC domains have been identified in a number of chromatin-associated proteins including Dnmt1 (42); methyl-DNA-binding protein 1 (Mbd1), a protein that binds methylated DNA and recruits a histone H3-Lys9 methyltransferase (12); Fbxl11, a histone demethylase specific for H3-Lys36 (56); leukemia-associated protein Lcx, found in Mll1 fusion proteins associated with acute myeloid leukemia (37); and Mll1, a histone H3-Lys4 methyltransferase (5). The MLL1 gene is a frequent target of chromosomal aberrations that are associated with leukemia (55), and the CXXC motif is essential for the transforming capacity of Mll1 fusion proteins (5). Similarly, mutation of the CXXC domain within Dnmt1 abolishes CXXC-mediated DNA binding and results in significant reduction of Dnmt1 catalytic activity and loss of genomic methylation of rDNA loci (42).
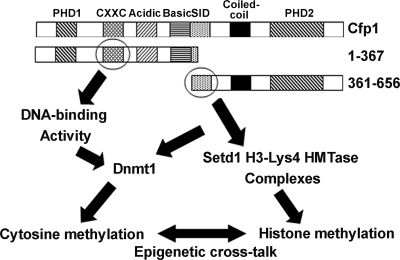
Remarkably, the presence of the CXXC DNA-binding domain is not required for Cfp1-mediated rescue of cytosine methylation, and interaction with the Setd1 complexes is not required for Cfp1-mediated rescue of appropriately regulated histone H3-Lys4 trimethylation. The finding of redundancy of Cfp1 functional domains suggests a model in which cross talk between epigenetic modifications permits ES cells to establish and maintain normal epigenetic function (Fig. 14). Thus, restoration of appropriate cytosine methylation might provide critical epigenetic marks to permit reestablishment and regulation of histone modifications, and restoration of appropriate regulation of histone H3-Lys4 methylation might permit subsequent restoration of normal patterns of cytosine methylation. It is now firmly established that cytosine methylation and histone modifications are intimately interconnected (14, 52, 53, 60). For example, Dnmt proteins associate with histone deacetylases, histone methyltransferases, and ATP-dependent chromatin remodeling enzymes (38), and deletion of the DNMT1 gene results in increased histone acetylation and decreased H3-Lys9 methylation (52). The histone H3-Lys27 methyltransferase Ezh2 is required for cytosine methylation and silencing of Ezh2-target promoters (58). In addition, depletion of Ezh2 leads to decreased recruitment of the Dnmt proteins to Ezh2-target genes, decreased H3-Lys4 methylation, and derepression of target gene expression (58). Loss of the catalytic SET domain of the histone H3-Lys4 methyltransferase Mll1 in mice causes defects in histone H3-Lys4 methylation and cytosine methylation at the promoters of several Hox genes (54). Also, the Suv39h1 H3-Lys9 methyltransferase is required for Dnmt3b-dependent methylation at pericentromeric repeats (16, 27). Hence, DNA methylation and histone modifications are highly integrated and mutually reinforcing mechanisms that establish and maintain heterochromatin. Cfp1 exhibits the interesting property of directly intersecting with both the cytosine methylation and histone methylation machinery. The results reported here identify Cfp1 as a possible integrator of epigenetic modifications.
FIG. 14.
Model for redundant Cfp1 functional domains. A schematic of the Cfp1 protein is presented to illustrate how the DNA-binding domain (CXXC) within the amino fragment of Cfp1 (aa 1 to 367) and the SID within the carboxyl fragment of Cfp1 (aa 361 to 656) may each support distinct epigenetic functions, which could lead to full rescue of the defects observed in CXXC1−/− ES cells due to cross talk between cytosine methylation and histone methylation.
Acknowledgments
This work was supported by the Riley Children's Foundation, the Lilly Endowment, and National Science Foundation grants NSF MCB-0344870 and MCB-0641851 (D.G.S.). C.M.T. was supported by a predoctoral fellowship from National Institutes of Health Grant T32 AI060519 and a Department of Education training grant for Graduate Assistance in Areas of National Need.
Footnotes
Published ahead of print on 11 May 2009.
REFERENCES
- 1.Aasland, R., T. J. Gibson, and F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 2056-59. [DOI] [PubMed] [Google Scholar]
- 2.Balaghi, M., and C. Wagner. 1993. DNA methylation in folate deficiency: use of CpG methylase. Biochem. Biophy. Res. Commun. 1931184-1190. [DOI] [PubMed] [Google Scholar]
- 3.Bestor, T. H. 2000. The DNA methyltransferases of mammals. Hum. Mol. Genet. 92395-2402. [DOI] [PubMed] [Google Scholar]
- 4.Bienz, M. 2006. The PHD finger, a nuclear protein-interaction domain. Trends Biochem. Sci. 3135-40. [DOI] [PubMed] [Google Scholar]
- 5.Birke, M., S. Schreiner, M. P. Garcia-Cuellar, K. Mahr, F. Titgemeyer, and R. K. Slany. 2002. The MT domain of the protooncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res. 30958-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 7.Butler, J. S., J. H. Lee, and D. G. Skalnik. 2008. CFP1 interacts with DNMT1 independently of association with the Setd1 histone H3K4 methyltransferase complexes. DNA Cell Biol. 27533-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caiafa, P., and M. Zampieri. 2005. DNA methylation and chromatin structure: the puzzling CpG islands. J. Biol. Chem. 94257-265. [DOI] [PubMed] [Google Scholar]
- 9.Carlone, D. L., J. H. Lee, S. R. L. Young, E. Dobrota, J. S. Butler, J. Ruiz, and D. G. Skalnik. 2005. Reduced genomic cytosine methylation and defective cellular differentiation in embryonic stem cells lacking CpG binding protein. Mol. Cell. Biol. 254881-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlone, D. L., and D. G. Skalnik. 2001. CpG binding protein is crucial for early embryonic development. Mol. Cell. Biol. 217601-7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan, M. F., R. Van Amerongen, T. Jijar, E. Cuppen, P. A. Jones, and P. W. Laird. 2001. Reduced rates of gene loss, gene silencing, and gene mutation in Dnmt1-deficient embryonic stem cells. Mol. Biol. Cell 217587-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cross, S. H., R. R. Meehan, X. Nan, and A. Bird. 1997. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat. Genet. 16256-259. [DOI] [PubMed] [Google Scholar]
- 13.Dillon, S. C., X. Zhang, R. C. Trievel, and X. Cheng. 2005. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuks, F., W. A. Burgers, A. Brehm, L. Hughes-Davies, and T. Kouzarides. 2000. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 2488-91. [DOI] [PubMed] [Google Scholar]
- 15.Glaser, S., J. Schaft, S. Lubitz, K. Vintersten, F. van der Hoeven, K. R. Tufteland, R. Aasland, K. Anastassiadis, S. L. Ang, and A. F. Stewart. 2006. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development 1331423-1432. [DOI] [PubMed] [Google Scholar]
- 16.Gopalakrishnan, S., B. O. Van Emburgh, and K. D. Robertson. 2008. DNA methylation in development and human disease. Mutat. Res. 130-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halbach, T., N. Scheer, and W. Werr. 2000. Transcriptional activation by the PHD finger is inhibited through an adjacent leucine zipper that binds 14-3-3 proteins. Nucleic Acids Res. 283542-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hattori, N., K. Nishino, Y. Ko, N. Hattori, J. Ohgane, S. Tanaka, and K. Shiota. 2004. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J. Biol. Chem. 27917063-17069. [DOI] [PubMed] [Google Scholar]
- 19.Jackson, M., A. Krassowska, N. Gilbert, T. Chevassut, L. Forrester, J. Ansell, and B. Ramsahoye. 2004. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol. Cell. Biol. 248862-8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 2931074-1080. [DOI] [PubMed] [Google Scholar]
- 21.Keller, G., M. Kennedy, T. Papayannopoulou, and M. Wiles. 1993. Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol. Cell. Biol. 13473-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, J. H., S. R. L. Hart, and D. G. Skalnik. 2004. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis 3832-38. [DOI] [PubMed] [Google Scholar]
- 23.Lee, J. H., and D. G. Skalnik. 2005. CpG-binding protein (CXXC Finger Protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J. Biol. Chem. 28041725-41731. [DOI] [PubMed] [Google Scholar]
- 24.Lee, J. H., and D. G. Skalnik. 2002. CpG-binding protein is a nuclear matrix- and euchromatin-associated protein localized to nuclear speckles containing human trithorax. J. Biol. Chem. 27742259-42267. [DOI] [PubMed] [Google Scholar]
- 25.Lee, J. H., C. M. Tate, J. Y. You, and D. G. Skalnik. 2007. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J. Biol. Chem. 28213419-13428. [DOI] [PubMed] [Google Scholar]
- 26.Lee, J. H., K. S. Voo, and D. G. Skalnik. 2001. Identification and characterization of the DNA binding domain of CpG-binding protein. J. Biol. Chem. 27644669-44676. [DOI] [PubMed] [Google Scholar]
- 27.Lehnertz, B., Y. Ueda, A. H. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 131192-1200. [DOI] [PubMed] [Google Scholar]
- 28.Li, E. 2002. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3662-673. [DOI] [PubMed] [Google Scholar]
- 29.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69915-926. [DOI] [PubMed] [Google Scholar]
- 30.Li, H., S. Ilin, W. Wqang, E. M. Dunchan, J. Wysocka, C. D. Allis, and D. J. Patel. 2006. Molecular basis for site-specific read-out of histone H3K4Me3 recognition by the BPTF PHD finger of NURF. Nature 44291-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loh, Y. H., Q. Wu, J. L. Chew, V. B. Vega, W. Zhang, X. Chen, G. Bourque, J. George, B. Leong, J. Liu, K. Y. Wong, K. W. Sung, C. W. Lee, X. D. Zhaoh, K. P. Chiu, L. Lipovich, V. A. Kuznetsov, P. Robson, L. W. Stanton, C. L. Wei, Y. Ruan, B. Lim, and H. H. Ng. 2006. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38431-440. [DOI] [PubMed] [Google Scholar]
- 32.Martin, D. G. E., K. Baetz, X. Shi, K. L. Walter, V. E. MacDonald, M. J. Wlodarski, O. Gozani, P. Hieter, and L. Howe. 2006. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol. Cell. Biol. 267871-7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClanahan, T., S. Dalrymple, M. Barkett, and F. Lee. 1993. Hematopoietic growth factor receptor genes as markers of lineage commitment during in vitro development of hematopoietic cells. Blood 812903-2915. [PubMed] [Google Scholar]
- 34.Miller, T., N. J. Krogan, J. Dover, H. Erdjument-Bromage, P. Tempst, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2001. COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. USA 2812902-12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niwa, H., J. Miyazaki, and A. G. Smith. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24372-376. [DOI] [PubMed] [Google Scholar]
- 36.Okano, M., and E. Li. 2002. Genetic analyses of DNA methyltransferase genes in mouse model system. Amer. Soc. Nutr. Sci. 1322462S-3465S. [DOI] [PubMed] [Google Scholar]
- 37.Ono, R., T. Take, T. Taketani, M. Taniwake, H. Kobayashi, and Y. Hayashi. 2002. LCX, leukemia-associated protein with a CXXC domain is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23). Cancer Res. 624075-4080. [PubMed] [Google Scholar]
- 38.Paulsen, M., and A. C. Ferguson-Smith. 2001. DNA methylation in genomic imprinting, development, and disease. J. Pathol. 19597-110. [DOI] [PubMed] [Google Scholar]
- 39.Pena, P. V., F. Davroaou, X. Shi, K. L. Walter, V. V. Verkhusha, O. Gozani, R. Zhao, and T. G. Kutatladze. 2006. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442100-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pesce, M., and H. R. Scholer. 2001. Oct-4: Gatekeeper in the beginnings of mammalian development. Stem Cells 19271-278. [DOI] [PubMed] [Google Scholar]
- 41.Peterson, C. L., and M. A. Laniel. 2004. Histones and histone modifications. Curr. Biol. 14R546-R551. [DOI] [PubMed] [Google Scholar]
- 42.Pradhan, M., P.-O. Esteve, H. G. Chin, M. Samaranayke, G.-D. Kim, and S. Pradhan. 2008. CXXC domain of human DNMT1 is essential for enzymatic activity. Biochemistry 4710000-10009. [DOI] [PubMed] [Google Scholar]
- 43.Puech, A., A. Dupressoir, M.-P. Loireau, M.-G. Mattei, and T. Heidmann. 1997. Characterization of two age-induced intracisternal A-particle-related transcripts in the mouse liver. J. Biol. Chem. 2725995-6003. [DOI] [PubMed] [Google Scholar]
- 44.Ragvin, A., H. Valvatne, S. Erdal, V. Arskog, K. R. Tufteland, K. Breen, A. M. Oyan, A. Eberharter, T. J. Gibson, P. B. Becker, and R. Aasland. 2004. Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. J. Mol. Biol. 337773-788. [DOI] [PubMed] [Google Scholar]
- 45.Rasmussen, T. P. 2003. Embryonic stem cell differentiation: a chromatin perspective. Reprod. Biol. Endocrinol. 1100-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruthenburg, A. J., H. Li, D. J. Patel, and C. D. Allis. 2007. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8983-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schindler, U., H. Beckmann, and A. R. Cashmore. 1993. HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 4137-150. [DOI] [PubMed] [Google Scholar]
- 48.Shi, X., T. Hong, K. L. Walter, M. Ewalt, E. Mischishita, T. Hung, D. Carney, D. Pena, P. Lan, M. R. Kaadige, N. Lacoste, C. Cayrou, F. Davrazou, A. Saha, B. R. Cairns, D. E. Ayer, T. G. Kutateladze, Y. Si, J. Cote, K. F. Chua, and O. Gozani. 2006. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 44296-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi, X., I. Kachirskaia, K. L. Walter, J.-H. A. Kuo, A. Lake, F. Davrazou, S. M. Chan, D. G. E. Martin, I. M. Fingerman, S. D. Briggs, L. Howe, P. J. Utz, T. G. Kutateladze, A. A. Lugovskoy, M. T. Bedford, and O. Gozani. 2007. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 2822450-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singal, R., and G. D. Ginder. 1999. DNA methylation. Blood 934059-4070. [PubMed] [Google Scholar]
- 51.Skalnik, D. G., E. C. Strauss, and S. H. Orkin. 1991. CCAAT displacement protein as a repressor of the myelomonocytic-specific gp91-phox gene promoter. J. Biol. Chem. 26616736-16744. [PubMed] [Google Scholar]
- 52.Szyf, M. 2005. DNA methylation and demethylation as targets for anticancer therapy. Biochemistry 70651-669. [DOI] [PubMed] [Google Scholar]
- 53.Tamaru, H., and E. U. Selker. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414277-283. [DOI] [PubMed] [Google Scholar]
- 54.Terranova, R., H. Agherbi, A. Boned, S. Meresse, and M. Djabali. 2006. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc. Natl. Acad. Sci. USA 1036629-6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tkachuk, D. C., S. Kohler, and M. L. Cleary. 1992. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 71691-700. [DOI] [PubMed] [Google Scholar]
- 56.Tsukada, Y., J. Fang, H. Erdjument-Bromage, M. E. Warren, C. H. Borchers, P. Tempst, and Y. Zhang. 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439811-816. [DOI] [PubMed] [Google Scholar]
- 57.Vannucchi, A. M., S. Linari, C. Cellai, M. J. Koury, and F. Paoletti. 1997. Constitutive and inducible expression of megakaryocyte-specific genes in Friend erythroleukaemia cells. Br. J. Haematol. 99500-508. [DOI] [PubMed] [Google Scholar]
- 58.Vire, E., C. Brenner, R. Deplus, L. Blanchon, M. Fraga, C. Didelot, L. Morey, A. Van Eynde, D. Bernard, and J. M. Vanderwinden. 2006. The polycomb group protein EZH2 directly controls DNA methylation. Nature 439871-874. [DOI] [PubMed] [Google Scholar]
- 59.Voo, K. S., D. L. Carlone, B. M. Jacobsen, A. Flodin, and D. G. Skalnik. 2000. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol. Cell. Biol. 202108-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weissmann, F., I. Muyrers-Chen, T. Musch, D. Stach, M. Wiessler, R. Paro, and F. Lyko. 2003. DNA hypermethylation in Drosophila melanogaster causes irregular chromosome condensation and dysregulation of epigenetic histone modifications. Mol. Cell. Biol. 232577-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wobus, A., G. Kaomei, J. Shan, M.-C. Wellner, J. Rohwedel, B. Guanju, H. A. Fleischmann, H. A. Katus, J. Hescheler, and W. M. Franz. 1997. Retinoic acid accelerates embryonic stem cell-derived cardiac differentiation and enhances development of ventricular cardiomyocytes. J. Mol. Cell. Cardiol. 291525-1539. [DOI] [PubMed] [Google Scholar]
- 62.Xu, C., G. Liguori, E. D. Adamson, and M. G. Persico. 1998. Specific arrest of cardiogenesis in cultured embryonic stem cells lacking Cripto-1. Dev. Biol. 196237-247. [DOI] [PubMed] [Google Scholar]
- 63.Yokochi, T., and K. D. Robertson. 2002. Preferential methylation of unmethylated DNA by mammalian de novo DNA methyltransferase Dnmt3a. J. Biol. Chem. 27711735-11745. [DOI] [PubMed] [Google Scholar]
- 64.Young, S. R. L., C. Mumaw, J. A. Marrs, and D. G. Skalnik. 2006. Antisense targeting of CXXC finger protein 1 inhibits genomic cytosine methylation and primitive hematopoiesis in zebrafish. J. Biol. Chem. 28137034-37044. [DOI] [PubMed] [Google Scholar]
- 65.Young, S. R. L., and D. G. Skalnik. 2007. CXXC finger protein 1 is required for normal proliferation and differentiation of the PLB-985 myeloid cell line. DNA Cell Biol. 2680-90. [DOI] [PubMed] [Google Scholar]